Abstract
Background:
Lipoxin A4 (LXA4) can alleviate lipopolysaccharide (LPS)-induced acute lung injury (ALI) and acute respiratory distress syndrome through promoting epithelial sodium channel (ENaC) expression in lung epithelial cells. However, how LXA4 promote ENaC expression is still largely elusive. The present study aimed to explore genes and signaling pathway involved in regulating ENaC expression induced by LXA4.
Methods:
A549 cells were incubated with LPS and LXA4, or in combination, and analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) of ENaC-α/γ. Candidate genes affected by LXA4 were explored by transcriptome sequencing of A549 cells. The critical candidate gene was validated by qRT-PCR and Western blot analysis of A549 cells treated with LPS and LXA4 at different concentrations and time intervals. LXA4 receptor (ALX) inhibitor BOC-2 was used to test induction of candidate gene by LXA4. Candidate gene siRNA was adopted to analyze its influence on A549 viability and ENaC-α expression. Phosphoinositide 3-kinase (PI3K) inhibitor LY294002 was utilized to probe whether the PI3K signaling pathway was involved in LXA4 induction of candidate gene expression.
Results:
The A549 cell models of ALI were constructed and subjected to transcriptome sequencing. Among candidate genes, N-myc downstream-regulated gene-1 (NDRG1) was validated by real-time-PCR and Western blot. NDRG1 mRNA was elevated in a dose-dependent manner of LXA4, whereas BOC-2 antagonized NDRG1 expression induced by LXA4. NDRG1 siRNA suppressed viability of LPS-treated A549 cells (treatment vs. control, 0.605 ± 0.063 vs. 0.878 ± 0.083, P = 0.040) and ENaC-α expression (treatment vs. control, 0.458 ± 0.038 vs. 0.711 ± 0.035, P = 0.008). LY294002 inhibited NDRG1 (treatment vs. control, 0.459 ± 0.023 vs. 0.726 ± 0.020, P = 0.001) and ENaC-α (treatment vs. control, 0.236 ± 0.021 vs. 0.814 ± 0.025, P < 0.001) expressions and serum- and glucocorticoid-inducible kinase 1 phosphorylation (treatment vs. control, 0.442 ± 0.024 vs. 1.046 ± 0.082, P = 0.002), indicating the PI3K signaling pathway was involved in regulating NDRG1 expression induced by LXA4.
Conclusion:
Our research uncovered a critical role of NDRG1 in LXA4 alleviation of LPS-induced A549 cell injury through mediating PI3K signaling to restore ENaC expression.
Keywords: Acute Lung Injury, Epithelial Sodium Channel, Lipopolysaccharide, Lipoxin A4, N-myc Downstream-Regulated Gene-1
摘要
背景:
脂氧素A4(LXA4)能够通过促进肺上皮细胞的钠离子通道(ENaC)表达而缓解脂多糖(LPS)诱导的急性肺损伤 (ALI)和急性呼吸窘迫综合征(ARDS)。然而,LXA4如何促进ENaC表达的主要机制依旧不明。本研究的目的是探索LXA4 诱导ENaC表达的相关基因和信号通路机制。
方法:
采用LPS和LXA4,或两者的组合,处理A549细胞,模拟ALI/ARDS治疗,通过real-time PCR分析ENaC-a/?转录水平。 对处理的A549细胞进行转录组测序,探索分析受LXA4影响的候选基因。对经不同浓度LPS和LXA4,以及不同时间间隔处理 的A549细胞进行real-time PCR和Western blot检测,验证关键的候选基因表达变化。利用LXA4 受体(ALX)抑制剂BOC-2验 证LXA4对候选基因的诱导表达作用。通过siRNA分析沉默候选基因对A549细胞存活率和ENaC-a表达的影响。利用PI3K抑制 剂LY294002分析PI3K信号通路是否参与调控LXA4诱导候选基因表达。
结果:
成功建立模拟ALI的A549细胞模型,并对其进行转录组测序。在多个候选基因中,通过real-time PCR和Western blot验 证了LXA4诱导NDRG1基因表达。NDRG1 mRNA水平随LXA4浓度增加而升高,而BOC-2则拮抗LXA4对NDRG1的诱导作 用。NDRG1 siRNA抑制LPS处理的A549细胞存活率(treatment vs. control, 0.605±0.063 vs. 0.878±0.083, P=0.040)和ENaC-a表 达(treatment vs. control, 0.458±0.038 vs. 0.711±0.035, P=0.008)。LY294002抑制NDRG1 (treatment vs. control, 0.459±0.023 vs. 0.726±0.020, P=0.001) 和ENaC-a (treatment vs. control, 0.236±0.021 vs. 0.814±0.025, P<0.001)的表达,以及SGK1的磷酸化水平 (treatment vs. control, 0.442±0.024 vs. 1.046±0.082, P=0.002),表明PI3K信号通路参与调控LXA4诱导NDRG1的表达。
结论:
本研究揭示了NDRG1在介导LXA4保护受LPS损伤的A549细胞关键作用。LXA4经PI3K信号通路上调NDRG1的表达水 平,增加ENaC的表达水平,缓解LPS诱导的A549细胞损伤。
INTRODUCTION
Acute lung injury (ALI) is a common clinical syndrome of acute respiratory failure that frequently occurs in severe patients, even with high morbidity. ALI and its exacerbated form, acute respiratory distress syndrome (ARDS), are categorized by their symptoms and severity according to the Berlin Definition.[1] ALI is characterized by increased permeability of the alveolar-capillary barrier, which consequently leads to lung edema with pulmonary infiltrates and hypoxemia.[2,3] ALI patients manifest significant pulmonary inflammation that causes flooding of the pulmonary alveoli, resulting in obstructed gas exchange and consequent hypoxemia. Lipopolysaccharide (LPS), a kind of endotoxin released from the outer membrane of Gram-negative bacteria, is one of the most common causes of ALI. LPS had been reported to induce expression of inflammatory genes in lung epithelial cells of ALI models, which impairs lung epithelial channel function.