Abstract
The human opportunistic pathogen Pseudomonas aeruginosa strain PA14 is a multihost pathogen that can infect Arabidopsis. We found that PA14 pathogenesis in Arabidopsis involves the following steps: attachment to the leaf surface, congregation of bacteria at and invasion through stomata or wounds, colonization of intercellular spaces, and concomitant disruption of plant cell wall and membrane structures, basipetal movement along the vascular parenchyma, and maceration and rotting of the petiole and central bud. Distinctive features of P. aeruginosa pathogenesis are that the surface of mesophyll cell walls adopt an unusual convoluted or undulated appearance, that PA14 cells orient themselves perpendicularly to the outer surface of mesophyll cell walls, and that PA14 cells make circular perforations, approximately equal to the diameter of P. aeruginosa, in mesophyll cell walls. Taken together, our data show that P. aeruginosa strain PA14 is a facultative pathogen of Arabidopsis that is capable of causing local and systemic infection, which can result in the death of the infected plant.
Pseudomonas aeruginosa, a ubiquitous gram-negative bacterium, has been intensively studied as an opportunistic human pathogen (Britigan et al., 1997) and as the dominant pathogen infecting the lungs of cystic fibrosis patients (Pier et al., 1996). There are also a few sporadic reports describing human clinical isolates of P. aeruginosa that are capable of eliciting soft-rot symptoms when infiltrated into a variety of plants including tomato, lettuce, onion, and tobacco (Elrod and Braun, 1942; Burkholder, 1950; Kominos et al., 1972). P. aeruginosa is also known to be a common colonizer of many fruits and green plants and can persist without causing disease symptoms (Cho et al., 1975).
Literature reports that specific strains of P. aeruginosa might function as a plant and animal pathogen led our laboratory to search for P. aeruginosa strains that elicited disease symptoms when infiltrated into Arabidopsis leaves. Among 75 P. aeruginosa strains tested (Rahme et al., 1995), several strains were identified that caused varying degrees of chlorosis and tissue maceration. A few caused severe soft-rot symptoms and at least two of these strains, UCBPP-PA14 (hereafter PA14), a human clinical isolate, and UCBPP-PA29 (hereafter PA29), a plant isolate, elicited severe soft-rot symptoms on some Arabidopsis ecotypes, but not on others. It is interesting that the most susceptible ecotypes were different for the two P. aeruginosa strains. This pattern of host-specific resistance is typical in plant-pathogen interactions and suggests that Arabidopsis may have evolved resistance to particular P. aeruginosa strains. Because PA14 was also shown to be highly infectious in a mouse burn model, it was selected for further studies and was subsequently shown to be a highly virulent pathogen of the insects greater wax moth (Galleria mellonella; Jander et al., 2000) and fruitfly (G. Lau, S. Mahajan-Miklos, F. Ausubel, L. Perkins, and L. Rahme, unpublished data) and to kill the nematode Caenorhabditis elegans (Mahajan-Miklos et al., 1999; Tan et al., 1999a, 1999b). It is important to point out that the ability of P. aeruginosa to be a multihost pathogen is not limited to PA14. The well-studied P. aeruginosa strains PAK and PAO1, which are virulent in a variety of animal models, elicit soft-rot symptoms when infiltrated into the mid-rib of lettuce leaf stems (Rahme et al., 1997). PA14 and PA29 are also pathogenic in lettuce leaves (Rahme et al., 1997; L. Rahme, unpublished data).
In previously published work our laboratory identified P. aeruginosa pathogenicity related genes by screening PA14 transposon insertion libraries for mutants that were less infectious in plants or nematodes (Rahme et al., 1997; Mahajan-Miklos et al., 1999; Tan et al., 1999b). It was remarkable that of the 13 pathogenicity related genes identified in nematode screens, 10 were also required for pathogenicity in plants (Mahajan-Miklos et al., 1999; Tan et al., 1999b). Moreover, out of a total of 21 genes identified in plant and nematode screens, 17 were required for mouse pathogenicity (Rahme et al., 1997; Mahajan-Miklos et al., 1999; Tan et al., 1999b). These studies suggested that there is a remarkable degree of conservation of the molecular basis of P. aeruginosa pathogenesis, regardless of the host, and that a detailed study of P. aeruginosa pathogenicity in plants would potentially be relevant to the study of P. aeruginosa pathogenesis in humans (Rahme et al., 1995).
The main goal of this study was the analysis of all of the stages of the P. aeruginosa infection process in Arabidopsis, including tissue penetration, distribution, and the effect of invading bacteria on plant cell walls and membrane structures.
RESULTS
Attachment of P. aeruginosa to Arabidopsis Leaves
In previously published work we showed that Arabidopsis ecotype Llagostera (Ll-0) is highly susceptible to infection by P. aeruginosa strain PA14, whereas ecotype Argentat (Ag-0) is resistant (Rahme et al., 1995). Figure 1 shows PA14 cells attached to various Ll-0 leaf surface structures after leaf discs were incubated at room temperature for 2 h in a suspension of PA14 cells and then incubated on the surface of 1.5% (w/v) water agar in closed Petri dishes. A minority of the bacterial cells are oriented perpendicular to the leaf surface (Fig. 1C) and as indicated by the white arrow in Figure 1A, some of the perpendicularly attached bacteria appeared to digest or degrade the outer layer of the epidermal wall. However, we never observed bacteria piercing through the entire thickness of the outer epidermal wall. Dense multilayered clusters of PA14 typically formed on trichomes as shown in Figure 1B, reminiscent of the structures of bacterial biofilm. Clusters of PA14 cells were also present above guard cells obscuring the stomatal openings (Fig. 1C). At higher magnifications, it was evident that most of these bacterial cells oriented perpendicular to the surfaces of the guard cells (not shown). Although ecotype Ag-0 is resistant to PA14 infection, PA14 also attached to the surface of Ag-0 leaves. However, the relative density of attached bacteria was 6 to 7-fold lower on Ag-0 leaves than on Ll-0 leaves (data not shown).
