Abstract
Background
Many arboviral outbreaks have occurred in various locations in Kenya. Entomological surveys are suitable methods for revealing information about circulating arboviruses before human outbreaks are recognized. Therefore, mosquitoes were collected in Kenya to determine the distribution of arboviruses.
Methods
Various species of mosquitoes were sampled from January to July 2012 using several collection methods. Mosquito homogenates were directly tested by reverse transcription-polymerase chain reaction (RT-PCR) using various arbovirus-targeted primer pairs.
Results
We collected 12,569 mosquitoes. Although no human-related arboviruses were detected, Culex flavivirus (CxFV), an insect-specific arbovirus, was detected in 54 pools of 324 Culex quinquefasciatus individuals collected during the rainy season. Of these 54 positive pools, 96.3% (52/54) of the mosquitoes were collected in Busia, on the border of western Kenya and Uganda. The remaining two CxFV-positive pools were collected in Mombasa and Kakamega, far from Busia. Phylogenetic analysis revealed minimal genetic diversity among the CxFVs collected in Mombasa, Kakamega, and Busia, even though these cities are in geographically different regions. Additionally, CxFV was detected in one mosquito pool collected in Mombasa during the dry season. In addition to Culex mosquitoes, Aedes (Stegomyia) and Anopheles mosquitoes were also positive for the Flavivirus genus. Cell fusing agent virus was detected in one pool of Aedes aegypti. Mosquito flavivirus was detected in three pools of Anopheles gambiae s.l. collected in the dry and rainy seasons.
Conclusions
Although no mosquitoes were positive for human-related arbovirus, insect-specific viruses were detected in various species of mosquitoes. The heterogeneity observed in the number of CxFVs in Culex mosquitoes in different locations in Kenya suggests that the abundance of human-related viruses might differ depending on the abundance of insect-specific viruses. We may have underestimated the circulation of any human-related arbovirus in Kenya, and the collection of larger samples may allow for determination of the presence of human-related arboviruses.
Keywords: Arbovirus, Insect-specific virus, Culex flavivirus, Aedes mosquito, Culex mosquito, Anopheles mosquito, Busia, Kakamega, Mombasa, Kenya
Background
Emergence and re-emergence of vector-borne diseases are crucial public health problems worldwide [1, 2]. In Kenya, many sporadic outbreaks have been reported in geographically different areas [3]. For example, an outbreak of dengue (DEN) fever occurred in the coastal towns of Malindi and Kilifi in 1982 [4], and in 1992–1993, an outbreak of yellow fever (YF) occurred in Rift Valley Province [5]. There were outbreaks of Rift Valley fever (RVF) in 1997 and 2006 [6–8], and an outbreak of chikungunya (CHIK) fever occurred in 2004 in the coastal area of Kenya [9, 10]. In Uganda, an epidemic of o’nyong’nyong (ONN) started in early 1959 and spread to Kenya [11, 12].
In general, febrile diseases caused by viruses are still confused with non-viral diseases, such as malaria [2]. Moreover, cases can remain unnoticed because some arboviral infections are mild and self-limiting during the early stage. Therefore, the number of human arboviral cases might be much higher than has been reported. Even in the absence of clinical outbreaks, historic serosurveys in Kenya can provide important clues about circulating arboviruses in various environments [13]. For instance, Mease et al. in [14] assessed the prevalence of IgG against yellow fever virus (YFV), West Nile virus (WNV), dengue virus (DENV), and chikungunya virus (CHIKV) using serum samples from healthy Kenyans. According to their data, 46.6% of the people in all study areas had antibodies against at least one of these arboviruses [14]. As historic serosurveys in Kenya have documented several arboviruses in geographically different areas [15], a large epidemic of arbovirus can occur anywhere at any time because, as demonstrated recently, many factors such as demographic, geographic environmental and climate change factors can complicate and worsen the situation [16]. Many studies have revealed that a threat of arboviral transmission is present throughout Kenya, regardless of the officially announced reports of outbreaks [17, 18].
Controlling arboviral diseases is difficult because of the complex environment and ecology, including relationships among viruses, vectors, and humans [2, 16, 19]. Multiple vector species are often involved in an arboviral disease, and a single vector can also transmit several diseases. Moreover, primary vectors vary among geographical areas, and the level of vector competence may also vary among species depending on each area [20]. Mosquitoes are known to carry not only human-related viruses but also insect-specific viruses, such as Culex flavivirus and Aedes flavivirus [21]. In addition, interactions between many types of viruses and many other organisms may affect vector competence inside the mosquito [22]. For example, the presence of co-infection with insect-specific virus and WNV has been reported [23]. In this case, co-infection might be considered a factor for the emergence of arbovirus, though the function of insect-specific viruses remains unclear. Assessing the potential for arbovirus outbreaks at the local level can be facilitated by identifying all patterns of relationships, including triangular relationships (human-vector-arbovirus environment), in each area [24]. Moreover, entomological baseline data may contribute to estimations of disease risk and allow precautionary measures to be taken against virus activity. In this study, we mainly selected collection sites where other researchers had previously found or suspected arbovirus activity. For example, border areas are suspected to be areas of potential arbovirus infection because busy transportation hubs may provide many opportunities for human-vector contact [25]. Although the presence of arboviruses has not yet been reported in some indigenous forests in Kenya, many species of mosquitoes can serve as bridge vectors of arboviruses, easily spreading sylvatic arboviruses such as sylvatic YF and sylvatic DENV from forests to human environments in these areas in Kenya [26]. We suspected that arboviruses were silently circulating, without outbreak detection. Therefore, an active survey was undertaken in border areas, including coastal boundaries and indigenous forests. The aim of this study was to obtain data regarding the presence of arboviruses in mosquitoes in selected areas of Kenya. Our additional goal was to recognize the main vector species of arboviruses.
Methods
Study areas
Mosquito sampling was performed in eastern (Mombasa and Kwale) and western (Kakamega and Busia) Kenya, which included a variety of areas, such as urban coastal border, land border, and rural areas next to a forest where there is suspected arbovirus activity (Fig. 1). The sampling was conducted in two different seasons: the rainy season and a season other than the rainy season; March to June in Kenya generally constitutes the rainy season. We initiated this study in January 2012, before the rainy season, which we conventionally termed the dry season. Between January 18 and 26, 2012 (representing dry-season sampling), we conducted a preliminary survey only in eastern Kenya. Between May 9 and June 8, 2012 (representing rainy-season sampling), we conducted the same survey in both eastern and western Kenya.
Fig. 1.
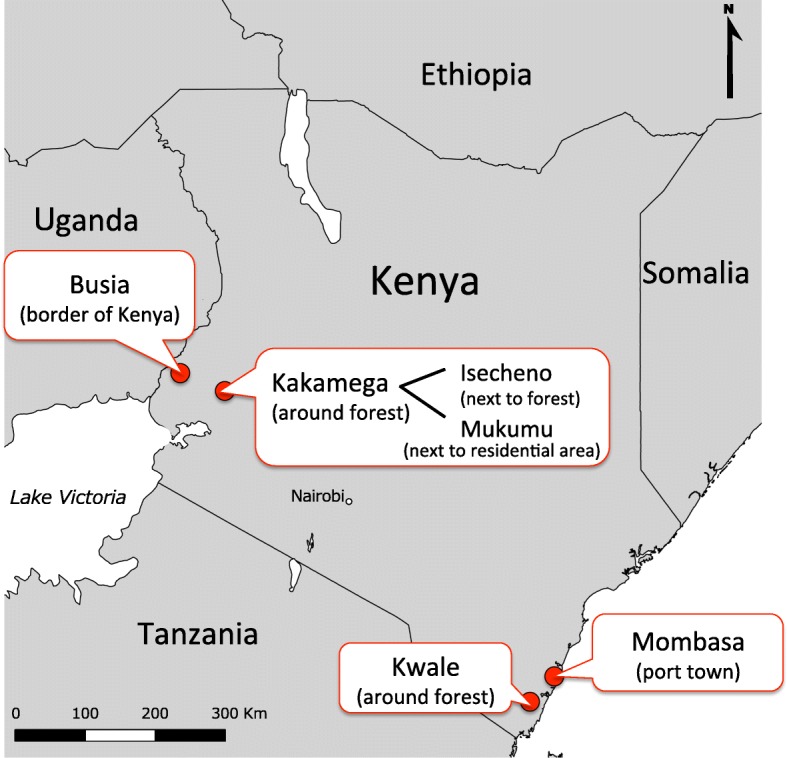
Map of study region. Location of mosquito sampling site in East Kenya; Kwale, Mombasa, and in West Kenya; Busia, Kakamega (Mukumu and Isecheno)
Eastern Kenya: Mombasa (the center: 4°3.509′S; 39°40.363′E)
This busy port town includes the urban coastal border with high levels of human activity. Dengue cases have been reported here for approximately the last 30 years [4]. We suspected that due to human activity, arboviral mosquitoes can be easily transported outside this area. Mosquitoes were collected in resident areas in the 2012 dry season (from January 24 to January 26) and in the 2012 rainy season (from May 15 to May 17).
