Nuclear mechanotransduction has been implicated in the control of chromatin organization and gene expression. Wang et al. show that, in Drosophila myofibers, the LINC complex is required for the regulation of DNA replication and synchronized cell-cycle progression in myonuclei.
Abstract
Nuclear mechanotransduction has been implicated in the control of chromatin organization; however, its impact on functional contractile myofibers is unclear. We found that deleting components of the linker of nucleoskeleton and cytoskeleton (LINC) complex in Drosophila melanogaster larval muscles abolishes the controlled and synchronized DNA endoreplication, typical of nuclei across myofibers, resulting in increased and variable DNA content in myonuclei of individual myofibers. Moreover, perturbation of LINC-independent mechanical input after knockdown of β-Integrin in larval muscles similarly led to increased DNA content in myonuclei. Genome-wide RNA-polymerase II occupancy analysis in myofibers of the LINC mutant klar indicated an altered binding profile, including a significant decrease in the chromatin regulator barrier-to-autointegration factor (BAF) and the contractile regulator Troponin C. Importantly, muscle-specific knockdown of BAF led to increased DNA content in myonuclei, phenocopying the LINC mutant phenotype. We propose that mechanical stimuli transmitted via the LINC complex act via BAF to regulate synchronized cell-cycle progression of myonuclei across single myofibers.
Introduction
Tissue mechanics has been linked to cellular and nuclear signals required for appropriate gene expression in differentiated cells (Discher et al., 2005; Hampoelz et al., 2011; Janmey and Miller, 2011; Li et al., 2011; Ho et al., 2013; Swift and Discher, 2014; Denais et al., 2016). However, the mechanisms transducing tissue mechanical signals into the nucleus and the way they impact the precise nuclear response have not been fully elucidated.
Striated muscle fibers are multinucleated cells with a highly ordered cytoplasmic architecture essential for muscle contraction. The myonuclei in each muscle fiber are exposed to both tissue mechanics, as well as to cytoplasmic forces promoted by waves of muscle contraction and relaxation. Whether these mechanical inputs are essential for controlling gene expression and whether and how the nuclear response is synchronized between the multiple nuclei of an individual muscle fiber are fundamental questions in muscle biology.
Cytoplasmic mechanical inputs are transmitted across the nuclear membrane by the linker of nucleoskeleton and cytoskeleton (LINC) complex, composed of cytoplasmic components of the Nesprin-family members (Wang et al., 2009; Rajgor and Shanahan, 2013; Chang et al., 2015; Meinke and Schirmer, 2015; Janota et al., 2017). These components are associated with the cytoplasmic cytoskeleton as well as with the outer nuclear membrane through their klarsicht, ANC-1, syne homology (KASH) domain. Subsequent mechanical input is transmitted through physical interaction of Nesprins with inner-nuclear-membrane Sad1/UNC-84 (SUN) domain proteins at the perinuclear space (Starr and Fridolfsson, 2010; Cain et al., 2014; Cain and Starr, 2015) and through their association with the nuclear lamina components lamin A/C and lamin B (Burke and Stewart, 2013, 2014; Osmanagic-Myers et al., 2015), as well as with various chromatin factors at the nucleoplasm (Dahl et al., 2008; Geyer et al., 2011; Barton et al., 2015). The LINC complex is therefore capable of transducing cytoplasmic mechanical input across the nuclear membrane, which can then be translated into chromatin binding and control of transcriptional output. Examples of transcriptional and epigenetic alterations induced by cellular mechanics have been recently described in stem cells (Chalut et al., 2012; Murphy et al., 2014; Le et al., 2016) and cultured cells under variable mechanical strains (Trappmann et al., 2012; Downing et al., 2013; Tajik et al., 2016). However, it is not clear how and whether cells within an intact tissue respond to mechanical stimuli and whether muscles, adapted to withstand contractile cytoplasmic mechanical inputs (induced by contraction/relaxation waves), are sensitive to such stimuli. The wide array of muscular dystrophies associated with mutations in LINC complex–associated genes emphasizes the functional significance of LINC-mediated mechanical inputs specifically for muscle tissue (Bossie and Sanders, 1993; Puckelwartz et al., 2009; Horn, 2014).
The Drosophila melanogaster genome contains two Nesprin-like genes, Msp300 (Volk, 1992) and Klarsicht (klar; Welte et al., 1998); a single SUN domain protein Klaroid (Koi; Kracklauer et al., 2007); and two lamin genes, lamC (Bossie and Sanders, 1993) and lam (Smith et al., 1987). To address myonuclear sensitivity to mechanical stimuli in muscle fibers, we analyzed fully differentiated striated muscles of Drosophila larvae mutant for the LINC complex genes klar and koi. Our previous studies indicated that myonuclear morphology is severely impaired in LINC complex mutants Msp300, klar, and koi, leading to defective larval movement. The myonuclei in these mutants are mispositioned, and their shape is altered (Elhanany-Tamir et al., 2012; Wang et al., 2015).
In the present study, we demonstrate that an observed variability in myonuclear size within individual myofibers of the LINC mutants results from deregulated, unsynchronized DNA endoreplication (endocycling) in myonuclei. Furthermore, impairment of LINC-independent mechanical input affected myonuclear DNA content in a similar manner, supporting a direct link between cytoplasmic mechanics and cell-cycle progression. Genomic analysis of LINC versus control muscles identified barrier-to-autointegration factor (BAF) as a potential transducer of LINC-dependent deregulated endoreplication. Our study thus demonstrates, for the first time to our knowledge, that the LINC complex is essential for regulation of cell-cycle progression in myonuclei sharing a common cytoplasm. Because mutations in LINC complex genes often lead to various muscular dystrophies and cardiomyopathies in humans, our study is likely to provide novel molecular insight into the way the loss of these genes leads to nonfunctional muscle tissue.
Results
LINC mutant myonuclei contain increased levels of DNA
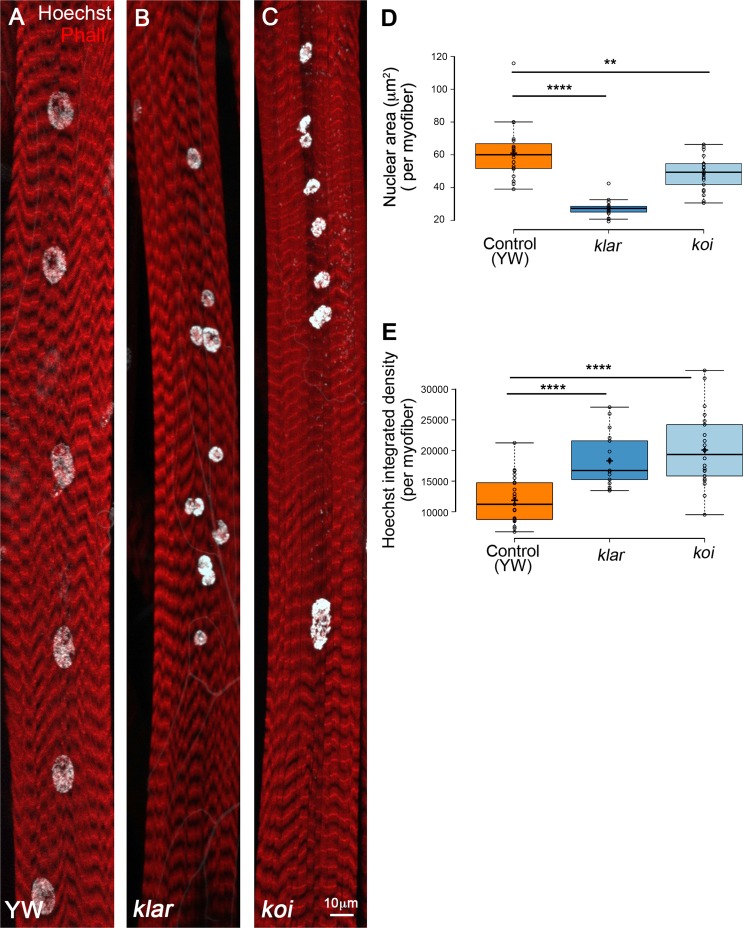
During larval growth, the skeletal muscles grow by ∼40-fold while maintaining a steady number of nuclei. Nuclear size scales with muscle growth by DNA endoreplication (Demontis and Perrimon, 2009). Interestingly, we found that in the LINC complex mutants, klar and koi, myonuclear growth did not scale with muscle growth resulting with variable nuclear size within individual myofibers. Fig. 1 (A–C) shows representative images of the myonuclei of muscle 7 in klar- and koi-null mutants demonstrating myonuclear position defects, described previously (Elhanany-Tamir et al., 2012), as well as size variability. Quantification of the nuclear area in klar and koi mutant myonuclei in muscle 7 (in four abdominal segments of six distinct larvae) indicated a smaller myonuclear size on average relative to control (Fig. 1 D).
Figure 1.
LINC mutant myonuclei are smaller and contain increased DNA content. Representative projected confocal images of muscle 7 from third-instar larvae of control (A; yw), klar (B), or koi (C) null mutants, labeled with Phalloidin (red) and Hoechst (white). Bar, 10 µm. (D) Quantitative analysis of the nuclear area of control, klar or koi myonuclei. Each dot represents the mean nuclear area value of all myonuclei in an individual myofiber. The myonuclear area of klar and koi are both significantly smaller than control (unpaired t test: ****, P = 10−10; **, P = 0.0039). (E) Quantitative analysis of Hoechst integrated density of control, klar, and koi myonuclei. Each data point represents the mean Hoechst integrated density value of all myonuclei of an individual myofiber 7. klar and koi myonuclei contain more DNA than control (unpaired t test: ****, P ≤ 10−6). Whiskers in D and E extend to data points less than 1.5 interquartile ranges from the first and third quartiles. Images were taken from six distinct larvae (control, n = 23; klar, n = 18; koi, n = 22).
