Abstract
Objectives
The aim of this study was to identify key pathological genes in osteoarthritis (OA).
Methods
We searched and downloaded mRNA expression data from the Gene Expression Omnibus database to identify differentially expressed genes (DEGs) of joint synovial tissues from OA and normal individuals. Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses were used to assess the function of identified DEGs. The protein-protein interaction (PPI) network and transcriptional factors (TFs) regulatory network were used to further explore the function of identified DEGs. The quantitative real-time polymerase chain reaction (qRT-PCR) was applied to validate the result of bioinformatics analysis. Electronic validation was performed to verify the expression of selected DEGs. The diagnosis value of identified DEGs was accessed by receiver operating characteristic (ROC) analysis.
Results
A total of 1085 DEGs were identified. KEGG pathway analysis displayed that Wnt was a significantly enriched signalling pathway. Some hub genes with high interactions such as USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, and ZNF423 were identified in the PPI and TFs network. The results of qRT-PCR showed that GADD45B, ADAMTS1, and TFAM were down-regulated in joint synovial tissues of OA, which was consistent with the bioinformatics analysis. The expression levels of USP46, CPVL, FOSL2, and PTGS1 in electronic validation were compatible with the bio-informatics result. CPVL and TFAM had a potential diagnostic value for OA based on the ROC analysis.
Conclusion
The deregulated genes including USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, ZNF423, ADAMTS1, and TFAM might be involved in the pathology of OA.
Cite this article: X. Zhang, Y. Bu, B. Zhu, Q. Zhao, Z. Lv, B. Li, J. Liu. Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis. Bone Joint Res 2018;7:298–307. DOI: 10.1302/2046-3758.74.BJR-2017-0245.R1.
Keywords: Osteoarthritis, Gene expression, Protein-protein interaction network, Transcriptional factors
Article focus
The objective of this study is identifying key pathological genes in osteoarthritis (OA).
Key messages
The deregulated genes including USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, ZNF423, ADAMTS1, and TFAM might be involved in the pathology of osteoarthritis.
Strengths and limitations
Bioinformatics analysis was used to identify the differentially expressed genes (DEGs) in osteoarthritis.
In vitro functional research is needed to study the function of identified DEGs in osteoarthritis.
Introduction
One of the bone joint structures, cartilage, is composed of two main extracellular matrix macromolecules: aggrecan and type II collagen.1,2 The aggrecan enables cartilage to resist compression, whereas the type II collagen ensures the tensile strength of cartilage. Generally, maintenance of normal joint structure and function depends on load adaptation of the bone and cartilage. The synovium encapsulates the joints and functions in providing structural support, lubricating the surfaces, and providing nutrients to the cartilage. Osteoarthritis (OA) is a common degenerative joint disease characterized by gradual thinning and eventual loss of articular cartilage, which can be accompanied by changes in other joint organs, including structural modifications of subchondral bone, pathological changes of the meniscus, and synovitis.3-6 It has been reported that there is a low degree of synovium inflammation in OA.7-11
OA is the most common chronic condition of the joints.12 The pharmacological treatment for OA is primarily focused on using analgesics and non-steroidal anti-inflammatory drugs. The pathological mechanism of OA is complex. It is reported that various inflammatory cytokines, including adipokines, interleukins, nerve growth factor, and tumour necrosis factor-alpha influence the progression of OA.13,14 The molecular mechanisms underlying OA are still poorly understood. Therefore, further understanding of the pathologica mechanisms and related genes in OA is needed.
By integrated analysis, Xiao et al15 found a number of rheumatoid arthritis associated genes. In view of this, we also sought to find differentially expressed genes (DEGs) in OA by similar integrated analysis. The mRNA expression data were downloaded from the Gene Expression Omnibus (GEO) database to identify differentially expressed genes (DEGs) by metaMA in R package (R Foundation for Statistical Computing, Vienna, Austria). Our study was helpful in understanding the pathogenic mechanism of OA.
Materials and Methods
Gene expression datasets
In this study, we searched datasets from the GEO database16 with the keywords "osteoarthritis" [MeSH Terms] OR osteoarthritis [All Fields] AND "knee"[MeSH Terms] OR "knee joint" [MeSH Terms] OR knee [All Fields] AND "Homo sapiens" [porgn] AND "gse" [Filter]. The study type was described as “expression profiling by array.” All selected data sets were genome-wide expression data in joint synovial tissues of OA and/or normal tissues.
Identification of DEGs
Raw expression data of OA patients in this study were obtained. The significant DEGs were identified between OA patients and normal controls through metaMA in R package (R Foundation for Statistical Computing). The false discovery rate (FDR) was performed for multiple testing corrections of the raw p-value through the Benjamin and Hochberg method.17,18 The threshold of DEGs was set as FDR < 0.05.
Functional enrichment analysis of DEGs
To obtain the biological function and signalling pathways of DEGs, the GeneCodis319-21 was used for GO annotation and KEGG pathway enrichment of DEGs.22,23 The threshold of GO function and KEGG pathway of DEGs was all set as FDR < 0.05.
PPI network construction
Studying the interactions between proteins can aid in the understanding of the molecular mechanism of osteoarthritis. In order to gain insights into the interaction between DEGs and proteins, the BioGRID database24 was used to retrieve the predicted interactions between top 40 proteins encoded by DEGs (20 up-regulated and 20 down-regulated) and other proteins. Then, PPI network was visualized by the Cytoscape Software25 A node in the PPI network denotes protein, and the edge denotes the interactions.