Eastern Kenya: Kwale (the center: 4°10.525′S; 39°27.087′E)
In this rural area, patches of indigenous forests (Shimba Hills National Reserve) exist next to the residential area. The edge of the indigenous forest can act as a border cross which arboviral mosquitoes can be transported from the forest to the residential area. Mosquitoes were sampled from houses in the 2012 dry season (from January 18 to January 20) and in the 2012 rainy season (from May 9 to May 12).
Western Kenya: Kakamega (the center: 0°16.923′N; 34°45.234′E)
Kakamega forest has a remarkable diversity of insects, birds and animals, which can serve as reservoir hosts of arboviruses [27]. We selected two areas: one exactly next to the indigenous forest (Isecheno), and another, a residential area (Mukumu) along the main road in this region. The edge of the indigenous forest is considered to be a dangerous border of arboviral activity, similar to Kwale. We suspect that the area is easily penetrable by arboviral mosquitoes from forests to residential areas and vice versa. The main road is also regarded as a border, which may encourage transmission of arboviruses. Mosquitoes were collected in Mukumu from June 2 to June 4 and in Isecheno from June 6 to June 8.
Western Kenya: Busia (the center: 0°27.914′N; 34°5.979′)
Busia is in the western land border (Kenya and Uganda) area, including a busy town with high human activity. Serological surveys were conducted and revealed a high positive rate of antibodies against arboviruses in healthy residents [14]. In this area, many residents may have already suffered from arboviral diseases, with or without symptoms. Transmission between humans and mosquitoes may have been underestimated due to the complicated human activity. Mosquito surveillance can provide other information to show the actual circulation of arboviruses. Mosquitoes were collected from May 25 to May 27.
Mosquito sampling
In each area, mosquitoes were collected for 3 consecutive days from 13 selected houses within approximately 0.5 km2 in each targeted area, except one area (for 4 consecutive days in Kwale in the rainy season). A systematic sampling method was applied for selecting study houses in each targeted area [28]. For example, in Kakamega, the main intersection (0°16.923′N; 34°45.234′E) on Kisumu-Kakamega Road was used as the starting point for the systematic sampling of houses. From this point, we established 13 sampling points at 250-m intervals. The nearest house from each point was then selected. The house belongs to a large family (> 5 people), and it was suggested because our study targets human-related arboviruses. Another house was selected if the household head or guardian was not willing to participate in the study. The same method was performed in other areas. Collection methods used the following traps: (i) CDC light traps, (ii) CDC gravid traps, and (iii) BG sentinel traps. Additionally, indoor resting mosquito collection with hand aspirators was performed in all houses. To use 20 traps effectively, we placed 2 types of traps randomly within each of the 13 study houses. We intended to collect as many mosquitoes as possible because arbovirus transmission is usually maintained at a low level in a mosquito population [29]. When the number of mosquitoes collected was insufficient, the position of the traps or type of traps was randomly changed. We used the most effective collection combination with positioning and type of traps at each study site.
In our study, the position of the traps depended on the structure of the house. CDC light traps were suspended > 1.5 m above the ground inside and outside of the houses but not near any other sources of artificial light. CDC gravid traps were placed in a stable area somewhere inside or outside the house where nothing could upset the medium in the pan, for example, under eaves. BG sentinel traps were placed in the house with enough space or outside of the house. CDC light traps were operated from dusk to dawn, whereas other traps were operated for 3–4 days continuously. Resting mosquito collection was performed using oral aspirators by three persons in all rooms of the selected houses in the early morning for 15 min each day; this occurred during all collection periods when the house was visited to remove the mosquito-sampling bags from the traps. To prevent RNA degradation, the captured mosquitoes were kept alive during transfer to the laboratory.
Mosquito identification
At the laboratory, the collected mosquitoes were killed at − 20 °C and placed on white filter paper in a Petri dish placed on a chill table and identified morphologically to the species level under a stereoscopic microscope using published keys [30–33]. For accurate identification, Aedes aegypti, Culex quinquefasciatus, Anopheles funestus, and An. rivulorum were confirmed by polymerase chain reaction (PCR) using specific primers (Table 1).
Table 1.
Primers used to detect and to sequence arbovirus from mosquito pools in Kenya
| Target | Primer name | Nucleotide sequence (5′ to 3′) | Polarity | Product (bp) | Cycle condition | Reference |
|---|---|---|---|---|---|---|
| Universal primers for flavivirus | MAMD | AACATGATGGGRAARAGRGARAA | Forward | 252 | 94°C, 2 min, 1 cycle; 94°C, 1 min, 53°C, 1 min, 72°C, 1 min, 35 cycles; 72°C, 5 min, 1 cycle | Scaramozzino et al. (2001) [48] |
| cFD2 | GTGTCCCAGCCGGCGGTGTCATCAGC | Reverse | ||||
| Universal primers for flavivirus | FLAVI-1 | AATGTACGCTGATGACACAGCTGGCTGGGACAC | Forward | 854–863 | 94°C, 5 min, 1 cycle; 94°C, 1 min, 58°C, 1 min, 72°C, 90 s, 45 cycles; 72°C, 10 min, 1 cycle | Ayers et al.(2006) [49] |
| FLAVI-2 | TCCAGACCTTCAGCATGTCTTCTGTTGTCATCCA | Reverse | ||||
| Universal primers for flavivirus (mainly YF) | YF-1 | GGTCTCCTCTAACCTCTAG | Forward | 675 | 94°C, 2 min, 1 cycle; 94°C, 30 s, 53°C, 30 s, 72°C, 1 min, 35 cycles; 72°C, 5 min, 1 cycle | Tanaka et al. (1993) [50] |
| YF-3 | GAGTGGATGACCACGGAAGACATGC | Reverse | ||||
| Universal primers for alpha viruses (mainly chikungunya and o’nyong’nyong viruses) | nsP1-S | TAGAGCAGGAAATTGATCC | Forward | 354 | 94°C, 2 min, 1 cycle; 94°C, 30 s, 53°C, 30 s, 72°C, 45 s, 35 cycles; 72°C, 5 min, 1 cycle | Hasebe et al. (2002) [51] |
| nsP1-C | CTTTAATCGCCTGGTGGTA | Reverse | ||||
| Universal primers for alpha viruses (mainly chikungunya and o’nyong’nyong viruses) | E1-S | TACCCATTCATGTGGGG | Forward | 294 | 94°C, 2 min, 1 cycle; 94°C, 30 s, 53°C, 30 s, 72°C, 45 s, 35 cycles; 72°C, 5 min, 1 cycle | Hasebe et al. (2002) [51] |
| E1-C | GCCTTTGTACACCACGAT | Reverse | ||||
| Rift Valley virus | RVF009 | CCAAATGACTACCAGTCAGC | Forward | 400–500 | 94°C, 2 min, 1 cycle; 94°C, 30 s, 50°C, 30 s, 72°C, 1 min, 35 cycles; 72°C, 5 min, 1 cycle | Jupp et al. (2000) [52] (modified) |
| RVF007 | GACAAATGAGTCTGGTAGCA | Reverse | ||||
| Mosquito RNA marker | Act-2F | ATGGTCGGYATGGGNCAGAAGGACTC | Forward | 683 | 94°C, 2 min, 1 cycle; 94°C, 30 s, 54°C, 30 s, 72°C, 45 s, 35 cycles; 72°C, 5 min, 1 cycle | Staley et al. (2010) [53] |
| Act-8R | GATTCCATACCCAGGAAGGADGG | Reverse | ||||
| Culex quinquefaciatus | ACEpip | GGAAACAACGACGTATGTACT | Forward | 610 | 94°C, 5 min, 1 cycle; 94°C, 30 s, 54°C, 30 s, 72°C, 1 min, 35 cycles; 72°C, 5 min, 1 cycle | Kasai et al. (2008) [39] |
| ACEquin | CCTTCTTGAATGGCTGTGGCA | Forward | 274 | |||
| B1246s | TGGAGCCTCCTCTTCACGG | Reverse | ||||
| Aedes aegypti | 18SFHIN | GTAAGCTTCCTTTGTACACACCGCCCGT | Forward | 550 | 97°C, 4 min, 1 cycle; 96°C, 30 s, 48°C, 30 s, 72°C, 2 min, 30 cycles; 72°C, 4 min, 1 cycle | Higa et al. (2010) [54] |
| aeg.r1 | TAACGGACACCGTTCTAGGCCCT | Reverse | ||||
| Anopheles funestus, Anopheles rivulorum | UV | TGTGAACTGCAGGACACAT | Forward | 94°C, 2 min, 1 cycle; 94°C, 30 s, 45°C, 30 s, 72°C, 40 s, 30 cycles; 72°C, 5 min, 1 cycle | Koekemoer et al. (2002) [55] | |
| FUN | GCATCGATGGGTTAATCATG | Reverse | 505 | |||
| RIV | CAAGCCGTTCGACCCTGATT | Reverse | 411 |
Note: Each 25 μl reaction mixture contained. (Accupower TM PCR PreMix kit with 2 μl template, 15.2 μl sterile water, and 1.4 μl of 100 pmol/μl each of primers)
Mosquito processing
A maximum of 30 individuals were pooled according to species, sex, physiological status, (i.e., unfed, blood fed, or gravid), and collection site and then were frozen in liquid nitrogen. For virus detection, we used all pools collected during the dry season, with each category (male, unfed, fed and gravid) examined separately. In contrast, for the pools collected during the rainy season, only unfed and gravid mosquito pools of Ae. aegypti, An. funestus, An. gambiae s.l., and Cx. quinquefasciatus were used. Moreover, both unfed and gravid mosquitoes were combined for some pools of each species. Blood-fed mosquitoes were excluded to prevent contamination of the virus contained in a blood meal, though we did utilize blood-fed mosquitoes collected during the dry season because of the small sample size. For the samples collected during the rainy season, we concentrated on detecting viruses in only female pools, excluding those that were blood-fed.
