Abstract
The foundation for the complex architecture of the brain is laid by means of a highly stereotyped pattern of proliferation and migration of neural progenitors during embryonic development. This process, termed early neurogenesis, is controlled by genetic mechanisms that have been studied in a number of experimentally amenable vertebrate and invertebrate model systems. Despite of the fact that many of these genetic mechanisms are highly conserved, fundamental structural aspects of early neurogenesis are different in the model systems used. The vertebrate brain is formed by layers of neurons that arise from large numbers of apical and basal progenitors embedded in an invaginated neuroepithelium (neural tube). By contrast, neurons in Drosophila or C. elegans descend from a relatively small number of invariant progenitors which divide in an asymmetric, stem cell-like pattern to create fixed lineages. In order to achieve a better understanding of neurogenesis it is beneficial to explore the evolution of this process. The use of molecular markers and experimental approaches spawned by the model systems has made it possible to study early neurogenesis in other animals, representing a variety of different clades, and our review attempts to provide a survey of this body of work. We divide the neurogenetic process into discrete elements, including origin, pattern, proliferation, and movement of neuronal progenitors, and compare these elements (the “toolkit” of early neurogenesis) in animals that represent the different clades. In cnidarians and many basal bilaterians the entire embryonic ectoderm produces neural precursors that differentiate within the epithelium or delaminate, and form a diffuse basiepithelial nerve net. In addition, one can distinguish in most basal bilaterians ectodermal subdomains (neuroectoderm), defined by conserved regulatory genes and signaling pathways, that contain neural progenitors at higher density, and with increased proliferatory activity. These neuroectodermal progenitors remain at the surface in the (few) basal lophotrochozoans (polychaetes) for which data exist; progenitors become internalized by a combination of delamination and invagination in basal ecdysozoans (onychophorans) and deuterostomes (hemichordates, cephalochordates). In more evolved bilaterians, larger nervous systems are realized by increasing the volume of invaginated neural progenitors (vertebrates, chelicerates), and/or advancing neural proliferation by switching to a mode of asymmetric, self-renewing mitosis (insects, crustaceans, derived annelids, vertebrates). In addition, the pattern of distribution and proliferation of neural progenitors is more precisely controlled, resulting in nervous systems with invariant neuronal architecture (annelids, arthropods, nematodes). Given their limited occurrence in derived clades, these aspects of neurogenesis have likely evolved independently multiple times.
Keywords: Nervous system, Development, Neural progenitor, Neural gene, Morphogenesis
Introduction
Many important aspects of the structure and function of the central nervous system (CNS), including the number and placement of neurons, as well as individual neuronal attributes, are ultimately controlled by processes that take place during development. The way in which neural progenitors are specified and spatially arranged, and in which they divide and migrate (events described as “early neurogenesis”), gives the immature embryonic CNS a certain shape and inner architecture, which in turn foreshadows the structure of the mature CNS. Furthermore, the phenotypic traits of neurons and glia that unfold during CNS differentiation (e.g., neurite growth and branching, synaptic connectivity, electric activity) are also strongly influenced by early neurogenetic events. Thus, neural progenitors express intrinsic determinants of neural fate (e.g., transcriptional regulators) and “forward” these factors by means of their particular mode of division to their progeny, where they control differentiation (Pearson and Doe, 2004; see Box 1). In addition, extrinsic signals can act upon neural progenitors at defined time points and change their mode of proliferation, or expression of intrinsic factors.
Box 1.
Terminology of Neurogenesis
With few exceptions, all cells of an embryo proliferate by mitotic cell division. Mitosis is defined as symmetric if the resulting daughter cells are identical in regard to size, shape, and position, and asymmetric if these aspects are not the same in the two daughter cells. Mitoses which are symmetric in regard to structure, but are known to produce daughter cells which receive different intrinsic molecular factors, are also usually called asymmetric; in order to distinguish them from the (structurally) overtly asymmetric division we will call them “molecularly asymmetric” divisions in the following.
Mitotically dividing cells in the developing organism are referred to as progenitors (progenitor cells). The term progenitor is often used interchangeably with another word, precursor. Another common use of “precursor” is to denote cells that have left the mitotic cycle, but are not yet differentiated. In this review, we will always use the term precursor in the latter sense, that is, to denote undifferentiated cells, which are either postmitotic or for which experimental evidence attesting to their postmitotic status is missing.
Whereas progenitor cells undergo a limited number of divisions before differentiating, stem cells are customarily defined as cells (usually studied in or extracted from mature organisms) that do not cease to proliferate. When talking about development, where the proliferative fate of a cell (unlimited or not?) is generally unknown, the terms are not used consistently in the literature. For example, the asymmetrically dividing neural progenitors (neuroblasts) of insects are classified as stem cells in many recent papers, even though their proliferatory activity is limited to the embryo and larva in most species. The distinction (just as in case of “precursor” vs “progenitor”) is semantic, depending on the criteria used to define a type of cell. In this review, which focuses on embryonic rather than adult neurogenesis, we will consistently use the term “neural progenitor”, rather than “stem cell”.
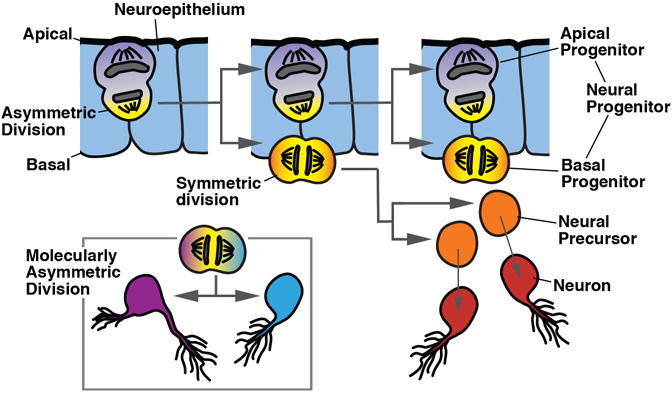
The relationship between cell division and the specification of cell fate is an important aspect of early neurogenesis. Neurons are typically specified (“born”) within growing, proliferating tissues. The step of neural determination is not necessarily linked to cell division. For example, cells distributed all over the early embryonic ectoderm, or restricted to specific domains (“growth zones”), divide and lead to an increase in ectodermal size. At the same time, mechanisms that are not involved in the pattern of mitosis act at specific locations to specify neural fate. In this instance, one may not call a dividing (ectodermal) cell a “neural progenitor”. On the other hand, proliferation and neurogenesis are often linked. Thus, progenitors located within specific regions of the ectoderm that have been specified as “neurogenic” (neuroectoderm), often increase their mitotic activity and produce exclusively neural cells (neurons or glia). These progenitors then represent neural progenitors. Neural progenitors may divide in a “stem cell-like”, asymmetric pattern, as shown for the “apical progenitors” of the vertebrate neural tube (Goetz and Huttner, 2007; Taverna et al., 2014), or the neuroblasts in insects. Alternatively, neural progenitors can divide symmetrically (e.g., basal or intermediate progenitors in vertebrates). Note that in many cases of non-model organisms that are surveyed in this review, the exact mode of division of neural progenitors has not yet been determined.
We would like to alert the reader to a difference in usage of the term “neuroblast” in the context of arthropod and vertebrate neurogenesis. As introduced above, the arthropod neuroblast denotes an asymmetrically dividing progenitor; in vertebrates, the term is often used for postmitotic neural precursors prior to differentiation (e.g., Yeo and Gautier, 2004).
Figure Box 1.
We can achieve a deeper understanding of the mechanisms controlling neural development by studying this process in different model systems, and trying to unravel the evolutionary changes that can be inferred. This review attempts to provide a comparative overview of neural progenitors found in the animal kingdom, and the neurogenetic mechanisms by which these cells frame the embryonic brain. For a few model systems (several vertebrate species, Drosophila melanogaster, and Caenorhabditis elegans), a great amount of detail is known, and in these cases we will narrowly focus on cell biological aspects of neurogenesis: where and when do neural progenitors appear, how do they divide and migrate. For selected species of some other groups, including arthropods, annelids, hemichordates, plathelminths and cnidarians (see Box 2), neural progenitors have been identified, and their proliferation, and/or expression of “neural” genes, has been studied to various extents. This allows one to draw a tentative outline describing the similarities and differences of the modes of early neurogenesis that are found in different clades, and to speculate about the phylogenetic relationships of these modes. Is it safe to conclude that asymmetrically dividing neural progenitors encountered in vertebrates and fruit flies are homologous? Is the process of neurulation, in the sense of an infolding of the proliferating neuroepithelium, an evolutionary novelty of members of the deuterostome clade? Before taking a look at what is known about early neurogenesis in the kingdom of metazoa, we will start out with a “dissection” of the neurogenetic process into discrete elements, or modules, including origin, pattern, proliferation, and movement of neuronal progenitors (see Box 1 for terminology). As presented in sections 1 and 2, these elements take on a variety of different forms. Using the modular approach as a guiding principle, we then compare the features of neurogenesis in metazoans, starting with cnidarians (section 3). For bilaterians (sections 4-6), our discussion is arranged according to the phylogenetic subdivision into the three major clades (Lophotrochozoa, Edysozoa, Deuterostomia; see Box 2). In the final section (7) we will assess possible scenarios of how the process of neurogenesis evolved during metazoan evolution.
Box 2.
Phylogeny
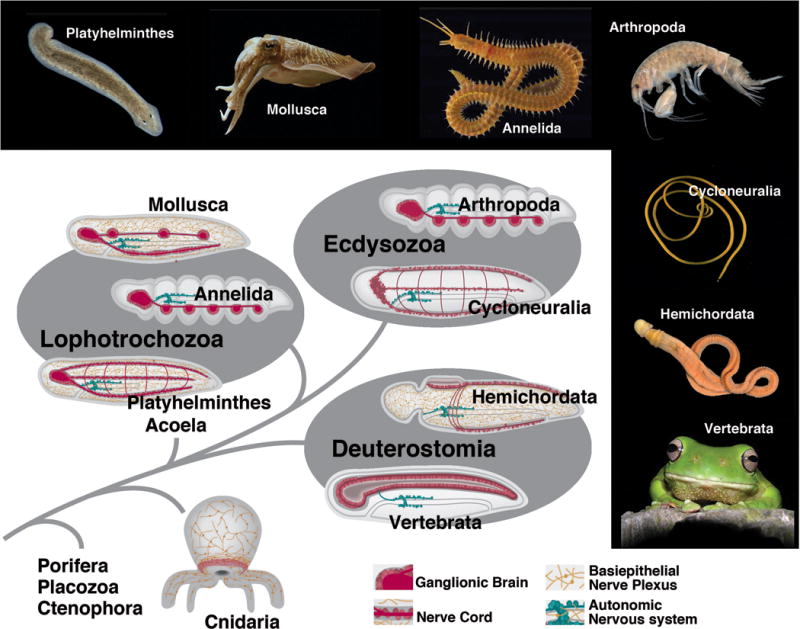
Based on fossil and molecular data, Metazoa (multicellular animals) evolved more than 800 Mio years ago (Erwin and Peterson, 2011). Two taxa that branched off the phylogenetic tree at an early stage (800-700 Mio years) are Placozoa and Porifera. Extant members of these groups do not possess a nervous system, although most elements of the molecular apparatus that mediates the function of a neuron (e.g., receptors for light and chemicals, ion channels, signal molecules stored in vesicles and released by docking of vesicles at membrane) were already present (Sakarya et al., 2007). Neurons, defined here as cells with specialized, elongated processes conducting electric impulses, are present in the next branch, Cnidaria (sea anemones, corals, jelly fishes), where sensory neurons and ganglion cells form a basiepithelial nerve plexus (nerve net), in addition to the first nerve centers associated with the mouth and tentacles (Sagaguchi et al., 1996; Mackie, 2004). The term “basiepithelial” refers to the fact that neurons and their processes form part of the epithelium (skin or gut); they are located in between the basal membrane of the epithelium and the basement membrane separating the epithelium from deeper tissue layers. Another type of jellies, Ctenophora, also possess a nerve net and were traditionally grouped close to the Cnidaria; based on recent findings, these animals split much earlier from the metazoan tree and evolved a nervous system independent of all other animals (Ryan et al., 2013; Moroz et al., 2014).
Bilaterian animals appeared on the scene between 700 and 650 Mio years ago. Bilateria are defined by their bilaterally symmetric body and a third tissue layer, the mesoderm, which gave rise to numerous specialized internal tissues and organs, including muscle, vascular, excretory and connective tissue/skeleton. Neurons of Bilateria are organized into central ganglia and peripheral sensory organs, although a subepidermal peripheral nerve net is still present in many basal bilaterian taxa (Fig. 2). Bilateria split into many diverse clades at the Ediacaran-Cambrian boundary (600-530 Mio years ago; Erwin et al., 2011). According to phylogenetic analysis started by the seminal analysis of Aguinaldo et al. (1997), and confirmed by most recent studies (e.g., Dunn et al., 2008), bilaterians comprise three major groups: deuterostomes and protostomes, the latter further subdivided into the “supertaxa” Ecdysozoa (molting animals; including arthropods, nematodes) and Lophotrochozoa (e.g., molluscs, annelids, platyhelminths, brachiopods).
The nervous system of basal deuterostomes [hemichordates (acorn worms), echinoderms (star fish)] forms a basiepithelial nerve plexus similar to that of cnidarians (for detailed review of the classical studies on invertebrate neuroanatomy, see Bullock and Horridge, 1965). In addition, local condensations of neurons occur in some regions where the neurogenic epithelium invaginates (e.g., the dorsal tube in the collar of hemichordates which is thought to foreshadow the vertebrate neural tube). In more highly derived deuterostomes [urochordates (sea squirts), vertebrates] a peripheral nerve plexus is absent. A neuroepithelium, represented by the dorsal ectoderm, invaginates as the neural tube. The central nervous system consists of layers of densely packed neurons and their processes that face outward, away from the basal surface of the invaginated neuroepithelium (Figure Box 2; see also Fig. 4B, C).