Assessment of the DNA content in the LINC mutant myonuclei was also performed. Fully differentiated larval myonuclei undergo DNA endoreplication, a process in which multiple cycles of G1 to S phases take place without progressing into G2 and M phases, leading to DNA polyploidy (Lee et al., 2009; Fox and Duronio, 2013; Frawley and Orr-Weaver, 2015; Orr-Weaver, 2015). In Drosophila, a wide variety of cell types undergo multiple rounds of endocycling during larval and adult stages leading to polyploid nuclei. Endoreplication is commonly associated with growth in cell size in response to physiological demands and was described in plants, mice, and humans (Bergmann et al., 2009; Lee et al., 2009; Liu et al., 2010; Zielke et al., 2013; Jamin and Wiebe, 2015; Cao et al., 2017). We measured the DNA content in individual myonuclei of LINC mutant and control muscle 7, by calculating Hoechst fluorescent integrated density, as described (Xiang et al., 2017). Whereas control myonuclei contained comparable DNA content in all myonuclei along a given myofiber, the LINC mutant myonuclei contained more DNA content overall, and a greater variability between myonuclei of individual myofibers was observed (Fig. 1 E). Because Msp300 mutant larvae are smaller, we did not perform this analysis on these mutants.
To address whether the aberrant nuclear position observed in all LINC mutants led to the variable DNA content, we measured the DNA content in larval muscles from the Oregon R strain, which exhibits myonuclear position defects, and could not find any correlation between nuclear position and DNA content (Fig. S1). Collectively, it was concluded that, despite their relatively smaller size, the LINC mutant myonuclei contain more DNA.
Enhanced, unsynchronized cell-cycle progression is observed in myonuclei of the LINC mutant klar
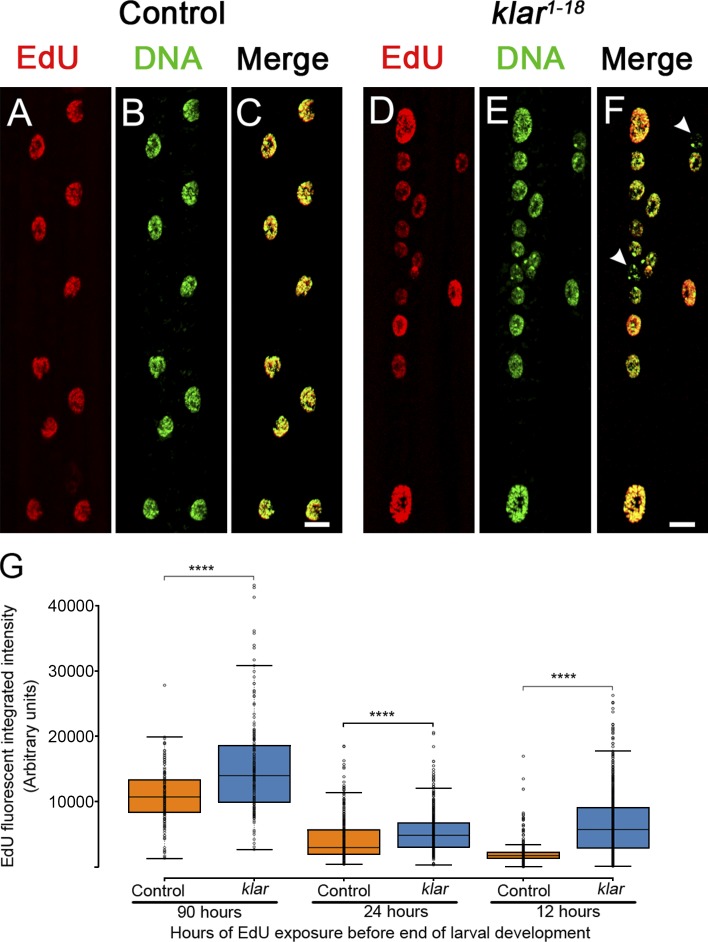
To further determine the basis for the elevated DNA content in LINC mutant myonuclei, DNA replication was analyzed in a LINC representative mutant klar. Larvae were fed with 5-ethynyl-2'-deoxyuridine (EdU)–enriched food, and the extent of EdU incorporation into DNA of myonuclei was monitored in the different syncytial myofibers. Feeding larvae during its growth phase for 48 h (from early-second to late-third instar) led to approximately equal incorporation of EdU into all myonuclei of control muscle fibers. However, klar mutant myonuclei incorporated variable EdU levels even within individual myofibers (Fig. 2, A–F). Quantification of the EdU incorporation after a wide window of EdU feeding (90 h, from first-instar stage), indicated a significantly higher EdU incorporation in klar mutant myonuclei (Fig. 2 G). This tendency was preserved when second-instar larvae were fed for 24 h with EdU (Fig. 2 G) or when third-instar larvae were fed for 12 h. In all three time phases the larvae underwent a recovery phase from the EdU containing food for 12 h before dissection. Notably, in the late developmental time point, control myonuclei showed very low incorporation of EdU, whereas klar mutant myonuclei continued to incorporate EdU into DNA, consistent with recurring endoreplication (Fig. 2 G). These results indicate that cell-cycle progression in klar myonuclei is dysregulated during larval growth. Because klar mutant larvae undergo pupariation at a time scale similar to the control, it is unlikely that the observed dysregulated endocycling occur because of developmental delay, favoring a direct impact of LINC-dependent function on cell-cycle progression.
Figure 2.
Impaired synchronization of DNA replication and dysregulated cell-cycle progression in klar mutant myonuclei. (A–F) Representative images of EdU incorporation (after EdU feeding for 48 h from early-second to the late-third instar larval stages) in WT (control) (A–C) or klar mutant muscle 6 (D–F) indicated by EdU labeling (red) and DAPI (green) labeling. All nuclei in a specific WT muscle fiber are approximately equally labeled, whereas in klar mutant myonuclei EdU labeling is variable (white arrowheads in F show an EdU-negative nucleus). Bars, 10 µm. (G) Quantification of EdU integrated density of EdU (fluorescence signal) in control (orange) and klar mutant (blue) myonuclei in muscle 7 after EdU feeding of 90, 24, or 12 h of larval development in staged larvae. Each data point represents a single nucleus. klar mutant myonuclei incorporated more EdU in all incubation periods (unpaired t test: ****, P ≤ 6 × 10−7). For 90 h: control, n = 158 and klar, n = 174 myonuclei; for 24 h: control, n = 377 and klar, n = 760; for 12 h: control, n = 568 and klar, n = 760; for all samples four muscles from six distinct larvae were analyzed. Whiskers extend to data points less than 1.5 interquartile ranges from the first and third quartiles.
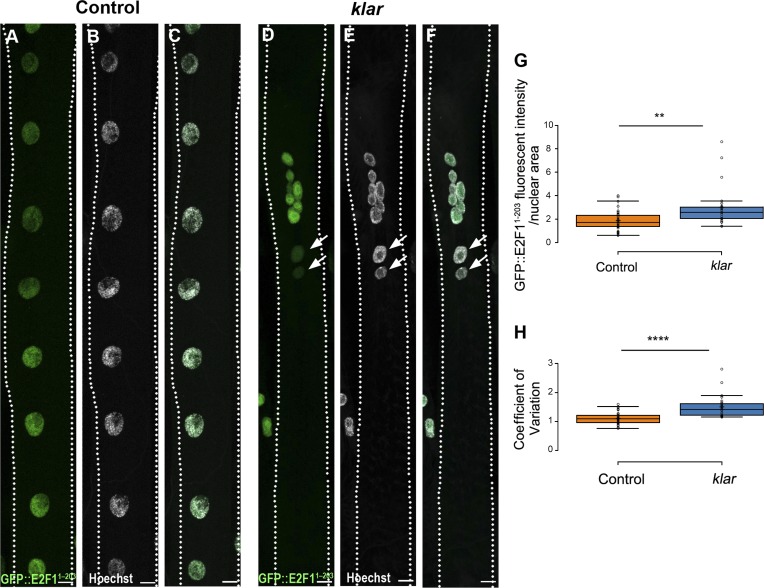
Cell-cycle progression in larval stages depends on E2F1 cycling (Edgar and Orr-Weaver, 2001; Edgar et al., 2014). We measured the rate of E2F1 degradation in myonuclei of individual myofibers. Toward this end, staged larvae expressing eGFP fused to E2F1 degron (eGFP::E2F11–203) under ubiquitous promoter (Zielke et al., 2014) were analyzed for GFP intensity in individual myofibers. Consistent with abrogated synchronization of cell-cycle progression in the klar mutant myofibers, we observed high degree of variability in eGFP::E2F11–203 fluorescent intensity in myonuclei of individual myofibers of klar mutant relative to control (Fig. 3, A–F and H). In addition, the GFP mean intensity (normalized to nuclear area) was higher in klar myonuclei, consistent with impaired G1 to S cell-cycle progression (Fig. 3 G). To evaluate whether endogenous E2F1 is expressed in larval muscles, we followed the levels of its direct target proliferating cell nuclear antigen (PCNA)–GFP in larval muscles and found a comparable level of GFP (Fig. S2). Collectively, it was concluded that cell-cycle progression is not synchronized between myonuclei of individual klar mutant myofibers, in contrast to control, and that dysregulation presumably takes place during the G1 to S phases.
Figure 3.