Construction of transcriptional regulatory networks
TFs regulate gene expression at the post-transcription,al level which can provide a better understanding of the underlying regulatory mechanisms in the development of OA. To provide a deeper knowledge about gene regulation underling OA, we extract information about TFs likely involved in regulating these DEGs. First, the promoter sequences of DEGs were obtained from the UCSC Genome Browser. Related TF information was then obtained via the matching tool in the TRANSFAC data set. Finally, transcriptional regulatory networks were visualized using Cytoscape.25
Validation of qRT-PCR
The quantitative real-time polymerase chain reaction (qRT-PCR) was applied to validate the result of bioinformatics analysis. In this study, six patients diagnosed as having OA and six unaffected individuals were enrolled in this study. Clinical information for these patients is shown in Table I. Both OA and corresponding normal knee joint synovial tissues were obtained and immediately frozen in liquid nitrogen. All participating individuals provided informed consent with the approval of the ethics committee of Tianjin Hospital (Tianjin, China).
Table I.
Qualitative PCR patient information
| Patient | Age (yrs) | Gender | BMI (kg/m2) | Duration of disease | Leukocyte count (cells/ml) | Platelet count (cells/ml) | The reference value of ESR (mm/1 hr) | CRP (mg/l) | Kellgren and Lawrence scores |
|---|---|---|---|---|---|---|---|---|---|
| Normal | 37 | Male | 23.1 | 4 mths | 6.85 × 106 | 2.15 × 108 | 4 | 5 | 0 |
| Normal | 43 | Male | 26.8 | 3 days | 6.31 × 106 | 1.79 × 108 | 12 | 5 | 0 |
| Normal | 26 | Male | 24.6 | 1 mth | 6.94 × 106 | 2.29 × 108 | 1 | 5 | 0 |
| Normal | 35 | Male | 25.4 | 2 mths | 7.64 × 106 | 1.99 × 108 | 2 | 5 | 0 |
| Normal | 50 | Female | 23.4 | 7 days | 8.33 × 106 | 2.51 × 108 | 5 | 7 | 0 |
| Normal | 31 | Male | 29.6 | 12 days | 4.92 × 106 | 1.66 × 108 | 5 | 5 | 0 |
| OA | 81 | Male | 24 | 2 yrs | 5.65 × 106 | 1.56 × 108 | 6 | 5 | 4 |
| OA | 60 | Female | 25.3 | 10 yrs | 6.68 × 106 | 2.95 × 108 | 42 | 5 | 4 |
| OA | 74 | Female | 25 | 2 yrs | 4.42 × 106 | 2.24 × 108 | 19 | 5 | 4 |
| OA | 65 | Female | 24 | 6 mths | 8.03 × 106 | 3.11 × 108 | 24 | 5 | 4 |
| OA | 67 | Male | 19.6 | 10 yrs | 3.94 × 106 | 1.90 × 108 | 8 | 5 | 4 |
| OA | 66 | Male | 24.7 | 2 yrs | 4.03 × 106 | 1.56 × 108 | 6 | 5 | 4 |
BMI, body mass index; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; OA, osteoarthritis; Kellgren and Lawrence scores, a grading method of OA severity. Generally, the grade from slight to severity is divided into 0, 1, 2, 3 and 4.
Total RNA of joint synovial tissues from OA patients and unaffected individuals was extracted using TRizol reagent (TFS, Foster City, California) according to the manual instructions. SuperScript III Reverse Transcription Kit (Invitrogen) was used to synthesize the cDNA. qRT-PCR reactions were performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, California) on Applied Biosystems 7,500 (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control for gene detection and the relative expression of genes was calculated using the log2 (fold change) equation.
Electronic validation of DEGs by GSE89408
The GSE89408 database (22 cases and 28 normal controls) was used to validate the expression of selected DEGs. We compared the expression levels of DEGs between OA cases and normal controls; the differences of expression levels were displayed by box-plots.
Receiver operating characteristic (ROC) analyses
Using pROC package in R language26, we performed receiver operating characteristic (ROC) analyses to assess the diagnostic value of selected DEGs. The area under the curve (AUC) under binomial exact confidence interval was calculated and the ROC curve was generated.
Results
DEGs in the gene expression data sets
After electronic search, a total of five expression profiling datasets met the inclusion criteria and were included, which are shown in Table II.27-30 Of these studies, 54 cases of OA and 43 cases of controls were included. By integrated analysis, 10 678 genes were obtained and a total of 1085 DEGs were identified in the OA datasets compared with normal tissues, including 604 up-regulated and 481 down-regulated DEGs. The top 20 DEGs are shown in Table III. In addition, the abbreviations of genes listed in the text and table listed in the supplementary Table I.
Table II.
Five expression profiling datasets of Gene Expression Omnibus (GEO)
| GEO ID | Normal | Case | Platform | Year | Country | Author |
|---|---|---|---|---|---|---|
| GSE55235 | 10 | 10 | GPL96[HG-U133A] Affymetrix Human Genome U133A Array | 2014 | Germany | Woetzel D27 |
| GSE55457 | 10 | 10 | GPL96[HG-U133A] Affymetrix Human Genome U133A Array | 2014 | Germany | Woetzel D27 |
| GSE46750 | 12 | 12 | GPL10558 Illumina HumanHT-12 V4.0 expression beadchip | 2013 | Belgium | Henrotin Y28 |
| GSE32317 | 7 | 19 | GPL570[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 2011 | USA | Wang Q29 |
| GSE41038 | 4 | 3 | GPL6883Illumina HumanRef-8 v3.0 expression beadchip | 2012 | Australia | Fischer R30 |
Table III.