Basal Lophotrochozoa [e.g., platyhelminths (flatworms), chaetognaths (arrow worms)] also retain a peripheral basiepithelial nerve plexus. The CNS takes the shape of an anterior ganglionic brain from which nerve cords extend posteriorly (Figure Box 2). A ganglion is formed by neurons that have separated from the neuroepithelium. Within a ganglion, neuronal cell bodies form the outer layer (cortex), and branched processes/synapses (neuropil) are at the core. Nerve cords are long axon bundles surrounded by neuronal cell bodies. The taxon Acoela (flatworms with a digestive syncytium instead of a gut), according to recent phylogenetic studies, resides at the vary base of Bilateria, either as the sister group to all Bilateria, or to Deuterostomia (Ruiz-Trillo et al., 1999; Philippe et al., 2011; Figure Box 2). The nervous system of acoels has many characteristics similar to those of platyhelminths (Ramachandra et al., 2002; Bery et al., 2010), featuring a ganglionic brain and multiple nerve cords at dorsal and ventral levels. More evolved Lophotrochozoa include the clades Annelida and Mollusca. Both have large ganglionic brains that process sensory information from complex sensory organs (eyes, tentacles). Other ganglia form a ventral, segmented chord (annelids), or are distributed throughout the body (molluscs).
Within the Ecdysozoa (molting animals), most taxa are assigned to two groups, Cycloneuralia and Arthropoda (Telford et al., 2008; Liu et al., 2014). Cycloneuralians are unsegmented, worm-like animals, including nematodes (round worms), nematomorphs (hair worms), priapulids (priaps worms) and multiple other small phyla. They possess an anterior ring-shaped brain that surrounds the pharynx (hence the name); nerve cords project from the brain posteriorly at all dorso-ventral levels. Arthropods include the terrestrial myriapods (e.g. millipedes, centipedes), chelicerates (e.g. spiders) and hexapods (e.g. insects), and the mainly aquatic crustaceans. Arthropods have large, complex sensory organs (e.g., eyes and antennae of insects) and ganglionic brains; segmentally arranged ganglia form a ventral nerve cord (Figure Box 2). A basiepithelial nerve plexus is absent in ecdysozoans.
Figure Box 2.
1. Elements of early neurogenesis: Ways to make a nervous system
Animals with a nervous system include all bilaterians, as well as a few pre-bilaterian clades (Cnidara, Ctenophora). Neural progenitors of most animals arise within the context of an epithelial layer, the ectoderm. The ectoderm is formed as the outer surface layer during gastrulation, when embryonic cells destined to produce the inner organs (endoderm and mesoderm) are internalized. The process of neurogenesis begins when ectodermal cells (in some groups, such as cnidarians, also endodermal cells) acquire the potential to form neural cells. This is followed by the separation of neural cells from the ectoderm, and then by their migration, proliferation, and differentiation. The process of neurogenesis can be dissected into a number of discrete steps, or modules, which can be compared between different clades. We have ordered these steps by respecting four different categories shown in Figure 1, and used throughout Figures 2–4.
Initial pattern of cells with neurogenic potential: The neurogenic potential can be spread out over the entire ectoderm (A.1), which in that case constitutes a generalized “neurogenic ectoderm”. Alternatively, neurogenesis becomes restricted to a particular domain, the neuroectoderm (A.2.). The former case, met in cnidarians (Richards and Rentzsch, 2014), but also lower deuterostomes (Cunningham and Casey, 2014), results in a nerve plexus; the latter case results in a central nervous system. In a number of clades, neurogenic potential rests with populations of subectodermal, mesenchymal cells [A.3; e.g., interstitial cells in some cnidarians (David, 2012), neoblasts of acoels and flatworms (Rink, 2013)].
Patterning of neural progenitors: Neural precursors or progenitors arise within the neuroectoderm as individual or groups of contiguous cells. The pattern of neural progenitors is variable (B.1) or strictly invariant (B.2) among different specimens.
Proliferation of neural progenitors (for nomenclature, see Box 1): Cells of the neuroectoderm directly become neural precursors that differentiate as neurons (C.1), or give rise to proliferating neural progenitors (C.2) which can have the capability of asymmetric, self-renewing mitosis (“stem cell-like progenitors”; C.3). Here, with each division, an asymmetrically dividing neural progenitor renews itself, and produces a second daughter cell that differentiates as neuron, or becomes an intermediate progenitor, which is defined as a dividing cell that does not self renew, but undergoes a fixed number of divisions before differentiating (Noctor et al., 2004; Kowalczyk et al., 2009). The pattern of proliferation of an asymmetrically dividing neural progenitor can be strictly invariant (C.4), which results in so called “fixed lineages”. Finally, in a number of presumably derived clades such as nematodes or leeches, a fixed lineage mechanism prevails from the beginning of development (C.5).
Movement of neural progenitors: Neural progenitors either remain integrated within the surface neuroepithelium (D.1), or become internalized by a process of delamination (D.2), ingression (D.3), or invagination (D.4). Delamination describes the movement by which an individual epithelial cell constricts apically, dislocates the nucleus basally, loses its contact to the junctional adhesive complex of neighboring epithelial cells, and slides to the basal surface of the epithelium. In ingression, the same cellular movements as in delamination occur in a group of contiguous cells. Invaginating cells start out by apical constriction and nuclear dislocation, but stay in junctional contact with each other. Interstitial cells in cnidarians and neoblasts in platyhelminths are motile, mesenchymal cells (D.5).
Figure 1.
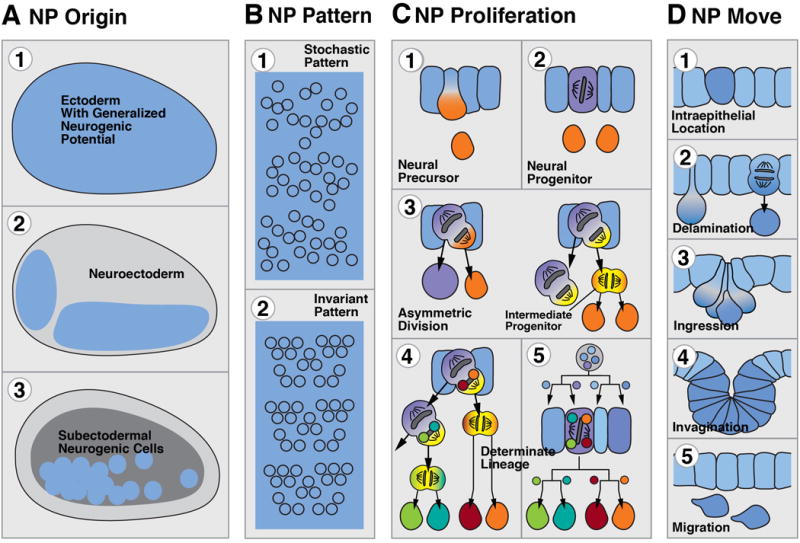
Modular representation of early neurogenesis, focusing on the origin (A), pattern (B), proliferation (C), and movement (D) of neural progenitor (NP) cells. The color coding that is applied in column (C), and maintained throughout the following figures 2–4, reflects the neurogenic potential and proliferative status of a cell: neuroepithelium: blue; actively dividing progenitor: purple; intermediate (basal) progenitors: yellow; undifferentiated precursor that is postmitotic or that has an undetermined, yet limited mitotic potential: orange. Small colored circles in cells C5 and C6 symbolize hypothetical intrinsic determinants that are channeled through asymmetric divisions into distinct neural cells (fixed lineages).
Figure 2.
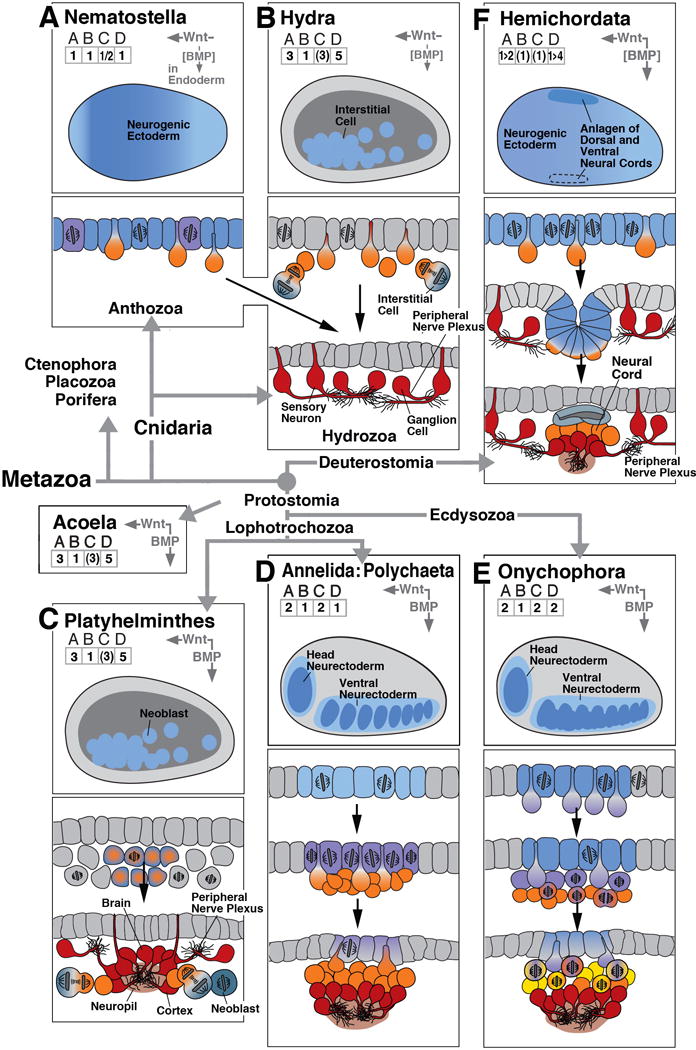
Early neurogenesis in cnidarians and basal bilaterians. In boxes (A)-(F), key events of early neurogenesis are depicted in a schematic, uniform manner for six different animal clades. Represented in the upper panel of each box is a “thumbnail” of an embryo of the corresponding clade at the onset of neurogenesis (side view; anterior to the left, dorsal up). Territories with neurogenic potential are outlined in blue. Role of BMP/BMP antagonist and Wnt signaling pathway, if present, is indicated by vertical and horizontal arrows at top right corner of upper panel. Arrows point in direction of decreasing activity (i.e., away from source) of these morphogens. Numbers in grids (ABCD) at top left capture the elements of early neurogenesis, as shown and explained in text section 1 and Figure 1. Drawings in lower panels of each box show schematic cross sections of embryos at sequential stages of development, capturing spatial characteristics and proliferatory behavior of neural progenitors. The drawings use the color code introduced in Box 1 and Figure 1. As an example, box (A) represents early neurogenesis in the cnidarian Nematostella. The Nematostella embryo shows generalized neurogenic potential all over the ectoderm (# 1 in grid A; colored blue in upper section). Neural cells are scattered stochastically over ectoderm (# 1 in grid B). Ectodermal cells form neural precursors (orange in upper section; #1 in grid C) and differentiate as epithelial, sensory neurons or delaminate to become ganglion cells (both red in bottom section). Ectoderm also contains dividing neural progenitors (# 2 in grid C; purple color in upper section). Based on published reports (Richards and Rentzsch, 2014) progenitors appear to divide in ectoderm (bracketed # 1 in grid D). Bracketing of numbers generally indicates that the implied aspect of neurogenesis is the most likely scenario, based on published data, but needs further confirmation. Bracketing of “BMP” indicates that morphogen is present but excerts no effect on neural organization. The sign “1>2” in grid A and “1>4” in grid D of box (F) signify that during an early embryonic phase of hemichordate neurogenesis, a generalized neurogenic ectoderm gives rise to neural precursors forming a nerve net; this is followed in the later embryo by a phase where the dorsal ectoderm invaginates as the dorsal neural cord, and the ventral ectoderm also gives rise to a ventral cord of higher neuronal density. Phylogenetic relationships between clades, in this and the following figures, are indicated by thick grey lines/arrows connecting the corresponding boxes. The remainder of the clades shown in this figure [(B)-(F)] and the following figures are composed in the manner explained for (A)
Figure 4.
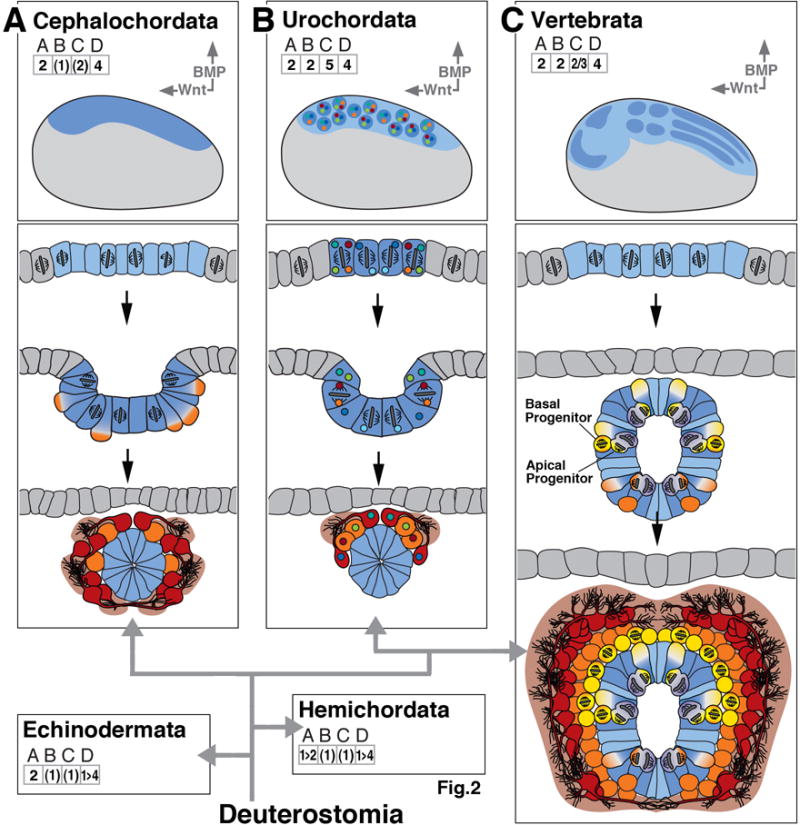
Early neurogenesis in deuterostomes; composition of figure as explained in legend of Figure 2. Deuterostomes include the basally branching echinoderms, hemichordates (represented in panel (F) of Figure 2), and cephalochordates [(A); lancelets], as well as more derived urochordates [(B); sea squirts] and vertebrates (C). Urochordates show fixed lineages with intrinsically specified neural fates.
