Abstract
Mdm2 is often overexpressed in tumors that retain wild-type TP53 but may affect therapeutic response independently of p53. Herein is shown that tumor cells with MDM2 amplification are selectively resistant to treatment with topoisomerase II poisons but not other DNA damaging agents. Tumor cells that overexpress Mdm2 have reduced DNA double-strand breaks in response to doxorubicin or etoposide. This latter result is not due to altered drug uptake. The selective attenuation of DNA damage in response to these agents is dependent on both Mdm2 levels and an intact ubiquitin ligase function. These findings reveal a novel, p53-independent activity of Mdm2 and have important implications for the choice of chemotherapeutic agents in the treatment of Mdm2-overexpressing tumors.
INTRODUCTION
Mdm2 is a well-characterized negative regulator of the p53 tumor suppressor protein. Mdm2 inhibits p53 transcriptional activity and ubiquitylates p53, targeting it for proteasomal degradation under normal conditions.1 In response to a variety of cell stressors, include genotoxic stress, p53 is stabilized by disruption of the Mdm2–p53 interaction and activates programs of tumor suppression that include cycle arrest, apoptosis and other downstream effects.2
Overexpression of Mdm2 has been documented in a variety of different malignancies and is often a consequence of gene amplification.3 Although a role for Mdm2 as an oncogene has focused on its role as an inhibitor of p53, there has been growing interest in p53-independent roles of Mdm2. Several studies have found that overexpression of Mdm2 is not mutually exclusive with loss or mutational inactivation of TP53, pointing to the possibility of p53-independent contributions.3–6 In addition, several studies have suggested that Mdm2 plays p53-independent roles in cell cycle progression, apoptosis, maintenance of genomic stability and response to DNA damage.7–9
Topoisomerase II poisons are among the most frequently employed chemotherapeutic agents in the treatment of many different cancers.10–14 Topoisomerase II enzymes catalyze the ATP-dependent passage of one intact DNA double helix through a double-strand DNA break produced in a second duplex, thereby relaxing DNA supercoiling and permitting such processes as DNA replication and transcription to occur.15 Topoisomerase II poisons bind to and trap the topoisomerase II holoenzyme in a so-called ‘cleavable complex’, at which point the enzyme is engaged in an otherwise transient covalent complex with the 5′ ends of cleaved DNA.16 This interaction inhibits re-ligation of the cleaved DNA duplex, and the lesion is interpreted by the cell as a double-stranded break.17 Doxorubicin and etoposide, members of the anthracycline and epidophyllotoxin subclasses of topoisomerase II poisons, respectively, achieve this outcome by different means. Whereas etoposide may promote double-strand breaks as a consequence of direct binding to topoisomerase II, doxorubicin intercalates into DNA and is thought to interfere with re-ligation at the topoisomerase II-DNA interface by perturbing the geometry of the DNA.16,18 In the present study, the effect of Mdm2 overexpression on the sensitivity of tumor cells to a panel of DNA damaging agents was investigated.
RESULTS
Tumor cells that overexpress Mdm2 as a result of gene amplification show selective resistance to topoisomerase II poisons
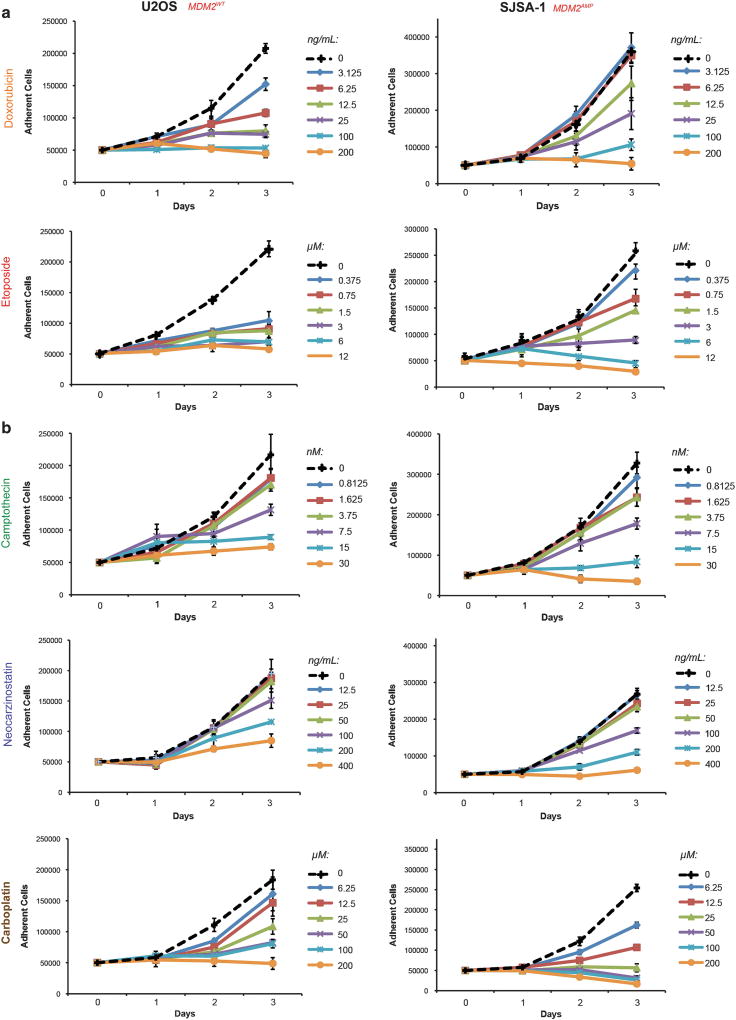
SJSA-1 osteosarcoma cells harbor a ~ 20-fold amplification of the MDM2 gene (Expanded View Figure 1a). The amplification of MDM2 in this cell line is associated with an approximately 50-fold elevation of MDM2 transcript over U2OS osteosarcoma cells and a correspondingly elevated level of Mdm2 protein (Supplementary Figures 1B–C). Both tumor lines are reported to be TP53WT; this was confirmed by sequencing19 (see Materials and methods). The effect of a range of doses of doxorubicin or etoposide on the proliferation of these cells was studied. The growth of U2OS cells was inhibited by all doses of doxorubicin or etoposide employed in growth curve assays relative to untreated controls (Figure 1a). In contrast, the growth of SJSA-1 cells was impaired only at higher doses of both drugs (Figure 1a). Low doses of doxorubicin or etoposide that inhibited U2OS cell proliferation failed to produce an antiproliferative response in SJSA-1 cells.
Figure 1.
Tumor cells overexpressing Mdm2 show selective resistance to topoisomerase II poisons. (a) U2OS and SJSA-1 cells were seeded into medium containing the indicated doses of either doxorubicin (DOX) or etoposide (VP16) and allowed to incubate for up to 3 days. Adherent cells were counted each day beginning at day 1 post-seeding. Data are mean ± s.e.m. (b) U2OS and SJSA-1 cells were seeded into medium containing the indicated doses of camptothecin (CPT), neocarzinostatin (NCS) or carboplatin (CBCDA). Cell proliferation was assessed as described in (a). Data are mean ± s.e.m.
To determine whether the difference in chemosensitivity of these tumor lines was unique to topoisomerase II poisons, the growth inhibitory effects of a panel of drugs that trigger DNA damage by other means were also investigated. SJSA-1 cells were not resistant to treatment with camptothecin, neocarzinostatin or carboplatin compared to U2OS cells (Figure 1b). U2OS and SJSA-1 cells were equally sensitive to camptothecin, while U2OS cells were more resistant to neocarzinostatin and carboplatin than SJSA-1 cells in the range of doses studied. These findings suggest that SJSA-1 cells are selectively resistant to topoisomerase II poisons but not to other DNA damaging agents.
