Summary
To initiate V(D)J recombination for generating the adaptive immune response of vertebrates, RAG1/2 recombinase cleaves DNA at a pair of recombination signal sequences, the 12- and 23-RSS. We have determined crystal and cryoEM structures of RAG1/2 with DNA in the pre-reaction and hairpin-forming complexes up to 2.75Å resolution. Both protein and DNA exhibit structural plasticity and undergo dramatic conformational changes. Coding-flank DNAs rotate, shift and deform extensively for nicking and hairpin formation. Two intertwined RAG1 subunits crisscross four times between the asymmetric pair of severely bent 12/23-RSS DNAs. Location-sensitive bending of 60° and 150° in 12- and 23-RSS spacers, respectively, must occur for RAG1/2 to capture the nonamers and pair the heptamers for symmetric double-strand breakage. DNA pairing is thus sequence-context dependent and structure specific, which partly explains the “beyond 12/23” restriction. Finally, catalysis in crystallo reveals the process of DNA hairpin formation and its stabilization by interleaved base stacking.
Keywords: DNA bending, CA/TG dinucleotides, polypurine tract, NBD, DNA hairpin, SCID
Introduction
The RAG1/2 recombinase initiates V(D)J recombination as an essential step in generating the adaptive immune systems of vertebrates (Gellert, 2002; Sakano et al., 1979). The antigen-binding surface of immunoglobulins or T-cell receptors is formed by a pair of variable domains encoded by V, D and J gene segments of heavy and light chains. Combinatorial assembly from 4–200 variations of each gene segment, imprecise rejoining of cleaved DNA, and stochastic pairing of heavy and light chains, result in the capability of the adaptive immunity system to recognize up to billions of different antigens (Schatz and Swanson, 2011). V, D and J segments are bordered by bipartite recombination signal sequences (RSS), which consist of conserved 7 and 9 bp sequences (heptamer and nonamer) separated by a relatively non-conserved spacer of 12 or 23 bp, thus known as 12- or 23-RSS (Akira et al., 1987; Ramsden et al., 1994). V(D)J recombination operates between pairs of sites with one 12RSS and one 23RSS (the 12/23 rule) thus joining V and J gene segments of light chains, and D and J or V and D of heavy chains. The 12/23 rule is strictly controlled by the RAG1/2 recombinase, which is necessary and sufficient for pairing one RSS DNA of each type and double-strand cleavage of both (Lapkouski et al., 2015; Ramsden et al., 1997). However, in the T-cell receptor (TCR) β and δ loci joining of V and J gene segments bordered by the 23- and 12-RSS, respectively, do not occur, and an intervening D segment has to be joined to J before V can be joined to DJ (Banerjee and Schatz, 2014; Bassing et al., 2000; Jung et al., 2003). This is described as the “beyond 12/23” restriction, for which the mechanism is unclear.
Of the 1040 and 527 residues of mouse RAG1 and RAG2, an essential catalytic core encompasses residues 384-1008 of RAG1 and 1-387 of RAG2 (Sadofsky et al., 1994; Sadofsky et al., 1993; Silver et al., 1993). The remaining N- and C-terminal regions serve regulatory roles and ensure that the potentially detrimental double-strand breaks occur only as programmed (Bellon et al., 1997; Grundy et al., 2010; Matthews et al., 2007; Ramon-Maiques et al., 2007; Schatz and Swanson, 2011; Singh and Gellert, 2015). With either the intact proteins or catalytic cores, RAG1/2 first hydrolyzes one strand to generate a nick with a 5´-phosphate on the RSS DNA and 3´-OH on the coding flank, and cleaves the second strand by strand transfer using the 3´-OH product of the first reaction as the nucleophile, thus producing a hairpin end on the coding flank and a blunt end on the RSS DNA (McBlane et al., 1995). These two reactions catalyzed by RAG1/2 occur consecutively in a single active site. After cleavage, RAG1/2 releases the coding segments for processing and ligation by non-homologous end joining (Boboila et al., 2012; Deriano and Roth, 2013).
The crystal structure of mouse core RAG1/2 revealed a Y-shaped dimer of RAG1/2 (Kim et al., 2015). Core RAG1 is structurally modular. From N- to C-terminus, it is made up of the NBD (nonamer binding domain), which is a domain-swapped dimer forming the stem of the Y, DDBD (dimerization and DNA binding domain), pre-RNH, RNH (Ribonuclease H-like), the Zn-binding ZnC2 and ZnH2 domains, and the C-terminal domain (CTD), which folds back to interact with the DDBD at the branch point of the “Y”. The catalytic residues, D600, D708 and E962 (Fugmann et al., 2000; Kim et al., 1999; Landree et al., 1999), reside in the RNH domain. The core of RAG2 is composed of a six-bladed β-propeller (Callebaut and Mornon, 1998), which sits at the top of each “Y” arm formed by the pre-RNH, RNH and Zn-binding domains of RAG1 (Grundy et al., 2009). Although the bulk of the RAG1/2 dimer follows a two-fold symmetry, the NBDs (aa 384-457) have a different dyad axis and are tilted toward one arm of the “Y” (Kim et al., 2015). Asymmetry of the RAG1/2 dimer with its off-centered NBDs is necessary to accommodate the 12- and 23-RSS pair of DNAs (Lapkouski et al., 2015).
Shortly after the crystal structure was determined, cryoEM structures of zebrafish RAG1/2 complexed with nicked or fully cleaved RSS DNAs were reported, reaching 3.4 Å resolution for symmetrized RAG1/2 dimer and 12/23-RSS DNAs that excluded the NBDs and the nonamers of the DNAs. These cryoEM structures revealed a large closing motion of the two RAG1/2 arms upon DNA binding, and severely distorted nicked RSS DNAs (Ru et al., 2015). However, the NBDs and nonamer DNAs were poorly defined in the non-symmetrized cryoEM structures, due to 7 Å resolution, and this region was modeled based on an earlier crystal structure of the NBD-nonamer complex, in which a crystallographic dyad axis symmetrizes the two halves of each nonamer segment (Yin et al., 2009). Consequently, the mechanism underlying the 12/23 rule that governs DNA pairing and cleavage in V(D)J recombination remained unresolved.
Here we report crystal structures up to 2.75 Å resolution of mouse core RAG1/2 in complex with both 12- and 23-RSS DNAs, either intact or with one or both DNAs nicked at the coding-heptamer border. These structures reveal the tracing of a complete RAG1/2 dimer and asymmetric recognition of 12- and 23-RSS DNAs. Dramatic DNA movement and three-part conformational changes of RAG1/2 occur from the apo form and from initial RSS-DNA binding to forming the hairpin products. Surprisingly, RAG1/2 uses the same protein surface to bind the major or minor groove of DNA equally well. We find that the conservation of the heptamer is largely determined by DNA deformability rather than hydrogen bonds with RAG1/2. Localized DNA kinks in the DNA spacers influence 12- and 23-RSS pairing and cleavage, and thus form a structural basis for the “beyond 12/23” restrictions. By cleaving DNA in crystallo (Nakamura et al., 2012; Samara et al., 2017), we have observed the process of DNA hairpin formation and also the hairpin structure with interleaved base stacking. Finally, we also have determined the cryoEM structure of mouse RAG1/2 complexed with nicked 12- and 23-RSS DNAs at 3.17 Å, validating the asymmetric crystal structure and showing that the 150° bend of the 23RSS spacer is facilitated by the connected A and B boxes of the accessory protein HMGB1, known to stimulate RAG cleavage (van Gent et al., 1997).
Results
Crystal and EM structures of mouse RAG1/2 complexed with 12/23-RSS DNAs
Our initial crystals of mouse RAG1/2 complexed with fully cleaved 12- and 23-RSS DNAs excluded the DNA due to lattice contacts formed by the proteins (Kim et al., 2015). By including the accessory DNA-binding protein HMGB1, and by varying DNA coding flank lengths on the 12- and 23-RSS DNAs, we obtained crystals of mouse RAG1/2 in complex with 12/23-RSS DNAs, either with both RSSs intact or with one or both nicked at the coding-heptamer border. These intact/intact, intact/nicked, and nicked/nicked forms, with different crystal lattices, diffract X-rays to 4.2 Å, 3.15 Å and 2.75 Å resolution, respectively (Table S1). The structures were determined by molecular replacement using the 3.2 Å apo-RAG1/2 structure (PDB: 4WWX) (Kim et al., 2015) (Fig. 1).
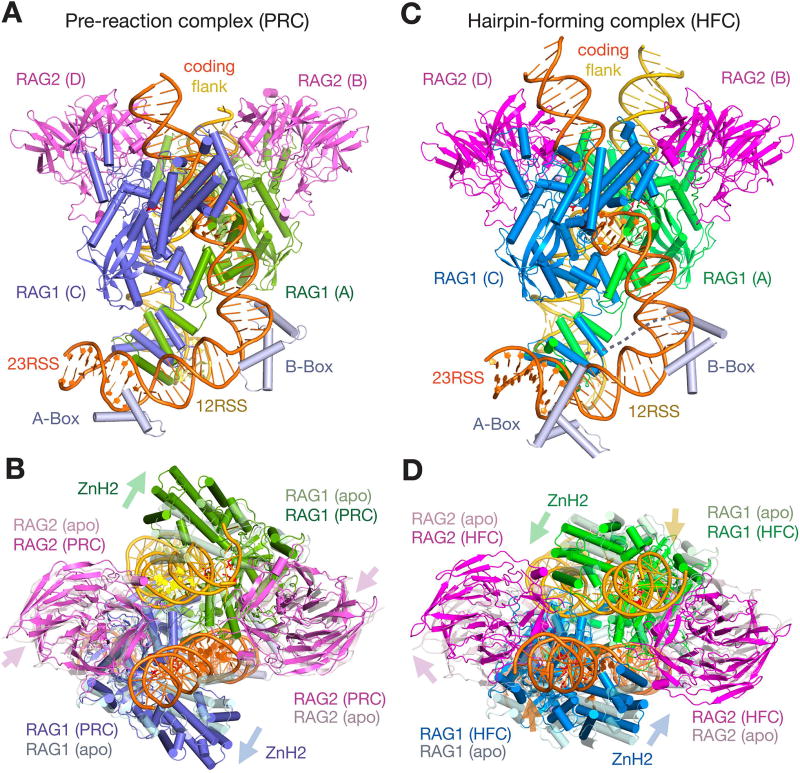
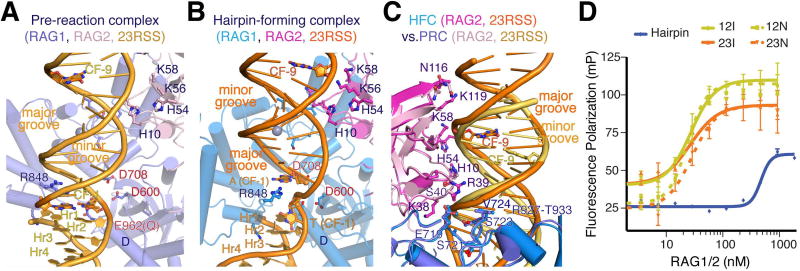
Figure 1.