Top 20 differentially expressed genes in osteoarthritis
| ID | Symbol | Combined effect size | p-value | False discovery rates | Up/down |
|---|---|---|---|---|---|
| 4616 | GADD45B | -2.30652 | 1.33E-15 | 1.42E-11 | Down |
| 55002 | TMCO3 | 1.860201 | 7.15E-14 | 3.82E-10 | Up |
| 5742 | PTGS1 | 1.772798 | 4.88E-13 | 1.74E-09 | Up |
| 53346 | TM6SF1 | 1.732943 | 1.14E-12 | 3.05E-09 | Up |
| 2355 | FOSL2 | -2.05783 | 5.63E-12 | 6.09E-09 | Down |
| 57018 | CCNL1 | -1.78228 | 5.71E-12 | 6.09E-09 | Down |
| 10560 | SLC19A2 | -2.1303 | 1.46E-11 | 1.20E-08 | Down |
| 64854 | USP46 | 1.671456 | 1.39E-11 | 1.20E-08 | Up |
| 2289 | FKBP5 | -1.65731 | 2.25E-11 | 1.60E-08 | Down |
| 1519 | CTSO | 1.590246 | 3.16E-11 | 2.11E-08 | Up |
| 25834 | MGAT4C | 1.652285 | 4.54E-11 | 2.85E-08 | Up |
| 29071 | C1GALT1C1 | 1.577996 | 4.91E-11 | 2.91E-08 | Up |
| 10797 | MTHFD2 | -1.68311 | 7.96E-11 | 4.09E-08 | Down |
| 79862 | ZNF669 | -1.54828 | 7.77E-11 | 4.09E-08 | Down |
| 25976 | TIPARP | -1.8764 | 1.02E-10 | 4.93E-08 | Down |
| 7538 | ZFP36 | -1.74104 | 1.11E-10 | 5.17E-08 | Down |
| 2628 | GATM | 1.510396 | 4.21E-10 | 1.73E-07 | Up |
| 23090 | ZNF423 | 1.489729 | 5.19E-10 | 1.85E-07 | Up |
| 2872 | MKNK2 | -1.44317 | 5.09E-10 | 1.85E-07 | Down |
| 54504 | CPVL | 1.439288 | 5.17E-10 | 1.85E-07 | Up |
Functional enrichment analysis
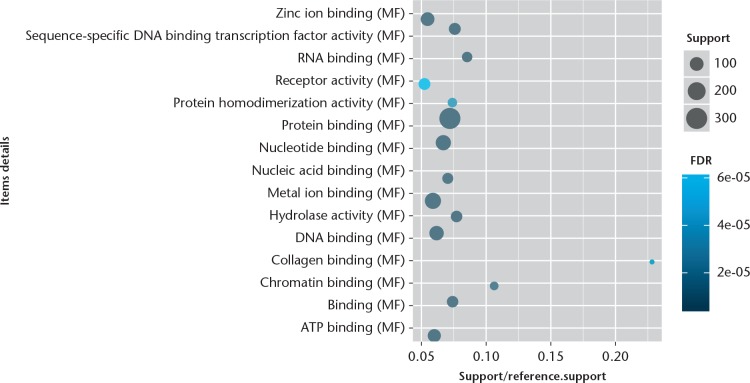
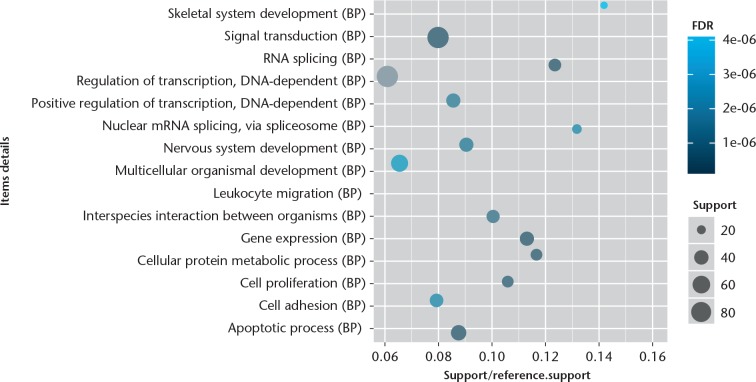
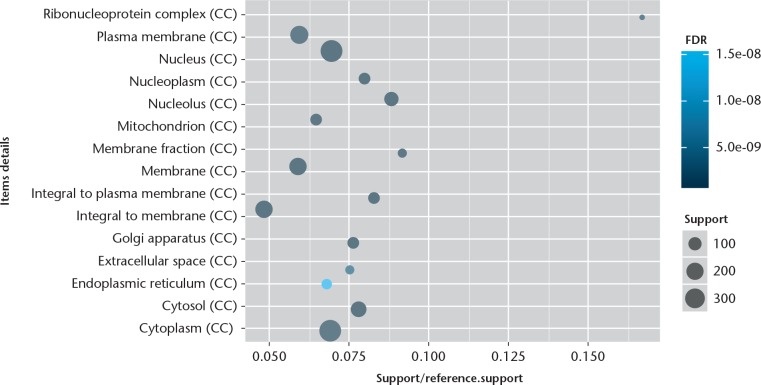
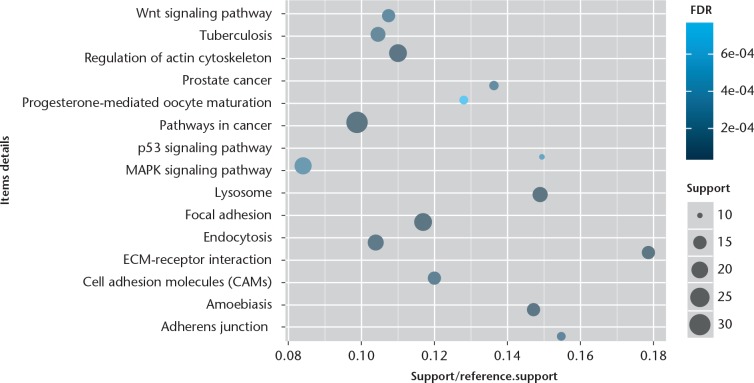
The GO enrichment analysis was performed to determine the biological function of DEGs. We found that the significantly enriched GO terms for molecular functions were metal ion binding, sequence-specific DNA binding transcription factor activity, and protein/nucleotide binding (Fig. 1), while those for biological processes were skeletal system development, signal transduction, RNA splicing, DNA-dependent regulation of transcription, apoptotic, and nervous system development (Fig. 2). Furthermore, the significantly enriched GO terms for cellular component were cytoplasm, nucleus, and plasma membrane (Fig. 3). The pathway enrichment analysis indicated that the most significantly enriched pathways were the Wnt signalling pathway, p53 signalling pathway, pathways in cancer, regulation of actin cytoskeleton, and focal adhesion (Fig. 4).