Tumor lines that overexpress Mdm2 show reduced DNA damage in response to topoisomerase II poisons
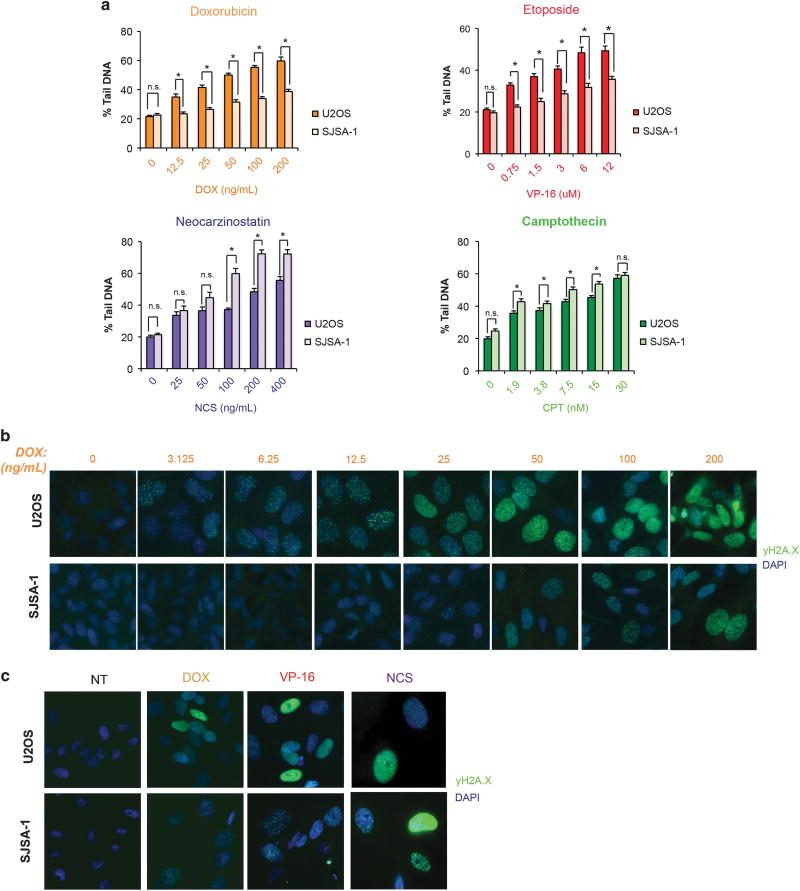
The mechanism of resistance of SJSA-1 cells to doxorubicin and etoposide was investigated. The induction of double-strand DNA breaks arising from treatment of either U2OS or SJSA-1 cells with these agents was assessed by neutral comet assay. Doxorubicin and etoposide produced significantly fewer DNA breaks in SJSA-1 than in U2OS cells across a range of relevant doses, as measured by percent tail DNA after treatment (Figure 2a, top, and Supplementary Figure 2A). SJSA-1 cells treated with either neocarzinostatin or camptothecin, however, did not develop fewer double-strand DNA breaks compared to U2OS cells (Figure 2a, bottom). The extent of DNA damage achieved by doxorubicin treatment of both cell lines was also determined by immunofluorescent detection of phospho-γH2A.X, a key mediator of DNA damage signaling and repair in response to double-strand breaks.20,21 In agreement with findings from the comet assays, whereas U2OS nuclei developed detectable phospho-γH2A.X foci at doses as low as 3.125 ng/ml doxorubicin, SJSA-1 cells did not demonstrate such foci at doses below 25 ng/ml (Figure 2b and Supplementary Figure 2B). Similar results were achieved with etoposide, while treatment with neocarzinostatin did not result in an appreciable difference in phospho-γH2A.X detection across either cell line, also in agreement with comet assay results (Figure 2c and Supplementary Figure 2c). Collectively, these results suggest that SJSA-1 cells are selectively resistant to induction of double-strand DNA breaks by topoisomerase II poisons.
Figure 2.
Tumor cells overexpressing Mdm2 show reduced DNA damage due to topoisomerase II inhibition. (a) The extent of double-stranded DNA breaks induced by doxorubicin (DOX), etoposide (VP16), neocarzinostatin (NCS) and camptothecin (CPT) in U2OS and SJSA-1 cells was determined by neutral comet assay following 48 h of treatment at the indicated doses of drug. Data are mean ± s.e.m. *P<0.05. ns=not significant. Results are representative. (b) U2OS and SJSA-1 cells were stained for phospho-γH2A.X after 48 h of treatment with the indicated doses of doxorubicin. Results are representative. (c) U2OS and SJSA-1 cells were stained for phospho-γH2A.X after 48 h of treatment with 100 ng/ml doxorubicin, 3 µM VP-16 and 200 ng/ml neocarzinostatin. Results are representative.
Resistance to the induction of DNA damage by topoisomerase II inhibition is Mdm2-dependent
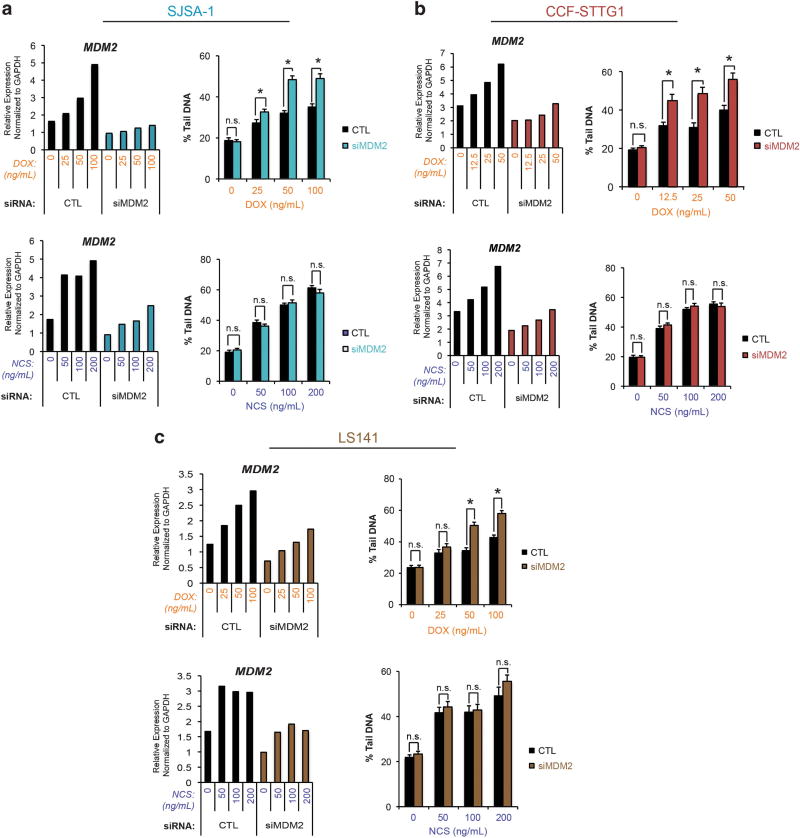
Given the striking difference in levels of MDM2 expression between U2OS and SJSA-1 cells (Expanded View Figures 1a–c), the influence of Mdm2 on the ability of topoisomerase II poisons to induce double-strand DNA breaks in SJSA-1 cells was investigated using an siRNA approach. SJSA-1 cells were transfected with either negative control or MDM2-directed siRNA oligonucleotides. Following transfection, SJSA-1 cells were treated with a range of doses of either doxorubicin or neocarzinostatin to allow for comparison of topoisomerase II-dependent and -independent means of double-strand DNA break induction. The extent of MDM2 knockdown by siRNA in each setting was quantified by qPCR and the degree of double-strand DNA breaks induced by treatment with these agents was measured by neutral comet assay. Mdm2 protein levels after MDM2 knockdown were determined by immunoblotting (Supplementary Figure 3A). Off-target effects of MDM2 siRNA oligonucleotides were ruled out by single oligonucleotide experiments (Supplementary Figure 3B). SJSA-1 cells transfected with MDM2 siRNA and treated with doxorubicin revealed significantly higher levels of double-strand DNA breaks than cells treated with control siRNA (Figure 3a, top). Conversely, SJSA-1 cells transfected with MDM2 siRNA and treated with neocarzinostatin did not show any detectable difference in induced double-strand DNA breaks when compared to control siRNA-transfectants (Figure 3a, bottom). No difference in DNA damage was detected in the untreated setting. Furthermore, knockdown of MDM2 was not shown to alter cell viability, as the fractions of hypodiploid cells after transfection with siRNA to MDM2 or negative control were comparable (Supplementary Figures 4A–C). These findings suggest that overexpression of Mdm2 blunts the ability of topoisomerase II poisons to generate double-strand DNA breaks in this cell line but does not affect the extent of topoisomerase II-independent double-strand DNA breaks generated by neocarzinostatin.
Figure 3.
Reduced DNA damage consequent to topoisomerase II inhibition is Mdm2-dependent. (a) MDM2 transcripts in SJSA-1 cell were targeted for degradation using an siRNA approach. Cells transfected with siRNA were treated for 48 h with either doxorubicin or neocarzinostatin at the indicated doses. A representative experiment is shown. Left: The extent of MDM2 knockdown at each point was quantified by qPCR. Right: The degree of double-stranded DNA breaks in control (CTL) or MDM2 siRNA-transfected cells treated with either doxorubicin or neocarzinostatin was determined by neutral comet assay. Data are mean ± s.e.m. (b) The experiment described in (a) was carried out in CCF-STTG1 cells. Data are mean ± s.e.m. Results are representative. (c) The experiment described in (a) was carried out using LS141 cells. Data are mean ± s.e.m. *P<0.05. ns=not significant. Results are representative.