Pooled specimens were placed in a 1.5-ml microcentrifuge tube with 300 μl of minimal essential medium (MEM) (minimum essential medium containing 10% foetal bovine serum, L-glutamine, penicillin, streptomycin, and amphotericin B). The mosquitoes were ground in MEM, and the homogenate was centrifuged; 200 μl of the supernatant was collected and kept at − 80 °C for future use (for cell culture). To maintain approximately 100 μl of the suspension, 75 μl of lysis buffer was added. The homogenates were prepared using sterile, RNase-free utensils.
Total RNA extraction and virus identification by reverse transcription-PCR
Total RNA was extracted from each pool of mosquitoes using an extraction kit (SV Total RNA Isolation System, Promega, Tokyo, Japan) according to the manufacturer’s instructions. RNA was eluted in 50 μl of sterile distilled water. Reverse transcription reactions were performed to synthesize first-strand cDNA using RNA to cDNA EcoDry Premix (Random Hexamers) (Clontech Laboratories, Inc., Mountain View, CA, USA). The cDNA was amplified by PCR using an AccuPower™ PCR Premix Kit (Bioneer Co., Daejon, Korea) with virus-specific primers (Table 1), and the products were evaluated by 1.5% agarose gel electrophoresis. For all positive samples, products of the expected size were extracted from the gel and were purified using a MonoFas DNA Purification Kit (GL Sciences, Tokyo, Japan). Purified amplicons were bidirectionally sequenced using a BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and analyzed with an ABI3130 Genetic Analyzer (Applied Biosystems). Nucleic acid sequences were compared with those in the GenBank database using the BLAST program.
The process was repeated for three universal primers for flavivirus (the main targets are DENV, YFV, and WNV), two universal primers for alpha viruses (the main targets are ONN virus and CHIKV) and single primer sets for RVFV (phlebovirus) (Table 1). For flaviviruses and alpha viruses, we prepared multiple primer sets to detect not only a well-known virus but also novel viruses. In the case of flavivirus detection, all pools were initially screened for flavivirus RNA by using universal flavivirus primer sets cFD2 and MAMD, which target the non-structural protein 5 (NS5) gene. To identify human-related flaviviruses, such as DENV, YFV, and WNV, all pools were screened with primer sets YF-1 and YF-3. To generate a larger NS5 cDNA segment for sequencing, putative positive samples detected using previous primer sets (cFD2 and MAMD) were again screened for flavivirus RNA using another universal flavivirus primer set (FLAVI1 and FLAVI2) targeting the NS5 gene. Confirmed bands of approximately 860 bp were sequenced as described above. In the case of alpha virus detection, primer sets (nsP1-S and nsP1-C; E1-S and E1-C) designed based on the genes non-structural protein 1 (nsP1) and glycoprotein E1 (E1) were used for amplification.
The following inactivated viruses available in the laboratory were used as positive controls: DEN-1 (Hawaii strain), YFV (17D strain, attenuated live vaccine strain), WNV (NY99 strain), CHIKV (S27 strain, African prototype), RVFV (Smithburn strain, attenuated live vaccine strain) (All positive controls were kindly provided by Dr. S Inoue). As a quality control for the detection step, each cDNA was checked by PCR using the mosquito β-actin primer.
Calculation of infection rates
We calculated the minimum infection rate (MIR) of arboviruses in each mosquito species at each site using the Poolscreen2 program [34]. MIR is expressed as the number of pools infected per 1000 mosquitoes tested, and it assumes that only one mosquito is positive in a pool. To determine the number of flavivirus-positive samples, the results using primer sets cFD2 and MAMD were employed. MIR was calculated when at least 100 mosquitoes were tested per species per site.
Phylogenetic analysis
For virus species identification, the collected sequences were confirmed by an alignment search in gene databases using MEGA6 with the ClustalW method [35]. Phylogenetic and molecular evolutionary analyses were conducted by using the p-distance option with the neighbor-joining (NJ) method. Bootstrap analyses were performed with 1000 replicates. Representative flavivirus sequences were used in the phylogenetic analysis as outgroup sequences.
Results
Mosquito collection
During the dry season in eastern Kenya (Table 2)
Table 2.
Summary of mosquitoes collected in the dry season in East Kenya
| Study site | Kwale | Mombasa | |||||
|---|---|---|---|---|---|---|---|
| Collection methods employed (number of trap sessions)# | As; 39, BG; 12, CDC; 15, GT; 12 | As; 39, BG; 12, CDC; 15, GT; 12 | |||||
| Methods collected mosquitoes (number of trap sessions)# | As; 7, BG; 3, CDC; 10, GT; 6 | As; 33, BG; 7, CDC; 14, GT; 12 | |||||
| Collection period | January 18–20, 2012 (3 days) | January 24–26, 2012 (3 days) | |||||
| Number of houses | 13 houses | 13 houses | |||||
| Species | Physiological status | No. collected | Pools | Positive pool | No. collected | Pools | Positive pool |
| Ae. aegypti | Fed | 2 | 1 | 0 | |||
| Unfed | 3 | 3 | 0 | 16 | 2 | 0 | |
| An. coustani | Fed | 1 | 1 | 0 | |||
| An. funestus | Fed | 1 | 1 | 0 | |||
| An. gambiae s.l. | Fed | 3 | 1 | 0 | |||
| Unfed | 8 | 1 | 1 | 2 | 1 | 1 | |
| An. longipulpis | Fed | 1 | 1 | 0 | |||
| An. rivulorum | Fed | 4 | 1 | 0 | |||
| Anopheles sp. | Male | 1 | 1 | 0 | |||
| Cx. cinereus | Gravid | 1 | 1 | 0 | |||
| Cx. decens | Unfed | 2 | 1 | 0 | |||
| Gravid | 4 | 1 | 0 | ||||
| Cx. quinquefasciatus | Male | 15 | 1 | 0 | 235 | 10 | 1 |
| Fed | 30 | 2 | 0 | 105 | 6 | 0 | |
| Unfed | 19 | 1 | 0 | 375 | 13 | 0 | |
| Gravid | 64 | 4 | 0 | 129 | 7 | 0 | |
| Cx. laticinctus | Gravid | 5 | 1 | 0 | |||
| Cx. simpsoni | Male | 1 | 1 | 0 | |||
| Unfed | 2 | 1 | 0 | ||||
| Cx. univiittetus | Unfed | 2 | 1 | 0 | |||
| Culex sp. | Male | 1 | 1 | 0 | |||
| Unfed | 1 | 1 | 0 | 3 | 1 | 0 | |
| Mansonia sp. | Fed | 2 | 1 | 0 | |||
| Unfed | 8 | 1 | 0 | ||||
| Others | Male | 2 | |||||
| Unfed | 3 | ||||||
| Total | 179 | 26 | 1 | 872 | 44 | 2 | |
#Abbreviations of collection methods are As aspirator, BG: BG sentinel trap, CDC:CDC light trap, GT CDC gravid trap
In Kwale (January 18–20, 2012), we employed a cumulative number of 39 trap sessions (per day per house) in 13 houses for 3 days (total numbers of each trap session per day per house were 12 BG sentinel, 15 CDC light, and 12 CDC gravid trap sessions) and a cumulative number of 39 aspirator catch sessions (per day per house) in 13 houses for 3 days using a 3-person aspirator catch team in each house. We collected 179 mosquitoes in the following subset of attempts: 3 BG sentinel trap sessions, 10 CDC light trap sessions, 6 CDC gravid trap sessions, and 7 aspirator catches. In Mombasa (January 24–26, 2012), we collected 872 mosquitoes by the same cumulative number of trap sessions as in Kwale. The collection methods entailed 7 BG sentinel trap sessions, 14 CDC light trap sessions, 12 CDC gravid trap sessions, and 33 aspirator catches. The total number of mosquitoes collected in Kwale and Mombasa was 1051. Of these mosquitoes, 796 (75.7%) were identified as females. For these samples collected during the dry season, all species were tested, including males of each species (70 pools) (Table 2). Only five mosquitoes were not identified and were excluded.