Fig. 1.
CapChart showing the top 15 significant enrichment molecular functions of differentially expressed genes (DEGs). The x-axis shows support/reference numbers and false discovery rate (FDR) value of DEGs; the y-axis shows different molecular functions.
Fig. 2.
Chart showing the top 15 significant enrichment biological processes of differentially expressed genes (DEGs). The x-axis shows support/reference numbers and false discovery rate (FDR) value of DEGs; the y-axis shows different biological processes.
Fig. 3.
Chart showing the top 15 significant enrichment cellular components of differentially expressed genes (DEGs). The x-axis shows support/reference numbers and false discovery rate (FDR) value of DEGs; the y-axis shows different cellular components.
Fig. 4.
Chart showing the top 15 Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathways of differentially expressed genes (DEGs). The x-axis shows support/reference numbers and false discovery rate (FDR) value of DEGs; the y-axis shows different signal pathways.
PPI network construction
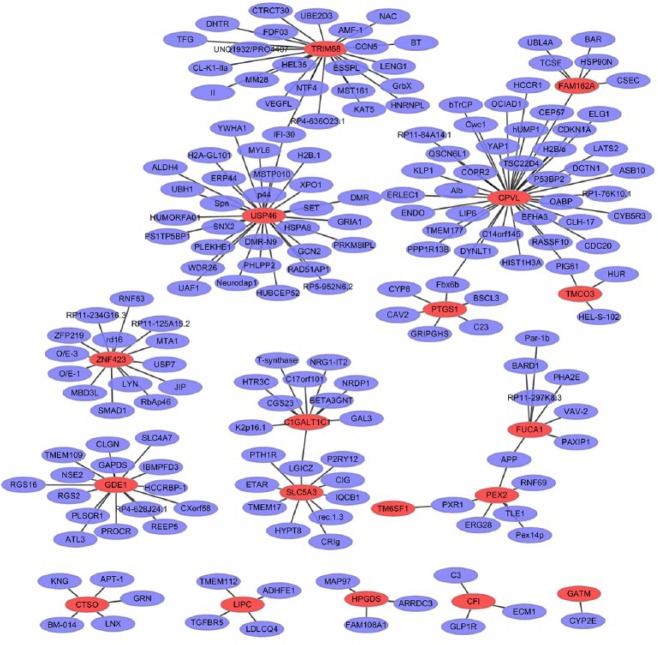
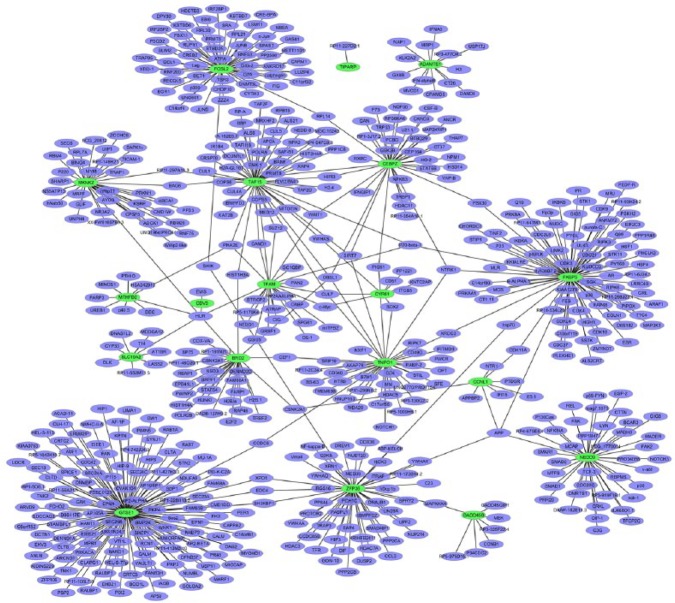
To obtain the interaction between the DEGs, the PPI network was explored and visualize by Cytoscape. PPI networks of top 20 upregulated and top 20 downregulated DEGs are presented in Figure 5 and Figure 6, respectively. The nodes with high interactions are described as the hub protein. The upregulated network consisted of 196 nodes and 185 edges. The top three significant hub proteins were USP46 (interaction = 30), CPVL (interaction = 37), and TRIM68 (interaction = 24) (Figure 5). The downregulated network consisted of 589 nodes and 618 edges in the network. The top three significant hub proteins were GTSE1 (interaction = 122), FKBP5 (interaction = 82), and FOSL2 (interaction = 55) (Figure 6).