Impaired synchronization of E2F1 degradation in klar mutant myonuclei. Representative confocal projection images of control (A–C) or klar mutant (D and E) muscle 7 in third-instar larvae, expressing GFP fused to E2F1 degron sequences (GFP::E2F11–203), and labeled with anti-GFP (A and D, green) and Hoechst (B and E, white). (C and F) Corresponding merged images. Bars, 10 µm. Arrows indicate negative GFP-labeled myonuclei in klar mutant. (G) Quantitative analysis of the ratio between GFP mean fluorescent intensity and nuclear area of muscle 7, indicating higher GFP intensity in klar mutant versus control myonuclei (t test: **, P = 0.0085). (H) Quantification of the variation in myonuclear GFP intensity per individual myofiber 7, in control and klar mutant myonuclei (control, n = 42; klar, n = 28). The graph presents the coefficient of variation (standard deviation divided by the mean GFP intensity in myonuclei of an individual myofiber) in klar versus control, indicating higher variability of GFP intensity in klar mutant myonuclei (t test: ****, P = 8.95 × 10−8). Whiskers in G and H extend to data points less than 1.5 interquartile ranges from the first and third quartiles.
Temporal disruption of the microtubule cytoskeleton (MT) in larval myofibers does not affect DNA content
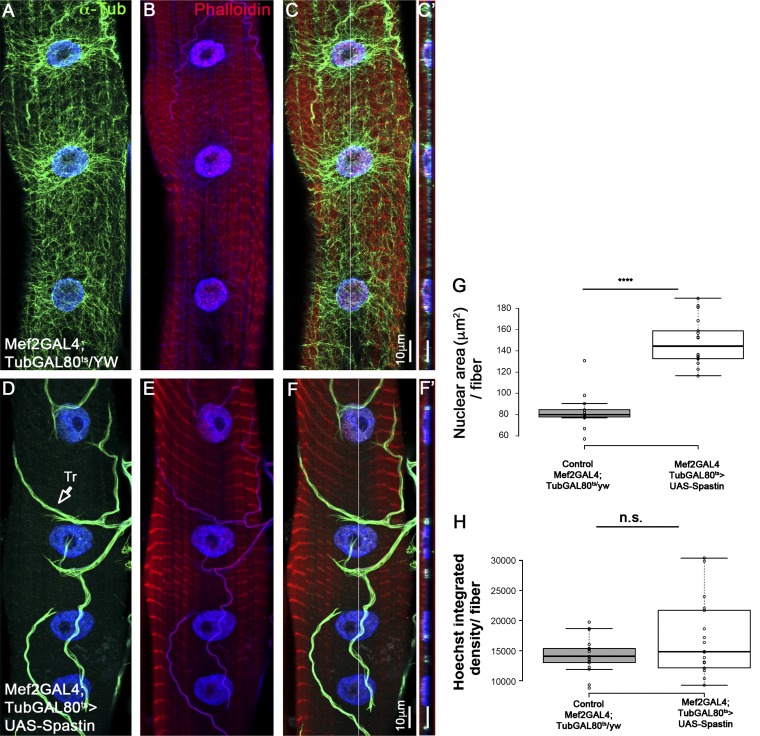
Previous studies indicated the essential contribution of the MT, MT-motor proteins, as well as MT-binding proteins, for myonuclear position (Elhanany-Tamir et al., 2012; Metzger et al., 2012; Folker et al., 2014; D’Alessandro et al., 2015; Wang et al., 2015; Wilson and Holzbaur, 2015; Gimpel et al., 2017). To address whether the MT network contributes to DNA endoreplication in larval myofibers, we induced temporal disruption of the MT network in second-instar larvae by expressing the MT-severing protein Spastin. Importantly, MT disruption was induced after myofiber differentiation and nuclear position were established but before the myofiber growth period, when DNA endoreplication takes place. Unexpectedly, we found that nuclear position was not significantly disrupted in myofibers lacking MT for 24–48 h (Fig. 4, A–F), and the DNA content was on average similar to that of control (Fig. 4 H). However, myonuclear area was greater relative to control (Fig. 4 G), implying a role for the MT network in providing nuclear constraints. Prolonged expression of Spastin (between 48 and 72 h) led to aberrant organization of the sarcomeres, as well as to defects in myonuclear position, possibly secondary for sarcomere disruption (unpublished data). These experiments imply that the nuclear-associated MT network does not primarily contribute to regulation of cell-cycle progression in larval myofibers.
Figure 4.
Myonuclear DNA content does not change in myofibers with disrupted microtubules. Representative single confocal stacks of third-instar larvae muscle 7 in control (A–C) or after muscle-specific, temporal expression of the MT-severing protein Spastin induced for 2 d during second to third-instar larval growth stages (D–F). Muscles were labeled with anti–α-Tubulin (green; A, C, D, and F), Hoechst (blue), and Phalloidin (red; B, C, E, and F). C and F are merged images. Bar, 10 µm. Note the deletion of α-Tubulin labeling in the Spastin-expressing muscles and its normal expression in the trachea (Tr., arrow; D). Ortho view shown in C′ and F′ corresponds to the white lines in C and in F. (G) Quantitative analysis of the mean myonuclear area in control (Mef2-GAL4;TubGAL80/yw) versus MT-depleted muscle 7 during second- to third-instar stage (Mef-GAL4;TubGAL80>UAS-Spastin) indicates higher nuclear area in the latter group (t test: ****, P = 6 × 10−12). (H) Quantitative analysis of the mean myonuclear Hoechst integrated density in control (Mef2-GAL4;TubGAL80/yw) versus MT-deleted muscle 7 during second- to third-instar stage (Mef-GAL4;TubGAL80>UAS-Spastin) indicates no significant difference between the two groups (t test: P = 0.13). Images were taken from five different larvae (control, n = 17; Spastin RNAi. n = 18). Whiskers in G and H extend to data points less than 1.5 interquartile ranges from the first and third quartiles.
Varying mechanical inputs within myofibers alters endoreplication in growing muscles
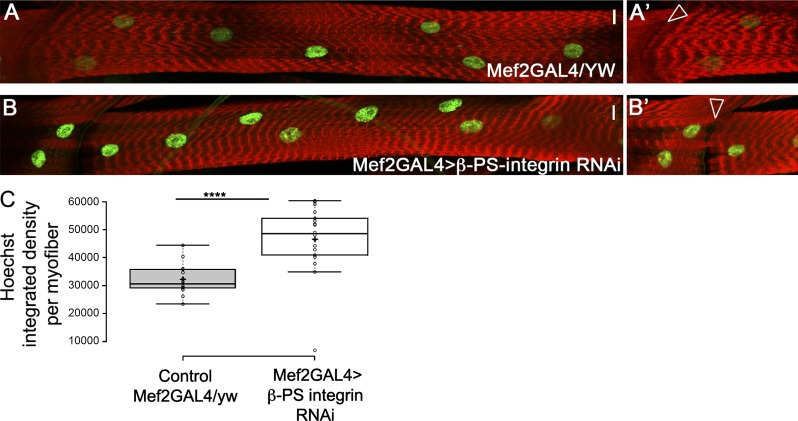
To further address the contribution of mechanical forces applied on myonuclei to DNA endoreplication, we knocked down β-PS-integrin specifically in muscles using RNAi. In this setup, muscle adhesion to tendons was partially inhibited, whereas sarcomere structure was preserved in third-instar larvae (Fig. 5, B and B′). We observed increased DNA content in the β-PS-integrin knocked-down myonuclei (Fig. 5 C) although nuclear position was not altered, relative to control. This result supports a direct link between altering the cytoplasmic mechanical inputs applied on myonuclei and regulation of the cell cycle. An additional attempt to alter nuclear mechanics was performed by a muscle-specific knockdown of Sallimus (Sls, Drosophila Titin). Sls is required for sarcomere organization (Hakeda et al., 2000) and for the recruitment of Msp300 to the Z-discs (Elhanany-Tamir et al., 2012). Assuming that myonuclei are tethered to the sarcomeres through Msp300 association with D-Titin/Sls, we expected an impact on myonuclear mechanics after knockdown of Sls by RNAi. Unfortunately, the Sls knockdown larvae were slightly smaller and exhibited significant impaired motility. Nevertheless, we observed abrogated nuclear position and decreased nuclear size in the Sls knockdown myofibers. Hoechst mean intensity was higher in these myonuclei; however, Hoechst integrated density was not significantly altered in the myonuclei (Fig. S3). We suspect that defects in larval movement and feeding could indirectly affect endoreplication. Interestingly, the decrease in myonuclear size correlates with that observed in the LINC mutants, supporting the possibility that myonuclear anchorage with the sarcomeres is essential for the maintenance of myonuclear size.
Figure 5.
Muscle-specific knockdown of β-PS-integrin promotes increased DNA content in myonuclei. Representative images of control (A and A′, Mef2-GAL4/yw) or β-PS-integrin knockdown (B and B′) muscle 7 labeled with phalloidin (red) and Hoechst (green). A′ and B′ are distinct focal planes at the level of muscle–muscle junction displaying similar magnification. Arrowheads demonstrate the adhesion defect induced by knockdown of β-PS-integrin (compare arrowheads in B′ to A′). Bars, 10 µm. (C) Quantification of the Hoechst integrated density of myonuclei per myofibers in muscle 7 indicates a significant increase in DNA content in β-PS-integrin knockdown versus control (t test: ****, P = 2.5 × 10−6). Images were taken from five distinct larvae (control, n = 14; integrin RNAi, n = 14). Whiskers extend to data points less than 1.5 interquartile ranges from the first and third quartiles.