To extend these studies to other cell lines harboring MDM2 amplification, similar experiments were carried out in CCF-STTG1 glioblastoma and LS141 liposarcoma cells, both of which possess in excess of 100 copies of MDM2 (Expanded View Figures 1a–c). As with SJSA-1 cells, both CCF-STTG1 and LS141 have been reported to be TP53WT; TP53 status was confirmed by sequencing (see Materials and methods).22–24 The relevant doses of doxorubicin for CCF-STTG1 cells were determined by proliferation assay (Supplementary Figure 5). Knockdown of MDM2 in CCF-STTG1 cells resulted in an increase in the extent of double-strand DNA breaks with doxorubicin treatment, but did not affect double-strand DNA break induction by neocarzinostatin treatment (Figure 3b). Likewise, LS141 cells transfected with MDM2 siRNA and treated with doxorubicin exhibited a higher degree of double-strand DNA breaks over cells transfected with control siRNA; this difference was not seen with neocarzinostatin treatment (Figure 3c). Taken together, these data support a role for Mdm2 in impairing the ability of topoisomerase II poisons to induce DNA damage.
Overexpression of Mdm2 in tumor cells does not affect doxorubicin uptake
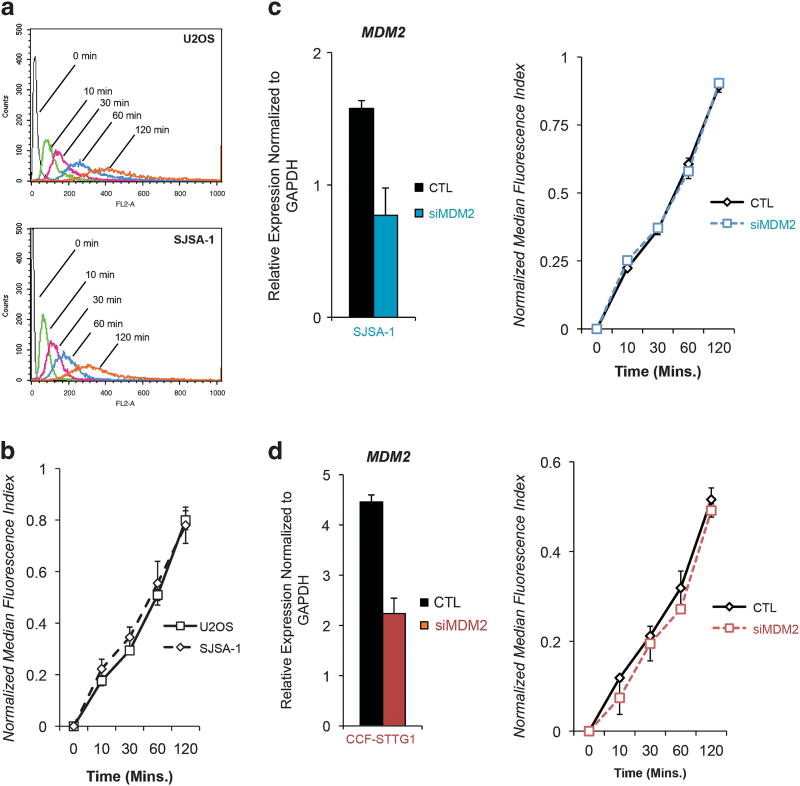
Previous work has reported that Mdm2 may induce expression of the P-glycoprotein drug efflux pump (P-gp).25 Furthermore, treatment of cells with the Mdm2 inhibitor Nutlin-3 has been shown to reduce P-gp activity.26 Therefore, the possibility that reduced DNA damage with topoisomerase II poisons in MDM2-amplified tumor lines may be a consequence of enhanced drug efflux was investigated. Uptake of doxorubicin was tracked in SJSA-1 and U2OS cells exposed to drug for varying periods of time and relative intracellular doxorubicin accumulation at each time point was estimated using an established flow cytometry approach.27,28 Both U2OS and SJSA-1 cells revealed comparable doxorubicin uptake profiles (Figures 4a and b). The absence of an effect of MDM2 knockdown by siRNA on intracellular doxorubicin accumulation was confirmed using this method in both SJSA-1 and CCF-STTG1 cells (Figures 4c and d). These results suggest that the difference in DNA damage observed with topoisomerase II poisons in cells that overexpress Mdm2 is not a consequence of reduced intracellular drug concentration.
Figure 4.
Mdm2 overexpression does not affect doxorubicin uptake. (a) U2OS and SJSA-1 cells were incubated in medium containing 2 µg/ml doxorubicin at 37 °C for up to 120 min and collected for flow cytometric analysis of doxorubicin uptake. Data are representative. (b) Relative intracellular doxorubicin accumulation was estimated by calculation of normalized median fluorescence index (NMFI). NMFI values were plotted over time. Data are mean ± s.e.m. (c) MDM2 transcripts in SJSA-1 cells were degraded using an siRNA approach and the effect of this manipulation on intracellular doxorubicin accumulation was studied. Left: The extent of Mdm2 knockdown by siRNA was quantified by qPCR. Error bars mean ± s.e.m. Right: NMFI values for control and Mdm2 siRNA-treated SJSA-1 cells exposed to 2 µg/ml doxorubicin for up to 120 min were calculated and plotted. Data are mean ± s.e.m. (d) The experiment described in (c) was performed using CCF-STTG1 cells. Data are mean ± s.e.m.
Mdm2 requires an intact RING finger domain to suppress topoisomerase II-dependent DNA damage
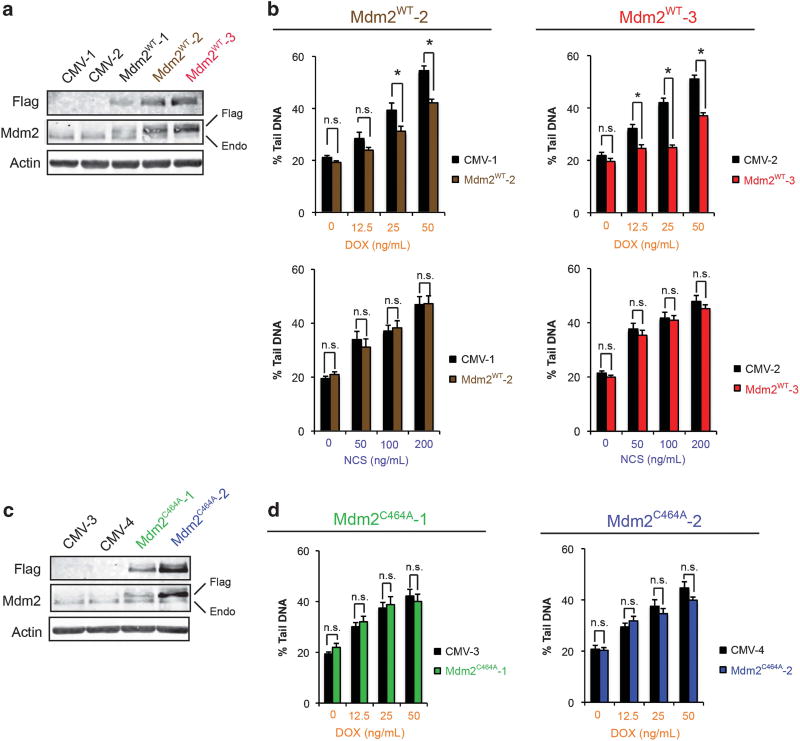
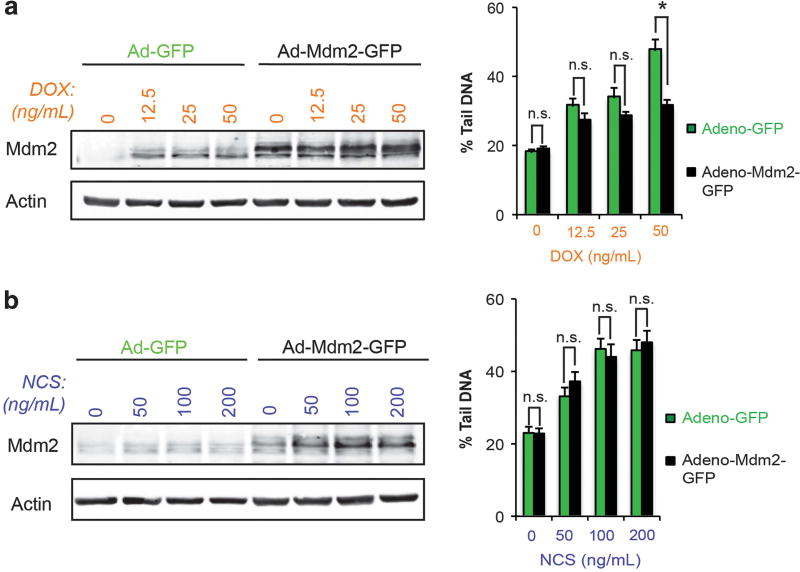
Mdm2 has been reported to ubiquitinate and regulate the levels of several target proteins in addition to p53.1,7,9 The contribution of the RING finger and associated ubiquitin ligase function of Mdm2 to the observed reduction in DNA damage with topoisomerase II poisons was studied. Stable clones of U2OS cells that overexpress either Flag-tagged, full-length wild-type Mdm2 (Mdm2WT) or a RING finger mutant (Mdm2C464A) were generated. Clones were screened for levels of Mdm2 expression by immunoblotting (Figures 5a and c). Doxorubicin-treated Mdm2WT clones revealed fewer double-strand DNA breaks compared to empty-vector controls, as measured by comet assay (Figure 5b, top). As expected, doxorubicin uptake in Mdm2WT and empty-vector controls was comparable (Supplementary Figure 6). In contrast, when treated with neocarzinostatin, the levels of DNA damage in Mdm2WT clones were indistinguishable from controls (Figure 5b, bottom). In agreement with these findings, transient overexpression of Mdm2 in U2OS cells using a viral transduction approach also reduced the extent of doxorubicin-induced double-strand DNA breaks compared to empty-vector controls (Figure 6a). No effect on the levels of DNA damage with neocarzinostatin was observed by this method (Figure 6b). Conversely, Mdm2C464A clones did not show reduced double-strand DNA breaks after doxorubicin treatment (Figure 5d). The findings from these studies are in agreement with those from MDM2-amplified tumor lines and further support a role for Mdm2 in selectively attenuating the ability of topoisomerase II poisons to induce double-strand DNA breaks. Moreover, given the failure of RING finger-mutant Mdm2 to reduce levels DNA damage achieved with doxorubicin, the ubiquitin ligase activity of Mdm2 is likely critical for this outcome.
