Saegusa et al. report that the SFT-4/Surf4 cargo receptor homologs mediate export of soluble proteins such as lipoproteins from the ER. Efficient export of yolk proteins in C. elegans intestinal cells, or apoliprotein trafficking in human hepatocytes, requires SFT-4/Surf4 so that they may enhance secretion by maintaining ER exit site organization.
Abstract
Lipoproteins regulate the overall lipid homeostasis in animals. However, the molecular mechanisms underlying lipoprotein trafficking remain poorly understood. Here, we show that SFT-4, a Caenorhabditis elegans homologue of the yeast Erv29p, is essential for the endoplasmic reticulum (ER) export of the yolk protein VIT-2, which is synthesized as a lipoprotein complex. SFT-4 loss strongly inhibits the ER exit of yolk proteins and certain soluble cargo proteins in intestinal cells. SFT-4 predominantly localizes at ER exit sites (ERES) and physically interacts with VIT-2 in vivo, which suggests that SFT-4 promotes the ER export of soluble proteins as a cargo receptor. Notably, Surf4, a mammalian SFT-4 homologue, physically interacts with apolipoprotein B, a very-low-density lipoprotein core protein, and its loss causes ER accumulation of apolipoprotein B in human hepatic HepG2 cells. Interestingly, loss of SFT-4 and Surf4 reduced the number of COPII-positive ERES. Thus, SFT-4 and Surf4 regulate the export of soluble proteins, including lipoproteins, from the ER and participate in ERES organization in animals.
Introduction
The biogenesis and secretion of lipoproteins such as very-low-density lipoproteins (VLDLs) and chylomicrons (CMs) are critical for regulating blood lipid levels and thus for overall lipid homeostasis, and defects in lipoprotein biogenesis/secretion are linked to the onset of dyslipidemia and related diseases. For example, excessive VLDL secretion increases the concentration of low-density lipoprotein (LDL), a metabolic product of VLDL, in the blood and results in subendothelial retention of atherogenic lipoproteins, which raises the risk of atherogenesis (Williams and Tabas, 1995; Tabas et al., 2007). Conversely, impaired VLDL secretion from hepatocytes results in the accumulation of neutral lipids in the liver, which leads to nonalcoholic fatty liver disease (Cefalù et al., 2013; Di Filippo et al., 2014).
Lipoproteins consist of large amphipathic glycoproteins, such as apolipoprotein B (ApoB), and lipids, including cholesterol, phospholipids, and neutral lipids. ApoB100 is an essential structural component of VLDLs, intermediate-density lipoproteins, and LDLs, and it is mainly expressed in the liver, whereas ApoB48, a core protein of CMs, is primarily produced by the intestine. These ApoBs are synthesized in the ER and form nascent lipoprotein particles by means of lipidation with neutral and polar lipids through a mechanism mediated by microsomal triglyceride-transfer protein (Hussain et al., 2003). Because nascent lipoproteins form considerably larger particles (diameter: VLDL, 30–80 nm; CM, 180–500 nm; Schekman and Mellman, 1997; Fisher and Ginsberg, 2002) as compared with canonical COPII-coated vesicles (diameter: 55–70 nm), the transport of nascent VLDLs and CMs from the ER is mediated by specialized vesicles called VLDL transport vesicles (VTVs; diameter: 100–120 nm) and preCM transport vesicles (PCTVs; diameter: 150–500 nm), respectively (Gusarova et al., 2003; Siddiqi, 2008). VTVs and PCTVs contain COPII components such as Sar1, a small GTPase, and Sec23 (Siddiqi, 2008), and thus these vesicles appear to share parts of the COPII coated-vesicle formation machinery. Intriguingly, Sar1 is required for VTV formation but is dispensable for PCTV formation in vitro, which suggests distinct requirements for the assembly of these vesicles (Siddiqi, 2008). Recently, human TANGO1, which was originally identified as a transmembrane cargo receptor for procollagen VII (Saito et al., 2009), was reported to be also involved in the ER export of bulky lipid particles (Santos et al., 2016). However, the molecular mechanisms underlying the formation of these massive vesicles and the sorting of nascent lipoproteins into the vesicles remain incompletely understood.
In Caenorhabditis elegans, yolk proteins are synthesized as lipoprotein particles in the intestine and secreted into the body cavity in adult hermaphrodites (Grant and Hirsh, 1999; Matyash et al., 2001). Subsequently, the secreted yolk proteins are taken up by growing oocytes through receptor-mediated endocytosis and used as the nutrient source during embryogenesis (Baker, 1988; Babin et al., 1999; Grant and Hirsh, 1999). This process is mediated by the yolk receptor RME-2, which is a member of the LDL-receptor superfamily (Grant and Hirsh, 1999), and this suggests that the mechanisms responsible for lipoprotein trafficking are well conserved from C. elegans to mammals.
Here, we report that C. elegans SFT-4 is required for the ER export of yolk proteins and certain soluble proteins in intestinal cells. SFT-4 is a C. elegans orthologue of cargo receptors of the Erv29p family, which binds soluble cargos and COPII components in yeast (Belden and Barlowe, 2001). SFT-4 was previously identified in genome-wide RNAi screening for genes whose knockdown affects VIT-2–GFP trafficking, but its role was not studied in detail (Balklava et al., 2007). We found that SFT-4 loss inhibits the export of certain soluble proteins, including yolk proteins, from the ER and causes the accumulation of these proteins in granular structures in the ER lumen of intestinal cells. SFT-4 predominantly localizes at ER exit sites (ERES) in intestinal cells and physically interacts with VIT-2 in vivo. Notably, we also determined that the depletion of Surf4, a mammalian homologue of SFT-4, inhibits efficient ER export of ApoB100 in human hepatic cell line HepG2 cells. We further found that loss of SFT-4/Surf4 causes a reduction of the number of COPII-positive ERES. Our findings suggest that SFT-4/Surf4 family proteins regulate the export of soluble proteins such as lipoproteins from the ER and are involved in the formation of COPII-positive ERES in animals.
Results
SFT-4 is required for secretion of yolk proteins from intestinal cells
To study the molecular mechanisms underlying lipoprotein trafficking, we used a model system for monitoring yolk protein transport in C. elegans. VIT-2, one of the yolk core proteins, is widely used for monitoring the trafficking of yolk proteins in C. elegans (Grant and Hirsh, 1999), and several important components involved in the secretion and receptor-mediated endocytosis of yolk proteins have been identified through genetic screening or genome-wide RNAi screening performed using fluorescent protein-tagged VIT-2 (Fig. 1 A and Fig. S1 A; Grant and Hirsh, 1999; Balklava et al., 2007). VIT-2 shares 22% identity with human ApoB in the N-terminal 1,000 amino acids (Babin et al., 1999). When a GFP fusion protein with VIT-2 (VIT-2–GFP) is expressed under the control of vit-2 promoter in WT animals, the protein is secreted from the intestine and mainly accumulated in growing oocytes and embryos (Fig. 1, A–A″, arrows; Grant and Hirsh, 1999). To identify the genes required for lipoprotein trafficking, we focused on C. elegans homologues of cargo receptors, which concentrate cargo proteins into ER-derived transport intermediates for efficient ER export. To date, several types of cargo receptors have been identified, such as a set of ER vesicle (Erv) proteins, L-type lectin family proteins, p24 proteins, and retrograde sorting receptors, and most of these are conserved in C. elegans (Table 1; Dancourt and Barlowe, 2010).
Figure 1.
