Abstract
Well-curated sets of pathology image features will be critical to clinical studies that aim to evaluate and predict treatment responses. Researchers require information synthesized across multiple biological scales, from the patient to the molecular scale, to more effectively study cancer. This article describes a suite of services and web applications that allow users to select regions of interest in whole slide tissue images, run a segmentation pipeline on the selected regions to extract nuclei and compute shape, size, intensity, and texture features, store and index images and analysis results, and visualize and explore images and computed features. All the services are deployed as containers and the user-facing interfaces as web-based applications. The set of containers and web applications presented in this article is used in cancer research studies of morphologic characteristics of tumor tissues. The software is free and open source.
Materials and Methods
Pathology data are employed in care guidelines and clinical settings for virtually all cancer disease sites. Historically, pathology interpretations for both research studies and clinical care have used microscopes and glass slides, but whole slide images are now widely employed in clinical research settings, with widespread clinical adoption of digital pathology platforms virtually certain to occur over the next 5 years. The combination of digital pathology platforms and maturing of image analysis and machine learning methodology will make possible adoption of image data-driven systems in research and clinical settings (1). Nuclear morphology plays a central role in the characterization of tumors. Morphologic descriptions of nuclei are crucial components of pathology classifications, and many groups, including our own, have linked nuclear features to cancer outcome and molecular classification (2–10).
We present in this article a software system that provides researchers with tools that support selection of whole slide regions of interest, segmentation of nuclei within one or more regions, and visualization of nuclear features. The software encompasses: (i) efficient and robust image analysis pipelines; (ii) databases to store and index large quantities of features and annotations computed by image analysis pipelines; (iii) user interfaces and applications to interact with and explore images and computed features. Users can interact with scatter plots depicting the distribution of nuclear feature values. Users can also select subsets of the feature plot and drill down to view images of nuclei.
We have employed state-of-the-art and emerging software technologies and frameworks in the design and implementation of the software. First, we have deployed all the core components and services as containers, more specifically as Docker containers. This decision is motivated by several factors: (i) containerization facilitates a modular design, which allows for collections of the services to be deployed as part of or interfaced with other software systems. (ii) Software to support tissue image analysis needs to leverage existing libraries and tools, which may have been developed using different programming languages and rely on different compilation, configuration, and service technologies. Indeed, our implementation spans a variety of software and computing technologies (C++, Java, Apache Tomcat, Node.js, cmake, etc.). Containerization makes the deployment process easier by providing self-contained, isolated components. (iii) Cloud computing has emerged as a means of scaling resources needed by a project or an institution. Containerization allows for deployment on a Cloud platform to speed up and to scale tissue image analyses and datasets as well as deployment on local server farms and on desktop machines. Processing a whole slide tissue image (WSI) can take a long time on a single CPU core depending on image resolution, tissue coverage, and the complexity of the analysis algorithm. This process can be accelerated by partitioning the WSI into tiles (e.g., 2048 × 2048 pixel tiles) and processing the tiles concurrently on multiple machines and CPU cores. In a recent analysis run with 130 images, it took on average 2 hours per image on a machine with 40 cores; please note that multiple images can also be processed concurrently on multiple machines.
Second, we have employed NoSQL document database technologies and the JavaScript Object Notation (JSON) format to devise data models and data management components. JSON and NoSQL systems together provide an agile data management environment by allowing for flexibility in document structures. The agility and scalability of the database is very important, because the specific set of features developed in a study often depends on scientific aims and evolves over the course of a study.
Third, we have developed web-based applications and user interfaces to interact with the software infrastructure, run image analysis pipelines, and query, visualize, and explore analysis results. Our web applications enable coordinated spatial and feature-based visual analytics. Interactive exploration and visual analysis of features are critical to augmenting the feature selection and curation processes. The user-facing components of our software are web-based applications instead of desktop applications, because browsers are becoming increasingly powerful as a computing and application platform. We expect that building our user-facing components on state-of-the-art web technologies will allow us to take advantage of browsers as an additional computational resource in future releases of our software.