Figure 5.
The ability of Mdm2 to suppress DNA damage due to topoisomerase II poisons is dependent on an intact RING finger domain. (a) Immunoblot analysis of stable clones of U2OS-derived cells transfected with empty vector (CMV) or Flag-tagged, full-length, wild-type Mdm2 (Mdm2WT). (b) The extent of DNA double-strand breaks due to doxorubicin (DOX) or neocarzinostatin (NCS) treatment in Mdm2WT clones versus empty vector controls was determined by neutral comet assay. A representative experiment is shown for two clones. Data are mean ± s.e.m. *P<0.05. (c) Immunoblot analysis of stable clones of U2OS-derived cells transfected with either empty vector (CMV) or Flag-tagged, full-length Mdm2 harboring a point mutation in the RING finger domain (Mdm2C464A). (d) Mdm2C464A clones were treated with doxorubicin as in (b) and the extent of DNA double-strand breaks was quantified by neutral comet assay. A representative experiment is shown for each clone. Data are mean ± s.e.m. ns=not significant.
Figure 6.
Expression of Mdm2 reduces DNA damage due to topoisomerase II inhibition. Left: Immunoblot analysis of U2OS cells transduced with an adenoviral vector driving expression of either GFP alone or in combination with full-length, wild-type Mdm2 and treated with the indicated doses of doxorubicin for 48 h. Right: The extent of DNA damage induced by doxorubicin in Adeno-GFP-versus Adeno-Mdm2-GFPtransduced U2OS cells was quantified by neutral comet assay. A representative experiment is shown. Data are mean ± s.e.m. (b) Left: Immunoblot analysis of U2OS cells transduced as described in (a) but treated with the indicated doses of neocarzinostatin for 48 h. Right: The extent of DNA damage induced by neocarzinostatin in Adeno-GFP- versus Adeno-Mdm2-GFP-transduced U2OS cells was quantified by neutral comet assay. A representative experiment is shown. Data are mean ± s.e.m. ns=not significant.
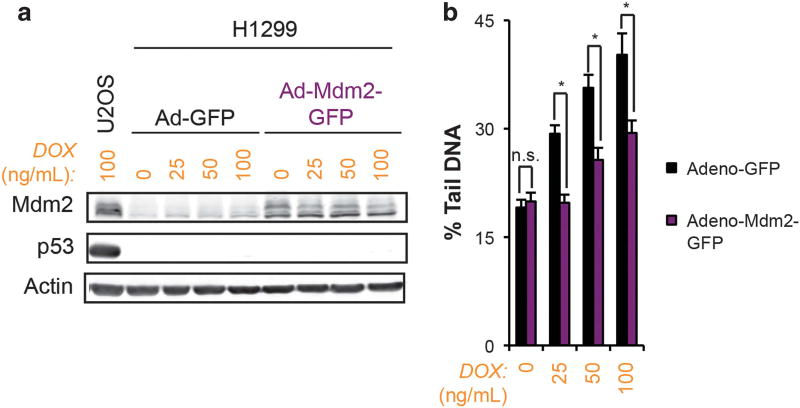
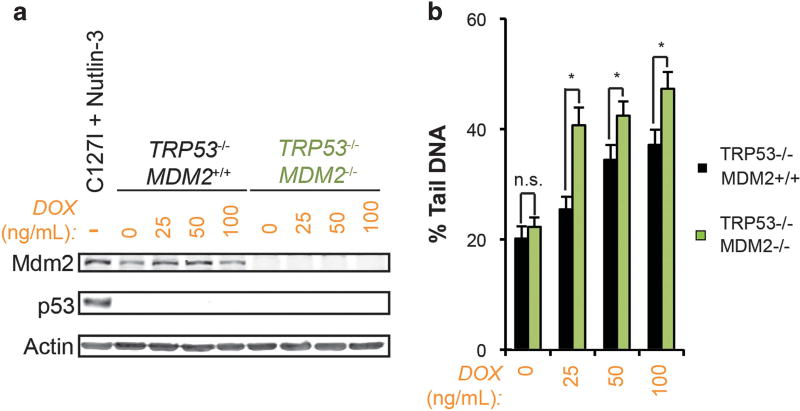
A role for Mdm2 regulation of p53 in mediating this effect was also studied. U2OS cells transfected with siRNA to TP53 and treated with doxorubicin did not demonstrate reduced double-strand DNA breaks by comet assay (Supplementary Figure 7). Additionally, SJSA-1 cells did not demonstrate further reduction in doxorubicin-induced double-strand DNA breaks after transfection with siRNA to TP53 (Supplementary Figure 8). In agreement with these findings, transient overexpression of Mdm2 in TP53−/− H1299 tumor cells using a viral transduction approach reduced the extent of doxorubicin-induced double-strand DNA breaks compared to empty-vector controls (Figure 7). To further validate these observations, TRP53−/−MDM2+/+ and TRP53−/−MDM2−/− mouse embryo fibroblasts (MEFs) were treated with doxorubicin and assessed for double-strand DNA breaks. TRP53−/−MDM2−/− MEFs demonstrated a significant increase in DNA double strand breaks over TRP53−/−MDM2+/+ controls (Figure 8). Taken together, these findings suggest that the effect of Mdm2 on DNA damage by topoisomerase II poisons is independent of Mdm2-mediated ubiquitination and degradation of p53. In agreement with published reports, co-treatment of SJSA-1 cells with Nutlin-3 and doxorubicin resulted in an increase in double-strand DNA breaks (Supplementary Figure 9).29 This finding has been observed by other investigators to be independent of p53, suggesting that the N-terminal hydrophobic region of Mdm2 may be contributing to the attenuation of DNA damage induced by topoisomerase II poisons through a separate mechanism. Finally, further knockdown of MDM2 in U2OS cells did not produce the effects seen with Mdm2 overexpression, arguing that the influence of Mdm2 on doxorubicin-dependent induction of double-strand DNA breaks may require threshold levels of protein (Supplementary Figure 7).
Figure 7.
Mdm2 suppression of DNA damage due to topoisomerase II inhibition is independent of p53. (a) Immunoblot analysis of H1299 cells transduced with an adenoviral vector driving expression of GFP alone or in combination with full-length, wild-type Mdm2 and treated with the indicated doses of doxorubicin for 48 h. (b) The extent of DNA damage induced by doxorubicin in Adeno-GFP versus Adeno-Mdm2-GFP-transduced H1299 cells was quantified by neutral comet assay. A representative experiment is shown. Data are mean ± s.e.m. ns=not significant.