Fig. 5.
The protein-protein interaction (PPI) networks of the top 20 upregulated differentially expressed genes (DEGs). All the nodes are proteins encoded by DEGs and the red borders represent proteins encoded by the top 20 upregulated DEGs; purple borders represent proteins encoded by other genes that interacted with proteins encoded by these DEGs.
Fig. 6.
The protein-protein interaction (PPI) networks of the top 20 downregulated differentially expressed genes (DEGs). All the nodes are proteins encoded by DEGs and the green borders represent proteins encoded by top 20 downregulated DEGs; purple borders represent proteins encoded by other genes interacted with proteins encoded by these DEGs.
Transcriptional regulatory networks
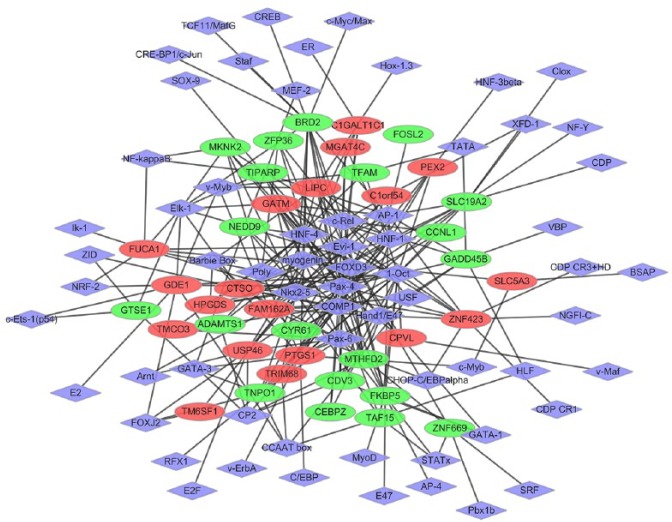
The regulatory network between DEG and TFs was constructed. Regulatory networks consisted of 817 TF-target interactions between 63 TFs and 39 DEGs in the context of OA (Fig. 7). The top five DEGs with high TFs interactions were PTGS1, FKBP5, ZNF423, USP46, and GADD45B. In addition, the top five TFs covering the most downstream DEGs were identified as crucial TFs: Pax-4, Oct-1, HNF-1, AP-1, and Evi-1, which are involved in the development of OA.
Fig. 7.
The established transcriptional regulatory network of osteoarthritis. Red- and green-coloured nodes represent products of up- and downregulated differentially expressed genes (DEGs), respectively. The purple nodes denote products of transcriptional factors (TFs) predicted to interact with the corresponding DEGs.
qRT-PCR
To verify the bioinformatics analyses, the expression level of DEGs was quantified by qRT-PCR in joint synovial tissues of six osteoarthritis patients and six normal individuals. Three DEGs (GADD45B, ADAMTS1, and TFAM) were selected as candidate genes based on bioinformatics results and our literature review.31-34 Our result showed that ADAMTS1 was significantly downregulated (p < 0.05) in osteoarthritis compared with those of normal controls. The expression level of GADD45B and TFAM was downregulated with no significant between OA and normal controls.
Electronic expression validation of genes
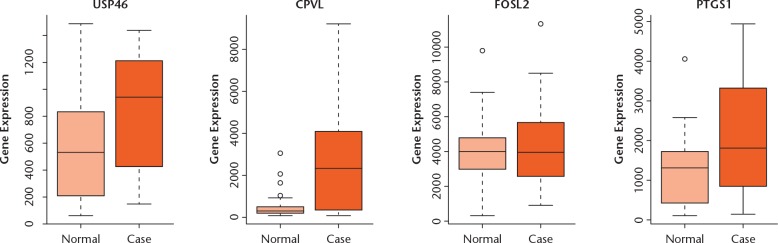
In this study, four DEGs (USP46, CPVL, FOSL2, and PTGS1) in OA were selected to perform expression validation (Fig. 8). Different expression levels of these DEGs between patients with OA and normal patients were analyzed and depicted through box-plots. These box-plots were shown as medians and interquartile ranges (IQR). The expression level of USP46, CPVL, and PTGS1 was significantly upregulated in the case group compared to the normal group. The expression level of FOSL2 was downregulated in the case group compared with the normal group. Compared with the normal group, the expression levels of them in the case group were consistent with our bioinformatics analysis result.
Fig. 8.
The validation of the expression levels of selected DEGs in OA based on GSE89408 database. The x-axis shows the case and normal groups and y-axis shows expression reads counts.