2. Conserved genetic modules of early neurogenesis
Many of the genetic factors that specify the neuroectoderm and, subsequently, guide neuroectodermal cells through their proliferative phase towards postmitotic neurons, appear to be highly conserved throughout the animal kingdom. Admittedly, we know specifics of these genes only from a few genetic model organims, but first “glimpses” into their expression in a wider array of animals is compatible with the conclusion of their conserved role.
Transcriptional regulators of the SoxB family are expressed in the ectoderm of the early embryo and specify populations of cells that have the potential to produce neurons. In many bilaterians, SoxB factors appear in the ectoderm around the stage of gastrulation. SoxB genes provide neurogenic potential, but at the same time inhibit neural differentiation, maintaining the neuroectoderm in a proliferative state (Sasai, 2001; Bylund et al., 2003; Elkouris et al., 2011). Expression of SoxB factors, and thereby the size and shape of the neuroectoderm, is controlled by several signaling pathways, notably the BMP/BMP antagonist pathway (Mizuseki et al., 1998), and the Wnt pathway (Niehrs, 2010). The role of these and other signaling pathways in shaping the neuroectoderm, and the body axes in general, is an intensely studied field (for review, see Arendt et al., 2008; Niehrs, 2010) in itself that falls outside the scope of this review.
The expression of basic Helix-Loop-Helix (bHLH) transcription factors of the Achaete Scute family (ASH) and Atonal family (ATO) family, known as “proneural genes”, is targeted to subdomains of the neuroectoderm (“proneural clusters”) where they define neurons or neural progenitors with more restricted fates (Quan and Hassan, 2005; Powell and Jarman, 2008). In cases where they have been studied in detail, including Drosophila and various vertebrates, proneural clusters form a highly invariant pattern within the neuroectoderm. Proneural genes oppose the effect of SoxB factors, moving cells out of the progenitor state and triggering neural differentiation.
The Notch signaling pathway acts locally within the proneural clusters to control the spatial and temporal pattern at which neurons/neural progenitors are born (Beatus and Lendahl, 1998; Hartenstein and Wodarz, 2013). Proneural genes trigger an inhibitory feed-back loop (“lateral inhibition”) within (or at the boundary of) the proneural clusters by transcriptionally activating Notch ligands (e.g., Delta). Cells receiving high levels of Notch activity turn down proneural genes and express another class of bHLH genes, the E(spl)/HES genes, which inhibit neural differentiation (Kageyama et al., 2008; Stigloher et al., 2008). HES-positive cells remain undifferentiated, while retaining their neurogenic potential for later rounds of neuroblast production. Cells with low levels of Notch activity progress towards a stage of commitment as neural progenitor or neural precursor.
Committed neural progenitors enter their phase of proliferation by expressing a set of zinc-finger transcription factors, among them Snail, which control the orientation and location of the mitotic spindle, and cell cycle genes (Ashraf and Ip, 2001; Zander et al., 2014). Conserved factors such as Prospero (Knoblich, 1997; Li and Vaessin, 2000; Kaltezioti et al., 2010) and Numb (Cayouette and Raff, 2002; Johnson, 2003; Zhong, 2003) trigger the exit from the cell cycle and initiate neural differentiation.
3. Neurogenesis before the rise of bilaterian animals
Cnidaria and Ctenophora are the first metazoan clades with neurons, even though the molecular machinery enabling a cell to sense external stimuli, and generate/conduct electric impulses evolved much earlier in single cell organisms (Cai, 2012; Liebeskind et al., 2011). Accordingly, in the first multicellular animals which still lacked a nervous system (e.g. sponges), one can detect different types of cells which already encapsulate many aspects of neurons. Examples are the photosensitive cilated cells of the pigmented ring (Leys and Degnan, 2001), or the flask cells found in larvae of the demosponge Amphimedon queenslandica (Sakarya et al., 2007; Renard et al., 2009). Flask cells contain secretory vesicles, and possess most of the post-synaptic scaffold proteins encountered in neurons. During embryonic development, proneural genes and the Notch signaling pathway controls the number and pattern of flask cells (Richards et al., 2008; Richards and Degnan, 2012). This or related cell types could have given rise to the neurons that occurred in the common ancestor of bilaterians and cnidarians; the origin of neurons is currently viewed as an event that has most likely occurred multiple types in metazoan evolution (Moroz, 2012)
Ctenophora and Cnidaria were traditionally linked together as Coelenterata; members of both clades possess a nervous system in the form of a basiepithelial nerve net (Jager et al., 2011; Satterlie and Eichinger, 2014; Eichinger and Satterlie, 2014). However, two independent studies of the ctenophore genome suggest a repositioning of the phylum to the very base of the Metazoa. Since placozoans and poriferans (sponges) do not possess a nervous system and branch off basal to the cnidarians (see Box 2), the parsimonious scenario is a convergent evolution of the nervous system in ctenophores and cnidiarians/bilaterians (Ryan et al., 2013; Moroz et al., 2014). This is supported by considerable differences in the composition of the neural genes. In particular, key genes/signal pathways of early neurogenesis present in other metazoans are either absent, modified or not expressed in the nervous system. For example, bHLH members of the achaete-scute family could not be detected in the ctenophore genome, and genes regulating Notch activity, such as Delta, Numb and Mindbomb, are either absent or considerably modified. Furthermore, Hox genes, many other neuronal patterning genes and neurotransmitters present in cnidarians and other metazoans are lacking in ctenophores.
The basiepithelial nerve plexus of cnidaria is first laid down during the embryonic stage for the larva; subsequently, during metamorphosis, the larval nerve net is supplanted by an adult nervous system (Nakanishi et al., 2008; Seipp et al., 2010). Embryonic development of cnidarians is characterized by cleavage leading to the formation of an epithelial blastula, followed by the generation of an inner and outer epithelial layer (ectoderm, endoderm) through gastrulation. The embryo gives rise to a motile larval stage, the planula, which seeks out a proper place for settlement and undergoes metamorphosis into a polyp. Polyps constitute the mature phenotype of several cnidarian phyla (e.g., most hydrozoa, anthozoa); in other phyla (e.g., Scyphozoa and Cubozoa) polyps cleave into multiple free-swimming medusae.
Differentiated epidermal cells and neurons, recognized by their expression of specific transmitters (e.g., FMRF) appear as early as the gastrula or even blastula stage in the cnidarian N. vectensis (Nakanishi et al., 2012; Richards and Rentzsch, 2014). In accordance with this premature differentiation, the expression of a SoxB orthologue in N. vectensis appears already at the blastula stage in small clusters of cells scattered all over the embryo (Magie et al., 2005; Richards and Rentzsch, 2014; Fig. 2A). These cell clusters represent neural progenitors (Fig. 2A, middle) that give rise to the three main neural lineages, epithelial (sensory) neurons, basiepithelial neurons (ganglion cells) (Fig. 2A, bottom), as well as nematocytes (stinging cells). By the gastrula/planula stage, the expression of NvSoxB, along with the expression of homologues of the proneural ASH/ATO transcription factors, is restricted to individual, differentiated cells whose axons form a basiepithelial plexus (Layden et al., 2012). In contrast to bilaterians, neural progenitors and neurons also form in the endoderm. Similar expressions of SoxB and proneural genes have been reported for other cnidaria (Grens et al., 2005; Hayakawa et al., 2004; Seipel et al., 2004; Shinzato et al., 2008). Members of the Notch pathway [Notch (N), Delta (Dl), Suppressor of Hairless (SuH), Hairy/Enhancer of Split (HES)] are expressed concomitantly with proneural genes, and pharmacological N knock-down largely increases the number of neurons in the planula (Marlow et al., 2012).
The planula larva exhibits a single axis with an anterior (aboral) pole and a posterior (oral pole), the latter defined by the blastopore. Neurons occur at all levels, but are concentrated closer to the anterior pole (Martin, 1992; Gröger and Schmid, 2001; Nakanishi et al., 2008; 2012; Seipp et al., 2010; Piraino et al., 2011). This gradient is established by a Wnt-secreting signaling center located at the oral pole (i.e., posterior end of the planula), and Wnt antagonists expressed further anteriorly in the domain where neurons are concentrated (Marlow et al., 2013; Sinigaglia et al., 2013; Fig. 2A, top). The Wnt/Wnt antagonist cassette and its role in neural patterning in cnidarians is comparable (and most likely homologous) to its counterpart in bilaterians, in particular vertebrates, where Wnt antagonists, expressed in the organizer, promote neural development (Baker et al., 1999). BMP signaling is active in cnidarians and establishes an axis perpendicular to the oral-aboral axis, but only in the endoderm; neurogenesis appears unaffected in experiments where BMP signaling is modulated (Saina et al., 2009).
Cnidarians enter a second phase of development after larval settling. The oral pole (posterior with respect to former larval movement) becomes the “head” of the polyp which develops tentacles and a new nervous system, consisting of a tentacular nerve net and a nerve ring surrounding the mouth and tentacle bases (Masuda-Nakagawa et al., 2000; Nakanishi et al., 2008; Seipp et al., 2010. This nerve ring, replicated in the medusae which (in some cnidarian phyla) form from the polyp, could be considered as the evolutionary origin of a brain (Koizumi, 2007). The nerve ring has a high density of neuronal processes and synapses, includes multiple circuits with separable functions in controlling feeding and swimming behaviors, and integrates sensory input from multimodal (visual, tactile, gravitational) sense organs, the rhopalia (Mackie and Meech, 2000; Mackie, 2004; Garm et al., 2006). The formation of this adult-specific nervous system is initiated with the de novo expression of SoxB and proneural genes in the oral ectoderm of the planula and young polyp (Magie et al., 2005; Shinzato et al., 2008; Galliot et al., 2009), followed by proliferation and neuronal differentiation.
A mode of neurogenesis which appears to be different from the above described has been documented in some hydrozoans (e.g., H. vulgaris). Here, sensory neurons and ganglion cells, as well as nematocytes and gland cells, are derived from migratory interstitial cells located in between the ectoderm and endoderm of the body column (Fig. 2B). Interstitial cells continuously proliferate and behave as asymmetrically dividing stem cells: they self renew, and at the same time give rise to a daughter cell that either directly, or following several additional (symmetric) divisions differentiates into neurons (Bode, 1996; David, 2012). However, interstitial cells do not only generate neural cells but also give rise to various secretory cell types and the germ line cells. Neuronal precursors and committed progenitors migrate basally and apically and insert themselves into the ectoderm as sensory neurons/nematocytes, or differentiate as basiepithelial ganglion cells (Fig. 2B middle, bottom). Proneural genes of the ASH family are expressed in subsets of interstitial cells and nematocytes/neurons they give rise to (Grens et al., 1995), and the Notch signaling pathway plays a role in neural cell commitment (Käsbauer et al., 2007).
In summary, many of the neurogenetic cellular mechanisms and most of the neurogenetically active gene cassettes can be found prior to the origin of bilaterians. Notably, there is evidence for committed neural progenitor cells (blastula of Nematostella) which can undergo asymmetric cell division. Neural precursors or differentiated neurons undergo delamination to form a basiepithelial nerve net. These cells are distributed widely over the embryo, rather than being restricted to a specific neuroectoderm, a feature that distinguishes cnidaria from virtually all bilaterian animals. Another difference to bilaterians can be seen in the expression of neural specification genes, including Sox and ASH genes, which are often maintained (or sometimes even switched on) in differentiated cells, rather than waning with the onset of differentiation.
4. Neurogenesis in Lophotrochozoa and Acoelomorpha
Platyhelminthes and Acoela
Platyhelminthes (flatworms) include a diverse assemblage of free-living or parasitic worms. The clade Acoelomorpha used to be grouped with Platyhelminthes, but several phylogenies position acoels now as the most basally branching bilaterian group (Ruiz-Trillo et al., 1999; Hejnol et al., 2009). A recent phlylogeny even suggests a sister group relationship of acoelomorphs and Xenoturbella within the deuterostomes (Philippe et al., 2011). Neurogenesis in both flatworms and acoels exhibits characteristics that resemble the “interstitial neural progenitor mode” summarized above for H. vulgaris. This mode of neurogenesis is unusual among bilaterians. Following cleavage, platyhelminthes embryos form a mesenchymal inner mass surrounded by ectoderm (Fig. 2C top, middle). We know little about the first appearance of neural cells; markers used so far indicate that a contiguous cluster within the mesenchymal mass gives rise to the brain (Younossi-Hartenstein et al., 2000; 2001; Younossi-Hartenstein and Hartenstein, 2000; Ramachandra et al., 2002; Morris et al., 2004). It is clear that from later embryonic stages onward, pluripotent stem cells (neoblasts), which, similar to hydrozoan interstitial cells, derive from the mesenchymal mass and then populate the interstitial spaces of the worm, produce most neurons of the central nervous system and nerve plexus. Neoblasts undergo asymmetric, self-renewing divisions, and, according to recent results (Nishimura et al., 2011; Rink, 2013; Scimone et al., 2014), give rise to committed neural progenitors (as well as other types of progenitors), which migrate towards the brain and body wall where they differentiate as neurons (Fig. 2C, bottom). Neoblasts also initiate expression of neural determination genes, and in some cases appear to maintain expression into the differentiated state (e.g., SoxB1 in the acoel S. roscoffensis; Semmler et al., 2010). As in Cnidaria, a dedicated tissue layer for neurogenesis (neuroectoderm) appears to be absent in platyhelminths.