Genomic analysis of RNA-Pol II occupancy in klar mutant larval muscles
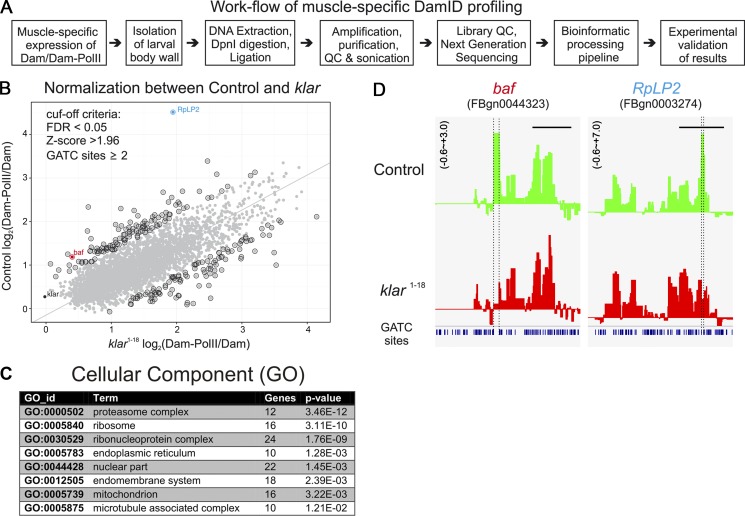
To identify genes that act downstream of the LINC complex, we adopted the targeted DamID approach, developed for tissue-specific expression in flies (Southall et al., 2013), to characterize genes with altered RNA-polymerase (Pol) II occupancy in klar mutant myofibers. This approach allows us to directly follow the profile of RNA-Pol II binding to DNA in klar and control mature myofibers, while circumventing the technical difficulty involved in isolating pure muscle tissue from the larvae as well as batch effects of RNA preparations, and thus compare directly the profile of RNA-Pol II binding to DNA in klar and control myofibers. In brief, muscle-specific RNA-Pol II (RpII215), fused to the DNA adenine methyltransferase from Escherichia coli, is driven by using a muscle-specific driver Mef2-GAL4, whose expression is activated only in mature larval muscles. A temperature-specific GAL4 inhibitor Gal80ts, was inactivated at the third-instar larval stage, allowing the Mef2-GAL4 to drive the Dam-Pol II in differentiated myofibers. The Dam-Pol II experiments included four independent repetitions for each of the four genotypes: (1) control, Dam only; (2) WT, Dam-Pol II; (3) klar, Dam only, and (4) klar, Dam-Pol II. The larvae of each genotype were dissected, and their DNA was extracted and processed with the specific DpnI enzyme that cuts the methylated GATC sites sensitive to the Dam activity. After an additional PCR amplification step and further processing of the DNA, we performed high-throughput DNA sequencing (Fig. 6, A and B). The results were further analyzed by a bioinformatics pipeline (Marshall and Brand, 2015), which essentially subtracts background of nonspecific Dam binding and further highlights the gene-specific loci.
Figure 6.
Profiling of Dam-RNA-Pol II genome occupancy in WT and klar mutant muscles. (A) Flow chart of the experimental procedures for the muscle-specific Dam-Pol II profiling. (B) Normalization of the high-throughput data by using a regression analysis between WT and the klar mutant. Gray dots in the plot represent single genes identified in the muscle-specific targeted Dam-Pol II occupancy. By setting the cut-off criteria false discovery rate (FDR) <0.05 and z-score >1.96 and focusing on more than one Dam-methylation site, the positive hits are highlighted by a black circle. The gene with highest z-score RpLP2 as well as klar are separately labeled. Principal component analysis was performed on a matrix composed of the x and y axis as features of the points. The axis was rotated to the principle components and derives the line that passes through the main axis of variation. This line puts equal weights to the x and y axes, but it is not restricted to pass through the origin. (C) GO analysis of the Dam-RNA-Pol II hits. (D) Two examples of the binding profiles of Dam-Pol II to baf and RpLP2 genes.
A regression analysis of the hits of the Dam-Pol II gene occupancy analysis in WT versus mutant muscles after normalizing each of them against Dam alone is shown in Fig. 6 B. A group of ∼200 genes was considered statistically different from that of the control (standard score, Z > 1.96, comparable to significance level, P < 0.05, or 95% confidential interval). Among these genes, a cluster of 12 genes coding for subunits of the 26S proteasome, a cluster of 16 ribosomal genes, and a cluster of 22 genes associated with the nucleus, as well as 10 genes coding for MT-associated proteins, were identified (Fig. 6 C).
High scores for RNA-Pol II occupancy of two gene loci, baf and RplP2, were of particular interest and are shown in Fig. 6 D. RplP2 encodes for ribosomal protein, and its down-regulation (also verified by quantitative PCR [qPCR]; Fig. S4) could lead to reduced protein synthesis in the klar mutant muscles, resulting in thinner muscles. baf encodes a protein that directly bridges between the chromatin and the nuclear lamina. It also binds to DNA, and its dimerization potentially mediates DNA–DNA interaction (Jamin and Wiebe, 2015). Baf locus was a strong hit for RNA-Pol II occupancy. A 2.5-fold reduction of baf mRNA was also observed by qPCR of klar versus control dissected larvae (Fig. S4), verifying the Dam-Pol II binding profile. Validation of the results obtained by the Dam-Pol II occupancy profiling was performed by qPCR analysis on RNA extracted from dissected WT and klar mutant larvae. Because it is technically infeasible to isolate the larval muscles from the rest of the tissue, the samples for the qPCR inevitably included the tissues of ectodermal origin and cells of the peripheral nervous system, in contrast to the highly tissue-specific Dam-Pol II experiment. A total of 28 genes that were either clearly up-regulated, down-regulated (Z score > 1.96), or unchanged by the Dam-Pol II analysis were analyzed by qPCR (normalized to the GAPDH mRNA) in WT versus klar mutant larvae (Fig. S4). Whereas we expected that genes exhibiting reduced Dam-Pol II binding in klar mutant muscles should consistently display down-regulation of their mRNA levels measured by qPCR, we speculate that genes with up-regulated Dam-Pol II binding profile could represent either transcriptionally active or stalled RNA-Pol II. The qPCR validation showed that 93% of the genes that exhibited reduced Dam-Pol II occupancy (15 of 16) were also decreased at their mRNA levels indicated by the qPCR analysis. However, eight of nine genes (88%) whose Dam-Pol II occupancy indicated elevated levels exhibited decreased mRNA levels by the qPCR analysis, possibly representing stalled RNA-Pol II at the promoter region, leading to inhibition of transcription (Zeitlinger et al., 2007; Nechaev et al., 2010). Some of these genes are highly enriched in salivary gland and trachea cells (the remnants of which might be present in the total RNA sample used for qPCR), possibly representing a different regulatory circuit. Importantly, genes that did not exhibit differential Dam-Pol II binding, including muscle genes such as myosin heavy chain (MHC) and myosin light chain or the nonmuscle gene CG5177, did not exhibit altered mRNA levels by the qPCR analysis (Fig. S4).
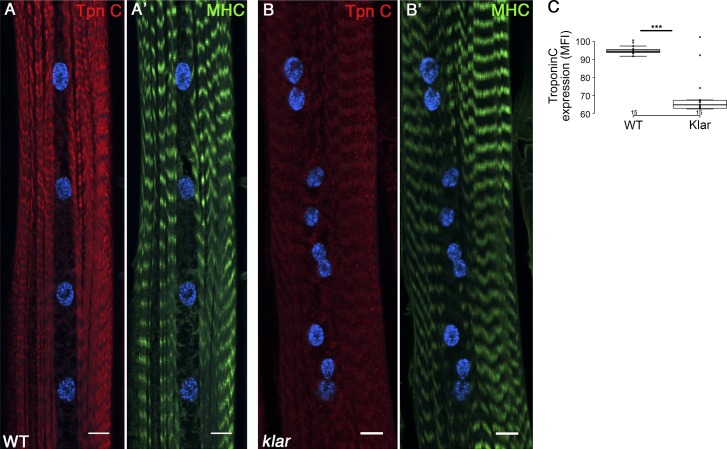
Reduced RNA-Pol II occupancy was also observed in two Troponin (Tpn) C loci, TpnC73F (Z score Dam-Pol II = −2.3; validated qPCR = −2.08) and TpnC47D (Z score = −2.1; validated qPCR = −1.1). Both Tpn C mRNAs are highly expressed in larval muscles (Fyrberg et al., 1994; Herranz et al., 2004). Staining with antibody recognizing both Tpn C proteins revealed a significant reduction in Tpn C protein levels in klar mutant muscles, relative to WT, whereas the overall fluorescent intensity of MHC did not change (Fig. 7, A–B′ and C′). Because Tpn C is essential for regulating Ca2+-dependent myosin activation, reduced levels of Tpn C in the klar mutant muscles are predicted to abolish proper larvae muscle contraction, consistent with our previous observation, where a significant slowing of larval and fly locomotion was reported in klar and Msp300 mutants (Elhanany-Tamir et al., 2012).
Figure 7.
Tpn C is down-regulated in klar mutant muscles. (A–B′) Representative single confocal stacks of WT (A and A′) and klar mutant (B and B′) muscle 7 labeled with anti–Tpn C (red), Dapi (blue), and MHC (green). A significant reduction in the protein levels of Tpn C is demonstrated, whereas MHC does not change. (C) Quantification of the mean fluorescent intensity of Tpn C in WT and klar mutant muscles is demonstrated. Bars, 10 µm. For control and klar statistics, n = 15 muscles taken from four distinct larvae; ***, P < 0.05. Whiskers extend to data points less than 1.5 interquartile ranges from the first and third quartile ranges.