During the rainy season in eastern and western Kenya (Table 3)
Table 3.
Summary of mosquitoes collected in the rainy season in East and West Kenya
| Study area | East Kenya | East Kenya | West Kenya | West Kenya | West Kenya | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study site | Kwale | Mombasa | Busia | Kakamega (Mukumu) | Kakamega (Isecheno) | |||||||||||
| Collection methods employed (number of trap sessions)# | As; 48, BG; 30, CDC; 16, GT; 22 | As; 30, BG; 13, CDC; 12, GT; 17 | As; 36, BG; 18, CDC; 12, GT; 15 | As; 39, BG; 15, CDC; 18, GT; 18 | As; 39, BG; 15, CDC; 21, GT; 21 | |||||||||||
| Methods collected mosquitoes (number of trap sessions)# | As; 42, BG; 25, CDC; 11, GT; 22 | As; 28, BG; 12, CDC; 11, GT; 17 | As; 36, BG; 17, CDC; 12, GT; 15 | As; 34, BG; 13, CDC; 16, GT; 15 | As; 20, BG; 8, CDC; 11, GT; 17 | |||||||||||
| Collection period (days) | May 9–12, 2012 (4 days) | May 15–17, 2012 (3 days) | May 25–27, 2012 (3 days) | June 2–4, 2012 (3 days) | June 6–8, 2012 (3 days) | |||||||||||
| Number of houses | 13 houses | 13 houses | 13 houses | 13 houses | 13 houses | |||||||||||
| Species | Physiological status | No. collected | Pool | Positive pool | No. collected | Pool | Positive pool | No. collected | Pool | Positive pool | No. collected | Pool | Positive pool | No. collected | Pool | Positive pool |
| Aedes sp. | Male | 2 | ||||||||||||||
| Unfed | 3 | 2 | 1 | 1 | ||||||||||||
| Ae. aegypti | Male | 2 | ||||||||||||||
| Unfed | 11 | 8 | 0 | 49 | 14 | 1* | 4 | 8 | 0 | 3 | 3 | 0 | 2 | 2 | 0 | |
| Gravid | 2 | 7 | 6 | 1 | 1 | |||||||||||
| An. brumripes | Unfed | 1 | 2 | |||||||||||||
| An. funestus | Male | 3 | ||||||||||||||
| Fed | 5 | |||||||||||||||
| Unfed | 59 | 7 | 0 | |||||||||||||
| Gravid | 2 | |||||||||||||||
| An. gambiae s.l. | Male | 22 | ||||||||||||||
| Fed | 2 | 37 | 3 | |||||||||||||
| Unfed | 18 | 3 | 0 | 402 | 34 | 1 | 3 | 4 | 0 | 3 | 3 | 0 | ||||
| Gravid | 9 | 1 | ||||||||||||||
| An. garnhami | Fed | 1 | ||||||||||||||
| Unfed | 6 | |||||||||||||||
| An. parensis | Unfed | 1 | ||||||||||||||
| An. rivulorum | Unfed | 4 | 3 | 0 | ||||||||||||
| Gravid | 1 | 1 | 0 | |||||||||||||
| Anopheles sp. | Male | 1 | ||||||||||||||
| Fed | 2 | |||||||||||||||
| Unfed | 6 | 2 | 1 | |||||||||||||
| Gravid | 1 | |||||||||||||||
| Cx. decens | Male | 1 | ||||||||||||||
| Gravid | 1 | |||||||||||||||
| Cx .quinquefasciatus | Male | 355 | 540 | 1554 | 332 | 38 | ||||||||||
| Fed | 202 | 351 | 331 | 840 | 79 | |||||||||||
| Unfed | 106 | 106 | 0 | 443 | 52 | 1* | 1243 | 113 | 52*** | 472 | 46 | 1** | 53 | 7 | 0 | |
| Gravid | 1867 | 554 | 906 | 431 | 79 | |||||||||||
| Cx. simpsoni | Unfed | 1 | ||||||||||||||
| Cx. univiittetus | Unfed | 1 | ||||||||||||||
| Culex sp. | Male | 3 | ||||||||||||||
| Fed | 3 | 1 | ||||||||||||||
| Unfed | 4 | 2 | 1 | 0 | ||||||||||||
| Gravid | 1 | 1 | 3 | |||||||||||||
| Lutzia | Gravid | 2 | ||||||||||||||
| Others | Male | 2 | ||||||||||||||
| Unfed | 1 | 15 | 3 | |||||||||||||
| Gravid | 1 | |||||||||||||||
| Total | 2592 | 117 | 0 | 1974 | 66 | 2 | 4598 | 165 | 53 | 2087 | 54 | 1 | 267 | 12 | 0 | |
Note: We used only unfed and gravid mosquitoes for the pools to detect arboviruses. Unfed and gravid mosquitoes were separated into each category, but some of them were combined into one pool
*The pool comprised only gravid mosquitoes
**The pool comprised only unfed mosquitoes
***Pools consisted of unfed and gravid mosquitoes
#Abbreviations of “collection methods” are As aspirator, BG BG sentinel trap, CDC CDC light trap, GT CDC gravid trap
In Kwale (May 9–12, 2012), we employed a cumulative number of 57 trap sessions (per day per house) in 13 houses for 4 days (total numbers of each trap session per day per house were 30 BG sentinel, 16 CDC light, and 22 CDC gravid trap sessions) and a cumulative number of 48 aspirator catches (per day per house) in 13 houses for 4 days using a 3-person aspirator catch team in each house. We collected 2592 mosquitoes in the following subset of attempts: 25 BG sentinel trap sessions, 11 CDC light trap sessions, 22 CDC gravid trap sessions, and 42 aspirator catches. In Mombasa (May 15–17, 2012), we employed a cumulative number of 42 trap sessions (per day per house) in 13 houses for 3 days (total numbers of trap sessions were 13 BG sentinel traps, 12 CDC light traps, and 17 CDC gravid traps) and a cumulative number of 30 aspirator catch sessions (per day per house) in 13 houses for 3 days using a 3-person aspirator catch team in each house. We collected 1974 mosquitoes in the following subset of attempts: 12 BG sentinel trap sessions, 11 CDC light trap sessions, 17 CDC gravid trap sessions, and 28 aspirator catches. In Busia (May 25–27, 2012), we employed a cumulative number of 45 trap sessions (per day per house) in 13 houses for 3 days (total numbers of trap sessions were 18 BG sentinel, 12 CDC light, and 15 CDC gravid trap sessions) and a cumulative number of 36 aspirator catch sessions (per day per house) in 13 houses for 3 days using 3-person aspirator catch team in each house. We collected 4598 mosquitoes in the following subset of attempts: 17 BG sentinel trap sessions, 12 CDC light trap sessions, 15 CDC gravid trap sessions, and 36 aspirator catches. In Kakamega (Mukumu) (June 2–4, 2012), we employed a cumulative number of 51 trap sessions (per day per house) in 13 houses for 3 days (total numbers of each trap sessions per day per house were 15 BG sentinel, 18 CDC light, and 18 CDC gravid trap sessions) and a cumulative number of 39 aspirator catches (per day per house) in 13 houses for 3 days using 3-person aspirator catch team in each house. We collected 2087 mosquitoes in the following subset of attempts: 13 BG sentinel trap sessions, 16 CDC light trap sessions, 15 CDC gravid trap sessions, and 34 aspirator catches. In Kakamega (Isecheno) (June 6–8, 2012), we employed a cumulative number of 57 trap sessions (per day per house) in 13 houses for 3 days (total numbers of trap sessions per day per house were 15 BG sentinel, 21 CDC light, and 21 CDC gravid trap sessions) and a cumulative number of 39 aspirator catch sessions (per day per house) in 13 houses for 3 days using a 3-person aspirator catch team in each house. We collected 267 mosquitoes in the following subset of attempts: 8 BG sentinel trap sessions, 11 CDC light trap sessions, 17 CDC gravid trap sessions, and 20 aspirator catches.
