Abstract
Objective
The purpose of this study is to describe the methodological innovations of a web-based system for storage, integration and computation of biomedical data, using a training imaging dataset to remotely compute a deep neural network classifier of temporomandibular joint osteoarthritis (TMJOA).
Methods
This study imaging dataset consisted of three-dimensional (3D) surface meshes of mandibular condyles constructed from cone beam computed tomography (CBCT) scans. The training dataset consisted of 259 condyles, 105 from control subjects and 154 from patients with diagnosis of TMJ OA. For the image analysis classification, 34 right and left condyles from 17 patients (39.9 ± 11.7 years), who experienced signs and symptoms of the disease for less than 5 years, were included as the testing dataset. For the integrative statistical model of clinical, biological and imaging markers, the sample consisted of the same 17 test OA subjects and 17 age and sex matched control subjects (39.4 ± 15.4 years), who did not show any sign or symptom of OA. For these 34 subjects, a standardized clinical questionnaire, blood and saliva samples were also collected. The technological methodologies in this study include a deep neural network classifier of 3D condylar morphology (ShapeVariationAnalyzer, SVA), and a flexible web-based system for data storage, computation and integration (DSCI) of high dimensional imaging, clinical, and biological data.
Results
The DSCI system trained and tested the neural network, indicating 5 stages of structural degenerative changes in condylar morphology in the TMJ with 91% close agreement between the clinician consensus and the SVA classifier. The DSCI remotely ran with a novel application of a statistical analysis, the Multivariate Functional Shape Data Analysis, that computed high dimensional correlations between shape 3D coordinates, clinical pain levels and levels of biological markers, and then graphically displayed the computation results.
Conclusions
The findings of this study demonstrate a comprehensive phenotypic characterization of TMJ health and disease at clinical, imaging and biological levels, using novel flexible and versatile open-source tools for a web-based system that provides advanced shape statistical analysis and a neural network based classification of temporomandibular joint osteoarthritis.
Keywords: neural network, web-based system, osteoarthritis
1. Introduction
No proven disease-modifying therapy exists for osteoarthritis (OA) and current treatment options for chronic arthritic pain are insufficient (Hunter et al., 2013). The NIH-funded categorization of OA includes imaging, clinical and molecular markers of inflammation, angiogenesis and bone resorption in arthritis initiation and progression. Variation in disease progression requires biomarkers that can reflect morphological and pathological changes in joints, beginning in the earliest stages of OA development and throughout the course of the disease. Biochemical markers may reflect ultrastructural changes in joint tissue metabolism very early in the disease process prior to any apparent change in imaging appearance. Not only local proteins in the synovial fluid, but also circulating levels of proteins, may play a role in the cross-talk among the different joint tissues. The ascertainment of variations between health and disease is essential information for detecting inflammatory and degenerative conditions of the tissues affected (Abramson and Attur, 2009).
Patient data in clinical research on Temporo-Mandibular Joint Osteoarthritis often includes large amounts of structured information, such as imaging data, biological marker levels, and clinical variables. The present study proposes improvement in the precision of the subjective radiological interpretation of morphological variability described in Su et al., (2014) the unsupervised statistical classification proposed by Gomes et al., (2015), and the shape statistical models proposed by Paniagua et al., (2017). Given the various sources of information, computerized methods can be a great help to clinicians to discover hidden patterns in the data. The computerized methods often employ data mining and machine learning algorithms, lending themselves as the computer-aided diagnosis tool that assists clinicians in making diagnostic decisions. State-of-the art methods to classify morphological variations include extreme learning machine, sparse representation-based classification and neural network deep learning (Ashinsky et al., 2017, Li, 2018). Neural network applications in computer-aided diagnosis represent the main stream of computational intelligence in medical imaging (Qian et al., 2007). Their application is generalizable to most medical problems due the adaptive and flexible nature of learning directly from input information. Given a suitable learning algorithm, the neural network can improve the algorithm performance in accordance with the variety and the change of input datasets. Neural networks have the capability of optimizing the relationship between the input and output via distributed computing, training, and processing, leading to reliable solutions to a specific clinical question. Diagnosis often relies on visual inspection of scans, and 3D imaging provides a most important tool for facilitating such inspection and visualization. Surpassing human-level performance on certain image recognition tasks, neural networks enable the incorporation of large training data sets, as well as the use of different shape analysis features within the same classification (Jiang et al., 2010).
The neural network deep learning proposed in this manuscript to classify morphological variability extract features from the mandibular condyle morphology to describe each patient 3D mesh. There is a compelling need for such more efficient software tools that facilitate the analyses of clinical, biological and imaging data of the complex heterogeneous conditions in TMJ OA. To answer the diagnostic and assessment of treatment effectiveness challenges, our innovative solutions include the development of a web-based system, which implements a broad set of statistics and an advanced neural network classifier that supports the analysis of shape variability based on a training dataset. Our DSCI (Data Storage for Computation and Integration) web system provides useful features for integrating databases, performing data quality control and sample selection. The purpose of this study is to describe the DSCI system methodological innovations in functionality and efficiency, using a training imaging data set to remotely compute a neural network classifier of temporomandibular joint osteoarthritis.
2. Materials and Methods
2.1. Materials
Data used in the preparation of this article were obtained from the Dental and Craniofacial Bionetwork of Image Analysis database (DCBIA). The DCBIA has performed an observational clinical study that collected imaging, biological, and clinical data to characterize TMJ OA. This study is in concordance with STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) guidelines for observational studies. The University of Michigan Institutional Review Board approved the data acquisition and analysis in this study.
This study included imaging, clinical and biological datasets. Three-dimensional (3D) surface representation (meshes) for 293 condyles were constructed from CBCT scans. The training dataset consisted of 259 condyles, 105 from control subjects and 154 from patients with diagnosis of TMJ OA. For the image analysis classification, 34 right and left condyles from 17 patients (39.9 ± 11.7 years), who experienced signs and symptoms of the disease for less than 5 years, were included as the testing dataset. For the integrative statistical model of clinical, biological and imaging markers, the sample consisted of the same 17 test OA subjects and 17 age and sex matched control subjects (39.4 ± 15.4 years), who did not show any sign or symptom of OA. For these 34 subjects, a standardized clinical questionnaire, blood and saliva samples were also collected. Subjects recruited from the university clinic and through advertisement, underwent a clinical exam by an orofacial pain specialist using the research diagnostic criteria for temporomandibular disorders (RDC/TMD) guidelines (Ahmad et al., 2009). Following clinical diagnosis of TMJ osteoarthritis or health, a Cone beam CT (CBCT) scan was taken on all participants, with 0.08 mm isotropic voxel size and 4cm × 4cm field of view, using the 3D Accuitomo 170, Morita Corp. Blood and saliva samples were collected on the same day by an experienced nurse at the Department of Oral and Maxillofacial Surgery. Subjects allowed the saliva to naturally drool down the funnel into the collection tube, until the amount of saliva collection reached around 3 ml or 15 min maximum. Immediately after collection, liquid saliva was aliquoted to exactly 2 ml, and inhibitor protease (Aprotinin + PMSF) was added. 400ul–500ul aliquots of the saliva + protease volume was placed into 4–5 Eppendorf tubes and stored at −80°C for future analysis. 4 ml of blood sample was collected in a unique EDTA tube. After collection, cells were removed from plasma by centrifugation for 20 minutes with 1000 revolutions per minute (rpm). Plasma and cells were placed into different Eppendorf tubes, 3 each one, and stored at −80°C.
