Activity of endogenous protein kinase A (PKA) could never be analyzed directly in the cellular environment. Isensee et al. used antibodies to quantify conformational changes leading to an open conformation of endogenous PKA-II holoenzymes, which allowed them to analyze and model its activation cycle in primary sensory neurons.
Abstract
Type II isoforms of cyclic adenosine monophosphate (cAMP)–dependent protein kinase A (PKA-II) contain a phosphorylatable epitope within the inhibitory domain of RII subunits (pRII) with still unclear function. In vitro, RII phosphorylation occurs in the absence of cAMP, whereas staining of cells with pRII-specific antibodies revealed a cAMP-dependent pattern. In sensory neurons, we found that increased pRII immunoreactivity reflects increased accessibility of the already phosphorylated RII epitope during cAMP-induced opening of the tetrameric RII2:C2 holoenzyme. Accordingly, induction of pRII by cAMP was sensitive to novel inhibitors of dissociation, whereas blocking catalytic activity was ineffective. Also in vitro, cAMP increased the binding of pRII antibodies to RII2:C2 holoenzymes. Identification of an antibody specific for the glycine-rich loop of catalytic subunits facing the pRII-epitope confirmed activity-dependent binding with similar kinetics, proving that the reassociation is rapid and precisely controlled. Mechanistic modeling further supported that RII phosphorylation precedes cAMP binding and controls the inactivation by modulating the reassociation involving the coordinated action of phosphodiesterases and phosphatases.
Introduction
cAMP-dependent protein kinase A (PKA) was discovered nearly 50 yr ago (Walsh et al., 1968). The PKA catalytic (C) subunit was the first protein kinase to be crystalized and has been studied in great detail as a prototypic serine/threonine kinase (Walsh et al., 1968; Knighton et al., 1991; Taylor et al., 2012). All PKA isoforms are tetramers composed of a regulatory (R) dimer (R2) and two C-subunits activated by cAMP. Nevertheless, key aspects such as the process of activation and inactivation in the complex cellular environment still remain unclear (Smith et al., 2017).
PKA isoforms are important regulators of cell biological functions such as cell differentiation, survival, memory formation, and pain sensitization. R2:C2 holoenzymes are classified according to the R-subunit isoform (RIα, RIβ, RIIα, and RIIβ) as PKA-I and PKA-II, respectively. Binding of the C-subunit to the inhibitory sites of the respective R-subunit renders the kinase inactive, whereas allosteric binding of cAMP to two C-terminal tandem cAMP-binding domains (CNB-A and CNB-B) of the R-subunits unleashes the catalytic activity of the holoenzyme (Taylor et al., 1990, 2012; Herberg et al., 1996). Numerous early in vitro experiments showed that cAMP causes the physical separation of C- and R-subunits (Tao et al., 1970; Reimann et al., 1971; Corbin et al., 1972; Rubin et al., 1972; Beavo et al., 1974). Studies under more physiological conditions, however, suggested that cAMP rather induces an isoform-specific conformational change that opens the holoenzyme and is modulated by substrates (Johnson et al., 1993; Yang et al., 1995; Vigil et al., 2004). Supporting this, even fusion proteins of R- and C-subunits can reconstitute PKA functionality in cells (Smith et al., 2017). Therefore, at least for PKA-II, the model of full dissociation may not completely reflect the reality in cells. It remains to be clarified whether endogenous PKA-II is activated by full separation or partial dissociation only involving a conformational change leading to a more open conformation.
The type I R-subunits inhibit C-subunits via nonphosphorylatable pseudosubstrates (RRxA/G). In contrast, inhibitory sites of RII-subunits are phosphorylatable substrates of the C-subunits (RRXS). Commonly, kinases quickly release their phosphorylated substrate to allow access to novel substrates. Indeed, phosphorylation of RII has been found to reduce the binding affinity to C-subunits (Erlichman et al., 1974; Zimmermann et al., 1999). Thus, it was assumed that activation of PKA-II occurred by (1) binding of cAMP to the R-subunits, followed by (2) phosphorylation of the inhibitory sites, leading to (3) the release of the then active C-subunits from the holoenzyme (Taylor et al., 1990). But recently, studies using cell homogenates and crystals of the PKA-RIIβ tetrameric holoenzyme (RIIβ2:C2) suggested that RII-subunits may skip the second step and are autophosphorylated already in the absence of cAMP (Manni et al., 2008; Zhang et al., 2012). Indeed, RIIβ-subunits are entirely phosphorylated while trapped in the inactive closed RIIβ2:C2 holoenzyme in RIIβ2:C2 crystals (Zhang et al., 2015). Thus, opening of the holoenzyme may be selectively controlled by cAMP and surrounding divalent ions but less, if at all, by phosphorylation of inhibitory sites. This suggests that the conserved phosphosite may be important for other aspects of PKA-II regulation, e.g., the process of inactivation during reassociation of RII- and C-subunits.
Whether instantaneous RII autophosphorylation occurs only in vitro or also in the cellular cytoplasm is currently unknown and challenging to investigate. Conflicting results have been reported. We have recently established an assay based on an antibody recognizing the phosphorylated RII inhibitory sites (pRII; Isensee et al., 2014a). In contrast to the recent biochemical data, cAMP analogues induced an immediate increase of pRII immunoreactivity in sensory neurons and glia cells, which we interpreted as a cAMP-induced increase of phosphorylated RII (Isensee et al., 2014a). However, the opposite has also been reported. Western blot analysis of cardiomyocyte lysates suggested a cAMP-induced reduction of RII phosphorylation (Manni et al., 2008). How could these conflicting data be reconciled? The decrease of RII phosphorylation in cardiomyocyte lysates could well be attributed to a lack of phosphatase inhibitors during cell lysis (Manni et al., 2008). The cAMP-dependent increase of the pRII immunoreactivity in intact cells, however, might indicate either increased phosphorylation of RII or alternatively increased accessibility of the otherwise hidden pRII epitope. An antibody sensitive for conformational changes resulting in opening of PKA-II holoenzymes would be an ideal tool to monitor the activity of endogenous PKA-II in fixed cells.
Therefore, we set out to identify the mechanism of endogenous PKA-II activation in nociceptors. These neurons initiate the sensation of painful stimuli and make up ∼60% of sensory neurons within the dorsal root ganglion (DRG). Nociceptors specifically express high levels of PKA-RIIβ and show pRII responses to various G protein–coupled receptor (GPCR) agonists (Isensee et al., 2014a,b, 2017a,b). In contrast, the PKA-RIIα isoform was detected at lower levels in all cells of the DRG. We found that RII phosphorylation is independent of cAMP-based activation and occurs already in the inactive PKA-II holoenzyme. Thus, the cAMP-dependent pRII increase in intact cells reflects the conformational state of the given holoenzyme rather than the degree of RII phosphorylation. To prove this, we characterized novel cell membrane–permeable cAMP antagonists and identified an antibody detecting preferentially the active C-subunit. The consistency of our data was tested by constructing a mechanistic mathematical model, which resembled all experimental data and could be applied to study the impact of phosphatase activity on the inactivation of PKA-II signaling.
Results
cAMP increases pRII immunoreactivity in intact cells, but not cell lysates
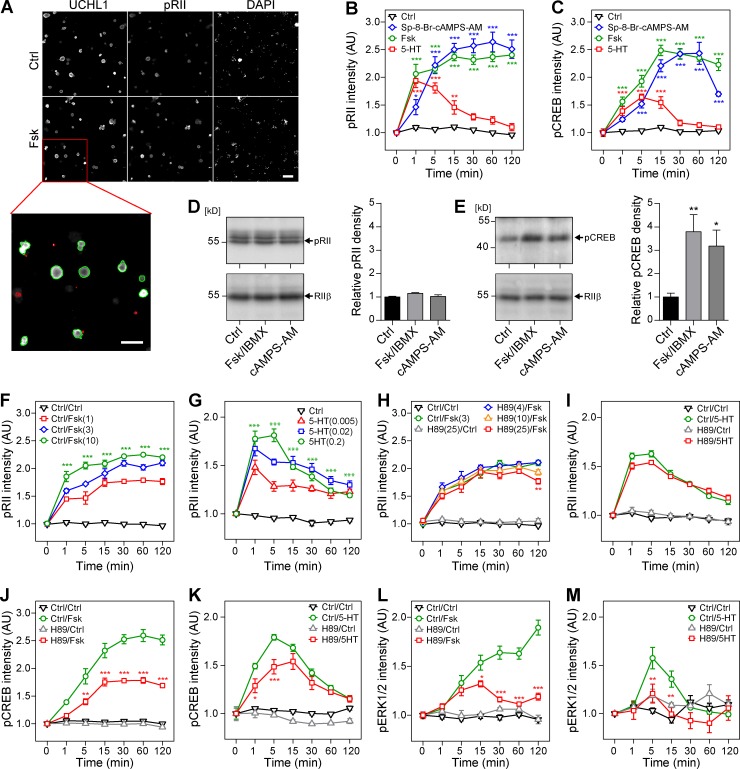
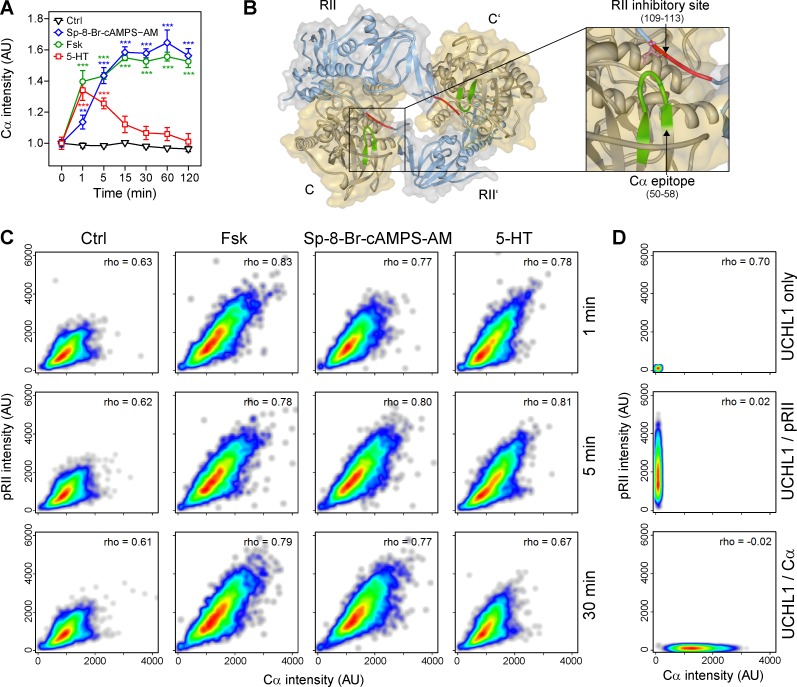
To detect the phosphorylated form of R-subunits RIIα and RIIβ (pRII), we recently introduced a high-content screening (HCS) microscopy approach to quantify endogenous cAMP/PKA dynamics in sensory neurons (Isensee et al., 2014a,b, 2017a). This system acquires thousands of images of labeled DRG neurons in multiwell plates. Neurons are then identified based on the expression of ubiquitin C-terminal hydrolase isozyme L1 (UCHL1) and selection parameters optimized for the sphere-like geometry of neurons after short-term culture (Fig. 1 A). Then, signal intensities (e.g., of pRII immunocytochemical stainings) are quantified within the neuronal regions. We reported previously that the pRII assay allows the analysis of stimulatory and inhibitory GPCR signaling at physiological concentrations (summarized in Table 1). Whereas stimulation with the adenylyl cyclase (AC) activator forskolin (Fsk; 10 µM) or the cell-permeable cAMP analogue Sp-8-Br-cAMPS-AM (10 µM) resulted in long-lasting pRII increase (Fig. 1, A and B), transient induction of cAMP formation by GPCR ligands such as serotonin (5-HT; 200 nM) led to a transient pRII response (Fig. 1 B).