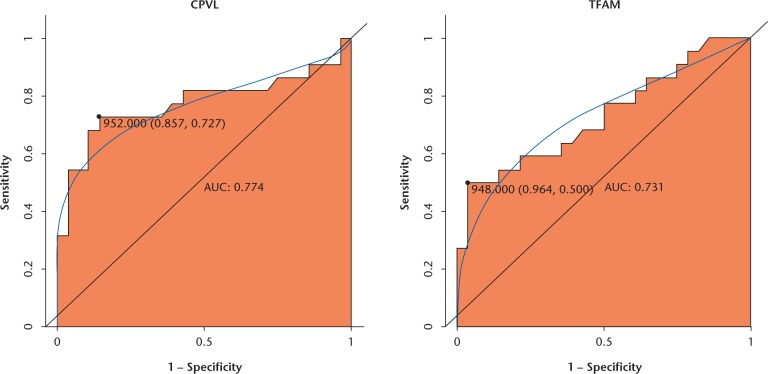
ROC curve analysis
We performed ROC curve analyses and calculated the AUC to assess the discriminatory ability of CPVL and TFAM from the GSE89408 data set (Fig. 9). The AUC for both was more than 0.7. For OA diagnosis, the values for 1-specificity (proportion of false positive) and sensitivity (proportion of true positive) of CPVL were 85.7% and 72.7%, respectively; the 1-specificity (proportion of false positive) and sensitivity (proportion of true positive) values of TFAM were 96.4% and 50%, respectively.
Fig. 9.
Receiver operator characteristic (ROC) curves of selected differentially expressed genes (DEGs) between osteoarthritic patients and healthy controls. The ROC curves were used to show the diagnostic ability of these selected DEGs with 1-specificity (the proportion of false positive) and sensitivity (the proportion of true positive).
Discussion
OA is known as the most prevalent chronic joint disease.12. In this study, we found a total of 1085 DEGs including 604 upregulated and 481 downregulated DEGs in synovial tissues of OA. Several genes with high interactions in the PPI and TF-target networks, such as USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, and ZNF423, were identified to play crucial roles in the presence of OA. Three candidate genes (GADD45B, ADAMTS1, and TFAM) were downregulated in qRT-PCR validation and gave results that were consistent with the bioinformatics analysis. Electronic validation expression of USP46, CPVL, FOSL2, and PTGS1 was also consistent with the bioinformatics analysis. CPVL and TFAM had a potential diagnostic value for OA.
Ubiquitin specific peptidase 46 (USP46) is involved in the ubiquitin proteolysis pathway. In this study, we first found upregulated expression of USP46 in the synovial tissues of OA. In addition, USP46 played an important regulatory role in both PPI and TFs target networks. This suggested that the overexpression of USP46 may be involved in the ubiquitin proteolysis pathway in the development of OA. Further research is needed to confirm this.
Carboxypeptidase, vitellogenic like (CPVL) is a hydrolase that is localized in the endoplasmic reticulum. It is noted that CPVL is differentially expressed in synovial membranes of patients with rheumatoid arthritis.35 In addition, compared with the neck of femur, the expression of CPVL is significantly upregulated in OA.36 Herein, we first found increased expression of CPVL in synovial tissues of OA. Moreover, it was one regulatory molecule in the PPI network. Significantly, CPVL had a potential diagnostic value for OA. Our results suggested that CPVL plays an important role in the process of OA.
FK506 binding protein 5 (FKBP5) belongs to the immunophilin protein family, which plays a key role in immunoregulation and cellular processes by affecting protein folding and transportation. Based on the gene expression profile in OA, the expression of FKBP5 is found to be downregulated in cartilage.37 In this study, we first found that FKBP5 was decreased in synovial tissues of OA and played regulatory roles in both PPI and TFs-target networks. This indicated that FKBP5 played a crucial role in the immune regulation of OA. Further study is needed to investigate the potential mechanism of FKBP5 in OA.
FOS like 2, AP-1 transcription factor subunit (FOSL2, also called FRA2), a FOS-related protein of the AP-1 family, is expressed in bone cells. It has been found that FOSL2 is downregulated in synovial tissue of rheumatoid arthritis.38 In this study, we first found that FOSL2 was downregulated in synovial tissues of OA and was one of top 20 DEGs with high interaction in the PPI network. Our result provided a new field in understanding of the pathology of OA.
Growth arrest and DNA damage inducible beta (GADD45B) is a member of the GADD45B family that is related to cell growth control and cell death.39 It has been demonstrated that GADD45B is significantly overexpressed in baseline synovial tissue of early rheumatoid arthritis patients.40 In this study, we found that the expression of GADD45B was downregulated and that the qRT-PCR validated the expression in synovial tissues of OA. We suggest that GADD45B may play roles in cell growth and death in OA.
Generally, prostaglandin-endoperoxide synthase 1 (PTGS1, also called COX-1) is an inflammatory molecule and is associated with pain. It has been found that the mRNA of PTGS1 is remarkably increased in the spine during OA pain.41 Knorth et al42 demonstrated the important role of synovial PTGS1 in patients with primary OA. Significantly, we found that PTGS1 was upregulated and involved in the TFs-target network in synovial tissues of OA. Therefore, we suggested that the upregulation of PTGS1 may be important in the inflammation of OA
Zinc finger protein 423 (ZNF423) has been demonstrated to be a molecule marker in the major cellular components of skeletal muscle.43 In this study, we first found that the expression of ZNF423 was increased and under the regulation of TFs in synovial tissues of OA. This indicated that ZNF423 played an important role in the process of OA.