2.2. Biological Samples Methods
Custom quantibody protein microarrays RayBiotech (Norcross, GA) were used to evaluate the saliva and serum samples for 17 biomarkers expressed in both synovial fluid and blood in our preliminary work (Cevidanes et al., 2014). This assay is array-based multiplex sandwich ELISA system for simultaneous, quantitative measurement the concentration of multiple proteins. Like an ELISA, it uses a pair of antigen-specific antibodies to capture the protein of interest. The use of biotinylated antibodies and a streptavidin-conjugated fluor allow detection levels for the specific proteins to be visualized using a fluorescence laser scanner (RayBiotech, 2017). For protein quantification, the reagent kit included protein standards, whose concentration had been predetermined, provided to generate a six-point standard curve of each protein. Standards and samples were assayed simultaneously. By comparing signals from unknown samples to the standard curve, the unknown protein concentrations in the samples were determined. Positive controls for each biomarker were included in each array and the array data obtained from densitometry were entered into the appropriate cells of the corresponding analysis tool, which plotted the standard curve for each analysis in addition to performing background subtraction/normalization. The biomarkers chosen were known to be associated with bone repair and degradation, inflammation or nociception, common processes seen in OA. Preprocessing steps for these samples were completed at the School of Dentistry and then shipped to RayBiotech for analysis. All samples were evaluated in duplicate for level of proteins 6ckine, ANG, BDNF, CXCL16, ENA-78, GM-CSF, IFNγ, IL-1α, IL-6, MMP-3, MMP-7, PAI-1, TGFβ1, TIMP-1, TNFα, VE-Cadherin and VEGF.
2.3. Image Analysis Methods
Correspondence of Condylar 3D Surface Meshes
The process of constructing surface models from the CBCTs is called segmentation, and was performed interactively with the open-source ITK-SNAP 2.4 software (Yushkevich et al., 2006). After generating all 3D surface models, left condyles were mirrored in the sagittal plane to be in the same orientation as the right condyles to facilitate bilateral comparisons. The regional superimposition technique used in the present study for a cross-subject comparisons was validated by Schilling et al., (2014). After registration, all condylar models were simultaneously cropped to define the condylar region of interest. SPHARM-PDM software (Paniagua et al., 2017) was used to generate a mesh with 1002 correspondent vertices, via spherical parameterization of the input segmentations. An average 3D condylar shape was generated for the TMJ OA groups and control group using the ShapeVariationAnalyzer (SVA) extension for the 3D Slicer open-source software (Dumast, 2017, Fedorov et al., 2012). In order to compute the group average and group variability, we established correspondence between each of the 1002 vertices in the condylar surface models across all subjects (Prieto et al., 2017). This homology is further optimized via group-wise correspondence (Lyu et al., 2013). For thirty condyles of other patients with chronic and advanced arthritic degeneration, the severe dismorphologies did not provide adequate, automatic establishment of homology; therefore, those condyles were not included in the training dataset.
Data Storage for Computational Integration of Biological, Clinical and Imaging Data
An encoded patient ID was used to de-identify subjects before storing in the DSCI web-system. No patient identifiers are stored. Data regarding specimen collection (blood and saliva samples), digital data from CBCT scans and the clinical survey data were recorded following ALCOA (Attributable, Legible, Contemporaneous, Original and Accurate) attributes. The DSCI allows the display of tables with all the data relative to the patients in a given study.
The architecture of the data storage is based on a NOSQL model using JavaScript Object Notation (JSON) files. The JSON files are text files with key-value pairs. Each JSON file represents a different object in the database with a 128-bit identification number, generated at creation, to identify them. Contrary to traditional SQL databases, in a NOSQL database, there is no requirement to define the relationships between objects in the database. Instead, the relationships are revealed using a map-reduce algorithm, where the fields in the JSON document serve as primary keys. We use the key-value pair notation to create lists of documents that are consumed by other applications written in various languages such as Python or C++.
The storage architecture has been designed with multiple levels. The top level is a project document; it can contain a name, a description, the number of patients, and one or multiple collections of data. The collection is the level below the project and is defined by a name and a list of de-identified/encoded patients. The lowest level is each patient document that can contain imaging, clinical and biological data for each project (Prieto et al., 2017).
On the DSCI website, a webpage has been implemented to ease selection of the display, creation and edits of different projects/collections. The dashboard and panels also allow management of projects, such as overall statistics of datasets, users activity or the degree of completion. The structure of the DSCI website and the database allows different types of collections:
-
–
The clinical and biological data collections were uploaded from Comma Separated Values (CSV) files or directly input on the website with a customized form for the study.
-
–
The morphological collections (imaging data) can be 3D surface models or volumes, that were uploaded and displayed directly on the website. To ease the manipulation of the morphological/imaging data, a plugin, DatabaseInteractor (Mirabel, 2017), has been developed for 3D Slicer. From the plugin, users can download data from and upload imaging data.
For this present study, the DSCI system trained and tested neural networks by constructing computational graphs of the imaging data. The DSCI also implemented an interface to run remotely the Multivariate Functional Shape Data Analysis (MFSDA) tool. The MFSDA computed statistical shape analysis models of TMJ OA, testing the integrating information of 3D mesh coordinates, clinical information and levels of biological markers. To run the code remotely, we set up a connection with Flux, a high-performance computing (HPC) Linux-based cluster provided by the Advanced Research Computing Technology Services (ARC-TS) at the University of Michigan (http://arc-ts.umich.edu/systems-and-services/flux/).