In total, we collected 11,518 mosquitoes at all sampling sites. Of these mosquitoes collected during the rainy season, 8663 (75.2%) were identified as female. Only unfed and gravid female mosquitoes (414 pools) were used for virus detection in samples collected during the rainy season (Table 4). The number of mosquitoes collected in Kakamega (Isecheno) was one order of magnitude lower than that collected at the other study sites.
Table 4.
Information of positive samples for insect specific arbovirus
| Places | Season | Species of mosquito | No. of mosquitoes | Physiological status of used pools | No. pools | No. positive pools | MIR* | MIR Lower-upper limits | Physiological status of positive pool |
|---|---|---|---|---|---|---|---|---|---|
| Ae. aegypti mosquito pools | |||||||||
| Kwale | Dry | Ae. aegypti | 3 | Unfed | 3 | 0 | NA | NA | |
| Kwale | Rain | Ae. aegypti | 13 | Unfed, gravid | 8 | 0 | NA | NA | |
| Mombasa | Dry | Ae. aegypti | 18 | Fed, unfed | 3 | 0 | NA | NA | |
| Mombasa | Rain | Ae. aegypti | 56 | Unfed, gravid (♂; excluded) | 14 | 1 | NA | NA | Female, gravid |
| Busia | Rain | Ae. aegypti | 10 | Unfed, gravid | 8 | 0 | NA | NA | |
| Kakamega (Mukumu) | Rain | Ae. aegypti | 4 | Unfed, gravid | 3 | 0 | NA | NA | |
| Kakamega (Isecheno) | Rain | Ae. aegypti | 3 | Unfed, gravid | 2 | 0 | NA | NA | |
| Cx. quinquefasciatus mosquito pools | |||||||||
| Kwale | Dry | Cx. quinquefasciatus | 128 | ♂, fed, unfed, gravid | 8 | 0 | NA | NA | |
| Kwale | Rain | Cx. quinquefasciatus | 1973 | Unfed, gravid (♂, fed; excluded) | 106 | 0 | NA | NA | |
| Mombasa | Dry | Cx. quinquefasciatus | 844 | ♂, fed, unfed, gravid | 36 | 1 | 1.18 | 0.07–5.75 | ♂ |
| Mombasa | Rain | Cx. quinquefasciatus | 997 | Unfed, gravid (♂, fed; excluded) | 52 | 1 | 1.01 | 0.06–4.89 | Female, gravid |
| Busia | Rain | Cx. quinquefasciatus | 2149 | Unfed, gravid (♂, fed; excluded) | 113 | 52 | 32.26 | 24.42–42.12 | Female, unfed + gravid |
| Kakamega (Mukumu) | Rain | Cx. quinquefasciatus | 903 | Unfed, gravid (♂, fed; excluded) | 46 | 1 | 1.11 | 0.06–5.37 | Female, unfed |
| Kakamega (Isecheno) | Rain | Cx. quinquefasciatus | 132 | Unfed, gravid (♂, fed; excluded) | 7 | 0 | NA | NA | |
| An. gambiae mosquito pools | |||||||||
| Kwale | Dry | An. gambiae | 11 | Fed, unfed | 2 | 1 | NA | NA | Female, unfed |
| Kwale | Rain | An. gambiae | 18 | Unfed, (fed; excluded) | 3 | 0 | NA | NA | |
| Mombasa | Dry | An. gambiae | 2 | Unfed | 1 | 1 | NA | NA | Female, unfed |
| Mombasa | Rain | An. gambiae | 0 | 0 | 0 | NA | NA | ||
| Busia | Rain | An. gambiae | 411 | Unfed, gravid (♂, fed; excluded) | 34 | 1 | 2.44 | 0.14–11.87 | Female, unfed |
| Kakamega (Mukumu) | Rain | An. gambiae | 4 | Unfed, gravid | 4 | 0 | NA | NA | |
| Kakamega (Isecheno) | Rain | An. gambiae | 3 | Unfed (fed; excluded) | 3 | 0 | NA | NA | |
*Minimum infection rate
Arbovirus detection
Overall, 484 pools consisting of 7788 mosquitoes were tested. The selected species collected in both seasons for the detection of arbovirus were Ae. aegypti (41 pools), An. funestus (8 pools), An. gambiae s.l. (47 pools), An. rivulorum (5 pools), and Cx. quinquefasciatus (368 pools). The following species of mosquitoes collected during only the dry season from East Kenya were also used for detection: An. coustani (1 pool), An. longipalpis (1 pool), Cx. cinereus (1 pool), Cx. decens (2 pools), Cx. laticinctus (1 pool), Cx. simpsoni (2 pool), Cx. univittatus (1 pool), Anopheles sp. (1 pool), Culex sp. (3 pools) and Mansonia sp. (2 pools). Although we collected 2 individuals of Cx. decens (1 male and 1 female from Isecheno), Cx. simpsoni (1 female from Busia), and Cx. univittatus (1 female from Busia) during the rainy season, we did not use these specimens for detection because of their small sample numbers compared to all other pools during the rainy season.
Human-related arboviruses from all mosquitoes
All pools were negative for human-related arboviruses, such as DENV, YFV, WNV, ONN, and CHINV.
Mosquito-related arboviruses from Culex quinquefasciatus
Using the primer sets cFD2 and MAMD, PCR bands were observed for 54 female Cx. quinquefasciatus pools during the rainy season and 1 male Cx. quinquefasciatus pool during the dry season in Mombasa (Tables 2 and 3). The nucleotide sequences for positive PCR reactions amplified using the primer sets cFD2 and MAMD from all these pools were compared with the GenBank database (BLAST), and sequencing results of all samples were 99% identical to the homologous region of Culex flavivirus (CxFV) strain Uganda08 (GQ165808.1). When we limited our analysis to female mosquitoes only, Busia yielded the most positive pools (52 pools) followed by Bamburi (1 pool) and Mukumu (1 pool).