Muscle-specific knockdown of BAF induces elevated DNA content in myonuclei
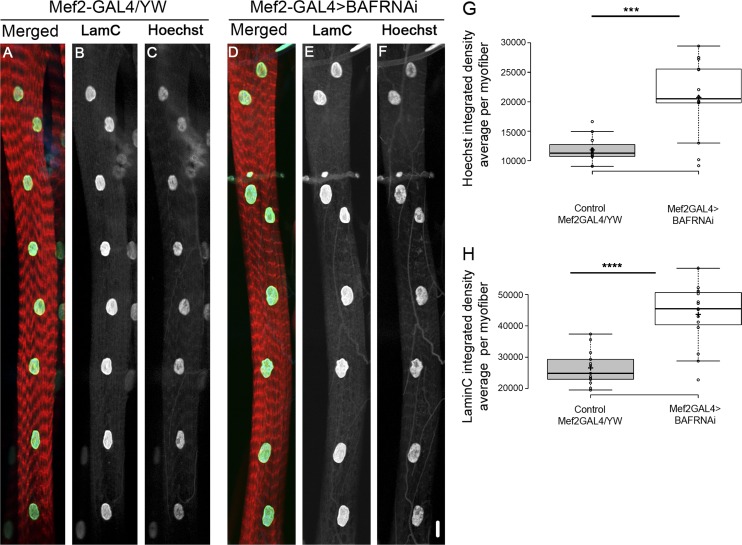
Next, the functional outcome of BAF down-regulation was examined by reducing BAF levels in myofibers using baf-RNAi and a muscle-specific driver. Notably, baf down-regulation led to a significant increase in the DNA content of the myonuclei relative to control (Fig. 8, A–G). Therefore, BAF decrease observed in klar myonuclei recapitulates LINC-induced elevated DNA endoreplication. Interestingly, lamin C levels were also increased in the BAF-knockdown myonuclei, suggesting a change in nuclear lamina stiffness in these nuclei. Nuclear position did not change (Fig. 8, D–F and H). These results imply BAF as a downstream component of LINC-mediated dysregulation of cell-cycle progression.
Figure 8.
Muscle-specific knockdown of BAF promotes increased DNA content in myonuclei. Representative image of muscle 7 of control (Mef2-GAL4/yw, A–C) or after muscle-specific knockdown of BAF (Mef2-GAL4>BAF RNAi, D–F), labeled with phalloidin (red, A and D), lamin C (green, A and D; white, B and E) and Hoechst (blue, A and D; white, C and F). (G) Quantification of Hoechst integrated density (averaged per myofiber) in the two groups indicates a significant increase of Hoechst integrated density after knockdown of BAF (***, P = 0.0001; control, n = 13; BAF RNAi, n = 15). (H) Quantification of lamin C integrated density (averaged per myofiber) in the two groups indicates a significant increase of lamin C fluorescence after knockdown of BAF (****, P = 7.4 × 10−6; control, n = 13; BAF RNAi, n = 15). Whiskers extend to data points less than 1.5 interquartile ranges from the first and third quartiles. Bar, 10 µm.
Discussion
In this manuscript we show, for the first time to our knowledge, that the LINC complex is required for regulation of DNA replication and synchronized cell-cycle progression within myonuclei of a given myofiber. This was primarily deduced from the variability in myonuclear DNA content, dysregulated EdU incorporation into DNA, and desynchronized E2F1 degradation observed in LINC mutant myofibers. We propose that cell-cycle progression and DNA endoreplication are functionally coupled to nuclear mechanotransduction, because mechanical perturbation similarly affected DNA replication in myonuclei. Moreover, our study implies that BAF, a regulator of chromatin organization, is a downstream component of the LINC complex–mediated cell cycle control.
Previous studies demonstrated contribution of the LINC complex to nuclear mechanotransduction (Lombardi et al., 2011; Arsenovic et al., 2016). We propose that the LINC complex contributes to proper regulation of cell-cycle progression and synchronized myonuclear response to mechanical stimulation, not only in Drosophila but also in vertebrate skeletal myofibers and cardiomyocytes. In support of this, recent study of heart regeneration in Zebrafish indicated that mechanical tension induces cell-cycle progression and endoreplication in epicardial cells after injury (Cao et al., 2017). Furthermore, our study might provide a molecular insight for the flawed muscle activity in Nesprin-related human diseases, such as Emery Dreifuss muscular dystrophy (Zhang et al., 2007), arthrogryposis multiplex congenital (Attali et al., 2009), dilated cardiomyopathy (Puckelwartz et al., 2010), and others.
Two lines of evidence support a direct link between myonuclear mechanical input and DNA replication: (a) Drosophila LINC mutants (klar and koi) reveal a similar phenotype of increased myonuclear DNA content. (b) Weakening adhesion between myofibers and tendons (revealed by β-PS-integrin knockdown) led to an increase in myonuclear DNA content.
The nature of the cytoskeletal elements that act upstream of the LINC complex in myofibers is not clear. The elaborate MT cytoskeleton that surrounds each myonucleus postulated previously to transmit mechanical signals into the nuclei (Hampoelz et al., 2011; Janota et al., 2017) appear to be less relevant to cell-cycle regulation. Moreover, our results imply that the MT cytoskeleton is not primarily involved in maintenance of nuclear position in mature myofibers, although extremely prolonged inhibition of the MT network in these cells (>3 d) did perturb both sarcomere organization and nuclear position in larval muscles (unpublished data). Based on previous experiments indicating the contribution of integrins to mechanical transmission (Li et al., 2012; Balasubramanian et al., 2013), we knocked down integrin β specifically in the larval muscles. Surprisingly, only a mild dissociation of the muscles from their tendon insertion sites was observed; however, this was sufficient to increase endoreplication in the myonuclei. It is therefore suggested that the major cytoskeletal element transmitting force into the myonuclei is the sarcomeres. Msp300 has been shown previously to bind D-Titin at the Z-discs (Elhanany-Tamir et al., 2012), might connect the myonuclei to the Z-discs, and thus provide mechanical support to the myonuclei.
The Dam-Pol II analysis proved to be highly consistent between replicates and significantly different between control and klar mutant. Because equal amounts of DNA from the dissected mutant and control larvae were taken at the first step of the analysis, it is assumed that the variations in DNA content produced because of endoreplication did not bias the outcome. An interesting and potentially relevant gene identified by the Dam-Pol II analysis is baf, which binds chromatin, Histone H3 (Montes de Oca et al., 2005), nuclear lamina constituents (Margalit et al., 2007; Amendola and van Steensel, 2014; Loi et al., 2016), Emerin (Berk et al., 2013), MAN1 (Mansharamani and Wilson, 2005), and LAP2b (Dechat et al., 2004; Bustin and Misteli, 2016). BAF dimerization is capable of inducing global changes in chromatin organization and therefore its reduction in klar and koi myofibers is expected to affect DNA condensation (Skoko et al., 2009). baf homozygous mutants exhibit cell-cycle defects (Furukawa et al., 2003); however, muscle-specific reduction of BAF caused increased levels of DNA in the myonuclei, phenocopying the LINC mutants. BAF is a phosphoprotein, whose nuclear translocation is regulated by dephosphorylation (Lancaster et al., 2007). Our experiments indicated an overall reduction in BAF mRNA levels in both klar and koi mutant myofibers; however, additional analysis is required to clarify whether its nuclear protein levels decreased in LINC mutants. Future availability of anti BAF antibody should clarify this point. Likewise, overexpression of BAF in myofibers is expected to rescue the enhanced DNA endoreplication in the myonuclei.
Few muscle-specific genes have been identified in the Dam-Pol II analysis. The observed decrease in Tpn C levels could be functionally relevant to the defective muscle activity and aberrant locomotion of the mutant larvae. Reduction in Tpn C levels would directly abrogate skeletal and cardiac muscle contractility in humans as well. Importantly, because human diseases associated with defective LINC complex exhibit a major muscle/cardiac weakness, fly muscles might represent a good model for explaining the pathogenesis of LINC-associated diseases (Zhang et al., 2007; Puckelwartz et al., 2009, 2010).
In summary, our analysis demonstrates for the first time that the LINC complex controls synchronized cell-cycle progression in myofibers and is tightly linked to comparable chromatin organization between myonuclei, affecting global gene expression levels.
Materials and methods
Fly stocks and husbandry
All crosses were performed at 25°C and raised on cornmeal yeast agar. The following stocks were distributed by the Bloomington Drosophila Stock Center: tubP-GAL80ts/TM2 (FBst0007017), GAL4-Mef2.R (FBst0027390), baf-RNAi (FBst0036108), mys RNAi (FBst0036108), and Sls RNAi (FBst0036108). klarΔ1–18 (FBal0277658) (Elhanany-Tamir et al., 2012; Wang et al., 2015), Df(2L)Msp300Δ3′(FBal0218044)/CyO, Kr-GAL4, UAS-GFP (Technau and Roth, 2008), and koi84(FBst0025105)/Cyo-dfd-eYfp (Kracklauer et al., 2007) have been described previously. UAS-LT3-Dm (FBtp0095492) and UAS-LT3-Dm-RpII215 (FBtp0095495) were received from A. Brand (The Gurdon Institute, University of Cambridge, Cambridge, England, UK; Southall et al., 2013); fly- FUCCI (FBst0055123) (Zielke et al., 2014) and PCNA-GFP were obtained from R. Duronio (University of North Carolina, Chapel Hill, NC; Swanhart et al., 2007); and UAS-Spastin flies were obtained from V. Brodu (Institute Jacques Monod, Paris, France).