Figure 1.
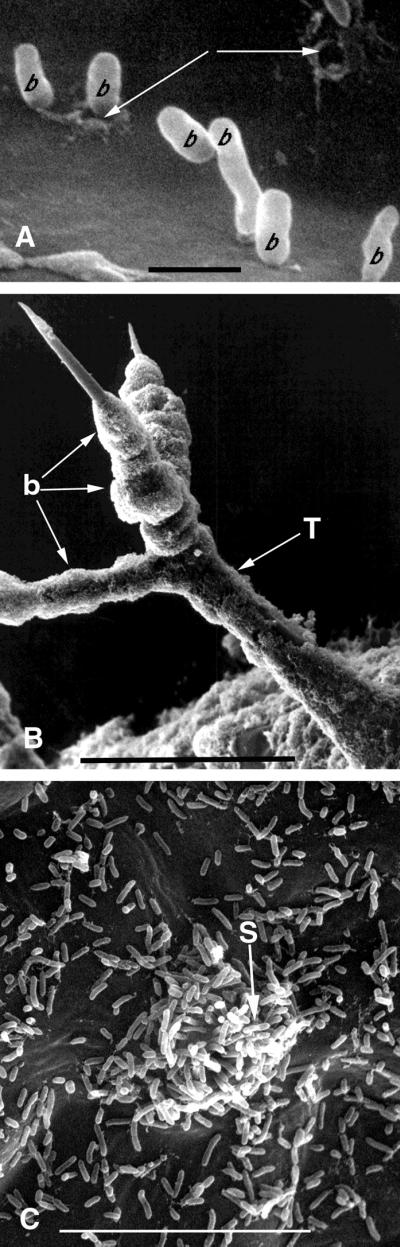
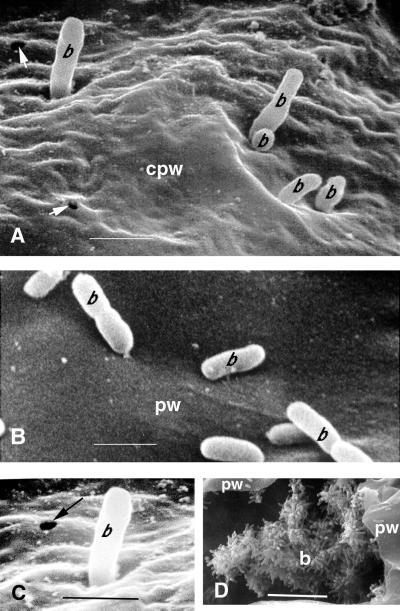
Scanning electron micrographs of P. aeruginosa PA14 interacting with the surface structures of an Arabidopsis Ll-0 leaf. Leaf discs were placed in a bacterial suspension for 2 h and then transferred to water agar for 48 h as described in “Materials and Methods.” A, PA14 cells attaching perpendicularly to the surface of the leaf and digestion of the leaf epidermis (arrows) at the site of bacterial attachment. Bar = 1 μm. B, PA14 cells concentrated on trichome surfaces. Bar = 100 μm. C, Concentration of PA14 cells at a stomatal opening. Bar = 10 μm. b, Bacterium; S, stoma; T, trichome.
Penetration of PA14 into Substomatal Cavities
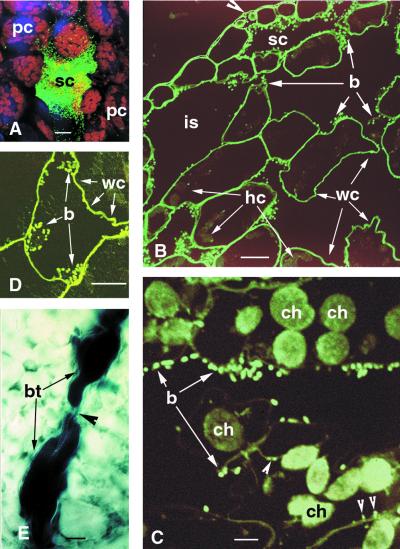
We studied the process by which P. aeruginosa PA14 penetrates Arabidopsis Ll-O leaves using PA14 expressing the green fluorescent protein (GFP). Detached leaves were incubated for 2 h in a bacterial suspension as described in “Materials and Methods.” Using a confocal scanning spectrophotometer (CSS), individual PA14 cells could be seen actively moving along the leaf surface toward open stomata, increasing their rate of movement as they approached the stomata (data not shown). Bacteria could also be readily seen entering into and then vanishing within the stomatal openings. As shown in Figure 2A, when the detached leaves that had been incubated in the bacterial suspension were subsequently placed on 1.5% (w/v) water agar, incubated for 24 h at room temperature, and optical sections inside the leaf were obtained with the CSS, GFP-labeled bacteria could be readily observed concentrated in the substomatal cavities. At 24 h, essentially all of the substomatal cavities were filled with bacteria. Accumulation of bacteria in substomatal cavities could also be seen in micrographs of cross sections of an Arabidopsis leaf stained with the fluorescent stain Syto 9 (Fig. 2B) and in transmission electron micrographs (Fig. 3A) at 48 h following incubation with PA14.
