High levels of autophagy exist in Leydig cells of the testis, but its physiological function is unknown. Gao et al. now show that autophagy promotes uptake of cholesterol, an essential precursor for testosterone synthesis, by removing the NHERF2 negative regulator of the high-density lipoprotein receptor SR-BI.
Abstract
Testosterone is indispensable for sexual development and maintaining male characteristics, and deficiency of this hormone results in primary or late-onset hypogonadism (LOH). Testosterone is primarily produced in Leydig cells, where autophagy has been reported to be extremely active. However, the functional role of autophagy in testosterone synthesis remains unknown. In this study, we show that steroidogenic cell–specific disruption of autophagy influenced the sexual behavior of aging male mice because of a reduction in serum testosterone, which is similar to the symptoms of LOH. The decline in testosterone was caused mainly by a defect in cholesterol uptake in autophagy-deficient Leydig cells. Further studies revealed that once autophagic flux was disrupted, Na+/H+ exchanger regulatory factor 2 (NHERF2) accumulated in Leydig cells, resulting in the down-regulation of scavenger receptor class B, type I (SR-BI) and eventually leading to insufficient cholesterol supply. Collectively, these results reveal that autophagy promotes cholesterol uptake into Leydig cells by eliminating NHERF2, suggesting that dysfunction of autophagy might be causal in the loss of testosterone production in some patients.
Introduction
Testosterone is an important adult male hormone that is needed for sexual development and for maintaining male characteristics (Isidori et al., 2005; Sinclair et al., 2015). A deficiency in serum testosterone levels is commonly associated with primary or late-onset hypogonadism (LOH; Bassil and Morley, 2010; Bassil, 2011), which is associated with not only male sexual dysfunction and decreased reproductive capacity but also with cardiovascular disease, diabetes, osteoporosis, and other diseases (Morales et al., 2010; Akishita and Yu, 2012; Wang et al., 2017).
In the testicular interstitium (Purvis et al., 1981), testosterone is primarily produced in Leydig cells, where autophagy has been reported to be extremely active (Tang, 1988; Tang and Zhang, 1990; Yi and Tang, 1991, 1995, 1999; Tang et al., 1992). Autophagy is a cellular metabolic process that uses lysosomal degradation of cellular components (such as organelles, nucleic acids, or proteins as well as other biological macromolecules) to provide raw materials to help cells survive under stress conditions (Rabinowitz and White, 2010; Goginashvili et al., 2015). Recent research shows that autophagy activity was decreased in aged rat Leydig cells (Li et al., 2011), and sex hormone levels reduced in autophagy-deficient mice with Atg5 expression in the brain (Yoshii et al., 2016). Because autophagy has been implicated in lipid metabolism via a process termed macrolipophagy to provide cells with sources of triglycerides (TGs) and cholesterol, we speculated that autophagy might be involved in testosterone synthesis by promoting lipid metabolism in Leydig cells.
To test this working hypothesis, we specifically disrupted autophagy by the conditional knockout of Atg7 or Atg5 in steroidogenic cells. Results showed that the disruption of autophagy affected male sexual behavior as a result of the sharp reduction in testosterone in serum, similar to the symptoms of LOH. In an effort to further address the relationship between autophagy and testosterone synthesis, we demonstrated that the decline in testosterone production resulted from the disruption of cholesterol uptake because of the down-regulation of the scavenger receptor class B, type I (SR-BI; gene name, Scarb1) in autophagy-deficient Leydig cells. Using a small-scale screen, a negative regulator of SR-BI, Na+/H+ exchanger regulatory factor 2 (NHERF2; gene name, Slc9a3r2), was found to be degraded via the autophagy–lysosome pathway. The accumulation of NHERF2 in Leydig cells of autophagy-deficient mice led to the down-regulation of SR-BI, thereby reducing testosterone synthesis. The cholesterol uptake defect could be partially rescued by Nherf2 knockdown in autophagy-deficient Leydig cells. In response to hormone stimulation, autophagic flux is induced in Leydig cells to promote testosterone synthesis by facilitating the degradation of NHERF2 and up-regulation of SR-BI. Thus, our research reveals a novel functional role for autophagy in testosterone synthesis through the regulation of cholesterol uptake via the degradation of NHERF2 in Leydig cells. These results hint that autophagy dysfunction might also play a role in the loss of testosterone production in some patients.