[4] Epithelial channel proteins, such as Na+-K+ ATPase, aquaporins, and epithelial sodium channel (ENaC), play important roles in effective removal of excessive alveolar edema, named alveolar fluid clearance (AFC).[5,6,7] Among these channels, ENaC is the primary drive of fluid transport to maintain the epithelial fluid layer of pulmonary alveoli. ENaC dysregulation caused by pro-inflammatory cytokines, such as tumor necrosis factor-α, transforming growth factor-β1, interleukin-1β (IL-1β), IL-4, and IL-13, is primarily responsible for lung edema.[7] Lipoxins (LXs), a group of endogenous lipids, can alleviate inflammation and show great therapeutic potential in lung edema. LXA4 ameliorates LPS-induced pulmonary edema and maintains lung airway epithelial cells and airway permeability.[8] LXA4 stimulates AFC through promoting alveolar ENaC subunits' expression and restores ENaC function against LPS.[9,10] The phosphoinositide 3-kinase (PI3K)/serine threonine kinase (AKT) signaling pathway is reported to be profoundly involved in regulating ENaC subunits' expression induced by LXA4.[10,11,12,13] However, how LXA4 enhance ENaC expression in lung alveolar epithelial cells is still largely elusive. This research aimed to explore the signaling pathway of how LXA4 promotes ENaC subunits' expression to preserve lung epithelial cell function inhibited by LPS. We adopted transcriptome sequencing of A549 cells treated with different doses of LPS and LXA4 and discovered N-myc downstream-regulated gene-1 (NDRG1) as a critical mediator of LXA4 and PI3K signaling to restore ENaC subunits' expression against LPS.
METHODS
Cell lines, reagents, and culture condition
The lung epithelial cell line and A549 cells were cultured in complete Dulbecco's Modified Eagle's medium (DMEM) (SH30022.01B, Hyclone, USA) containing 10% (v/v) fetal bovine serum (FBS) (10100147, Gibco Sera, USA), supplemented with penicillin-streptomycin Solution (SV30010, ×100, HyClone, USA). For experimental tests, A549 cells were inoculated into 10 cm dishes or 6-well plates and cultured in humidified atmosphere containing 5% CO2 at 37°C for 24 h. As cell confluence reached 80%, A549 cells were further cultured with FBS-free DMEM for 12 h. Then, A549 cells were treated with LPS (Sigma, USA), LXA4 (Sigma), LXA4 receptor (ALX) inhibitor (BOC-2, GenScript, USA), or PI3K inhibitor LY-294002 (Sigma) for 6 h or more time.
Quantitative real-time polymerase chain reaction
Total RNAs were extracted from A549 cells treated with LPS, LXA4, or siRNA in 6-well culture plates, according to the manufacturer's protocol (MiniBEST Universal RNA Extraction Kit 9767, Takara, Japan). The RNA concentrations of each sample were determined by NanoDrop2000 (Thermo, USA) detection. The cDNAs (10 μl) were synthesized from 2 μg RNA of each sample by utilizing reverse transcription kit (PrimeScript™ RT reagent Kit with gDNA Eraser RR047Q, Takara). The cDNAs were diluted ×20 (200 μl), and 0.5–1.0 μl of the diluted cDNAs were analyzed by quantitative real-time-polymerase chain reaction (qRT-PCR) based on SYBR Green kit (SYBR® Premix Ex Taq™ II [Tli RNaseH Plus] RR820A, Takara, Japan). The relative mRNA levels were calculated by CFX Manager software (Bio-Rad Laboratories, USA) and the comparative Ct (ΔΔCt) method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were deployed as a reference gene. The primer sequences were as the followings: ENaC-α forward: 5'TGACATCCCAGGAATGGGTC3', reverse: 5'CCAGCAGGTCAAAGACGAGC3'; ENaC-γ forward: 5'GTGCTTGTCCATCAG-CAGAA3', reverse: 5'AAGGCAGATCTGGAGGGAAT3'; GAPDH forward: 5'GGT-GGTCTCCTCTGACTTCAACA3', reverse: 5'GTTGCTGTAGCCAAATTCGTTG-T3'; β-actin forward: 5'CATGTACGTTGCTATCCAGGC3', reverse: 5'CTCCTT-AATGTCACGCACGAT3'; NDRG1 forward: 5'CAAAGACCACTCTCCTCAAGAT-G3', reverse: 5'TGCCATCCAGAGAAGTGACG3'. The PCR program was set for incubation at 95°C for 30 s, followed by 35 times of PCR cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. After the cycles were finished, the melting curves of primers were detected from 65°C to 95°C at an interval of 0.5°C. The Ct values of β-actin or GAPDH were utilized as sample internal references.