Figure 2.
Distribution of P. aeruginosa PA14 in Arabidopsis Ll-0 leaves. As described in “Materials and Methods,” detached leaves were incubated in a bacterial suspension for 2 h and then transferred to water agar (A–D) or intact plants were infiltrated with a bacterial suspension using a syringe (E). A, Confocal scanning micrograph of GFP-labeled PA14 concentrated in substomatal cavity 24 hpi as seen in the same focal plane as the parenchyma cells. Strong red autofluorescence from chloroplasts is readily apparent. Bar = 10 μm. B, Syto 9-stained leaf cross section depicting PA14 localization in the intercellular space 48 hpi. Partial degradation of host cytoplasm (hc), enlargement of intercellular space (is), decrease of cell volume, and convolution of plant walls (wc) are apparent. Arrowhead indicates stomatal opening. Bar = 10 μm. C, Syto 9-stained leaf cross section depicting PA14 attachment to the outer (arrows) and inner (arrowheads) surfaces of mesophyll walls at 48 hpi. Redistribution and degradation of host organelles is apparent. Bar = 1 μm. D, Syto 9-stained cross section depicting a mesophyll cell with broken wall, cell wall convolutions (wc), and bacteria concentrated around cell organelles. Bar = 10 μm. E, Trypan blue staining of bacterial-filled vessel parenchyma cells 48 hpi. Xylem vessels do not contain bacteria (arrowheads). Bar = 10 μm. b, Bacterium; bt, bacterial thread; ch, chloroplast; hc, host cytoplasm; is, intercellular space; pc, parenchyma cell; sc, substomatal cavity; wc, wall convolution.
Figure 3.
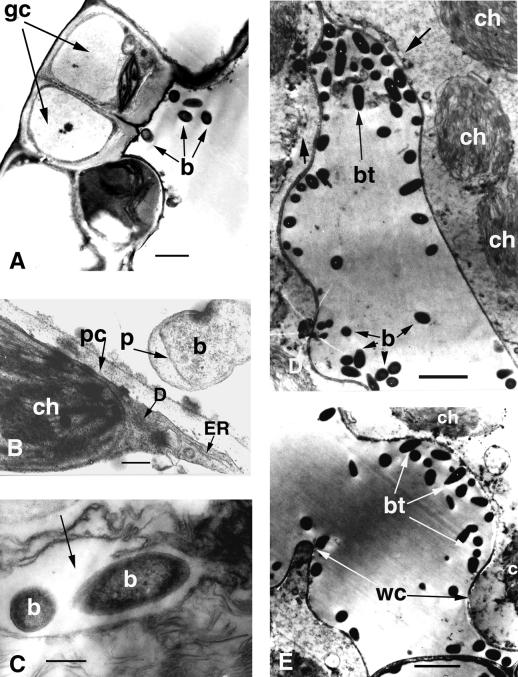
Transmission electron microscopy of Arabidopsis Ll-0 leaves infected with P. aeruginosa PA14. Detached leaves were incubated in a bacterial suspension OD600 = 0.02 in 10 mm MgSO4 for 2 h under vacuum and then transferred to water agar plates. A, Cross section of a stoma and bacteria inside the substomatal cavity 24 hpi. Bar = 10 μm. B, Interaction of a PA14 cell with an Arabidopsis cell illustrating concentration of host organelles at the site of interaction with the bacteria at 24 hpi. Bar = 1 μm. C, Two bacteria shown inside host cytoplasm (arrow). Bar = 1 μm. D, Advanced stage of infection illustrating disrupted host membranes at 72 hpi. The bacterial thread has digested the middle lamella and separated the mesophyll cells from each other. The host plasmalemma (short large arrow) is highly undulated (left cell) or partly destroyed (right cell). The outer membranes of the chloroplasts are at various stages of degradation. Bar = 10 μm. E, Advanced stage of infection illustrating thinning of the cell wall and cell wall convolutions. The plasmalemma and chloroplasts are severely degraded. Bar = 10 μm. b, Bacterium; bt, bacterial thread; ch, chloroplast; D, dictyosomes; ER, endoplasmic reticulum; gc, guard cells; p, bacterial periplasmic space, pc, plant periplasmic space; wc, wall convolution.
Development of the Bacterial Infection
In the experiments described in the previous section Ll-0 leaves were incubated in PA14 and then transferred to water agar plates at room temperature. After PA14 cells entered the substomatal cavity, they began multiplying and rapidly spread through the leaf mesophyll. As in the case of other well-studied phytopathogenic bacteria, PA14 formed “bacterial threads” (dense populations of bacterial cells that adopt an elongated and branched structure) that colonized the intercellular space, presumably by digesting the middle lamellae and separating the plant cells from each other (Figs. 2, B and C and 3D). At relatively early stages of the infection (2 d) almost all the bacteria were found in intercellular spaces attached to Arabidopsis mesophyll walls (Fig. 2, B–D), although some bacterial cells could also be observed attached to the inner surface of cell walls (Fig. 2C). After proliferation in the intercellular space at the site of infection, PA14 bacterial cells entered vascular parenchyma cells and then spread from one parenchyma or companion cell to the next, moving in a basipetal direction (not shown, but refer to Fig. 2E described below). At later stages of infection (3–4 d), many of the mesophyll cells were severely damaged and contained intracellular bacteria (Fig. 5D).