Figure 1.
cAMP induces pRII immunoreactivity in intact sensory neurons, but not cell lysates. (A) Representative HCS microscopy images of control (Ctrl; 0.1% DMSO) and 10 µM Fsk-stimulated (15 min) rat sensory neurons. Cultures were stained with UCHL1 to identify the neurons and pRII to quantify PKA-II signaling. Green/red encircled neurons indicate automatically selected/rejected objects. Bar, 100 µm. (B) Time course of pRII intensity in sensory neurons after stimulation with Ctrl (0.1% DMSO), 10 µM Fsk, 10 µM Sp-8-Br-cAMPS-AM, or 200 nM 5-HT. (C) Time course of pCREB (Ser133) intensity. (D) Immunoblot of sensory neuron lysates probed with pRII and RIIβ antibodies (left) and densitometry results (right). The pRII antibody recognized phosphorylated RIIα and RIIβ. Stimulation with 10/100 µM Fsk/IBMX or 10 µM Sp-8-Br-cAMPS-AM for 15 min did not increase the pRII density. (E) Immunoblot and densitometry results of the same cell lysates probed with the pCREB antibody. (F and G) Time course of pRII intensity after stimulation with different doses of 1–10 µM Fsk or 5–200 nM 5-HT. (H and I) Pretreatment of sensory neurons with 4–25 µM H89 (30 min) did not inhibit the pRII response to 3 µM Fsk (H) or 200 nM 5-HT (I). (J and K) Pretreatment with 25 µM H89 (30 min) significantly inhibited the induction of CREB phosphorylation by 10 µM Fsk (H) or 200 nM 5-HT (K). (L and M) Pretreatment with H89 also inhibited the induction of ERK1/2 phosphorylation. HCS results are means ± SEM; n = 3–4; >1,000 neurons/condition; two-way ANOVA with Bonferroni’s test. Immunoblot results are means ± SEM; n = 6; one-way ANOVA with Dunnett’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Kinetic experiments are plotted in nonlinear scale.
Table 1. Summary of GPCR agonists and cAMP analogs analyzed using the pRII assay in rat or mouse sensory neurons.
| Compound | Action | Lipophilicity (log K′g)a | EC50 or IC50 for ratb | EC50 or IC50 for mouseb | Reference |
|---|---|---|---|---|---|
| 5-HT | Gs-GPCR agonist | 14 nM | 47 nM | Isensee et al., 2014a, 2017a | |
| Prostaglandin I2 | Gs-GPCR agonist | 74 nM | 27 nM | Isensee et al., 2014a, 2017a | |
| Prostaglandin D2 | Gs-GPCR agonist | 377 nM | Isensee et al., 2014b | ||
| DAMGOc | Gi-GPCR agonist | 321 nM | Isensee et al., 2017a | ||
| Fentanyl | Gi-GPCR agonist | 97 nM | 36 nM | Isensee et al., 2017a | |
| Fsk | Activator | 1.7 µM | 2.6 µM | Isensee et al., 2014a, 2017a | |
| Sp-8-Br-cAMPS | Activator | 1.66 | 203 µM | This study | |
| Sp-8-Br-cAMPS-AM | Activator | 1.3 µM | 6.8 µM | Isensee et al., 2014a, 2017a | |
| Rp-cAMPS | Inhibitor | 1.21 | Schwede et al., 2015 | ||
| Rp-cAMPS-pAB | Inhibitor | ∼10 µM | This study | ||
| Rp-8-pCPT-cAMPS | Inhibitor | 2.72 | This study | ||
| Rp-8-pCPT-cAMPS-pAB | Inhibitor | ∼4 µM | This study | ||
| Rp-8-Br-cAMPS | Inhibitor | 1.47 | This study | ||
| Rp-8-Br-cAMPS-pAB | Inhibitor | ∼3 µM | This study |
Lipophilicity (log K′g values) according to reversed-phase HPLC protocol (Krass et al., 1997).
Half maximal effective concentration (EC50) or half maximal inhibitory concentration (IC50) determined using the pRII assay in rat or mouse sensory neurons.
DAMGO, [D-Ala2, N-MePhe4, Gly-ol]-enkephalin.
To corroborate that the pRII increase reflects activation of PKA, we analyzed changes of a prototypical PKA downstream substrate, i.e., phosphorylation of the cAMP response-element–binding protein at Ser133 (pCREB). We have previously shown that the induction of pCREB by 5-HT is sensitive to the PKA inhibitor H89, but not to inhibitors of the ERK1/2 cascade, supporting that Ser133 phosphorylation is PKA dependent in sensory neurons (Isensee et al., 2014a). As kinases require time to perform substrate phosphorylation, the increase of pCREB intensity was slightly delayed compared with the pRII kinetic (Fig. 1 C).
These results suggest that changes in pRII intensity follow changes in cytoplasmic cAMP levels and thus reflect PKA-II activity. Nevertheless, the detected change in pRII intensity could have two underlying mechanisms: (1) the phosphorylation of the R-subunit occurs during the process of PKA activation, or (2) the R-subunit is constitutively phosphorylated and only accessible to the detecting antibody after PKA activation. To differentiate between these two mechanisms, we analyzed cell lysates for activation-induced changes in RII phosphorylation by immunoblotting. As denatured proteins were separated, this allowed us to evaluate for changes of phosphorylated RII independent of potentially masking protein–protein interactions. To induce substantial cellular cAMP accumulation, sensory neurons were stimulated for 15 min either with 10 µM Fsk while inhibiting cAMP degradation with the nonspecific phosphodiesterase (PDE) inhibitor 3-isobutyl-1-methylxanthine (IBMX; 100 µM, 10 min pretreatment) or with the PDE-resistant cAMP analogue Sp-8-Br-cAMPS-AM (10 µM), respectively. To prevent dephosphorylation of RII-subunits, cell lysis was performed using a buffer containing high concentrations of urea, sodium dodecylsulfate, and phosphatase inhibitors. In Western blots for pRII, we observed two bands with the expected molecular weights of RIIα and RIIβ (Fig. 1 D). In contrast to the analysis of pRII intensities in intact cells, however, densitometry of the Western blot signals did not reveal increased RII phosphorylation (Fig. 1 D), even though PKA was activated, as shown by induction of CREB phosphorylation (Fig. 1 E).
ATP-site inhibitors of PKA fail to block increase in pRII immunoreactivity but inhibit PKA downstream signaling
To explain the inconsistency between immunocytochemistry and Western blotting, we hypothesized that RII subunits are fully phosphorylated in the inactive RII2:C2 complex but the accessibility of the pRII epitope for the detecting antibody increased after cAMP-induced dissociation. To test this hypothesis, we first determined the dynamic range of the assay. The pRII increase by stimulation with Fsk or 5-HT was dose dependent and reached maximal values approximately twofold higher than baseline (Fig. 1, F and G). Inhibition of cAMP degradation with 100 µM IBMX (10 min) led to elevated basal pRII intensity but did not increase the response to Fsk, indicating that the maximal response is not limited by PDE activity (Fig. S1 A). The transient response to 5-HT, however, was substantially prolonged by IBMX, showing that the pRII decline depends on PDE activity after rapid deactivation of 5-HT4 receptors (Fig. S1 B; Isensee et al., 2017b).
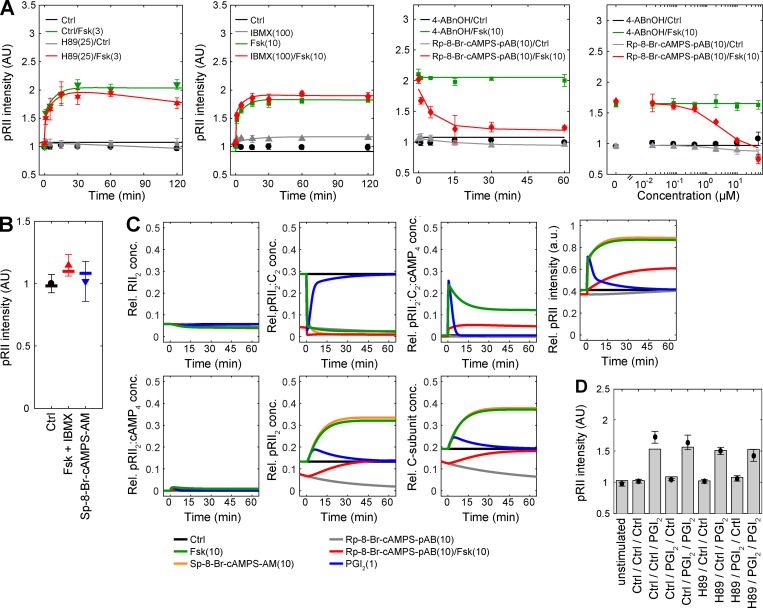
Assuming that RII-subunits are fully phosphorylated in the inactive holoenzyme (Zhang et al., 2015), basal and acute induction of pRII intensity should be rather resistant against inhibitors of catalytic activity such as H89. To test this, we pretreated the sensory neurons for 30 min with 4–25 µM H89 followed by stimulation with a low dose of Fsk (3 µM). Indeed, H89 inhibited neither basal nor induced pRII intensity (Fig. 1 H). Also the transient response to 200 nM 5-HT was unchanged by H89 (Fig. 1 I). To confirm that H89 inhibited PKA, we analyzed the effect of H89 on PKA-dependent downstream signaling such as phosphorylation of CREB and cross talk to the MAPK cascade (Isensee et al., 2014a, 2017b). As expected, H89 substantially reduced CREB and ERK1/2 phosphorylation induced by Fsk or 5-HT (Fig. 1, J–M). As H89 represents a rather unspecific ATP-site inhibitor (Murray, 2008), we tried to verify this using the structurally unrelated ATP-site inhibitor KT5720 and a peptide derived from the endogenous protein kinase inhibitor (myristoylated PKI 14–22). Both compounds, however, failed to inhibit not only the pRII increase but also PKA downstream signaling, most likely because of low cell permeability or rapid degradation (Fig. S1, D–K).
Cell-permeable Rp-isomers inhibit the opening of PKA isoenzymes
To further corroborate that phosphorylated RII-subunits are not fully accessible for antibodies in the inactive PKA-II holoenzyme, we intended to inhibit cAMP-dependent conformational changes rather than inhibition of the catalytic activity. Rp-isomers of adenosine-3′, 5′-cyclic monophosphorothioate (cAMPS) have sulfur substitutions in the equatorial position of the cyclophosphate ring and stabilize the inactive state of the PKA holoenzyme (Rothermel et al., 1983; Bacskai et al., 1993; Gjertsen et al., 1995; Prinz et al., 2006a; Badireddy et al., 2011). They therefore act as competitive cAMP antagonists. Several Rp-isomers, however, did not inhibit Fsk-induced pRII intensity in sensory neurons, probably because of limited cell-permeability (unpublished data). Supporting this, 135-times higher concentrations of the PKA agonist Sp-8-Br-cAMPS were required to reach the same response if compared with the highly cell-permeable acetoxymethyl-ester Sp-8-Br-cAMPS-AM (EC50 = 203 vs. 1.5 µM; Fig. S1 C).
To overcome this limitation of unconjugated Rp-isomers, we were aiming to use novel highly cell-permeable para-acetoxybenzyl (pAB) ester prodrugs of Rp-isomers (Chepurny et al., 2013; Schwede et al., 2015) to block PKA-II in sensory neurons. As the pRII assay, however, specifically detects changes only of PKA-II, but not of PKA-I, we first used bioluminescence resonance energy transfer (BRET) sensors to test the potency of pAB-conjugated Rp-cAMPS, Rp-8-CPT-cAMPS, and Rp-8-Br-cAMPS on all PKA-I and PKA-II isoforms in transfected HEK293 cells. BRET sensors consist of a mutant Renilla luciferase–conjugated R-subunits as bioluminescent donor proteins and GFP-tagged Cα-subunit as acceptor protein (Prinz et al., 2006a,b; Diskar et al., 2007; Schwede et al., 2015). In the inactive PKA complex, close proximity of luciferase and GFP results in energy transfer and a strong BRET signal, which declines after cAMP-induced dissociation.