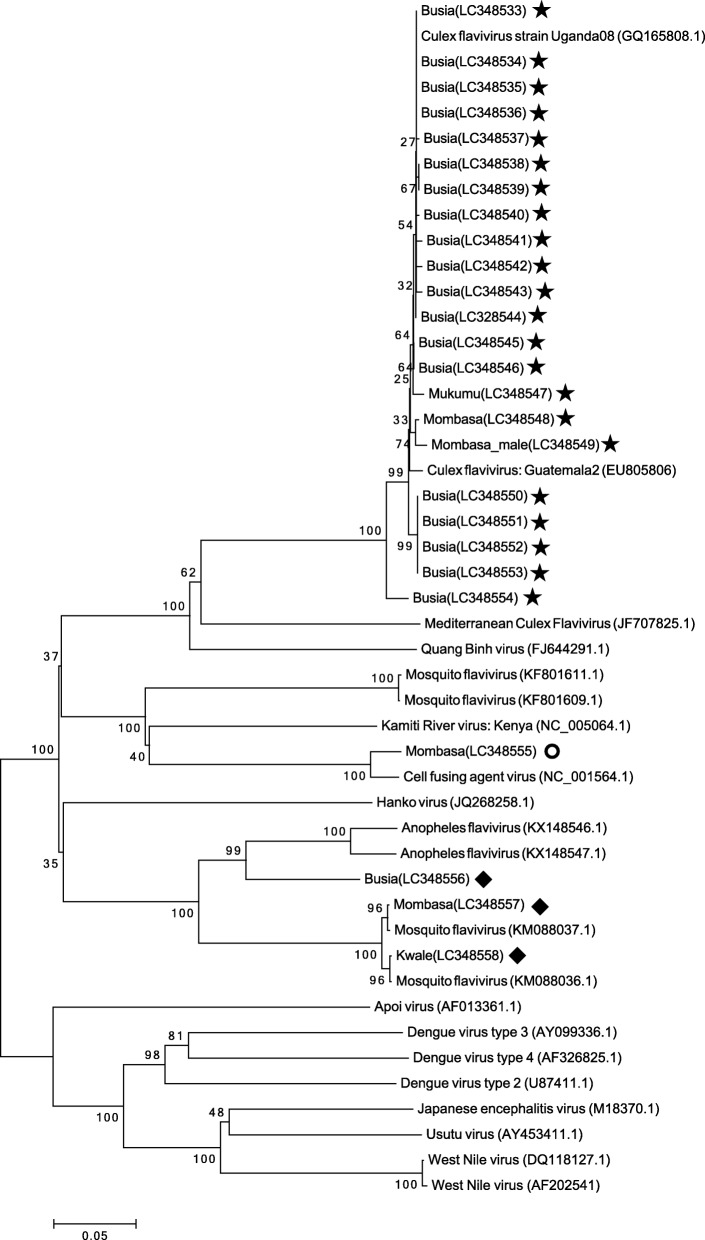
To generate a larger NS5 cDNA segment for sequencing to be used in phylogenetic analyses, only pools that were positive for flavivirus using the primer sets cFD2 and MAMD were amplified with the primer sets FLAVI1 and FLAVI2. Bands of approximately 860 nt (597 nt was used) were observed, and nucleotide sequencing was successful for 22 pools of Cx. quinquefasciatus (21 female pools and 1 male pool) among 55 pools (54 female pools and 1 male pool). The genomic sequences obtained using both primer sets (FLAVI1 and FLAVI2) share similar nucleotide sequence identity (99%) with CxFV from Uganda (GenBank: GQ165808.1). This result was the same as that using the primer sets cFD2 and MAMD. A phylogenetic tree was constructed with the NJ method using NS5 gene sequences of 22 CxFV strains by adding CxFV NS5 gene sequences from Uganda (GenBank: GQ165808.1) and Guatemala (GenBank: EU805806) obtained from BLAST. Additionally, NS5 gene sequences of human-related flaviviruses, such as WNV (GenBank: DQ118127.1, GenBank: AF202541), DNV (GenBank: AY099336.1, GenBank: AF326825.1, GenBank: U87411.1), and Japanese encephalitis virus (GenBank: M18370.1), were included as outgroup sequences. The NS5 gene sequences of our samples from Kenya clustered with CxFV NS5 gene sequences from Uganda and Guatemala. Although Busia, Kakamega, and Mombasa are in completely different regions of Kenya, the phylogenetic tree shows sequence similarity (Fig. 2).
Fig. 2.
Phylogenetic tree of the positive sequences based on the 597 nucleotides of the NS5 gene. The tree was constructed by employing the program MEGA 6, using the neighbor-joining method and distance-p model with 1000 bootstrap replicates. GenBank accession numbers are indicated in the parenthesis in the tree. Numbers on internal branches indicate bootstrap values for 1000 replicates. Our samples are marked with star (Cx. quinquefasciatus), with circle (Ae. aegypti), and with diamond shape (An. gambiae)
Mosquito-related arboviruses from Ae. aegypti and An. gambiae
The PCR products using the primer sets FLAVI1 and FLAVI2 for one pool of Ae. aegypti were shown to correspond to cell-fusing agent virus (CFAV) (NC_001564.1, 96% BLAST identity). In terms of An. gambiae s.l. pools, PCR products using the same primer sets as above were observed for three female pools, consisting of one pool from Kwale and one pool from Mombasa (both collected during the dry season) and one pool from Busia (collected during the rainy season). The nucleotide sequencing results of the two samples collected in Kwale and Mombasa were similar to mosquito flavivirus sequences (KM088036.1 and KM088037.1, 99% BLAST identity) reported from Kenya. The sequence of the sample collected from one pool from Busia was moderately divergent from the other two, being most similar a sequence of Anopheles flavivirus (KX148546.1, 85% BLAST identity) reported from Liberia. According to Kuno et al., a viral species is defined as the same group of viruses with > 84% nucleotide sequence identity among them [36]. Our sequence analysis demonstrated slightly higher nucleotide sequence identity than this cut-off. Therefore, the viruses from An. gambiae s.l. collected in Busia represent a variant of the closely related Anopheles flavivirus. The phylogenetic analyses including arboviruses from Cx. quinquefasciatus are presented in Fig. 2.
Minimum infection rate (MIR)
Although our study sites were geographically limited, MIR for Cx. quinquefasciatus showed a heterogeneous distribution for this species among the selected sites. Busia was the region with the highest MIR among all Cx. quinquefasciatus pools collected in Kenya (Table 4). Other Cx. quinquefasciatus pools revealed only one positive pool, with an MIR of approximately 1.0 (Table 4). Furthermore, taking into account differences in sampling efficiency among the study sites, seasons and traps, the Cx. quinquefasciatus specimens collected in Busia showed a higher MIR (MIR = 32.26; 95% CI = 24.42–42.12) than those collected in Mombasa during the rainy season (MIR = 1.01; 95% CI = 0.06–4.89) and during the dry season (MIR = 1.18; 95% CI = 0.07–5.75), and those collected in Kakamega during the rainy season (MIR = 1.11; 95% CI = 0.06–5.37). CxFV was detected in Mombasa during the dry season in a male pool as well as in female pools; however, there were no positive samples found in female pools during the dry season. No differences in MIR were found between the dry and rainy seasons in Mombasa, even though the pools of male and fed mosquitoes collected in the rainy season were not tested. Because of the limited number of samples, it is uncertain whether heterogeneity exists among Ae. aegypti and An. gambiae MIRs.
Discussion
In this study in Kenya, we did not detect any human-related arboviruses, and the main vector species of arboviruses were not found. Instead, we did detect mosquito-specific arboviruses from many types of mosquitoes. In particular, high prevalence of CxFV is Cx. quinquefasciatus was found in Busia, and this strain of CxFV is similar to one reported in Uganda by Cook et al. [37, 38]. Additionally, a similar CxFV was detected in each female pool from Mombasa and Kakamega. These areas in Kenya are separated by great distances. Additional sampling in the area between Busia and Kakamega in western Kenya and in the area between Kakamega and Mombasa in middle to eastern Kenya will likely increase the precision of the data regarding CxFV prevalence and geographic variation in Kenya. At present, the consequences of this geographic variation in Kenya are not clear. Moreover, we detected CxFV in one male pool collected in Mombasa. This result suggests that vertical maintenance may be common, even though Mombasa is an area with a lower positive rate compared to Busia.
Although many studies have reported mosquito-specific flavivirus detection in Culex and Aedes [39], there is little information about flaviviruses from anopheline mosquitoes, except for a few recent reports from Africa [40, 41]. In addition to Ae. aegypti, we also obtained flavivirus sequences from An. gambiae s.l. Our phylogenetic data using flavivirus NS5 gene sequences suggest that the sequences from Ae. aegypti are related to CFAV and that the sequences from An. gambiae s.l. are most closely related to mosquito flaviviruses (KM088037.1 and KM088036.1) from An. gambiae s.l. in West Africa and Kenya [40, 41]. Overall, reports of mosquito-specific flaviviruses are increasing.
Our results are based on partial sequences (NS5) of flaviviruses directly detected in mosquitoes. However, other regions of flavivirus nucleotide sequences (such as a region of NS3) were not determined, and there is a possibility that these sequences differ. Thus, further sequence information might be required, especially for a novel mosquito flavivirus, to establish the detailed taxonomic status of arboviruses. None of the mosquitoes in our samples were infected with human-related flaviviruses, though the detection rate might have been slightly higher if we had performed cell culture. Another limitation is the small sample size, and the number of mosquito species was also small. Larger studies are needed to provide a more accurate view of the prevalence of arboviruses.
Additionally, the abundance of Ae. aegypti, one of the most effective arboviral vectors in the human environment, obtained was relatively smaller than we expected. This mosquito is thought to have originated from Africa and to have been introduced to other continents such as Asia and South America through maritime trade [1]. Because this mosquito can easily adapt to urban areas on these continents, DENV transmitted by Ae. aegypti has become a major threat to humans. In this study, there were no positive pools of arboviruses, including DEN and CHIK, among 107 female Ae. aegypti samples. It is clear that this small sample size is insufficient. Additionally, due to this small sample size, the existence of another important vector, Aedes albopictus, cannot be determined, even though the distribution of this Asian-based mosquito has already been extended throughout the world, including West and Central Africa [42, 43]. Currently, this mosquito is not reported in Kenya. However, methods of collecting Aedes mosquitoes in Kenya remain an issue. We recognize that the effectiveness of the BG sentinel trap is quite low in certain areas such as Africa [44], though we did not analyze the effectiveness of each trap.