siRNA transfection
A459 cells were cultured in 6-well plates until 50% confluence and transfected with 50 nmol/L of small interfering RNA (siRNA) targeting NDRG1 (5'GCUGAUCCAGUUUCCGGAAtt3') or scrambled siRNA (siScr) for 24 h, according to the siRNA kit instructions (X-tremeGENE™ siRNA transfection reagent, Roche, Switzerland). Twenty-four hours after cell transfection, the treated A549 cells were digested and seeded in 96-well plates for Cell Counting Kit-8 (CCK-8) analysis or cultured for another 24 h in 10 cm dishes for Western blot analysis.
Cell Counting Kit-8 assay
A549 cells cultured in 6-well plate were transfected with siScr or NDRG1 siRNA. After 24 h, A549 cells were digested and suspended in complete medium at 0.5 × 105/ml. 100 μl aliquots of A549 cells was transferred to 96-well plate and cultured for 24 h and incubated with serum-free medium for 12 h before treatment with 1 μg/ml LPS or 100 nmol/L LXA4, or in combination, for 6 h. Then, 10 μl of CCK-8 (CK04, Dojindo Laboratories, Japan) substrate was directly added into each well of the 96-well plate and incubated at 37°C for 2 h. The 450 nm absorbance of each well was detected by Multiskan™ GO device (Thermo Scientific, USA).
Western blot analysis
Protein expressions and phosphorylation were detected by Western blot. A549 cells cultured in 10 cm dish or 6-well plate were treated with LPS, LXA4, or in combination, or 10 μmol/L BOC-2 and 25 μmol/L LY294002 for 6 h. Total proteins were extracted from the treated A549 cells lysed with 50–300 μl cold radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, China) supplemented with ×100 diluted Halt™ protease and phosphatase inhibitor cocktail (Thermo Scientific Pierce, USA). The cell pellets were vortexed every 5 min for 9 times and incubated on ice. The cell lysates were centrifuged at 14,000 ×g for 10 min at 4°C to discard fragment pellets. Total protein concentrations of the supernatant were determined by Pierce™ BCA Protein Assay Kit (Thermo Scientific Pierce). An equal amount of total protein (20–50 μg) was loaded into sodium dodecyl sulphate-polyacrylamide gel electrophoresis and separated by electrophoresis under 80–100 V. After electrophoresis, the protein bands were transferred to polyvinylidene difluoride (PVDF) membrane under 100 V for 1 h in cold water bath. Then, the PVDF membranes were blocked with phosphate-buffered saline (PBS) containing 5% nonfat milk or BSA for 1 h and incubated with the primary antibodies (×1000 dilution) overnight at 4°C. The PVDF membrane was washed 3 times with phosphate-buffered saline containing 0.05% Tween-20 (PBST) and incubated with the horseradish peroxidase (HRP)-conjugated secondary antibody (×10,000 dilution) at 37°C for 1 h. The PVDF membranes were washed 3 times with PBST again and labeled with the enhanced Pierce™ Fast Western blot Kit (Thermo Scientific Pierce, USA) for 1 min at room temperature in dark. Finally, the PVDF membranes were exposed to the imaging system (Bio-Rad, USA) to capture the light signals of protein bands.
For Western blot analysis, the adopted antibodies included ENaC-α antibody (70R-13299, Fitzgerald, USA); serum- and glucocorticoid-inducible kinase 1 (SGK1) antibody (ab59337, Abcam, UK), SGK1-phosphor-S422 antibody (ab55281, Abcam), β-actin antibody AC-15 (ab6276 Abcam), goat anti-rabbit IgG H&L HRP-conjugated antibody (ab6721, Abcam), and goat anti-mouse IgG-Fc fragment HRP-conjugated antibody (Bethyl, USA).
Statistical analysis
All experiments were performed in triplicate. Data were analyzed by the statistical software GraphPad Prism 5 (GraphPad Software Inc., USA). Data were presented as a mean ± standard deviation (SD). Statistical analysis was executed by one-way analysis of variance (ANOVA), and P < 0.05 was considered to be statistically significant.