Structures of RAG1/2 PRC and HFC complexes with 12/23-RSS DNAs. A, B. The front (A) and top (B) views of pre-reaction complex (PRC). C, D. The front (C) and top (D) views of hairpin-forming complex (HFC) are shown after superposition of their DDBD domains. Protein and DNA are color coded. Zn2+ ions are shown as grey spheres, and the catalytic carboxylates are shown as red sticks. Both PRC and HFC are compared to the apo structure (shown in light and semi-transparent colors) in B and D, after superposition of DDBD domains. Movement of RAG2 and the ZnH2 domains relative to the apo form is indicated by color-coded arrows. Compared to the PRC, the coding flank DNAs in HFC also move as indicated by yellow (12RSS) and orange (23RSS) arrows.
The resulting electron density map of the nicked/nicked DNA complex allowed unequivocal tracing of 28 and 39 bp of the 12- and 23-RSSs and 9–15 bp of each coding flank DNA, and structure refinement of two RAG1 (aa 385-1007 and 391-1008), two RAG2 (aa 1-350), and one HMGB1 chain in each asymmetric unit to R and Rfree of 20.0 and 23.4% (Table S1). We have also determined the equivalent cryoEM structure at 3.17 Å (see Methods, Table S2, Fig. S1). Compared to the EM structure, the crystal structure is slightly more compact as if dehydrated, and NBDs and DNA ends are better defined in the crystal structure due to reduced mobility (Fig. S2A). In both the crystal and EM structure, the nicked RSS DNAs are poised in the active sites for the second strand cleavage and hairpin formation. Therefore, these structures are referred to as the “hairpin-forming complex” (HFC) from here on. Compared to the structure of zebrafish RAG1/2-RSS complexes (Ru et al., 2015), both NBDs and nonamers are finally resolved at 2.75 Å resolution and the curvatures of the spacer DNAs are unequivocally defined. In addition, details of DNA local structures and protein-DNA interactions deviate from the cryoEM structure of the corresponding zebrafish complex, probably due to the improved resolution (Fig. S3).
The crystal structure of an intact/intact RAG1/2-RSS complex was obtained with 10 and 14 bp coding flanks on 12- and 23-RSS DNA, respectively. Although this structure is resolved only to 4.2 Å (Table S1), it is clear that the RAG1/2 protein, particularly the ZnH2 domain, is much more expanded from the core than in the HFC (Fig. 1). But the scissile phosphates for hydrolysis are ∼20 Å away from the catalytic centers, and severe distortions of DNA will be necessary to engage them for the cleavage reactions. Therefore, this structure represents DNA captured before chemical reaction and is hereafter referred to as a “pre-reaction complex” (PRC). Even when a nick was introduced to either 12- or 23-RSS DNA (forming a nicked/intact or intact/nicked pair, respectively), the resulting complexes have essentially the same structures as the intact/intact DNA complex, although with better diffracting crystals and improved resolution extending to 3.15 Å (Table S1, Fig. S4). Moreover, when the complexes of RAG1/2 with intact/intact 12/23 RSS DNA were examined using cryoEM, the majority of particles belonged to this pre-reaction state (data not shown), which must be prevalent and stable. The highest resolution PRC structure contains intact/nicked 12/23-RSS DNAs and is used for structural analysis hereafter.
Overall structures of pre-reaction and hairpin-forming complexes
In both PRC and HFC structures, the RAG1/2 dimer retains the overall Y shape of the apo protein (Fig. 1). Binding of RSS DNA induces the RAG1/2 arms of the Y to pivot, and the NBDs at the stem of the Y to tilt significantly toward one arm (Fig. 2A). Moreover, each RAG1/2 arm is hinged into two parts, as the ZnH2 domain (aa 794-927 of RAG1) in each arm swings out in the pre-reaction state and then swings back in the hairpin-forming complex (Fig. 1B, 1D, 2). Compared to the “open” apo-protein structure, where the two RAG1/2 arms are separated by a gap, the gap appears even wider in the PRC, and disappears in the HFC, with the two RAG1/2 arms hugging each other (Movies 1–4).
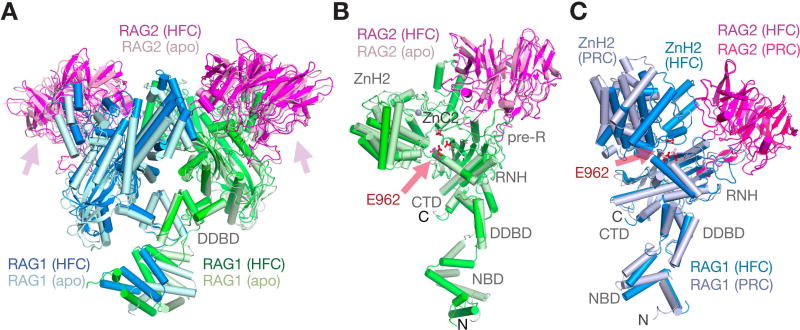
Figure 2.
Structural rearrangement of RAG1/2. A. Superposition of the Y-shaped RAG1/2 dimer in apo (light colors) and HFC (darker colors) by their DDBD domains shows the large rotational movement of NBD domains at the stem and also movement in both arms. B. Structure comparison of a single RAG1/2 heterodimer (A and B subunit as an example) between the apo form and HFC. After superposition of RAG2 (B subunit), the pre-RNH, RNH, ZnC2 domains of RAG1 are also superimposable, but ZnH2 rotates by 9° and shifts by 2.6 Å, and the NBD rotates by ∼40° and shifts by 10Å. C. Large movement of ZnH2 between HRC and HFC. The RAG1/2 (C and D subunit as an example) is shown after superimposing RAG2. Movement of helix D with E962 (marked by a pink arrow) of RNH is obvious.
The 28 and 39 bp of the recombination signal regions of the 12- and 23-RSS DNA substrates are nearly superimposable in the PRC and HFC, except for the flipping out of the first C and an increased kink between the last two nucleotides (TG) of the heptamers in the HFC (Fig. 3). These 12- and 23-RSS DNAs bind to a RAG1 dimer on the outside of the stem and lower one third of the Y arms similarly, except that the extra 11 bp of the 23RSS spacer are bulged out (Fig. 1A, C). RAG1 acts like a zipper and crisscrosses between the two RSS DNAs, so that each RSS is bound by both RAG1 subunits (Fig. 4A, B). The two nonamers bound by the NBDs are almost anti-parallel, but the heptamers bound near the RAG1 active sites are nearly parallel (with a mere ∼30° crossing angle) (Fig. 3). The DNA directional change is accomplished by the 90° bend in the middle of the 23RSS spacer that is aided by the HMGB1 protein (see below), and ∼60° bending in each RSS spacer adjacent to the nonamer. The dyad axis relating the NBDs is at a ∼60° angle to the dyad relating the two RAG1/2 arms.
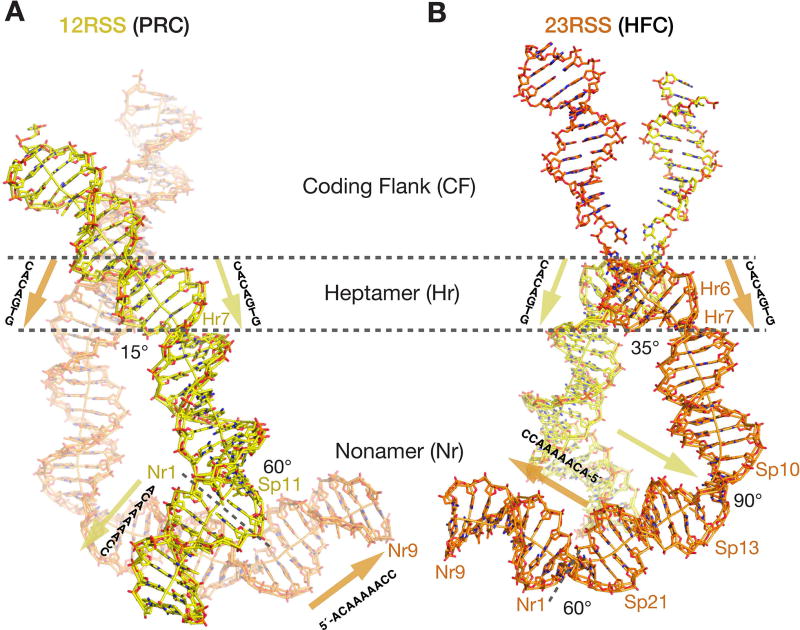
Figure 3.
The overall curvature of 12/23-RSS DNAs. A, B. Stick representation of the RSS DNAs in the PRC with 12RSS (yellow) in the front (A) and the HFC with 23RSS (orange) in the front (B). The borders separating the coding flank (CF), heptamer (Hr1 to Hr7), spacer (Sp) and nonamer regions (Nr1 to Nr9) are marked by dashed lines. The DNA helical axes calculated by Curves5 (Lavery et al., 2009) are also shown. Angles of sharp bends are marked, and locations of kinks are labeled (Hr7, Sp11 on 12RSS, and Sp10-13 and Sp21 on 23RSS). The conventional 5´ to 3´ directions of the heptamers (nearly parallel) and nonamers (nearly antiparallel) are marked by yellow and orange arrowheads, respectively.