Figure 8.
Loss of MDM2 in mouse embryo fibroblasts is associated with increased sensitivity to DNA damage due to topoisomerase II inhibition independent of p53. (a) Immunoblot analysis of TRP53−/−MDM2− +/+ and TRP53−/−MDM2−/− mouse embryo fibroblasts (MEFs) treated with the indicated doses of DOX for 48 h. C127I mouse mammary epithelial cells treated with 10 µm Nutlin-3 for 24 h are a positive control for Trp53 and Mdm2. (b) Neutral comet assay quantification of double-strand DNA breaks arising from DOX treatment at the indicated dose for 48 h in TRP53−/−MDM2− +/+ versus TRP53−/−MDM2−/− MEFs. A representative experiment is shown. Data are mean ± s.e.m. ns=not significant.
The extent of DNA damage induced by topoisomerase II poisons is not dependent on levels of topoisomerase IIα
Although a variety of toxicity mechanisms have been described using different doses of doxorubicin in different model systems, the topoisomerase IIα isoform of topoisomerase II is frequently considered to be the molecular target of this drug responsible for the generations of double-strand DNA breaks.30,31 Several studies have suggested a direct correlation between the levels of topoisomerase IIα and sensitivity to both doxorubicin and etoposide.32,33 To elucidate a possible mechanism by which Mdm2 may be mediating resistance to topoisomerase II poisons, the influence of topoisomerase IIα levels on the extent of doxorubicin-induced DNA damage was studied. U2OS cells transfected with siRNA to TOP2A and treated with doxorubicin showed no discernible difference in phospho-γH2A.X staining compared to cells transfected with control siRNA (Supplementary Figure 10A). Similarly, SJSA-1 cells were neither sensitized nor rendered more resistant to DNA damage by doxorubicin after TOP2A knockdown (Supplementary Figure 10B). Moreover, despite the differences in the ability of doxorubicin to induce DNA damage in U2OS and SJSA-1 cells, a difference in topoisomerase IIα levels between these lines, either under basal conditions or during doxorubicin treatment, was not seen by immunoblotting (Supplementary Figure 10C). Taken together, these findings do not support a role for levels of expression of topoisomerase IIα in the ability of Mdm2 to attenuate the DNA damage induced by topoisomerase II poisons.
DISCUSSION
Despite attempts to correlate Mdm2 levels with response to chemotherapy, a prognostic role for Mdm2 has been elusive. Although Mdm2 overexpression has been reported in a variety of different malignancies, studies have produced contradictory conclusions.3 In osteosarcoma, for example, it was suggested that MDM2 amplification was associated with a more aggressive disease course owing to detection predominantly in metastatic or recurrent tumor specimens.34,35 Another study found MDM2 to be more frequently amplified in parosteal osteosarcomas, which are typically lower in pathologic grade than the conventional subtype and associated with better outcomes.36 More recently, copy number gain of MDM2 was associated with poor response to chemotherapy regardless of pathologic subtype.37 Attempts to draw conclusions from these few studies are complicated not only by differences in methods used to detect MDM2 amplification but also by the absence of treatment data accompanying the samples analyzed. As the management of osteosarcoma frequently involves treatment with any combination of cisplatin, doxorubicin, methotrexate, ifosfamide and related agents, the challenge of identifying a possible influence of Mdm2 expression on tumor responsiveness to individual drugs in this relatively rare disease is enormous.10,38 These obstacles have been noted by others attempting to pool data from analyses of soft tissue sarcomas, gliomas and other cancers.39,40
There is an important question as to whether Mdm2 is reducing the level of DNA damage or is enhancing DNA repair. Studies from the laboratory of Christine Eischen have established a clear role for Mdm2 in disrupting DNA double-strand break repair. Mdm2 has been found to associate with Nbs1, a member of the MRN (Mre11-Rad50-Nbs1) complex that is essential for DNA repair, cell cycle checkpoint signaling and telomere maintenance.41,42 Expression of wild-type Mdm2 was found to delay the recovery of DNA damage after ionizing radiation, but this effect was abolished if an Mdm2 mutant incapable of binding Nbs1 was used.41 The delay in double-strand DNA break resolution was determined to be p53-independent. These studies demonstrate that Mdm2 plays a role in inhibiting the repair of double-strand DNA breaks via a mechanism involving this interaction with Nbs1. Further, the repair of the DNA damage that is caused by doxorubicin (as a topoisomerase II inhibitor) versus that of neocarzinostatin (acting as an radiomimetic) is unlikely to be occurring by distinct pathways. Yet, the effects seen with Mdm2 are specific for the former. Taken together, the findings reported here thus are more likely explained by Mdm2 exerting an effect at the level of the DNA damage itself, rather than subsequent steps in the cellular response to that damage.
The work presented here has not examined the effect of ionizing radiation in the setting of Mdm2 overexpression, although the radiomimetic compound neocarzinostatin was used.43 An effect of Mdm2 on neocarzinostatin-induced double-strand DNA breaks was not identified by comet assay, neither after MDM2 siRNA transfection in MDM2-amplified cell lines nor with ectopic expression of Mdm2 in U2OS cells. In addition, neocarzinostatin treatment in these experiments was continuous, limiting the ability to infer a role for Mdm2 in the repair of these DNA lesions. Similarly, although phosphoγH2A.X has been described, as an early mediator of the DNA damage response to double-strand DNA breaks, no difference in staining between U2OS and SJSA-1 cells treated with neocarzinostatin was detected. Whether overexpression of Mdm2 delays double-strand DNA break repair after drug withdrawal remains to be seen, although at least one study has contended that Mdm2 may be necessary for effective DNA repair.29
An intriguing possibility was recently proposed by Yeo et al. which involves a potential role for p53 in preventing interference between transcription and replication thereby influencing cellular responses to inhibitors of topoisomerase II.44 This effect was shown to be influenced by levels of TOP2A, whereas the effect in Mdm2-overexpressing SJSA-cells was not influenced by TOP2A knockdown (Supplementary Figure 10). Further, the Mdm2-dependent outcomes described herein have been shown to be p53-independent. Nevertheless, a possible contribution of Mdm2 in regulating DNA damage generated by an interplay between transcription and replication cannot be ruled out.
Nayak et al. reported reduced sensitivity to an array of topoisomerase II poisons in a panel of SNP309G/G cell lines relative to SNP309T/T cells.45 The T→G transversion in SNP309, located in the first intron of the MDM2 gene, is associated with enhanced binding of the Sp1 transcription factor and a consequent increase in Mdm2 expression.46 Consistent with the findings in the present work, while G/G cell lines were less sensitive to topoisomerase II poisons than their T/T counterparts, this was not true of treatment with camptothecin, cisplatin and non-DNA damaging drugs such as taxanes. MDM2 knockdown in G/G lines increased sensitivity to etoposide, while no change in sensitivity was observed with MDM2 knockdown in T/T lines. Interestingly, the authors reported an indirect correlation between Mdm2 and topoisomerase IIα levels and identified an interaction between topoisomerase IIα and Mdm2 by co-immunoprecipitation. The authors concluded that elevated Mdm2 levels promote the ubiquitin-mediated degradation of topoisomerase IIα during treatment with topoisomerase II poisons, thereby reducing the availability of topoisomerase IIα for inhibition and rendering cells resistant to this class of drug. Other reports have failed to demonstrate a relationship between topoisomerase IIα levels and cell sensitivity to doxorubicin, and an interaction between Mdm2 and topoisomerase IIα has not been consistently observed, likely owing to differences in reagents and conditions47,48. Nayak et al. used the SMP14 antibody to immunoprecipitate Mdm2 in their cell extracts following treatment with etoposide. There is evidence that the epitope of this antibody is masked by Mdm2 phosphorylation after DNA damage that reduces the ability to detect Mdm2 by immunoblotting.49 Additionally, the Sepharose beads employed in their co-IP were washed in phosphate-buffered saline (PBS), whereas a more stringent wash buffer containing a higher salt concentration and detergent was used in the studies presented here.