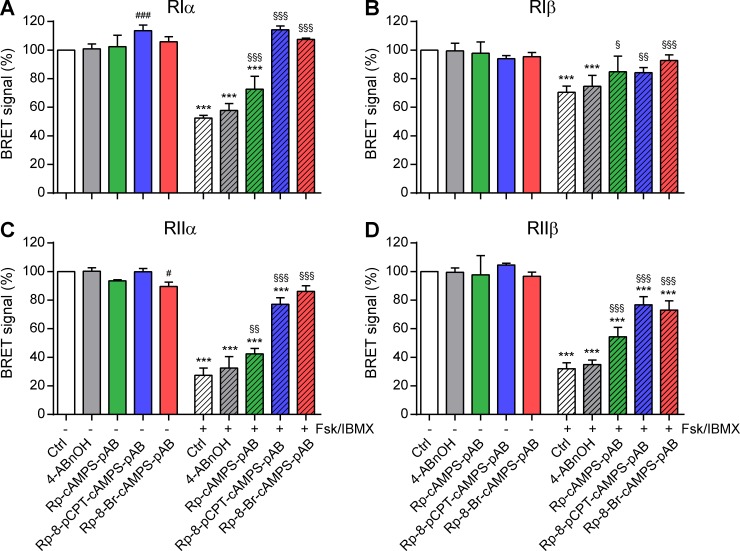
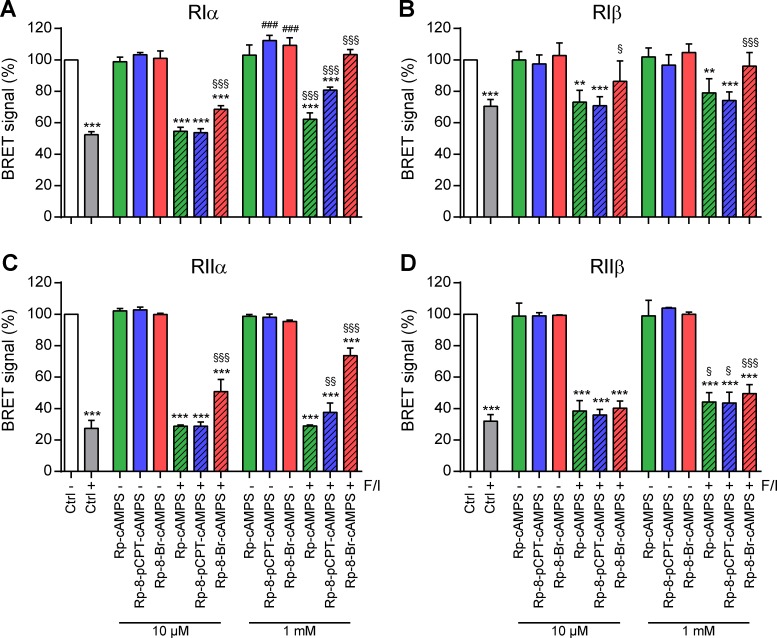
As expected, Fsk/IBMX reduced the BRET signal of PKA-Iα, PKA-Iβ, PKA-IIα, and PKA-IIβ sensors to 51 ± 3%, 75 ± 2%, 25 ± 1%, and 28 ± 3%, respectively (Fig. 2, A–D). Pretreatment with 10 µM Rp-cAMPS-pAB, Rp-8-pCPT-cAMPS-pAB, or Rp-8-Br-cAMPS-pAB effectively inhibited the dissociation of all PKA sensors, whereas the control reagent, pAB alcohol (4-ABnOH), was ineffective. Compared with Rp-cAMPS, modification of the adenine moiety at position eight in Rp-8-pCPT-cAMPS-pAB and Rp-8-Br-cAMPS-pAB increased their potency to inhibit PKA-I and PKA-II isoenzymes, respectively (Fig. 2, A–D, blue/red bars vs. green bar). Verifying that pAB-ester prodrugs crossed the cell membrane more efficiently, much higher doses of their unconjugated parent compounds were required to inhibit the dissociation of PKA sensors (Fig. 3). Even at concentrations of 1 mM, none of the unconjugated Rp-isomers effectively inhibited all PKA isoforms. Substantial inhibition of the PKA-RIIβ isoform was not achieved with any of the unconjugated Rp-isomers (Fig. 3 D). As observed for the pAB-conjugated compounds, modification of the adenine moiety at position eight in Rp-8-pCPT-cAMPS and Rp-8-Br-cAMPS increased their potency.
Figure 2.
pAB ester prodrugs of Rp-isomers are effective inhibitors of PKA. Modifications at position 8 of the adenine nucleobase increases their efficiency. (A–D) HEK293 cells were transfected with isoform-specific BRET sensors composed of mutant Renilla luciferase–conjugated R-subunits hRIα-RLuc8 (A), RIβ-RLuc8 (B), hRIIα-RLuc8 (C), and hRIIβ-RLuc8 (D) as bioluminescent donor proteins and GFP-tagged Cα-subunits as acceptor proteins. Close proximity of luciferase and GFP in the inactive PKA complex results in BRET signal (white bars), which is reduced after activation by Fsk/IBMX (striped white bars, 50/100 µM; 20 min). Pretreatment with 10 µM Rp-8-Br-cAMPS-pAB (green), Rp-8-pCPT-cAMPS-pAB (blue), or Rp-8-Br-cAMPS-pAB (red; 15 min) inhibited the reduction in BRET signals, whereas the control, 4-ABnOH (gray), was ineffective. Values are means ± SD; n = 3; one-way ANOVA with Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 indicate significance between control and Fsk-stimulated conditions; §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 between Fsk stimulations in the absence/presence of Rp-isomers; #, P < 0.05; ##, P < 0.01; ###, P < 0.001 between the unstimulated controls in the absence/presence of Rp-isomers.
Figure 3.
High doses of unconjugated Rp-cAMPS isomers are required to inhibit dissociation of PKA isoforms. (A–D) HEK293 cells were transfected with BRET sensors composed of mutant Renilla luciferase–conjugated R-subunits hRIα-RLuc8 (A), RIβ-RLuc8 (B), hRIIα-RLuc8 (C), and hRIIβ-RLuc8 (D) as bioluminescent donor proteins and GFP-tagged Cα-subunits as acceptor proteins. Close proximity of luciferase and GFP in the inactive PKA complex results in BRET signal (white), which is reduced after activation by Fsk/IBMX (gray; 50/100 µM, 20 min). Rp-8-Br-cAMPS (green), Rp-8-pCPT-cAMPS (blue), or Rp-8-Br-cAMPS (red) was applied 15 min before stimulation. Values are means ± SD; n = 3; one-way ANOVA with Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 indicate significance levels between control and Fsk-stimulated conditions; §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 between Fsk-stimulations in the absence/presence of Rp-isomers; #, P < 0.05; ##, P < 0.01; ###, P < 0.001 between the unstimulated controls in the absence/presence of Rp-isomers.
Cell-permeable Rp-isomers inhibit the pRII response in sensory neurons
We have demonstrated that the amount of phosphorylated RII-subunits is unchanged in the presence of cAMP in lysates of the DRG and that the cAMP-dependent increase of pRII immunoreactivity is resistant to inhibition of catalytic activity in intact neurons (Fig. 1). This suggests that increased accessibility of the pRII epitope caused by conformational changes of the holoenzyme is key for the pRII assay. Therefore, inhibiting the open conformation of PKA-II using the pAB-conjugated Rp-isomers should block the pRII increase.
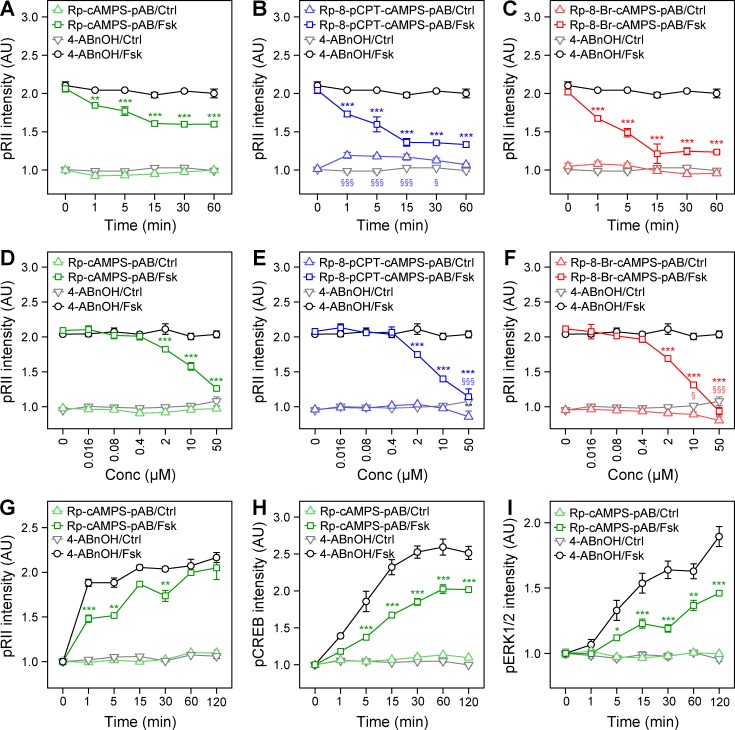
Indeed, pretreating sensory neurons with each of the pAB-conjugated Rp-isomers (10 µM) inhibited the response to 10 µM Fsk (5 min) within 15 min, whereas the control reagent (4-ABnOH) was ineffective (Fig. 4, A–C). Rp-isomers alone (without Fsk treatment) had no impact on the basal pRII intensity, although we observed slight pRII induction by Rp-8-pCPT-cAMPS-pAB (Fig. 4 B). This could indicate trace amounts of the PKA agonist Sp-8-pCPT-cAMPS or unspecific off-target effects (Herfindal et al., 2014). The inhibition of Fsk-induced pRII immunoreactivity by Rp-isomers was dose dependent (Fig. 4, D–F). In line with the BRET experiments, modifications at position eight of the adenine residue increased their potency. Estimated IC50 values were 10, 4, and 3 µM for Rp-cAMPS-pAB, Rp-8-pCPT-cAMPS-pAB, and Rp-8-Br-cAMPS-pAB, respectively.
Figure 4.
Cell-permeable Rp-isomers inhibit the induction of pRII intensity and PKA downstream signaling in sensory neurons. (A–C) HCS microscopy analyses of Fsk-induced (10 µM, 5 min) pRII intensity after pretreatment of sensory neurons for up to 60 min with 10 µM Rp-cAMPS-pAB (A), Rp-8-pCPT-cAMPS-pAB (B), or Rp-8-Br-cAMPS-pAB (C). 10 µM 4-ABnOH served as negative control. (D–F) Dose–response experiments after 30 min pretreatment with Rp-isomers followed by stimulation with 10 µM Fsk (5 min). (G–I) Pretreatment with 10 µM Rp-cAMPS-pAB (30 min) effectively inhibited the pRII increase (G) and phosphorylation of CREB (H) and ERK1/2 (I). Values are means ± SEM; n = 3–4; >1,000 neurons/condition; two-way ANOVA with Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 indicate significance levels between Fsk-stimulated conditions; §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 indicate significance levels between basal conditions.
We also measured phosphorylation of ERK1/2 in the very same cells to analyze whether Rp-isomers also block PKA-dependent cross talk to the MAP cascade (Isensee et al., 2017b). Surprisingly, Rp-8-pCPT-cAMPS-pAB and Rp-8-Br-cAMPS-pAB with modifications at position eight of the adenine residue strongly induced ERK1/2 phosphorylation, whereas Rp-cAMPS-pAB had an inhibitory effect to concentrations up to 10 µM (Fig. S4, A–F). This induction of ERK activity was accompanied by a significant cell loss, indicating rapid detachment of sensory neurons (Fig. S4, G–I). We therefore applied Rp-cAMPS-pAB to study the inhibition of PKA downstream signaling. In contrast to H89 but in line with its role in maintaining the PKA holoenzyme in a closed conformation, pretreatment with 10 µM Rp-cAMPS-pAB (30 min) caused long-lasting inhibition of Fsk-induced pRII immunoreactivity (Fig. 4 G). As expected, phosphorylation of CREB and ERK1/2 was also inhibited by Rp-cAMPS-pAB (Fig. 4, H and I).