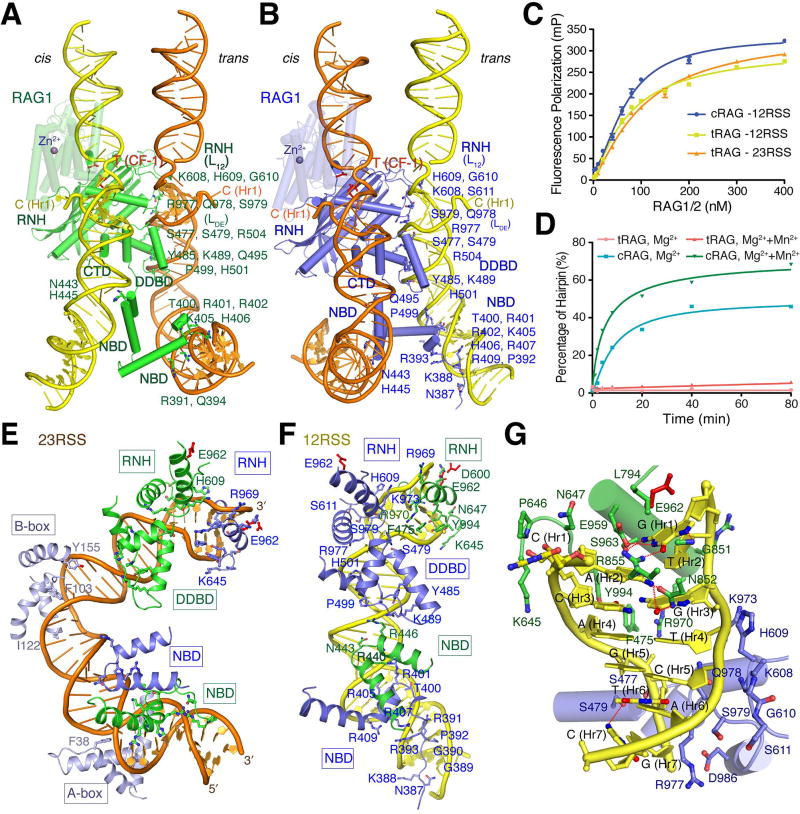
Figure 4.
RSS DNA recognition. (A and B) 23RSS is dominantly bound by NBD, DDBD and RNH of RAG1-A (green) (shown in A), but cleaved by RAG1-C (blue), while 12RSS is dominantly bound by RAG1-C (blue) (shown in B) and cleaved by RAG1-A (green). ZnH2 domain is shown in semi-transparency. The flipped-out C (H1) and T (CF-1) are labeled. C. DNA binding by tRAG1/2 is similar to cRAG1/2 and unaffected by removing the NBD domain. Pre-nicked 12- or 23-RSS DNAs were fluorescence-labeled (6-FAM). D. Hairpin formation by tRAG1/2 that lacks the NBD is abolished, and an addition of 2 mM Mn2+ cannot rescue the tRAG1/2 recombinase activity. (E and F) Recognition of 23RSS (orange) (E) and 12RSS (yellow) (F). RAG1 domains are color coded according to subunits A (green) and C (blue). Heptamers and nonamers are shown with sugar and base rings, while the spacer DNAs are shown as ladders only. G. A close-up view of the 12RSS heptamer recognition.
The coding flank DNAs occupy the upper two thirds of the Y arms and are surrounded by both RAG1 and RAG2 (Fig. 1). In transforming from the pre-reaction to the hairpin forming complex, each coding flank DNA moves dramatically, rotating ∼180° and translating ∼8Å upward around the helical axis, with the bases adjacent to the heptamers now poised to form a hairpin loop (Fig. 3). As a result of this DNA transformation, the same RAG1/2 surface contacts the major groove in one complex but the minor groove in the other.
Structural changes in RAG1/2
Three parts of the RAG1/2 dimer are mobile. Relative to the DDBD domains, which form the pivot point in the center of the Y, the main part of each Y arm consisting of the catalytic regions of RAG1 (pre-RNH, RNH and ZnC2) together with the entire RAG2 rotates en bloc by 5 to 11° in different directions in the two DNA complexes compared to the apo form (Fig. 1B, D). In addition, the NBDs at the stem of the “Y” rotate ∼40° between the apo and DNA-bound forms with the centroid shifting by over 10 Å. The NBDs become much more tilted in both DNA complexes (Fig. 2A). Although only six residues in length (aa 457-462), the linker between NBD and DDBD can span 6.5 Å to 15.6 Å when bound to 12- or 23-RSS DNA, respectively, to accommodate the asymmetry of RAG1/2-RSS complexes. These flexible linkers also result in slightly different tilt of the Y stem among different RAG1/2-DNA structures.
NBDs and nonamer regions easily become blurred or even “disappear” in the EM structures (Ru et al., 2015) due to their concerted flexibility. However, nonamer recognition by the NBD is absolutely necessary for RAG1/2 function (Fig. S5). We found that when the NBD (aa 383-458) is deleted from the RAG1 core, the truncated RAG1/2 (tRAG1/2) binds to 12/23-RSS DNAs with an affinity similar to the core RAG1/2, but fails to cleave the DNAs at the heptamer and coding flank junction in the presence of Mg2+ (Fig. 4C, D). Even with an addition of Mn2+, nicking and hairpin formation by tRAG1/2 is barely detectable (Fig. S5C).
In addition to the en bloc movements, each helical ZnH2 domain, which binds the coding flank and heptamer surrounding the DNA cleavage sites, opens by a 25° rotation and 13 Å centroid movement within the Y arm (Fig. 1B, D) to allow initial binding of undistorted RSS DNAs, and then closes by rotating ∼16° to bring the pair of nicked coding flank DNAs closer, with significant DNA rotation, translation and deformation in the hairpin-forming complex (Fig. 3). Even between the apo form and the HFC, each ZnH2 domain rotates by 9–10° outwards with a 2.6 to 2.9 Å shift of the centroid relative to the main part of each Y-arm to accommodate the DNAs (Fig. 2B–C). These ZnH2 movements, which are necessary for binding and orienting the DNA substrates for the cleavage reactions, were not noted in the previous zebrafish RAG1/2 structures (Ru et al., 2015).
Structure of the 12/23-RSS DNAs and HMGB1 protein
The 12- and 23-RSS DNAs are generally in B-form with local distortions around the nicked cleavage sites, and a ∼60° kink in the 12RSS and a total of ∼150° curvature in the 23RSS DNA (Fig. 3). DNA bending is necessary because RAG1/2 binding shortens the end-to-end distance by 15Å and 55Å on the 12RSS (28 bp) and 23RSS (39 bp), respectively. In all RAG1/2-DNA complexes, the 23RSS is bent nearly 90° towards the major groove at 10–13 bp from the heptamer border (in the middle of the spacer) and stabilized by the B-box of HMGB1 protein, which approaches the RAG1/2 complex from outside (Fig. 1A, C). In the HFC, the A-box of HMGB1 binds the 23RSS DNA one helical turn away at the border with the nonamer, where the DNA is bent an additional ∼60°. In the PRC, the A-box is shifted by 4 Å along the DNA minor groove due to crystal lattice contacts with a neighboring RAG1, but the sharp bend 1 bp into the spacer from the nonamer border remains unchanged (Fig. 3). A similar 60° bend is also observed in the 12RSS DNA 1 bp into the spacer (Fig. 3). Adjacent to the bent 12RSS, weak electron density for a low-occupancy second HMGB1 is also observed, but it is insufficient to build a reliable model. The 12- and 23-RSS DNAs are superimposable from the beginning of the nonamer to 5 bp into the spacer regions (Fig. S6). The 60° bend in the 12RSS is mainly stabilized by the contiguous protein-DNA interaction made by the NBDs, DDBDs and RNHs (Fig. 4F) and probably occurs without the aid of HMGB1 in solution (Grundy et al., 2009; van Gent et al., 1997).
To distinguish the roles of the A- and B-box of HMGB1 on the 23RSS, hairpin formation catalyzed by RAG1/2 was assayed with each DNA-binding box of HMGB1 alone, or two linked boxes at the molar ratio of 1:1 for a 12/23-RSS pair of DNAs (Fig. S2C). The presence of an A- or B-box alone barely affects RAG1/2 catalysis, but the two linked boxes of HMGB1 stimulate hairpin formation. This is likely due to the increased avidity of joined A- and B-boxes and the necessity for the B-box to stabilize the 90° bend in 23RSS, which has no contact with RAG1/2. The ∼50 Å distance between the end of the A-box (K76) and the beginning of the B-box (R97) can be readily spanned by the 20-residue linker between them. Although electron density resulting from X-ray diffraction for the linker is weak, in the cryoEM map at 3.9Å resolution, the linker threads through the inside of the DNA bulge, between RAG1 and the 23RSS (Fig. S2B), which is reminiscent of the binding of TFAM (an HMGB homolog) to the promoter DNA in the mitochondrial transcription initiation complex (Hillen et al., 2017).
RAG1 dimer crisscrosses between 12/23-RSS DNAs
Each RSS DNA is bound by both RAG1 subunits, and each RAG1 crosses between the DNA pair four times. DNA recognition and cleavage occur in trans, as a RAG1 subunit predominantly binds one RSS but cleaves the other (Fig. 4A–B). The conserved heptamer and nonamer regions of the 12- and 23-RSS have the same interactions with RAG1, and even the 12RSS spacer interacts with RAG1 similarly to the first 7 and last 5 bp of the 23RSS spacer (Fig. 4E–F, S6). Thus, the interactions between RAG1/2 and 12RSS that are detailed below are also applicable to the 23RSS. The interactions with the nonamer and spacers are also conserved in the pre-reaction and hairpin-forming complexes.
Nonamer recognition starts from tight minor groove binding by the N-terminal extended peptide of NBD-C (aa 383-393, C subunit of RAG1 as labeled in Fig. 1) (Fig. 4F). The bases 5´-AAC-3´ at the end of the nonamer are recognized by the mainchain polar atoms of K388, G389, G390 and R391. The entire sidechain of R391 is buried deeply in the characteristically narrow minor groove of the five consecutive A/T pairs in the middle of the nonamer. After tracking the phosphosugar backbone of the T5 strand from R393 to T400, the NBD adopts a long and kinked helical structure (aa 400-424), which recognizes the A/T and C/G pairs at the beginning of the nonamer in the major groove. R402 forms the typical bifurcated hydrogen bonds with the guanine base, and the sidechain of H406 forms van der Waals contacts with the thymine adjacent to the guanine. The major groove between the nonamer and the spacer is compressed where R407 contacts the backbone of the T5 strand, and R401, K405 and K412 interact with three consecutive phosphate groups of the spacer on the other side.