Shape Variation Analysis and Classification
The classification system in this study was trained to distinguish different degrees of shape deformation in TMJ OA. A consensus visualization and interpretation of 3D surface morphology by two expert clinicians (MSY and ACR) was used to classify subjects with clinical diagnosis of TMJ OA. The clinicians classified the condylar morphology into 5 sub-groups with different degrees of condylar degeneration that were later compared to SVA’s automatic classification (Dumast, 2017). The SVA module of the Slicer software was used for automatic classification of morphological variation in TMJ OA, based on a neural network. The training of the neural network can be performed in the DSCI system, where the classifier learns from features extracted from the 3D meshes of the condyles. The SVA module computes the average shape of each group of condylar dysmorphology, as well as geometric features at each vertex of the mesh. The features are stored into arrays and linked to their corresponding vertices in the 3D meshes. Those vertex-wise features (Koenderink, 1990) are:
-
–
Normal vector: 3 scalars for the x, y, and z coordinates
-
–
Curvatures: 4 scalars for mean, minimum, maximum, and Gaussian curvature.
-
–
Distances: As many components as classes
-
–
Shape Index: 1 scalar
-
–
Curvedness: 1 scalar
-
–
Position: 3 scalars for the x, y, and z coordinates
The shape index and curvedness (Koenderink and Vandoorn, 1992) were computed using the principal curvatures (κ1, κ2) at every point in the surface. The shape index described local surface topology in terms of the principal curvatures, calculated as follows:
The curvedness was calculated as a measurement of the amount or ‘intensity’ of the surface curvature as follows:
Neural network architecture: The neural network learns tasks by considering examples. It is based on a collection of connected units called neurons organized in layers. We use a soft-max layer with one output per class. The output vector will be the probability for each shape to belong to a class. The algorithm extracts shape features to classify each sample in the training data in a class and can then classify new samples thanks to this probabilistic function (the softmax function). The TensorFlow open-source library (Abadi et al., 2015) was used to train and test the neural network by constructing computational graphs. The neural network was trained to classify a given shape into one of the 5 groups indicating the severity of the disease. The input data to train the network was stored in a matrix with dimensions [number (nbr) of subjects, nbr of vertices, nbr of features]. By training a neural network we sought to identify discriminative patterns of these features and encoded them in the network (deep learning). The neural network architecture has two hidden layers and follow the geometric pyramid rule (Masters, 1993). The output of the first layer is connected to a rectified linear unit and the second layer generates the scores or probabilities of belonging to a class. The deeper the neural network, ie, if we added more hidden layers, 3 or 4, more abstraction from the data would be learned. At the stage of development of our deep learning network, we chose 2 hidden layers, because more hidden layers could cause overfitting with our current sample size (Zagoruyko et al., 2016).
In order to identify patterns that generalize well and create a working classifier of shape variability, it is necessary to train the network using as many samples as possible. Moreover, it is important to have the same number of samples for each class. As our database contains fewer samples for some of the disease groups, the condylar database was not used in its entirety during the training phase. Having unbalanced datasets in the machine-learning field is common and is known as the ‘class imbalance problem. In order not to over-train the network for one of the groups, the training procedure requires the same number of meshes in each training group (LeCun et al., 2015). To increase the number of datasets in each training group, we simulated data by adding perlin noise (vtk.org) of small magnitude to each coordinate in the shape, and then the features were recomputed. Perlin noise is an algorithm to generate smooth noise in the X, Y, Z coordinates of the meshes 1002 points, creating different shapes from our sample. The simulated data was visually inspected to assess if the shape looked realistic. For training the neural network, we simulated data to ensure that 74 meshes were available per group. For groups that had more than 74 meshes such as the control group, the preprocessing step randomly selected 74 condyles.
Validation of the neural network: Before the training phase, 3 meshes per each group were removed from the training dataset. The validation dataset was used during the training, in order to monitor the progression of the network. Thus, the training dataset consisted of (74-3) × 6 groups= 426 meshes.
The testing dataset consisted of another 34 condyles. The SVA classification of the testing dataset provides the estimated classification group for the input meshes (34 condylar surface meshes in this study). The SVA classification was then compared to the clinical expert classification.
3. Statistical Analysis
For statistical analysis of the classification of condylar 3D meshes variability, a confusion matrix, also known as error or contingency matrix (Stehman, 1997), allowed visualization of the performance of the supervised learning SVA algorithm.
We considered the Multivariate Functional Shape Data Analysis (MFSDA) on the 3D meshes data. In MFSDA, the Multivariate Varying Coefficient Model (MVCM) (Huang et al., 2017, Zhu et al., 2012) is introduced to build the relationship between the 3D meshes data and other variables of interest. In particular, the different groups of variables are taken into account in MVCM, i.e., demographic variables (age, gender), clinical variables (diagnosis information, clinical markers), and biological markers. In order to select the variables in each group we used Principal Component Analysis (PCA). Specifically, variables highly correlated with the first principal components are selected from each group of variables.
To estimate the parameters in MFSDA, we employ a weighted least squares (WLS) method based on the multivariate local polynomial kernel smoothing technique (Fan and Gijbels, 1996). In our shape data analysis, we are interested in testing whether there is significant morphological difference caused by the variables we are interested in. We introduce both local and global Wald-type test statistics (Zhu et al., 2012) to investigate this hypothesis testing problem. In particular, the global p values are calculated by wild Bootstrap method (Zhu et al., 2007) and the local p values are corrected via the false discovery rate (FDR) method (Benjamini, 2010) to deal with the multiple comparison issues. The package MFSDA, along with its documentation, is accessible from the website https://github.com/BIG-S2/MFSDA.
3.1. Software
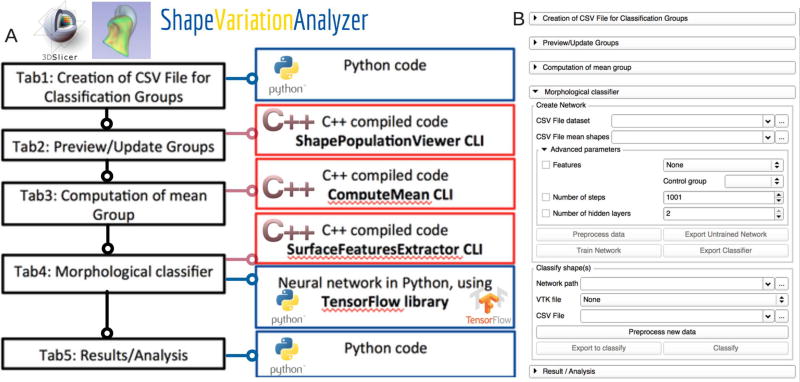
ShapeVariationAnalyzer module (SVA): is a 3D Slicer plug-in to compute average morphologies and classify morphological variability using a neural network. The tool architecture uses Python, C++ code and VTK C++ libraries. The neural network is developed using TensorFlow (Abadi et al., 2015), an open-source library for machine learning. The source code is free and available on GitHub (Figure 1) (Dumast, 2017).