Figure 5.
Scanning electron micrographic visualization of freeze-fractured infected Arabidopsis Ll-0 leaves depicting P. aeruginosa PA14 cells attaching to and penetrating the convoluted cell walls of vessel parenchyma cells at 96 hpi. The leaves of intact Ll-0 plants were infiltrated with a bacterial suspension (OD600 = 0.02) under vacuum and detached leaves were incubated in Petri dishes on 1.5% (w/v) water agar as described in “Materials and Methods” for 72 h. A, PA14 cells on the surface of a highly convoluted cell wall of a parenchyma vessel cell. Holes in the plant cell wall with a diameter similar to that of the bacteria are apparent (arrowheads). Bar = 1 μm. B, P. syringae cells on the surface of a nonundulated cell wall of a parenchyma vessel cell. Bar = 1 μm. C, Detail of Figure 5A to highlight a hole in the plant cell wall. D, PA14 cells attached to the degrading cytoplasm in the lumen of a vessel parenchyma cell. Bar = 10 μm. b, Bacterium, cpw, convoluted plant wall; pw, plant wall.
We also followed the systemic spread of PA14 infections at 30°C in intact plants after infiltration of the top portion of leaves with a syringe at a dose of approximately 5 × 103 cfu/cm2 leaf area. By 24 h post-infection (hpi), PA14 had propagated to high titers in the intercellular space at the loci of infection. In a similar manner to the experiment described in the preceding paragraph, by 48 hpi bacterial cells filled parenchyma and companion cells surrounding small vessels close to the initial inoculation site (Fig. 2E) and as above, the infection spread in a basipetal direction from one parenchyma or companion cell to the next. By 72 hpi the propagating bacteria had spread along the major Arabidopsis leaf vein to the petiole, filling essentially all of the vessel parenchyma cells and resulting in complete maceration of the petiolar tissue by 96 hpi. In contrast, we did not find any evidence that P. aeruginosa were present in substantial numbers in phloem or xylem.
P. aeruginosa proliferated faster in old leaves (lower leaves of 6-week-old plants) than in young ones (fully expanded leaves of 6-week-old plants). In young leaves it took about 4 d before the bacteria started spreading along the leaf veins in a basipetal direction. The time course of the infection in these experiments is summarized in Table I. PA14 also produced spreading soft-rot lesions in infiltrated plants at room temperature. In contrast to P. aeruginosa strain PA14, P. syringae pv maculicola strain ES4326, which is considered to be a virulent Arabidopsis pathogen (Schott et al., 1990; Dong et al., 1991), did not exhibit the systemic distribution characteristic of PA14, but only caused localized water-soaked lesions without spreading along the vessel elements.
Table I.
Spreading of P. aeruginosa infection in intact infiltrated Arabidopsis leaves
| Incubation Time | Progression of Disease
Symptomsa
|
|||
|---|---|---|---|---|
| Young
Leaves
|
Old Leaves
|
|||
| Portion of leaf infected | Portion of leaf infected | |||
| 1/4 | 1/2 | 1/4 | 1/2 | |
| 1 | 0 | 0 | 0 | 0 |
| 2 | 1 | 1 | 1 | 1 |
| 3 | 2 | 2 | 3 | 3 |
| 4 | 2 | 2 | 3 | 3 |
0, No visible bacterial clusters; 1, inter- and intracellular bacteria in the locus of inoculation; 2, spreading bacteria along the leaf blade; 3, appearance of bacterial clusters (bacteria fill the whole cell volume) in the petiole.
Effect of PA14 Infection on Plant Ultrastructure
In this section we describe experiments that show that infection of Arabidopsis leaves with PA14 has a dramatic effect on the structures of the host cell walls and membranes and on the location and organization of host cell organelles. Uninfected Arabidopsis mesophyll cells have a large central vacuole surrounded by a thin layer of cytoplasm containing a nucleus and organelles. The first sign of host cell degeneration following PA14 infection was slight plasmolysis and concentration of host membrane structures including chloroplasts, endoplasmic reticulum, and dictyosomes at the site of bacterial contact (Fig. 3B). At this stage a limited number of single bacteria appeared to be able to penetrate into metabolically active plant cells (Fig. 3C).
Proliferation of bacteria in the intercellular space resulted in a redistribution and alteration of host organelles 72 hpi. Host plasmalemma became highly undulated and disrupted (Fig. 3D), there was swelling and disruption of outer chloroplast membranes and thylakoids (Fig. 3, D and E), and there was destruction of mitochondrial cristae. In addition, cell organelles including chloroplasts became redistributed in the cell and were no longer found only at the periphery (Fig. 2, C and D). Figure 3E shows severely damaged Arabidopsis cells in contact with the bacterial cells. Cell organelles are degraded and the cell walls are thin and highly convoluted. This unusual convolution of cell walls is also readily apparent in Figure 2, B through D. Host cell collapse was the final step of the bacterial infection.