How, then, might overexpression of Mdm2 confer resistance to DNA damage induction by topoisomerase II poisons? In the absence of uniform support for topoisomerase IIα levels mediating sensitivity to these drugs, it is reasonable to propose that post-translational modification of topoisomerase IIα is playing a critical role. The phosphorylation, acetylation, ubiquitination and sumoylation of topoisomerase IIα have all been reported, but the consequences of these modifications are a matter of dispute.50 Nevertheless, it is conceivable that a reduction in the catalytic activity topoisomerase IIα may reduce sensitivity to topoisomerase II poisons without altering cell viability in the absence of drug. Although one study has suggested that topoisomerase IIα catalytic activity may not affect the sensitivity of cells to topoisomerase II poisons, the authors conceded that differences in drug uptake, extractability of nuclear topoisomerase IIα for activity assays and overall susceptibility of cells to apoptosis were not rigorously assessed and may have influenced their results.48
Several lines of evidence suggest that a reduction in topoisomerase IIα activity may attenuate the ability of topoisomerase II poisons to stabilize cleavage complexes and induce double-strand DNA breaks. In yeast, a temperature-sensitive top2 strain of Saccharomyces cerevisiae grown at semi-permissive temperature showed reduced topoisomerase II enzyme activity and was resistant to topoisomerase II poisons amsacrine and etoposide but hypersensitive to camptothecin.51 In human cells, overexpression of the atypical ζ isoform of protein kinase C (PKCζ) resulted in phosphorylation of topoisomerase IIα and a concomitant reduction in catalytic activity, DNA-protein crosslinks and etoposide-induced double-strand DNA breaks.52 Consistent with this observation, blockade of PKCζ activation by wortmannin treatment or stable expression of a dominant-negative PKCζ enhanced topoisomerase IIα catalytic activity and increased the extent of etoposide-induced double-strand DNA breaks.53 Additionally, a role for calcium-regulated phosphorylation of topoisomerase IIα by casein kinase I δ/ε with similar effects on catalytic activity has also been described.54,55 Mdm2 may be modifying the levels or activity of a number of upstream kinases in a ubiquitin-dependent manner. The consequent downregulation of topoisomerase IIα catalytic activity may confer resistance to topoisomerase II poisons by limiting the ability of these drugs to generate double-strand DNA breaks.
Apart from post-translational modification, protein–protein interactions may also influence topoisomerase II poison-induced double-strand DNA breaks, either by affecting topoisomerase IIα activity or accessibility. For example, Nayak et al. observed relocalization of topoisomerase IIα from the nucleus to the cytoplasm during treatment of SNP309G/G cells with etoposide.45 If Mdm2 were to promote the shuttling of topoisomerase IIα out of the nucleus, then the ability of etoposide to poison the enzyme in the nucleus would be limited, leading to fewer double-strand DNA breaks. Although evidence in support of a direct interaction with topoisomerase IIα has proven elusive, Mdm2 may still act on this enzyme through an intermediary protein or pathway. Previous work has identified an interaction between ERK and topoisomerase IIα that enhanced topoisomerase IIα catalytic activity but was not dependent on phosphorylation.56
Finally, although the work presented here has not directly addressed DNA repair, sumoylation of topoisomerase IIα has been observed after the enzyme is trapped in a cleavage complex with DNA by topoisomerase II poisons.50 There is increasing evidence that hydrolysis of 5′-phosphotyrosyl linkages between the SUMO-1-conjugated enzyme and DNA by tyrosyl phosphodiesterase 2 is critical for clearance of DNA-enzyme adducts and subsequent repair of etoposide-induced DNA damage.57–59 Thus, It will be important to address whether elevated levels of Mdm2 affect this step of the DNA damage response.
These studies have shown that overexpression of Mdm2 selectively suppresses the levels of DNA double-strand breaks achieved by known inhibitors of topoisomerase IIα and is associated with selective resistance of MDM2-amplified tumor lines to this class of drugs. As MDM2 amplification and overexpression have been documented across a spectrum of malignancies at varying frequency, the prognostic value of Mdm2 levels on tumor response in patients treated with these agents warrants further investigation. In patients undergoing treatment, the effects of Mdm2 both on induction and repair of DNA damage by topoisomerase II poisons are likely to be critical and concurrent determinants of tumor response to these drugs.
MATERIALS AND METHODS
Cells and vectors
U2OS, SJSA-1, H1299 and CCF-STTG1 cells were purchased from ATCC. LS141 cells were a generous gift from Dr Jonathan Fletcher. Trp53−/− and Trp53−/−Mdm2−/− MEFs were a generous gift from Carol A. Prives. Experiments in MEFs were carried out at matched cell passage numbers. The pCMV-Flag-Mdm2WT expression plasmid expresses Flag-tagged, full-length human Mdm2 under the control of the CMV promoter. The pCMV-Flag-Mdm2C464A expression plasmid was generated by site-directed mutagenesis using the pCMV-Flag-Mdm2WT construct as a template. The recombinant, bicistronic Adeno-Mdm2-GFP adenovirus encodes human Mdm2 and green fluorescent protein, both of which are expressed by their respective CMV promoters. For establishment of stable Mdm2-overexpressing clones, U2OS cells were co-transfected with either empty pCDNA3, pCMV-Flag-Mdm2WT, or pCMV-Flag-Mdm2C464A and pBabe-Puro. Clones were selected in 1 µg/ml puromycin (Sigma, St. Louis, MO, USA).
mRNA and protein analysis
RT–PCR as performed as previously described60 with the following primers.
| Target | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| MDM2 | CATTCAGGTGATTGGTTGGA | CACAGTAACTTGATATACCTCATCATC |
| TP53 | CTGCCCTCAACAAGATGTTTTG | CTATCTGAGCAGCGCTCATGG |
| TOP2A | CCTGTAAATGAAAATATGCAAGTCA | CCACATTTGCTGGGTCAC |
| GAPDH | CAATGACCCCTTCATTGACC | GATCTCGCTCCTGGAAGATG |
Immunoblotting was performed as described.60 The following antibodies were used anti-Mdm2 (EMD Millipore, Ab-1), anti-Flag (Sigma, F1804-M2), anti-topoisomerase IIα (Abcam, Cambridge, UK, EP1102Y) and anti-β-actin (Sigma A2066). Immunofluorescence was performed as described60 using anti-phospho-γH2A.X (Millipore, St Louis, MO, USA, JBW301).
Fluorescence in situ hybridization
Cell lines were processed for fluorescence in situ hybridization analysis using standard cytogenetic preparation methods. Interphase fluorescence in situ hybridization analysis was performed using two probes, MDM2 labeled in Spectrum Orange and Centromere Enumeration Probe 12 labeled in Spectrum Green (Abbott Molecular, Abbott Park, IL, USA). Two hundred interphase cells were analyzed per cell line.
TP53 sequencing
RNA was isolated from cell lines using an RNeasy kit (Qiagen, Hilden, Germany). Following isolation, 1µg of RNA was used for cDNA synthesis using a qScript cDNA Synthesis kit (Quanta Biosciences, Beverly, MA, USA) according to the manufacturer’s protocol. The p53 cDNA sequence was amplified by PCR (DreamTaq Green PCR Master Mix, Thermo Scientific, Waltham, MA, USA), purified by agarose gel electrophoresis, and sequenced by Sanger sequencing (Genewiz, South Plainfield, NJ, USA).
The primers used for PCR amplification of the p53 cDNA sequence were 5′-AGTCTAGAGCCACCGTCCA-3′ and 5′-TCTGACGCACACCTATTGCAAGC-3′. The primers used for sequencing were 5′-GTGCTTTCCACGACGGTGAC-3′, 5′-CTGTGACTTGCACGTACTCC-3′, 5′-AGTGGTAATCTACTGGGACGGAAC-3′, 5′-TCTGGCATTCTGGGAGCTTC-3′, 5′-TAGGGCACCACCACACTATGTC-3′, and 5′-GGAACAAGAAGTGGAGAATGTC-3′.
siRNA transfections
Individual and pooled siRNA oligonucleotides were purchased directly from Dharmacon/Thermo Scientific Molecular Biology (Lafayette, CO, USA) and used according to the manufacturer’s instructions. Oligonucleotide sequences are as follows.