RESULTS
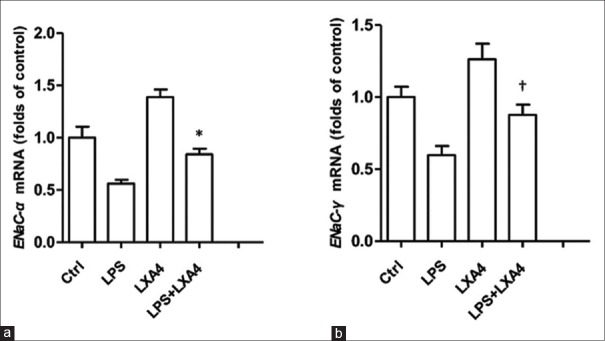
To construct lung epithelial cell model of LXA4 alleviation of LPS-induced ALI, we treated A549 cells with LPS, LXA4, or in combination In vitro. As well known, ENaC complex consists of three subunits, α, β, and γ. LPS could suppress ENaC function, whereas LXA4 induce ENaC subunits' expression. To our expect, real-time-PCR analysis results showed that LPS suppressed ENaC-α mRNA expression, whereas LXA4 antagonized LPS inhibition and enhanced ENaC-α mRNA expression [Figure 1a] (treatment vs. control, P = 0.006). LXA4 also alleviated inhibition of ENaC-γ mRNA expression by LPS [Figure 1b] (treatment vs. control, P = 0.026). These results indicated that LXA4 could promote ENaC subunits' transcription to restore ENaC function impaired by LPS. It also meant that the A549 cell model treated with LPS and LXA4 was successfully constructed. Through treating A549 cells with different concentrations of LPS and LXA4 (data not shown), the eligible treatments were validated as 1 μg/ml LPS, 100 nmol/L LXA4, or in combination, and adopted for further experimental tests.
Figure 1.
LXA4 restored ENaC subunits' mRNA expressions in A549 cells challenged by LPS. (a) LXA4 rescued ENaC-α subunit mRNA expression in A549 cells treated with LPS. The mRNA levels were determined by real-time PCR analysis. The treated groups were Control, LPS (1 μg/ml), LXA4 (100 nmol/L), and in combination. *P = 0.006, LPS versus LPS + LXA4. (b) LXA4 restored EnaC-γ subunit mRNA expression in A549 cells treated with LPS. †P = 0.026, LPS versus LPS + LXA4. LXA4: Lipoxin A4; LPS: Lipopolysaccharide; NDRG1: N-myc downstream-regulated gene 1; ENaC-α: Epithelial sodium channel α subunit; ENaC-γ: Epithelial sodium channel γ subunit; PCR: Polymerase chain reaction.
To investigate candidate genes' expression affected by LXA4, we employed transcriptome sequencing of A549 cell models. A549 cells in 6-well plate were treated with 1 μg/ml LPS, 100 nmol/L LXA4, or in combination, for 6 h. Then, 20 μg of total RNAs extracted from each A549 cell group were subjected to transcriptome analysis. Comparisons of mRNA expression profile between A549 cell groups were made to explore potential candidate genes affected by LXA4 [Figure 2]. The mRNA levels of some candidate genes changed significantly upon LXA4 stimulation, compared with that of the LPS-treated group [Table 1]. To verify these candidate genes, we performed real-time PCR to detect mRNA change dynamics of ANGPTL4, NDRG1, LRP1, PLIN2, and so on. Among these genes, NDRG1 mRNA was preliminarily proved to be positively correlated with LXA4 (data not shown).
Figure 2.

Transcriptome sequencing and comparison of the treated A549 cells. Transcriptome array analysis of A549 cells treated with LPS (1 μg/ml), LXA4 (100 nmol/L), and in combination, respectively. LXA4: Lipoxin A4; LPS: Lipopolysaccharide.
Table 1.
The mRNA level changes of candidate genes
| Gene | LPS | LXA4 | LPS + LXA4 |
|---|---|---|---|
| IGFBP3 | 1 | 2.59 | 2.30 |
| ANGPTL4 | 1 | 1.73 | 1.85 |
| NDRG1 | 1 | 1.53 | 1.35 |
| LRP1 | 1 | 1.48 | 1.28 |
| PLIN2 | 1 | 1.21 | 1.03 |
| BLOC1S5 | 1 | 7.43 | 5.70 |
| CCDC180 | 1 | −6.09 | −6.09 |
| FSBP | 1 | −6.13 | −6.13 |
| SCHIP1 | 1 | −7.19 | −7.19 |
The mRNA levels of candidate genes of the LPS treated group were define as “1”, and that of the LXA4 and LPS + XA4 treated groups were calculated by comparison with the LPS treated group. IGFBP3: Insulin-like growth factor binding protein 3; ANGPTL4: Angiopoietin-like 4; NDRG1: N-myc downstream regulated 1; LRP1: LDL receptor-related protein 1; PLIN2: Perilipin 2; BLOC1S5: Biogenesis of lysosomal organelles complex 1 subunit 5; CCDC180: Coiled-coil domain containing 180; FSBP: Fibrinogen silencer binding protein; SCHIP1: Schwannomin interacting protein 1; LXA4: Lipoxin A4; LPS: Lipopolysaccharide; LDL: Low-density lipoprotein.