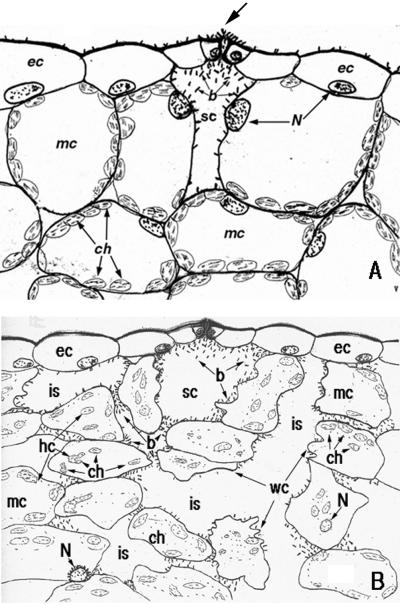
The spread and location of PA14 cells in a susceptible Arabidopsis host and the effect of the infection on the structure of host cells is summarized in the drawings shown in Figure 4, A and B. Figure 4A shows the initial steps of PA14 penetration through stomata and the concentration of bacteria in a substomatal cavity and their attachment to host cell walls. At this early stage of the infection (4–5 hpi) most host cells are intact except for the ones bordering on the substomatal cavity. These cells show degradation of organelles adjacent to the cavity. Figure 4B shows that at later stages (48 hpi) of the infection bacterial cells have spread in the intercellular spaces and are attached to the outer walls of epidermal and mesophyll cells. The mesophyll cells become separated due to the destruction of the middle lamella and the volume of the intercellular space increases with a concomitant decrease in the plant cell volume. The shrinkage of mesophyll cells is most likely a consequence of the degradation of mesophyll cell walls and plasmalemma resulting in the leakage of cytoplasmic contents, as well as the absorption of host cell nutrients by the proliferating bacteria.
Figure 4.
Drawing illustrating the course of P. aeruginosa PA14 infection in an Arabidopsis Ll-0 leaf. Detached leaves were incubated in a suspension of GFP-labeled PA14 for 2 h followed by transfer to water agar plates. At various times leaves were examined by confocal laser microscopy as described in “Materials and Methods.” A, Penetration of PA14 through a stoma (arrow) and accumulation in a substomatal cavity at 4 to 5 hpi. Redistribution and degradation of host organelles is seen only in cells in immediate contact with the bacteria. B, Distribution of PA14 in intercellular space at 48 hpi. Disruption of intercellular contacts and separation of mesophyll cells from each other, redistribution and degradation of host organelles; and convolution of plant cell walls are depicted. b, Bacteria; ch, chloroplast; ec, epidermal cells; is, intercellular space; mc, mesophyll cell; N, nucleus; sc, substomatal cavity; WC, wall convolution.
Attachment of PA14 to Plant Cell Walls
As described above in Figure 2C, when PA14 penetrates in the intercellular space between Arabidopsis cells, the bacteria attach perpendicularly to plant cell walls. In addition, as shown in Figure 5A, scanning electron microscopy of freeze-fractured Ll-O vessel parenchyma showed that PA14 appears to be able to penetrate the cell walls. Figure 5A shows what appears to be various stages of bacterial penetration through a region of cell wall that has the unusual convoluted or undulated surface described above. Moreover, Figure 5, A and C, shows holes in the plant cell wall with a diameter similar to that of PA14. These holes are presumably formed as a consequence of bacterial attachment and/or penetration. As described above, during systemic infection most vessel parenchyma cells were found to be invaded by large numbers of PA14 cells (Fig. 5D). In these cells PA14 was perpendicularly attached to the plasmolyzed host cytoplasm or organelles. Bacterial attachment to nuclei and chloroplasts can also be seen in the confocal micrograph shown in Figure 2D. As illustrated in Figure 5B, in contrast to P. aeruginosa, P. syringae pv maculicola ES4326 cells were found oriented parallel to the plant cell wall and no convolutions, undulations, or holes were ever observed.
Variation in Susceptibility of Arabidopsis Ecotypes to PA14
To assess differences in susceptibility of various Arabidopsis ecotypes to PA14 we determined the depth of PA14-GFP penetration in detached Ll-0, Columbia (Col-0), Wassilevskija (Ws-0), and Ag-0 leaf discs following a 2-h exposure to PA14 cell suspension (OD600 = .002) followed by incubation at room temperature on water agar. The depths at which PA14/GFP fluorescence was detectable starting from the surface of the leaves was measured using a CSS. PA14 penetrated through the entire thickness of the Ll-0 and Ws-0 leaves. In contrast, PA14 penetrated through approximately 50% and 20% of Col-0 and Ag-0 leaves, respectively. PA14 also penetrated all the way through a npr1-1 mutant leaf (in the Col-0 genetic background). The npr1-1 mutant was included in these studies because our laboratory had previously shown that npr1 mutants, which appear to be blocked in salicylic acid signaling and which exhibit enhanced susceptibility to a wide variety of pathogens including P. syringae, Erysiphe orontii, and Peronospora parasitica (Cao et al., 1994; Reuber et al., 1998), are also more susceptible to P. aeruginosa (Volko et al., 1998).
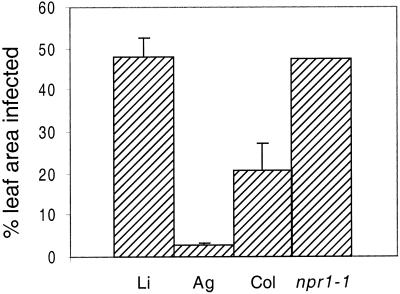
We also evaluated the differences in susceptibility of the same Arabidopsis accessions (and the npr1-1 mutant) by measuring the percentage of the leaf displaying disease symptoms when agar cylinders containing a lawn of PA14 were placed bacterial side down on detached leaves and incubated for 4 d at room temperature on 1.5% (w/v) water agar plates. The results showed that Ll-0 is more susceptible than Col-0 and Ag-0 and that npr1-1 is more susceptible than its wild-type Col-0 parent (Fig. 6).