Annelids
By contrast to acoels and platyhelminths, all other members of the Lophotrochozoa have a neuroectoderm that is clearly delineated both structurally and genetically. In annelids, molluscs, and chaetognaths, spatially restricted domains within the ectoderm take on “placoideal” characters, with high cylindrical epithelial cells and increased proliferatory activity. Early neurogenesis has been described in greatest detail in recent papers on the polychaete annelids Platynereis dumerilii (Denes et al., 2007) and Capitella sp. (Meyer and Seaver, 2009), as well as the leeches Helobdella robusta and Theromyzon rude (Stuart et al., 1989; Wedeen and Weisblat, 1991; Ramirez et al., 1995; Shankland, 1995; Shain et al., 1998; Zhang and Weisblat, 2005). In P. dumerilii, an anterior and a posterior-ventral subdomain within the ectoderm, which give rise to the brain and ventral nerve cord, respectively, express SoxB (Kerner et al., 2009). Smaller, segmentally arranged expression domains of the proneural genes ASH, ATO and Olig are nested within the neuroectoderm, which consists of rapidly proliferating neural precursors (Simionato et al., 2008; Demilly et al., 2013; Fig. 2D, top). The restriction of these neural specification genes to the ventral neuroectoderm is dependent on the BMP signaling pathway (Denes et al., 2007). Postmitotic cells delaminate from the neuroectoderm and form a second, deeper layer of neural precursors (Fig. 2D, middle). These cells lose expression of specification genes, and turn on genes, including prospero and elav, that promote neural differentiation (Simionato et al., 2008). We see here the characteristic three-layered architecture of the neural primordium encountered in derived bilateria (e.g., vertebrates and arthropods): an apical, epithelial layer of neural precursors, an intermediate layer of delaminated neural precursors and/or intermediate precursors, and an inner layer of differentiated neural cells (Fig. 2D, bottom).
The mature nervous system of annelids and molluscs, similar to that of insects and other arthropods discussed below, features many large, invariant, and often uniquely identifiable cells (Bullock and Horridge, 1965; Koester and Kandel, 1965; Zipser, 1982). One developmental mechanism to achieve such precision in neuronal architecture is by fixed neural lineages. Here, intrinsic determinants are expressed in progenitors that divide according to an asymmetric, fixed pattern, and are thereby channeled into specific daughter cells; based on exactly which determinant it inherits, a daughter cell adopts a specific cell type. It is currently not yet clear whether the neural progenitors studied in P. dumerilii or other polychaetes do indeed follow this mode of proliferation. However, asymmetric and fixed lineages form a conspicuous attribute of early (cleavage) divisions in many lophotrochozoans, and do extend into the phase of neurogenesis in at least one derived group of annelids, the Hirudinea (leeches). In the early leech embryos, a bilateral set of four, posteriorly located blastomeres, called teloblasts (Stent, 1985; Weisblat, 2007), generate the entire trunk of the worm (Fig. 3C, top). One of the teloblasts (N) forms most of the neurons of the ventral nerve cord. The N-teloblast buds off, one after another, pairs of so called N-blast cells, which are assigned to one segment of the ventral nerve cord (Fig. 3C, top, middle). The blast cells (Np and Nf) continue to divide in an asymmetric, fixed pattern to form neurons (Zhang and Weisblat, 2005; Fig. 3C, middle). Little is known about genetic mechanisms controlling leech neurogenesis. It is clear that members of the Notch signaling pathway are expressed in blast cells and teloblasts, and that inhibition of Notch signaling disrupts the formation of discrete ganglion priomordia (Rivera and Weisblat, 2009).
Figure 3.
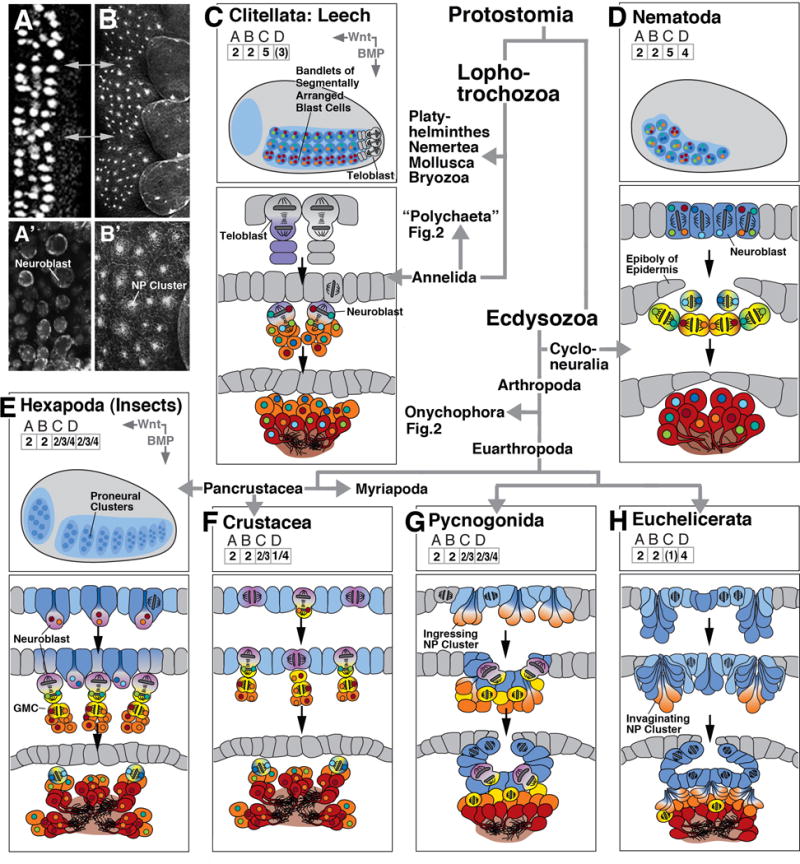
Early neurogenesis in derived protostome clades; composition of figure as explained in legend of Figure 2. Lophotrochozoa represented by leeches [Hirudinea, (C)]), a derived branch of the annelids. Within the Ecdysozoa, the branch Cycloneuralia is represented by the nematodes (D). The Arthropoda (E-H) comprise Pancrustacea, including Hexapoda (insects; E) and crustaceans, Myriapoda (centipedes; F), and Checlicerata, including Pycnogonida (sea spiders, G) and Euchelicerata (H). Thumbnail of embryo shown for Hexapoda (E) applies to all other arthropod clades.
Early neurogenesis in leech (C) and nematode (D) is characterized by fixed lineages, generated by asymmetric cell division. Positional fate (antero-posterior and dorsoventral) is controlled by intrinsic determinants and local cell-cell interactions, rather than by long range BMP/Wnt gradients.
Panels at upper left show horizontal confocal sections of the ventral neuroectoderm of an insect (A: D. melanogaster; A’: T. castaneum) and a spider (B/B’: C. salei), illustrating the similarity between the pattern of individual neuroblasts (insect) and invaginating NEURAL PROGENITOR clusters (spider). Grey arrows in A/B indicate neuromere boundaries.
Molluscs, chaetognaths, bryozoans
The ganglia of molluscs are derived from ectodermal placode that contain densely packed, proliferating neural progenitors (Jacob, 1984, for Aplysia californica). Cells that delaminate or ingress from the placodes are mostly postmitotic neural precursors, similar to what has been described for annelids. This also holds true for neurogenesis in chaetognaths (arrow worms), where mitotic, symmetrically dividing progenitors form a highly regular array within the ventral neuroectoderm of the embryo, whereas postmitotic precursors and differentiated neurons appear basally in the neuroectoderm (Rieger et al., 2011; Perez et al., 2013, for Spadella cephaloptera). No experimental or molecular-genetic data exist for the steps of early neurogenesis, such as the establishment of proneural clusters, or the control of proliferation. Bryozoa are members of the superclade Lophophorata, sessile and mostly colonial marine organisms that have in common a tentacular organ used for feeding. Even though nothing is known about the process of early neurogenesis in these animals, they deserve mentioning because the mature ganglia in several species are epithelial: differentiated neurons are located basally within internalized epithelial vesicles (Gruhl and Bartolomaeus, 2008, for Fredericella sultana and Plumatella emarginata). This implies that invagination of (part of) the neuroectoderm is an event during bryozoan (and thereby lophotrochozoan) neurogenesis. As discussed below, neuroectodermal invaginations represent a standard module in the repertoire of neurogenetic mechanisms in deuterostomes and ecdysozoans.
5. Neurogenesis in Ecdysozoa
Arthropods
Arthropods (insects, crustaceans, myriapods, chelicerates and onychophorans) show a great variety of neural precursors and mechanisms of neurogenesis, some of them surprisingly similar to the mechanisms observed in chordates (see below), others quite different. This raises the question of how these different mechanisms are phylogenetically related: which features can be considered as plesiomorphic, and which are derived. Recent large-scale molecular phylogenies have greatly improved the resolution of arthropod relationships (Regier et al., 2010) and thus facilitate the investigation of the evolution of arthropod neurogenesis. It is now widely accepted that the paraphyletic crustaceans group with the insects, forming the pancrustacean clade (Fig. 3). The pancrustaceans are a sister group of the myriapods and are united with the latter in the Mandibulata group (Regier et al., 2010). The chelicerates represent a basal branch and are a sister group of the Mandibulata. Insects, crustaceans, myriapods and chelicerates together compose the euarthropods. The onychophorans (velvet worms) are the sister group of the euarthropods and together with the latter form the phylum Arthropoda.
In all arthropods, a large neuroectodermal domain, recognizeable by characteristic structural and molecular hallmarks [e.g., expression of the SoxB gene in insects (Buescher et al., 2002) and myriapods (Pioro and Stollewerk, 2006)], is specified around the stage of gastrulation. This domain encompasses an anterior (procephalic or head) region that gives rise to the brain, and a ventral region from which the ventral nerve cord arises (Fig. 2E, top). In onychophorans, the earliest branch of the arthropods, large numbers of neural progenitors delaminate from the neuroectoderm (Eriksson and Stollewerk, 2010). The pattern of these cells shows considerable variability, in contrast to euarthropods, where neural progenitors are highly invariant (see below). Delaminated progenitors appear to divide symmetrically into intermediate neural progenitors; these in turn divide again, generating postmitotic neural cells (Fig. 2E, middle)(Eriksson and Stollewerk, 2010; Mayer and Whitington, 2009b). The different morphological mechanisms are reflected in the gene expression patterns of the proneural and neurogenic genes. In contrast to euarthropods, members of both classes of genes are not upregulated in a fixed spatio-temporal pattern; rather, they are expressed homogeneously in the neuroectoderm (Fig. 2E, top). Strong expression of the achaete-scute homologue is only seen in delaminated neural precursors. Following the delamination of neural progenitors, large parts of the neuroectoderm thicken or, in the head region, invaginate to form the so called “ventral organs”. Whereas the ventral organs of the trunk appear to undergo cell death (Mayer and Whitington, 2009a), those of the head are incorporated into the developing brain (Eriksson et al., 2003). In the adult they are named hypocerebral organs and it is assumed that they function as glands (Eriksson et al., 2013).
In insects and crustaceans most neurons are generated from stem cell-like neural progenitors, called neuroblasts (Fig. 3E, middle). Each insect neuroblasts is uniquely identifiable by its expression of a characteristic combination of transcriptional regulators (Doe, 1992; Urbach and Technau, 2003; Biffar and Stollewerk, 2014). Neuroblasts delaminate from the neuroectoderm, similar to the neural progenitors of onychophorans. However, following delamination, insect neuroblasts divide asymmetrically with a spindle that is oriented perpendicular to the plane of the neuroectoderm (Fig. 3E, middle). The apical daughter cell remains active as a neuroblast (self-renewal), the basal daughter–called ganglion mother cell–divides once to generate neurons and/or glial cells (Goodman and Doe, 1993). The presence and fixed arrangement of neuroblasts has been demonstrated in basal and derived insects and thus seems to belong to the ground pattern of insect neurogenesis (Bate, 1976; Doe and Goodman, 1985; Truman and Ball, 1998). In crustaceans (where early neurogenesis has been studied in detail for two groups, the malacostracans and the branchiopods; (Gerberding, 1997; Harzsch, 2001; Scholtz, 1992; Ungerer et al., 2011; Ungerer et al., 2012; Ungerer and Scholtz, 2008; Wheeler and Skeath, 2005) neuroblasts show a similar arrangement as in insects, and there is strong evidence for homology of individual malacostracan and insect neuroblasts based on cell lineage studies. In contrast to insects where neuroblasts delaminate from the neuroectoderm, crustacean neuroblasts remain epithelial and produce ganglion mother cells that are pushed inside by directed mitosis.
The genetic control of early neurogenesis is well established in insects. Neuroblasts are specified by transcription factors, notably the bHLH proteins encoded by the proneural genes, which are expressed in small groups of contiguous cells (“proneural clusters”) forming an invariant pattern in the neuroectoderm (Fig. 3E, top)(Bertrand et al., 2002; Cabrera et al., 1987; Ungerer et al., 2011; Wheeler et al., 2003). Proneural genes activate Notch/Delta-mediated cell-cell interactions (lateral inhibition) that result in the specification and delamination of one neuroblast per proneural cluster (Heitzler et al., 1996). Delaminated neuroblasts enter their phase of proliferation by expressing a set of zinc-finger transcription factors, among them Snail and Escargot, which control the orientation and location of the mitotic spindle, and the basal trafficking of the proteins Prospero, Numb and Brat (Cai et al., 2001; Knoblich et al., 1995). These factors end up in the ganglion mother cells where they terminate mitosis and trigger neural differentiation. Cells that had originally been part of the proneural clusters left behind in the neuroectoderm express bHLH genes of the E(spl)/HES family; once all neuroblasts have delaminated, HES expression fades, and ectodermal cells lose their neurogenic potential to become epidermal precursors (Heitzler and Simpson, 1991).
In crustaceans, the expression and function of neural genes (investigated for the branchiopod Daphnia magna; (Ungerer et al., 2011; Ungerer et al., 2012) shows several significant differences from insects which, in part, may reflect the epithelial position of the proliferative neuroblasts. In contrast to insects, Notch signaling antagonizes achaete-scute homologue (ASH) expression within neuroblasts and keeps them in an immature state. Up-regulation of ASH in individual neuroblasts results in prospero expression and asymmetric division. This mechanism resembles neural progenitor regulation in vertebrates (Kageyama et al., 2009).
It should be emphasized that additional neural precursors types and modes of neurogenesis exist in the developing insect and crustacean brain. The stomatogastric nervous system and the neuroendocrine system of D. melanogaster, as well as the large optic lobe in insects, are formed by groups of neuroectodermal cells that invaginate, closely resembling the prevalent mode of neurogenesis in chelicerates and myriapods (see below; Green et al., 1993; Hartenstein et al., 1994; de Velasco et al., 2007).