Results: Software Description
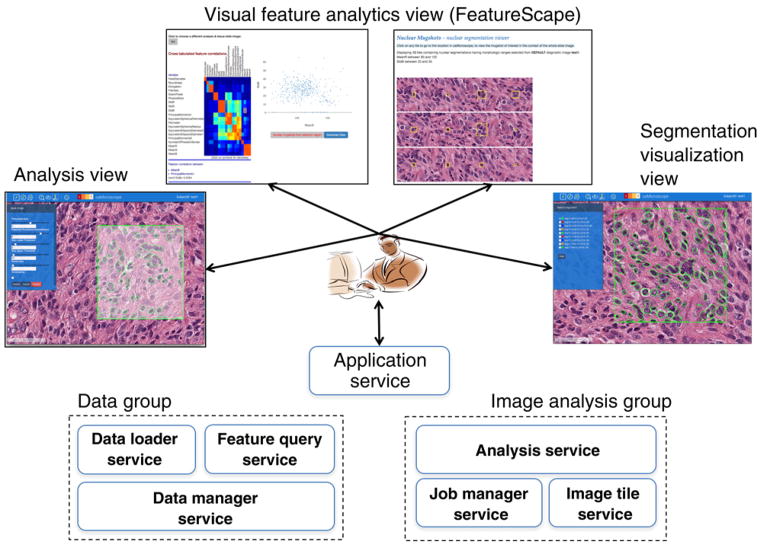
Figure 1 shows the architecture of our software. It consists of three core service groups. The application service group is a single container that hosts the application home page and web applications. The user interacts with the application service to view images, execute image analyses, explore feature sets generated from analyses, and visualize analysis results (i.e., segmentation results) overlaid on images. The image analysis group is made up of three containers. They collectively execute image analysis requests submitted by the user using the web applications. The third group is the data group. It is responsible for data loading, data management, and query processing. It is implemented as a set of three containers. In the following sections, we describe each of the groups and their containers.
Figure 1.
Software architecture. There are three groups of services: application service, data group, and analysis group. These groups are implemented as a collection of containers that interact with each other through REST and file system interfaces. The user interacts with the application service, which hosts the home page and the web applications. The web applications present several graphical user interfaces to the user for viewing and analyzing images, interacting with image features, and visualizing analysis results (segmentation results) along with images.
The application service
This container hosts the web applications to interact with the software infrastructure, submit analysis requests, and view images and analysis results. The component that facilitates the interactive exploration of WSIs and nuclear segmentation results (overlaid on images as polygons) is caMicroscope (http://camicroscope.org; ref. 11). caMicroscope provides graphical user interfaces and application programming interfaces (API), which allow the programmatic creation of a presentation state (i.e., a representation of the data for interaction). This is particularly useful when interfacing with the visual feature analytics component, referred to here as FeatureScape. Users can select a group of nuclear segmentations in a certain area and launch FeatureScape to explore these features. FeatureScape supports visualizations such as scatter plots to allow one to take a deep dive into nucleus-level features. Users can subsequently select a subregion in the scatter plot to generate a list of image patches that are representative of areas of the image. The middle of each image patch contains a segmented nucleus, the feature values of which are within the bounds of the subregion selected in the scatter plot. If the user clicks on an image patch, FeatureScape then takes the user back into caMicroscope interface with the source WSI. Such an interactive back-and-forth between a visual image-based exploration in caMicroscope and a more quantitative feature-based exploration in FeatureScape provides novel and unique insight into the significance and quality of features, thereby enabling the development of robust quantitative feature sets. Please see Supplementary Video S1, which shows an example use of the web applications and interfaces.
The image analysis group
This group of containers manages the execution of image analyses on image tiles selected by the user using the application web interfaces. The analysis service container encapsulates one or more analysis pipelines. In our current implementation, we use a level set–based nuclear segmentation algorithm developed by our group (12). This algorithm segments tiles in hematoxylin and eosin–stained tissue images to extract nuclei and computes a set of size, shape, and intensity features. The job manager service container keeps track of analysis jobs submitted by the user via the web application. The image tile service extracts user-selected image tiles from WSIs and serves them to the analysis service for processing. The analysis service subscribes to the job manager service and is triggered when an analysis job is submitted. For an analysis job, it gets the corresponding image tile from the image tile service, executes the analysis pipeline, and posts the analysis results to the data loader service, which loads them to the database.
The data group
The data management components of our infrastructure are deployed as three containers. The data manager service implements the database, referred to as FeatureDB, for storing image metadata, analysis results, and metadata about analyses (e.g., analysis parameters). To support the management of analysis results, we have created a data model called μAIM, which borrows elements from our prior work with the Pathology Analytical Imaging Standards (13) and the Annotation and Image Markup (AIM; ref. 14) data models and organizes them using a GeoJSON compliant specification. GeoJSON is a widely used format for encoding geospatial data as JSON documents (15). μAIM expresses segmentation results as polygons and size, shape, intensity, and texture features, such as area, mean intensity, for each segmented object as key-value pairs. These documents are in a MongoDB database hosted in the data manager service container. The data loader service container implements the functions for loading image metadata and image analysis results, which are output from the analysis service as image masks and csv files, which contain the computed features and boundaries of segmented objects. The feature query service container hosts a REST API for querying feature results by the FeatureScape web application. Each object (i.e., segmented nucleus) stored in the database is assigned a randomly generated index (randval) to facilitate “statistical zooming” into the search space. This index can be used to sample from a large set of nuclei extracted from thousands of images. This process speeds up queries into the database when there are hundreds of thousands or millions of nuclei per image.