Figure 6.
Susceptibility of Arabidopsis ecotypes and the Arabidopsis npr1 mutant to P. aeruginosa PA14. Detached leaves were infected using the agar cylinder procedure described in “Materials and Methods.” The bar graph depicts the percentage of total leaf area that displayed disease symptoms at 96 hpi. The percentage of leaf area infected is reported instead of lesion size because the lesions were irregularly shaped and elongated in the basipetal direction and because different accessions had significantly different sized leaves.
DISCUSSION
P. aeruginosa Is a Facultative Pathogen of Arabidopsis
The data presented in this paper indicate that P. aeruginosa strain PA14 is a facultative (as opposed to an obligate) pathogen of Arabidopsis. P. aeruginosa parasitism in Arabidopsis starts after bacterial attachment to the plant leaf surface and entry into Arabidopsis tissues via stomatal openings, followed by proliferation in the substomatal cavity and intercellular space. As the bacteria proliferate Arabidopsis cell walls become undulated and at least some bacteria appear to penetrate through the walls. Aggressive PA14 propagation in the intercellular space results in maceration and autolysis of plant cells. A systemic infection results from infection of parenchyma vessel cells and basipetal movement of P. aeruginosa along the vessels, apparently from one vessel parenchyma cell to the next. These data argue strongly against the possibility that the soft-rot symptoms elicited by P. aeruginosa on Arabidopsis and other plants that we reported in previous publications (Rahme et al., 1995, 1997; Mahajan-Miklos et al., 1999; Tan et al., 1999b) are merely the consequence of the infiltrating of a relatively large number of bacterial cells that are secreting a variety of potent degradative enzymes.
Attachment of P. aeruginosa to Plant Cell Surfaces
A distinct feature of P. aeruginosa infection in Arabidopsis is the perpendicular attachment of P. aeruginosa cells to mesophyll cell walls. The vast majority of P. aeruginosa cells were adherent to plant walls in the intercellular space during the parasitic stages of the infection. Although perpendicular attachment of Ralstonia solanacearum has also been observed to tobacco tissue culture cells (Van Gijsegem et al., 2000) and to tomato xylem cell walls (Vasse et al., 2000), most bacterial phytopathogens do not appear to attach to plant walls in this manner (Romantschuk, 1992). For example we found that P. syringae orients randomly or parallel to the plant cell walls in an infected Arabidopsis leaf. On the other hand, P. aeruginosa strain PAK A549 is also capable of attaching perpendicularly to the surface of human respiratory epithelial cells (S. Lory, personal communication), indicating that P. aeruginosa may utilize a similar mode of attachment to human and plant tissues.
Arabidopsis ecotypes exhibit a significant range in their susceptibility to PA14 infection. For example, ecotypes Ll-0 and Ag-0 are susceptible and resistant to PA14 infection, respectively. We previously interpreted this difference in susceptibility as an indication that some Arabidopsis ecotypes may have evolved resistance to particular P. aeruginosa strains. It is interesting to note and we show here that P. aeruginosa PA14 not only penetrates further into and forms larger lesions on the leaves of the susceptible Ll-0 ecotype than on resistant Ag-0 leaves, but that PA14 also attaches at least 5-fold more efficiently to the epidermis of Ll-0 leaves than to Ag-0 leaves.
Colonization of Arabidopsis Leaves
Following attachment to the leaf epidermis P. aeruginosa cells accumulate in the substomatal cavities and attach to the walls of the mesophyll cells lining the cavities. As the bacteria replicate, they spread in the intercellular space, presumably by digesting the middle lamellae. At this stage of the infection, the bacterial cells move to leaf vessels where they accumulate in vessel companion and parenchyma cells. In these experiments we did not obtain any direct evidence that P. aeruginosa can actively invade intact parenchyma vessel cells. The bacteria then move in a basipetal direction, apparently traveling through the contiguous vessel companion and parenchyma cells. These latter cells accumulate and transport photosynthates from photosynthesizing mesophyll cells to phloem sieve elements. In some cases these contiguous cells can also unload the sieve elements at sites of photosynthate utilization. Thus P. aeruginosa parasitism in vessel parenchyma allows the bacteria access to photosynthates in the phloem vessels as well. In contrast to the accumulation of bacteria in the vessel companion and parenchyma cells, we did not detect any bacteria in xylem vessels, unlike other pathogens such as Xanthomonas phaseoli and X. campestris. P. aeruginosa movement along minor veins was the prelude to systemic infection via maceration of the whole petiole and central bud.
Formation of Holes in Mesophyll Cell Walls
An interesting feature of P. aeruginosa pathogenesis in Arabidopsis is the perforation of mesophyll cell walls generating permanent holes. These holes only appear to form on cell walls that have a convoluted appearance, suggesting that the walls have been subjected to a significant degree of overall enzymatic digestion by hydrolytic enzymes prior to the formation of the holes. It is interesting that the holes, which are probably the result of localized enzymatic activity of bacterial cells, are 0.15 to 0.30 μm in diameter, approximately the same size as the bacterial cells that have a diameter of 0.2 to 0.4 μm. No holes were observed in mesophyll cell walls in P. syringae-infected leaves. Although P. aeruginosa cells can be observed directly penetrating through mesophyll cell walls, since we obtained no evidence of intracellular PA14 proliferation, it is most likely that P. aeruginosa forms the holes to gain access to nutrients.