Staging of the larvae was performed by 4- to 6-h embryo collection. Flies were caged on agar plates and further grown on yeast paste for the appropriate length of time.
Immunofluorescence
Immunofluorescent staining was performed as previously described (Wang et al., 2015). For fixation, paraformaldehyde (4% from 16% stock of EM grade; 15710; Electron Microscopy Sciences) was used without methanol to avoid damage of native F-actin or chromosomal morphology. Specimens were fixed for ∼30 min and subsequently washed several times in PBS with 0.1% Triton X-100 on a horizontal shaker with gentle agitation. Image analyses were consistently performed on muscle 7. All specimens were mounted in Shandon ImmuMount for microscopy (Thermo Fisher Scientific).
Antibodies and synthetic dyes
The following antibodies, created by the National Institute of Child and Health and Human Development of the National Institutes of Health and maintained at The University of Iowa, were used: mouse–anti lamin C (LC28.26-c; DSHB), rat–anti Tpn C (ab51106; Abcam), rat–anti Tubulin α (MCA78G; Bio-Rad), chicken–anti GFP (13970; Abcam), and rabbit–anti MHC (provided by P. Fisher, State University of New York at Stony Brook, Stony Brook, NY). Alexa Fluor 488–, 532–, and 546–conjugated secondary antibodies against rat, chick, rabbit, and mouse were purchased from The Jackson Laboratory and Thermo Fisher Scientific.
For labeling of the chromatin we used different dyes: DAPI (1 µg/ml; Sigma-Aldrich) specifically binds the minor groove of AT rich sequences of heterochromatin and displays a distinct banding pattern in polytene chromosomes. Hoechst 33258 (0.2 µg/ml; Thermo Fisher Scientific) was sometimes used as well. TRITC-Phalloidin (P1951; Sigma-Aldrich) was used for labeling F-actin.
To avoid antibody cross-reactions and fluorescent spectral cross talks, either single labeling or fluorophore swaps were performed to examine the labeling specificity.
Microscopy and image analysis
Microscopic images were acquired at 23°C on confocal microscopes Zeiss LSM 800 with the following lenses: Zeiss C-Apochromat 40×/1.20-W Korr M27, 20× PlanApochromat 20/0.8. The microscopic samples were embedded with Coverslip High Precision 1.5H ± 5 µm (Marienfeld-Superior). Immersion medium Immersol W 2010 (ne = 1.3339) and Immersion oil Immersol 518 F (ne = 1.518) were used, respectively. Images were analyzed with Fiji including plug-ins and adapted scripts. Figure panels were finally assembled by using Photoshop CS5. The acquisition software was Zen 2.3 (blue edition).
Automatic data collection
A macro in the IJ1 ImageJ programming language was used as a nonbiased and fast method to select all myonuclei within an image of a given myofiber automatically over many image files. A threshold for lamin C antibody intensity was set, above which shapes (nuclei) with a circularity >0.6 and a size >25 pixels were selected (the selected region being the region of interest [ROI]). All slices through the selected nucleus within the ROI were summed to create a projection image. Within ROIs on the projection image, the antibody stains were measured for the desired metrics, namely mean intensity across the selected region, area (μm2), and integrated density (the product of mean intensity and area).
Statistical analysis
Statistical analysis was performed using SPSS software (version 19; IBM SPSS for Windows) and Microsoft Excel 2013. Measurements were first evaluated for normality by using the Shapiro-Wilk test and subsequently analyzed by using a two-independent samples t test. P-values <0.05 were considered statistically significant. BoxPlotR was used to generate box plots (Spitzer et al., 2014), in which the center lines represent the medians, box limits indicate 25th and 75th percentiles, crosses represent sample means, and outliers (data points beyond the 95% confidence intervals) are represented by dots. Furthermore, whiskers, determined by using the Tukey method, extended to data points <1.5 interquartile ranges from the first and third quartiles as determined by the BoxPlotR software. All experiments were performed at least twice independently and in at least five randomly selected larvae from which muscle 7 was monitored in at least four abdominal segments per experimental group.
Detection of de novo DNA synthesis in Drosophila larva by EdU labeling
We applied a feeding protocol enabling the subsequent detection of EdU in situ. The labeling was performed by using Invitrogen Click-iT EdU imaging kits: 10 µl stock solution (20 mM stock solution in DMSO) and a few granules of Bromophenol Blue (114391-5G; Sigma-Aldrich) were mixed with 1 ml fly medium to obtain a final EdU concentration of 0.2 mM.
When they had reached the desired stage, larvae were dissected, rinsed, and fixed immediately in 4% paraformaldehyde using standard protocols. PBT with 0.1% Triton X-100 was used to permeabilize the samples for up to 1 h at room temperature. After removal of PBT, samples were incubated with freshly prepared Click-iT reaction cocktail for at least 30 min or longer at room temperature, protected from light. The specimens were subsequently antibody-stained according to standard procedures after the EdU detection.
MefGAL4 (control) and klar/TM6 lines were laid for 6 h on agar laying plates with ample yeast paste at 25°C. Three separate timing regimens were used to label the DNA, namely 12, 24, and 90 h. In the first group, developing larvae were starved for 10 h beginning from 3 d and 12 h ± 3 h after egg laying. They were subsequently fed with yeast paste containing 8% EdU and bromophenol blue for 12 h. In the second group, developing larvae were starved for 6 h beginning from 3 d ± 3 h after egg laying. They were subsequently fed with yeast paste containing 8% EdU and bromophenol blue for 24 h. In the third group, developing larvae were placed on yeast paste containing 8% EdU and bromophenol blue at 24 h ± 3 h after egg laying. Larva were subsequently fed for 90 h until they reached the late-third-instar stage. After feeding with EdU-yeast paste, all groups were given normal yeast paste for a short time until larvae developed into the late-third-instar stage. Larvae were dissected at 5 d after egg laying, were fixed, and underwent the Click-iT EdU detection reaction for 3 h by using the Click-iT EdU Alexa Fluor 594 imaging kit (Thermo Fisher Scientific). Larvae were subsequently antibody stained for lamin C, H3 phospho-Ser10, and Hoechst 33258.
DamID
DamID was performed as described previously (Marshall et al., 2016). We drove the expression of Dam-Pol II in muscles of WT and klarΔ–-18 Drosophila larvae using Mef2-GAL4 driver. A temperature-sensitive GAL80 was ubiquitously expressed to allow temporal control of GAL4 expression. The Dam-Pol II experiments comprise five independent replicates for each of the following four genotypes: (1) WT, Dam only; (2) WT, Dam-Pol II; (3) klar, Dam only, and (4) klar, Dam-Pol II. 20 third-instar larvae of each genotype were dissected, and their genomic DNA was extracted and processed with methylation-specific DpnI enzymes that cleave at GATC and generated a pool of blunt-ended DNA fragments. A double-stranded oligonucleotide adaptor with a 32-bp 5′ overhang (comprising oligos AdRt and AdRb) was ligated to the blunt ends to ensure directional ligation. The ligation was followed by digestion with DpnII, which is also a methylation-sensitive enzyme that cuts only unmethylated GATCs. Finally, a PCR primer identical to the 15 most 3′ nucleotides of the AdRt oligonucleotide sequence and the 5′ TC nucleotides on the gDNA fragment was used to amplify adaptor-ligated sequences. These amplified sequences were deep sequenced, the results were further analyzed by a bioinformatic pipeline (Marshall and Brand, 2015), and reads were normalized to filter out nonspecific Dam binding.
Library production
20 ng fragmented DNA was processed as previously described (Blecher-Gonen et al., 2013). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of 20 samples on two lanes. Between 6 and 12 million single-end 60-bp reads were obtained per sample on the Illumina HiSeq 2500 instrument. Sequencing was performed by using a custom-made specific primer with an addition of GATC in the 3′ end of the Illumina RD1 primer.
DamID-seq pipeline
We used the DamID-seq pipeline20 to process the data. The pipeline generates a BedGraph file containing normalized ratios for each pair of samples: DAM-POL and DAM control. This was followed by a script for calculating mean RNA-Pol II occupancy per gene (https://github.com/owenjm/polii.gene.call).
Prebuilt GATC fragment files were downloaded from the pipeline’s website (https://github.com/owenjm/damidseq_pipeline/raw/gh-pages/pipeline_gatc_files/Dmel_BDGP6.GATC.gff.gz).
Reference FASTA file and GFF were downloaded from the FlyBase repository: ftp://ftp.flybase.net/releases/FB2016_02/dmel_r6.10/fasta/dmel-all-chromosome-r6.10.fasta.gz and ftp://ftp.flybase.net/releases/FB2016_02/dmel_r6.10/gff/dmel-all-filtered-r6.10.gff.gz.
Initially, each replicate was processed by the pipeline separately, and a sample correlation matrix was calculated based on the per-gene Pol II occupancy. After outlier detection, all remaining replicates were pooled together and processed again by the pipeline. A regression of control on klarΔ1–18 was established, and a z-score for each of the points around the regression line was calculated. A threshold for the z-score was set (i.e., Z > 2). In the present analysis, cut-off criteria GATC sites >2, false discovery rate <0.01, and Z >1.96 (2-tailed) were used to filter the results, which gives approximate 95% confidence intervals equal to a significance level of 0.05. Then all the candidate genes were clustered, and among these genes, 11 were related to muscle development or function. Additionally, Log2(DamPolII/Dam) binding profiles of control versus klarΔ1–18 were examined independently by using IGV browser (Broad Institute of Massachusetts Institute of Technology and Harvard) to identify potential candidates for validation.