| Target | Oligonucleotide # | Sequence |
|---|---|---|
| MDM2 | 9 | 5′-GCCAGUAUAUUAUGACUAA-3′ |
| 10 | 5′-GAUGAGAAGCAACAACAUA-3′ | |
| 25 | 5′-CCCUAGGAAUUUAGACAAC-3′ | |
| 26 | 5′-AAAGUCUGUUGGUGCACAA-3′ | |
| TP53 | 14 | 5′-GAAAUUUGCGUGUGGAGUA-3′ |
| 15 | 5′-GUGCAGCUGUGGGUUGAUU-3′ | |
| 16 | 5′-GCAGUCAGAUCCUAGCGUC-3′ | |
| 17 | 5′-GGAGAAUAUUUCACCCUUCC-3′ | |
| TOP2A | 06 | 5′-CGAAAGGAAUGGUUAACUA-3′ |
| 07 | 5′-GAUGAACUCUGCAGGCUAA-3′ | |
| 08 | 5′-GGAGAAGAUUAUACAUGUA-3′ | |
| 09 | 5′-GGUAACUCCUUGAAAGUAA-3′ | |
| Negative control | 5′-UUUGUAAUCGUCGAUACCCUG-3′ |
Neutral comet assay of double-strand DNA breaks
Single-cell comet electrophoresis was performed using the Trevigen CometAssay kit (Trevigen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. Cells were treated with the agents described for the durations indicated and collected by trypsinization. Cells were pelleted at 2000 r.p.m. × 5 min and washed once in 1 × PBS. Approximately 2×105 cells were resuspended in 0.1% low-melting point agarose in 1 × PBS. 50 µl of the cell suspension was pipetted onto designated areas of CometSlides and allowed to solidify for 10 min at 4 °C. Cells were lysed in CometAssay Lysis Solution for 1 h in the dark at 4 °C. Samples were immersed in 1 × Neutral Electrophoresis Buffer (NED; Tris-acetate pH 9.0) for 30 min at 4 °C and then transferred to an electrophoresis unit. Slides were aligned equidistant from electrodes. The electrophoresis unit was filled with 1 × NEB to a height of 0.5 cm above the slides, and a constant current of 1.0 V/cm, measured between electrodes, was applied for 60 min at 4 °C. Slides were removed from the unit and placed flat in DNA precipitation solution (1M NH4Ac in 95% ethanol) for 30 min at room temperature in the dark. Afterward, slides were transferred into 70% ethanol for an additional 30 min at room temperature in the dark. Samples were subsequently dried at room temperature, in the dark, overnight. Samples were stained the following day with 1 × SYBR Green I stain (Invitrogen) diluted 1:10 000 in Tris-EDTA buffer pH 7.5. Comets were visualized at × 10 magnification using an Axioplan 2 IE fluorescent microscope equipped with an AxioCam MRm digital camera (Zeiss). Images were captured using AxioVision 4.8 software (Zeiss). Percent tail DNA for 50 nuclei per experimental point was determined using ImageJ software (NIH) with the Comet Assay plug-in (original macro from Herbert M. Geller, NIH, 1997, later development by Robert Bagnell, 2011, UNC-CH). Statistical significance of comet assay results was determined using a nonparametric Mann–Whitney U test.
Doxorubicin uptake analysis
doxorubicin (2 µg/ml) in the appropriate cell culture medium was added to each well of a 12-well cell culture dish. 5 × 105 exponentially growing cells were added to three wells containing medium and drug at 120, 60, 30 and 10 min before harvest. All cells were collected and centrifuged at 2000 r.p.m. for 5 min. The cell pellets were washed once with ice-cold 1 × PBS and centrifuged a second time. The cells were resuspended in 300 µl of ice-cold 1 × PBS and fluorescence data were acquired immediately on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) as described by others.27,28 Briefly, laser excitation was set to 488 nm and all fluorescence greater than 530 nm (IGFL, integrated fluorescence) was captured for analysis. Relative intracellular doxorubicin concentration was estimated using normalized median fluorescence index (NMFI), in which cell volume is monitored by forward area light scatter (FALS) in parallel with IGFL: NMFI = (IGFL/FALS) − (IGFL0/FALS0).
Propidium iodide staining and cell cycle analysis
Cells were collected by trypsinization and centrifuged at 2000 r.p.m. for 5 min. Cell pellets were washed once in 1 × PBS and spun again. Approximately 1 × 106 cells were fixed in 70% ethanol at − 20 °C for a minimum of 12 h before analysis. Samples were centrifuged at 2000 r.p.m. for 5 min, resuspended in 1 × PBS, pelleted, and resuspended a second time in 1 × PBS containing 20 µg/ml propidium iodide and 1 mg/ml RNAse A (Sigma). Samples were transferred to 5 ml flow cytometry tubes and incubated for 12 h the dark at 4 °C. Samples data were acquired on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using CellQuest Pro software (BD Biosciences).
Immunofluorescence
Cells were split 1:5–1:10 (depending on cell type) into 100 mm cell culture dishes and grown on sterile glass coverslips for 24 h before treatment. Untreated cells were collected and fixed the following day and maintained at 4 °C until further processing. All treated cells were collected at the timepoints indicated. Upon collection, cells were washed twice in 1 × PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. After fixing, cells were washed twice more in 1 × PBS. Cells were subsequently permeabilized and blocked for 2 h at room temperature in a blocking solution comprising 0.1% Triton-X-100 and 2% bovine serum albumin in 1 × PBS. Primary anti-phosphpo-γH2A.X (Millipore, JBW301) was diluted 1:500 in blocking solution. Samples were incubated in primary antibody overnight. The coverslips were subsequently washed three times in blocking solution and incubated with 1:500 Alexa Fluor 488-conjugated goat anti-mouse for1 h at room temperature. Afterward, samples were washed twice in blocking solution and a third time in 1 × PBS. Samples were mounted onto slides and nuclei were counterstained using VECTASHIELD HardSet mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA). All imaging was performed at × 40 magnification using an Axioplan 2 IE fluorescent microscope equipped with an AxioCam MRm digital camera (Zeiss). Exposure time was kept constant for all samples. Images were captured using AxioVision 4.8 software (Zeiss). yH2A.X foci were manually counted using ImageJ software.
Supplementary Material
Acknowledgments
We wish to thank the members of the Manfredi laboratory—Lois Resnick-Silverman, Luis Carvajal, Pierre-Jacques Hamard, Crystal Tonnessen and Nicolas Bartelery—as well as Matthew O’Connell, Miguel Gama-Sosa, Stuart Aaronson and Robert Maki for helpful discussions. We also wish to thank Jonathan Fletcher (Brigham and Women’s Hospital) for the LS141 liposarcoma cell line, Ze’ev Ronai (Sanford-Burnham Medical Research Institute) for the expression vector for Flag-tagged Mdm2WT and Shohreh Varmeh for the recombinant Adeno-Mdm2-GFP adenovirus. We wish to thank Carol Prives (Columbia University) for Trp53−/−MDM2+/+ and Trp53−/−Mdm2−/− MEFs. Joseph Tripodi and Vesna Najfeld are gratefully thanked for performing the FISH analysis. These studies were supported by National Cancer Institute grants T32CA078207 to JCS and R03CA216466 to JJM and developmental support from P30CA196521.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 4.Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794. [PubMed] [Google Scholar]
- 5.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Hotta T, Ichikawa A, Kinoshita T, Nagai H, Uchida T, et al. The MDM2 oncogene overexpression in chronic lymphocytic leukemia and low-grade lymphoma of B-cell origin. Blood. 1994;84:3158. [PubMed] [Google Scholar]
- 7.Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci. 2009;34:279–286. doi: 10.1016/j.tibs.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 10.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 11.Khasraw M, Bell R, Dang C. Epirubicin: is it like doxorubicin in breast cancer? A clinical review. Breast. 2012;21:142–149. doi: 10.1016/j.breast.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Movva S, Verschraegen C. Systemic management strategies for metastatic soft tissue sarcoma. Drugs. 2011;71:2115–2129. doi: 10.2165/11594500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 13.Neal JW, Gubens MA, Wakelee HA. Current management of small cell lung cancer. Clin Chest Med. 2011;32:853–863. doi: 10.1016/j.ccm.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106:1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 15.Chikamori K, Grozav AG, Kozuki T, Grabowski D, Ganapathi R, Ganapathi MK. DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy. Curr Cancer Drug Targets. 2010;10:758–771. doi: 10.2174/156800910793605785. [DOI] [PubMed] [Google Scholar]
- 16.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 19.Ottaviano L, Schaefer KL, Gajewski M, Huckenbeck W, Baldus S, Rogel U, et al. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer. 2010;49:40–51. doi: 10.1002/gcc.20717. [DOI] [PubMed] [Google Scholar]