Penetration of PA14 into Plant Cells
When P. aeruginosa was present in individual plant cells, the bacteria were found attached to the inner surface of the plant walls, as well as to the plasmolyzed host cytoplasmic structures. However, we have no direct evidence that PA14 is capable of penetrating or propagating in live plant cells. At later stages of the infectious process, most plant cells that contained internalized bacteria had highly undulated cells walls. In the case of mammalian infections, P. aeruginosa is capable of invading host cells and in some cases host cell invasion has been shown to be an important component of virulence (Fleiszig et al., 1995; Zaidi et al., 1999)
The Arabidopsis npr1 Mutant Is More Susceptible to PA14
In a previous publication we reported that the Arabidopsis npr1-1 mutant supports higher levels of PA14 growth than wild-type plants following the forced infiltration of PA14 into Arabidopsis leaves (Volko et al., 1998). The npr1-1 mutant was initially isolated on the basis that it failed to express the pathogenesis related PR-1 gene following treatment with salicylic acid (Cao et al., 1994), an important secondary messenger involved in the activation of a variety of plant defense-related responses. The data presented here confirm and extend our previous observations. The npr1-1 mutant was more susceptible to P. aeruginosa than wild-type Col-0. In experiments involving either the incubation of leaves in a suspension of PA14 or the placement of agar blocks containing a bacterial lawn on the surface of leaves, PA14 penetrated deeper into the leaf interior and formed significantly larger lesions than on wild-type plants.
MATERIALS AND METHODS
Bacterial Strains and Growth of Bacteria
Pseudomonas aeruginosa strain UCBPP-PA14 was propagated on solid or liquid Luria-Bertani medium containing 50 μg/mL of rifampicin at 37°C. Plasmid pSMC21 expressing a derivative of the Aequorea victoria GFP has been described (Bloemberg et al., 1997).
Plant Material and Growth of Plants
Arabidopsis ecotypes Col-0, Ll-0, Ws-0, and Ag-0 were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Arabidopsis plants were grown in Metro-Mix 2000 in either a climate-controlled greenhouse at 19°C under a 12-h light/dark cycle with supplemental fluorescent illumination or in a Percival AR-60L growth chamber at 20°C.
Arabidopsis Pathogenicity Assays
Six- to 8-week-old intact or detached Arabidopsis rosette leaves were used for pathogenicity assays. Intact leaves were infiltrated with a bacterial suspension as previously described (Rahme et al., 1995) using a 1-mL disposable syringe without a needle at a dose of approximately 103 cfu/cm2 leaf area. Infiltrated plants were incubated at 30°C and approximately 80% relative humidity under a 12-h light/dark cycle and analyzed 1, 2, 3, and 4 d post-infiltration. In some experiments whole leaves or 4-mm diameter leaf discs cut from the central portion of a leaf from each side of the central vein with a cork borer were immersed in a PA14 suspension at OD600 = 0.02 in 10 mm MgSO4 and incubated at room temperature. Detached leaves were placed on a 1.5% (w/v) water agar surface with their petioles embedded into the agar. The detached leaves were subsequently infected by placing in the middle of the leaf two 3-mm diameter agar cylinders cut from a Luria agar plate containing an overnight lawn of PA14. The agar cylinders were placed bacterial-side down on either side of the central vein.
Light Microscopy and Histochemistry
For trypan blue staining infected leaves were cleared in a solution of lactophenol:ethanol (1 vol of lactophenol:2 vol ethanol) for a period of 12 to 24 h with one change of solution. Lactophenol was prepared by mixing equal volumes of phenol, lactic acid, glycerol, and water. Cleared Arabidopsis leaves were moved to a fresh lactophenol solution containing 1 mg/mL of trypan blue. Leaves were stained for 10 min before mounting on slides and examination with an Axioscope (Zeiss, Jena, Germany) with bright field and Nomarski optics. Photomicrographs were taken with an MC 80 automatic camera.
For Syto 9 staining, small pieces (1 × 3 mm) of leaf tissue were fixed first in a 4% (w/v) glutaraldehyde solution in 0.1 n cacodilate buffer at pH 7.2 for 3 h and then in 2% (w/v) osmium tetroxide solution in the same buffer, dehydrated in an alcohol series (30%, 70%, 96%, and 100%), and embedded in Spurr medium at 60°C for 16 h. Thin sections of the polymerized leaf material were stained with the fluorescent dye Syto 9 (Molecular Probes, Eugene, OR) and analyzed with a confocal laser spectrophotometer (TCS NT, Leica Microsystems, Wetzlar, Germany) by excitation at 504 nm and emission at 523 nm. SYTO 9 is a non-specific cell-permeant cyanine dye that stains nucleic acids, bacteria, and a variety of subcellular structures, including plant cell walls.
The attachment of PA14/pSMC21 (expressing GFP) to leaf surface structures and the depth of PA14/pSMC21 penetration in Arabidopsis leaves was visualized with a confocal laser spectrophotometer (TCS NT, Leica) by excitation at 488 nm and monitoring emission intensity at 511 nm.