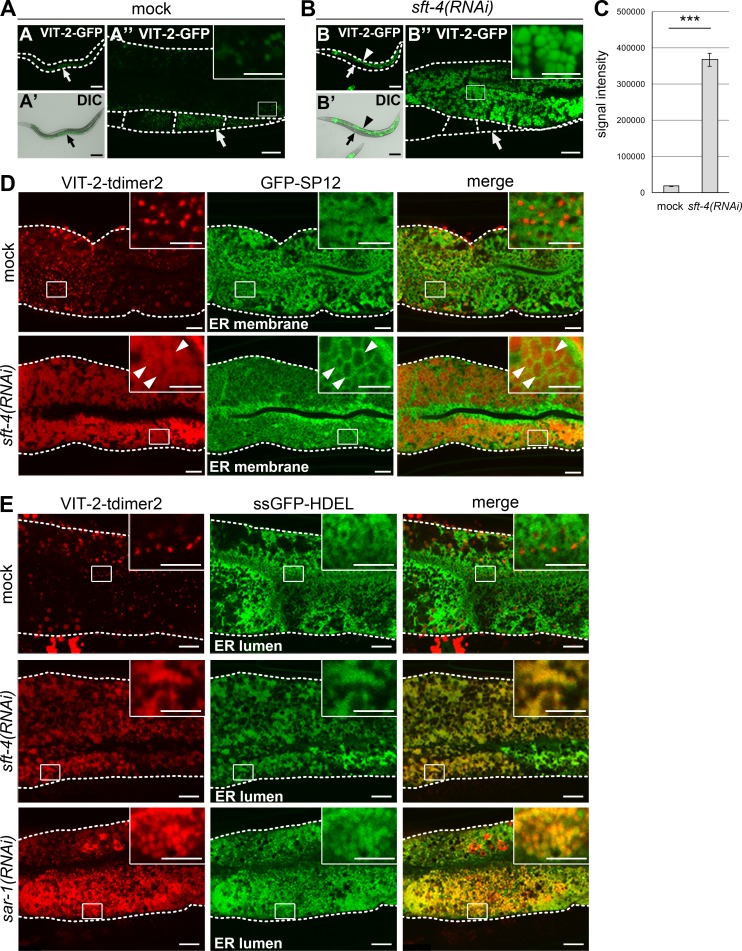
SFT-4 is required for efficient ER export of VIT-2 from intestinal cells. (A–C) Loss of sft-4 caused high accumulation of VIT-2 in intestinal cells. (A–A″) In mock-treated animals, VIT-2–GFP is detected on certain small punctate structures in intestinal cells and is prominently accumulated in oocytes (arrows). (B–B″) VIT-2–GFP is highly accumulated in sft-4(RNAi) intestinal cells (arrowheads) but is almost undetectable in oocytes (arrows). (C) The amount of VIT-2–GFP in intestinal cells was significantly increased after knockdown of sft-4. Fluorescence signal intensities per unit area were measured and statistically analyzed using Student’s t test; ***, P < 0.001; error bars: SEM (n = 34 and 31 intestines from mock and sft-4(RNAi) animals, respectively). Dotted lines indicate the outlines of worm bodies (A and B) or intestines and oocytes (other panels). Regions surrounded by squares are enlarged (16×) in insets. Bars: (A, A′, B, and B′) 50 µm; (A″ and B″) 10 µm; (insets) 5 µm. (D and E) Subcellular localization of VIT-2 in mock and sft-4(RNAi) intestinal cells. Intestines of transgenic animals coexpressing VIT-2–tdimer2 and GFP-SP12 (D) or ssGFP-HDEL (E) are shown. L3 larvae were treated with RNAi for 2 d in the case of sar-1(RNAi). VIT-2–tdimer2 localizes to granular structures in the ER lumen labeled with ssGFP-HDEL in sft-4(RNAi) or sar-1(RNAi) intestinal cells. Dotted lines indicate the outlines of intestines. Regions surrounded by squares are enlarged (16×) in insets. Bars: 10 µm; (insets) 5 µm.
Table 1. Effects of RNAi knockdown of C. elegans cargo receptor homologs in VIT-2 secretion from the intestine.
| C. elegans | S. cerevisiae | Mammals | Identified cargo proteins (species) | VIT-2 accumulation in the intestine |
|---|---|---|---|---|
| ile-1 | Emp46p/Emp47p | ERGIC-53 | Coagulation factors V and VIII, Cathepsin C and Z, α1-antitrypsin (Hs) | − |
| sel-9 | Emp24p | p24β | Gas1p (Sc) | − |
| rer-1 | Rer1p | Rer1 | Sec12p, Sec63p, Sec71p, Mns1p (Sc) | − |
| sft-4 | Erv29p | Surf4 | gp-α-factor, CPY, proteinase A (Sc) | +++ |
Hs, Homo sapiens; Sc, Saccharomyces cerevisiae.
We found that the loss of SFT-4, one of the aforementioned C. elegans homologues, caused severe accumulation of VIT-2–GFP in large granule-like structures in intestinal cells, whereas the knockdown of the other cargo receptor homologues did not (Fig. 1, B–B″ and C; and Table 1). SFT-4 was previously identified as a candidate regulator of VIT-2 trafficking by means of a genome-wide RNAi screening, but SFT-4 has not yet been studied in detail (Balklava et al., 2007). We confirmed the accumulation of endogenous VIT-2 in sft-4(RNAi) intestinal cells by immunostaining with an anti–VIT-2 antibody (Fig. S1 B), which indicated that SFT-4 is required for the secretion of yolk proteins from the intestine.
The gene sft-4 (C54H2.5) encodes a homologue of yeast Erv29p and mammalian Surf4. In yeast, Erv29p functions as a cargo receptor, which is required for the ER export of a yeast sex-pheromone precursor, pro-α-factor, and a vacuolar protease, carboxypeptidase Y (CPY; Belden and Barlowe, 2001). The C. elegans genome contains one paralog of sft-4, T02E1.7, which encodes a protein that shares 33% identity with SFT-4 (Henricson et al., 2004). Because T02E1.7 RNAi did not noticeably hinder VIT-2–GFP secretion (unpublished data), we focused on sft-4 in further experiments. RNAi of sft-4 caused severe larval lethality (49% lethality at the L3 stage), and the deletion mutant sft-4(gk301) also showed a high rate of embryonic lethality (Fig. S1 D). Some of the sft-4(gk301) homozygotes reached adulthood but were sterile. The embryonic lethal phenotype of sft-4(gk301) was rescued by the expression of GFP–SFT-4. These results demonstrated that SFT-4 is essential for animal viability and reproduction in C. elegans.
Loss of SFT-4 results in VIT-2 accumulation in the ER lumen
Because yeast Erv29p is required for the COPII-dependent ER export of cargo proteins, we suspected that the ER export of yolk proteins would be blocked by the depletion of SFT-4. Accordingly, the loss of SAR-1, a small GTPase, or SEC-23, a coat protein of COPII inner layer, also led to the accumulation of VIT-2–GFP in granule-like structures as observed in the intestine of sft-4(RNAi) animals (Fig. S1 C), which suggests that SFT-4 and COPII function at a similar step in VIT-2–GFP transport. This result also confirmed that yolk proteins are transported from the ER in a COPII-dependent manner as reported previously (Grant and Hirsh, 1999). We further sought to demonstrate that VIT-2 is accumulated in the ER in sft-4(RNAi) animals. To distinguish between the ER membrane and lumen, we used GFP-SP12 and signal-sequence-conjugated GFP (ssGFP)–HDEL as markers: SP12 is a 12-kD subunit of ER signal peptidase and GFP-SP12 localizes to the ER membrane (Fig. 1 D), whereas ssGFP-HDEL is a ssGFP fusion that contains the ER retrieval signal HDEL at its C terminus and localizes to the ER lumen (Fig. 1 E). We found that VIT-2–tdimer2 (a red fluorescent protein) did not exhibit overlapping distribution with GFP-SP12, but was accumulated in a region that was surrounded by GFP-SP12–positive mesh-like structures in sft-4(RNAi) intestinal cells (Fig. 1 D, arrowheads). In contrast, VIT-2–tdimer2 mostly colocalized with ssGFP-HDEL in sft-4(RNAi) intestinal cells, which suggests that VIT-2 mainly accumulates in the ER lumen in the absence of SFT-4 (Fig. 1 E). VIT-2–GFP did not colocalize with the late-Golgi glucuronyl transferase SQV-8 or the lysosomal membrane protein LMP-1 in sft-4(RNAi) intestinal cells (Fig. S2, A and B). Moreover, we examined whether VIT-2–tdimer2 colocalizes with GFP-tagged PLIN-1, a C. elegans perilipin homologue that localizes to cytosolic lipid droplets (LDs), in sft-4(RNAi) animals. No clear colocalization of these proteins was detected (Fig. S2 C), which suggests that VIT-2 is not mistargeted to cytosolic LDs.
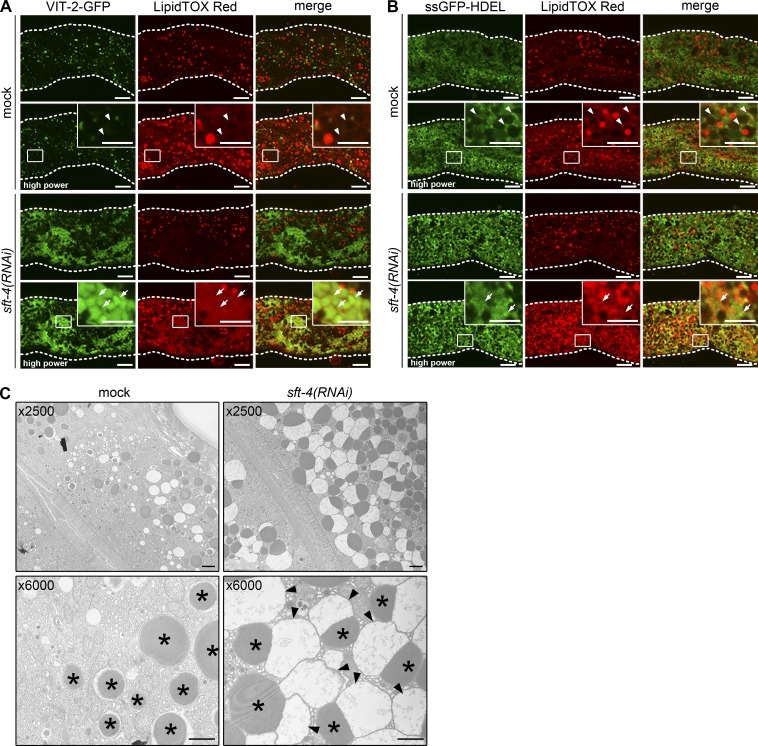
We also determined whether SFT-4 loss affects neutral-lipid distribution in intestinal cells. In mock-treated animals, LipidTOX Red neutral-lipid stain (LTR) labeled numerous granular structures, which probably represent cytosolic LDs, and also weakly stained reticular structures (Fig. 2 A, arrowheads). The LTR-positive reticular structures appear to represent neutral lipids in the ER lumen, because these structures were labeled with ssGFP-HDEL (Fig. 2 B, arrowheads). In sft-4(RNAi) animals, LTR stained the expanded ER lumen, wherein VIT-2–GFP was heavily accumulated, as well as the LDs, which suggests that SFT-4 loss leads to the accumulation of neutral lipids in the ER lumen (Fig. 2, A and B, arrows).