Presence of cAMP increases the binding of pRII antibodies to PKA-II
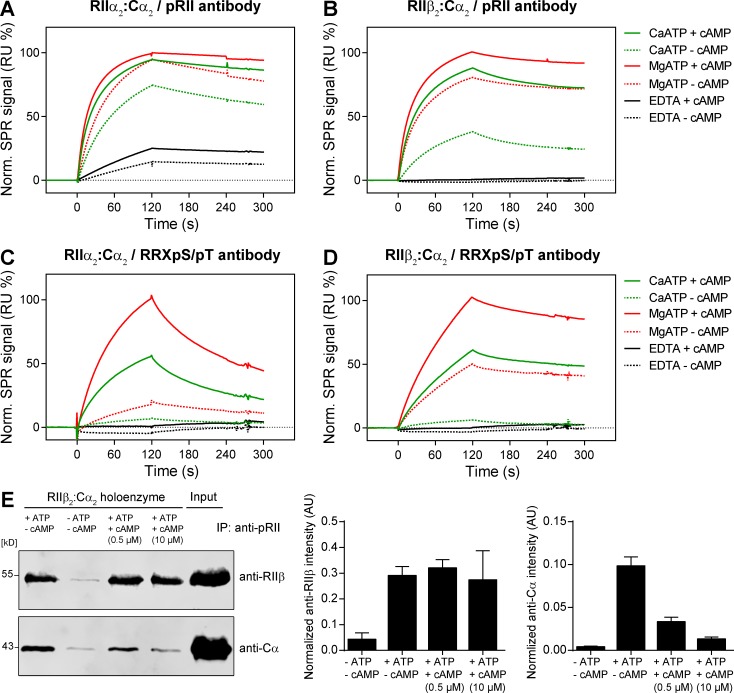
To prove that pRII antibody binding is increased by cAMP-induced conformational changes of the PKA holoenzyme, we applied surface plasmon resonance (SPR) spectroscopy and quantified the effect of cAMP on the binding of PKA-II holoenzymes to pRII antibodies. Therefore, we immobilized pRII antibodies covalently to a sensor chip and assessed the binding of preformed recombinant RIIα2:C2 and RIIβ2:C2 holoenzymes in buffer containing 1 mM MgCl2 and 0.5 mM ATP. Dissociation of holoenzymes was induced by adding cAMP (10 µM) to the preformed RII2:C2 holoenzymes before SPR measurements.
We found that cAMP increased the binding of both PKA-II isoenzymes to immobilized pRII antibodies (Fig. 5, A and B). The cAMP-induced change in binding signal was stronger for RIIβ2:Cα2 than for RIIα2:Cα2, which may indicate that the pRII assay is more sensitive for the activation of RIIβ2:Cα2 holoenzymes. In contrast, nonphosphorylated holoenzymes, preincubated in buffer without ATP, showed no (RIIβ) or weak (RIIα) binding, proving the phosphospecificity of the antibody (Fig. 5, dashed black lines). The binding of inactive holoenzymes to pRII antibodies reveals a certain degree of conformational flexibility at the active site even in the absence of cAMP, making the inhibitory sequence of RII accessible for the antibody. We therefore analyzed the effect of Ca2+ instead of Mg2+ ions on the binding efficiency, as previous results indicated that Ca2+ stabilizes the inactive state and thereby slows down cAMP-induced dissociation (Knape et al., 2015; Zhang et al., 2015). Supporting that Ca2+ promotes stabilization of an inactive state and in line with a reduced accessibility of the phosphoepitope, we observed decreased binding to the antibody in the presence of Ca2+. However, Ca2+ predominantly affected cAMP-free holoenzymes, reflected in an even stronger reduction of the binding signal. SPR measurements using another antibody specific for the PKA consensus site RRXpS/pT verified these results. Again, increased binding was observed in presence of cAMP and Ca2+ stabilized the respective holoenzyme (Fig. 5, C and D).
Figure 5.
Presence of cAMP increases the binding of pRII antibodies to PKA-II holoenzymes. (A–D) SPR analysis using pRII- (A-B) or RRXpS/pT-specific antibodies (C and D) covalently immobilized on sensor chips. Binding of preformed RIIα2:Cα2 (A and C) and RIIβ2:Cα2 holoenzymes (B and D) was assessed in buffer containing 1 mM MgCl2 or 1 mM CaCl2 and 0.5 mM ATP. Dissociation of holoenzymes was induced by adding 10 µM cAMP before SPR analysis. (E) Coimmunoprecipitation of RIIβ and Cα using pRII antibodies captured on protein G functionalized beads. Pull-downs of preformed RIIβ2:C2 holoenzymes were performed in the absence or presence of 1 mM ATP or 0.5 and 10 µM cAMP. Representative immunoblots (left) and densitometry results are shown (right). Values are means ± SD; n = 3. IP, immunoprecipitation; RU, resonance units.
The SPR measurements suggest that conformational flexibility of the inactive holoenzyme causes binding of pRII- and RRXpS/pT-specific antibodies to inhibitory sites in the absence of cAMP. To prove this hypothesis, we performed coimmunoprecipitation experiments and incubated preformed RIIβ2:Cα2 holoenzymes with pRII-specific antibodies immobilized on protein G–conjugated beads. As expected, pull-down of RIIβ was ATP dependent (Fig. 5 E). Supporting that conformational flexibility in the inactive RIIβ2:Cα2 holoenzyme allows binding of pRII antibodies, we were able to pull down C-subunits in the absence of cAMP (Fig. 5 E). The presence of cAMP led to dose-dependent reduction of C-subunits in the precipitate, indicating dissociation of the holoenzyme.
C-subunit–specific antibodies against the glycine-rich loop show a cAMP-dependent intensity pattern
The results above indicate that the pRII epitope is not fully accessible for antibodies in the inactive PKA-II holoenzyme. Therefore, epitopes of C-subunits, which are localized face to face with pRII sites, should also be masked in the inactive PKA-II holoenzyme. Antibodies specific for such masked epitopes on the C-subunit are currently unknown but would confirm our results and potentially allow to measure activation of PKA-I isoforms, which lack the pRII epitope. Therefore, we screened several commercially available antibodies specific for C-subunits for changes in signal intensity after activation of the cAMP–PKA pathway. Indeed, a commercial mouse monoclonal antibody, which was generated using the full-length Cα protein as antigen, showed a cAMP-dependent intensity pattern almost identical to the pRII response (Fig. 6 A). In addition, signal intensities by both the pRII- and the Cα-specific antibody strongly correlated on a single-cell level (Fig. 6 C). This correlation was not caused by spillover between fluorescence channels, which we rigorously compensated for (Fig. 6 D; see Materials and methods for details).
Figure 6.
Antibodies binding the glycine-rich loop of C-subunits present a cAMP-dependent intensity pattern in sensory neurons. (A) Time course of Cα intensity after stimulation with Ctrl (0.1% DMSO), 10 µM Fsk, 10 µM Sp-8-Br-cAMPS-AM, or 200 nM 5-HT. (B) Nano–LC-MS/MS was applied to map the epitope of the Cα-specific antibody. The antibody binds to the sequence GTGSFGRVM corresponding to aa 50–58 of C-subunits. This region contains the glycine-rich loop and is localized face to face with the pRII site in the holoenzyme structure (Protein Data Bank accession no. 3TNP; Zhang et al., 2012). (C) Cell density plots showing single-cell data of pRII/Cα–labeled neurons stimulated for 1, 5, or 30 min with 10 µM Fsk, 10 µM Sp-8-Br-cAMPS-AM, or 0.2 µM 5-HT. The Spearman’s rank correlation coefficient is shown. (D) Controls indicating that spillover between fluorescence channels was properly compensated to avoid false correlations. HCS results in A show means ± SEM; n = 3–4; >1,000 neurons/condition; two-way ANOVA with Bonferroni’s test. ***, P < 0.001. Kinetic experiments are plotted in nonlinear scale.
To identify the antibody-binding epitope, we applied a nanoscale liquid chromatography coupled to tandem mass spectrometry (nano–LC-MS/MS) approach and followed two strategies. The first strategy was to use limited proteolysis of the antibody-bound protein. The intrinsically stable antibody remains almost completely intact, whereas the antibody-bound protein is digested, such that only the epitope should remain associated (Fig. S3). For this purpose we made use of the broad-specificity enzyme subtilisin, as it generates peptides of similar length covering the whole protein and enables a fast and reproducible digestion. Cα subunits were incubated with anti–Cα antibodies, digested with subtilisin, filtrated to remove unbound peptides, and treated with trifluoroacetic acid (TFA) to elute the antibody-bound peptides. After LC-MS analysis of the eluate, we found that 44% (479) of all Cα peptide-spectrum matches (PSMs) belonged to the peptide GTGSFGRVM, which is a strong indication that this indeed represents the bound epitope (Fig. S4 A). Analyzing subtilisin-digested PKA-Cα without antibody enrichment, GTGSFGRVM amounted to only 4% of PSMs (Fig. S4 C). As a second strategy to validate the potential epitope, we incubated subtilisin-digested PKA-Cα with the Cα-specific antibody to enrich the epitope-containing peptide. Again, the analysis of the eluate confirmed our previous results, because 80% of the three identified sequences represented the peptide GTGSFGRVM (161 PSMs; Fig. S4 B).
Mapping the peptide to the Cα protein sequence revealed amino acids 50–58 within the N-lobe of the catalytic core (Fig. 6 B). This region contains the glycine-rich loop (GxGxxG) that is highly conserved in eukaryotic protein kinases and essential for binding the phosphoryl groups of ATP (Hemmer et al., 1997). Interestingly, this epitope of the C-subunit is localized face to face with the pRII epitope of RII-subunits within the holoenzyme (Fig. 6 B).
Mechanistic modeling reveals an essential role of phosphatases for regulating PKA-II activity
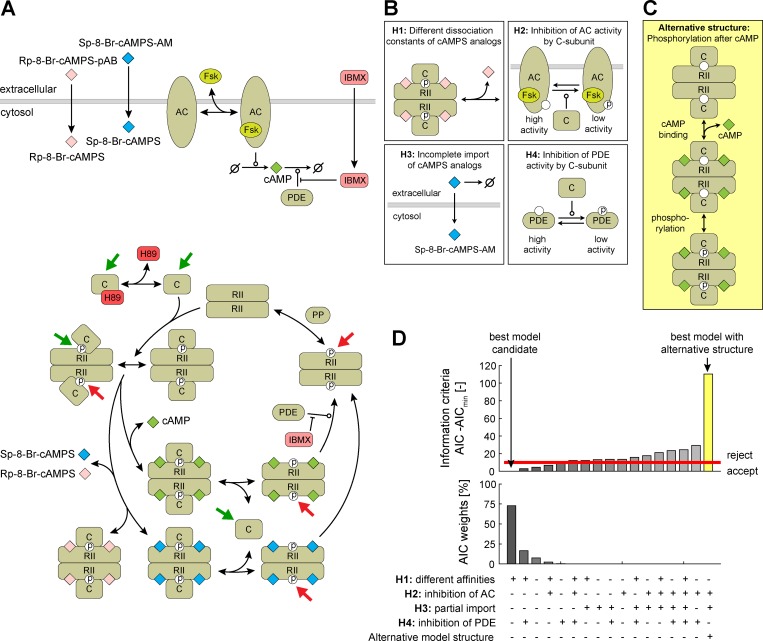
Our experimental results extend the recent biochemical data suggesting that the phosphorylation of the RII epitope precedes cAMP binding also in primary neurons. To verify the consistency of our hypotheses, further dissect specific roles of individual processes, and assess competing hypotheses, we implemented a mechanistic mathematical model of PKA-II activity in response to treatment with Fsk, IBMX, H89, and cAMP analogues (Fig. 7 A). The model was optimized to integrate more than 750 data points observed in more than 60 experimental conditions (see Materials and methods). The experimental data to train the model, the model implementation, best parameters found for each model, and plots of the fitting results are provided (see the online supplemental material modeling file). The model accounts for the reversible binding of Fsk to AC, yielding the complex AC–Fsk with increased cAMP production rate (Fig. 7 A). The passive transport of cAMP analogues across the membrane and the subsequent processing is modeled as a single reaction. Based on our experimental data we assumed that conformational flexibility in the inactive holoenzyme causes switching between a closed and partially open conformation, allowing pRII and Cα antibodies to bind in the absence of cAMP. Free cAMP then binds to the pRII2:C2 complex, resulting in pRII2:C2:cAMP4, which is prone to dissociate into pRIIβ2:cAMP4 and two C-subunits. PDEs degrade free but also pRII2-bound cAMP with different degradation rates, as recently suggested (Moorthy et al., 2011), and are inhibited by IBMX. As binding of the C-subunit to phosphorylated RIIβ2 (pRIIβ2) is weaker (Zhang et al., 2015), we modeled a dephosphorylation event mediated by a phosphatase. C-subunits bind with high affinity only to unphosphorylated RII2. Consistent with the observation that RIIβ2 is quickly phosphorylated after binding of the C-subunit, we assumed that this binding yields pRII2:C2. Released C-subunits are considered as catalytically active if not bound by H89. In addition to cAMP, cAMP analogues can reversibly bind pRII2:C2. Depending on whether these analogues are Sp or Rp analogues acting as agonist or antagonists of PKA, respectively, we assumed the dissociation to occur or to be negligible.