The 12RSS spacer from nonamer to heptamer contacts NBD-C (detailed above), NBD-A (A subunit), and DDBD-C in the major, minor and major groove, respectively (Fig. 4F). The minor groove of the spacer region is bound by the domain-swapped NBD-A (aa 426-457), and N443 on the turn between two α-helices interacts with a DNA base. Next to it, the three-helix bundle of DDBD-C (aa 463-517) interacts with the major groove framed by the spacer and heptamer. The third α-helix of DDBD roughly parallel to the DNA bases in the major groove is anchored by H501 and R504, which interact with the surrounding phosphate backbones. The preceding α-helix pushes the heptamer backbone with S477 and S479 on one end, and reaches to the spacer backbone on the other side with H485, K489 and Q495 (Fig. 4E–F). Two intertwined RAG1 subunits are sandwiched between 12- and 23-RSS DNAs (Fig. 4A–B). The NBD and DDBD bound to 12RSS contact each other, but not those bound to 23RSS (Fig. 4E–F). Although not sequence specific, the extended interactions with DNA backbones lead to expanded minor grooves and a compressed major groove, forming the 60° bend in the 12RSS spacer immediately next to the nonamer.
Each heptamer is contacted by both RAG1 subunits mostly in the minor groove (Fig. 4G) and undergoes a slight rotation between the PRC and HFC, concurrent with the ZnH2 movement (Fig. 1, 2). The 4 bp of the heptamer adjacent to the 12RSS spacer region (AGTG) are contacted across the minor groove by S477 and S479 of the DDBD (see above) and two loops coming from the trans subunit, K973-S979 (after the catalytic residue E962) and K608-S611 (after D600). R977, supported by salt bridges with D986 and π-cation stacking with Y982, is buried deep in the minor groove of the 6th and 7th bp of the heptamer (TG consensus), and causes a large roll angle between them and a 15° and 35° bend in PRC and HFC, respectively (Fig. 3, 4E–G).
The first 3 bp of the heptamer (CAC) adjacent to the coding flank interact with only the cis RAG1 subunit that cleaves it, and have very different interactions in the two RAG1/2 complexes. In the PRC, the CAC nucleotides are continuous with the coding flank without noticeable helical distortion, and all interactions with RAG1, including the R848-R855 loop of ZnH2 and the catalytic residue E962 and N965 on helix D (aa 958-975), occur in the minor groove with little base contact (Fig. 5A). In the HFC, both CACs are severely distorted; the first C is unpaired with the base completely flipped out of the helix (Fig. 4G). The RAG1 cleavage subunit straddles the nicked heptamer. The ZnH2 domain presents G851-R855 for major groove interactions with the CAC. The mainchain of G851 and sidechains of R855, E959 and S963 interact with the unpaired first G, and N852 makes bifurcated hydrogen bonds with the next two base pairs (AC) across the duplex (Fig. 4G). RAG1 also contacts the minor groove with E962, S963, K966, R969 and R970 along helix D and the loop of K645 to E649. The flipped-out C is stabilized by the loop of K890 to C902.
Figure 5.
Coding flank recognition. A, B. Coding flanks in the PRC (A) and HFC (B) are bound by RAG1-C (blue) and RAG2 (pink) in the same Y-arm. The coding flank in the HFC is rotated by ∼180° and translocated by ∼ 8Å compared to the PRC, driven by the movement of ZnH2 and RAG2. 23RSS DNA, RAG1-C and RAG2-D are shown after superimposing RAG2-Ds. R848 is buried in the minor groove in the PRC (A), but interacts extensively with the beginning of the hairpin forming coding flank (CF-1) nucleotides in the major groove in the HFC. (B). Both CF-1 and one CF-9 are shown as sticks for reference. C. Superimposed structures of PRC and HFC reveal that the same protein surface contacts the major groove in the HFC (orange) and the widened minor groove in the PRC (yellow-orange). D. Fluorescence anisotropy-based DNA-binding analysis of RAG1/2. RAG1/2 binds intact and nicked 12- and 23-RSS DNAs equally well, but binds about 20-fold weaker to the hairpin product.
In summary, the heptamer structure is markedly distorted in the hairpin-forming complex, with under winding, reduced base stacking, large roll angles, and close contact between adjacent base pairs in the major groove (Fig. 3, 4). With limited protein-DNA base interactions, the strong sequence conservation in the heptamer appears to be due to the ability of CA and TG dinucleotides to maintain the distorted duplex structures.
Dramatically different coding flank interactions
Coding-flank DNAs bind the inside of the Y-shape RAG1/2. The first 5 bp nearest the heptamer are bound by RAG1 only, and the rest contact RAG2 (Fig. 5). In the PRC, the ZnH2 domain on each Y-arm twists open to accommodate the coding-flank DNA (Fig. 1B, Movies 1–2), while the rest of the RAG1/2 arm remains similar to the apo structure. The non-sequence-specific interactions between the DNA and RAG1/2 leave the DNA in a relaxed B-form. In contrast, in the HFC, concurrently with the Y-arm’s en bloc rotation and the closing movement of ZnH2 subdomains, each coding flank is rotated ∼180° in the unwinding direction and translocated 8Å away from the heptamer (Fig. 5A–B, Movies 3–4). The dramatic conformational changes of DNA together with flipping out the first base in the coding flank, which is attached to the scissile phosphate, provides the space for the 3´-OH of the nicked strand to bend toward the scissile phosphate for hairpin formation in the active site (Fig. 5B). The requirement of separating the first base pair in the coding flank for hairpin formation favors T (thymine) for flipping out with little protein contact, and the unpaired A for strong base stacking with the rest of the coding flank. The distal end of the coding flank leans on RAG2 (N116 to K119) (Fig. 5C), while RAG2 is supported by ZnH2 on the other Y-arm as the two Y-arms close the gap between them (Fig. 1D).
As a result, a single patch of RAG1/2 surface, if it contacts the major groove of coding-flank DNA in the PRC (or HFC), interacts with the minor groove in the HFC (or PRC), and vice versa (Fig. 5A–C). In the HFC, R848 approaches DNA in the major groove by forming π-cation stacking with the base of the attacking nucleophile (3´-OH) and charge-charge interactions with the scissile phosphate opposite the two divalent cations in the active site (Fig. 5B), thus playing a crucial role in orienting the substrate for catalysis. In the PRC, R848 adopts two alternative conformations and instead is inserted deeply in the minor groove (Fig. 5A). At the same time, two RAG1 loops (aa 720-726 and 927-933) bind the first 5 bp of the coding-flank in the major groove (in PRC) or the minor groove (in HFC) (Fig. 5C).
Beyond the first 5 bp, RAG2 dominates the coding flank binding. Whether it is the major groove in the HFC or the slightly widened minor groove in the PRC, R39 together with K38 and S40 in a 310 helical turn contacts the DNA backbone 6–8 bp from the heptamer, and H10 and H54 to K58 contact the other DNA backbone at 8–10 bp from the heptamer (Fig. 5C). The role of R39 in supporting the conformational changes of the coding flank explains why the R39G mutation and the nearby C41W lead to human SCID (Piirila et al., 2006).
Despite the dramatically different protein-DNA interactions in the PRC and HFC, the binding constants of core RAG1/2 for the intact and pairwise-nicked RSS DNAs are both in the 25nM range (Fig. 5D). This is probably because in the severely distorted nicked form, the coding flank interacts more extensively with RAG1/2, involving more than 12 bps.
Hairpin forming and strand transfer reaction
Not only do the NBD domains and Y-arms of RAG1/2 rearrange upon binding DNA, but the three catalytic residues undergo significant conformational changes (Fig. 6, S7). In the PRC, without the scissile phosphate captured in the active site, the catalytic residue E962 is dislocated as in the apo form (E962Q mutant was used in the PRC) (Movie 2), while D600 and D708 adopt the reaction-ready rotamer conformations as in the HFC. To become cleavage competent after pairing nicked 12/23-RSS DNAs asymmetrically, E962 undergoes a random loop to helix conformational change and shifts by > 2 Å (Fig. 2B–C). The helix (D), to which E962 belongs, extends from one active site to binding the heptamer in the other catalytic site with K973, R977 and Q978 on the loop LDE (Fig. 6A–B, 4G). In addition, the loop L12 (K608-S611) after D600, which is disordered in the apo form and PRC, become ordered in the HFC and forms trans interactions with the heptamer of the other RSS. These crosstalks between the two catalytic centers orchestrate the two active sites into coupled action. The ability of one active site to catalyze two different reactions using either a water molecule or a 3´-OH as the nucleophile probably entails the flexibility of the catalytic residues and a requirement of substrate-assisted catalytic center formation.
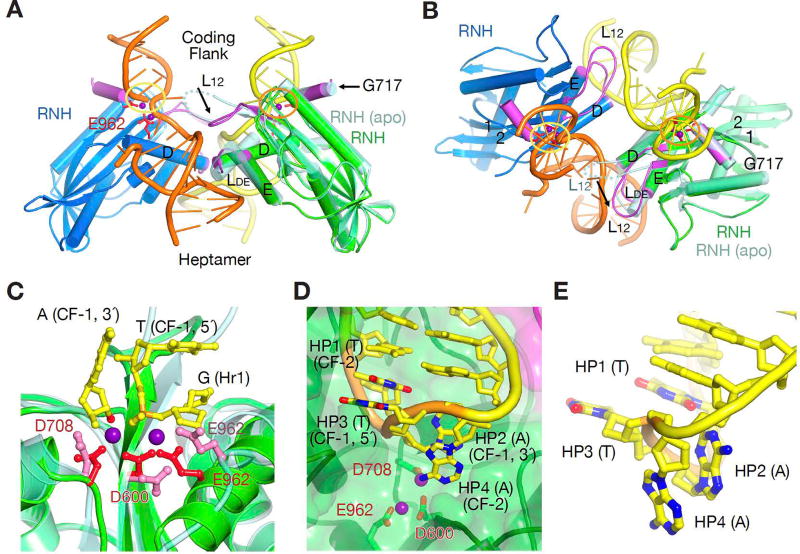
Figure 6.