Scanning Electron Microscopy
Pieces of Arabidopsis leaves were fixed in 4% (w/v) paraformaldehyde and passed through an ethanol series (30%, 50%, 70%, 96%, and 100%). The fixed plant leaves, including those freeze-fractured in liquid nitrogen as described (Andreev and Plotnikova, 1989), were dried in a critical point drying apparatus (Samdri-PVT-3B, Tousimis, Rockville, MD), mounted on stubs, coated with a 20- to 25-μm layer of gold-palladium in a Hummer II Sputter Coater (Technics, Alexandria, VA), and studied using a scanning electron microscope (AMRAY 1000, Bedford, MA). Fixation, dehydration, drying at critical point, coating with gold, and scanning electron microscopy analysis was performed as described above.
Transmission Electron Microscopy
Small pieces of infected or uninfected Arabidopsis leaves (1 × 3 mm) were fixed overnight in 3% (w/v) glutaraldehyde in 0.1 n cacodilate buffer at pH 7.2, washed in the same buffer, post-fixed in 2% (w/v) osmium tetroxide, dehydrated in an alcohol series (30%, 70%, 96%, and 100%), and embedded in Spurr medium at 60°C for 16 h. After polymerization, ultra-thin sections were cut with an LKB 8 800 Ultratome, stained in 2% (w/v) lead citrate and 2% (w/v) uranyl acetate (Reynolds, 1963), and analyzed using an electron microscope (401, Philips, Eindhoven, The Netherlands).
ACKNOWLEDGMENTS
We would like to thank Livingston Van De Water and Bob Crowther (Shriners' Burns Institute, Boston) for the use of the electron microscope.
Footnotes
This work was supported by the National Institutes of Health (grant no. GM48707 to F.M.A.) and by a grant from Aventis SA to Massachusetts General Hospital.
LITERATURE CITED
- Andreev LN, Plotnikova JM. Structure and ultrastructure of rust fungi. In: Gorlenko MV, editor. Wheat Rust: Cytology and Physiology. Moscow: Nauka; 1989. pp. 5–84. [Google Scholar]
- Bloemberg GV, O'Toole GA, Lugtenberg BJJ, Colter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britigan BE, Rasmussen GT, Cox CD. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun. 1997;63:1071–1076. doi: 10.1128/iai.65.3.1071-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder WH. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117. [Google Scholar]
- Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is non-responsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JJ, Schroth MN, Kominos SD, Green SK. Ornamental plants as carriers of Pseudomonas aeruginosa. Phytopathology. 1975;65:425–431. [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod RP, Braun AC. Pseudomonas aeruginosa: its role as a plant pathogen. J Bacteriol. 1942;44:633–645. doi: 10.1128/jb.44.6.633-645.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiszig SMJ, Zaidi TS, Pier GB. Pseudomonas aeruginosa invasion of and multiplication within corneal epithelial cells in vitro. Infect Immun. 1995;63:4072–4077. doi: 10.1128/iai.63.10.4072-4077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominos SD, Copeland CE, Grosiak B, Postic B. Introduction of Pseudomonas aeruginosa into a hospital via vegetables. Appl Microbiol. 1972;24:567–570. doi: 10.1128/am.24.4.567-570.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan M-W, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Pier GB, Grout M, Zaidi TS, Olsen JC, Johnson LG, Yankaskas JR, Goldberg JB. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Tan M-W, Le L, Wong SM, Tompkins RG, Calderwood SB, Ausubel FM. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM. Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 1998;16:473–485. doi: 10.1046/j.1365-313x.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantschuk M. Attachment of plant pathogenic bacteria to plant surfaces. Annu Rev Phytopathol. 1992;30:225–243. doi: 10.1146/annurev.py.30.090192.001301. [DOI] [PubMed] [Google Scholar]
- Schott EJ, Davis KR, Dong X, Mindrinos M, Guevara P, Ausubel FM. Pseudomonas syringae infection of Arabidopsis thaliana as a model system for studying plant-bacterial interactions. In: Silver S, Chakrabarty AM, Iglewski B, Kaplan S, editors. Pseudomonas: Biotransformation, Pathogenesis, and Evolving Biotechnology. Washington, DC: American Society of Microbiology; 1990. pp. 82–90. [Google Scholar]
- Tan M-W, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999a;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-W, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999b;96:2408–2499. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijsegem F, Vasse J, Camus JC, Marenda M, Boucher C. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol Microbiol. 2000;36:249–60. doi: 10.1046/j.1365-2958.2000.01851.x. [DOI] [PubMed] [Google Scholar]
- Vasse J, Genin S, Frey P, Boucher C, Brito B. The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol Plant-Microbe Interact. 2000;13:259–267. doi: 10.1094/MPMI.2000.13.3.259. [DOI] [PubMed] [Google Scholar]
- Volko SM, Boller T, Ausubel FM. Isolation of new Arabidopsis mutants with enhanced disease susceptibility. Genetics. 1998;149:537–548. doi: 10.1093/genetics/149.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi TS, Lyczak J, Preston M, Pier GB. Cystic fibrosis transmembrane conductance regulator-mediated corneal epithelial cell ingestion of Pseudomonas aeruginosa is a key component in the pathogenesis of experimental murine keratitis. Infect Immun. 1999;67:1481–1492. doi: 10.1128/iai.67.3.1481-1492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]