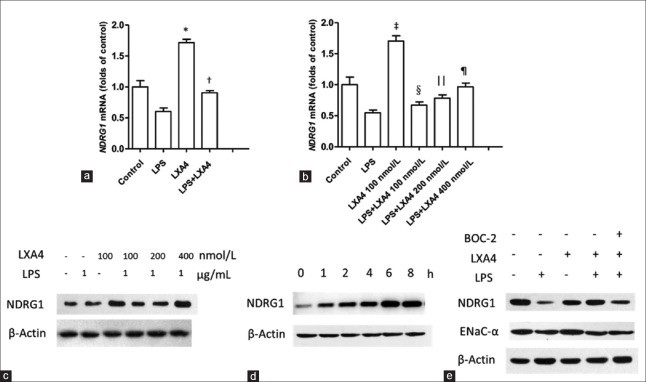
Detection of NDRG1 mRNA levels by real-time-PCR confirmed that LPS treatment suppressed NDRG1 transcription, whereas LXA4 heightened NDRG1 mRNA levels and alleviated LPS inhibition effects on NDRG1 transcription [Figure 3a] (treatment vs. control, P < 0.001 and P = 0.004, respectively). To probe the relationship between NDRG1 mRNA transcription and LXA4 concentration, we treated A549 cells with LXA4 at 100, 200, and 400 nmol/L, respectively, in the presence of LPS. The real-time-PCR results showed that NDRG1 mRNA levels were elevated in a dose-dependent manner of LXA4 [Figure 3b] (LPS vs. LXA4 [100 nmol/L], P < 0.001; LPS vs. LPS + LXA4 [100 nmol/L], P = 0.122, non-significant; LPS vs. LPS + LXA4 [200 nmol/L], P = 0.013;LPS vs. LPS + LXA4 [400 nmol/L], P = 0.001). Accordingly, it was proved by Western blot analysis that NDRG1 protein levels increased in a positive correlation with LXA4 concentrations [Figure 3c]. In addition, NDRG1 protein expression was induced by LXA4 in a time-dependent manner as its level increased until 6 h after LXA4 stimulation [Figure 3d]. On the contrary, ALX inhibitor BOC-2 antagonized expressions of NDRG1 and ENaC-α protein induced by LXA4 [Figure 3e]. These results revealed an important role of LXA4 in promoting NDRG1 protein expression through potentiation of mRNA transcription.
Figure 3.
LXA4 promoted NDRG1 expression in A549 cells. (a) LXA4 induced NDRG1 expression and alleviated inhibition of LPS. *P < 0.001, LPS versus LXA4; †P = 0.004, LPS versus LPS + LXA4. (b) LXA4 (100, 200, and 400 nmol/L) restored NDRG1 mRNA expressions suppressed by LPS. ‡P < 0.001, LPS versus LXA4 (100 nmol/L); §P = 0.122, NS, LPS versus LPS + LXA4 (100 nmol/L); ||P = 0.013, LPS versus LPS + LXA4 (200 nmol/L); ¶P = 0.001, LPS versus LPS + LXA4 (400 nmol/L). (c) LXA4 enhanced NDRG1 protein expression inhibited by LPS. (d) LXA4-induced NDRG1 expression in a time-dependent manner. (e) BOC-2 suppressed NDRG1 and ENaC-α expression induced by LXA4. LXA4: Lipoxin A4; LPS: Lipopolysaccharide; NDRG1: N-myc downstream-regulated gene 1; ENaC-α: Epithelial sodium channel α subunit; NS: Non-significant.
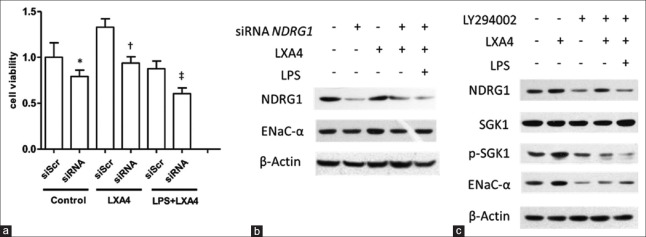
To further explore the function of NDRG1 in improving A549 cell viability, we employed siRNA to knockdown NDRG1 expression. Downregulation of NDRG1 protein expression resulted in marked reduction of A549 cell viability (treatment vs. control, 0.605 ± 0.063 vs. 0.878 ± 0.083, P = 0.040) and attenuated protection function of LXA4 against LPS [Figure 4a]. NDRG1 siRNA caused obvious decline in ENaC-α expression, which accounted for impaired cell viability challenged by LPS [Figure 4b] (treatment vs. control, 0.458 ± 0.038 vs. 0.711 ± 0.035, P = 0.008). It has been reported that PI3K signaling was intimately involved in regulating ENaC subunits expression, and NDRG1 was a substrate of SGK1. In accordance with previous reports, the PI3K inhibitor LY294002 could efficiently repress SGK1 phosphorylation (treatment vs. control, 0.442 ± 0.024 vs. 1.046 ± 0.082, P = 0.002) and downregulate expressions of NDRG1 (treatment vs. control, 0.459 ± 0.023 vs. 0.726 ± 0.020, P = 0.001) and ENaC-α (treatment vs. control, 0.236 ± 0.021 vs. 0.814 ± 0.025, P < 0.001), indicating that the PI3K signaling pathway was profoundly involved in regulating NDRG1 and ENaC expressions stimulated by LXA4 [Figure 4c] (treatment vs. control, P = 0.002). In conclusion, our research uncovered a critical role of NDRG1 in mediating LXA4 protection of A549 cells from LPS lesion in which PI3K signaling was involved [Figure 4].