The active site and hairpin formation. (A and B) Formation of the active catalytic centers in the HFC (darker green and blue RNH domains) depends on coupling of two RAG1 subunits, the correct DNA conformation, and metal ions (Mn2+ or Mg2+). Orthogonal views of the dimeric interface are shown with the dyad in plane (A) and perpendicular to the view plane (B). RAG1 structural changes (highlighted in purple and black arrows) occur after D600 (L12), D708 (G717 helix) and E962 (LDE). For comparison, one subunit of the apo form is shown in light green. C. A zoom-in view of the two-Mn2+-bound active site in the HFC (green protein, red carboxylates and purple Mn2+) superimposed with the apo form structure (lighter green and pink). The first nucleotide in the heptamer (H1), the scissile phosphorus (orange, on the flipped-out T (CF-1)) and the 3´-OH (red) are shown as ball-and-sticks. D. The active site after hairpin formation. The 4 nucleotides forming the loop of the hairpin are highlighted in orange (backbone), yellow (carbon), blue (nitrogen) and red (oxygen) and labeled sequentially as HP1 to HP4. E. A close-up view of the base stacking in the hairpin loop, HP1 with HP3 and HP2 with HP4.
The HFC complexes can be formed with an E962Q mutant or 3´-dideoxy nucleotides at the nicked ends to prevent reaction. WT RAG1/2 protein can also be crystallized with nicked DNA substrate with 3´-OH ends if the catalytic Mg2+ is replaced with Ca2+, which has been shown to support protein-DNA complex formation but not catalysis (Grundy et al., 2009; Nowotny et al., 2007). We find that RAG1/2 can catalyze hairpin formation in crystallo when the WT crystals are transferred and soaked in 2–5 mM Mn2+ stabilization buffer for 30 s to 10 min. Surprisingly, with WT protein, native DNA and Ca2+, the active sites are not fully formed and devoid of the second metal ion (Fig. S7). It requires soaking in 2 mM Mn2+ for the metal ions to occupy the catalytic site and coordinate the scissile phosphate and the 3´-OH for the reaction to occur (Fig. 6C). At 5 mM Mn2+, reaction was nearly complete in 2 min at 4°C (Fig. 6D). The electron density of the hairpin DNA became weaker during the reaction process, indicating the release of the hairpin DNA product. Solution analysis showed that RAG1/2 binds the hairpin product ∼20-fold more weakly than the intact or nicked RSS DNAs (Fig. 5D).
The in crystallo reaction also afforded a glimpse of the hairpin structures of the coding flank DNA, which are fully base paired in the PRC. The 2 bp that form the hairpin loop are completely unpaired to allow the phosphosugar backbone to be connected, and the loop structure is stabilized in two alternate conformations, each with two pairs of base stackings. The major conformation is formed by interleaving base stacking, 1–3 and 2–4 (Fig. 6D–E), and the minor one has 1–2 and 3–4 stacking with the 3rd and 4th bases solvent exposed. In both cases, the first base in the loop is more stacked with the duplex portion of the hairpin.
Discussion
DNA sequence context that govern 12/23-RSS pairing
The heptamer sequence in 12- and 23-RSS is highly conserved (Ramsden et al., 1996). However, in both PRC and HFC, there are few protein-base specific hydrogen bonds; they are limited to the first 3 bp. This is unexpected and different from the usual sequence-specific protein-DNA associations. With the consensus heptamer sequence, backbone distortion and DNA kinking are pronounced between the 6th and 7th bp (Fig. 3, 4G). The 2.75 Å resolution crystal structure of the HFC suggests that the particular heptamer sequences, rich in CA and TG dinucleotides, are chosen for the high deformability of the DNA in unwinding, reduced base stacking, and increased base rolling and tilting (thus resulting in a bent helix), so that each heptamer can form cis and trans interactions with both RAG1 subunits and allow communication across two catalytic centers (Fig. 6).
Coupled recognition of a pair of 12/23-RSS DNAs is essential for the catalytic center formation. In the well-defined HFC structure, DNA kinks between the 10th and 11th, 12th and 13th, and 22nd and 23rd bp of the 23RSS spacer, and between 11th and 12th bp of the 12RSS spacer (Fig. 3). Although HMGB1 can facilitate DNA bending, the location of kinks is determined not by HMGB1 but by the local DNA sequence context and macromolecular assembly. When DNA is kinked as in the RSS spacers, adjacent base pairs contact each other closely in the major groove, and TG and CA dinucleotides favor such contacts (Fig. 4G). A polypurine tract, particularly an A run, as in the center of the conserved nonamer, makes the DNA duplex rather stiff and straight, but it accentuates DNA kinks in its vicinity (Rice et al., 1996). The 23RSS spacer needs to bend ∼150° to bind RAG1/2. Exact locations of kinks in the 12- and 23-RSS may vary, but they have to be matched so that the heptamers and nonamers can be bound by NBDs and RNHs for productive DNA cleavage (see discussion below). Therefore, the sequence context of the spacers, instead of conservation of individual nucleotides, is important for pairing 12- and 23-RSS DNAs.
A survey of the human and mouse germline TCR and Ig (immunoglobulin) sequences reveals that (1) the consensus TG dinucleotide in positions 6 and 7 of the heptamer is often absent in TCR V and J segments (Glusman et al., 2001; Ramsden et al., 1994), (2) CA dinucleotides often occur between the 10th and 13th positions of the 23RSS spacer on Ig gene segments, (3) CA or TG is often found at the end of the spacer bordering the nonamers of both 12- and 23-RSS DNAs, (4) polypurine tracts are prevalent in spacer sequences and placed uniquely in different RSS DNAs, and (5) consecutive CA and TG dinucleotides are found in other spacer regions. Collectively these distinct features influence the locations of straight and bent portions in each RSS as well as favored and disfavored pairing of certain 12- and 23-RSS DNAs.
A particular RSS sequence can result in enhanced RAG1/2-DNA complex formation and can also dramatically bias V(D)J joining. For example, a single C to T change at the 10th bp of the 23RSS bordering the human TCR Vβ3, thus making a TG dinucleotide, increases the V(D)J recombination product by 8 fold (Posnett et al., 1994). In another example, the 23RSS bordering the D segment of the TCRβ locus, which has the consensus heptamer and nonamer sequence and pro-bending sequence TATG at the 10th to 13th bp of the spacer, promotes D to J joining, and even allows V to be joined with J when placed at the border of the V segment (Wu et al., 2003). In contrast, the native 23RSS bordering the V segment has non-consensus heptamer and nonamer and the anti-bending GGGC at the 10th to 13th bp of the spacer. A 23RSS as good as the one bordering TCR Dβ1 can even pair with off-target DNA within a chromatin domain, as long as CAC (the first three nucleotides of the heptamer) is present to mark the cleavage site (Hu et al., 2015). The “beyond 12/23” restriction can thus be explained in part by the requirement of location-sensitive DNA kinks and bends for productive RSS pairing and cleavage by RAG1/2.
Sequence context of DNA coding flanks also plays a critical role in V(D)J recombination (Gerstein and Lieber, 1993). Because base flipping out at the heptamer-coding flank junction is necessary for the hairpin formation (Fig. 6), an A/T pair is favored immediately adjacent to the heptamer, and unpaired pyrimidines (T and C) are preferentially flipped out and their partners (A and G) are stabilized by base stacking with adjacent duplexes. Because the DNA hairpin product depends on base pairing to maintain a duplex structure beyond the hairpin loop, GCs are preferred in the coding flank after the first 2 bp (Gerstein and Lieber, 1993).
HMGB1 and its preference for AC/TG dinucleotides
Since the discovery of HMG proteins in 1970s, no structure of the two-box HMGB-DNA complex has been reported before its participation in the PRC and HFC, where its complex with RAG1/2 establishes 60° and 90° bends in the 23-RSS spacer (Fig. 1). In contrast, structures of TFAM, which also contains two HMG boxes, have been determined in complex with a transcription initiation site, where the DNA is bent 90° by each of the two HMG boxes in phase, making a 180° U-turn (Hillen et al., 2017; Rubio-Cosials et al., 2011). We notice that wherever TFAM bends the DNA, there is a CA or TG dinucleotide. It is likely that strategically located pro-bending CA/TG dinucleotides in the PRC and HFC would stabilize otherwise unstable HMGB1-bent DNA complexes as they influence V(D)J recombination efficiency and partner choice (Posnett et al., 1994; Wu et al., 2003). If a DNA kink is shifted by 1 bp, the direction of DNA curvature would be changed by ∼36°. Given the flexible linker between the two HMG boxes, HMGB1 can bend DNA into different directions according to the functional requirement of particular macromolecular complexes. The threading of HMGB1 under 23RSS also presents a topological puzzle, as whether HMGB1 wraps around the DNA prior to RAG1/2 binding or a RAG1/2-DNA complex partially dissociates for HMGB1 to bind fully.
Synchronized DNA pairing by NBD and RNH domains
Formation of each active site in the RNH domain critically depends on its precise contact with the other heptamer in trans (Fig. 4G, 6–7), and the proper communication between RNH domains depends on the NBD domains binding to nonamers. Acquisition of the NBD domain by RAG1 differentiates the chordates that carry out V(D)J recombination from those that do not (Huang et al., 2016). It may be surprising that the truncated RAG1/2 without NBD binds RSS DNAs as well as core RAG1/2 but fails to cleave them (Fig. 4C–D, S5C). The sequence-specific DNA binding by NBDs apparently is not utilized to increase RSS affinity, but to increase the diversity and selectivity of DNA substrates. To coordinate binding of a pair of antiparallel nonamers by the intertwined NBDs and a pair of nearly parallel heptamers by the RNH domains requires the asymmetric 12- and 23-RSS spacer DNAs to bend differently but synchronously (Fig. 7). The requirement of communication between the NBD and RNH domains and structural synchrony between 12- and 23-RSS DNAs makes RAG1/2 recombinase far more versatile and tunable, and substrate selection more stringent, than any other DNA transposases.