ADAM metallopeptidase with thrombospondin type 1 motif 1 (ADAMTS1) is an aggrecanase. It is found that ADAMTS1 is downregulated in synovium tissues of OA.44 Herein, we found downregulated expression of ADAMTS1 in synovium tissues of OA, which was in line with previous reports. Our result further demonstrated the crucial role of ADAMTS1 in the development of OA.
Transcription factor A, mitochondrial (TFAM) is a mitochondrial derived key transcription factor in maintaining healthy skeletal muscle and is associated with osteoblast differentiation.45,46 Decreased TFAM expression in OA chondrocytes is related to mitochondrial biogenesis capacity.47 In this study, we found that TFAM was downregulated in synovium tissues of OA. In addition, TFAM had a potential diagnostic value for OA. This suggested that TFAM played an important role in the bone differentiation of OA.
According to KEGG enrichment analysis, we found that Wnt was a significantly enriched signalling pathway. The Wnt signalling pathway is inhibited under normal circumstances but promotes the development of OA once activated.48 Activation of Wnt signalling pathway in chondrocytes induces cartilage damage.49 Furthermore, overactivation of Wnt signalling pathways stimulates the expression of chondrocyte hypertrophic markers and promotes apoptosis.50 Therefore, understanding the mechanism that regulates Wnt signalling in OA can provide new keys to control cartilage formation. Additionally, modifying the WNT pathway has been a target of therapeutic development for OA.51
There are limitations to this study. First, the effective mechanism of identified DEGs in OA is needed for further research. Second, larger numbers of OA samples are needed to further demonstrate the expression of identified genes.
In summary, we found a series of DEGs (USP46, CPVL, FKBP5, FOSL2, GADD45B, PTGS1, ZNF423, ADAMTS1, and TFAM) that played an important role in the pathology of OA. Interestingly, CPVL and TFAM had an important role in the diagnosis of OA. Our findings can better help understanding regulatory mechanisms of OA.
Footnotes
Author Contributions: X. Zhang: Manuscript writing.
Y. Bu: Experiment conduction and data analysis.
B. Zhu: Experiment conduction and data analysis.
Q. Zhao: Experiment conduction and data analysis.
Z. Lv: Experiment conduction and data analysis.
B. Li: Experiment conduction and data analysis.
J. Liu: Manuscript subject design.
Conflict of Interest Statement: None declared
Follow us @BoneJointRes
Supplementary Material
A full list of abbreviations used in the paper is available alongside the online version of this paper.
Funding Statement
None declared
References
- 1. Roughley PJ. Articular cartilage and changes in arthritis: noncollagenous proteins and proteoglycans in the extracellular matrix of cartilage. Arthritis Res 2001;3:342-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eyre D. Collagen of articular cartilage. Arthritis Res 2002;4:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuettner KE, Cole AA. Cartilage degeneration in different human joints. Osteoarthritis Cartilage 2005;13:93-103. [DOI] [PubMed] [Google Scholar]
- 4. Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet 2005;365:965-973. [DOI] [PubMed] [Google Scholar]
- 5. Grynpas MD, Alpert B, Katz I, Lieberman I, Pritzker KP. Subchondral bone in osteoarthritis. Calcif Tissue Int 1991;49:20-26. [DOI] [PubMed] [Google Scholar]
- 6. Brandt KD, Radin EL, Dieppe PA, van de, Putte L. Yet more evidence that osteoarthritis is not a cartilage disease. Ann Rheum Dis 2006;65:1261-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pearle AD, Scanzello CR, George S, et al. Elevated high-sensitivity C-reactive protein levels are associated with local inflammatory findings in patients with osteoarthritis. Osteoarthritis Cartilage 2007;15:516-523. [DOI] [PubMed] [Google Scholar]
- 8. Benito MJ, Veale DJ, FitzGerald O, van den Berg WB, Bresnihan B. Synovial tissue inflammation in early and late osteoarthritis. Ann Rheum Dis 2005;64:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith MD, Triantafillou S, Parker A, Youssef PP, Coleman M. Synovial membrane inflammation and cytokine production in patients with early osteoarthritis. J Rheumatol 1997;24:365-371. [PubMed] [Google Scholar]
- 10. Saito I, Koshino T, Nakashima K, Uesugi M, Saito T. Increased cellular infiltrate in inflammatory synovia of osteoarthritic knees. Osteoarthritis Cartilage 2002;10:156-162. [DOI] [PubMed] [Google Scholar]
- 11. Haynes MK, Hume EL, Smith JB. Phenotypic characterization of inflammatory cells from osteoarthritic synovium and synovial fluids. Clin Immunol 2002;105:315-325. [DOI] [PubMed] [Google Scholar]
- 12. Tatebe M, Iwatsuki K, Hirata H, et al. Effects of depression and inflammatory factors on chronic conditions of the wrist. Bone Joint J 2016;98-B:961-968. [DOI] [PubMed] [Google Scholar]
- 13. Hulejová H, Baresová V, Klézl Z, et al. Increased level of cytokines and matrix metalloproteinases in osteoarthritic subchondral bone. Cytokine 2007;38:151-156. [DOI] [PubMed] [Google Scholar]
- 14. Gómez R, Conde J, Scotece M, et al. What’s new in our understanding of the role of adipokines in rheumatic diseases? Nat Rev Rheumatol 2011;7:528-536. [DOI] [PubMed] [Google Scholar]
- 15. Xiao X, Hao J, Wen Y, et al. Genome-wide association studies and gene expression profiles of rheumatoid arthritis: an analysis. Bone Joint Res 2016;5:314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. No authors listed. Geo Database. http://www.ncbi.nlm.nih.gov/geo/ (date last accessed 22 March 2018).