Figure 4.
The PI3K signaling pathway was involved in the induction of NDRG1 expression by LXA4. (a) NDRG1 siRNA resulted in decreased viability of A549 cells. A549 cell viability was detected by CCK-8 kit. *P = 0.278, ns, siScr versus siRNA of control; †P = 0.014, siScr versus siRNA of LXA4; ‡P = 0.040, siScr versus siRNA of LPS + LXA4. (b) NDRG1 siRNA caused ENaC-α decreased in A549 cells (P = 0.008, lane 3 vs. lane 4). (c) The PI3K inhibitor LY294002 (25 μmol/L) antagonized SGK1 phosphorylation and ENaC-α expression induced by LXA4 (100 nmol/L) (P = 0.002, lane 2 vs. lane 4). LXA4: Lipoxin A4; LPS: Lipopolysaccharide; NDRG1: N-myc downstream-regulated gene 1; ENaC-α: Epithelial sodium channel α subunit; siScr: Scramble small interfering RNA.
DISCUSSION
ALI/ARDS is a common clinical syndrome of acute respiratory failure that is characterized by lung edema with pulmonary infiltrates and hypoxemia. LPS, a kind of pro-inflammatory endotoxin from Gram-negative bacteria, can initiate inflammatory genes' expression and impair channel proteins' function in lung epithelial cells, resulting in pulmonary edema in ALI patients.[2,14] ENaC is the primary executor for effective removal of excessive alveolar edema through active Na+ transport across the alveolar epithelium.[9,15] LPS is reported to activate nuclear factor-κB signaling to inhibit SGK1 and ENaC expression in alveolar epithelial cells.[16] In contrast, LXA4, an endogenously synthesized lipid, can effectively alleviate LPS-caused inflammation through priming AKT signaling and promoting ENaC expression.[9,10] Great efforts have been devoted to the investigation of LXA4 protecting lung epithelial cells from LPS damage. However, the mechanism of how LXA4 promote ENaC expression and alleviate LPS-induced impairment is still not well understood. The present study aimed to explore genes and signaling pathways involved in regulating ENaC expression by LXA4.
As well known, ENaC expression is inversely affected by LPS and LXA4. In accordance with previous reports, A549 cells treated with LPS showed decreased ENaC subunits' mRNA levels, while LAX4 alleviated LPS inhibition of ENaC subunits' mRNA expression. Through transcriptome sequencing of A549 cell models treated with different concentrations of LPS and LXA4, or in combination, we found that mRNA levels of some candidate genes changed dramatically upon LPS and LXA4 stimulation. Among these candidate genes, we proved a critical role of NDRG1 in mediating LXA4 protection of A549 cell injury from LPS lesion. Our results showed that NDRG1 mRNA downregulated by LPS could be restored by LXA4. NDRG1 expression was elevated in a concentration-dependent manner of LXA4. Our results also uncovered an important role of NDRG1 in mediating LXA4 stimulation to promote ENaC expression. Interestingly, NDRG1 was reported to maintain the integrity of airway epithelial barrier as NDRG1 knockdown significantly decreased claudin-9 expression and impaired the barrier function.[17] However, there were controversial issues about NDRG1 functions. NDRG1 was recognized as a potent tumor suppressor, while its ectopic expression promoted tumor cell survival.[18,19,20] NDRG1 promoted the stem-like properties and survival of lung cancer cells.[21] Suppression of NDRG1 significantly counteracted hepatocellular carcinoma cell growth due to extensive cellular senescence.[22] NDRG1 deficiency obstructed fetal growth and the intrauterine response to hypoxic injury.[23] These reports mainly supported the pro-survival function of NDRG1. In our experiment, NDRG1 siRNA dampened the protection effects of LXA4 on A549 cell viability under LPS challenge. As expected, NDRG1 siRNA inhibited ENaC-α expression induced by LXA4.