Figure 1.
ShapeVariationAnalyzer (SVA) Slicer plug-in. A, SVA architecture; B, User interface.
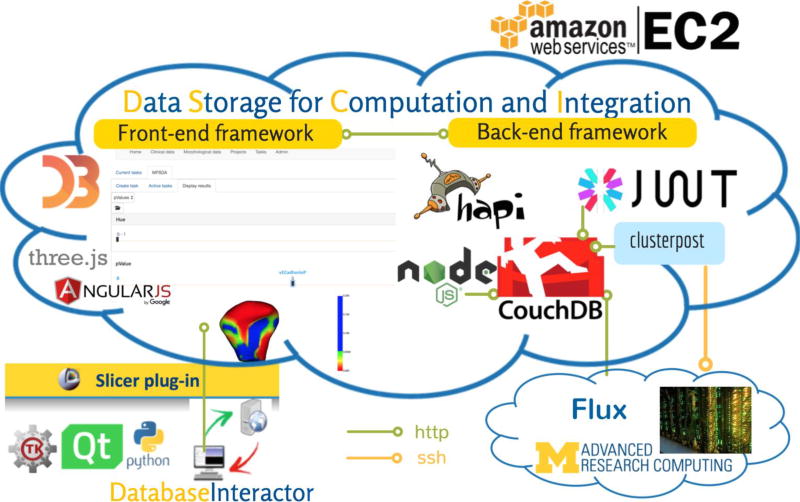
DatabaseInteractor module: is a 3D Slicer plug-in to manage patient data, upload and download imaging data in the web-based system, and create and manage image processing tasks. The tool architecture has been developed in Python. The source code is free and available on GitHub (Figure 2) (Mirabel, 2017).
Figure 2.
DSCI web-based system architecture based on plug-ins for a distributed application. The framework allows easy integration of a variety of plug-ins. The authentication system is based on JSON Web Tokens. The clusterpost plug-in allows submitting heavy computational tasks to remote computing grids, such as Flux, Linux-based high-performance computing at the University of Michigan.
Data Storage for Computation and Integration (DSCI) web-based system: the web-based system utilizes Node and Hapi for the back-end framework, Couchdb for storage, and JSON Web Token (JWT) for the authentication system. A plug-in was developed for Node that allows storage and retrieval of user information, as well as JWT encryption upon user login. We also integrated the Clusterpost plug-in into the system to allow submitting tasks to remote computing grids, using the data stored in the system. The front end of the application is based on Angular.js. The visualizations utilize D3.js and Threejs. The application is hosted using Amazon web services or the Elastic Computing Cloud (EC2). The computation tasks, such as MFSDA, use 4 cores, with 4 GB of RAM per core, stored on Flux (Figure 2). Flux consists of approximately 27,000 cores – including 1,372 compute nodes composed of multiple CPU cores, with at least 4 GB of RAM per core, interconnected with InfiniBand networking.
4. Results
4.1. Classification of Condylar Morphology
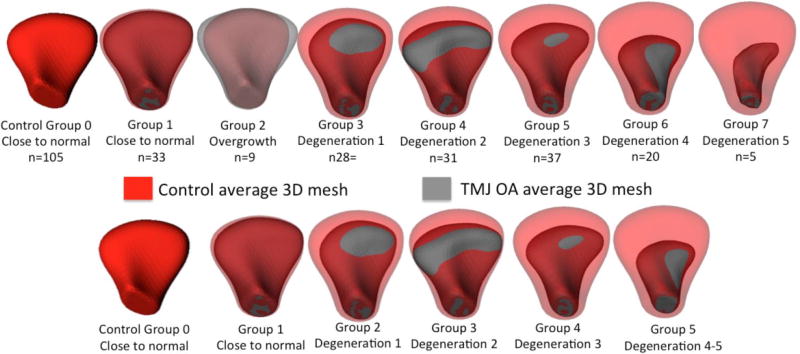
The classification of 3D morphology performed by experienced clinicians resulted in 8 groups of morphological variability shown in Figure 3. The 8 subgroups of condylar morphology had marked differences in size in the partition of the data. The smallest group (Degeneration 5) had only 5 condyles, whereas the largest one (Control group) had more than 100 condyles. The two small groups with the most severe phenotypes of bone destruction were merged (Table 1 and Figure 3). The bone proliferations and overgrowth group were not included in the training database as hyperplastic/ hypertrophic conditions with bone proliferations do not seem to have a defined, common morphological pattern (Figure 4), and are a different clinical condition compared to progressive degenerative joint conditions.
Figure 3.
Average 3D meshes for each group of morphology variability. Eight subgroups of condylar morphology variability are shown in the top row. For the neural network training data, the overgrowth group was not included and two subgroups with the most severe phenotypes of bone destruction were merged.
Table 1.
Distribution of the training and testing datasets into 5 subgroups of OA following the clinician visual classification. Note that with the addition of simulated data the number of the condylar meshes included in the training network were 74 meshes per group. For groups that had more than 74 meshes, such as the control group, the preprocessing step randomly selected 74 condyles.