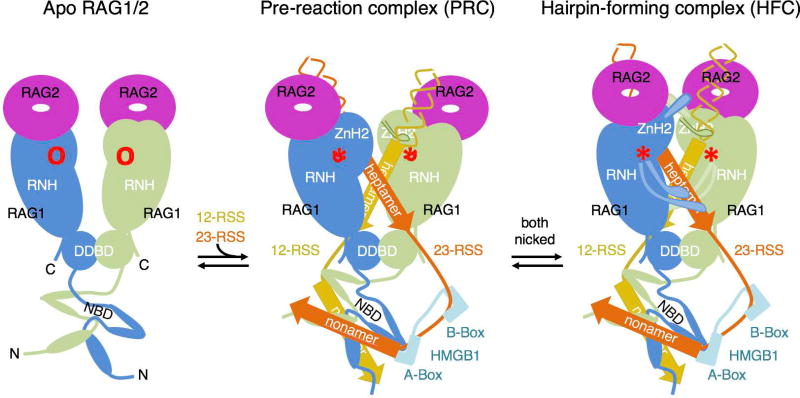
Figure 7.
Cartoon diagrams of the reaction process of RAG1/2 from the apo form, to PRC and HFC.
RAG2 and ZnH2 may provide another layer of 12/23-RSS selection
Comparison of the apo, PRC and HFC structures reveals the previously unappreciated large domain movement of ZnH2. ZnH2 is tethered to the RAG1/2 arm by the Zn2+ coordination between H937 and H942 of ZnH2 and C727 and C730 of ZnC2, which form extensive interactions with RAG2 to make each RAG1/2 arm move as one unit. The mobility of the helix carrying H937 and H942 and the hinge region (aa 928-933) allows the bulk of ZnH2 (aa 794-927) to undergo an opening and closing movement from apo to PRC and from PRC to HFC (Fig. 7). The deformation of the coding flank at the heptamer junction in the HFC is accompanied by the dramatic domain rotation of ZnH2 supported by the smaller movement of the remaining RAG1/2 arm. The ZnH2 domain, particularly the loop containing R848-N852, which undergoes minimal structural changes, needs to hop over the DNA backbone and change from contacting the minor groove in the PRC to reading the nicked heptamer in the major groove in the HFC (Fig. 7).
RAG2, which works with ZnH2 to embrace coding flank DNAs, is unique in RAG1/2 and absent in other DNA transposases, which undergo a similar two-step DNA double-strand cleavage (Hickman et al., 2014). The role of RAG2 appears to be to equalize DNA binding in the PRC and HFC and support this dramatic DNA transformation. Our observation of singly nicked RSS DNA reverting to the pre-reaction state rather than committing to the nick-forming state may indicate that asynchronous nicking of one DNA, e.g. as a result of incorrect pairing of non-complementary 12/23-RSS DNAs, could contribute to the “beyond 12/23” restriction by reversing the process and DNA release (Drejer-Teel et al., 2007).
The X-ray crystallography versus cryoEM approaches
To the best of our knowledge, the mouse and zebrafish HFC represent the first example of a macromolecular structure determined first by cryoEM and then by X-ray crystallography at near atomic resolutions. The differences in these structures reveal interesting aspects of the pros and cons of each method. The obvious advantage of cryoEM not needing crystals is well established, but the precise alignment of trillions of molecules in crystals alleviates the intrinsic errors of aligning individual molecules picked from noisy micrographs produced by cryoEM. The outcome is that crystal structure is more precise in determining the hydrogen bonding patterns. For molecules such as RAG1/2-DNA complexes, which have discrete functional states, crystallography remains a powerful tool for high-precision analysis.
Moreover, Brownian motion in solution introduces heterogeneity into structurally and functionally identical molecules, with the result that structure variations in cryoEM images are expected, even from the same conformational species. For example, the NBD domains and nonamer regions are blurry and of relatively low resolution in all cryoEM maps, particularly when those maps are viewed after B-value sharpening. Lattice contacts in crystals, however, selectively stabilize a certain macromolecular conformation and allow precise model building of otherwise intractable structures. Unambiguous characterization of the DNA deformations and locations of DNA kinks revealed in the crystal structure of HFC provide a structural basis for understanding the 12/23 rule and the “beyond 12/23” restriction.
Concluding remarks
By now, many structures of the RAG1/2 recombinase have been determined, including the apo protein, PRC and HFC, hairpin-product complex, and a cleaved complex variant (Ru et al., 2015). These structures illuminate the asymmetric 12/23-RSS DNA and RAG1/2 protein configurations involved in achieving the sophisticated site-specific DNA manipulations required to form our highly diverse immune system. The atomic details explain why many naturally occurring mutations in RAG1 or RAG2 cause human SCID or Omenn syndrome and shed light on restricted pairing of 12/23-RSS DNAs. However, how RAG1/2 makes the first nick in each RSS DNA, how RAG1/2 recombinase activity is regulated by modifications of histones and the RAG proteins themselves, and whether we can use the molecular understanding to improve functions of genetically defective RAG1/2 and fight cancers, remain to be addressed.
STAR methods
Contact for Reagent and Resource Sharing
Email contact for further information, reagent and resource sharing: weiy@niddk.nih.gov.
Data and Software Availability
The accession codes for the crystal structures of the HFC and PRC reported in this paper are PDB: 5ZDZ, 5ZE0-2, and 6CIK-M (Table S1). The cryoEM density maps and the atomic models derived from these maps are deposited in the EMDB: 7470 and 7480 and PDB: 6CG0 and 6CIJ, respectively (Table S2).
Protein and DNA preparation
The mouse core RAG1/2 (wildtype or E962Q mutant RAG1 aa 384-1008 and RAG2 aa 1-359, tRAG1/2 (RAG1 aa 459-1008) and extended RAG1/2 (RAG1 aa 265-1040 and RAG2 aa 1-520 with a T490A mutation) proteins were expressed as N-terminal His6-MBP fusions in HEK293T cells and purified as previously described (Kim et al., 2015), except that one extra step of Mono Q anion exchange chromatography was added after amylose affinity purification to improve protein purity and eliminate a trace amount of random DNA. The salt concentration in the protein eluted from amylose column was lowered before purification on a Mono Q 10/100 GL anion exchange column (GE Healthcare). Mono Q was pre-equilibrated with 50 mM HEPES (pH 7.3), 100 mM KCl, 5% glycerol, 2 mM DTT, 0.1 mM EDTA. RAG1/2 protein was eluted by a linear gradient of 100–500 mM KCl. The purified RAG1/2 protein was buffer exchanged into a storage buffer containing 25 mM HEPES (pH 7.3), 500 mM KCl, 20% glycerol, 0.1 mM EDTA, 2 mM DTT, concentrated to 4–6 mg/ml, and stored at −80°C.
Human HMGB1 (1-163 amino acids) was prepared as reported previously (Grundy et al., 2009). The coding sequences of Human HMGB1-A box (aa 1-78) and HMGB1-B box (aa 88-163 amino acids) were PCR amplified and subcloned into the pET15 vector with an N-terminal MBP-tag. Proteins were expressed in BL21(DE3) cells by IPTG induction. After harvesting, cells were lysed by sonication in lysis buffer (20 mM TRIS (pH 8.0), 0.5 M KCl, 1 mM TCEP (pH7.0), 1 mM EDTA and protease inhibitor cocktail (Roche)). All purification steps were carried out at 4°C. The lysate was cleared by centrifugation at 35,000 rpm for 1 h and loaded onto amylose resin (NEB). After washing with 100X column volumes of lysis buffer, protein was eluted with lysis buffer supplemented with 40 mM maltose. Eluted protein was treated with PreScission protease to cleave off the His6-MBP-tag at 4°C overnight. Further purification included HiTrap Heparin, HiTrap SP, and a HiLoad 16/60 Superdex 75 column (GE Healthcare) in the final buffer containing 20 mM Tris (pH 8.0), 0.1 M KCl, and 1 mM TCEP. After adding glycerol to 20% final concentration, the protein was stored at −80°C before use.
12- and 23-RSS DNAs used for structural analyses and binding and cleavage assays (Table S3) were synthesized as ssDNA (Integrated DNA Technologies). Long oligonucleotides (>36 nucleotides) were purified by 8–15% TBE-Urea PAGE in a small gel cassette (Life Technologies). Gel purified oligonucleotides were then loaded onto a Glen Gel-Pak column (Glen Research) and eluted in TE buffer. dsDNA was annealed in a Thermocycler in annealing buffer containing 20 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 50 mM NaCl.
To generate 3´-dideoxy oligos, 14 or 15 nucleotides of coding DNA were annealed with a complementary oligo with a 5´ overhang of 10 nucleotides (to create length difference) and incubated with ddATP, Klenow polymerase (NEB) in Klenow reaction buffer at 37°C for 2 hours. The reaction was stopped by addition of 2X volume of stop solution (90% v/v formamide, 0.025% SDS, and 50 mM EDTA). After heat denaturation, the 3´-dideoxy oligo was purified by TBE-urea PAGE.
The procedure for assembly and purification of the PRC and HFC was similar to that of the SEC complex described previously (Kim et al., 2015). Pure wildtype or E962Q-RAG1/2 tetramer, intact or pre-nicked 12RSS, intact or pre-nicked 23RSS, and HMGB1 (aa 1-163) were mixed at 1:1.2:1.2:2 molar ratio in buffer containing 25 mM HEPES (pH 7.3), 100 mM KCl, 5 µM ZnCl2, 1 mM TCEP and 5 mM divalent cation (Ca2+ for WT enzyme-substrate complex or Mg2+ for 3´-dideoxy pre-nicked 12/23-RSS DNAs or E962Q mutant RAG1/2) and incubated at 37°C for 20 min. To remove the His6-MBP fusion tags from the PRC and HFC complexes, PreScission protease was added at a 1:100 mass ratio (protease to RAG1/2) and incubated overnight at 4°C. The non-tagged RAG1/2:12RSS:23RSS:HMGB1 complex (1:1:1:1 ratio) was separated from free-MBP, PreScission protease, excess DNA and HMGB1 by size exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) in buffer containing 25 mM HEPES (pH 7.3), 100 mM KCl, 5% glycerol, 0.1 mM EDTA, 1 mM TCEP, 5 mM divalent cation.