Figure 2.
Loss of sft-4 results in VIT-2 accumulation in the ER lumen. (A and B) The intestines of transgenic worms expressing VIT-2–GFP or ssGFP-HDEL were dissected and stained with LipidTOX Red, which labels neutral lipids. In mock animals, LipidTOX Red mainly stained cytosolic LDs (A and B) and weakly stained the ER lumen (arrowheads). In sft-4(RNAi) animals, the accumulated VIT-2–GFP and ssGFP-HDEL overlapped with LipidTOX Red–positive regions (A and B, arrows). Cytosolic LDs, which appeared as multiple punctate structures, were superficially normal. Dotted lines indicate the outlines of intestines. Regions surrounded by squares are enlarged (16×) in insets. Bars: 10 µm; (insets) 5 µm. (C) Electron microscopy analysis of intestinal cross sections from WT and sft-4(RNAi) animals. Mock-treated animals showed normal intestinal organelle distribution, such as the tubular structures of the ER, orderly alignment of microvilli, and several vesicles and granules. The large electron-dense granules (asterisks) likely contain yolk components. In contrast, sft-4(RNAi) animals showed increased numbers of electron-dense granules juxtaposed to electron-lucent granules (arrowheads) in the cytoplasm. Notably, these structures appear to be surrounded by a membrane. Bars, 1 µm.
We further examined the effect of SFT-4 depletion on organelle morphology by performing electron microscopy. In mock-treated intestinal cells, we detected large electron-dense granules, which are considered to contain yolk components (Fig. 2 C, asterisks; Paupard et al., 2001). Notably, granular structures with the similar electron density were drastically increased in number and size in sft-4(RNAi) cells (Fig. 2 C, asterisks), and most of these electron-dense structures were accompanied by electron-lucent structures resembling structures observed when protein export from the ER is blocked (Fig. 2 C, arrowheads). We tentatively refer to these structures as dalma (electron-dense and lucent membrane–associated) bodies. Dalma bodies were surrounded by a membrane, implying that they are formed in the expanded ER lumen. We assume that yolk components and other soluble cargos are separately retained in the ER lumen, resulting in formation of dalma bodies. Collectively with the results of fluorescent microscopy, our data suggest that sft-4(RNAi) causes massive expansion of the ER lumen.
SFT-4 loss strongly impairs ER export of soluble proteins but not that of transmembrane proteins
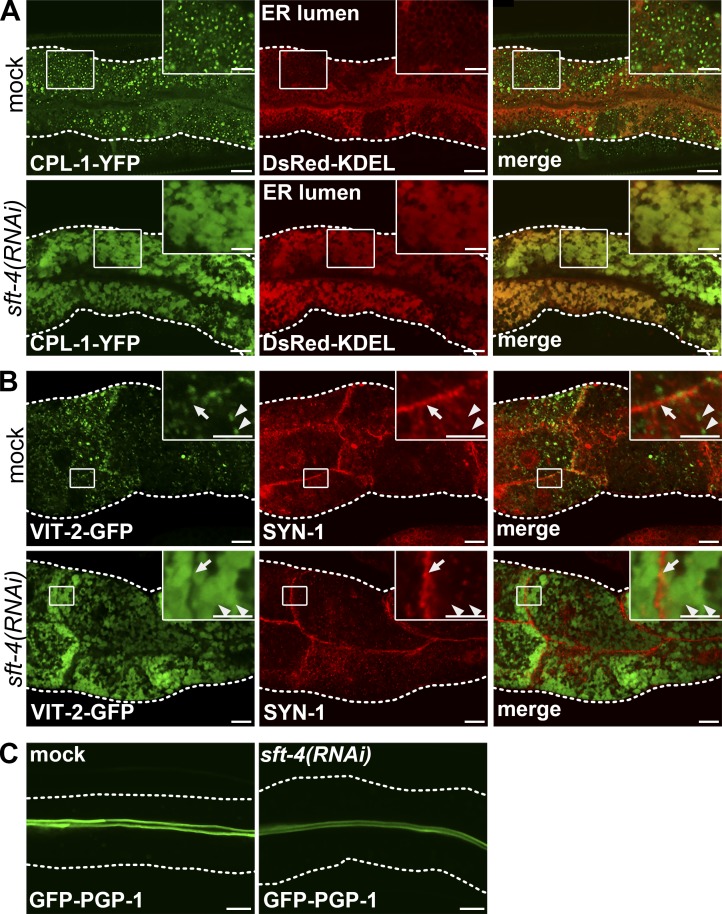
Because SFT-4 was found to be required for the efficient export of VIT-2 from the ER, we hypothesized that SFT-4 functions as a cargo receptor for soluble cargos, including VIT-2. To test this, we examined how sft-4 RNAi affected the ER export of other cargo proteins. C. elegans CPL-1 is a lysosomal cathepsin L–like cysteine protease, which is an orthologue of yeast CPY (Miedel et al., 2012). We found that sft-4 RNAi resulted in the accumulation of CPL-1–YFP in the ER lumen (labeled with DsRed-KDEL), which indicates that SFT-4 is also required for the ER exit of CPL-1 (Fig. 3 A).
Figure 3.
SFT-4 is required for transport from the ER of CPL-1, but not SYN-1 or PGP-1. (A) ER export of CPL-1–YFP is impaired by RNAi of sft-4. Subcellular localization of CPL-1–YFP and DsRed-KDEL was examined in the intestines of mock- and sft-4(RNAi)–treated animals. (B) Intestines of transgenic animals expressing VIT-2–GFP were stained with an antibody against SYN-1. When sft-4 was depleted, SYN-1 was not markedly affected: although VIT-2–GFP was strongly accumulated in the ER (arrowheads), SYN-1 mainly localized to the basolateral membrane (arrows). (C) Subcellular localization of GFP–PGP-1 in the intestinal cells of mock- or sft-4(RNAi)–treated animals. GFP–PGP-1 was mostly transported to the apical plasma membrane even in sft-4(RNAi) animals. Dotted lines indicate the outlines of intestines. Regions surrounded by squares are enlarged (4× in A and 16× in B) in insets. Bars: 10 µm; (insets) 5 µm.
We next tested whether the loss of SFT-4 affects the ER export of transmembrane proteins. We found that VIT-2–GFP was strongly accumulated in the ER by the loss of SFT-4, but SQV-8 and LAMP-1 were not (Fig. S2, A and B). To monitor the transport of plasma membrane proteins in intestinal cells, we selected SYN-1 and PGP-1 (Sato et al., 2011). SYN-1, a C. elegans homologue of syntaxin-1, is transported to the basolateral membrane in WT intestinal cells (Fig. 3 B, arrows), whereas PGP-1, a P-glycoprotein homologue, localizes to the apical region of the plasma membrane (Fig. 3 C). Immunostaining using an anti–SYN-1 antibody labeled the basolateral plasma membrane of intestinal cells, and this signal was abolished by RNAi of syn-1 gene, confirming the specificity of this antibody (Fig. S3 A). We found that endogenous SYN-1 still localized to the basolateral membrane in sft-4(RNAi) cells, in which VIT-2–GFP was largely accumulated in the ER (Fig. 3 B, arrowheads), and further that GFP-tagged PGP-1 was transported to the apical membrane in the sft-4(RNAi) intestinal cells (Fig. 3 C). Notably, when we knocked down sar-1, GFP–PGP-1 was partly accumulated in the reticular ER and also in large punctate structures near the basal membrane, suggesting COPII-dependent trafficking of membrane proteins such as PGP-1 (Fig. S3 B). These observations suggest differential requirements of SFT-4 for the transport of soluble cargos and transmembrane proteins.