Figure 7.
Mechanistic mathematical modeling supports that RII phosphorylation occurs before opening of PKA-II holoenzymes. (A) Illustration of the core PKA-II model. Colored arrows indicate accessible epitopes recognized by pRII (red) and Cα (green) antibodies, respectively. (B) Illustration of model extensions allowing for differences between cAMP analogues (H1 and H3) and feedback regulation (H2 and H4). (C) Illustration of an alternative model structure assuming that cAMP induces RII phosphorylation. (D) Ranking of model hypotheses according to the AIC value.
We used the mechanistic model to (1) corroborate the experimental results that the pRII epitope is only accessible in an open holoenzyme conformation, (2) analyze differences between cAMP analogues and feedback regulation mechanisms (Fig. 7 B), and (3) test an alternative model structure in which binding of cAMP to RII-subunits induces RII phosphorylation (Fig. 7 C). Regarding the cAMP analogues, we asked whether the dissociation constants of cAMP analogues differ (Fig. 7 B, hypothesis 1 [H1]) or whether the imported fraction differs because of different import rates (H3). Furthermore, we considered negative feedback loops reported in other contexts such as phosphorylation and inhibition of AC and AC–Fsk by active C-subunits (H2; Iwami et al., 1995) and activation of PDEs by active C-subunits (H4; Sette and Conti, 1996). These hypotheses were selected from a large set of possible model extensions, e.g., further modes of regulation (Calejo and Taskén, 2015; Torres-Quesada et al., 2017), as they appeared to be supported by the observed experimental data. The hypotheses were implemented in the model, the parameters of the resulting models were estimated, and the models were compared using the Akaike information criterion (AIC).
Model simulation and evaluation indicates that the pRII antibody indeed predominately binds the open holoenzyme conformation and that the best model candidate accounts for differences in binding affinities of cAMPS analogues (Fig. 7 D). This model provides a good quantitative description of the experimental data generated by immunostaining (Fig. 8 A; see supplemental modeling file) and Western blotting (Fig. 8 B). The alternative model structure assuming that binding of cAMP induces RII phosphorylation (Figs. 7 C and S5 A) led to much larger AIC values and thus could be rejected (Fig. 7 D, yellow bar). Indeed, although the alternative model was able to describe the pRII immunostainings (Fig. S5 C), it failed to fit the Western blot data (Fig. S5 B).
Figure 8.
Mechanistic mathematical modeling describes the experimental data for PKA-II activation. (A) Comparison of experimental data obtained by immunostaining of intact neurons with simulation results for the best model. Experimental data for H89 are also included in Fig. 1 H, for IBMX in Fig. S1, and for Rp-8-Br-cAMPS-pAB in Fig. 4 (C and F). The good agreement indicates the consistency of the best model assumptions. (B) Comparison of experimental data obtained by Western blotting with simulation results. (C) Prediction of the dynamics of selected internal states of the best model. (D) Experimental data for repetitive PGI2 treatment (black data points) and the corresponding model predictions (gray bars) for an assumed PGI2 half-life time of 90 s. Sensory neurons were pretreated for 30 min with 25 µM H89 or solvent (DMSO, 0.1%) followed by stimulation with 1 µM PGI2 for 60 min and then again for 2 min. HCS results in A represent means ± SEM; n = 3–4; >1,000 neurons/condition. Immunoblot results in B represent mean ± SD; n = 6.
We explored the properties of the best model candidate by simulating a series of experimental conditions (Fig. 8 C). These simulations provide predictions for the internal states of the model, which cannot be assessed experimentally. Consistent with our experimental data, the internal states of the model indicate that the concentration of free pRII2 indeed resembles the concentration of free active C-subunits unless bound by H89 (Fig. 8 C).
To validate the mechanistic model, we predicted the response to double perturbation experiments. We pretreated cells with H89 or solvent control and followed the response to PGI2, an agonist of the Gs-coupled IP1 receptor and thereby activator of AC (Fig. 8 D). PGI2 is stable in basic buffers but rapidly hydrolyzed to 6-keto PGF1α at physiological pH (Stehle, 1982). Because the half-life ranges from 30 s to a few minutes, we implemented it in the model as a transient maximal production of cAMP, circumventing the introduction of additional parameters. Although no data for PGI2 or double perturbations were used to train the model, it provided for a range of plausible PGI2 half-lives an accurate prediction of the experimental data verifying the predictive power of the model.
Next, we used the validated model to study the impact of phosphatase activity on the pRII response. At baseline, we found that increased phosphatase activity elevates the level of pRII2:C2 holoenzymes, resulting in decreased pRII and C-subunit concentrations (Fig. 9, Ctrl). As rapid dephosphorylation of pRII by increased phosphatase activity results in faster reassociation of RII and C-subunits, this finding suggests that phosphatase activity controls the basal level of dissociated PKA-II. Indeed, in previous experiments application of the phosphatase inhibitor calyculin A resulted in ever increasing basal pRII intensities in the absence cAMP stimulation (Isensee et al., 2014a). Using the modeling, we now found that an increase of phosphatase-activity also decreased the maximal amplitude of dissociated pRII2 and C-subunit concentrations after cAMP-stimulation (Fig. 9, Fsk). Increased phosphatase activity modulated also the kinetic of the response resulting in faster establishment of equilibrium between the association states (Fig. 9, Fsk). To study the impact of phosphatase-activity on PKA-II deactivation, we modeled the response to the IP1 receptor agonist PGI2. PGI2 induces a rapid but transient production of cAMP (Fig. 9, PGI2). The result of the modeling indicated that increased phosphatase activity also leads to more rapid termination of PKA-II activity.
Figure 9.
Phosphatase activity decreases dissociation of PKA-II at baseline and after stimulation. The effect of increasing phosphatase activity on the internal states of the best model after stimulation with Fsk or PGI2 is shown. Different colors indicate phosphatase activities ranging from a 100-fold reduction (blue lines) to a 100-fold increase (red lines) compared with the estimated phosphatase activity (green lines).
Discussion
Despite the plethora of powerful methods, endogenous PKA activity could never be measured directly in the cellular environment. PKA activity in cells was mainly deduced indirectly from measuring its activator, cAMP, phosphorylation of downstream substrates, or using ectopically expressed reporter proteins (Adams et al., 1991; Bacskai et al., 1993; Zhang et al., 2001; Prinz et al., 2006a; Martin et al., 2007; Smith et al., 2017). PKA is the first kinase, which gets regulated after ligand-induced activation of the huge group of stimulatory and inhibitory G protein–coupled receptors. Thus, it is essential to understand the molecular changes of PKA regulation and its isoform-specific activity in distinct primary cells models, in which PKA isoforms control various rather different biological processes (Taylor et al., 2005, 2012; Pidoux and Taskén, 2010). Although the C-subunit has been studied in great detail to reveal the catalytic mechanism of the AGC kinase family (Taylor and Kornev, 2011; Srivastava et al., 2014; Meharena et al., 2016), its isoform-specific regulation by R-subunits in cells is less investigated. Already the fundamental question of the time point and the function of the phosphorylation of RII inhibitory sites remained to be established in vivo (Zhang et al., 2015).
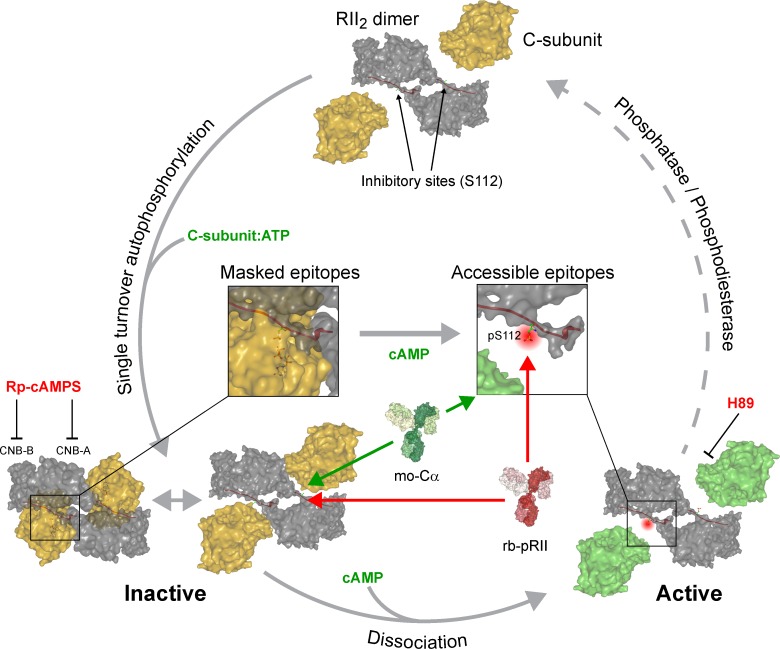
We recently found that pRII intensities follow changes in cAMP levels and correlate with PKA substrate phosphorylation (Isensee et al., 2014a, 2017a,b). This indicated a classical cAMP-induced activation and subsequent phosphorylation of RII. Nevertheless, these data are in disagreement with biochemical data, which argue for a cAMP-independent phosphorylation of RII by C-subunits (Zhang et al., 2012, 2015). We now show that full binding of the pRII-specific antibodies requires cAMP-dependent conformational changes, which increase the accessibility of the pRII epitope (Fig. 10). Therefore, cAMP-dependent changes in pRII intensities are not based on regulation of RII phosphorylation itself but reflect the conformational state of the holoenzyme, which has constitutively phosphorylated RII-subunits.
Figure 10.
Proposed model of PKA-II activation based on quantifying pRII and Cα signal intensities in intact sensory neurons. Our data support that the RII dimer associates with ATP-loaded C-subunits, resulting in a single-turnover autophosphorylation event (Zhang et al., 2015). Phosphorylated RII-subunits remain associated with C-subunits in the inactive holoenzyme. This complex is prone for conformational flexibility at the active site even in the absence of cAMP, making a fraction of RII inhibitor sites (pS112) as well as residues in the catalytic cleft of C-subunits accessible for antibodies. The presence of cAMP then induces full opening of holoenzymes, resulting in increased accessibility for antibodies recognizing pS112 of RII (red) or the glycine-rich loop of C-subunits (green). Reassociation of RII and C-subunits requires the concerted hydrolysis of cAMP by phosphodiesterases and dephosphorylation of S112 by phosphatases and occur immediately after dephosphorylation in a precisely coordinated manner. CNB-A/B, cyclic nucleotide binding domains.
To prove this concept, we applied SPR and found that cAMP indeed increases the binding of RII2:C2 holoenzymes to immobilized pRII antibodies (Fig. 5). However, also the inactive cAMP-free RII2:C2 holoenzyme binds to the pRII antibody to some extent, which we also verified by coimmunoprecipitation experiments (Fig. 5 E). These observations suggest yet-unexpected dynamic conformational changes of PKA-II holoenzymes in the absence of cAMP. This could well explain why we observed substantial basal pRII immunoreactivity in unstimulated intact neurons (Fig. 1 A). The conformational flexibility of PKA holoenzymes is likely constrained after fixation, and the epitope accessibility depends on the captured conformation. The doubling of pRII immunoreactivity after maximal stimulation (Fig. 1 B) would then indicate full opening of holoenzymes in the presence of cAMP.
To study the PKA-II activation cycle in intact neurons, we used novel cell-permeable Rp-isomers of cAMPS, which stabilize the inactive holoenzyme conformation and therefore act upstream compared with classical ATP-site inhibitors (Rothermel et al., 1983; Bacskai et al., 1993; Gjertsen et al., 1995; Prinz et al., 2006a; Badireddy et al., 2011). Using a BRET approach, we demonstrate that esterification of Rp-isomers to pAB-conjugates results in enhanced cell permeability and thereby more effective inhibition of all PKA isoforms in intact HEK293 cells (Figs. 2 and 3). Indeed, pAB-conjugated Rp-isomers also allowed the study of PKA-II signaling in sensory neurons, further demonstrating their value to study cyclic nucleotide signaling in difficult-to-access cell models (Fig. 4; Chepurny et al., 2013; Schwede et al., 2015). The pAB-conjugated Rp-isomers, but not H89, effectively inhibited the pRII increase, whereas both decreased CREB phosphorylation and cross talk to the MAPK cascade (Fig. 1, H–M). This pharmacological approach again corroborates that RII-subunits are phosphorylated in the inactive PKA-II holoenzyme and cAMP-dependent dissociation leads to enhanced accessibility of the pRII epitope.