Crystallization and diffraction data collection
Crystals were grown by hanging drop vapor diffusion at 4°C. The HFC crystals were produced by mixing equal volumes of the complex (at ∼5 mg/ml protein) and reservoir solution containing 100 mM MES (pH 6.8), 10–15 % PEG 3350 and 200 mM potassium formate in each droplet. Crystals appeared in 3 days and reached a maximum size in 3 to 6 weeks. Crystals were cryo-protected in reservoir solution supplemented with 25 % ethylene glycol and flash cooled in liquid nitrogen. HFC crystallized in the P21 space group with one RAG1/2 heterotetramer, one 12RSS, one 23RSS, and one HMGB1 in each asymmetric unit.
Intact/intact, intact/nicked, and nicked/intact PRC complexes were grown in similar crystallization conditions containing 12–18% PEG3350, 200 mM NaNO3 or KNO3, or NaCl, and 50–100 mM HEPES (pH 6.9) or MES (pH 7.0). Crystals of intact/intact, intact/nicked, and nicked/intact PRC complexes, which initially diffracted X-rays to 7Å resolution, were dehydrated at 4°C in the crystallization droplets against well solutions of crystallization buffer with 4% stepwise increments of PEG3350 up to 35% (v/v), for 30 min at each step. Mn2+ (5mM) was included at a final step of dehydration of the inactivated mutant (E962Q) RAG1/2 and incubated overnight. The harvested crystals were cryo-protected in the final dehydration solution supplemented with 5%, 10% and 15% (v/v) glycerol stepwise and then flash cooled in liquid nitrogen. All X-ray diffraction data were collected at 100K on beamline 22ID (SER-CAT) of the Advanced Photon Source at Argonne National Laboratory. All data were processed and scaled by XDS (Kabsch, 2010) or HKL2000 (Otwinowski and Minor, 1997).
Crystal structure determination and refinement
Both HFC and PRC structures were determined by molecular replacement using Phaser-MR in PHENIX (Adams et al., 2010) with the apo RAG1/2 structure (PDB: 4WWX) as a search model. The initial model derived from Phaser-MR consisted of one RAG1/2 heterotetramer per asymmetric unit (chains A-D). Electron density for the 12- and 23-RSS DNAs was obvious in the HFC, but in the PRC the end of the coding flank of 23RSS and some base pairs at and beyond the nonamer ends on both RSSs were not as well defined. The DNAs, HMGB1, and flexible regions in RAG1/2 were manually built iteratively in COOT (Emsley et al., 2010), and all structures were refined using PHENIX. Highly flexible regions of protein and DNA with ambiguous density were not included in the model. The intact/nicked structure was used as a template to determine structures of intact/intact and nicked/intact complexes.
Secondary structure restraints and non-crystallographic two-fold symmetry averaging restraints were used throughout the refinement. The final HFC model contains amino acids 385-1008 of RAG1 and amino acids 1-351 of RAG2, and one Zn2+ ion in each RAG1 subunit. The N-terminal residues 384-390 are disordered in one RAG1 subunit. Due to poor electron densities, residues 82-87 and 336-340 of RAG2 were not included in the final model. The final refinement statistics are shown in Table S1. Restraints on base pairs and base stacking of DNA were also applied. The Ramachandran plot and model quality were calculated with MolProbity. All structural figures were prepared with PyMOL (www.pymol.org).
CryoEM sample preparation and data collection
To obtain cryoEM images of the HFC, an extended form of RAG1/2 with RING finger and PHD domains included was used rather than core RAG1/2. With core RAG1/2, a severe problem of preferential orientation was always encountered, regardless of what kind of RSS DNAs were used, and only a biased map could be obtained. By using extended RAG1/2, we overcame the preferential orientation problem. HFC containing extended RAG1/2 (0.2 mg/ml) was loaded on C-flat CF-1.2/1.3-;4C holey carbon grids (3µl each), blotted for 4 s, and flash-frozen in liquid ethane in a Vitrobot at 100% humidity. The frozen grids were stored in liquid nitrogen and loaded into a Titan Krios electron microscope operated at 300 kV for automated image acquisition with Leginon (Suloway et al., 2005). Movies were recorded on a Gatan K2 Summit direct electron detector, using the super-resolution mode at 130K nominal magnification (calibrated pixel size of 1.07Å on the sample level, corresponding to 0.535Å in super-resolution mode) and defocus values ranging from −1.4 to −3.0 µm. The dose rate at the detector was set to about 5.5 electrons per physical pixel per second. The total exposure time for each movie was 12s, which was fractionated into 60 frames of sub-images, with 0.2s per frame. The first and last two frames were removed during motion correction, and the remaining frames were aligned and summed to generate a dose-weighted micrograph using Motioncorr2 (Zheng et al., 2017). Micrographs without dose weighting were also generated and used for defocus determination and particle picking. The micrographs with dose weighting were phase-flipped using Bsoft (Heymann, 2001). Out of 2300 dose-weighted micrographs obtained in a continuous session, 1688 micrographs were selected by manual screening, and 693,167 particles were initially picked from these selected micrographs using Gautomatch (developed by Dr. K. Zhang; http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) and extracted in RELION-2.1 (Fernandez-Leiro and Scheres, 2017). Using the extracted particles, an initial map at ∼3.5Å resolution was obtained with cryoSPARC (Punjani et al., 2017) and served as the reference for 3D structure classification with RELION (Scheres, 2012). A low-pass (15 Å) filtered map generated from the coordinates of our crystal structure of HFC using EMAN2 (Ludtke, 2016) worked equally well as the reference for 3D classification with RELION. Two cycles of 3D classification were performed to classify the extracted particles. In the first cycle, 3 classes were assigned, and one of them showed good structural features and was thus selected for the second cycle of classification. In the second cycle, 4 classes were assigned, 3 of which showed a clear NBD domain and the fourth contained partial structural features. One of the 3 good classes showed the partial linker between A- and B-boxes of HMGB1 bound to the 23RSS. 139,781 particles belonging to the 3 classes and 49,624 in the class with the partial linker were then selected to refine the models and generate the 3.17 Å and 3.90 Å maps in C1 symmetry using 3D auto-refine in RELION, and the maps were sharpened using post-processing also in RELION. All reported resolutions are based on the “gold standard” refinement procedures and the 0.143 Fourier Shell Correlation (FSC) criterion (Swint-Kruse and Brown, 2005). Local resolution was estimated using Resmap (Kucukelbir et al., 2014). For model building, we first fit our 2.75Å HFC crystal structure into the cryoEM map using Chimera, and then manually adjusted and re-built the model according to the cryoEM density in COOT (Emsley et al., 2010). Phenix real-space refinement was used to refine the model. MolProbity and EMRinger (Barad et al., 2015) were used to validate the final model. The refinement statistics are shown in Table S2.
HMGB1 requirement in the hairpin formation reaction
For hairpin formation, 55 nM pre-nicked 12- and 23- RSS DNAs, of which 12RSS was 5´- labeled with 6-FAM on the 16-bp coding flank (Table S3) were incubated with 100 nM RAG1/2 in the reaction buffer (20 mM HEPES (pH 7.3), 80 mM KCl, 1 mM CaCl2, 1 µM ZnCl2, and 0.1 mg/ml BSA) in the presence of 50 nM to 100 nM HMGB1, or a single A- or B-box, at 22 °C for 5 min. Reaction was initiated by addition of 5 mM MgCl2 and terminated at the indicated time points by mixing with 2X volume of stop solution (90% v/v formamide, 0.025% SDS, and 50 mM EDTA). After heat denaturation, reaction products were resolve on 15% polyacrylamide TBE-urea gels, visualized on a Typhoon PhosphorImager, and quantified using ImageQuant software.
Fluorescence-based substrate, intermediate and product DNA binding assays
The fluorescence-based DNA binding assays were performed in a binding buffer containing 20 mM HEPES (pH 7.5), 100 mM KCl, 2 mM DTT, 0.1 mg/ml BSA, 1 mM MgCl2, 0.1 mM EDTA, and 1% glycerol. For binding of intact or nicked RSS DNA substrates with 16 bp coding ends, 20 nM 5´-Cy5-labeled 12- or 23-RSS and an equal amount of unlabeled 23- or 12-RSS were incubated with 40 nM HMGB1 and mouse cRAG1/2 E962Q tetrameric protein at concentrations from 2 to 1024 nM at 25°C for 5 min. For binding of 16 bp hairpin products, 40 nM cleaved 12RSS and 40 nM 23RSS without coding flank, and 80 nM 5´-Cy5-labeled hairpin coding-flank products were incubated with HMGB1 and cRAG1/2 as described above. Fluorescence polarization values were measured on a CLARIOstar instrument, and the binding curves and Kd values were plotted and calculated with Graphpad Prism software (version 7.0). Plots of biochemical data are shown with the mean ± SD from three independent experiments.
Comparison of DNA binding and cleavage by tRAG1/2 and cRAG1/2
DNA binding assays were carried out in a binding buffer containing 25 mM HEPES (pH 7.5), 60 mM KCl, 1 mM DTT, 0.1 mg/ml BSA, 1 mM CaCl2 and 0.1 mM EDTA, with either wildtype cRAG1/2 or tRAG1/2. Either pre-nicked 12- or 23-RSS DNA (each 20 nM) was FAM-labeled on the 5´-coding flank and incubated with WT tRAG1/2 at concentrations from 2 to 400 nM at 25°C for 5 min. DNA binding by WT cRAG1/2 to pre-nicked 12/23-RSS DNA with the FAM-labeled 12RSS was also measured in the presence of 20 nM HMGB1 as a reference. The Kd values were generated in the same way as described above.
Cleavage assays were performed in the reaction buffer containing 25 mM HEPES (pH 7.5), 60 mM KCl, 1 mM DTT, 0.1 mg/ml BSA, and 5 mM MgCl2 or 5 mM MgCl2 plus 2 mM MnCl2. Pre-nicked 12- and 23-RSS DNA each with a 16 bp coding flank and both FAM-labeled at 60 nM were incubated with 50 nM (based on the heterotetrameric MW) WT cRAG1/2 or tRAG1/2 and 120 nM HMGB1 at 37°C for 0–80 min. Reactions were stopped by adding an equal volume of the formamide buffer (95% (v/v) formamide and 12 mM EDTA and 0.3% bromophenol blue) and heating at 95°C for 10 min. Cleavage products were separated by 15% TBE-urea PAGE, visualized and quantified using a Typhoon PhosphorImager (GE Healthcare).