SFT-4 is enriched at ERES, and its loss affects the ERES organization
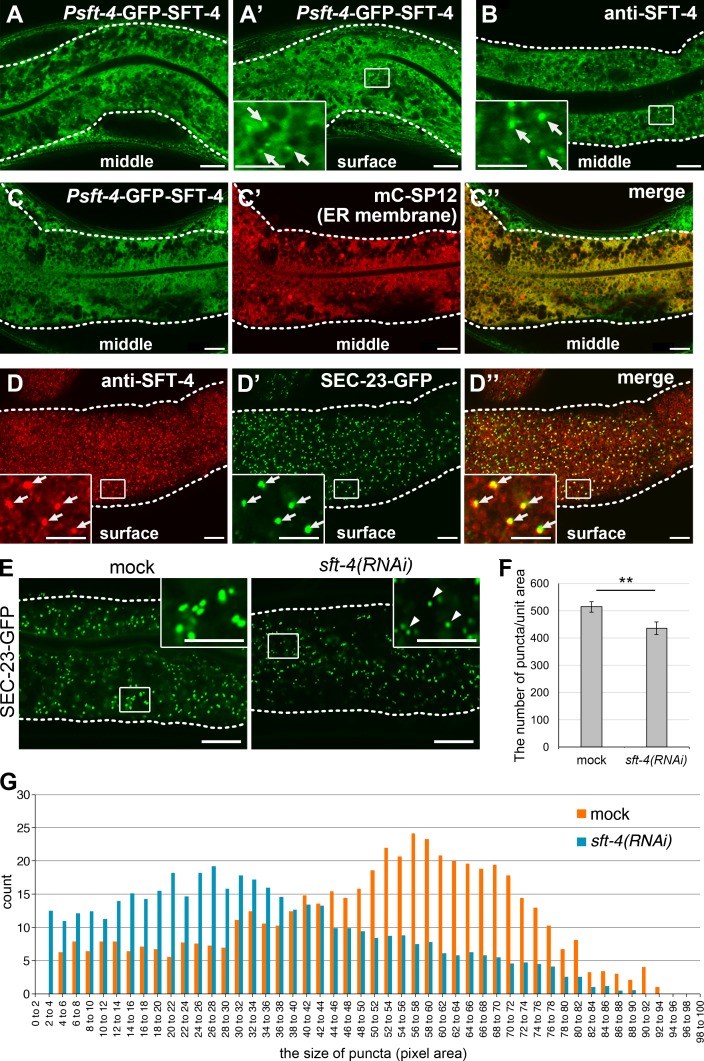
Next, to determine the expression and subcellular localization of SFT-4 in live animals, we generated transgenic animals in which the expression of GFP-tagged SFT-4 was driven by sft-4 promoter; we found that GFP–SFT-4 was strongly expressed in the intestine, body wall muscle, and spermatheca (Fig. S4, arrowheads). In intestinal cells, GFP–SFT-4 localized to typical ER reticular structures (Fig. 4 A) and punctate structures (Fig. 4 A′, arrows). Anti–SFT-4 antibody staining of endogenous SFT-4 also exhibited a similar pattern (Fig. 4 B). Notably, punctate structures tended to be observed in the region underneath the plasma membrane. This signal was abolished by RNAi of sft-4, confirming the specificity of this antibody (Fig. S4 B). The reticular distribution of GFP–SFT-4 overlapped with that of an ER marker, mCherry-SP12 (mC-SP12), which indicates that SFT-4 mainly localizes to the ER (Fig. 4, C–C″). We also compared the localization of SFT-4 with that of SEC-23–GFP, which labels ERES in intestinal cells. Most punctate structures stained by anti–SFT-4 antibody colocalized with SEC-23–GFP–labeled puncta, which indicates that a population of SFT-4 is concentrated at ERES (Fig. 4, D–D″, arrows).
Figure 4.
SFT-4 localizes at ERES and affects the ERES organization. (A and A′) GFP–SFT-4 localizes to reticular network structures in the middle of intestinal cells (A) and punctate structures beneath the cell surface (A′; arrows). GFP–SFT-4 expression was driven by sft-4 promoter. (B) Immunostaining of endogenous SFT-4 shows reticular and punctate (arrows) structures as observed with GFP–SFT-4. (C–C″) GFP–SFT-4 localizes to the ER membrane, labeled here with mC-SP12. GFP–SFT-4 and mCherry-SP12 (mC-SP12) were coexpressed in the intestine and examined. (D–D″) SFT-4 localizes at ERES. The intestines of transgenic animals expressing SEC-23–GFP were stained with anti–SFT-4 antibody. SFT-4 colocalizes with SEC-23–GFP (D″; arrows). (E–G) Loss of SFT-4 reduces the number and size of SEC-23–GFP–positive ERES. SEC-23–GFP was localized to punctate structures, which are presumably ERES in mock animals (E). The number and size of SEC-23–GFP–positive ERES was reduced in sft-4(RNAi) intestinal cells (E, arrowheads). The number (F) and size (G) of SEC-23–GFP–positive ERES in mock and sft-4(RNAi) animals were measured (n = 7 and 10 intestines from mock and sft-4(RNAi) animals, respectively). **, P < 0.05 (Student’s t test); error bars indicate SEM (F). Dotted lines indicate the outlines of intestines. Regions surrounded by squares are enlarged (16× in A–D″ and 9× in E) in insets. Bars: 10 µm; (insets) 5 µm.
We further examined whether loss of SFT-4 affects the ERES formation. When sft-4 was knocked down, SEC-23–GFP–positive ERES were reduced in the number and size, suggesting that SFT-4 is involved in the organization of COPII-positive ERES (Fig. 4, E–G).
SFT-4 specifically recognizes VIT-2 in vivo
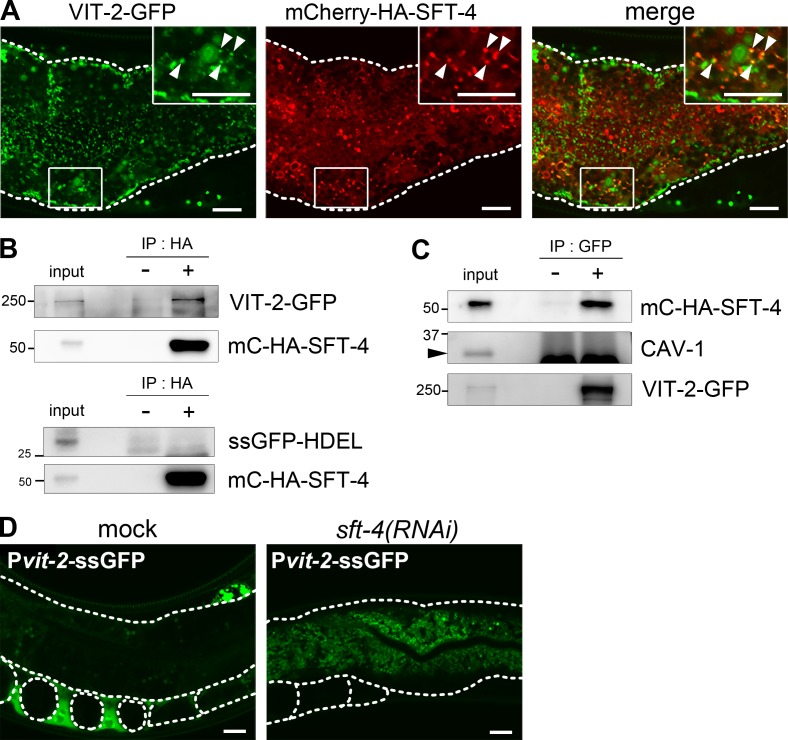
If SFT-4 functions as a sorting receptor for specific substrates, SFT-4 would physically interact with these substrates in the ER. We examined this by generating transgenic animals coexpressing mCherry–HA–SFT-4 (mC–HA–SFT-4) and VIT-2–GFP in the intestine, and found that mC–HA–SFT-4–positive puncta partly overlapped with a population of VIT-2–GFP, which suggests that SFT-4 transiently colocalizes with a population of VIT-2 at ERES (Fig. 5 A, arrowheads). We next performed coimmunoprecipitation experiments to examine the interaction between SFT-4 and VIT-2; here, as negative controls, we used ssGFP-HDEL, which is retained in the ER lumen at steady-state, and CAV-1, a plasma membrane protein (Sato et al., 2006). We found that mC–HA–SFT-4 coimmunoprecipitated with VIT-2–GFP but not ssGFP-HDEL (Fig. 5, B and C), which suggests that SFT-4 specifically interacts with VIT-2 in vivo.
Figure 5.
SFT-4 interacts with VIT-2 in vivo. (A) Subcellular localization of VIT-2 and SFT-4 in intestinal cells. VIT-2–GFP partially colocalized with mC–HA–SFT-4 on punctate structures (arrowheads). (B and C) SFT-4 interacts with VIT-2. Lysates of whole animals coexpressing VIT-2–GFP and mC–HA–SFT-4 were immunoprecipitated with anti-HA (B; upper panel) and anti-GFP (C) antibodies. The precipitates and 0.3% of the total lysate were immunoblotted with anti-HA and anti-GFP antibodies. SFT-4 does not interact with ssGFP-HDEL when lysates of whole animals coexpressing ssGFP-HDEL and mC–HA–SFT-4 were immunoprecipitated with anti-HA antibody (B; lower panel). CAV-1 (C; arrowheads) was not coimmunoprecipitated with VIT-2–GFP. (D) Secretion of ssGFP from the intestine into the body cavity was impaired in sft-4(RNAi) animals. Dotted lines indicate the outlines of intestines, oocytes, and embryos. Regions surrounded by squares are enlarged (4×) in insets. Bars, 10 µm.
We further tested whether SFT-4 is selectively required for a subset of soluble cargos, or whether SFT-4 regulates the transport of soluble cargos in general. For these studies, we created transgenic animals expressing ssGFP under the control of vit-2 promoter; this protein should contain no export signal. ssGFP was synthesized in the intestine and secreted into the body cavity as efficiently as VIT-2, but accumulated in the body cavity and coelomocytes (scavenger cells in the body cavity; Fig. 5 D, left). Unexpectedly, ssGFP was also accumulated in the ER and not secreted into the body cavity in sft-4(RNAi) intestinal cells (Fig. 5 D, right). This result raises the possibility that SFT-4 performs a general function of regulating the ER export of cargo proteins in addition to cargo sorting in animals.