| Number of 3D condylar meshes |
Group 0 Control |
Group 1 Close to normal |
Group 2 Degeneration 1 |
Group 3 Degeneration 2 |
Group 4 Degeneration 3 |
Group 5 Degeneration 4–5 |
|---|---|---|---|---|---|---|
| Available for the training =259 | 105 | 33 | 28 | 31 | 37 | 25 |
| Available + simulated = 530 | 105 | 99 | 84 | 93 | 74 | 75 |
| Testing = 34 | 0 | 3 | 9 | 16 | 6 | 0 |
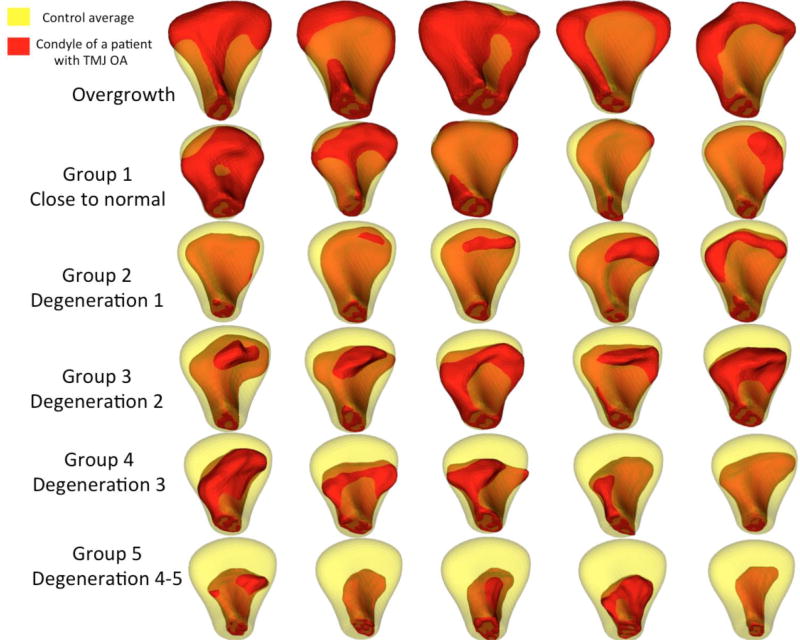
Figure 4.
Examples of condyles in each subgroup with semitransparency overlay of each condyle (red) and the average control condyle (transparent yellow). Condyles with overgrowth did not present a common morphology pattern. The degeneration groups show progressive degeneration that start with flattening of the articular surface in the degree 1 of degeneration; more marked resorption of the lateral pole and bone proliferations anteriorly were observed in the degree 2 of degeneration; further resorption of the articular surface and bone proliferation anteriorly led to changed condylar torque in the degree 3 of degeneration; advanced degenerative stages presented loss of overall condylar structure in the degree 4 of degeneration.
From the different combinations of features that were used to train the network, the features that led to higher accuracy of the morphological classification, compared to the clinician expert assessments, were normal vectors, mean curvature, and the distances to the average meshes at each mesh vertex. The results reported in this paper were run using those features, with neural network architecture of 1 hidden layer, 2001 iterations, 50 epochs (number of times the entire dataset was used in the training). The SVA performance is shown in the confusion matrix (Table 2), where each column represents the instances in the SVA classification group, while each row represents the group instances as assessed by the consensus between two clinician experts. Agreement between the clinician consensus and the SVA classification are located in the main diagonal of the table. Cells adjacent to the main diagonal (1 diagonal to the right and 1 diagonal to the left) indicate the classification of the degree of degeneration was within 1 group difference. The main and adjacent diagonals include 91% of the testing dataset. The cells outside the 3 middle diagonals show that 3 out of 34 condyles were classified with 2 or more groups difference.
Table 2.
Confusion matrix: columns show the SVA classification of condylar morphology; rows show the clinician experts consensus classification. The main diagonal cells show when the group was classified by the SVA exactly the same as the clinicians. The cells in 1 diagonal to the right and 1 diagonal to the left show number of condyles where the prediction differed from clinicians by only one group and still indicates an acceptable estimate of severity/risk level of disease staging. A challenge is that clinician expert assessment is limited by their visual perception of the 3D morphology. Note that as the testing dataset is from patients with less than 5 years of diagnosis of the disease, SVA did not classify any group 4 or 5 of more advanced disease stag in the testing sample, as expected.in such disease condition that the degeneration progresses in a chronic process through many years.
| SVA | 0 | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| ClinE | ||||||
| 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 1 | 7 | 1 | 0 | 0 | 0 | 0 |
| 2 | 2 | 2 | 1 | 4 | 0 | 0 |
| 3 | 1 | 0 | 1 | 10 | 0 | 0 |
| 4 | 0 | 0 | 0 | 2 | 2 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 |
4.2. Multivariate Functional Shape Data Analysis (MFSDA)
The MFSDA statistics included a subset of the above dataset: mandibular condyle 3D meshes from 34 subjects (17 TMJOA patients and 17 asymptomatic controls). Each subject has 2 TMJ condyles, left and right, but the biological and clinical data refer to on each subject without being side specific. For this reason, the left or right condyle of choice for the MSFDA statistics was the side with most severe symptoms in the OA group, and the matching control condyle. The demographic information included were age and sex. Seven pain–related clinical variables were tested: current facial pain intensity, worst facial pain over the last 6 months, average facial pain intensity over the last 6 months, pain duration in years, distress by headaches during last month and distress by muscle soreness during the last month (Table 3). The MSFDA model also tested 8 biological markers that were expressed at the best confidence levels in both in saliva and plasma samples (ANG, CXCL16, ENA-78, MMP-3, MMP-7, TIMP-1, TNFα, VE-Cadherin and VEGF, Table 4). The statistical analysis of pain-related clinical variables revealed that the variable “current pain intensity” in OA subjects was significantly correlated with “worst pain intensity in last 6 months” and “average facial pain intensity over the last 6 months”. Principal Component Analysis (PCA) showed that the percentage of the total variance explained by first two principal components is over 82%. The variable “worst pain intensity in last 6 months” was highly correlated with the first PC score (pearson correlation = 0.905, p-value=6.08E-7); and the variable “pain duration in years” was highly correlated with the second PC score (pearson correlation = 0.666, p-value=0.0035). Thus, these two-clinical painrelated variables were included in the MSFDA model (results reported in Table 3 and Figure 5).
Table 3.
Pearson correlation among 7 pain related clinical variables.
| Pearson Correlation |
Current pain |
Worst pain | Average pain in 6 months |
Pain duration |
Last month headaches |
Last month muscle soreness |
Pain Location |
|---|---|---|---|---|---|---|---|
| P-value | |||||||
| Current pain | 1 | 0.57 | 0.61 | 0.2 | 0.17 | 0.37 | 0.11 |
| Worst pain | 0.02* | 1 | 0.9 | 0.46 | 0.39 | 0.19 | −0.06 |
| Average pain in 6 months | 0.01* | 0.00* | 1 | 0.4 | 0.23 | 0.29 | −0.05 |
| Pain duration | 0.43 | 0.06 | 0.11 | 1 | 0.44 | 0.46 | 0 |
| Last month headaches | 0.51 | 0.12 | 0.36 | 0.08 | 1 | 0.31 | −0.29 |
| Last month muscle soreness | 0.14 | 0.46 | 0.25 | 0.06 | 0.23 | 1 | 0.13 |
| Pain Location | 0.67 | 0.83 | 0.85 | 0.99 | 0.26 | 0.62 | 1 |
Table 4.