- 17. Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J 2007;49:107-126. [DOI] [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc. 1995;57:289-300. [Google Scholar]
- 19. Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Research 2012;40:478-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nogales-Cadenas R, Carmona-Saez P, Vazquez M, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 2009;37:317-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 2007;8:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. No authors listed. Gene Ontology Annotation. http://www.geneontology.org/ (date last accessed 17 January 2018).
- 23. No authors listed. Kyoto Encyclopedia of Genes and Genomes pathway analysis. http://www.genome.jp/kegg/pathway.html (date last accessed 17 January 2018).
- 24. No authors listed. BioGRID. http://thebiogrid.org (date last accessed 17 January 2018).
- 25. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. No authors listed. ExPASy Bioinformatics Resource Portal, http://web.expasy.org/pROC/ (date last accessed 22 March 2018).
- 27. Woetzel D, Huber R, Kupfer P, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther 2014;16:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henrotin Y, Lambert C. Chondroitin and glucosamine in the management of osteoarthritis: an update. Curr Rheumatol Rep 2013;15:361. [DOI] [PubMed] [Google Scholar]
- 29. Wang Q, Rozelle AL, Lepus CM, et al. Identification of a central role for complement in osteoarthritis. Nat Med 2011;17:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fischer R, Trudgian DC, Wright C, et al. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Molecular & cellular proteomics 2012;11:M111013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimada H, Otero M, Tsuchimochi K, et al. CCAAT/enhancer binding protein β (C/EBPβ) regulates the transcription of growth arrest and DNA damage-inducible protein 45 β (GADD45β) in articular chondrocytes. Pathol Res Pract 2016;212:302-309. [DOI] [PubMed] [Google Scholar]
- 32. Ijiri K, Zerbini LF, Peng H, et al. Differential expression of GADD45beta in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum 2008;58:2075-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burleigh A, Chanalaris A, Gardiner MD, et al. Joint immobilization prevents murine osteoarthritis and reveals the highly mechanosensitive nature of protease expression in vivo. Arthritis Rheum 2012;64:2278-2288. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu-Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor γ coactivator 1α. Arthritis Rheumatol 2015;67:2141-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harnett MM, Melendez AJ, Harnett W. The therapeutic potential of the filarial nematode-derived immunodulator, ES-62 in inflammatory disease. Clin Exp Immunol 2010;159:256-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swingler TE, Waters JG, Davidson RK, et al. Degradome expression profiling in human articular cartilage. Arthritis Res Ther 2009;11:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thakur M, Dawes JM, McMahon SB. Genomics of pain in osteoarthritis. Osteoarthritis Cartilage 2013;21:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dey P, Panga V, Raghunathan S. A cytokine signalling network for the regulation of inducible nitric oxide synthase expression in rheumatoid arthritis. PLoS One 2016;11:e0161306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Smaele E, Zazzeroni F, Papa S, et al. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature 2001;414:308-313. [DOI] [PubMed] [Google Scholar]
- 40. De Groof A, Ducreux J, Humby F, et al. Higher expression of TNFα-induced genes in the synovium of patients with early rheumatoid arthritis correlates with disease activity, and predicts absence of response to first line therapy. Arthritis Res Ther 2016;18:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prochazkova M, Zanvit P, Dolezal T, Prokesova L, Krsiak M. Increased gene expression and production of spinal cyclooxygenase 1 and 2 during experimental osteoarthritis pain. Physiol Res 2009;58:419-425. [DOI] [PubMed] [Google Scholar]
- 42. Knorth H, Dorfmüller P, Lebert R, et al. Participation of cyclooxygenase-1 in prostaglandin E2 release from synovitis tissue in primary osteoarthritis in vitro. Osteoarthritis Cartilage 2004;12:658-666. [DOI] [PubMed] [Google Scholar]
- 43. Guo B, Greenwood PL, Cafe LM, et al. Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC Genomics 2015;16:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davidson RK, Waters JG, Kevorkian L, et al. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther 2006;8:R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goljanek-Whysall K, Iwanejko LA, Vasilaki A, Pekovic-Vaughan V, McDonagh B. Ageing in relation to skeletal muscle dysfunction: redox homoeostasis to regulation of gene expression. Mamm Genome 2016;27:341-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Twine NA, Harkness L, Kassem M, Wilkins MR. Transcription factor ZNF25 is associated with osteoblast differentiation of human skeletal stem cells. BMC Genomics 2016;17:872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu-Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor γ coactivator 1α. Arthritis Rheumatol 2015;67:2141-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Corr M. Wnt-beta-catenin signaling in the pathogenesis of osteoarthritis. Nat Clin Pract Rheumatol 2008;4:550-556. [DOI] [PubMed] [Google Scholar]
- 49. Burr DB, Gallant MA. Bone remodelling in osteoarthritis. Nat Rev Rheumatol 2012;8:665-673. [DOI] [PubMed] [Google Scholar]
- 50. Goldring MB. Articular cartilage degradation in osteoarthritis. HSS J 2012;8:7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beyer C, Schett G. Pharmacotherapy: concepts of pathogenesis and emerging treatments. Novel targets in bone and cartilage. Best Pract Res Clin Rheumatol 2010;24:489-496. [DOI] [PubMed] [Google Scholar]