Depletion of mammalian Surf4 impairs ER export of ApoB and affects ERES organization in HepG2 cells
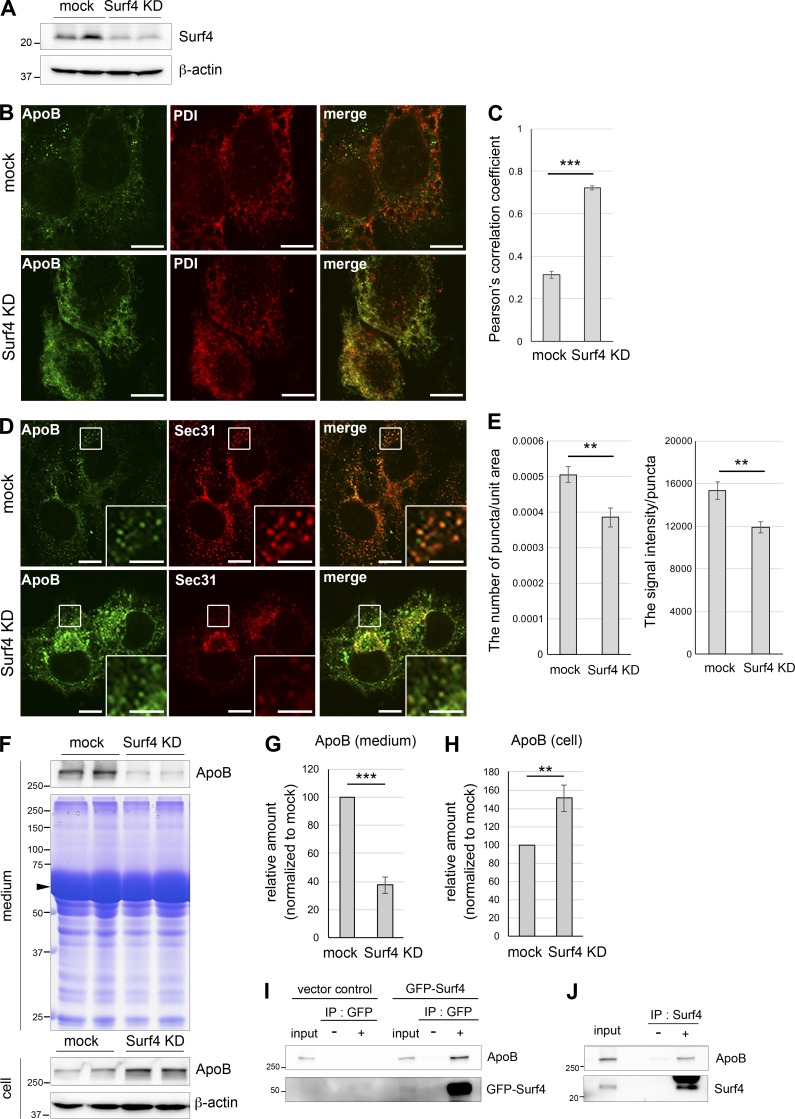
Last, we investigated whether a mammalian homologue of SFT-4, Surf4, also functions similarly to SFT-4 in lipoprotein trafficking. Because C. elegans VIT-2 is a lipid-carrier lipoprotein similar to human ApoB, we examined the requirement of human Surf4 for ApoB secretion from human hepatocellular carcinoma HepG2 cells. We cultured HepG2 cells in the presence of oleic acid (0.1 mM) to induce expression of the ApoB gene (Santos et al., 2016). We then knocked down Surf4 expression by introducing siRNAs into HepG2 cells (Fig. 6 A), and examined the intracellular trafficking of ApoB by using immunofluorescence microscopy. In mock-treated cells, ApoB was mainly detected on punctate structures at steady-state (Fig. 6 B). In contrast, when Surf4 was depleted, ApoB was accumulated in reticular structures and largely colocalized with the ER chaperone protein disulfide isomerase (PDI; Fig. 6, B and C). These results suggest that Surf4 is required for the export of ApoB from the ER. We also found that Surf4 depletion reduced the number of Sec31-positive puncta spread in the cytoplasm, although perinuclear clusters of Sec31-positive puncta were still observed. The signal intensity of each Sec31-positive punctate structure spread in the cytoplasm was also reduced by Surf4 knockdown. This result suggests that Surf4 is involved in the formation or distribution of ERES (Fig. 6, D and E).
Figure 6.
Surf4, a mammalian homologue of SFT-4, is required for ER export of ApoB in HepG2 cells. (A) Whole-cell lysates were immunoblotted with antibodies against Surf4 and β-actin. Surf4 expression was efficiently silenced. (B) Surf4 loss causes ApoB accumulation in the ER. HepG2 cells were transiently transfected for 3 d with control (mock) or surf4 siRNA (Surf4 KD) and then fixed and stained with anti-ApoB and anti-PDI antibodies. In Surf4 KD cells, ApoB was accumulated in the ER and was colocalized with PDI. Bars, 10 µm. (C) Colocalization quantifications were calculated by using Pearson’s correlation coefficient and statistically analyzed using Student’s t test; ***, P < 0.001; error bars: SEM (n = 96 and 87 of mock and Surf4 KD cells, respectively). (D) Loss of Surf4 reduced the number and size of Sec31-positive ERES. HepG2 cells were transiently transfected for 3 d with control (mock) or surf4 siRNA (Surf4 KD) and then fixed and stained with anti-ApoB and anti-Sec31 antibodies. Regions surrounded by squares are enlarged (9×) in insets. Bars: 10 µm; (insets) 5 µm. (E) The quantifications of the number and signal intensity of Sec31-positive punctate structures spread in the cytoplasm were measured and statistically analyzed using Student’s t test; **, P < 0.05; error bars: SEM (left panel, n = 70 and 50 of mock and Surf4 KD cells for the number of puncta/unit area; right panel, n = 62 and 66 of mock and Surf4 KD cells for the signal intensity/puncta, respectively). (F) The pattern of total secreted proteins. Whole-cell lysates and culture medium were immunoblotted with anti-ApoB. Coomassie Brilliant Blue staining of culture medium is also shown in middle panel. The secretion was not generally inhibited by the loss of Surf4. The arrowhead presumably indicates BSA conjugated with oleic acids. (G and H) ApoB amount was quantified through densitometric scanning of band intensities, and the relative amount was determined. The amount of ApoB secreted from Surf4-depleted cells was significantly decreased as compared with that secreted from mock-treated cells (G), whereas the amount of ApoB in whole cell lysates was significantly increased (H). Results were analyzed using Student’s t test; **, P < 0.05; ***, P < 0.001; error bars: SEM, n = 6 (G) or 4 (H) are shown. (I) Lysates from vector control or GFP-Surf4 transfected HepG2 cells were immunoprecipitated with anti-GFP antibody. The precipitates and 1% of the total lysate were immunoblotted with anti-ApoB antibody. (J) HepG2 cell lysates were immunoprecipitated with anti-Surf4 antibody. The precipitates and 1% of the total lysate were immunoblotted with anti-ApoB antibody.
We next analyzed the amount of ApoB protein in the cell lysate and the medium by immunoblotting with an anti-ApoB antibody: Surf4 siRNA-treated cells showed a reduction in the amount of secreted ApoB compared with mock-treated cells, whereas the amount of ApoB retained in the cells was increased in knocked-down cells. These results again suggest that Surf4 is necessary for the efficient secretion of ApoB in HepG2 cells (Fig. 6, F–H). The patterns of total secreted proteins were comparable between mock and Surf4 siRNA-treated cells, suggesting that the secretion still in part occurs in the absence of Surf4 (Fig. 6 F; Mitrovic et al., 2008).
We also examined whether Surf4 physically interacts with ApoB by coimmunoprecipitation experiments. HepG2 cells expressing GFP-Surf4 were subjected to coimmunoprecipitation experiments using an anti-GFP antibody. We found that ApoB was coimmunoprecipitated with GFP-Surf4 (Fig. 6 I). We further tested this interaction by using an anti-Surf4 antibody and found that endogenous ApoB was coimmunoprecipitated with endogenous Surf4, suggesting that a part of Surf 4 exists in a complex containing ApoB (Fig. 6 J). Collectively, these results indicate that Surf4 physically interacts with ApoB and is required for efficient ER export of lipoproteins.
Discussion
In this study, we identified SFT-4/Surf4 family proteins as key regulators of the efficient ER export of soluble proteins such as lipoproteins in animals. In C. elegans, SFT-4 is required for the ER export of yolk proteins in intestinal cells. SFT-4 is also critical for the ER exit of the soluble lysosomal protein CPL-1, but not for the trafficking of transmembrane proteins such as SYN-1 and PGP-1, which suggests that SFT-4 is essential for the ER exit of soluble cargo proteins. Moreover, SFT-4 interacts physically with VIT-2–GFP, which raises the possibility that SFT-4 regulates the ER export of soluble cargo proteins, including lipoproteins, by acting as a cargo receptor in C. elegans as Erv29p does in yeast. We further revealed that mammalian Surf4 physically interacts with ApoB and is necessary for the secretion of human ApoB from HepG2 cells. These findings suggest that the roles of SFT-4/Surf4 family proteins in the ER export of soluble cargo proteins including lipoproteins are widely conserved from worms to mammals.