- 20.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 22.Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK. Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene. 2007;26:3473–3481. doi: 10.1038/sj.onc.1210136. [DOI] [PubMed] [Google Scholar]
- 23.Dimitriadi M, Poulogiannis G, Liu L, Backlund LM, Pearson DM, Ichimura K, et al. p53-independent mechanisms regulate the P2-MDM2 promoter in adult astrocytic tumours. Br J Cancer. 2008;99:1144–1152. doi: 10.1038/sj.bjc.6604643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Reifenberger G, Liu L, Collins VP, James CD. Analysis of glioma cell lines for amplification and overexpression of MDM2. Genes Chromosomes Cancer. 1994;11:91–96. doi: 10.1002/gcc.2870110205. [DOI] [PubMed] [Google Scholar]
- 25.Kondo S, Kondo Y, Hara H, Kaakaji R, Peterson JW, Morimura T, et al. mdm2 gene mediates the expression of mdr1 gene and P-glycoprotein in a human glioblastoma cell line. Br J Cancer. 1996;74:1263–1268. doi: 10.1038/bjc.1996.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaelis M, Rothweiler F, Klassert D, von Deimling A, Weber K, Fehse B, et al. Reversal of P-glycoprotein-mediated multidrug resistance by the murine double minute 2 antagonist nutlin-3. Cancer Res. 2009;69:416–421. doi: 10.1158/0008-5472.CAN-08-1856. [DOI] [PubMed] [Google Scholar]
- 27.Durand RE, Olive PL. Flow cytometry studies of intracellular adriamycin in single cells in vitro. Cancer Res. 1981;41:3489–3494. [PubMed] [Google Scholar]
- 28.Luk CK, Tannock IF. Flow cytometric analysis of doxorubicin accumulation in cells from human and rodent cell lines. J Natl Cancer Inst. 1989;81:55–59. doi: 10.1093/jnci/81.1.55. [DOI] [PubMed] [Google Scholar]
- 29.Conradt L, Henrich A, Wirth M, Reichert M, Lesina M, Algul H, et al. Mdm2 inhibitors synergize with topoisomerase II inhibitors to induce p53-independent pancreatic cancer cell death. Int J Cancer. 2013;132:2248–2257. doi: 10.1002/ijc.27916. [DOI] [PubMed] [Google Scholar]
- 30.Chen W, Qiu J, Shen YM. Topoisomerase IIalpha, rather than IIbeta, is a promising target in development of anti-cancer drugs. Drug Discov Ther. 2012;6:230–237. [PubMed] [Google Scholar]
- 31.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 32.Davies SM, Robson CN, Davies SL, Hickson ID. Nuclear topoisomerase II levels correlate with the sensitivity of mammalian cells to intercalating agents and epipodophyllotoxins. J Biol Chem. 1988;263:17724–17729. [PubMed] [Google Scholar]
- 33.Friche E, Danks MK, Schmidt CA, Beck WT. Decreased DNA topoisomerase II in daunorubicin-resistant Ehrlich ascites tumor cells. Cancer Res. 1991;51:4213–4218. [PubMed] [Google Scholar]
- 34.Ladanyi M, Cha C, Lewis R, Jhanwar SC, Huvos AG, Healey JH. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res. 1993;53:16–18. [PubMed] [Google Scholar]
- 35.Miller CW, Aslo A, Won A, Tan M, Lampkin B, Koeffler HP. Alterations of the p53, Rb and MDM2 genes in osteosarcoma. J Cancer Res Clin Oncol. 1996;122:559–565. doi: 10.1007/BF01213553. [DOI] [PubMed] [Google Scholar]
- 36.Wunder JS, Eppert K, Burrow SR, Gokgoz N, Bell RS, Andrulis IL. Co-amplification and overexpression of CDK4, SAS and MDM2 occurs frequently in human parosteal osteosarcomas. Oncogene. 1999;18:783–788. doi: 10.1038/sj.onc.1202346. [DOI] [PubMed] [Google Scholar]
- 37.Duhamel LA, Ye H, Halai D, Idowu BD, Presneau N, Tirabosco R, et al. Frequency of Mouse Double Minute 2 (MDM2) and Mouse Double Minute 4 (MDM4) amplification in parosteal and conventional osteosarcoma subtypes. Histopathology. 2012;60:357–359. doi: 10.1111/j.1365-2559.2011.04023.x. [DOI] [PubMed] [Google Scholar]
- 38.Lamoureux F, Trichet V, Chipoy C, Blanchard F, Gouin F, Redini F. Recent advances in the management of osteosarcoma and forthcoming therapeutic strategies. Expert Rev Anticancer Ther. 2007;7:169–181. doi: 10.1586/14737140.7.2.169. [DOI] [PubMed] [Google Scholar]
- 39.Ohnstad HO, Castro R, Sun J, Heintz KM, Vassilev LT, Bjerkehagen B, et al. Correlation of TP53 and MDM2 genotypes with response to therapy in sarcoma. Cancer. 2013;119:1013–1022. doi: 10.1002/cncr.27837. [DOI] [PubMed] [Google Scholar]
- 40.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1–8. [PubMed] [Google Scholar]
- 41.Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 42.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28:4862. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Issell BF, Prestayko AW, Comis RL, Crooke ST. Zinostatin (neocarzinostatin) Cancer Treat Rev. 1979;6:239–249. doi: 10.1016/s0305-7372(79)80040-7. [DOI] [PubMed] [Google Scholar]
- 44.Yeo CQX, Alexander I, Lin Z, Lim S, Aning OA, Kumar R, et al. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016;15:132–146. doi: 10.1016/j.celrep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Nayak MS, Yang JM, Hait WN. Effect of a single nucleotide polymorphism in the murine double minute 2 promoter (SNP309) on the sensitivity to topoisomerase II-targeting drugs. Cancer Res. 2007;67:5831–5839. doi: 10.1158/0008-5472.CAN-06-4533. [DOI] [PubMed] [Google Scholar]
- 46.Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005;65:5481–5484. doi: 10.1158/0008-5472.CAN-05-0825. [DOI] [PubMed] [Google Scholar]
- 47.Binaschi M, Farinosi R, Austin CA, Fisher LM, Zunino F, Capranico G. Human DNA topoisomerase IIalpha-dependent DNA cleavage and yeast cell killing by anthracycline analogues. Cancer Res. 1998;58:1886–1892. [PubMed] [Google Scholar]
- 48.Yamazaki K, Isobe H, Hanada T, Betsuyaku T, Hasegawa A, Hizawa N, et al. Topoisomerase II alpha content and topoisomerase II catalytic activity cannot explain drug sensitivities to topoisomerase II inhibitors in lung cancer cell lines. Cancer Chemother Pharmacol. 1997;39:192–198. doi: 10.1007/s002800050559. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Q, Chen J. The phenotype of MDM2 auto-degradation after DNA damage is due to epitope masking by phosphorylation. Cell Cycle. 2011;10:1162–1166. doi: 10.4161/cc.10.7.15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishida R, Hamatake M, Wasserman RA, Nitiss JL, Wang JC, Andoh T. DNA topoisomerase II is the molecular target of bisdioxopiperazine derivatives ICRF-159 and ICRF-193 in Saccharomyces cerevisiae. Cancer Res. 1995;55:2299–2303. [PubMed] [Google Scholar]
- 52.Plo I, Hernandez H, Kohlhagen G, Lautier D, Pommier Y, Laurent G. Overexpression of the atypical protein kinase C zeta reduces topoisomerase II catalytic activity, cleavable complexes formation, and drug-induced cytotoxicity in monocytic U937 leukemia cells. J Biol Chem. 2002;277:31407–31415. doi: 10.1074/jbc.M204654200. [DOI] [PubMed] [Google Scholar]
- 53.Reis C, Giocanti N, Hennequin C, Megnin-Chanet F, Fernet M, Filomenko R, et al. A role for PKCzeta in potentiation of the topoisomerase II activity and etoposide cytotoxicity by wortmannin. Mol Cancer Ther. 2005;4:1457–1464. doi: 10.1158/1535-7163.MCT-05-0156. [DOI] [PubMed] [Google Scholar]
- 54.Chikamori K, Grabowski DR, Kinter M, Willard BB, Yadav S, Aebersold RH, et al. Phosphorylation of serine 1106 in the catalytic domain of topoisomerase II alpha regulates enzymatic activity and drug sensitivity. J Biol Chem. 2003;278:12696–12702. doi: 10.1074/jbc.M300837200. [DOI] [PubMed] [Google Scholar]
- 55.Grozav AG, Chikamori K, Kozuki T, Grabowski DR, Bukowski RM, Willard B, et al. Casein kinase I delta/epsilon phosphorylates topoisomerase IIalpha at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res. 2009;37:382–392. doi: 10.1093/nar/gkn934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritke MK, Murray NR, Allan WP, Fields AP, Yalowich JC. Hypophosphorylation of topoisomerase II in etoposide (VP-16)-resistant human leukemia K562 cells associated with reduced levels of beta II protein kinase C. Mol Pharmacol. 1995;48:798–805. [PubMed] [Google Scholar]
- 57.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilon A, Quintero C, Ju L, et al. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng Z, Cortes-Ledesma F, El Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5'-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamard PJ, Lukin DJ, Manfredi JJ. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J Biol Chem. 2012;287:22397–22407. doi: 10.1074/jbc.M111.331298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.