Supplementary Material
Figure S1. Related to Figure 1. CryoEM structure of HFC.
Figure S2. Related to Figure 1. Requirement of HMGB1 in RSS-binding and cleavage by RAG1/2.
Figure S3. Related to Figure 1. Comparison of the crystal structure of mouse HFC and the cryoEM structure of zebrafish HFC.
Figure S4. Related to Figure 2. Superimposable crystal structures of PRC complexes with intact/intact, intact/nicked and nicked/intact 12/23-RSS DNAs.
Figure S5. Related to Figure 3. NBD domain and nonamer binding are necessary for DNA cleavage by RAG1/2.
Figure S6. Related to Figure 4. Similar protein interactions made by 12- and 23-RSS DNAs.
Figure S7. Related to Figure 6. Formation of the catalytically competent active sites requires both DNA substrate and correct divalent cations.
Table S1. Related to Figure 1 & 2. X-ray diffraction data and structure refinement Statistics of HFC and PRC.
Table S2. Related to Figure 1. Statistics of cryoEM data collection and structure refinement of HFC.
Table S3. Related to Figure 1, 4 & 5. Oligos used in structural and biochemical studies.
Movie 1. Related to Figure 1. Morphing of structural transition from apo to PRC complex. A front view of RAG1/2 dimer with RAG1 subunits shown as blue and green and RAG2 in magenta. The NBD domains tilt more severely in the DNA bound form than in the apo protein. On the Y-arms, each RAG2 moves slightly, and ZnH2 of RAG1 significantly.
Movie 2. Related to Figure 1. Morphing of structural transition from apo to PRC complex. A top view of RAG1/2 dimer reveals the slight movement of RAG2 towards RAG1 on the same Y-arm as ZnH2 opens out significantly for DNA binding.
Movie 3. Related to Figure 2. Morphing of structural transition from PRC to HFC. A front view of RAG1/2 dimer with RAG1 subunits shown as blue and green and RAG2 in magenta. The 12RSS DNA is shown in yellow and 23RSS in orange. The NBD domains move a little, probably because of different crystal lattice contacts. The A-box bound to the 23RSS nonamer-spacer junction slides more than 4Å along the minor groove because of crystal lattice contacts in the PRC structure. The Y-arms together with the nicked DNAs extend outwards to make space for hairpin formation.
Movie 4. Related to Figure 2. Morphing of structural transition from PRC to HFC. A top view of RAG1/2 dimer reveal the ‘hugging” movement of RAG2 and ZnH2 brings the two coding flank DNAs tilted toward RAG2 and closer to each other. Maneuvering the coding flank DNA is important for positioning the scissile phosphate and 3´-OH nucleophile for hairpin formation.
Acknowledgments
We thank D. Leahy for critical reading of the manuscript, H. Wang (NIDDK) for help with EM grid preparation, and W. Jiang (Purdue) for help with using cryoSPARC. This research was supported by the National Key R&D Program (2017YFA0504803) and Fundamental Research Funds for the Central Universities of China to XZ, the National Institutes of Health Intramural AIDS targeted Antiviral Program (IATAP), and the National Institute of Diabetes and Digestive and Kidney Diseases to W. Y. (DK036147 and DK036144) and M. G. (DK036167). The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057, 1S10OD018111 and GM071940), NSF (DBI-1338135) and CNSI at UCLA.
Footnotes
Author contributions
M.S.K. determined the HFC crystal structures and performed the assays of HMGB1 requirement; W.C. and X.C. determined the PRC crystal structures; X.C., Y.C. and X.Z. determined the cryoEM structures; X.C. performed all biochemical assays except for the HMGB1 requirement; W.Y. and M.G. conceived and supervised the project; and H.Z. supervised the cryoEM analysis. All authors participated in data analysis and manuscript preparation.
Declaration of Interests
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- Banerjee JK, Schatz DG. Synapsis alters RAG-mediated nicking at Tcrb recombination signal sequences: implications for the “beyond 12/23” rule. Mol Cell Biol. 2014;34:2566–2580. doi: 10.1128/MCB.00411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Alt FW, Hughes MM, D’Auteuil M, Wehrly TD, Woodman BB, Gartner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six- bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Drejer-Teel AH, Fugmann SD, Schatz DG. The beyond 12/23 restriction is imposed at the nicking and pairing steps of DNA cleavage during V(D)J recombination. Mol Cell Biol. 2007;27:6288–6299. doi: 10.1128/MCB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, Wang K, Koop BF, Hood L. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–349. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- Grundy GJ, Ramon-Maiques S, Dimitriadis EK, Kotova S, Biertumpfel C, Heymann JB, Steven AC, Gellert M, Yang W. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol Cell. 2009;35:217–227. doi: 10.1016/j.molcel.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB. Bsoft: image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ewis HE, Li X, Knapp JA, Laver T, Doss AL, Tolun G, Steven AC, Grishaev A, Bax A, et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell. 2014;158:353–367. doi: 10.1016/j.cell.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen HS, Morozov YI, Sarfallah A, Temiakov D, Cramer P. Structural Basis of Mitochondrial Transcription Initiation. Cell. 2017;171:1072–1081. doi: 10.1016/j.cell.2017.10.036. e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18:65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapkouski M, Chuenchor W, Kim MS, Gellert M, Yang W. Assembly Pathway and Characterization of the RAG1/2-DNA Paired and Signal-end Complexes. J Biol Chem. 2015;290:14618–14625. doi: 10.1074/jbc.M115.641787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ. Single-Particle Refinement and Variability Analysis in EMAN2.1. Methods Enzymol. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Piirila H, Valiaho J, Vihinen M. Immunodeficiency mutation databases (IDbases) Hum Mutat. 2006;27:1200–1208. doi: 10.1002/humu.20405. [DOI] [PubMed] [Google Scholar]
- Posnett DN, Vissinga CS, Pambuccian C, Wei S, Robinson MA, Kostyu D, Concannon P. Level of human TCRBV3S1 (V beta 3) expression correlates with allelic polymorphism in the spacer region of the recombination signal sequence. J Exp Med. 1994;179:1707–1711. doi: 10.1084/jem.179.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, van Gent DC, Gellert M. Specificity in V(D)J recombination: new lessons from biochemistry and genetics. Curr Opin Immunol. 1997;9:114–120. doi: 10.1016/s0952-7915(97)80167-7. [DOI] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Samara NL, Gao Y, Wu J, Yang W. Detection of Reaction Intermediates in Mg(2+)-Dependent DNA Synthesis and RNA Degradation by Time-Resolved X-Ray Crystallography. Methods Enzymol. 2017;592:283–327. doi: 10.1016/bs.mie.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Gellert M. Role of RAG1 autoubiquitination in V(D)J recombination. Proc Natl Acad Sci U S A. 2015;112:8579–8583. doi: 10.1073/pnas.1510464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Swint-Kruse L, Brown CS. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics. 2005;21:3327–3328. doi: 10.1093/bioinformatics/bti511. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bassing CH, Jung D, Woodman BB, Foy D, Alt FW. Dramatically increased rearrangement and peripheral representation of Vbeta14 driven by the 3’Dbeta1 recombination signal sequence. Immunity. 2003;18:75–85. doi: 10.1016/s1074-7613(02)00515-0. [DOI] [PubMed] [Google Scholar]
- Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1. CryoEM structure of HFC.
Figure S2. Related to Figure 1. Requirement of HMGB1 in RSS-binding and cleavage by RAG1/2.
Figure S3. Related to Figure 1. Comparison of the crystal structure of mouse HFC and the cryoEM structure of zebrafish HFC.
Figure S4. Related to Figure 2. Superimposable crystal structures of PRC complexes with intact/intact, intact/nicked and nicked/intact 12/23-RSS DNAs.
Figure S5. Related to Figure 3. NBD domain and nonamer binding are necessary for DNA cleavage by RAG1/2.
Figure S6. Related to Figure 4. Similar protein interactions made by 12- and 23-RSS DNAs.
Figure S7. Related to Figure 6. Formation of the catalytically competent active sites requires both DNA substrate and correct divalent cations.
Table S1. Related to Figure 1 & 2. X-ray diffraction data and structure refinement Statistics of HFC and PRC.
Table S2. Related to Figure 1. Statistics of cryoEM data collection and structure refinement of HFC.
Table S3. Related to Figure 1, 4 & 5. Oligos used in structural and biochemical studies.
Movie 1. Related to Figure 1. Morphing of structural transition from apo to PRC complex. A front view of RAG1/2 dimer with RAG1 subunits shown as blue and green and RAG2 in magenta. The NBD domains tilt more severely in the DNA bound form than in the apo protein. On the Y-arms, each RAG2 moves slightly, and ZnH2 of RAG1 significantly.
Movie 2. Related to Figure 1. Morphing of structural transition from apo to PRC complex. A top view of RAG1/2 dimer reveals the slight movement of RAG2 towards RAG1 on the same Y-arm as ZnH2 opens out significantly for DNA binding.
Movie 3. Related to Figure 2. Morphing of structural transition from PRC to HFC. A front view of RAG1/2 dimer with RAG1 subunits shown as blue and green and RAG2 in magenta. The 12RSS DNA is shown in yellow and 23RSS in orange. The NBD domains move a little, probably because of different crystal lattice contacts. The A-box bound to the 23RSS nonamer-spacer junction slides more than 4Å along the minor groove because of crystal lattice contacts in the PRC structure. The Y-arms together with the nicked DNAs extend outwards to make space for hairpin formation.
Movie 4. Related to Figure 2. Morphing of structural transition from PRC to HFC. A top view of RAG1/2 dimer reveal the ‘hugging” movement of RAG2 and ZnH2 brings the two coding flank DNAs tilted toward RAG2 and closer to each other. Maneuvering the coding flank DNA is important for positioning the scissile phosphate and 3´-OH nucleophile for hairpin formation.