We showed that the loss of SFT-4 results in the ER accumulation of soluble secretory proteins, including VIT-2 and CPL-1. Unexpectedly, we also found that the secretion of an artificial secreted protein, ssGFP, was inhibited in sft-4(RNAi) intestinal cells. Because mC–HA–SFT-4 was coimmunoprecipitated with VIT-2–GFP but not ssGFP-HDEL, the ER export of ssGFP might be inhibited nonspecifically by the excessive accumulation of authentic cargo proteins in the ER. Supporting this possibility, a large amount of yolk proteins was accumulated in the ER lumen in sft-4(RNAi) intestinal cells. Moreover, the expression level of hsp-4, the gene encoding one of two C. elegans heat shock proteins homologous to mammalian grp78/BiP, was increased in sft-4(RNAi) intestinal cells (unpublished data), which suggests that ER stress was induced by the loss of SFT-4. Thus, the ER export of the artificial secretory protein could be inhibited indirectly by the accumulation of a large amount of cargo proteins in the ER. Alternatively, SFT-4 could be required for the general processes operating in the early secretory pathway, as well as for cargo concentration because loss of SFT-4 and Surf4 reduced the number of COPII-positive ERES in C. elegans and mammalian HepG2 cells, respectively. Yeast Erv29p molecules are known to form a prebudding complex with Sec23p/Sec24p (Miller et al., 2002). A recent study reported that TANGO1, which was originally identified as a cargo receptor for procollagen VII, is also involved in the general process of ER export of secretory proteins: TANGO1 interacts with and recruits Sec16 to ERES and functions as a scaffold protein and acts in a coordinated manner with Sec16 in organizing the formation of ERES (Maeda et al., 2017). Such cargo receptors might play critical roles in the formation of ER-derived vesicles in addition to functioning in the concentration of soluble cargo proteins in animal cells. Surf4 was also reported to interact with ER–Golgi intermediate compartment (ERGIC)–53 and p24 proteins and regulate the structure of ERGIC and Golgi by modulating COPI recruitment in mammalian cells (Mitrovic et al., 2008), and this suggests additional roles of SFT-4/Surf4 proteins in the early secretory pathway, although how SFT-4 works in any of these events is still unclear.
We also showed that Surf4 loss in HepG2 cells causes ApoB accumulation in the ER, which leads to impaired secretion of ApoB. However, the secretion was not generally inhibited by the loss of Surf4 (this study; Mitrovic et al., 2008), which suggests that Surf4 is specifically required for the secretion of a subset of soluble secretory proteins, including ApoB. Recently, the TANGO1/cTAGE5 complex was shown to necessary for ApoB secretion (Santos et al., 2016). The C. elegans genome does not contain genes encoding homologues of TANGO1 or cTAGE5, but in mammals, Surf4 might function redundantly or in a coordinated manner with the TANGO1/cTAGE5 complex in the process of cargo enrichment and loading into VLDL particles.
The secretion of yolk proteins from the intestine was strongly impaired in sft-4–deficient animals; thus, we expected the number of LDs to be increased in intestinal cells. However, we did not observe any noticeable increase of LTR-labeled LDs in sft-4(RNAi) intestinal cells. Instead of an effect on LDs, we detected the accumulation of neutral lipids together with yolk proteins in the ER lumen in sft-4(RNAi) intestinal cells. Furthermore, electron microscopy analysis revealed the accumulation of characteristic electron-dense spherical structures juxtaposed to electron-lucent structures (dalma bodies) in sft-4–deficient intestinal cells. Intriguingly, similar structures have been observed in intestinal cells derived from patients with Anderson’s disease/CM retention disease (AD/CMRD), a rare autosomal recessive disorder associated with mutations in SAR1B (Dannoura et al., 1999; Georges et al., 2011; Okada et al., 2011). AD/CMRD patients present hypocholesterolemia caused by the lack of ApoB-containing lipoproteins and high accumulation of neutral lipids in the small intestinal epithelium. Notably, in the small intestinal epithelium of AD/CMRD patients, numerous lipoprotein-like particles are accumulated in membrane-bound compartments (Georges et al., 2011), and these membrane structures closely resemble the dalma bodies observed in sft-4(RNAi) intestinal cells, which suggests that defective lipoprotein trafficking results in a similar phenotype in C. elegans and mammals. Thus, our findings not only provide insights into the mechanisms underlying the biogenesis and secretion of VLDLs and CMs but also indicate that C. elegans could serve as a highly favorable model system for identifying new targets for the treatment of dyslipidemia, including AD/CMRD and hyperlipidemia.
Materials and methods
General methods and strains
Methods for C. elegans handling and culturing have been described previously (Brenner, 1974). Strains expressing GFP- or mCherry-tagged transgenes were grown at 20°C. The WT parent for all strains was C. elegans var. Bristol N2. The unc-119(ed3) mutant (Maduro and Pilgrim, 1995), VK1256(vkEx1256[Pnhx-2::cpl-1::YFP; Pnhx-2::DsRed::KDEL]), and the deletion allele of sft-4(gk301) balanced with szT1[lon-2(e678)](I;X), were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). Heterozygotes of the sft-4(gk301)/szT1 strain produce WT-like heterozygotes, arrested szT1 aneuploids/homozygotes, Lon males, and dead embryos or arrested larvae lacking szT1, which indicates that sft-4(gk301) homozygosity is mostly embryonic lethal. The transgenic strains used were the following: pwIs23[Pvit-2::vit-2::GFP; unc-119(+)] (Balklava et al., 2007), pwIs98[Pvit-2::vit-2::tdimer2; unc-119(+)] (Sato et al., 2008b), and dkIs37[Pact-5::GFP::pgp-1; unc-119(+)] (Sato et al., 2011). The following transgenic strains were constructed for this study: dkIs778[Psft-4::GFP::sft-4; unc-119(+)], dkIs759[Pact-5::mCherry::HA::sft-4; unc-119(+)], dkIs315[Pact-5::GFP::SP12; unc-119(+)], dkIs116[Pact-5::mCherry::SP12; unc-119(+)], dkIs570[Pact-5::ssGFP::HDEL; unc-119(+)], dkIs808[Pact-5::sec-23::GFP; unc-119(+)], dkIs850[Pplin-1::plin-1::GFP; unc-119(+)], and dkIs738[Pvit-2::ssGFP; unc-119(+)].
Plasmids and transgenic C. elegans
To construct Psft-4::GFP::sft-4, a genomic fragment containing the sft-4 coding region, the 3ʹ UTR of sft-4 (687 bp), and the 5ʹ UTR of sft-4 (2696 bp), and the GFP coding sequence were PCR-amplified, and then these fragments were joined together by means of PCR performed using each fragment as a template. Next, the GFP-sft-4 fragment containing the 5ʹ and 3ʹ UTR of sft-4 was cloned into the Entry vector pDONR221 and transferred into the destination vector pMS20.2 (Sato et al., 2018) by using Gateway recombinational cloning technology (Invitrogen). pMS20.2 containing WT unc-119 was created by cloning a Gateway cassette A into the XbaI site in pDP#MM016b (Praitis et al., 2001). To construct Pplin-1::plin-1::GFP, a genomic fragment containing the 5ʹ UTR of plin-1 (1,042 bp) and the coding region of plin-1, the GFP sequence, and the 3ʹ UTR of plin-1 (1,000 bp) were PCR-amplified, after which the fragments were joined together by performing PCR by using each fragment as a template. The plin-1-GFP fragment containing the 5ʹ and 3ʹ UTR of plin-1 was cloned into pDONR221 and transferred into the destination vector pMS20.2. To construct Pact-5::mCherry::HA::sft-4, Pact-5::GFP::SP12, Pact-5::mCherry::SP12, and Pact-5::sec-23::GFP, genomic fragments containing the coding regions of sft-4, C34B2.10 (SP12), and sec-23 were PCR amplified and cloned into pDONR221 and transferred into the destination vectors pMS6.3, pKS18, pMS6.2 (Sato et al., 2011), and pKS17 (Saegusa et al., 2014), respectively. To construct Pvit-2::ssGFP, a stop codon was cloned into pDONR221 and transferred into the destination vector pMS4.1 containing the promoter region of vit-2 and the DNA fragments encoding the vit-2 signal sequence and GFP. These plasmids were introduced into unc-119(ed3) through microparticle bombardment as described previously (Praitis et al., 2001).
RNAi
We conducted RNAi experiments by using a feeding method (Timmons et al., 2001). L1 larvae were placed on nematode growth medium agar plates containing 5 mM isopropyl β-d-thiogalactopyranoside and Escherichia coli [strain HT115(DE3)] carrying double-stranded RNA expression constructs, and were allowed to grow at 20°C for 3 d. To observe sar-1(RNAi) and sec-23(RNAi) animals, L3 larvae were incubated on RNAi plates at 20°C for 2 d and allowed to grow to the adult stage. The RNAi constructs fed to the animals contained genomic DNA fragments of the genes sft-4, sar-1, sec-23, ile-1, sel-9, rer-1, and syn-1, obtained from the Ahringer Genomic RNAi Library (Kamath and Ahringer, 2003). As a negative control, we used L4440 harboring a cDNA encoding the human transferrin receptor (Sato et al., 2006).