RT-qPCR
Gene expression quantification was performed by RT-qPCR. In brief, total muscle RNA was isolated from larval body walls of 30 dissected Drosophila larvae including the somatic muscles of WT and klarΔ1–18 by using RNeasy Protect Mini kit (Qiagen). 2 µg of total RNA was used for first-strand cDNA synthesis by RT by using SuperScript Reverse Transcription IV (Invitrogen) with oligonucleotide dT primers. qPCR was performed by using gene-specific primers on the ABI 7500 Real-Time PCR System (Applied Biosystems) with Fast SYBR Green Master Mix (Applied Biosystems) for detection. Each sample was run in triplicate. The primer sequences are listed in Table S2. The difference in gene expression was calculated by using the fold change (ΔΔCt method; Schmittgen and Livak, 2008). ΔCt is the Ct value for the gene of interest normalized to the Ct value of the respective GAPDH control in both WT and klarΔ1–18 Drosophila larvae. ΔΔCt values were calculated as a relative change in ΔCt of the target gene in klarΔ1–18 with respect to WT. Fold changes were expressed as 2−ΔΔCt for up-regulated genes and the negative reciprocal of the fold change for down-regulated genes (where 2−ΔΔCt < 1).
Online supplemental material
Fig. S1 shows that the distance between nuclei does not correlate with DNA content in Oregon R myonuclei. Fig. S2 shows that PCNA-GFP, a direct target of E2F1, is expressed in larval myofibers. Fig. S3 shows that muscle-specific knockdown of D-Titin (Sls) reveals smaller nuclei with similar DNA content. Fig. S4 shows a comparison of the muscle-specific Dam-Pol II occupancy gene hits and qPCR analysis in klar and in koi mutant dissected larvae. Table S1 shows a summary of the actual values obtained by the Dam-ID pipeline. Table S2 shows a summary of the major hits identified by the DamID pipeline. Table S3 shows the primer sequences used in this study. Supplemental data file shows a macro designed to allow the user to specify a range of conditions in which he or she can select desired nuclei, flatten the images, and obtain a range of different measurements for all nuclei in a given cell; the macro uses IJ1 Macro language.
Supplementary Material
Acknowledgments
We thank A. Brand for the Dam-Pol II fly lines and O.J. Marshall for advice on the DamID protocol and pipeline. We thank the Bloomington Drosophila Stock Centre for various fly lines, the DSHB for antibodies, and FlyBase for important genomic data. S. Motola and S. Gilad from the Institute for Personalized Medicine provided critical input and advice on the DamID experiments, and Y. Gruenbaum and Z. Paroush critically commented on the manuscript. We thank J. Silvermetz for his useful input and advice on the data acquisition and data analysis.
This study was supported by grants from the Minerva Foundation (711743 to T. Volk), the German-Israeli Foundation (1305 to T. Volk), Weizmann UK joint research program (to T. Volk), and The French Muscular Dystrophy Association (AFM-Téléthon grant 18982).
The authors declare no competing financial interests.
Author contributions: S. Wang conceptualized and conducted the experiments. E. Stoops conducted the experiments and performed formal analysis on the images. U. CP conducted the experiments. B. Markus performed formal analysis on the Dam-RNA-Pol II data. A. Reuveny and E. Ordan conducted the experiments. and T. Volk conceptualized, wrote the manuscript, and provided the funding.
References
- Amendola M., and van Steensel B.. 2014. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr. Opin. Cell Biol. 28:61–68. 10.1016/j.ceb.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Arsenovic P.T., Ramachandran I., Bathula K., Zhu R., Narang J.D., Noll N.A., Lemmon C.A., Gundersen G.G., and Conway D.E.. 2016. Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys. J. 110:34–43. 10.1016/j.bpj.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attali R., Warwar N., Israel A., Gurt I., McNally E., Puckelwartz M., Glick B., Nevo Y., Ben-Neriah Z., and Melki J.. 2009. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum. Mol. Genet. 18:3462–3469. 10.1093/hmg/ddp290 [DOI] [PubMed] [Google Scholar]
- Balasubramanian L., Lo C.M., Sham J.S., and Yip K.P.. 2013. Remanent cell traction force in renal vascular smooth muscle cells induced by integrin-mediated mechanotransduction. Am. J. Physiol. Cell Physiol. 304:C382–C391. 10.1152/ajpcell.00234.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L.J., Soshnev A.A., and Geyer P.K.. 2015. Networking in the nucleus: a spotlight on LEM-domain proteins. Curr. Opin. Cell Biol. 34:1–8. 10.1016/j.ceb.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H., et al. . 2009. Evidence for cardiomyocyte renewal in humans. Science. 324:98–102. 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk J.M., Tifft K.E., and Wilson K.L.. 2013. The nuclear envelope LEM-domain protein emerin. Nucleus. 4:298–314. 10.4161/nucl.25751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., and Amit I.. 2013. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8:539–554. 10.1038/nprot.2013.023 [DOI] [PubMed] [Google Scholar]
- Bossie C.A., and Sanders M.M.. 1993. A cDNA from Drosophila melanogaster encodes a lamin C-like intermediate filament protein. J. Cell Sci. 104:1263–1272. [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2013. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 14:13–24. 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2014. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 109:1–52. 10.1016/B978-0-12-397920-9.00006-8 [DOI] [PubMed] [Google Scholar]
- Bustin M., and Misteli T.. 2016. Nongenetic functions of the genome. Science. 352:aad6933 10.1126/science.aad6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain N.E., and Starr D.A.. 2015. SUN proteins and nuclear envelope spacing. Nucleus. 6:2–7. 10.4161/19491034.2014.990857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain N., Tapley E.C., McDonald K., Cain B., and Starr D.A.. 2014. The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J. Cell Biol. 206:163–172. 10.1083/jcb.201405081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wang J., Jackman C.P., Cox A.H., Trembley M.A., Balowski J.J., Cox B.D., De Simone A., Dickson A.L., Di Talia S., et al. . 2017. Tension creates an endoreplication wavefront that leads regeneration of epicardial tissue. Dev. Cell. 42:600–615. 10.1016/j.devcel.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut K.J., Höpfler M., Lautenschläger F., Boyde L., Chan C.J., Ekpenyong A., Martinez-Arias A., and Guck J.. 2012. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J. 103:2060–2070. 10.1016/j.bpj.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Worman H.J., and Gundersen G.G.. 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro M., Hnia K., Gache V., Koch C., Gavriilidis C., Rodriguez D., Nicot A.S., Romero N.B., Schwab Y., Gomes E., et al. . 2015. Amphiphysin 2 Orchestrates Nucleus Positioning and Shape by Linking the Nuclear Envelope to the Actin and Microtubule Cytoskeleton. Dev. Cell. 35:186–198. 10.1016/j.devcel.2015.09.018 [DOI] [PubMed] [Google Scholar]
- Dahl K.N., Ribeiro A.J., and Lammerding J.. 2008. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102:1307–1318. 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Gajewski A., Korbei B., Gerlich D., Daigle N., Haraguchi T., Furukawa K., Ellenberg J., and Foisner R.. 2004. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117:6117–6128. 10.1242/jcs.01529 [DOI] [PubMed] [Google Scholar]
- Demontis F., and Perrimon N.. 2009. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development. 136:983–993. 10.1242/dev.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D.E., Janmey P., and Wang Y.L.. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science. 310:1139–1143. 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- Downing T.L., Soto J., Morez C., Houssin T., Fritz A., Yuan F., Chu J., Patel S., Schaffer D.V., and Li S.. 2013. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 12:1154–1162. 10.1038/nmat3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., and Orr-Weaver T.L.. 2001. Endoreplication cell cycles: More for less. Cell. 105:297–306. 10.1016/S0092-8674(01)00334-8 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., and Gutierrez C.. 2014. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15:197–210. 10.1038/nrm3756 [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H., Yu Y.V., Shnayder M., Jain A., Welte M., and Volk T.. 2012. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 198:833–846. 10.1083/jcb.201204102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., Schulman V.K., and Baylies M.K.. 2014. Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development. 141:355–366. 10.1242/dev.095612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D.T., and Duronio R.J.. 2013. Endoreplication and polyploidy: insights into development and disease. Development. 140:3–12. 10.1242/dev.080531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frawley L.E., and Orr-Weaver T.L.. 2015. Polyploidy. Curr. Biol. 25:R353–R358. 10.1016/j.cub.2015.03.037 [DOI] [PubMed] [Google Scholar]
- Furukawa K., Sugiyama S., Osouda S., Goto H., Inagaki M., Horigome T., Omata S., McConnell M., Fisher P.A., and Nishida Y.. 2003. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116:3811–3823. 10.1242/jcs.00682 [DOI] [PubMed] [Google Scholar]
- Fyrberg C., Parker H., Hutchison B., and Fyrberg E.. 1994. Drosophila melanogaster genes encoding three troponin-C isoforms and a calmodulin-related protein. Biochem. Genet. 32:119–135. 10.1007/BF00554420 [DOI] [PubMed] [Google Scholar]
- Geyer P.K., Vitalini M.W., and Wallrath L.L.. 2011. Nuclear organization: Taking a position on gene expression. Curr. Opin. Cell Biol. 23:354–359. 10.1016/j.ceb.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel P., Lee Y.L., Sobota R.M., Calvi A., Koullourou V., Patel R., Mamchaoui K., Nedelec F., Shackleton S., Schmoranzer J., et al. . 2017. Nesprin-1alpha-dependent microtubule nucleation from the nuclear envelope via Akap450 is necessary for nuclear positioning in muscle cells. Curr. Biol. 27:2999–3009. 10.1016/j.cub.2017.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda S., Endo S., and Saigo K.. 2000. Requirements of Kettin, a giant muscle protein highly conserved in overall structure in evolution, for normal muscle function, viability, and flight activity of Drosophila. J. Cell Biol. 148:101–114. 10.1083/jcb.148.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B., Azou-Gros Y., Fabre R., Markova O., Puech P.H., and Lecuit T.. 2011. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development. 138:3377–3386. 10.1242/dev.065706 [DOI] [PubMed] [Google Scholar]