Among the clades Chelicerata and Myriapoda, neurogenesis has been investigated for a number of spiders (Linne and Stollewerk, 2011; Stollewerk, 2004; Stollewerk et al., 2001), horseshoe crab (Mittmann, 2002), sea spiders (Brenneis et al., 2013), millipedes (Dove and Stollewerk, 2003; Pioro and Stollewerk, 2006) and centipedes (Chipman and Stollewerk, 2006; Kadner and Stollewerk, 2004). In all species analyzed, the CNS is formed from clusters of contiguous neuroectodermal cells that resemble insect proneural clusters in size and pattern, but invaginate or ingress in toto, rather than giving rise to single neuroblasts (Fig. 3A, F-H)). In euchelicerates, one can distinguish an early phase of neurogenesis where a dense array of small clusters (5 to 9 cells) invaginates from the ventral neuroectoderm (Fig. 3H, middle)(Stollewerk et al., 2001); in a later phase, large groups of up to 40 cells appear that seem to increase in size and invaginate; these invaginates appear to persist into larval stages (Stollewerk, 2004)(Fig. 3H, bottom). The phase of proliferation mainly seems to take place prior to invagination; in other words, in contrast to insects/crustaceans where a numerically small neuroectoderm gives rise to a certain number of individual progenitors which then divide asymmetrically, we see in spiders a long phase of ectodermal growth (presumably by means of symmetric epithelial division), resulting in a large neuroectoderm from which then groups of neural precursors separate. Pycnogonids (sea spiders; representing an early branching group of chelicerates, or their own clade within the arthropods) even show a mixture of ingressing neural precursor groups and asymmetrically dividing neural progenitors (Fig. 3G, middle)(Brenneis et al., 2013). At a later phase, the entire neuroectoderm, encompassing neural precursor clusters and associated progenitors, invaginate to form segmental structures called “ventral organs” in the classical literature (Morgan, 1891); Fig. 3G, bottom). Similarly, in the euchelicerate Cupiennius salei, the formation of the brain involves a mechanism by which arrays of small neural precursor invaginations that had appeared at an early stage of neurogenesis are later folded inside as part of large grooves and vesicles (Döffinger et al., 2010).
Neurogenesis in myriapods resembles that in sea spiders to a large degree. Small clusters of neural precursors ingress from the neuroectoderm (Fig. 3F, middle), and at a later stage, at least in some classes, the neuroectoderm as a whole invaginates (Fig. 3F, bottom; Dove and Stollewerk, 2003). Mitotically active epithelial cells are associated with each precursor group, which could either indicate that the neural precursors of a group are related by lineage, or that additional cells are recruited to existing neural precursor groups. The exact pattern of proliferation and neural lineage relationships in chelicerates and myriapods awaits further investigation.
The molecular mechanisms of neural precursor selection seem to be similar in chelicerates and myriapods (Dove and Stollewerk, 2003; Stollewerk et al., 2001). The achaete-scute homologues show expression in large areas out of which neural precursor groups are selected subsequently. Functional studies in the spider Cupiennius salei demonstrate that the achaete-scute homologues are required for neural precursor formation and that Notch signaling restricts the expression of achaete-scute homologues to neural precursor groups (Stollewerk, 2002). However, Notch signaling has an additional function in keeping neural precursors in the epithelium over a period of several days. In Notch loss-of-function experiments neural precursors differentiate prematurely (Gold et al., 2009; Stollewerk, 2002).
Cycloneuralia
Basally branching cycloneuralia (e.g., Priapulida, Kinorhyncha) comprises multiple clades of mostly marine, unsegmented worms. The larval nervous system of these animals resembles that of other basal bilaterian groups, including scattered basal epithelial neurons and an anterior nerve center (Bullock and Horridge, 1965; Herranz et al., 2013). Nothing is known about the early developmental stages in neurogenesis of marine cycloneuralia. A cycloneuralian clade with presumably many highly derived characters are the nematodes (round worms), mostly microscopic, free living or parasitic worms. Numerous nematodes, in particular Caenorhabditis elegans, are well studied model systems. The nervous system of the nematode C. elegans consists of a fixed number of 302 neurons, which are produced by invariant cell lineages. The neuronal lineages originate from neuroblasts located in the ventral ectoderm (Fig. 3D, top) which, similar to arthropods, also generates epidermal precursors (Sulston et al., 1983; Wadsworth and Hedgecock, 1992). Nematode neuroblasts are internalized by epiboly of the dorsal ectoderm as it gives rise to the epidermis (Fig. 3D, middle). Following internalization, neuroblasts undergo 1-3 additional, molecularly asymmetric divisions before differentiating as neurons (Fig. 3D, bottom).
Nematode neurogenesis involves many different classes of intrinsic determinants that are channeled into the proper neurons through the process of molecularly asymmetric divisions. As in the (similarly derived) leeches, a stage where a neuroectoderm is subdivided into distinct groups of “generic” potential neural progenitors (i.e., proneural clusters, defined by proneural genes) which are then acted upon by signaling mechanisms (i.e., Notch signaling) can not be discerned. However, proneural genes of the achaete-scute and atonal family are required for the neural fate in some of the C. elegans neural lineages. Loss of function of the achaete-scute homologue (hlh-14) or the atonal homologue (lin-32) leads to a transformation of several types of neuroblasts into epidermal cells (Zhao and Emmons, 1995; Frank et al., 2003). Interestingly, the reverse transformation occurs in the epidermal lineages V1-4 if the homolog of hairy/Enhancer of Split (which suppresses in other anim systems), lin-22, is mutated. In the absence of lin-22, the atonal homologue lin-32 is upregulated in V1-4 leading to the formation of neuroblasts (Wrischnik and Kenyon, 1997). Furthermore, the requirement of Notch activity (glp-1) for the formation of non-neuronal cells has been demonstrated in the ABara lineage (Moskowitz et al., 1994).
6. Neurogenesis in Deuterostomia
Basal deuterostomes: hemichordates and echinoderms
CNS architecture and early neurogenesis in echinoderms and hemichordates, animal clades at the base of the deuterostome tree, bears many of the plesiomorphic characteristics present already in cnidarians. Both clades have a basiepithelial nerve plexus with specialized domains where neurons are packed at higher density and form cords or ganglia (Bullock and Horridge, 1965). In early echinoderm and hemichordate embryos, transcriptional regulators defining neurogenic potential, notably SoxB genes, are expressed early and ubiquitously in the blastula (Lowe et al., 2003; Cunningham and Casey, 2014, for Saccoglossus kowalevskii; Fig. 2F, top). Wnt signaling at the blastopore/posterior pole defines the endomesoderm, and initiates a gradient along the primary, antero-posterior axis, which concentrates neurogenic potential near the anterior pole (Darras et al., 2011). This is reflected in the expression of Wnt-inhibited genes like six3/6, which (as in Nematostella vectensis planulae) becomes restricted to the anterior ectoderm (Lowe et al., 2003). In echinoderm larvae, this domain produces the “brain-like” apical organ, which (aside from a higher neuronal density) contains several “brain specific” cell types, including serotonergic neurons. In hemichordates, the anterior ectoderm forms the proboscis, which also shows highest neuronal density along the anterior-posterior axis, and contains serotonergic neurons. A secondary, dorso-ventral axis forms under the control of BMP signaling in echinoderm embryos, and restricts neurogenic ectoderm in the trunk to a specific domain, the “ciliary band ectoderm” (Angerer et al., 2011; Burke et al., 2014, for Strongylocentrotus purpuratus). In hemichordates, neurogenesis is initially not restricted along the dorsoventral axis and is insensitive to an existing BMP gradient, allowing for the formation of neurons at all dorso-ventral levels (Lowe et al., 2006; Cunningham and Casey, 2014; Fig. 2F, top, middle). Secondarily, a restricted dorsal portion of the ectoderm (dorsal collar) gives rise to an exclusively neural territory, which invaginates to form the dorsal neural cord, considered as a forerunner of the chordate neural tube (Fig. 2F, middle). The cord consists of a strand of ependymal cells flanked ventrally by a band of neuronal cell bodies and sparse neuropil (Nomaksteinsky et al., 2009; Kaul and Stach, 2010; Fig. 2F, bottom).
Based on the few reports that exist it appears as if neuronal differentiation, similar to the situation in cnidarian embryos, takes place within the ectoderm (Fig. 2F, middle). Markers for postmitotic neurons (Elav, Synaptotagmin) are expressed as early as the gastrula stage in scattered cells within the ectoderm, where they overlap with the widespread, diffuse expression of early neural specification genes (Cunningham and Casey, 2014). The relationship between neural specification and proliferation has not been investigated so far. Thus, it is not clear whether dedicated neural progenitors exist that undergo symmetric or asymmetric divisions prior to neural differentiation.
Urochordates, cephalochordates, and vertebrates
Urochordates (=tunicates, ascidians), cephalochordates and vertebrates form a neuroectoderm restricted to the dorsal side of the ectoderm (Fig. 4A-C, top). Expression of SoxB genes, as well as the involvement of the BMP-BMP antagonist cassette and Wnt signaling in setting up the neuroectoderm, has been shown in all three clades [cephalochordates (Branchiostoma floridae): Holland et al., 2000, 2005; Lu et al., 2012; Onai et al., 2012; urochordates (Halocynthia roretzi): Miya and Nishida, 2003; Darras and Nishida, 2001; vertebrates: reviewed in Altmann and Brivanlou, 2001; Niehrs, 2010; Schmidt et al., 2013]. The dorsal neuroectoderm invaginates to form a hollow epithelial cylinder, the neural tube. All cells of the neural tube possess neurogenic potential and undergo diverse patterns of proliferation to produce neurons and glia.
In the ascidian Ciona intestinalis, neurogenesis follows a determinate lineage pattern that, similar to that found in C. elegans and in leech, is considered highly derived. Ciona larvae are freely swimming organisms shaped similar to a vertebrate tadpole and possess a small number of cells which, from the beginning of cleavage onward, are all produced in a fixed lineage mechanism (Nicol and Meinertzhagen, 1988a, b; Lemaire, 2009). The neural plate consists of 48 uniquely identifiable neural progenitors that are arranged in a regular orthogonal pattern, forming four columns and six rows. Following neurulation (Fig. 4B, middle), these progenitors undergo another 2-3 rounds of mitosis before a subset of them delaminates to form the invariant, small set of larval neurons (Fig. 4B, bottom). Proneural genes of the bHLH family are expressed in distinct neuronal progenitors, and the Notch signaling cassette, in addition to other signaling pathways (FGFR, Nodal), control the pattern of neurogenesis by mediating cell-cell interactions between cells belonging to different columns within the neuroepithelium (Hudson et al., 2007). The larval nervous system undergoes major, not yet characterized changes during metamorphosis, leading up to the cerebral ganglion of the mature, sessile sea squirt.
Neural development in cephalochordates follows a similar pathway as in urochordates, aside from the fact that determinate lineages do not seem to exist. The neural plate expresses the proneural gene neurogenin in segmentally organized clusters (Holland et al., 2000). The Notch ligand Dl appears in dispersed epithelial cells of the neural plate, but is not expressed in the neural tube of late embryos (Rasmussen et al., 2007). This finding, along with the fact that the gene Elav (typically found in postmitotic neural precursors) is widely expressed in the neural plate (Satoh et al., 2001), suggests that the birth of larval neurons occurs at an early stage within the neural plate and during neurulation (Fig. 4A, middle). Proliferation follows a distinct pattern within the neural plate and neural tube; no asymmetric mitoses have been described (Holland and Holland, 2006).
The neural plate and neural tube of vertebrates is considerably larger than that one of protochordates, and goes through an extended phase of proliferative neurogenesis. Proliferating cells constitute part of the neuroepithelium (apical progenitors), or have delaminated and cover the basal surface of the neuroepithelium (basal progenitors; also called intermediate progenitors; Noctor et al., 2004; Götz and Huttner, 2005; Taverna et al., 2014; Fig. 4C middle, bottom). In addition, various other types of dividing progenitors have been described for specialized domains within the neural tube (e.g., the progenitors of the cerebellar rhombic lip; Wingate, 2001). At an early stage during and shortly after neurulation, apical progenitors undergo symmetric division, resulting in an expansion of the neural tube. At later stages, apical progenitors increasingly switch to an asymmetric, self-renewing mode of division. One cell remains mitotically active, the other one delaminates and differentiates as a neuron, oligodendrocyte, or basal progenitor. At this point (i.e., the onset of neuronal and glial birth), apical progenitors have transitioned molecularly into a different state, expressing markers of astroglial cells, and are called radial glial cells (Götz and Huttner, 2005). Their name notwithstanding, radial glia continues to produce mostly neurons until a late stage in embryogenesis when most of these cells differentiate as astroglia; a smaller fraction becomes the ependymal lining of the ventricle, and some radial glia remain undifferentiated and give rise to adult neural stem cells (Tramontin et al., 2003).
Unlike apical progenitors, basal progenitors do not self renew, but undergo a limited number (typically just one; Miyata et al., 2004; Kowalczyk et al., 2009) of equal divisions before differentiating. Basal progenitors are found predominantly in the anterior part of the neural tube, in particular the telencephalon, where they form from late embryonic stages onward (E14 in mouse) the massive subventricular zone (SVZ; Fig. 4C, bottom). The formation and size of the basal progenitor population is correlated with the expansion of the dorsal telencephalon, which gives rise to the cerebral cortex in mammals.
Many of the molecular pathways involved in vertebrate neurogenesis show all the conserved features sketched in section 2 above. Conserved factors such as SoxB transcription factors (Xenopus: Mizuseki et al., 1998; chicken and mouse: Pevny and Plczek, 2005; zebrafish: Schmidt et al., 2013) and Snail zinc finger proteins (Zander et al., 2014; for mouse) play a role in the neuroepithelium to promote proliferation and prevent cell death. Another member of the Snail family of transcription factors, Scratch, is required for delamination of neural precursors by downregulating E-cadherin (Itoh et al., 2013; for mouse); this function could be very similar to the role of Drosophila Snail, which comes on in neuroblasts about to lose contact with the apical surface (Ashraf and Ip, 2001). Snail also downregulates epithelial cadherin in gastrulation, and epithelial-to-mesenchymal transitions in general (Schäfer et al., 2014).