Pearson correlation among 8 biological variables.
| Pearson Correl. |
Saliva | Plasma | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| AN G |
MMP 3 |
MMP 7 |
TIMP 1 |
VE- cadheri n |
CXCL1 6 |
ENA7 8 |
VEG F |
AN G |
MMP 3 |
MMP 7 |
TIMP 1 |
VE- cadheri n |
CXCL1 6 |
ENA7 8 |
VEG F |
|
|
|
||||||||||||||||
| p-value | ||||||||||||||||
| ANG | 1.00 | 0.22 | 0.60 | 0.45 | 0.16 | 0.16 | 0.05 | 0.49 | 1.00 | 0.39 | 0.19 | 0.04 | −0.36 | 0.56 | 0.41 | −0.06 |
| MMP3 | 0.39 | 1.00 | 0.60 | 0.22 | 0.99 | 0.30 | 0.08 | 0.18 | 0.13 | 1.00 | 0.60 | −0.10 | −0.03 | 0.71 | 0.02 | 0.06 |
| MMP7 | 0.01* | 0.01* | 1.00 | 0.15 | 0.60 | 0.38 | 0.29 | 0.41 | 0.46 | 0.01* | 1.00 | 0.28 | 0.41 | 0.55 | 0.38 | 0.09 |
| TIMP1 | 0.07 | 0.40 | 0.55 | 1.00 | 0.18 | 0.39 | 0.09 | 0.70 | 0.87 | 0.71 | 0.28 | 1.00 | 0.21 | −0.13 | 0.18 | −0.12 |
| VE-cadherin | 0.54 | 0.00* | 0.01* | 0.48 | 1.00 | 0.30 | 0.08 | 0.13 | 0.15 | 0.91 | 0.10 | 0.41 | 1.00 | −0.14 | 0.21 | 0.52 |
| CXCL1 6 | 0.53 | 0.25 | 0.13 | 0.12 | 0.24 | 1.00 | 0.56 | 0.69 | 0.02* | 0.00* | 0.02* | 0.61 | 0.59 | 1.00 | 0.47 | 0.07 |
| ENA78 | 0.85 | 0.77 | 0.27 | 0.74 | 0.77 | 0.02* | 1.00 | 0.43 | 0.10 | 0.95 | 0.13 | 0.50 | 0.43 | 0.06 | 1.00 | 0.35 |
| VEGF | 0.05 | 0.50 | 0.11 | 0.00* | 0.61 | 0.00* | 0.08 | 1.00 | 0.82 | 0.83 | 0.72 | 0.65 | 0.03* | 0.78 | 0.17 | 1.00 |
Figure 5.
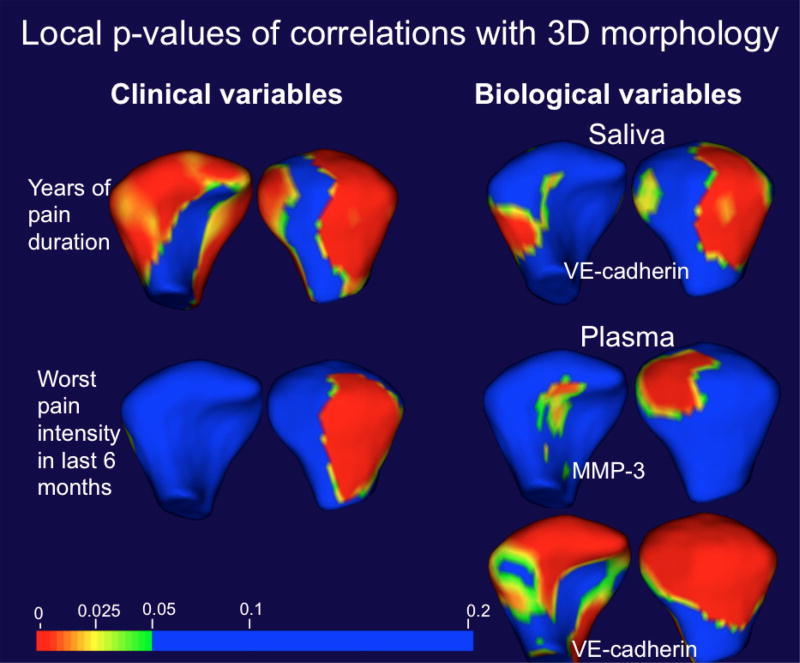
MSFDA statistics showing the local p-value maps of significant correlations of clinical and biological co-variates with morphological variability in the testing dataset.
The statistical analysis of the biological variables, measured by levels of specific proteins in plasma and saliva, revealed that saliva levels of MMP-7 in OA subjects were significantly correlated with ANG and MMP-3, VE-cadherin levels correlated with MMP-3 and MMP-7, ENA-78 levels correlated with CXCL16 and VEGF levels correlated with TIMP-1 and CXCL16. Plasma levels of MMP-7 in OA subjects were significantly correlated with MMP-3; CXCL16 levels were correlated with ANG, MMP-3 and MMP-7; and VEGF levels were correlated with VE-cadherin (Table 4). PCA showed that the percentage of the total variance of protein levels in saliva explained by the first principal component was over 99%. As VE-cadherin levels in saliva (VE-cadherin_S) were highly correlated with the first PC score (pearson correlation = 1.00, p-value=1.71E-41), this variable was included in the MSFDA model. PCA also showed that the percentage of the total variance of protein levels in plasma explained by first two principal components was over 99%. VE-cadherin levels in plasma (VEcadherin_ P) were highly correlated with the first PC score (pearson correlation = 1.000, p-value=5.84E-36); MMP-3 levels in plasma (MMP-3_P) were highly correlated with the second PC score (pearson correlation = 0.758, p-value=5.35E-23). Thus, both VE-cadherin_P and MMP-3_P were included in the MSFDA model and results for global and clustering p-values shown in Table 5, and local–values for clinical and biological variables are shown in Figure 5.
Table 5.
Demographic, clinical and biological co-variates in MSFDA model of correlations with 1002 vertices in condylar 3D meshes
| Co-variates in MSFDA model | global p-value | clustering p-value |
|---|---|---|
| Age | 0.133 | 0.092 |
| Sex | 0.031 | 0.024 |
| worst pain | 0.031 | 0.084 |
| duration of pain in years | 0.04 | 0.054 |
| VE-cadherin levels in saliva | 0.027 | 0.05 |
| MMP-3 levels in plasma | 0.107 | 0.068 |
| VE-cadherin levels in plasma | 0.001 | 0.018 |
5. Discussion
The present study incorporates novel approaches to temporomandibular joint osteoarthritis, conditions that were described by the RDC/TMD consortium in 2010 (Look et al., 2010a, 2010b). To our knowledge, this is the first study to compare phenotypic findings across different degrees of 3D joint degeneration using a neural network classifier that may be augmented and also modified to input diverse patient data into the training database. The DSCI web-based system may aid researchers gain insight into biomarkers to help guide treatment or to predict risk factors of patient specific outcomes. This system stores, runs computing-intensive tasks, and integrates data from different sources. The classification of 3D morphological variation presented in this paper addresses a clinical problem applicable to any research on morphological pathology and can be applied and generalized to multiple projects across institutions.