Antibodies
Keyhole limpet hemocyanin-conjugated peptides corresponding to the N terminus of C. elegans SFT-4 (NH2-C+DAAEDFFRKTRTYLPHIAR-COOH), SYN-1 (NH2-C+PRDRLKELQEKATVNTIHA-COOH), and CAV-1 (NH2-C+MSTEQDIKTEEQI-COOH) were produced, and the purified peptides were used for generating polyclonal antibodies in rabbits at Sigma-Aldrich, and Lab Frontier Co Ltd. Mouse monoclonal anti–VIT-2 antibody was provided by B.D. Grant (Rutgers University, New Brunswick, NJ; Grant and Hirsh, 1999). Mouse monoclonal anti–SQV-8 and anti–LMP-1 antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health and maintained at the Department of Biology, University of Iowa (Iowa City, IA). These antibodies were purchased: mouse monoclonal anti–pan-actin antibody (C4; Millipore), goat polyclonal anti-GFP antibody conjugated to HRP (Research Diagnostics), mouse monoclonal anti-GFP antibody (3E6; Q-BIOgene), mouse monoclonal anti-HA antibody (16B12; Covance Research Products), rat monoclonal anti-HA antibody conjugated to peroxidase (3F10; Roche Applied Science), goat polyclonal anti-human ApoB antibody (Rockland), goat polyclonal anti-Surf4 antibody (S-12; Santa Cruz Biotechnology), rabbit monoclonal anti-PDI antibody (C81H6; Cell Signaling Technology), and mouse monoclonal anti-SEC31 antibody (32/Sec31A; BD Biosciences).
Immunoprecipitation from worm lysates
For immunoprecipitation assays, ∼0.5 ml of young adults was collected and washed with M9 buffer, resuspended in a homogenization buffer (1% Triton X-100, 1% sodium deoxycholate, 25 mM Tris-HCl, pH 7.5, and 150 mM NaCl, supplemented with protease inhibitors), and homogenized in a stainless-steel homogenizer (Wheaton). Homogenates were centrifuged at 15,000 g at 4°C, and the collected supernatants were incubated with mouse anti-GFP (3E6) or mouse anti-HA (16B12) antibodies for 1 h at 4°C. Then the immunoprecipitates were collected with Magnetic DynaBeads (Invitrogen). The precipitated beads were washed and eluted according to the manufacturer’s instructions, and the eluates were immunoblotted with a goat anti-GFP antibody conjugated to HRP, a rat anti-HA antibody conjugated to peroxidase (3F10), or a rabbit anti–CAV-1 polyclonal antibody.
Immunostaining and microscopy of the worm intestine
To observe live worms expressing transgenes, worms were mounted on agarose pads containing 20 mM levamisole in M9 buffer (Sato et al., 2008a). Dissected intestines were stained as described previously (Grant and Hirsh, 1999; Sato et al., 2006). For VIT-2 and SYN-1 staining, dissected intestines were fixed in 1.25% PFA/egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 25 mM Hepes, pH 7.3) for 10 min at room temperature. For SQV-8 and LMP-1 staining, dissected intestines were fixed in 3.7% formaldehyde/egg buffer and postfixed in precooled (−20°C) methanol for 5 min. For SFT-4 staining, dissected intestines were fixed in precooled (−20°C) methanol for 5 min. After fixation, the dissected intestines were washed four times in 1 ml of PTB buffer (1% BSA, 1× PBS, 0.1% Tween-20, 0.05% NaN3, and 1 mM EDTA) for 2 h, and then incubated with anti–VIT-2 (1:100), anti–SYN-1 (1:100), anti–SQV-8 (1:100), anti–LMP-1 (1:100), or anti–SFT-4 (1:200) antibodies overnight at 4°C. Next, the samples were washed 4 times with PTC buffer (0.1% BSA, 1× PBS, 0.1% Tween-20, 0.05% NaN3, and 1 mM EDTA) and incubated (overnight, 4°C) with appropriate secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 555 (1:1000; Life Technologies). For LD staining, dissected intestines were fixed in 1.25% PFA/M9 buffer for 10 min and washed in PTB buffer, and then incubated with HCS LTR (Thermo Fisher Scientific) for 30 min at room temperature. Images were obtained using an FV1000 or FV1200 confocal microscopy system (Olympus Corp.). Electron microscopy of C. elegans samples was conducted as described previously (Sato et al., 2011). Analysis of SEC-23–GFP puncta was performed using MetaMorph (Molecular Devices), and the number and size of SEC-23–GFP–positive puncta in the intestinal cells were analyzed (n = 7 and 10 intestines from mock and sft-4(RNAi) animals, respectively).
Cell culture, transfection, and immunostaining of HepG2 cells
HepG2 cells were cultured in DMEM/high glucose (Wako Pure Chemical) supplemented with 10% (vol/vol) FBS at 37°C in a CO2 incubator. Cells were transiently transfected with RNAi oligonucleotides by using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions, and cultured for 2 d. RNAi oligonucleotides (Dharmacon) were used as a mixed siRNA reagent containing four oligonucleotides: human Surf4 siRNAs 09 5′-CCACAAGGGUAGUCGAACA-3′, 10 5′-CGAAUAUUGGUAAGAUCGA-3′, 11 5′-GCUCCCUGUUAGUGCCGUA-3′, and 12 5′-ACGUAUAUUUCAACGCCUU-3′. AllStars Negative Control siRNA (Dharmacon) was used as a control. The cells were incubated with fresh medium without FBS for 1 d and further incubated with fresh medium without FBS but containing 0.1 mM oleic acid conjugated with BSA (Sigma-Aldrich) for 5 h. The cells were washed three times with 1× PBS and fixed with 4% PFA for 15 min. To observe endogenous ApoB, PDI, and Sec31, the cells were permeabilized and blocked with 0.05% saponin and 5% FBS in 1× PBS containing 0.3% BSA for 1 h. Subsequently, the cells were incubated with goat anti-ApoB and anti-PDI or anti-Sec31A antibodies for 1 h at room temperature, washed in 1× PBS, and then treated with appropriate secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 555 (1:1,000; Life Technologies) for 1 h at room temperature. Images were obtained using an FV1000 or FV1200 confocal microscopy system (Olympus Corp.). Colocalization analysis was performed using Fiji software (Schindelin et al., 2012). We cropped each cell as regions of interest, and Pearson’s correlation coefficiency was determined using colocalization analysis with Coloc 2 plugin. In some cells, we observed few spots showing a very bright signal for anti-PDI staining. We avoided such regions to be included in regions of interest because saturated signal hinders colocalization analysis. The number and fluorescent intensity of Sec31-positive puncta in the cytoplasm were quantified using MetaMorph. Perinuclear clusters of Sec31-positive puncta were not counted in this analysis.
Immunoblotting and immunoprecipitation in HepG2 cells
HepG2 cells were treated with siRNA and stimulated with oleic acids for 5 h as described above. To examine the secreted proteins, the culture media were collected and precipitated with 10% TCA; the pellets were washed with ice-cold acetone, dissolved in Laemmli sample buffer, and analyzed by immunoblotting with an anti-ApoB antibody and Coomassie Brilliant Blue staining. To prepare the total cell lysates, cells were washed in 1× PBS and resuspended in a lysis buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, and 1 mM EDTA, supplemented with protease inhibitors), and after incubation for 30 min at 4°C, the cell lysates were centrifuged for 15 min at 15,000 g at 4°C. The supernatants were mixed with Laemmli sample buffer and analyzed by immunoblotting with anti-Surf4 or anti-ApoB antibodies.
For immunoprecipitation assays, pcDNA3.1(+) (vector control) or pcDNA3.1(+)-GFP-Surf4 was transfected into HepG2 cells by using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions, and incubated for 72 h. Cells were collected and washed with 1× PBS and resuspended in a lysis buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, and 1 mM EDTA, supplemented with protease inhibitors), and after incubation for 1 h at 4°C, the cell lysates were centrifuged for 15 min at 15,000 g at 4°C. The supernatants were incubated with anti-GFP (3E6) or anti-Surf4 (S-12) antibodies for 4 h at 4°C. The immune complex was precipitated with protein G Sepharose beads (GE Healthcare). The precipitated beads were washed and eluted according to the manufacturer’s instructions, and the eluates were immunoblotted with an anti-ApoB antibody.
Online supplemental material
Fig. S1 examines the VIT-2 accumulation in sft-4(RNAi), sar-1(RNAi), and sec-23(RNAi) intestinal cells and the brood size of the sft-4 deletion mutants. Fig. S2 examines the morphology of late-Golgi, lysosome, and cytosolic LDs in sft-4(RNAi) intestinal cells. Fig. S3 examines the specificity of anti–SYN-1 antibody and the localization of GFP–PGP-1 in sar-1(RNAi) intestinal cells. Fig. S4 examines the tissue-expression pattern of SFT-4 and the specificity of anti–SFT-4 antibody.