Discussion and Software Availability
The software is open source and is freely available for use by the public. An instance of the software is deployed at http://quip1.bmi.stonybrook.edu. It hosts a database of about 1.9 billion segmented nuclei and their features (17 shape, size, and intensity features per segmented nucleus) and allows for users to visualize images, features, and segmentation results. Instructions and Linux scripts to install and run a local instance of the software are available at https://github.com/SBU-BMI/quip_distro. The software distribution repository has links to a video that shows the basic use of the software (also see Supplementary Video S1) as well as to the code repositories that make up the software code base. The software has been tested on Linux systems and with the Google Chrome web browser.
Supplementary Material
Acknowledgments
Grant Support
This work was supported in part by 1U24CA180924-01A1 from the NCI, and R01LM011119-01 and R01LM009239 from the NLM and by NCIP/Leidos 14 × 138.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: J. Saltz, A. Sharma, E. Bremer, J.S. Almeida, Y. Gao, M. Saltz, T. Kurc
Development of methodology: J. Saltz, A. Sharma, G. Iyer, E. Bremer, F. Wang, A. Jasniewski, J.S. Almeida, Y. Gao, M. Saltz, T. Kurc
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A. Sharma, F. Wang, T. Zhao
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Sharma, F. Wang, J.S. Almeida, Y. Gao
Writing, review, and/or revision of the manuscript: J. Saltz, A. Sharma, J.S. Almeida, T. Kurc
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A. Sharma, G. Iyer, E. Bremer, A. Jasniewski, M. Saltz
Study supervision: A. Sharma, T. Kurc
Other (software developer): T. DiPrima
Other (provided pathology expertise and user interface feedback): T. Zhao
References
- 1.Colen R, Foster I, Gatenby R, Giger ME, Gillies R, Gutman D, et al. NCI workshop report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. 2014;7:556–69. doi: 10.1016/j.tranon.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang H, Fontenay GV, Han J, Cong G, Baehner FL, Gray JW, et al. Morphometric analysis of TCGA glioblastoma multiforme. BMC Bioinformatics. 2011;12:484. doi: 10.1186/1471-2105-12-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu KH, Zhang C, Berry GJ, Altman RB, Re C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurcan MN, Pan T, Shimada H, Saltz J. Image analysis for neuroblastoma classification: segmentation of cell nuclei. Conf Proc IEEE Eng Med Biol Soc. 2006;1:4844–7. doi: 10.1109/IEMBS.2006.260837. [DOI] [PubMed] [Google Scholar]
- 5.Foran DJ, Yang L, Chen W, Hu J, Goodell LA, Reiss M, et al. ImageMiner: a software system for comparative analysis of tissue microarrays using content-based image retrieval, high-performance computing, and grid technology. J Am Med Inform Assoc. 2011;18:403–15. doi: 10.1136/amiajnl-2011-000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang K, Mosaliganti K, Cooper L, Machiraju R. Quantitative phenotyping using microscopic images. In: Rittscher J, Machiraju R, Wong STC, editors. Microscopic Image Analysis for Life Science Applications. Boston, MA: Artech House; 2008. [Google Scholar]
- 7.Lu C, Mahmood M, Jha N, Mandal M. Automated segmentation of the melanocytes in skin histopathological images. IEEE J Biomed Health Inform. 2013;17:284–96. doi: 10.1109/TITB.2012.2199595. [DOI] [PubMed] [Google Scholar]
- 8.Dong F, Irshad H, Oh EY, Lerwill MF, Brachtel EF, Jones NC, et al. Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS One. 2014;9:e114885. doi: 10.1371/journal.pone.0114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:642–53. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 10.Cooper LA, Kong J, Gutman DA, Wang F, Gao J, Appin C, et al. Integrated morphologic analysis for the identification and characterization of disease subtypes. J Am Med Inform Assoc. 2012;19:317–23. doi: 10.1136/amiajnl-2011-000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A, Kazerouni A, Saghar N, Commean P, Tarbox L, Prior F. Framework for Data Management and Visualization of The National Lung Screening Trial Pathology Images. Pittsburgh, PA: Pathology Informatics Summit; 2014. May 13–16, [Google Scholar]
- 12.Gao Y, Ratner V, Zhu LJ, Diprima T, Kurc T, Tannenbaum A, et al. Hierarchical nucleus segmentation in digital pathology images. Proc SPIE Int Soc Opt Eng. 2016:9791, 979117. doi: 10.1117/12.2217029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL. The cabig annotation and image markup project. J Digit Imaging. 2010;23:217–25. doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Kong J, Cooper L, Pan T, Kurc T, Chen W, et al. A data model and database for high-resolution pathology analytical image informatics. J Pathol Inform. 2011;2:32. doi: 10.4103/2153-3539.83192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler H, Daly M, Doyle A, Gillies S, Schaub T, Schmidt C. The GeoJSON format specification. Rapport technique. 2008:67. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.