The conformation dependency of the pRII antibody makes it a powerful tool. It allows for the investigation of the regulation of PKA-II, even in the complex environment of a cell, without the need to disturb the cellular organization (e.g., by transfection of a reporter protein). The characterization of a second conformation-dependent antibody corroborates and extends the possibility of this assay even further. The antibody against the glycine-rich loop potentially expands the assay to PKA-I, because the C-subunit (isoform α or β) is shared by all PKA isoforms. The response to cAMP obtained with pRII- and Cα-specific antibodies shows temporal similarity and correlates at the single-cell level, suggesting predominant activation of PKA-II in sensory neurons (Fig. 6). Because of the almost identical response kinetic, dephosphorylation of RII subunits and reassociation with C-subunits must occur almost simultaneously in vivo. Supporting a local and highly dynamic interplay among ACs, PDEs, and phosphatases, a recent study showed that PKA activation at physiological cAMP concentrations does not lead to full dissociation of PKA-II holoenzymes, yet intact PKA-II tetramers are able to phosphorylate nearby substrates (Smith et al., 2017).
Markov state models were recently applied to gain mechanistic understanding of the activation of the PKA-RIα isoform by cAMP (Boras et al., 2014). With the help of our novel tools, we have now established the first computational model of PKA-II activity in primary sensory neurons (Figs. 7, 8, and 9). The computational analysis not only validated the consistency of our data and hypotheses but also further helped to suggest a role for the phosphorylation site. Although phosphorylation may also play a role in the activation process of PKA-II, the dephosphorylation dynamic defines the amplitude and duration of the PKA-II activity. Thereby, PKA-II integrates not only over the changes of cAMP and its catabolizing enzymes, the phosphodiesterases, but also over the activity of the phosphatase as well as changes of divalent cations. To verify these model predictions, the application of phosphatase inhibitors would be required. Indeed, application of the PP2A/B inhibitor calyculin A resulted in ever increasing basal pRII immunoreactivity (Isensee et al., 2014a). Calyculin A, however, was cytotoxic, leading to rapid detachment of sensory neurons and preventing its use in kinetic experiments. The other phosphatase inhibitors tested so far have been ineffective, and further research is required to verify our model predictions in the future.
In a broader view, our study demonstrates that conformational changes in kinases, especially within the glycine-rich loop and at the inhibitory site, facilitate the quantification of kinase activity in intact cells using classical immunocytochemistry. The development of antibodies specific for homologous epitopes of other kinases may further prove this concept in the future.
Materials and methods
Antibodies
The following antibodies were used in this study: chicken polyclonal anti-UCHL1 (1:2,000; NB110-58872; Novus), rabbit monoclonal anti–phospho-RII (S99; 1:1,000; clone E151; ab32390; Abcam), rabbit monoclonal anti–PKA substrate (RRXpS/pT; lone 100G7E; 9624; Cell Signaling), rabbit monoclonal anti–phospho-CREB pS133 (1:500; clone F.959.4; MA5-11192; Thermo Fisher Scientific), mouse monoclonal anti–Cα (1:500, clone 5B; 610980; BD Transduction Laboratories), mouse monoclonal anti–PKA RIIβ (1:2,000; 610625; BD Transduction Laboratories), and highly cross-adsorbed Alexa Fluor 647–, 594–, and 488–conjugated secondary antibodies (Invitrogen).
Drugs
10 mM 5-HT (in dH2O) was purchased from Sigma-Aldrich. 10 mM fentanyl (in dH2O) and 10 mM Fsk (in DMSO) were from Tocris. cAMP, PO4-AM3, Sp-8-Br-cAMPS, Sp-8-Br-cAMPS-AM, Rp-cAMPS, Rp-8-Br-cAMPS, and Rp-8-pCPT-cAMPS (all 10 mM in DMSO) were from BioLog. Rp-cAMPS-pAB, Rp-8-pCPT-cAMPS-pAB, Rp-8-Br-cAMPS-pAB, and 4-ABnOH (all 10 mM in DMSO) were synthesized and provided by F. Schwede (BioLog).
Animals
Male Sprague–Dawley rats (200–225 g, aged 8–10 wk) were obtained from Harlan. Rats were kept on a 12-h light/dark cycle and provided with food and water ad libitum. All animal experiments were performed in accordance with the German animal welfare law and approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen. Animals were sacrificed between 9 and 12 am by CO2 intoxication. DRGs of rats (L1–L6) were removed within 30 min per animal.
DRG neuron cultures
DRGs were desheathed, pooled, and incubated in Neurobasal A/B27 medium (Invitrogen) containing 10 U/ml collagenase P (1 h, 37°C, 5% CO2; Roche). DRGs were dissociated by trituration with fire-polished Pasteur pipettes. Axon stumps and disrupted cells were removed by BSA gradient centrifugation (15% BSA, 120 g, 8 min). Viable cells were resuspended in NeurobasalA/B27 medium, plated in 0.1 mg/ml poly-l-ornithine/5 µg/ml laminin–precoated 96-well imaging plates (Greiner) and incubated overnight (37°C, 5% CO2). Neuron density was 1,500 neurons/cm2.
Stimulation of DRG neurons
DRG neurons were stimulated 24 h after isolation in 96-well imaging plates. Compounds were dissolved in 12.5 µl PBS in 96-well V-bottom plates, mixed with 50 µl medium from the culture wells, and added back to the same wells. Stimulations were performed with automated 8-channel pipettes (Eppendorf) at low dispense speed on heated blocks; stimulated cells were placed back in the incubator. The cells were fixed for 10 min at RT by adding 100 µl 8% PFA, resulting in a final concentration of 4%.
Immunofluorescence staining
PFA-fixed cells were treated with goat serum blocking (2% goat serum, 1% BSA, 0.1% Triton X-100, and 0.05% Tween 20; 1 h, RT) and incubated with respective primary antibodies diluted in 1% BSA in PBS at 4°C overnight. Subsequent to three washes with PBS (30 min, RT), cells were incubated with secondary Alexa Fluor dye–coupled antibodies (1:1,000, 1 h, RT). After three final washes (30 min, RT), wells of 96-well plates were filled with PBS, sealed, and stored at 4°C until scanning.
Quantitative microscopy
We used a Cellomics ArrayScan XTI microscope equipped with an X1 CCD camera and LED light source to scan stained cultures of sensory neurons in 96-well plates. 2 × 2 binned images (1,104 × 1,104 pixels) were acquired with a 10× (NA = 0.3) EC Plan NeoFluar objective (Zeiss) and analyzed using the Cellomics software package. In brief, images of UCHL1 stainings were background corrected (low-pass filtration), converted to binary image masks (fixed threshold), and segmented (geometric method), and neurons were identified by the object selection parameters size 80–7,500 µm2, circularity (perimeter 2/4π area) 1–3, length-to-width ratio 1–2, mean intensity 800–12,000, and total intensity 2 × 105–5 × 107. The image masks were then used to quantify signals in other channels. Three respective controls were prepared for each triple staining: (1) UCHL1 alone, (2) UCHL1 + antibody 1, and (3) UCHL1 + antibody 2. Raw fluorescence data of the controls were used to calculate the slope of best fit straight lines by linear regression, which was then used to compensate spill-over as described previously (Roederer, 2002). Compensated data were scaled to a mean value of 1 (or 1,000) for the unstimulated cells to adjust for variability between experimental days. One- and two-dimensional probability density plots were generated using R packages (R Development Core Team, 2011). Gating of subpopulations was performed by setting thresholds at local minima of probability density plots.
BRET assay
HEK293 cells (DSMZ) were cotransfected with expression vectors for GFP-hCα and hRIα-Rluc8, hRIβ-Rluc8, hRIIα-Rluc8, and hRIIβ-Rluc8, respectively (Prinz et al., 2006a). BRET experiments were performed as described previously (Chepurny et al., 2013; Isensee et al., 2014a; Schwede et al., 2015). In brief, HEK293 cells were seeded on 96-well Nunc plates (Thermo Fisher Scientific) at a density of 2 × 104 cells/well and cultured in DMEM high glucose (GE Healthcare) supplemented with 10% fetal calf serum gold (PAA Laboratories) at 37°C and 5% CO2. After 24 h, cells were transfected using polyethyleneimine (25 kD, linear; Polysciences Europe) with GFP-hCα and the desired R-subunit in a 1:1 ratio. Cells were rinsed with HBSS 2 d after transfection, treated with the respective compound for 20 min, and before adding the substrate coelenterazine 400A (BIOTREND Chem.) at a final concentration of 5 µM in a total volume of 30 µl HBSS. After 15 min, BRET measurements at wavelengths 410 ± 80 nm for the donor and 515 ± 30 nm for the acceptor were performed using a POLARstar Omega microplate reader (BMG Labtech). Emission values of untransfected cells (n.t.) were subtracted, and BRET signals were calculated according to the following formula: [emission (515 nm) − n.t. (515 nm)]/[emission (410 nm) − n.t. (410 nm)]. Control measurements with cells expressing RLuc8 alone were included in each experiment.
Protein expression, purification, and holoenzyme formation
Recombinant PKA regulatory isoforms (human isoforms Iα, Iβ, IIα, and IIβ) were expressed in Escherichia coli BL21 DE3 RIL and purified by cAMP affinity chromatography using Sp8-AEA-cAMPS agarose (BioLog) and 20 mM cyclic guanosine monophosphate for elution as described before (Bertinetti et al., 2009; Zhang et al., 2015). PKA C-subunits (human and murine isoform α1) were expressed in E. coli BL21 DE3 and purified using an IP20-resin according to Knape et al. (2015) and Olsen and Uhler (1989). Holoenzyme formation was performed by mixing PKA-Cα and PKA-RII in a 1.2:1 molar ratio and subsequent dialysis against buffer A (20 mM MOPS, pH 7, 150 mM NaCl, and 2 mM β-mercaptoethanol).
SPR analysis
A Biacore T200 SPR instrument (GE Healthcare) was used to analyze the binding of the anti–phospho-RII and anti–PKA substrate (Cell Signaling) to PKA-II. For this, both antibodies were immobilized on parallel flow cells (5,000–8,000 resonance units) on a CM5 sensor chip (GE Healthcare) using standard N-hydroxysuccinimide (NHS)/1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) chemistry. A reference flow cell without antibodies was generated by the activation of carboxy groups (NHS/EDC) and subsequent deactivation (ethanolamine). Binding analysis was performed in running buffer (20 mM MOPS, pH 7, 150 mM NaCl, and 0.01% P20 surfactant) by injecting 500 nM recombinant type IIα and 250 nM type IIβ holoenzymes under different buffer conditions (i.e., buffer alone or supplemented with 0.5 mM ATP and 1 mM MgCl2 or CaCl2) for 120 s at 30 µl/min at 25°C. As shown before, the addition of ATP and divalent metal ions results in an instantaneous phosphorylation of RII-subunits (Zhang et al., 2012, 2015). Dissociation of PKA holoenzymes was induced by the addition of 10 µM cAMP. To ensure dissociation as well as phosphorylation of RII-subunits all samples were incubated at 25°C for at least 10 min before run. Regeneration of the sensor chip surface was achieved by injecting 10 mM glycine, pH 1.9, for 30 s. Unspecific binding was removed by subtracting binding signals of the reference surface as well as control runs with buffer only (double referencing). BIAevaluation 4.1.1 software (GE Healthcare) was used for data processing.