- Herranz R., Díaz-Castillo C., Nguyen T.P., Lovato T.L., Cripps R.M., and Marco R.. 2004. Expression patterns of the whole troponin C gene repertoire during Drosophila development. Gene Expr. Patterns. 4:183–190. 10.1016/j.modgep.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Ho C.Y., Jaalouk D.E., Vartiainen M.K., and Lammerding J.. 2013. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 497:507–511. 10.1038/nature12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F. 2014. LINC complex proteins in development and disease. Curr. Top. Dev. Biol. 109:287–321. 10.1016/B978-0-12-397920-9.00004-4 [DOI] [PubMed] [Google Scholar]
- Jamin A., and Wiebe M.S.. 2015. Barrier to Autointegration Factor (BANF1): interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol. 34:61–68. 10.1016/j.ceb.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P.A., and Miller R.T.. 2011. Mechanisms of mechanical signaling in development and disease. J. Cell Sci. 124:9–18. 10.1242/jcs.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janota C.S., Calero-Cuenca F.J., Costa J., and Gomes E.R.. 2017. SnapShot: nucleo-cytoskeletal interactions. Cell. 169:970 10.1016/j.cell.2017.05.014 [DOI] [PubMed] [Google Scholar]
- Kracklauer M.P., Banks S.M., Xie X., Wu Y., and Fischer J.A.. 2007. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin). 1:75–85. 10.4161/fly.4254 [DOI] [PubMed] [Google Scholar]
- Lancaster O.M., Cullen C.F., and Ohkura H.. 2007. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J. Cell Biol. 179:817–824. 10.1083/jcb.200706067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H.Q., Ghatak S., Yeung C.Y.C., Tellkamp F., Günschmann C., Dieterich C., Yeroslaviz A., Habermann B., Pombo A., Niessen C.M., and Wickström S.A.. 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18:864–875. 10.1038/ncb3387 [DOI] [PubMed] [Google Scholar]
- Lee H.O., Davidson J.M., and Duronio R.J.. 2009. Endoreplication: polyploidy with purpose. Genes Dev. 23:2461–2477. 10.1101/gad.1829209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Wu Y., Manso A.M., Gu Y., Liao P., Israeli S., Yajima T., Nguyen U., Huang M.S., Dalton N.D., et al. . 2012. β1 integrin gene excision in the adult murine cardiac myocyte causes defective mechanical and signaling responses. Am. J. Pathol. 180:952–962. 10.1016/j.ajpath.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chu J.S., Kurpinski K., Li X., Bautista D.M., Yang L., Sung K.L., and Li S.. 2011. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys. J. 100:1902–1909. 10.1016/j.bpj.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yue S., Chen X., Kubin T., and Braun T.. 2010. Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ. Res. 106:1498–1506. 10.1161/CIRCRESAHA.109.211888 [DOI] [PubMed] [Google Scholar]
- Loi M., Cenni V., Duchi S., Squarzoni S., Lopez-Otin C., Foisner R., Lattanzi G., and Capanni C.. 2016. Barrier-to-autointegration factor (BAF) involvement in prelamin A-related chromatin organization changes. Oncotarget. 7:15662–15677. 10.18632/oncotarget.6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M.L., Jaalouk D.E., Shanahan C.M., Burke B., Roux K.J., and Lammerding J.. 2011. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286:26743–26753. 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansharamani M., and Wilson K.L.. 2005. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J. Biol. Chem. 280:13863–13870. 10.1074/jbc.M413020200 [DOI] [PubMed] [Google Scholar]
- Margalit A., Brachner A., Gotzmann J., Foisner R., and Gruenbaum Y.. 2007. Barrier-to-autointegration factor–a BAFfling little protein. Trends Cell Biol. 17:202–208. 10.1016/j.tcb.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Marshall O.J., and Brand A.H.. 2015. damidseq_pipeline: An automated pipeline for processing DamID sequencing datasets. Bioinformatics. 31:3371–3373. 10.1093/bioinformatics/btv386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Southall T.D., Cheetham S.W., and Brand A.H.. 2016. Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat. Protoc. 11:1586–1598. 10.1038/nprot.2016.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P., and Schirmer E.C.. 2015. LINC’ing form and function at the nuclear envelope. FEBS Lett. 589(19PartA, 19 Pt A):2514–2521. 10.1016/j.febslet.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., and Baylies M.K.. 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 484:120–124. 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Lee K.K., and Wilson K.L.. 2005. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 280:42252–42262. 10.1074/jbc.M509917200 [DOI] [PubMed] [Google Scholar]
- Murphy W.L., McDevitt T.C., and Engler A.J.. 2014. Materials as stem cell regulators. Nat. Mater. 13:547–557. 10.1038/nmat3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., and Adelman K.. 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 327:335–338. 10.1126/science.1181421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L. 2015. When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 31:307–315. 10.1016/j.tig.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Dechat T., and Foisner R.. 2015. Lamins at the crossroads of mechanosignaling. Genes Dev. 29:225–237. 10.1101/gad.255968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz M.J., Kessler E., Zhang Y., Hodzic D., Randles K.N., Morris G., Earley J.U., Hadhazy M., Holaska J.M., Mewborn S.K., et al. . 2009. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 18:607–620. 10.1093/hmg/ddn386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz M.J., Kessler E.J., Kim G., Dewitt M.M., Zhang Y., Earley J.U., Depreux F.F., Holaska J., Mewborn S.K., Pytel P., and McNally E.M.. 2010. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol. 48:600–608. 10.1016/j.yjmcc.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D., and Shanahan C.M.. 2013. Nesprins: From the nuclear envelope and beyond. Expert Rev. Mol. Med. 15:e5 10.1017/erm.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., and Livak K.J.. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Skoko D., Li M., Huang Y., Mizuuchi M., Cai M., Bradley C.M., Pease P.J., Xiao B., Marko J.F., Craigie R., and Mizuuchi K.. 2009. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc. Natl. Acad. Sci. USA. 106:16610–16615. 10.1073/pnas.0909077106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.E., Gruenbaum Y., Berrios M., and Fisher P.A.. 1987. Biosynthesis and interconversion of Drosophila nuclear lamin isoforms during normal growth and in response to heat shock. J. Cell Biol. 105:771–790. 10.1083/jcb.105.2.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T.D., Gold K.S., Egger B., Davidson C.M., Caygill E.E., Marshall O.J., and Brand A.H.. 2013. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell. 26:101–112. 10.1016/j.devcel.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M., Wildenhain J., Rappsilber J., and Tyers M.. 2014. BoxPlotR: a web tool for generation of box plots. Nat. Methods. 11:121–122. 10.1038/nmeth.2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., and Fridolfsson H.N.. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanhart L.M., Sanders A.N., and Duronio R.J.. 2007. Normal regulation of Rbf1/E2f1 target genes in Drosophila type 1 protein phosphatase mutants. Dev. Dyn. 236:2567–2577. 10.1002/dvdy.21265 [DOI] [PubMed] [Google Scholar]
- Swift J., and Discher D.E.. 2014. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 127:3005–3015. 10.1242/jcs.149203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A.S., and Wang N.. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15:1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M., and Roth S.. 2008. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly (Austin). 2:82–91. 10.4161/fly.6288 [DOI] [PubMed] [Google Scholar]
- Trappmann B., Gautrot J.E., Connelly J.T., Strange D.G.T., Li Y., Oyen M.L., Cohen Stuart M.A., Boehm H., Li B., Vogel V., et al. . 2012. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 11:642–649. 10.1038/nmat3339 [DOI] [PubMed] [Google Scholar]
- Volk T. 1992. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 116:721–730. [DOI] [PubMed] [Google Scholar]
- Wang N., Tytell J.D., and Ingber D.E.. 2009. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10:75–82. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- Wang S., Reuveny A., and Volk T.. 2015. Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. J. Cell Biol. 209:529–538. 10.1083/jcb.201408098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A., Gross S.P., Postner M., Block S.M., and Wieschaus E.F.. 1998. Developmental regulation of vesicle transport in Drosophila embryos: Forces and kinetics. Cell. 92:547–557. 10.1016/S0092-8674(00)80947-2 [DOI] [PubMed] [Google Scholar]
- Wilson M.H., and Holzbaur E.L.. 2015. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development. 142:218–228. 10.1242/dev.114769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Bandura J., Zhang P., Jin Y., Reuter H., and Edgar B.A.. 2017. EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat. Commun. 8:15125 10.1038/ncomms15125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., and Young R.A.. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39:1512–1516. 10.1038/ng.2007.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T., et al. . 2007. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 16:2816–2833. 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- Zielke N., Edgar B.A., and DePamphilis M.L.. 2013. Endoreplication. Cold Spring Harb. Perspect. Biol. 5:a012948 10.1101/cshperspect.a012948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Korzelius J., van Straaten M., Bender K., Schuhknecht G.F., Dutta D., Xiang J., and Edgar B.A.. 2014. Fly-FUCCI: A versatile tool for studying cell proliferation in complex tissues. Cell Reports. 7:588–598. 10.1016/j.celrep.2014.03.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.