The PI3K signaling pathway is implicated in the regulation of ENaC expression to control AFC and relieve pulmonary edema.[10,11,12,24] We sought to test whether NDRG1 was influenced by the PI3K signaling activated by LXA4. Our results proved that NDRG1 was a downstream target of PI3K as the PI3K inhibitor LY294002 downregulated NDRG1 expression and SGK1 phosphorylation, indicating the PIK3-SGK1 signaling pathway was involved in regulating NDRG1 expression induced by LXA4. These results were consistent with the previous report that NDRG family members were substrates of SGK1 and GSK3.[25] SGK1 significantly increases phosphorylation of NDRG1 to confer cell proliferation.[26] In this study, we proposed NDRG1 as a critical mediator of LXA4 to promote ENaC expression, which was at downstream of the PI3K-SGK1 signaling.
In summary, our research uncovered an important role of NDRG1 in the LXA4 alleviation of LPS-induced A549 cell injury. NDRG1 was upregulated by LXA4 and subsequently mediated PI3K signaling to restore ENaC expression and promoted A549 cell survival.
Financial support and sponsorship
This work was supported by a grant from the Shenzhen Municipal Science and Technology Innovation Committee (No. JCYJ20150403102020225).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Qiang Shi
REFERENCES
- 1.Ferguson ND, Fan E, Camporota L, Antonelli M, Anzueto A, Beale R, et al. The berlin definition of ARDS: An expanded rationale, justification, and supplementary material. Intensive Care Med. 2012;38:1573–82. doi: 10.1007/s00134-012-2682-1. doi: 10.1007/s00134-012-2682-1. [DOI] [PubMed] [Google Scholar]
- 2.Grommes J, Soehnlein O. Contribution of neutrophils to acute lung injury. Mol Med. 2011;17:293–307. doi: 10.2119/molmed.2010.00138. doi: 10.2119/molmed.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiland JE, Davis WB, Holter JF, Mohammed JR, Dorinsky PM, Gadek JE, et al. Lung neutrophils in the adult respiratory distress syndrome. Clinical and pathophysiologic significance. Am Rev Respir Dis. 1986;133:218–25. doi: 10.1164/arrd.1986.133.2.218. doi: 10.1164/arrd.1986.133.2.218. [DOI] [PubMed] [Google Scholar]
- 4.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun. 2004;72:7247–56. doi: 10.1128/IAI.72.12.7247-7256.2004. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adir Y, Welch LC, Dumasius V, Factor P, Sznajder JI, Ridge KM, et al. Overexpression of the Na-K-ATPase alpha2-subunit improves lung liquid clearance during ventilation-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1233–7. doi: 10.1152/ajplung.00076.2007. doi: 10.1152/ajplung.00076.2007. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Xu M, Fan Q, Xie X, Zhang Y, Mu D, et al. Tanshinone IIA ameliorates seawater exposure-induced lung injury by inhibiting aquaporins (AQP) 1 and AQP5 expression in lung. Respir Physiol Neurobiol. 2011;176:39–49. doi: 10.1016/j.resp.2011.01.005. doi: 10.1016/j.resp.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Wynne BM, Zou L, Linck V, Hoover RS, Ma HP, Eaton DC, et al. Regulation of lung epithelial sodium channels by cytokines and chemokines. Front Immunol. 2017;8:766. doi: 10.3389/fimmu.2017.00766. doi: 10.3389/fimmu.2017.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng X, He S, Yuan J, Miao S, Gao H, Zhang J, et al. Lipoxin A4 attenuates LPS-induced mouse acute lung injury via Nrf2-mediated E-cadherin expression in airway epithelial cells. Free Radic Biol Med. 2016;93:52–66. doi: 10.1016/j.freeradbiomed.2016.01.026. doi: 10.1016/j.freeradbiomed.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Lian QQ, Li R, Ying BY, He Q, Chen F, et al. Lipoxin A(4) activates alveolar epithelial sodium channel, Na, K-ATPase, and increases alveolar fluid clearance. Am J Respir Cell Mol Biol. 2013;48:610–8. doi: 10.1165/rcmb.2012-0274OC. doi: 10.1165/rcmb.2012-0274OC. [DOI] [PubMed] [Google Scholar]
- 10.Qi W, Li H, Cai XH, Gu JQ, Meng J, Xie HQ, et al. Lipoxin A4 activates alveolar epithelial sodium channel gamma via the microRNA-21/PTEN/AKT pathway in lipopolysaccharide-induced inflammatory lung injury. Lab Invest. 2015;95:1258–68. doi: 10.1038/labinvest.2015.109. doi: 10.1038/labinvest.2015.109. [DOI] [PubMed] [Google Scholar]