At the onset of neurulation, the expression of proneural genes defines extensive “proneural domains” from which precursors of early born neurons (primary neurons) are selected (Fig. 4C, top). These domains are surrounded by territories where primary neurogenesis is inhibited by expression of the E(spl)/HES genes (Stigloher et al., 2008; Schmidt et al., 2013, for zebrafish); these domains become active postembryonically as a source for secondary neurons. Within the embryonic proneural domains, Notch signaling defines the differential fate of the asymmetrically dividing apical progenitors; the daughter cell ending up with lower Notch activity loses its progenitor fate and differentiates as neuron, or becomes an intermediate progenitor. Interestingly, several different mechanisms seem to exist that regulate the level of Notch activity. One mechanism, described in mouse (Bultje et al., 2009), is directly tied to the unequal distribution of the apical Par3 complex resulting from an asymmetric division. Due to a tilted plane of division, only the more apical daughter cell inherits Par3; this cell upregulates Notch and continues as an epithelial apical progenitor, whereas the other daughter cell downregulates Notch and delaminates. Asymmetric distribution of the Notch inhibitor Numb may also be involved in promoting differentiation of the delaminating cell (Shen et al., 2002; Petersen et al., 2004). In zebrafish, Par3 inherited by the apical daughter cell also regulates Notch, yet in the opposite direction as in mouse: by activating the factor Mindbomb (Mib), which increases the activity of the Notch ligand Delta, the level of Notch increases in the neighboring, basal cell; this cell re-inserts into the epithelium as a progenitor, whereas the apical cell delaminates to become the neuron (Alexandre et al., 2010). Following delamination, high proneural gene activity in neural precursors activates Prospero. As in Drosophila, Prospero initiates a program of neuronal differentiation and blocks continued proliferation; there is a direct feedback between the expression of Prospero and decreased Notch activity in neural precursors (mouse: Misra et al., 2008; Kaltezioti et al., 2010).
7. The evolution of neurogenetic mechanisms in Bilateria
The comparison of neural architectures and the underlying developmental mechanisms among bilaterian clades surveyed in the previous sections suggests that the last common ancestor of bilaterian animals (“Urbilateria”) must have possessed a well-stocked “toolkit” of neurogenesis, comprising the following elements:
Specialized ectodermal domains of Urbilateria were designated as neuroectoderm, destined to produce a centralized nervous system, by means of signaling pathways (Wnt/BMP), and the transcription factors promoting and maintaining neurogenic potential (e.g., SoxB genes). This condition has now been well confirmed for most bilaterian animals, and therefore most likely constitutes a plesiomorphy inherited from the last common ancestor.
Given that many of the clades considered basal among bilaterians have retained a basiepithelial nerve plexus, it seems likely that the ectoderm of Urbilateria had widespread neurogenic potential outside the central neuroectoderm. The condition in extant hemichordates (generalized expression of SoxB and proneural genes, followed secondarily by restricted expression in domains that invaginate) may serve as a model for the structure of such an ancestral combination of central and generalized neurogenesis.
Cells of the generalized neurogenic ectoderm of Urbilateria differentiated directly into epithelial, sensory neurons, as well as delaminating ganglion cells; these cells then constituted the basiepithelial nerve plexus.
The central neuroectoderm was able to produce neurons in higher numbers than a generalized neurogenic ectoderm by two mechanisms, invagination of (parts of) the neuroepithelium, and formation of dedicated neural progenitors. In many extant clades (e.g., arthropods, chordates) both mechanisms coexist, and it is therefore possible that both are plesiomorphic neurogenetic features handed down from Urbilateria.
The “neural gene cassette”, whose central elements are the proneural genes controlled by Notch signaling, specified the number and pattern of neural precursors and progenitors in the generalized neurogenic ectoderm and the central neuroectoderm. High levels of Notch activity maintain neural progenitors in the neuroepithelium, and prevent neural differentiation, by transcriptional repression of the proneural genes. Small, random shifts in the expression levels of proneural genes and a Notch ligand (e.g., Delta) could lead to a reduction of Notch signaling in single neural precursors/progenitors, which then would allow them to delaminate. Such a mechanism would result in a stochastic pattern of neural precursor/progenitor segregation, which most likely represents the ancestral pattern.
Factors like Prospero or Numb, which have been confirmed in members of all three super clades (Deuterostomia, Ecdysozoa, Lophotrochozoa), most likely formed already integral parts of the “neural gene cassette” in Urbilateria and controlled the transition of neural precursors from proliferation to differentiation.
In conclusion, most of the modules of early neurogenesis formed already part of the “toolkit” that was at the disposal of Urbilateria. On the other hand, it is likely that at least two elements of neurogenesis encountered in many derived bilaterians, the high degree of determinacy of the positioning of neural progenitors, and intrinsically fixed lineages, are derived features that could have evolved independently multiple times.
Patterning neural progenitors: what we can learn from arthropods
Based on the comparison of neurogenetic mechanisms observed among the extant euarthropod clades the hypothesis can be put forward that the versatility of Notch function and its interactions with the proneural genes could be the basis for the diversity of neural precursor patterns. When considering the outgroup, Onychophora, we may expect a variable pattern of individual neural precursors delaminating from the neuroectoderm at the base of the arthropods (Eriksson and Stollewerk, 2010). In the last common ancestor of euarthropods, the formation of neural precursors became spatio-temporally fixed. This could have been achieved by two events: (1) changes in the regulatory regions of the proneural genes that led to an up-regulation in domains/patches within the neuroectoderm, (2) the inclusion of additional regulatory mechanism of Notch signaling. Biological and mathematical models show that the pattern of neural precursor selection can change from domains to groups of cells to single cells depending on the regulation of the Notch pathway (Wang et al., 2011). The selection mechanisms are linked to pre-patterning mechanisms that determine the anterior-posterior and dorso-ventral polarity within segments, setting up the precise areas of proneural gene expression and thus the fixed arrangement of neural precursors (Skeath, 1999; Skeath et al., 1992).
Clusters of contiguous neural precursor distributed in an invariant pattern most likely represented the ancestral feature of arthropod neurogenesis; we see this condition in extant chelicerates and myriapods (Fig. 3C, D, E). Subsequently, asymmetrically dividing neural progenitors evolved from these clusters, which, among others, is supported by the strikingly similar arrangement and number of neural precursor groups and neuroblasts, respectively, in the ventral neuromeres of euarthropods (Fig. 3A, B)(Döffinger and Stollewerk, 2010; Eriksson and Stollewerk, 2010). The function of Notch signaling in maintaining cells in an epithelial state and controlling binary cell fate decisions might have culminated in singling out individual cells from the precursors groups. The difference in the final position of the neuroblasts observed in insects and crustaceans (i.e. delamination in insects/maintenance in the epithelium in crustaceans) could also be explained by variations in the regulation of Notch signaling.
Asymmetric division and fixed lineage patterns of neural progenitors
An invariant pattern of proliferation that is “fixed” by intrinsic, asymmetrically distributed factors, is often correlated with an invariant neuronal architecture of the mature brain. In such brains, one encounters conserved arrays of unique cells that can be recognized in every specimen. The connection between fixed lineage and invariant neuronal pattern is plausible: intrinsic determinants are expressed in progenitors that divide according to an invariant, asymmetric pattern, and are thereby channeled into specific daughter cells; based on exactly which determinant it inherits, a daughter cell adopts a specific cell fate (Pearson and Doe, 2004).
As described above, well documented cases of asymmetrically dividing neural progenitors producing fixed lineages are observed in several derived clades of Ecdysozoa (insects, nematodes), Lophotrochozoa (leech) and Deuterostomia (ascidians). The fixed lineage mechanism (in the nervous system) has not been described in clades considered basal, and has therefore likely evolved independently, using molecular modules that existed as preadaptations. Thus, asymmetric, fixed divisions are widespread throughout all clades during early embryogenesis. For example, in all spirally cleavaging embryos (e.g., molluscs, annelids, some platyhelminths), and in many cycloneuralians, the cleavage divisions are asymmetric and invariant, giving rise to blastula stage embryos with several hundred cells each of which has a predetermined fate (for review, see relevant chapters in Gilbert and Raunio, 1997). The molecular machinery that controls asymmetric fixed divisions is probably conserved. For example, the Par3 complex (discovered through its function during C. elegans cleavage) plays a role in positioning the mitotic spindle, and in distributing certain determinants (like Prospero or Numb), in asymmetrically dividing cells in C. elegans, Drosophila, and vertebrates (Doe, 2001; Suzuki and Ohno, 2006). Thus, a cellular mechanism for generic asymmetric divisions was available as a Urbilaterian plesiomorphy, so that, once the “necessity arose” for asymmetric neural progenitor divisions, this mechanism could be utilized towards that purpose. Such then could be the evolutionary relationship between asymmetrically dividing NPs in the vertebrate and insect embryo: their presence does not imply that Urbilateria possessed asymmetrically dividing neural progenitors, let alone neuroblasts with fixed lineages; the link is the conserved genetic cassette (Par3 complex, Insc, Numb) that was present in all bilaterians, and that was extracted from the toolkit whenever asymmetric division coupled to self renewal became advantageous during neurogenesis, independently in the vertebrate lineage (to generate more neurons), and the insect/crustacean lineage (to efficiently distribute intrinsic determinants for fixed lineages).
A final point that deserves mentioning is the question how, if at all, an invariant neuronal architecture can be generated without fixed lineages. In polychaetes or molluscs, which have stereotyped patterns of neurons in many ganglia (Hulsebosch and Bittner, 1981; Koester and Kandel, 1977), fixed lineages have not been reported so far, but might come to light once techniques like live imaging and clonal analysis become available. Invariant neural architecture in the apparent absence of fixed lineages also occurs in many arthropods, including spiders and myriapods discussed above. One might speculate that here, local cell-cell interaction within the individual neural precursor groups/invaginations that produce the ganglia could generate an invariant neural architecture. As described above, neural precursor clusters are similar in pattern and overall cell number (per cluster) to the primary lineages of insects (approximately 30 lineages with 10-15 cells each per hemisegment). It is conceivable that in the spider, similar to what happens in the Drosophila eye disc during the specification of different ommatidial cell types (Kumar, 2012), one cell is selected first in each cluster cluster, which then acts as a signaling center to specify in a fixed temporal order the other cell types. Just as in the compound eye, such a local signaling mechanism, given the small number of cells that are involved, could account for an invariant neural architecture.