Coimmunoprecipitation
Protein G functionalized Dynabeads (Thermo Fisher Scientific) were used to probe binding of PKA-II holoenzymes to the phospho-RII antibodies. Anti–phospho-RII (S99) antibodies (∼2 µg input; Abcam) were captured following the manufacturer’s instructions, washed twice in HBS-P buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.01% P20 surfactant), and incubated for 20 min with purified, preformed PKA-II holoenzyme (Cα:RIIβ, 3.6 µM:3 µM) in HBS-P buffer. The buffer was supplemented with 1 mM ATP, 10 mM MgCl2, and 0.5 or 10 µM cAMP. The beads were washed four times and transferred into new tubes. Elution was performed using 10 mM glycine, pH 2.2, and SDS sample buffer. The samples were subjected to SDS-PAGE and proteins were detected by Western blotting using Cα (sc-28315; Santa Cruz) and RIIβ (610625; BD Transduction Laboratories) on separate Western blots. Detection and signal quantification was performed using the Odyssey Fc Imager (Li-Cor) with the fluorescently labeled anti–mouse IRDye800 (Li-Cor) secondary antibody or mouse IgG κ-binding protein CFL790 (sc-516181; Santa Cruz).
Nano–LC-MS/MS
For sample preparation, ∼15 µg PKA-Cα was resuspended in 50 mM ammonium bicarbonate (ABC; Sigma-Aldrich), pH 7.8. The solution was filtrated using a 30-kD Amicon Ultra-0.5 ml centrifugal filter (Merck Millipore) by centrifugation for 10–15 min at 10,000 g and 4°C to remove degradation products. To identify the antibody-binding epitope, we followed two alternative strategies. First, PKA-Cα was incubated with anti–PKA-Cα for 1 h at 37°C in 50 mM ABC buffer, pH 7.8, followed by digestion with the broad-specificity enzyme subtilisin (protease/protein ratio of 1:20) for 5 min at 56°C (Gonczarowska-Jorge et al., 2016). The sample was filtrated as above to remove PKA-Cα peptides not bound (flow-through) by anti–PKA-Cα. The concentrate was incubated with 0.1% TFA (Biosolve) for 5 min at 4°C to elute the antibody-bound peptides from anti-PKA-Cα, followed by another ultrafiltration as above to remove intact anti-PKA-Cα (eluate). The eluate and flow-through were dried under vacuum and resuspended in 20 µl of 0.1% TFA. PKA-Cα, anti–PKA-Cα as well as coincubated PKA-Cα + anti–PKA-Cα were separated on a monolithic HPLC system before and after digestion (Fig. S1; Burkhart et al., 2012). Second, PKA-Cα was digested with subtilisin (1:20) for 5 min at 56°C in 50 mM ABC, followed by ultrafiltration as above. The filtrate containing proteolytic peptides was incubated with anti–PKA-Cα for 1 h at 37°C in 50 mM ABC, pH 7.8, followed by ultrafiltration as above to remove peptides not bound (flow-through) to anti–PKA-Cα. The concentrate was treated as above.
Samples were analyzed using an Ultimate 3000 nano RSLC (Thermo Fisher Scientific) coupled to an Orbitrap Fusion Lumos (Thermo Fisher Scientific) in data-dependent acquisition mode. Peptides were preconcentrated on a 75 µm × 2 cm C18 Acclaim Pepmap trapping column (Thermo Fisher Scientific) in 0.1% TFA for 10 min using a flow rate of 20 µl/min, followed by separation in a on a 75 µm × 50 cm Acclaim Pepmap C18 main column using a binary gradient (A, 0.1% formic acid; B, 84% acetonitrile, 0.1% formic acid) ranging from 3% to 35% B in 55 min at a flow rate of 250 nl/min. MS survey scans were acquired in the Orbitrap from 300 to 1,500 m/z at a resolution of 120,000 using the polysiloxane ion at 371.1012 m/z as lock mass (Gonczarowska-Jorge et al., 2016). The AGC target value was 5 × 103 ions, and the maximum injection time was 50 ms. For MS/MS, precursors were selected using the top speed option (3 s) without dynamic exclusion. Peptides were fragmented using higher-energy collisional dissociation at 32%, and fragment ion spectra were acquired in the Orbitrap with a resolution of 30,000, an AGC target value of 5 × 103 ions, and a maximum injection time of 150 ms.
For data analysis MS raw files were searched with the help of Proteome Discoverer 1.4 (Thermo Fisher Scientific), using Mascot 2.4 against a Mus musculus Uniprot database (downloaded July 2015; 16,684 target sequences). The following search settings were used: Enzyme specificity was set to “none” and oxidation of Met was set as variable modification. MS and MS/MS tolerances were set to 10 ppm and 0.02 D, respectively. Percolator (Käll et al., 2007) was used for false discovery rate estimation, and data were filtered at 1% false discovery rate on the PSM level.
Statistical analysis
Statistical analyses were performed with one- or two-way ANOVA with respective post hoc tests as indicated in the figure legends. P < 0.05 was considered as statistically significant. HCS microscopy experiments were analyzed with R (R Development Core Team, 2011) using ordinary two-way ANOVA. Bonferroni’s post hoc analysis was applied to determine p-values of selected pairs defined in a contrast matrix using the R library multcomp. Error bars represent the SEM of n = 3–5 independent replicate experiments using cells of different animals. Dose–response curves from HCS microscopy in Fig. S1 C were generated using nonlinear regression curve-fitting (three parameters, standard Hill slope) with Prism (GraphPad). We assumed that mean values of all neurons analyzed per experiment (usually >500 per well) are normally distributed, which we proved on a larger datasets using normality tests such as the Shapiro–Wilk, Anderson–Darling, or Lillifors test. Similar variance of values before and after stimulation was analyzed using the Fligner–Killeen and Bartlett test.
Molecular modeling of holoenzyme dissociation
For mechanistic mathematical modeling of PKA signaling and hypotheses testing, we used the open-source MATLAB toolbox Data2Dynamics (Raue et al., 2015). The parameters of all model alternatives were determined using Bayesian parameter estimation, taking into account >750 data points observed under >60 experimental conditions and prior knowledge for dissociation constants (e.g., AC and Fsk). The measurement noise was assumed normally distributed with unknown SD. The maximum a posterior estimate was computed using multistart local optimization. For local optimization, we used the MATLAB routine lsqnonlin.m with gradients computed using forward sensitivity equations. For each model, 1,000 local optimizations were performed with starting values drawn from a log-uniform distribution between 10−5 and 103. Using the optimization results, model section was performed using the AIC. A difference of 10 in the AIC of two models was considered substantial. In addition, we computed the AIC weights (Burnham and Anderson, 2004), an approximation for the posteriori probability of the model given the data. The experimental data, Data2Dynamics implementation, visualization of the fitting results, and best parameters found for each model are provided in the online supplemental material.
Online supplemental material
Fig. S1 shows that KT5720 and Myr-PKI cannot inhibit the induction of pRII intensity as well as PKA downstream signaling in sensory neurons. Fig. S2 demonstrates that high doses of Rp-isomers induce the phosphorylation of ERK1/2 and cell loss depending on their modification at position 8 of the adenine nucleobase. Fig. S3 provides monolithic HPLC pre- and postdigestion controls for the anti–Cα antibody, PKA-Cα subunits, and coincubations thereof. Fig. S4 shows the distribution of PSMs identified by LC-MS analysis as a measure of peptide abundance indicating that the sequence GTGSFGRVM binds to the anti–Cα antibody. Fig. S5 depicts an alternative model structure, which assumes that cAMP induces RII phosphorylation. The supplemental modeling archive includes the used experimental data, Data2Dynamics implementation, visualization of the fitting results, and best parameters found for each model. The modeling archive is also available at https://doi.org/10.6084/m9.figshare.5991136.v2.
Supplementary Material
Acknowledgments
We thank Stephanie Brosig and Maike Siobal for excellent technical support.
J. Isensee, M. Kaufholz, F.W. Herberg, and T. Hucho were supported by grants from the Federal Ministry of Education and Research, Germany (NoPain, FKZ0316177A, and FKZ0316177F). M.J. Knape was supported by The Michael J. Fox Foundation for Parkinson’s Research (grant 11425). R.P. Zahedi and H. Gonczarowska-Jorge received financial support by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, and the Federal Ministry of Education and Research. H. Gonczarowska-Jorge furthermore thanks the CAPES Foundation for funding. J. Hasenauer was supported by the Postdoctoral Fellowship Program of the German Research Center for Environmental Health.
F. Schwede is head of research and development at BIOLOG LSI, which sells some of the cAMP analogs that were used in this study.
The authors declare no further conflict of interest.
Author contributions: J. Isensee, F.W. Herberg, and T. Hucho designed the study and wrote the manuscript. J. Isensee and H. Hammerich performed cellular experiments using HCS microscopy. F. Schwede synthesized and provided Rp-cAMPS-pAB prodrugs. M. Kaufholz designed and performed the BRET experiments. M.J. Knape designed and performed SPR experiments. R.P. Zahedi and H. Gonczarowska-Jorge performed epitope mapping by nano–LC-MS/MS. J. Hasenauer generated the mathematical model of PKA-II activation. M. Kaufholz, M.J. Knape, R.P. Zahedi, and J. Hasenauer contributed to writing the manuscript.