- 11.Qi D, He J, Wang D, Deng W, Zhao Y, Ye Y, et al. 17β-estradiol suppresses lipopolysaccharide-induced acute lung injury through PI3K/Akt/SGK1 mediated up-regulation of epithelial sodium channel (ENaC) in vivo and In vitro. Respir Res. 2014;15:159. doi: 10.1186/s12931-014-0159-1. doi: 10.1186/s12931-014-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JL, Zhuo XJ, Lin J, Luo LC, Ying WY, Xie X, et al. Maresin1 stimulates alveolar fluid clearance through the alveolar epithelial sodium channel Na, K-ATPase via the ALX/PI3K/Nedd4-2 pathway. Lab Invest. 2017;97:543–54. doi: 10.1038/labinvest.2016.150. doi: 10.1038/labinvest.2016.150. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Zheng X, Cheng Y, Zhang YL, Wen HX, Tao Z, et al. Resolvin D1 stimulates alveolar fluid clearance through alveolar epithelial sodium channel, Na, K-ATPase via ALX/cAMP/PI3K pathway in lipopolysaccharide-induced acute lung injury. J Immunol. 2014;192:3765–77. doi: 10.4049/jimmunol.1302421. doi: 10.4049/jimmunol.1302421. [DOI] [PubMed] [Google Scholar]
- 14.Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L379–99. doi: 10.1152/ajplung.00010.2008. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morty RE, Eickelberg O, Seeger W. Alveolar fluid clearance in acute lung injury: What have we learned from animal models and clinical studies? Intensive Care Med. 2007;33:1229–40. doi: 10.1007/s00134-007-0662-7. doi: 10.1007/s00134-007-0662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Seigneux S, Leroy V, Ghzili H, Rousselot M, Nielsen S, Rossier BC, et al. NF-kappaB inhibits sodium transport via down-regulation of SGK1 in renal collecting duct principal cells. J Biol Chem. 2008;283:25671–81. doi: 10.1074/jbc.M803812200. doi: 10.1074/jbc.M803812200. [DOI] [PubMed] [Google Scholar]
- 17.Gon Y, Maruoka S, Kishi H, Kozu Y, Kazumichi K, Nomura Y, et al. NDRG1 is important to maintain the integrity of airway epithelial barrier through claudin-9 expression. Cell Biol Int. 2017;41:716–25. doi: 10.1002/cbin.10741. doi: 10.1002/cbin.10741. [DOI] [PubMed] [Google Scholar]
- 18.Akiba J, Murakami Y, Noda M, Watari K, Ogasawara S, Yoshida T, et al. N-myc downstream regulated gene1/Cap43 overexpression suppresses tumor growth by hepatic cancer cells through cell cycle arrest at the G0/G1 phase. Cancer Lett. 2011;310:25–34. doi: 10.1016/j.canlet.2011.05.034. doi: 10.1016/j.canlet.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 19.Song Y, Oda Y, Hori M, Kuroiwa K, Ono M, Hosoi F, et al. N-myc downstream regulated gene-1/Cap43 may play an important role in malignant progression of prostate cancer, in its close association with E-cadherin. Hum Pathol. 2010;41:214–22. doi: 10.1016/j.humpath.2009.07.011. doi: 10.1016/j.humpath.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang AH, Rao JN, Zou T, Liu L, Marasa BS, Xiao L, et al. P53-dependent NDRG1 expression induces inhibition of intestinal epithelial cell proliferation but not apoptosis after polyamine depletion. Am J Physiol Cell Physiol. 2007;293:C379–89. doi: 10.1152/ajpcell.00547.2006. doi: 10.1152/ajpcell.00547.2006. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Zhou Y, Tao F, Chai S, Xu X, Yang Y, et al. N-myc downstream regulated gene 1(NDRG1) promotes the stem-like properties of lung cancer cells through stabilized c-Myc. Cancer Lett. 2017;401:53–62. doi: 10.1016/j.canlet.2017.04.031. doi: 10.1016/j.canlet.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Lu WJ, Chua MS, So SK. Suppressing N-Myc downstream regulated gene 1 reactivates senescence signaling and inhibits tumor growth in hepatocellular carcinoma. Carcinogenesis. 2014;35:915–22. doi: 10.1093/carcin/bgt401. doi: 10.1093/carcin/bgt401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin J, Chen B, Shi XH, Mishima T, Kokame K, Barak Y, et al. NDRG1 deficiency attenuates fetal growth and the intrauterine response to hypoxic injury. Endocrinology. 2014;155:1099–106. doi: 10.1210/en.2013-1425. doi: 10.1210/en.2013-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng W, Li CY, Tong J, Zhang W, Wang DX. Regulation of ENaC-mediated alveolar fluid clearance by insulin via PI3K/Akt pathway in LPS-induced acute lung injury. Respir Res. 2012;13:29. doi: 10.1186/1465-9921-13-29. doi: 10.1186/1465-9921-13- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384(Pt 3):477–88. doi: 10.1042/BJ20041057. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer EM, Dry H, Cross D, Guichard S, Davies BR, Alessi DR, et al. Elevated SGK1 predicts resistance of breast cancer cells to Akt inhibitors. Biochem J. 2013;452:499–508. doi: 10.1042/BJ20130342. doi: 10.1042/BJ20130342. [DOI] [PMC free article] [PubMed] [Google Scholar]