References
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Altmann CR, Brivanlou AH. Neural patterning in the vertebrate embryo. Int Rev Cytol. 2001;203:447–482. doi: 10.1016/s0074-7696(01)03013-3. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: insights from sea urchin development. Development. 2011;138:3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Denes AS, Jékely G, Tessmar-Raible K. The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci. 2008;363:1523–1528. doi: 10.1098/rstb.2007.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Ip YT. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development. 2001;128:4757–4767. doi: 10.1242/dev.128.23.4757. [DOI] [PubMed] [Google Scholar]
- Bailly X, Reichert H, Hartenstein V. The urbilaterian brain revisited: novel insights into old questions from new flatworm clades. Dev Genes Evol. 2013;223:149–157. doi: 10.1007/s00427-012-0423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999;13:3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J Embryol Exp Morphol. 1976;35:107–123. [PubMed] [Google Scholar]
- Beatus P, Lendahl U. Notch and neurogenesis. J Neurosci Res. 1998;54:125–136. doi: 10.1002/(SICI)1097-4547(19981015)54:2<125::AID-JNR1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bery A, Cardona A, Martinez P, Hartenstein V. Structure of the central nervous system of a juvenile acoel, Symsagittifera roscoffensis. Dev Genes Evol. 2010;220:61–76. doi: 10.1007/s00427-010-0328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffar L, Stollewerk A. Conservation and evolutionary modifications of neuroblast expression patterns in insects. Dev Biol. 2014;388:103–116. doi: 10.1016/j.ydbio.2014.01.028. [DOI] [PubMed] [Google Scholar]
- Bode HR. The interstitial cell lineage of hydra: a stem cell system that arose early in evolution. J Cell Sci. 1996;109:1155–1164. doi: 10.1242/jcs.109.6.1155. [DOI] [PubMed] [Google Scholar]
- Brenneis G, Stollewerk A, Scholtz G. Embryonic neurogenesis in Pseudopallene sp (Arthropoda, Pycnogonida) includes two subsequent phases with similarities to different arthropod groups. Evodevo. 2013;4:32. doi: 10.1186/2041-9139-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd GE, Telford MJ. The origin and evolution of arthropods. Nature. 2009;457:812–817. doi: 10.1038/nature07890. [DOI] [PubMed] [Google Scholar]
- Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- Bullock TH, Horridge GA. Structure and Function in the Nervous System of Invertebrates. San Francisco: Freeman; 1965. [Google Scholar]
- Burke RD, Moller DJ, Krupke OA, Taylor VJ. Sea urchin neural development and the metazoan paradigm of neurogenesis. Genesis. 2014;52:208–221. doi: 10.1002/dvg.22750. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cabrera CV, Martinez-Arias A, Bate M. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell. 1987;50:425–433. doi: 10.1016/0092-8674(87)90496-x. [DOI] [PubMed] [Google Scholar]
- Cai X. Evolutionary genomics reveals the premetazoan origin of opposite gating polarity in animal-type voltage-gated ion channels. Genomics. 2012;99:241–245. doi: 10.1016/j.ygeno.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chia W, Yang X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 2001;20:1704–1714. doi: 10.1093/emboj/20.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat Neurosci. 2002;5:1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Casey ES. Spatiotemporal development of the embryonic nervous system of Saccoglossus kowalevskii. Dev Biol. 2014;386:252–263. doi: 10.1016/j.ydbio.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ. b-catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development. 2011;138:959–970. doi: 10.1242/dev.059493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CN. Interstitial stem cells in Hydra: multipotency and decisionmaking. Int J Dev Biol. 2012;56:489–497. doi: 10.1387/ijdb.113476cd. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302:309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Demilly A, Steinmetz P, Gazave E, Marchand L, Vervoort M. Involvement of the Wnt/b-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat Commun. 2013;4:1915. doi: 10.1038/ncomms2915. [DOI] [PubMed] [Google Scholar]
- Denes AS, Jékely G, Steinmetz PR, Raible F, Snyman H, Prud’homme B, Ferrier DE, Balavoine G, Arendt D. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Cell polarity: the PARty expands. Nat Cell Biol. 2001;3:E7–E9. doi: 10.1038/35050684. [DOI] [PubMed] [Google Scholar]
- Doeffinger C, Hartenstein V, Stollewerk A. Compartmentalization of the precheliceral neuroectoderm in the spider Cupiennius salei: development of the arcuate body, optic ganglia, and mushroom body. J Comp Neurol. 2010;518:2612–2632. doi: 10.1002/cne.22355. [DOI] [PubMed] [Google Scholar]
- Doeffinger C, Stollewerk A. How can conserved gene expression allow for variation? Lessons from the dorso-ventral patterning gene muscle segment homeobox. Dev Biol. 2010;345:105–116. doi: 10.1016/j.ydbio.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Dove H, Stollewerk A. Comparative analysis of neurogenesis in the myriapod Glomeris marginata (Diplopoda) suggests more similarities to chelicerates than to insects. Development. 2003;130:2161–2171. doi: 10.1242/dev.00442. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Eichinger JM, Satterlie RA. Organization of the ectodermal nervous structures in medusae: cubomedusae. Biol Bull. 2014;226:41–55. doi: 10.1086/BBLv226n1p41. [DOI] [PubMed] [Google Scholar]
- Elkouris M, Balaskas N, Poulou M, Politis PK, Panayiotou E, Malas S, Thomaidou D, Remboutsika E. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells. 2011;29:89–98. doi: 10.1002/stem.554. [DOI] [PubMed] [Google Scholar]
- Eriksson BJ, Stollewerk A. Expression patterns of neural genes in Euperipatoides kanangrensis suggest divergent evolution of onychophoran and euarthropod neurogenesis. Proc Natl Acad Sci USA. 2010;107:22576–22581. doi: 10.1073/pnas.1008822108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson BJ, Tait NN, Budd GE. Head development in the onychophoran Euperipatoides kanangrensis with particular reference to the central nervous system. J Morphol. 2003;255:1–23. doi: 10.1002/jmor.10034. [DOI] [PubMed] [Google Scholar]
- Eriksson BJ, Samadi L, Schmid A. The expression pattern of the genes engrailed, pax6, otd and six3 with special respect to head and eye development in Euperipatoides kanangrensis Reid 1996 (Onychophora: Peripatopsidae) Dev Genes Evol. 2013;223:237–246. doi: 10.1007/s00427-013-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- Frank CA, Baum PD, Garriga G. HLH-14 is a C. elegans achaete-scute protein that promotes neurogenesis through asymmetric cell division. Development. 2003;130:6507–6518. doi: 10.1242/dev.00894. [DOI] [PubMed] [Google Scholar]
- Gerberding M. Germ band formation and early neurogenesis of Leptodora kindti (Cladocera): first evidence for neuroblasts in the entomostracan crustaceans. Invertebr Reprod Dev. 1997;32:63–73. [Google Scholar]
- Gilbert SF, Raunio AM. Embryology: Constructing the Organism. Sunderland: Sinauer Associates; 1997. [Google Scholar]
- Gold K, Cotton JA, Stollewerk A. The role of Notch signaling and numb function in mechanosensory organ formation in the spider Cupiennius salei. Dev Biol. 2009;327:121–131. doi: 10.1016/j.ydbio.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Goodman CS, Doe CQ. In: Embryonic development of the Drosophila central nervous system In The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1131–1206. [Google Scholar]
- Goetz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993;273:583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- Grens A, Mason E, Marsh JL, Bode HR. Evolutionary conservation of a cell fate specification gene: the Hydra achaete-scute homolog has proneural activity in Drosophila. Development. 1995;121:4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]
- Groeger H, Schmid V. Larval development in Cnidaria: a connection to Bilateria? Genesis. 2001;29:110–114. doi: 10.1002/gene.1013. [DOI] [PubMed] [Google Scholar]
- Gruhl A, Bartolomaeus T. Ganglion ultrastructure in phylactolaemate Bryozoa: evidence for a neuroepithelium. J Morphol. 2008;269:594–603. doi: 10.1002/jmor.10607. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Wodarz A. Initial neurogenesis in Drosophila. Wiley Interdiscip Rev Dev Biol. 2013;2:701–721. doi: 10.1002/wdev.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Tepass U, Gruszynski-Defeo E. Embryonic development of the stomatogastric nervous system in Drosophila. J Comp Neurol. 1994;350:367–381. doi: 10.1002/cne.903500304. [DOI] [PubMed] [Google Scholar]
- Harzsch S. Neurogenesis in the crustacean ventral nerve cord: homology of neuronal stem cells in Malacostraca and Branchiopoda? Evol Dev. 2001;3:154–169. doi: 10.1046/j.1525-142x.2001.003003154.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa E, Fujisawa C, Fujisawa T. Involvement of Hydra achaete-scute gene CnASH in the differentiation pathway of sensory neurons in the tentacles. Dev Genes Evol. 2004;214:486–492. doi: 10.1007/s00427-004-0430-4. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguñà J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz M, Pardos F, Boyle MJ. Comparative morphology of serotonergic-like immunoreactive elements in the central nervous system of kinorhynchs (Kinorhyncha, Cyclorhagida) J Morphol. 2013;274:258–274. doi: 10.1002/jmor.20089. [DOI] [PubMed] [Google Scholar]
- Holland ND, Holland LZ. Stage- and tissue-specific patterns of cell division in embryonic and larval tissues of amphioxus during normal development. Evol Dev. 2006;8:142–149. doi: 10.1111/j.1525-142X.2006.00085.x. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Schubert M, Holland ND, Neuman T. Evolutionary conservation of the presumptive neural plate markers AmphiSox1/2/3 and AmphiNeurogenin in the invertebrate chordate amphioxus. Dev Biol. 2000;226:18–33. doi: 10.1006/dbio.2000.9810. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Panfilio KA, Chastain R, Schubert M, Holland ND. Nuclear beta-catenin promotes non-neural ectoderm and posterior cell fates in amphioxus embryos. Dev Dyn. 2005;233:1430–1443. doi: 10.1002/dvdy.20473. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lotito S, Yasuo H. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development. 2007;134:3527–3537. doi: 10.1242/dev.002352. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Moriyama Y, Hasegawa T, Endo TA, Toyoda T, Gotoh Y. Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nat Neurosci. 2013;16:416–425. doi: 10.1038/nn.3336. [DOI] [PubMed] [Google Scholar]
- Jacob MH. Neurogenesis in Aplysia californica resembles nervous system formation in vertebrates. J Neurosci. 1984;4:1225–1239. doi: 10.1523/JNEUROSCI.04-05-01225.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager M, Chiori R, Alié A, Dayraud C, Quéinnec E, Manuel M. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Müller, 1776) J Exp Zoolog B Mol Dev Evol. 2011;316B:171–187. doi: 10.1002/jez.b.21386. [DOI] [PubMed] [Google Scholar]
- Johnson JE. Numb and Numblike control cell number during vertebrate neurogenesis. Trends Neurosci. 2003;26:395–396. doi: 10.1016/S0166-2236(03)00166-8. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr Opin Cell Biol. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Kaltezioti V, Kouroupi G, Oikonomaki M, Mantouvalou E, Stergiopoulos A, Charonis A, Rohrer H, Matsas R, Politis PK. Prox1 regulates the notch1-mediated inhibition of neurogenesis. PLoS Biol. 2010;8:e1000565. doi: 10.1371/journal.pbio.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaesbauer T, Towb P, Alexandrova O, David CN, Dall’armi E, Staudigl A, Stiening B, Böttger A. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 2007;303:376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Kaul S, Stach T. Ontogeny of the collar cord: neurulation in the hemichordate Saccoglossus kowalevskii. J Morphol. 2010;271:1240–1259. doi: 10.1002/jmor.10868. [DOI] [PubMed] [Google Scholar]
- Kerner P, Simionato E, Le Gouar M, Vervoort M. Orthologs of key vertebrate neural genes are expressed during neurogenesis in the annelid Platynereis dumerilii. Evol Dev. 2009;11:513–524. doi: 10.1111/j.1525-142X.2009.00359.x. [DOI] [PubMed] [Google Scholar]
- Khalturin K, Anton-Erxleben F, Milde S, Ploetz C, Wittlieb J, Hemmrich G, Bosch TC. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol. 2007;309:32–44. doi: 10.1016/j.ydbio.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric cell division during animal development. Curr Opin Cell Biol. 1997;9:833–841. doi: 10.1016/s0955-0674(97)80085-3. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Koester J, Kandel ER. Further identification of neurons in the abdominal ganglion of Aplysia using behavioral criteria. Brain Res. 1977;121:1–20. doi: 10.1016/0006-8993(77)90435-8. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo DH, Weisblat DA. A new molecular logic for BMP-mediated dorsoventral patterning in the leech Helobdella. Curr Biol. 2011;21:1282–1288. doi: 10.1016/j.cub.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layden MJ, Boekhout M, Martindale MQ. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development. 2012;139:1013–1022. doi: 10.1242/dev.073221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol. 2009;332:48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biol Bull. 2001;201:323–338. doi: 10.2307/1543611. [DOI] [PubMed] [Google Scholar]
- Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci USA. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiao S, Shao T, Broce J, Zhang H. The oldest known priapulid-like scalidophoran animal and its implications for the early evolution of cycloneuralians and ecdysozoans. Evol Dev. 2014;16:155–165. doi: 10.1111/ede.12076. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GO. Central neural circuitry in the jellyfish Aglantha: a model ‘simple nervous system’. Neurosignals. 2004;13:5–19. doi: 10.1159/000076155. [DOI] [PubMed] [Google Scholar]
- Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev Genes Evol. 2005;215:618–630. doi: 10.1007/s00427-005-0022-y. [DOI] [PubMed] [Google Scholar]
- Marlow H, Arendt D. Evolution: ctenophore genomes and the origin of neurons. Curr Biol. 2014;24:R757–R761. doi: 10.1016/j.cub.2014.06.057. [DOI] [PubMed] [Google Scholar]
- Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev Biol. 2012;362:295–308. doi: 10.1016/j.ydbio.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev Biol. 2013;380:324–334. doi: 10.1016/j.ydbio.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJ. Characterization of a RFamide-positive subset of ganglionic cells in the hydrozoan planular nerve net. Cell Tissue Res. 1992;269:431–438. doi: 10.1007/BF00353898. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Copley RR, Dunn CW, Hejnol A, Eccleston H, Halanych KM, Martindale MQ, Telford MJ. Broad taxon and gene sampling indicate that chaetognaths are protostomes. Curr Biol. 2006;16:R575–R576. doi: 10.1016/j.cub.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Mayer G, Whitington PM. Neural development in Onychophora (velvet worms) suggests a step-wise evolution of segmentation in the nervous system of Panarthropoda. Dev Biol. 2009a;335:263–275. doi: 10.1016/j.ydbio.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Mayer G, Whitington PM. Velvet worm development links myriapods with chelicerates. Proc Biol Sci. 2009b;276:3571–3579. doi: 10.1098/rspb.2009.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer NP, Seaver EC. Neurogenesis in an annelid: characterization of brain neural precursors in the polychaete Capitella sp I. Dev Biol. 2009;335:237–252. doi: 10.1016/j.ydbio.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Mittmann B. Early neurogenesis in the horseshoe crab Limulus polyphemus and its implication for arthropod relationships. Biol Bull. 2002;203:221–222. doi: 10.2307/1543407. [DOI] [PubMed] [Google Scholar]