The patient testing data in the present study was stored in the DSCI web-based system in a structured manner. The imaging variables consisted of geometric features computed at each of the 3D condylar meshes 1002 vertices that are stored into arrays and linked to their corresponding vertices in the 3D meshes. Clinicians use diverse information from initial records for diagnosis and treatment planning. Variability in patient symptoms and imaging findings pose challenges in diagnosing, what leads to frequent disagreement among clinicians. Gaining better insight into patient data improves diagnostic accuracy, and helps clinicians assist in making decisions. Therefore, we need to develop algorithms to discover hidden information in diverse data sources. Data mining and machine learning methods are valuable tools in this regard. Over the past several decades, many algorithms have been proposed and have demonstrated good performance in mining medical data. Decision tree and support vector machines (SVM) are well-known algorithms (Wu et al., 2008). Deep learning has been regarded as the new generation of the neural networks since 2006, and now data analysis faces a new exciting era, in which big data, deep learning, and significant computational power meet (Li, 2018). The deep learning architecture chosen for this study was able to capture complex morphology patterns.
Important advances of the present work compared to our previous Diagnostic Index (Paniagua et al., 2017) were the flexibility of use of different shape features in the SVA classifier and improved shape correspondence by running rigid alignment prior to SPHARM. The differences in classification between the clinicians assessment and the SVA classifier, may be due to the need to increase sample size to improve SVA classification, limitations in the clinicians visual perception, or the fact the registration and correspondence/homology of vertices in the surface meshes affect the computation of shape features in the SVA classifier. Moreover, the addition of simulated data does not increase the diversity inside a group. For the SVA classifier, future increase in training data can add more actual individual patient variability. Due to the number of 3D surface meshes in each group of this study training database, it could have been possible to add more simulated data, but such simulations would not improve the assessments of individual variability. Even if the simulated meshes are not exactly the same as the actual ones, they still have very similar morphologies. Training the neural network with too many similar meshes would over fit those specific meshes that would always be well classified. Then, the neural network would not have been able to properly classify meshes that are variably different but may have a similar stage of condylar degeneration. Our continued goal is to increase the training database sample size to extensively represent the variability possible in a certain group or degree of condylar degeneration. Once this variability is further represented, the SVA precision in classification of meshes will improve with deeper learning of the classification criteria.
Assessment of any new methodology includes testing the precision of its performance, compared to a gold standard. Recently, artificial intelligence (AI) has been touted as a new way to increase productivity by replacing clinician subjective interpretation. But long before AI or machines may replace clinicians, they will be helping us to make smart clinical decisions, so we can provide more precise and personalized treatment and become more productive. This use of technology is called “intelligence augmentation” and because of its imminent and extensive impact, it deserves a closer look (Lavenda, 2017). Whether we call it neural network, artificial intelligence (AI) or intelligence augmentation (IA), these cognitive systems are neither autonomous nor sentient, but they form a new kind of intelligence that has nothing artificial about it. They augment our capacity to understand what is happening in the complex world around us (Rometty, 2017). While artificial intelligence can improve efficiency by replacing humans for focused tasks, it is in the application of machine intelligence to augment human decision-making that the real advances in precise and personalized health care may occur. Understanding the roles machine learning can play is the key to maximizing both artificial and human intelligence in diagnosis and planning of treatment, as well as in assessment of treatment outcomes.
Our long-term goal is to create a repository for Osteoarthritis of the TMJ. Such repository requires maintaining the data in a distributed computational environment to allow contributions to the database from multi-clinical centers and to share trained models for TMJ classification. Both the novel shape statistics computation and the neural network training and classification require the advanced computational power provided by the DCSI web-system in this study. The distributed computation allows executing these computing intensive tasks outside the clinical centers. The web-based system in this study is not only an improved way to display complex data in any databases. It has also been designed to run programs, such as neural networks and advanced statistical programs, using data stored in the DSCI database. Subsets can be created from datasets in projects and the website allows users to create jobs to be run on a remote computing grid, to check the status of job execution, and get the outputs. One of the challenges in this study was to integrate different types of data in the DSCI web-based system. The results of this study demonstrate the feasibility of testing comprehensive phenotypic characterization, including clinical symptoms, 3D morphology and molecular levels of patient specific information. This versatile and robust web-system allows managing many different projects, where each dataset remains independent, so the data can be shared between projects with the same patient ID.
6. Conclusion
The findings of this study demonstrate a comprehensive phenotypic characterization of TMJ health and disease at clinical, imaging and biological levels, using novel flexible and versatile open-source tools for a web-based system that provides advanced shape statistical analysis and a neural network based classification of temporomandibular joint osteoarthritis.
Highlights.
Web-based system for storage, integration and computation of biomedical data.
Deep neural network to classify temporomandibular joint osteoarthritis.
Web-based system provides advanced shape statistical analysis.
Acknowledgments
The authors acknowledge the financial support received from National Institutes of Health (NIH) (grant numbers R01EB021391, R01DE024450, R21DE025306).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
References
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. [July 23, 2017];TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. 2015 http://download.tensorflow.org/paper/whitepaper2015.pdf.
- Abramson SB, Attur M. Developments in the scientific understanding of osteoarthritis. Arthritis Res Ther. 2009;11(3):227. doi: 10.1186/ar2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Hollender L, Anderson Q, Kartha K, Ohrbach R, Truelove EL, et al. Research diagnostic criteria for temporomandibular disorders (RDC/TMD): development of image analysis criteria and examiner reliability for image analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107(6):844–860. doi: 10.1016/j.tripleo.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashinsky BG, Bouhrara M, Coletta CE, Lehallier B, Urish KL, Lin PC, et al. Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J Orthop Res. 2017;35(10):2243–2250. doi: 10.1002/jor.23519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. Discovering the false discovery rate. J R Stat Soc B. 2010:72405–416. [Google Scholar]
- Cevidanes LH, Walker D, Schilling J, Sugai J, Giannobile W, Paniagua B, et al. 3D osteoarthritic changes in TMJ condylar morphology correlates with specific systemic and local biomarkers of disease. Osteoarthritis Cartilage. 2014;22(10):1657–1667. doi: 10.1016/j.joca.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumast PD. [July 13m, 2017];ShapeVariationAnalyzer. 2017 https://github.com/DCBIA-OrthoLab/ShapeVariationAnalyzer.