Here, we report the detection of CxFV from Cx. quinquefasciatus, CFAV from Ae. aegypti, mosquito flavivirus from An. gambiae s.l., and a new virus from An. gambiae s.l. However, we did not detect any arboviruses that are responsible for human disease. Many individuals might be exposed to a considerable risk of arbovirus infection in Kenya. Muyeku et al. reported the seroprevalence of CHIKV, YFV, and WNV in children at a hospital in Busia. According to their data for 2010, the virus with the highest positive rate was WNV (31% of 296 tested) followed by YFV (17% of 310 tested) and CHIV (11% of 298 tested) [45]. Moreover, there is an anecdotal report that the WNV infection rate might be higher than that reported because many infections are not obvious or are mild among those who live on the border of Kenya and Uganda, where this virus was first isolated in 1937 [46]. Regardless, the detection of human-related arboviruses in mosquitoes is very difficult in the absence of an outbreak. Our results, which indicate relatively high CxFV positivity among Cx. quinquefasciatus mosquitoes in Busia, might support risk prediction for future patterns of epidemics of arboviral infection. One previous study reported a positive association between insect-specific flaviviruses and human-related arboviruses, such as WNV [23]. Interestingly, Bolling et al. [47] identified early suppression of WNV infection in Culex pipiens naturally infected with CxFV. This suppression is one of the possible explanations for the lack of arbovirus detection, despite the high prevalence of CxFV in Cx. quinquefasciatus in our study. Thus, it is important to determine whether mosquitoes infected with mosquito-specific flaviviruses are resistant or susceptible to infection with other human-related flaviviruses. Future research on these viruses and their potential interactions with other flaviviruses in arthropod vectors will provide important new insight into not only virological but also public health aspects.
Conclusions
Insect-specific viruses were detected in various species of mosquitoes. In particular, the abundance of CxFV in Culex mosquitoes in Busia is higher than in other areas of Kenya. We suspect that this heterogeneity in various areas of Kenya may reflect the heterogeneity of the abundance of human-related virus vectors. These results, together with the absence of positive pools of human-related arbovirus, can be used as a baseline for future studies of human arboviruses. Future efforts to detect the circulation of arboviruses will help clarify the relationship between human-related arboviruses and various arboviruses, including insect-specific viruses. Detection methods that are more sensitive, such as next-generation sequencing (NGS), will facilitate obtaining real data about the presence of arboviruses.
Acknowledgements
We are deeply grateful to Dr. Kouichi Morita, Dr. Yoshio Ichinose, Dr. Masaaki Shimada, Dr. Charles Mwandawiro, Dr. Futoshi Hasebe, Dr. Shingo Inoue, Dr. Kazuhiko Moji, Dr. Matilu Mwau, and Mr. Haruki Kazama for technical support and Ms. Yukie Saito and Ms. Junko Sakemoto for providing administrative support. Special thanks go to Mr. Matthew Munyao, Mr. Johnstone Muyodi, Ms. Jecinta Odeo Lumumba, Mr. James Omondi Kongere, Ms. Mercy Syombua Mwania, Ms. Scholastica Achieng Wagalla, and Dr. Yuki Takamatsu, who devoted themselves to the fieldwork and experiments. Finally, we wish to express our gratitude to the residents of Kwale, Mombasa, Busia, and Kakamega who participated in this study.
Funding
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Young Researcher Overseas Visits Program for Vitalizing Brain Circulation, MEXT, Japan; the Global COE program, MEXT, Japan; the Japan Initiative for Global Network on Infectious Diseases (J-GRID), MEXT, Japan; and a Grant-in-Aid for Scientific Research from the Ministry of Health, Labour, and Welfare, Japan, Nagasaki University Kenya Research Station NUITM-KEMRI Project.
Availability of data and materials
Raw data can be obtained from the corresponding author upon request.
Abbreviations
- CFAV
Cell-fusing agent virus
- CHIKV
Chikungunya virus
- CxFV
Culex flavivirus
- DEN
Dengue
- DENV
Dengue virus
- MIR
Minimum infection rate
- NJ
Neighbor-joining
- ONN
O’nyong’nyong
- PCR
Polymerase chain reaction
- RVF
Rift Valley fever
- WNV
West Nile virus
- YF
Yellow fever
- YFV
Yellow fever virus
Authors’ contributions
HI, YH, KF, and NM conceived and designed this study. SN helped design and plan the study in Kenya. HI, YH, KF, and PA collected the field data, and HI, YH, and KF organized and conducted the laboratory work. HI and TN performed the data analyses. HI drafted the first manuscript, and HI and NM finalized the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All residents of the houses we visited to collect mosquitoes were informed about the study and agreed to participate. This study was approved by the Ethics Committee of Kenya Medical Research Institute (KEMRI) (SSC No. 2420), Kenya.
Consent for publication
This paper is published with the permission of the Director of KEMRI.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hanako Iwashita, Email: hanako@med.u-ryukyu.ac.jp.
Yukiko Higa, Email: yukko@nagasaki-u.ac.jp.
Kyoko Futami, Email: futami@nagasaki-u.ac.jp.
Peter A. Lutiali, Email: pakweywa@yahoo.com
Sammy M. Njenga, Email: sammynjenga@gmail.com
Takeshi Nabeshima, Email: mtmikami@tm.nagasaki-u.ac.jp.
Noboru Minakawa, Email: minakawa@nagasaki-u.ac.jp.
References
- 1.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/S0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, Reisen WK. Present and future arboviral threats. Antivir Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutherland LJ, Cash AA, Huang YJ, Sang RC, Malhotra I, Moormann AM, et al. Serologic evidence of arboviral infections among humans in Kenya. Am J Trop Med Hyg. 2011;85:158–161. doi: 10.4269/ajtmh.2011.10-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson BK, Ocheng D, Gichogo A, Okiro M, Libondo D, Kinyanjui P, et al. Epidemic dengue fever caused by dengue type 2 virus in Kenya: preliminary results of human virological and serological studies. East Afr Med J. 1982;59:781–784. [PubMed] [Google Scholar]
- 5.Sanders EJ, Marfin AA, Tukei PM, Kuria G, Ademba G, Agata NN, et al. First recorded outbreak of yellow fever in Kenya, 1992-1993. I. Epidemiologic investigations. Am J Trop Med Hyg. 1998;59:644–649. doi: 10.4269/ajtmh.1998.59.644. [DOI] [PubMed] [Google Scholar]
- 6.Nguku PM, Sharif S, Mutonga D, Amwayi S, Omolo J, Mohammed O, et al. An investigation of a major outbreak of Rift Valley fever in Kenya: 2006–2007. Am J Trop Med Hyg. 2010;83(2 Suppl):05–13. doi: 10.4269/ajtmh.2010.09-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods CW, Karpati AM, Grein T, McCarthy N, Gaturuku P, Muchiri E, et al. An outbreak of Rift Valley fever in Northeastern Kenya, 1997-98. Emerg Infect Dis. 2002;8:138–144. doi: 10.3201/eid0802.010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabtree M, Sang R, Lutomiah J, Richardson J, Miller B. Arbovirus surveillance of mosquitoes collected at sites of active Rift Valley fever virus transmission: Kenya, 2006–2007. J Med Entomol. 2009;46(4):961–964. doi: 10.1603/033.046.0431. [DOI] [PubMed] [Google Scholar]
- 9.Sergon K, Njuguna C, Kalani R, Ofula V, Onyango C, Konongoi LS, et al. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg. 2008;78:333–337. [PubMed] [Google Scholar]
- 10.Charrel RN, De Lamballerie X, Raoult D. Chikungunya outbreaks - the globalization of vectorborne diseases. N Engl J Med. 2007;356:769–771. doi: 10.1056/NEJMp078013. [DOI] [PubMed] [Google Scholar]
- 11.Haddow AJ, Davies CW, Walter AJ. O’nyong-nyong fever: an epidemic virus disease in East Africa - introduction. Trans R Soc TropMed Hyg. 1960;54:517–522. doi: 10.1016/0035-9203(60)90025-0. [DOI] [Google Scholar]
- 12.Williams MC, Woodall JP, Corbet PS, Gillett JD. O’nyong-nyong fever: an epidemic virus disease in East Africa. 8. Virus isolations from anopheles mosquitoes. Trans R Soc Trop Med Hyg. 1965;59:300–306. doi: 10.1016/0035-9203(65)90012-X. [DOI] [PubMed] [Google Scholar]
- 13.Geser A, Henderson BE, Christensen S. A multipurpose serological survey in Kenya. 2. Results of arbovirus serological tests. Bull World Health Organ. 1970;43:539–552. [PMC free article] [PubMed] [Google Scholar]