- Miya T, Nishida H. Expression pattern and transcriptional control of SoxB1 in embryos of the ascidian Halocynthia roretzi. Zoolog Sci. 2003;20:59–67. doi: 10.2108/zsj.20.59. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998;21:77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Monjo F, Romero R. Embryonic development of the nervous system in the planarian Schmidtea polychroa. Dev Biol. 2015;397:305–319. doi: 10.1016/j.ydbio.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Stud Biol Lab. Vol. 5. Johns Hopkins Univ; Baltimore: 1891. A contribution to the embryology and phylogeny of the pycnogonids; pp. 1–76. [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J, Nallur R, Ladurner P, Egger B, Rieger R, Hartenstein V. The embryonic development of the flatworm Macrostomum sp. Dev Genes Evol. 2004;214:220–239. doi: 10.1007/s00427-004-0406-4. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Yuan D, Jacobs DK, Hartenstein V. Early development, pattern, and reorganization of the planula nervous system in Aurelia (Cnidaria, Scyphozoa) Dev Genes Evol. 2008;218:511–524. doi: 10.1007/s00427-008-0239-7. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Renfer E, Technau U, Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development. 2012;139:347–357. doi: 10.1242/dev.071902. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev Biol. 1988a;130:737–766. doi: 10.1016/0012-1606(88)90364-8. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. I. The early lineages of the neural plate. Dev Biol. 1988b;130:721–736. doi: 10.1016/0012-1606(88)90363-6. [DOI] [PubMed] [Google Scholar]
- Niehrs C. On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development. 2010;137:845–857. doi: 10.1242/dev.039651. [DOI] [PubMed] [Google Scholar]
- Nielsen C, Martinez P. Patterns of gene expression: homology or homocracy? Dev Genes Evol. 2003;213:149–154. doi: 10.1007/s00427-003-0301-4. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Inoue T, Yoshimoto K, Taniguchi T, Kitamura Y, Agata K. Regeneration of dopaminergic neurons after 6-hydroxydopamineinduced lesion in planarian brain. J Neurochem. 2011;119:1217–1231. doi: 10.1111/j.1471-4159.2011.07518.x. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nomaksteinsky M, Röttinger E, Dufour HD, Chettouh Z, Lowe CJ, Martindale MQ, Brunet JF. Centralization of the deuterostome nervous system predates chordates. Curr Biol. 2009;19:1264–1269. doi: 10.1016/j.cub.2009.05.063. [DOI] [PubMed] [Google Scholar]
- Onai T, Takai A, Setiamarga DH, Holland LZ. Essential role of Dkk3 for head formation by inhibiting Wnt/b-catenin and Nodal/Vg1 signaling pathways in the basal chordate amphioxus. Evol Dev. 2012;14:338–350. doi: 10.1111/j.1525-142X.2012.00552.x. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Perez Y, Rieger V, Martin E, Müller CH, Harzsch S. Neurogenesis in an early protostome relative: progenitor cells in the ventral nerve center of chaetognath hatchlings are arranged in a highly organized geometrical pattern. J Exp Zoolog B Mol Dev Evol. 2013;320:179–193. doi: 10.1002/jez.b.22493. [DOI] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioro HL, Stollewerk A. The expression pattern of genes involved in early neurogenesis suggests distinct and conserved functions in the diplopod Glomeris marginata. Dev Genes Evol. 2006;216:417–430. doi: 10.1007/s00427-006-0078-3. [DOI] [PubMed] [Google Scholar]
- Piraino S, Zega G, Di Benedetto C, Leone A, Dell’Anna A, Pennati R, Carnevali DC, Schmid V, Reichert H. Complex neural architecture in the diploblastic larva of Clava multicornis (Hydrozoa, Cnidaria) J Comp Neurol. 2011;519:1931–1951. doi: 10.1002/cne.22614. [DOI] [PubMed] [Google Scholar]
- Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra NB, Gates RD, Ladurner P, Jacobs DK, Hartenstein V. Embryonic development in the primitive bilaterian Neochildia fusca: normal morphogenesis and isolation of POU genes Brn-1 and Brn-3. Dev Genes Evol. 2002;212:55–69. doi: 10.1007/s00427-001-0207-y. [DOI] [PubMed] [Google Scholar]
- Ramírez FA, Wedeen CJ, Stuart DK, Lans D, Weisblat DA. Identification of a neurogenic sublineage required for CNS segmentation in an Annelid. Development. 1995;121:2091–2097. doi: 10.1242/dev.121.7.2091. [DOI] [PubMed] [Google Scholar]
- Rasmussen SL, Holland LZ, Schubert M, Beaster-Jones L, Holland ND. Amphioxus AmphiDelta: evolution of Delta protein structure, segmentation, and neurogenesis. Genesis. 2007;45:113–122. doi: 10.1002/dvg.20278. [DOI] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Renard E, Vacelet J, Gazave E, Lapébie P, Borchiellini C, Ereskovsky AV. Origin of the neuro-sensory system: new and expected insights from sponges. Integr Zool. 2009;4:294–308. doi: 10.1111/j.1749-4877.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- Richards GS, Degnan BM. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. Evodevo. 2012;3:15. doi: 10.1186/2041-9139-3-15. http://dx.doi.org/10.1186/2041-9139-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GS, Rentzsch F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development. 2014;141:4681–4689. doi: 10.1242/dev.112029. [DOI] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Rieger V, Perez Y, Mueller CH, Lacalli T, Hansson BS, Harzsch S. Development of the nervous system in hatchlings of Spadella cephaloptera (Chaetognatha), and implications for nervous system evolution in Bilateria. Dev Growth Differ. 2011;53:740–759. doi: 10.1111/j.1440-169X.2011.01283.x. [DOI] [PubMed] [Google Scholar]
- Rink JC. Stem cell systems and regeneration in planaria. Dev Genes Evol. 2013;223:67–84. doi: 10.1007/s00427-012-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, Weisblat DA. And Lophotrochozoa makes three: Notch/Hes signaling in annelid segmentation. Dev Genes Evol. 2009;219:37–43. doi: 10.1007/s00427-008-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DT, Herniou EA, Baguña J. Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, et al. NISC Comparative Sequencing Program The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saina M, Genikhovich G, Renfer E, Technau U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc Natl Acad Sci USA. 2009;106:18592–18597. doi: 10.1073/pnas.0900151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M, Mizusina A, Kobayakawa Y. Structure, development, and maintenance of the nerve net of the body column in Hydra. J Comp Neurol. 1996;373:41–54. doi: 10.1002/(SICI)1096-9861(19960909)373:1<41::AID-CNE4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Roles of Sox factors in neural determination: conserved signaling in evolution? Int J Dev Biol. 2001;45:321–326. [PubMed] [Google Scholar]
- Satoh G, Wang Y, Zhang P, Satoh N. Early development of amphioxus nervous system with special reference to segmental cell organization and putative sensory cell precursors: a study based on the expression of pan-neuronal marker gene Hu/elav. J Exp Zool. 2001;291:354–364. doi: 10.1002/jez.1134. [DOI] [PubMed] [Google Scholar]
- Satterlie RA, Eichinger JM. Organization of the ectodermal nervous structures in jellyfish: scyphomedusae. Biol Bull. 2014;226:29–40. doi: 10.1086/BBLv226n1p29. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Straehle U, Scholpp S. Neurogenesis in zebrafish -from embryo to adult. Neural Dev. 2013;8:3. doi: 10.1186/1749-8104-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz G. Cell lineage studies in the crayfish Cherax destructor (Crustacea, Decapoda): germ band formation, segmentation and early neurogenesis. Rouxs Arch Dev Biol. 1992;202:36–48. doi: 10.1007/BF00364595. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. Developmental and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Dev Biol. 2004;269:331–345. doi: 10.1016/j.ydbio.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Seipp S, Schmich J, Will B, Schetter E, Plickert G, Leitz T. Neuronal cell death during metamorphosis of Hydractina echinata (Cnidaria, Hydrozoa) Invert Neurosci. 2010;10:77–91. doi: 10.1007/s10158-010-0109-7. [DOI] [PubMed] [Google Scholar]
- Semmler H, Chiodin M, Bailly X, Martinez P, Wanninger A. Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis. Dev Growth Differ. 2010;52:701–713. doi: 10.1111/j.1440-169X.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Shain DH, Ramırez-Weber FA, Hsu J, Weisblat DA. Gangliogenesis in leech: morphogenetic processes leading to segmentation in the central nervous system. Dev Genes Evol. 1998;208:28–36. doi: 10.1007/s004270050150. [DOI] [PubMed] [Google Scholar]
- Shankland M. Formation and specification of neurons during the development of the leech central nervous system. J Neurobiol. 1995;27:294–309. doi: 10.1002/neu.480270304. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Iguchi A, Hayward DC, Technau U, Ball EE, Miller DJ. Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol. 2008;8:311. doi: 10.1186/1471-2148-8-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Kerner P, Dray N, Le Gouar M, Ledent V, Arendt D, Vervoort M. atonal- and achaete-scute-related genes in the annelid Platynereis dumerilii: insights into the evolution of neural basic-Helix-Loop-Helix genes. BMC Evol Biol. 2008;8:170. doi: 10.1186/1471-2148-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 2013;11:e1001488. doi: 10.1371/journal.pbio.1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB. At the nexus between pattern formation and cell-type specification: the generation of individual neuroblast fates in the Drosophila embryonic central nervous system. BioEssays. 1999;21:922–931. doi: 10.1002/(SICI)1521-1878(199911)21:11<922::AID-BIES4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Panganiban G, Selegue J, Carroll SB. Gene regulation in two dimensions: the proneural achaete and scute genes are controlled by combinations of axis-patterning genes through a common intergenic control region. Genes Dev. 1992;6(12B):2606–2619. doi: 10.1101/gad.6.12b.2606. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Mazza-Curll KL, van Wolfswinkel JC, Reddien PW. Whole-body acoel regeneration is controlled by Wnt and Bmp-Admp signaling. Curr Biol. 2014;24:1107–1113. doi: 10.1016/j.cub.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Stent GS. The role of cell lineage in development. Philos Trans R Soc Lond B Biol Sci. 1985;312:3–19. doi: 10.1098/rstb.1985.0174. [DOI] [PubMed] [Google Scholar]
- Stigloher C, Chapouton P, Adolf B, Bally-Cuif L. Identification of neural progenitor pools by E(Spl) factors in the embryonic and adult brain. Brain Res Bull. 2008;75:266–273. doi: 10.1016/j.brainresbull.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Stollewerk A. Recruitment of cell groups through Delta/Notch signaling during spider neurogenesis. Development. 2002;129:5339–5348. doi: 10.1242/dev.00109. [DOI] [PubMed] [Google Scholar]
- Stollewerk A. Secondary neurons are arrested in an immature state by formation of epithelial vesicles during neurogenesis of the spider Cupiennius salei. Front Zool. 2004;1:3. doi: 10.1186/1742-9994-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollewerk A, Weller M, Tautz D. Neurogenesis in the spider Cupiennius salei. Development. 2001;128:2673–2688. doi: 10.1242/dev.128.14.2673. [DOI] [PubMed] [Google Scholar]
- Stuart DK, Torrence SA, Law MI. Leech neurogenesis. I. Positional commitment of neural precursor cells. Dev Biol. 1989;136:17–39. doi: 10.1016/0012-1606(89)90128-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- Taverna E, Goetz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Bourlat SJ, Economou A, Papillon D, Rota-Stabelli O. The evolution of the Ecdysozoa. Philos Trans R Soc Lond B Biol Sci. 2008;363:1529–1537. doi: 10.1098/rstb.2007.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontin AD, García-Verdugo JM, Lim DA, Alvarez-Buylla A. Postnatal development of radial glia and the ventricular zone (VZ): a continuum of the neural stem cell compartment. Cereb Cortex. 2003;13:580–587. doi: 10.1093/cercor/13.6.580. [DOI] [PubMed] [Google Scholar]
- Ungerer P, Scholtz G. Filling the gap between identified neuroblasts and neurons in crustaceans adds new support for Tetraconata. Proc Biol Sci. 2008;275:369–376. doi: 10.1098/rspb.2007.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer P, Eriksson BJ, Stollewerk A. Neurogenesis in the water flea Daphnia magna (Crustacea, Branchiopoda) suggests different mechanisms of neuroblast formation in insects and crustaceans. Dev Biol. 2011;357:42–52. doi: 10.1016/j.ydbio.2011.05.662. [DOI] [PubMed] [Google Scholar]
- Ungerer P, Eriksson BJ, Stollewerk A. Unravelling the evolution of neural stem cells in arthropods: notch signalling in neural stem cell development in the crustacean Daphnia magna. Dev Biol. 2012;371:302–311. doi: 10.1016/j.ydbio.2012.08.025. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Wadsworth WG, Hedgecock EM. Guidance of neuroblast migrations and axonal projections in Caenorhabditis elegans. Curr Opin Neurobiol. 1992;2:36–41. doi: 10.1016/0959-4388(92)90159-i. [DOI] [PubMed] [Google Scholar]
- Wang R, Liu K, Chen L, Aihara K. Neural fate decisions mediated by trans-activation and cis-inhibition in Notch signaling. Bioinformatics. 2011;27:3158–3165. doi: 10.1093/bioinformatics/btr551. [DOI] [PubMed] [Google Scholar]
- Wedeen CJ, Weisblat DA. Segmental expression of an engrailed-class gene during early development and neurogenesis in an annelid. Development. 1991;113:805–814. doi: 10.1242/dev.113.3.805. [DOI] [PubMed] [Google Scholar]
- Weisblat DA. Asymmetric cell divisions in the early embryo of the leech Helobdella robusta. Prog Mol Subcell Biol. 2007;45:79–95. doi: 10.1007/978-3-540-69161-7_4. [DOI] [PubMed] [Google Scholar]
- Weller M, Tautz D. Prospero and Snail expression during spider neurogenesis. Dev Genes Evol. 2003;213:554–566. doi: 10.1007/s00427-003-0362-4. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Skeath JB. The identification and expression of achaete-scute genes in the branchiopod crustacean Triops longicaudatus. Gene Expr Patterns. 2005;5:695–700. doi: 10.1016/j.modgep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Carrico ML, Wilson BA, Brown SJ, Skeath JB. The expression and function of the achaete-scute genes in Tribolium castaneum reveals conservation and variation in neural pattern formation and cell fate specification. Development. 2003;130:4373–4381. doi: 10.1242/dev.00646. [DOI] [PubMed] [Google Scholar]
- Wingate RJ. The rhombic lip and early cerebellar development. Curr Opin Neurobiol. 2001;11:82–88. doi: 10.1016/s0959-4388(00)00177-x. [DOI] [PubMed] [Google Scholar]
- Yeo W, Gautier J. Early neural cell death: dying to become neurons. Dev Biol. 2004;274:233–244. doi: 10.1016/j.ydbio.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Hartenstein V. The embryonic development of the polyclad flatworm Imogine mcgrathi. Dev Genes Evol. 2000;210:383–398. doi: 10.1007/s004270000086. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Ehlers U, Hartenstein V. Embryonic development of the nervous system of the rhabdocoel flatworm Mesostoma lingua (Abilgaard, 1789) J Comp Neurol. 2000;416:461–474. doi: 10.1002/(sici)1096-9861(20000124)416:4<461::aid-cne4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Jones M, Hartenstein V. Embryonic development of the nervous system of the temnocephalid flatworm Craspedella pedum. J Comp Neurol. 2001;434:56–68. doi: 10.1002/cne.1164. [DOI] [PubMed] [Google Scholar]
- Zander MA, Burns SE, Yang G, Kaplan DR, Miller FD. Snail coordinately regulates downstream pathways to control multiple aspects of mammalian neural precursor development. J Neurosci. 2014;34:5164–5175. doi: 10.1523/JNEUROSCI.0370-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Weisblat DA. Applications of mRNA injections for analyzing cell lineage and asymmetric cell divisions during segmentation in the leech Helobdella robusta. Development. 2005;132:2103–2113. doi: 10.1242/dev.01802. [DOI] [PubMed] [Google Scholar]
- Zhao C, Emmons SW. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]
- Zhong W. Diversifying neural cells through order of birth and asymmetry of division. Neuron. 2003;37:11–14. [Google Scholar]
- Zipser B. Complete distribution patterns of neurons with characteristic antigens in the leech central nervous system. J Neurosci. 1982;2:1453–1464. doi: 10.1523/JNEUROSCI.02-10-01453.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