- Fan J, Gijbels I. Local Polynomial Modelling and Its Applications: Monographs on Statistics and Applied Probability. Vol. 66. Chapman and Hall; London: 1996. [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magnetic Resonance Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LR, Gomes M, Jung B, Paniagua B, Ruellas AC, Goncalves JR, et al. Diagnostic index of 3D osteoarthritic changes in TMJ condylar morphology. Proceedings of SPIE--the International Society for Optical Engineering. 2015;9414 doi: 10.1117/12.2082226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Thompson P, Wang Y, Yu Y, Zhang J, Kong D, et al. FGWAS: Functional genome wide association analysis. Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Eckstein F, Kraus VB, Losina E, Sandell L, Guermazi A. Imaging biomarker validation and qualification report: sixth OARSI Workshop on Imaging in Osteoarthritis combined with third OA Biomarkers Workshop. Osteoarthritis Cartilage. 2013;21(7):939–942. doi: 10.1016/j.joca.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Trundle P, Ren J. Medical image analysis with artificial neural networks. Comput Med Imaging Graph. 2010;34(8):617–631. doi: 10.1016/j.compmedimag.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Koenderink JJ. Solid Shape. MIT Press; Massachusetts: 1990. [Google Scholar]
- Koenderink JJ, Vandoorn AJ. Surface Shape and Curvature Scales. Image Vision Comput. 1992;10(8):557–564. doi: 10.1016/0262-8856(92)90076-F. [DOI] [Google Scholar]
- Lavenda D. How machine learming influences your productivity. [July 13, 2017];Venturebeat. 2017 https://venturebeat.com/2017/05/07/how-machine-learning-influences-your-productivity/
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li R. Data Mining and Machine Learning Methods for Dementia Research. Methods Mol Biol. 2018 doi: 10.1007/978-1-4939-7704-8_25. 1750363-370. [DOI] [PubMed] [Google Scholar]
- Look JO, John MT, Tai F, Huggins KH, Lenton PA, Truelove EL, et al. The Research Diagnostic Criteria For Temporomandibular Disorders. II: reliability of Axis I diagnoses and selected clinical measures. J Orofac Pain. 2010a;24(1):25–34. [PMC free article] [PubMed] [Google Scholar]
- Look JO, Schiffman EL, Truelove EL, Ahmad M. Reliability and validity of Axis I of the Research Diagnostic Criteria for Temporomandibular Disorders (RDC/TMD) with proposed revisions. J Oral Rehabil. 2010b;37(10):744–759. doi: 10.1111/j.1365-2842.2010.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu I, Kim SH, Seong JK, Yoo SW, Evans AC, Shi Y, et al. Group-wise cortical correspondence via sulcal curve-constrained entropy minimization. Inf Process Med Imaging. 2013:23364–375. doi: 10.1007/978-3-642-38868-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters T. Practical neural network recipes in C. Academic Press; Boston: 1993. [Google Scholar]
- Mirabel C. [July 12, 2017];DatabaseInteractorExtension. 2017 https://github.com/DCBIA-OrthoLab/DatabaseInteractorExtension.
- Paniagua B, Pascal L, Prieto J, Vimort JB, Gomes L, Yatabe M, et al. Diagnostic Index: An open-source tool to classify TMJ OA condyles. Proc SPIE Int Soc Opt Eng. 2017:10137. doi: 10.1117/12.2254070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JC, Paniagua B, Yatabe MS, Ruellas ACO, Fattori L, Muniz L, et al. Federating Heterogeneous Datasets to Enhance Data Sharing and Experiment Reproducibility. Proc SPIE Int Soc Opt Eng. 2017:10137. doi: 10.1117/12.2254689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhukov T, Song D, Tockman MS. Computerized analysis of cellular features and biomarkers for cytologic diagnosis of early lung cancer. Anal Quant Cytol Histol. 2007;29(2):103–111. [PubMed] [Google Scholar]
- RayBiotech. [July 12, 2017];Quantibody multiplex Elisa array. 2017 https://www.raybiotech.com/quantibody-multiplex-elisa-array/
- Rometty G. IBM CEO: AI will be man and machine, not man vs. machine. [july 18, 2017];Wall Street Journal. 2017 http://www.wsj.com/video/ibm-ceo-ai-will-be-man-and-machine-not-man-vs-machine/74769095-C3C1-4173-9269-AAE812B0D340.html.
- Schilling J, Gomes LC, Benavides E, Nguyen T, Paniagua B, Styner M, et al. Regional 3D superimposition to assess temporomandibular joint condylar morphology. Dentomaxillofac Radiol. 2014;43(1) doi: 10.1259/dmfr.20130273. 20130273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman SV. Selecting and interpreting measures of thematic classification accuracy. Remote Sens Environ. 1997;62(1):77–89. doi: 10.1016/S0034-4257(97)00083-7. [DOI] [Google Scholar]
- Su N, Liu Y, Yang X, Luo Z, Shi Z. Correlation between bony changes measured with cone beam computed tomography and clinical dysfunction index in patients with temporomandibular joint osteoarthritis. J Craniomaxillofac Surg. 2014;42(7):1402–1407. doi: 10.1016/j.jcms.2014.04.001. [DOI] [PubMed] [Google Scholar]
- vtk.org. [July 24, 2017];vtkPerlinNoise Class Reference. http://www.vtk.org/doc/nightly/html/classvtkPerlinNoise.html.
- Wu XD, Kumar V, Quinlan JR, Ghosh J, Yang Q, Motoda H, et al. Top 10 algorithms in data mining. Knowl Inf Syst. 2008;14(1):1–37. doi: 10.1007/s10115-007-0114-2. [DOI] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zhu H, Ibrahim JG, Tang N, Rowe DB, Hao X, Bansal R, et al. A Statistical Analysis of Brain Morphology Using Wild Bootstrapping. Ieee Transactions on Medical Imaging. 2007;26(7):954–966. doi: 10.1109/TMI.2007.897396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Li R, Kong L. Multivariate Varying Coefficient Model for Functional Responses. Ann Stat. 2012;40(5):2634–2666. doi: 10.1214/12-AOS1045SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]