- 14.Mease LE, Coldren RL, Musila LA, Prosser T, Ogolla F, Ofula VO, et al. Seroprevalence and distribution of arboviral infections among rural Kenyan adults: a cross-sectional study. Virol J. 2011;8:371. doi: 10.1186/1743-422X-8-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sang RC, Dunster LM. The growing threat of arbovirus transmission and outbreaks in Kenya: a review. East Afr Med J. 2001;78:655–661. doi: 10.4314/eamj.v78i12.8936. [DOI] [PubMed] [Google Scholar]
- 16.Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutomiah J, Bast J, Clark J, Richardson J, Yalwala S, Oullo D, et al. Abundance, diversity, and distribution of mosquito vectors in selected ecological regions of Kenya: public health implications. J Vector Ecol. 2013;38:134–142. doi: 10.1111/j.1948-7134.2013.12019.x. [DOI] [PubMed] [Google Scholar]
- 18.Labeaud AD, Sutherland LJ, Muiruri S, Muchiri EM, Gray LR, Zimmerman PA, et al. Arbovirus prevalence in mosquitoes, kenya. Emerg Infect Dis. 2011;17:233–241. doi: 10.3201/eid1702.091666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould EA, Higgs S. Impact of climate change and other factors on emerging arbovirus diseases. Trans R Soc Trop Med Hyg. 2009;103(2):109–121. doi: 10.1016/j.trstmh.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, et al. Two chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One. 2007;2(11):e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshino K, Isawa H, Tsuda Y, Sawabe K, Kobayashi M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology. 2009;391:119–129. doi: 10.1016/j.virol.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Ciota AT, Kramer LD. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses. 2013;5(12):3021–3047. doi: 10.3390/v5123021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman CM, Cerutti F, Anderson TK, Hamer GL, Walker ED, Kitron UD, et al. Culex Flavivirus and west Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis. 2011;11:1099–1105. doi: 10.1089/vbz.2010.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W, Novak RJ. Short report: detection probability of arbovirus infection in mosquito populations. Am J Trop Med Hyg. 2004;71:636–638. [PubMed] [Google Scholar]
- 25.Weaver SC. Urbanization and geographic expansion of zoonotic arboviral diseases: mechanisms and potential strategies for prevention. Trends Microbiol. 2013;21(8):360–363. doi: 10.1016/j.tim.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanley KA, Monath TP, Weaver SC, Rossi SL, Richman RL, Vasilakis N. Fever versus fever: the role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infect Genet Evol. 2013;19:292–311. doi: 10.1016/j.meegid.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berens DG, Farwig N, Schaab G, Boehning-Gaese K. Exotic guavas are foci of forest regeneration in Kenyan farmland. Biotropica. 2008;40:104–112. [Google Scholar]
- 28.Southwood TRE, ECOLOGICAL METHODS second Edition 1977.
- 29.Gu W, Unnasch TR, Katholi CR, Lampman R, Novak RJ. Fundamental issues in mosquito surveillance for arboviral transmission. Trans R Soc Trop Med Hyg. 2008;102:817–822. doi: 10.1016/j.trstmh.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbach RE. Pictorial keys to the genera of mosquitoes, subgenera of Culex and the species of Culex (Culex) occurring in southwestern Asia and Egypt, with a note on the subgeneric placement of Culex deserticola (Diptera: Culicidae) Mosq Syst. 1985;17:83–107. [Google Scholar]
- 31.Huang YM. A pictorial key to the mosquito genera of the world, including subgenera of Aedes and Ochlerotatus (Diptera: Culicidae) Ins Koreana. 2002;19:1–130. [Google Scholar]
- 32.Reinert JF. Descriptions of Zavortinkius, a new subgenus of Aedes, and the eleven included species from the Afrotropical region (Diptera: Culicidae) 1999. pp. 1–105. [Google Scholar]
- 33.Rueda LM. Pictorial keys for the identification of mosquitoes (Diptera:Culicidae) associated with dengue virus transmission. Zootaxa. 2004;589:1–60. doi: 10.11646/zootaxa.589.1.1. [DOI] [Google Scholar]
- 34.Biggerstaff BJ. PooledInfRate, version 3.0: a Microsoft excel add-in to compute prevalence estimates from pooled samples. Ft. Collins, CO: Centers for Disease Control and Prevention; 2006. [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook S, Moureau G, Harbach RE, Mukwaya L, Goodger K, Ssenfuka F, et al. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol. 2009;90:2669–2678. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mwangangi JM, Midega J, Kahindi S, Njoroge L, Nzovu J, Githure J, et al. Mosquito species abundance and diversity in Malindi, Kenya and their potential implication in pathogen transmission. Parasitol Res. 2012;110:61–71. doi: 10.1007/s00436-011-2449-6. [DOI] [PubMed] [Google Scholar]
- 39.Kasai S, Komagata O, Tomita T, Sawabe K, Tsuda Y, Kurahashi H, et al. PCR-based identification of Culex pipiens complex collected in Japan. Jpn J Infect Dis. 2008;61:184–191. [PubMed] [Google Scholar]
- 40.Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, Fakoli LS, 3rd, et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288e99. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Villinger J, Mbaya MK, Ouso D, Kipanga PN, Lutomiah J, Masiga DK. Arbovirus and insect-specific virus discovery in Kenya by novel six genera multiplex high resolution melting analysis. Mol Ecol Resour. 2017;17:466–480. doi: 10.1111/1755-0998.12584. [DOI] [PubMed] [Google Scholar]
- 42.Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect. 2009;11:1177–1185. doi: 10.1016/j.micinf.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. elife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivagnaname N, Gunasekaran K. Need for an efficient adult trap for the surveillance of dengue vectors. Indian J Med Res. 2012;136:739–749. [PMC free article] [PubMed] [Google Scholar]
- 45.Muyeku MI, Seroprevalence of chikungunya, yellow fever and West Nile viruses in children at the Alupe District Hospital in Western Kenya. http://erepository.uonbi.ac.ke/bitstream/handle/11295/3785/Muyeku_Seroprevalence%20of%20Chikungunya%2c%20Yellow%20fever%20and%20West%20Nile%20Viruses%20in%20Children.pdf?sequence=1&isAllowed=y (2011). Accessed 5 Dec 2017.
- 46.Smithburn KC, Hughes TP, Burke AW, Paul JH. A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med Hyg. 1940;20:471–473. doi: 10.4269/ajtmh.1940.s1-20.471. [DOI] [Google Scholar]
- 47.Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, Blair CD. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology. 2012;427:90–97. doi: 10.1016/j.virol.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription–PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol. 2001;39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayers M, Adachi D, Johnson G, Andonova M, Drebot M, Tellier R. A single tube RT-PCR assay for the detection of mosquito-borne flaviviruses. J Virol Methods. 2006;135:235–239. doi: 10.1016/j.jviromet.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka M. Rapid identification of flavivirus using the polymerase chain reaction. J Virol Methods. 1993;41:311–322. doi: 10.1016/0166-0934(93)90020-R. [DOI] [PubMed] [Google Scholar]
- 51.Hasebe F, Parquet MC, Pandey BD, Mathenge EG, Morita K, Balasubramaniam V, et al. Combined detection and genotyping of chikungunya virus by a specific reverse transcription-polymerase chain reaction. J Med Virol. 2002;67:370–374. doi: 10.1002/jmv.10085. [DOI] [PubMed] [Google Scholar]
- 52.Jupp PG, Grobbelaar AA, Leman PA, Kemp A, Dunton RF, Burkot TR, et al. Experimental detection of Rift Valley fever by reverse transcription–polymerase chain reaction assay in large samples of mosquitoes. J Med Entomol. 2000;37:467–471. doi: 10.1093/jmedent/37.3.467. [DOI] [PubMed] [Google Scholar]
- 53.Staley M, Dorman KS, Bartholomay LC, Fernández-Salas I, Farfan-Ale JA, Loroño-Pino MA, et al. Universal primers for the amplification and sequence analysis of actin-1 from diverse mosquito species. J Am Mosq Control Assoc. 2010;26:214–218. doi: 10.2987/09-5968.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higa Y, Toma T, Tsuda Y, Miyagi IA. Multiplex PCR-based molecular identification of five morphologically related, medically important subgenus Stegomyia mosquitoes from the genus Aedes(Diptera: Culicidae) found in the Ryukyu Archipelagon Japan. Jpn J Infect Dis. 2010;63:312–316. [PubMed] [Google Scholar]
- 55.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. Cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data can be obtained from the corresponding author upon request.