Supplementary Material
Acknowledgments
We thank Dr. Barth D. Grant for supplying reagents. Certain strains used in this study were provided by the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (grant P40 OD010440). We also thank Katsuya Sato, Dr. Rika Hirai, and the other members of the Sato laboratory, Drs. Aisa Sakaguchi (Osaka University), Kota Saito (University of Tokyo), Toyoshi Fujimoto (Nagoya University), Satoshi Yoshida (Waseda University), and Ken Sato (University of Tokyo), for technical assistance and discussions.
K. Sato was supported by the Japan Society for the Promotion of Science KAKENHI (grants 26291036, 17K19377, and 17H03669), the Uehara Memorial Foundation, the Ono Medical Research Foundation, and the Takeda Science Foundation. M. Sato was supported by the Ministry of Education, Culture, Sports, Science and Technology KAKENHI (grant 16H01191). K. Saegusa was supported by a Grant-in-Aid for Japan Society for the Promotion of Science Fellows (grant 16J11698). This work was supported by the Joint Research Program of the Institute for Molecular and Cellular Regulation at Gunma University.
The authors declare no competing financial interests.
K. Saegusa, M. Sato, and K. Sato conceived and designed the experiments. K. Saegusa, M. Sato, N. Morooka, and T. Hara performed the experiments. K. Saegusa, M. Sato, and K. Sato analyzed the data and wrote the manuscript.
References
- Babin P.J., Bogerd J., Kooiman F.P., Van Marrewijk W.J., and Van der Horst D.J.. 1999. Apolipophorin II/I, apolipoprotein B, vitellogenin, and microsomal triglyceride transfer protein genes are derived from a common ancestor. J. Mol. Evol. 49:150–160. 10.1007/PL00006528 [DOI] [PubMed] [Google Scholar]
- Baker M.E. 1988. Is vitellogenin an ancestor of apolipoprotein B-100 of human low-density lipoprotein and human lipoprotein lipase? Biochem. J. 255:1057–1060. 10.1042/bj2551057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Belden W.J., and Barlowe C.. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 294:1528–1531. 10.1126/science.1065224 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cefalù A.B., Pirruccello J.P., Noto D., Gabriel S., Valenti V., Gupta N., Spina R., Tarugi P., Kathiresan S., and Averna M.R.. 2013. A novel APOB mutation identified by exome sequencing cosegregates with steatosis, liver cancer, and hypocholesterolemia. Arterioscler. Thromb. Vasc. Biol. 33:2021–2025. 10.1161/ATVBAHA.112.301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J., and Barlowe C.. 2010. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79:777–802. 10.1146/annurev-biochem-061608-091319 [DOI] [PubMed] [Google Scholar]
- Dannoura A.H., Berriot-Varoqueaux N., Amati P., Abadie V., Verthier N., Schmitz J., Wetterau J.R., Samson-Bouma M.E., and Aggerbeck L.P.. 1999. Anderson’s disease: exclusion of apolipoprotein and intracellular lipid transport genes. Arterioscler. Thromb. Vasc. Biol. 19:2494–2508. 10.1161/01.ATV.19.10.2494 [DOI] [PubMed] [Google Scholar]
- Di Filippo M., Moulin P., Roy P., Samson-Bouma M.E., Collardeau-Frachon S., Chebel-Dumont S., Peretti N., Dumortier J., Zoulim F., Fontanges T., et al. 2014. Homozygous MTTP and APOB mutations may lead to hepatic steatosis and fibrosis despite metabolic differences in congenital hypocholesterolemia. J. Hepatol. 61:891–902. 10.1016/j.jhep.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Fisher E.A., and Ginsberg H.N.. 2002. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem. 277:17377–17380. 10.1074/jbc.R100068200 [DOI] [PubMed] [Google Scholar]
- Georges A., Bonneau J., Bonnefont-Rousselot D., Champigneulle J., Rabès J.P., Abifadel M., Aparicio T., Guenedet J.C., Bruckert E., Boileau C., et al. 2011. Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson’s disease (Chylomicron retention disease). Orphanet J. Rare Dis. 6:1 10.1186/1750-1172-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., and Hirsh D.. 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 10:4311–4326. 10.1091/mbc.10.12.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova V., Brodsky J.L., and Fisher E.A.. 2003. Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J. Biol. Chem. 278:48051–48058. 10.1074/jbc.M306898200 [DOI] [PubMed] [Google Scholar]
- Henricson A., Sonnhammer E.L., Baillie D.L., and Gomes A.V.. 2004. Functional characterization in Caenorhabditis elegans of transmembrane worm-human orthologs. BMC Genomics. 5:85 10.1186/1471-2164-5-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M.M., Shi J., and Dreizen P.. 2003. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J. Lipid Res. 44:22–32. 10.1194/jlr.R200014-JLR200 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Maduro M., and Pilgrim D.. 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 141:977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M., Katada T., and Saito K.. 2017. TANGO1 recruits Sec16 to coordinately organize ER exit sites for efficient secretion. J. Cell Biol. 216:1731–1743. 10.1083/jcb.201703084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Geier C., Henske A., Mukherjee S., Hirsh D., Thiele C., Grant B., Maxfield F.R., and Kurzchalia T.V.. 2001. Distribution and transport of cholesterol in Caenorhabditis elegans. Mol. Biol. Cell. 12:1725–1736. 10.1091/mbc.12.6.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedel M.T., Graf N.J., Stephen K.E., Long O.S., Pak S.C., Perlmutter D.H., Silverman G.A., and Luke C.J.. 2012. A pro-cathepsin L mutant is a luminal substrate for endoplasmic-reticulum-associated degradation in C. elegans. PLoS One. 7:e40145 10.1371/journal.pone.0040145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E., Antonny B., Hamamoto S., and Schekman R.. 2002. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 21:6105–6113. 10.1093/emboj/cdf605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic S., Ben-Tekaya H., Koegler E., Gruenberg J., and Hauri H.P.. 2008. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell. 19:1976–1990. 10.1091/mbc.E07-10-0989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Miyashita M., Fukuhara J., Sugitani M., Ueno T., Samson-Bouma M.E., and Aggerbeck L.P.. 2011. Anderson’s disease/chylomicron retention disease in a Japanese patient with uniparental disomy 7 and a normal SAR1B gene protein coding sequence. Orphanet J. Rare Dis. 6:78 10.1186/1750-1172-6-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupard M.C., Miller A., Grant B., Hirsh D., and Hall D.H.. 2001. Immuno-EM localization of GFP-tagged yolk proteins in C. elegans using microwave fixation. J. Histochem. Cytochem. 49:949–956. 10.1177/002215540104900803 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., and Austin J.. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa K., Sato M., Sato K., Nakajima-Shimada J., Harada A., and Sato K.. 2014. Caenorhabditis elegans chaperonin CCT/TRiC is required for actin and tubulin biogenesis and microvillus formation in intestinal epithelial cells. Mol. Biol. Cell. 25:3095–3104. 10.1091/mbc.E13-09-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Chen M., Bard F., Chen S., Zhou H., Woodley D., Polischuk R., Schekman R., and Malhotra V.. 2009. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 136:891–902. 10.1016/j.cell.2008.12.025 [DOI] [PubMed] [Google Scholar]
- Santos A.J., Nogueira C., Ortega-Bellido M., and Malhotra V.. 2016. TANGO1 and Mia2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J. Cell Biol. 213:343–354. 10.1083/jcb.201603072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M., Audhya A., Oegema K., Schweinsberg P., and Grant B.D.. 2006. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol. Biol. Cell. 17:3085–3094. 10.1091/mbc.E06-03-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Grant B.D., Harada A., and Sato K.. 2008a Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J. Cell Sci. 121:3177–3186. 10.1242/jcs.034678 [DOI] [PubMed] [Google Scholar]
- Sato M., Sato K., Liou W., Pant S., Harada A., and Grant B.D.. 2008b Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27:1183–1196. 10.1038/emboj.2008.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Saegusa K., Sato K., Hara T., Harada A., and Sato K.. 2011. Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol. Biol. Cell. 22:2579–2587. 10.1091/mbc.E11-04-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K., Tomura K., Kosako H., and Sato K.. 2018. The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat. Cell Biol. 20:81–91. 10.1038/s41556-017-0008-9 [DOI] [PubMed] [Google Scholar]
- Schekman R., and Mellman I.. 1997. Does COPI go both ways? Cell. 90:197–200. 10.1016/S0092-8674(00)80326-8 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S.A. 2008. VLDL exits from the endoplasmic reticulum in a specialized vesicle, the VLDL transport vesicle, in rat primary hepatocytes. Biochem. J. 413:333–342. 10.1042/BJ20071469 [DOI] [PubMed] [Google Scholar]
- Tabas I., Williams K.J., and Borén J.. 2007. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 116:1832–1844. 10.1161/CIRCULATIONAHA.106.676890 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D.L., and Fire A.. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Williams K.J., and Tabas I.. 1995. The response-to-retention hypothesis of early atherogenesis. Arterioscler. Thromb. Vasc. Biol. 15:551–561. 10.1161/01.ATV.15.5.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.