References
- Adams S.R., Harootunian A.T., Buechler Y.J., Taylor S.S., and Tsien R.Y.. 1991. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 349:694–697. 10.1038/349694a0 [DOI] [PubMed] [Google Scholar]
- Bacskai B.J., Hochner B., Mahaut-Smith M., Adams S.R., Kaang B.K., Kandel E.R., and Tsien R.Y.. 1993. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 260:222–226. 10.1126/science.7682336 [DOI] [PubMed] [Google Scholar]
- Badireddy S., Yunfeng G., Ritchie M., Akamine P., Wu J., Kim C.W., Taylor S.S., Qingsong L., Swaminathan K., and Anand G.S.. 2011. Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol. Cell. Proteomics. 10:M110.004390 10.1074/mcp.M110.004390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavo J.A., Bechtel P.J., and Krebs E.G.. 1974. Preparation of homogeneous cyclic AMP-dependent protein kinase(s) and its subunits from rabbit skeletal muscle. Methods Enzymol. 38:299–308. 10.1016/0076-6879(74)38046-9 [DOI] [PubMed] [Google Scholar]
- Bertinetti D., Schweinsberg S., Hanke S.E., Schwede F., Bertinetti O., Drewianka S., Genieser H.G., and Herberg F.W.. 2009. Chemical tools selectively target components of the PKA system. BMC Chem. Biol. 9:3 10.1186/1472-6769-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras B.W., Kornev A., Taylor S.S., and McCulloch A.D.. 2014. Using Markov state models to develop a mechanistic understanding of protein kinase A regulatory subunit RIα activation in response to cAMP binding. J. Biol. Chem. 289:30040–30051. 10.1074/jbc.M114.568907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart J.M., Schumbrutzki C., Wortelkamp S., Sickmann A., and Zahedi R.P.. 2012. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteomics. 75:1454–1462. 10.1016/j.jprot.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Burnham K.P., and Anderson D.R.. 2004. Multimodel inference - understanding AIC and BIC in model selection. Sociol. Methods Res. 33:261–304. 10.1177/0049124104268644 [DOI] [Google Scholar]
- Calejo A.I., and Taskén K.. 2015. Targeting protein-protein interactions in complexes organized by A kinase anchoring proteins. Front. Pharmacol. 6:192 10.3389/fphar.2015.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurny O.G., Bertinetti D., Diskar M., Leech C.A., Afshari P., Tsalkova T., Cheng X., Schwede F., Genieser H.G., Herberg F.W., and Holz G.G.. 2013. Stimulation of proglucagon gene expression by human GPR119 in enteroendocrine L-cell line GLUTag. Mol. Endocrinol. 27:1267–1282. 10.1210/me.2013-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin J.D., Brostrom C.O., Alexander R.L., and Krebs E.G.. 1972. Adenosine 3′,5′-monophosphate-dependent protein kinase from adipose tissue. J. Biol. Chem. 247:3736–3743. [PubMed] [Google Scholar]
- Diskar M., Zenn H.M., Kaupisch A., Prinz A., and Herberg F.W.. 2007. Molecular basis for isoform-specific autoregulation of protein kinase A. Cell. Signal. 19:2024–2034. 10.1016/j.cellsig.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Erlichman J., Rosenfeld R., and Rosen O.M.. 1974. Phosphorylation of a cyclic adenosine 3′:5′-monophosphate-dependent protein kinase from bovine cardiac muscle. J. Biol. Chem. 249:5000–5003. [PubMed] [Google Scholar]
- Gjertsen B.T., Mellgren G., Otten A., Maronde E., Genieser H.G., Jastorff B., Vintermyr O.K., McKnight G.S., and Døskeland S.O.. 1995. Novel (Rp)-cAMPS analogs as tools for inhibition of cAMP-kinase in cell culture. Basal cAMP-kinase activity modulates interleukin-1 beta action. J. Biol. Chem. 270:20599–20607. 10.1074/jbc.270.35.20599 [DOI] [PubMed] [Google Scholar]
- Gonczarowska-Jorge H., Dell’Aica M., Dickhut C., and Zahedi R.P.. 2016. Variable Digestion Strategies for Phosphoproteomics Analysis. Methods Mol. Biol. 1355:225–239. 10.1007/978-1-4939-3049-4_15 [DOI] [PubMed] [Google Scholar]
- Hemmer W., McGlone M., Tsigelny I., and Taylor S.S.. 1997. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 272:16946–16954. 10.1074/jbc.272.27.16946 [DOI] [PubMed] [Google Scholar]
- Herberg F.W., Taylor S.S., and Dostmann W.R.. 1996. Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase. Biochemistry. 35:2934–2942. 10.1021/bi951647c [DOI] [PubMed] [Google Scholar]
- Herfindal L., Krakstad C., Myhren L., Hagland H., Kopperud R., Teigen K., Schwede F., Kleppe R., and Døskeland S.O.. 2014. Introduction of aromatic ring-containing substituents in cyclic nucleotides is associated with inhibition of toxin uptake by the hepatocyte transporters OATP 1B1 and 1B3. PLoS One. 9:e94926 10.1371/journal.pone.0094926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J., Diskar M., Waldherr S., Buschow R., Hasenauer J., Prinz A., Allgöwer F., Herberg F.W., and Hucho T.. 2014a Pain modulators regulate the dynamics of PKA-RII phosphorylation in subgroups of sensory neurons. J. Cell Sci. 127:216–229. 10.1242/jcs.136580 [DOI] [PubMed] [Google Scholar]
- Isensee J., Wenzel C., Buschow R., Weissmann R., Kuss A.W., and Hucho T.. 2014b Subgroup-elimination transcriptomics identifies signaling proteins that define subclasses of TRPV1-positive neurons and a novel paracrine circuit. PLoS One. 9:e115731 10.1371/journal.pone.0115731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J., Krahé L., Moeller K., Pereira V., Sexton J.E., Sun X., Emery E., Wood J.N., and Hucho T.. 2017a Synergistic regulation of serotonin and opioid signaling contributes to pain insensitivity in Nav1.7 knockout mice. Sci. Signal. 10:10 10.1126/scisignal.aah4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J., Schild C., Schwede F., and Hucho T.. 2017b Crosstalk from cAMP to ERK1/2 emerges during postnatal maturation of nociceptive neurons and is maintained during aging. J. Cell Sci. 130:2134–2146. 10.1242/jcs.197327 [DOI] [PubMed] [Google Scholar]
- Iwami G., Kawabe J., Ebina T., Cannon P.J., Homcy C.J., and Ishikawa Y.. 1995. Regulation of adenylyl cyclase by protein kinase A. J. Biol. Chem. 270:12481–12484. 10.1074/jbc.270.21.12481 [DOI] [PubMed] [Google Scholar]
- Johnson D.A., Leathers V.L., Martinez A.M., Walsh D.A., and Fletcher W.H.. 1993. Fluorescence resonance energy transfer within a heterochromatic cAMP-dependent protein kinase holoenzyme under equilibrium conditions: new insights into the conformational changes that result in cAMP-dependent activation. Biochemistry. 32:6402–6410. 10.1021/bi00076a013 [DOI] [PubMed] [Google Scholar]
- Käll L., Canterbury J.D., Weston J., Noble W.S., and MacCoss M.J.. 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 4:923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- Knape M.J., Ahuja L.G., Bertinetti D., Burghardt N.C., Zimmermann B., Taylor S.S., and Herberg F.W.. 2015. Divalent Metal Ions Mg2+ and Ca2+ Have Distinct Effects on Protein Kinase A Activity and Regulation. ACS Chem. Biol. 10:2303–2315. 10.1021/acschembio.5b00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., and Sowadski J.M.. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 253:407–414. 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- Krass J.D., Jastorff B., and Genieser H.G.. 1997. Determination of lipophilicity by gradient elution high-performance liquid chromatography. Anal. Chem. 69:2575–2581. 10.1021/ac961246i [DOI] [PubMed] [Google Scholar]
- Manni S., Mauban J.H., Ward C.W., and Bond M.. 2008. Phosphorylation of the cAMP-dependent protein kinase (PKA) regulatory subunit modulates PKA-AKAP interaction, substrate phosphorylation, and calcium signaling in cardiac cells. J. Biol. Chem. 283:24145–24154. 10.1074/jbc.M802278200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B.R., Deerinck T.J., Ellisman M.H., Taylor S.S., and Tsien R.Y.. 2007. Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1alpha-mediated localization in living cells. Chem. Biol. 14:1031–1042. 10.1016/j.chembiol.2007.07.017 [DOI] [PubMed] [Google Scholar]
- Meharena H.S., Fan X., Ahuja L.G., Keshwani M.M., McClendon C.L., Chen A.M., Adams J.A., and Taylor S.S.. 2016. Decoding the Interactions Regulating the Active State Mechanics of Eukaryotic Protein Kinases. PLoS Biol. 14:e2000127 10.1371/journal.pbio.2000127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy B.S., Gao Y., and Anand G.S.. 2011. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol. Cell. Proteomics. 10:M110.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.J. 2008. Pharmacological PKA inhibition: all may not be what it seems. Sci. Signal. 1:re4 10.1126/scisignal.122re4 [DOI] [PubMed] [Google Scholar]
- Olsen S.R., and Uhler M.D.. 1989. Affinity purification of the C alpha and C beta isoforms of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 264:18662–18666. [PubMed] [Google Scholar]
- Pidoux G., and Taskén K.. 2010. Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J. Mol. Endocrinol. 44:271–284. 10.1677/JME-10-0010 [DOI] [PubMed] [Google Scholar]
- Prinz A., Diskar M., Erlbruch A., and Herberg F.W.. 2006a Novel, isotype-specific sensors for protein kinase A subunit interaction based on bioluminescence resonance energy transfer (BRET). Cell. Signal. 18:1616–1625. 10.1016/j.cellsig.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Prinz A., Diskar M., and Herberg F.W.. 2006b Application of bioluminescence resonance energy transfer (BRET) for biomolecular interaction studies. ChemBioChem. 7:1007–1012. 10.1002/cbic.200600048 [DOI] [PubMed] [Google Scholar]
- Raue A., Steiert B., Schelker M., Kreutz C., Maiwald T., Hass H., Vanlier J., Tönsing C., Adlung L., Engesser R., et al. . 2015. Data2Dynamics: a modeling environment tailored to parameter estimation in dynamical systems. Bioinformatics. 31:3558–3560. 10.1093/bioinformatics/btv405 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reimann E.M., Walsh D.A., and Krebs E.G.. 1971. Purification and properties of rabbit skeletal muscle adenosine 3′,5′-monophosphate-dependent protein kinases. J. Biol. Chem. 246:1986–1995. [PubMed] [Google Scholar]
- Roederer M. Compensation in flow cytometry. Curr Protoc Cytom. 2002;Chapter 1:Unit 1.14. 10.1002/0471142956.cy0114s22 [DOI] [PubMed] [Google Scholar]
- Rothermel J.D., Stec W.J., Baraniak J., Jastorff B., and Botelho L.H.. 1983. Inhibition of glycogenolysis in isolated rat hepatocytes by the Rp diastereomer of adenosine cyclic 3′,5′-phosphorothioate. J. Biol. Chem. 258:12125–12128. [PubMed] [Google Scholar]
- Rubin C.S., Erlichman J., and Rosen O.M.. 1972. Molecular forms and subunit composition of a cyclic adenosine 3′,5′-monophosphate-dependent protein kinase purified from bovine heart muscle. J. Biol. Chem. 247:36–44. [PubMed] [Google Scholar]
- Schwede F., Chepurny O.G., Kaufholz M., Bertinetti D., Leech C.A., Cabrera O., Zhu Y., Mei F., Cheng X., Manning Fox J.E., et al. . 2015. Rp-cAMPS Prodrugs Reveal the cAMP Dependence of First-Phase Glucose-Stimulated Insulin Secretion. Mol. Endocrinol. 29:988–1005. 10.1210/me.2014-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette C., and Conti M.. 1996. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J. Biol. Chem. 271:16526–16534. 10.1074/jbc.271.28.16526 [DOI] [PubMed] [Google Scholar]
- Smith F.D., Esseltine J.L., Nygren P.J., Veesler D., Byrne D.P., Vonderach M., Strashnov I., Eyers C.E., Eyers P.A., Langeberg L.K., and Scott J.D.. 2017. Local protein kinase A action proceeds through intact holoenzymes. Science. 356:1288–1293. 10.1126/science.aaj1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.K., McDonald L.R., Cembran A., Kim J., Masterson L.R., McClendon C.L., Taylor S.S., and Veglia G.. 2014. Synchronous opening and closing motions are essential for cAMP-dependent protein kinase A signaling. Structure. 22:1735–1743. 10.1016/j.str.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle R.G. 1982. Physical-Chemistry, Stability, and Handling of Prostaglandins-E2, Prostaglandins-F2-Alpha, Prostaglandins-D2, and Prostaglandins-I2 - a Critical Summary. Methods Enzymol. 86:436–458. 10.1016/0076-6879(82)86216-2 [DOI] [Google Scholar]
- Tao M., Salas M.L., and Lipmann F.. 1970. Mechanism of activation by adenosine 3′:5′-cyclic monophosphate of a protein phosphokinase from rabbit reticulocytes. Proc. Natl. Acad. Sci. USA. 67:408–414. 10.1073/pnas.67.1.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., and Kornev A.P.. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36:65–77. 10.1016/j.tibs.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., Buechler J.A., and Yonemoto W.. 1990. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59:971–1005. 10.1146/annurev.bi.59.070190.004543 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Kim C., Vigil D., Haste N.M., Yang J., Wu J., and Anand G.S.. 2005. Dynamics of signaling by PKA. Biochim. Biophys. Acta. 1754:25–37. 10.1016/j.bbapap.2005.08.024 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ilouz R., Zhang P., and Kornev A.P.. 2012. Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 13:646–658. 10.1038/nrm3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Quesada O., Mayrhofer J.E., and Stefan E.. 2017. The many faces of compartmentalized PKA signalosomes. Cell. Signal. 37:1–11. 10.1016/j.cellsig.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Vigil D., Blumenthal D.K., Brown S., Taylor S.S., and Trewhella J.. 2004. Differential effects of substrate on type I and type II PKA holoenzyme dissociation. Biochemistry. 43:5629–5636. 10.1021/bi0499157 [DOI] [PubMed] [Google Scholar]
- Walsh D.A., Perkins J.P., and Krebs E.G.. 1968. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243:3763–3765. [PubMed] [Google Scholar]
- Yang S., Fletcher W.H., and Johnson D.A.. 1995. Regulation of cAMP-dependent protein kinase: enzyme activation without dissociation. Biochemistry. 34:6267–6271. 10.1021/bi00019a002 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ma Y., Taylor S.S., and Tsien R.Y.. 2001. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc. Natl. Acad. Sci. USA. 98:14997–15002. 10.1073/pnas.211566798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Smith-Nguyen E.V., Keshwani M.M., Deal M.S., Kornev A.P., and Taylor S.S.. 2012. Structure and allostery of the PKA RIIβ tetrameric holoenzyme. Science. 335:712–716. 10.1126/science.1213979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Knape M.J., Ahuja L.G., Keshwani M.M., King C.C., Sastri M., Herberg F.W., and Taylor S.S.. 2015. Single Turnover Autophosphorylation Cycle of the PKA RIIβ Holoenzyme. PLoS Biol. 13:e1002192 10.1371/journal.pbio.1002192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann B., Chiorini J.A., Ma Y., Kotin R.M., and Herberg F.W.. 1999. PrKX is a novel catalytic subunit of the cAMP-dependent protein kinase regulated by the regulatory subunit type I. J. Biol. Chem. 274:5370–5378. 10.1074/jbc.274.9.5370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.