Results
Impaired autophagy in low-testosterone patients
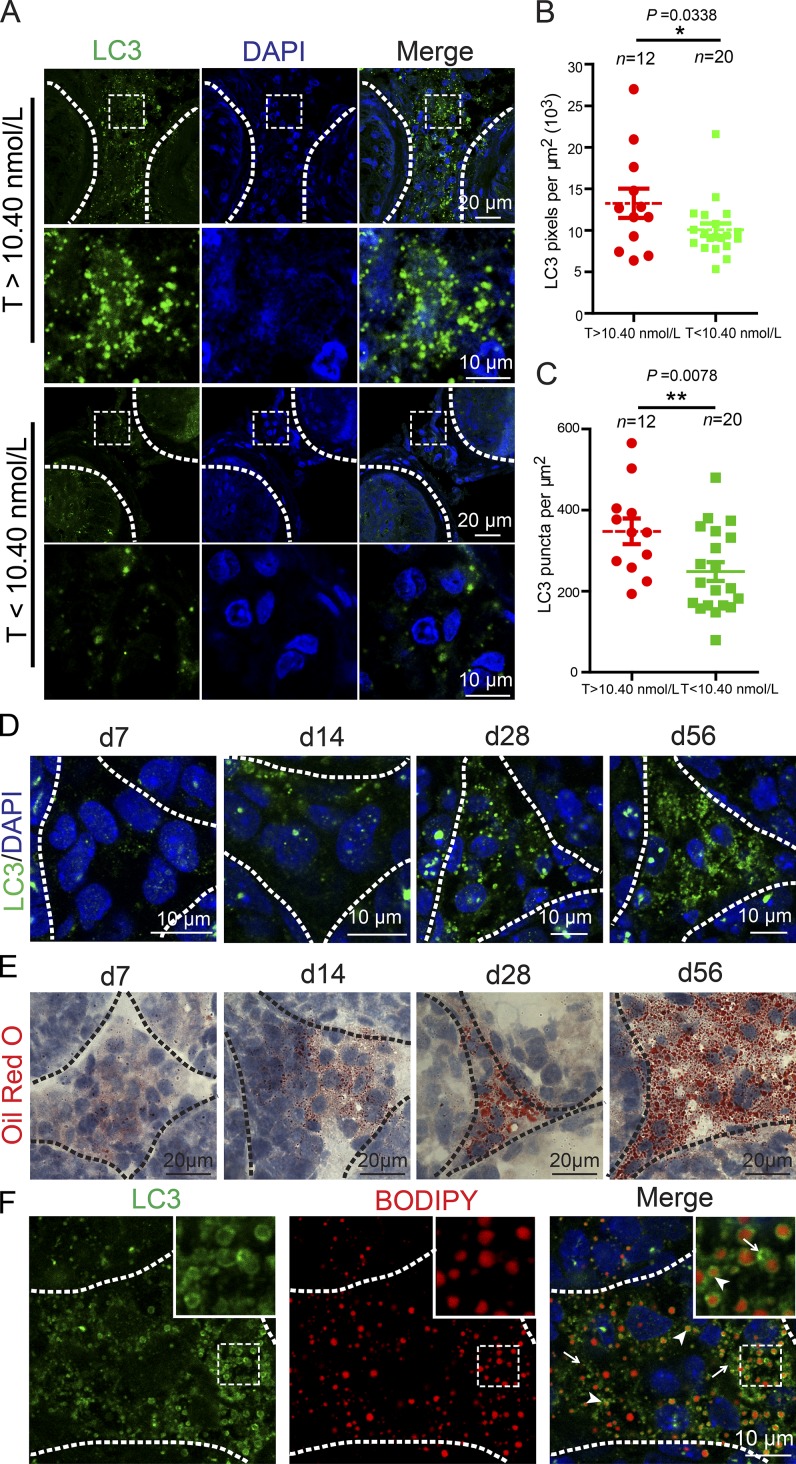
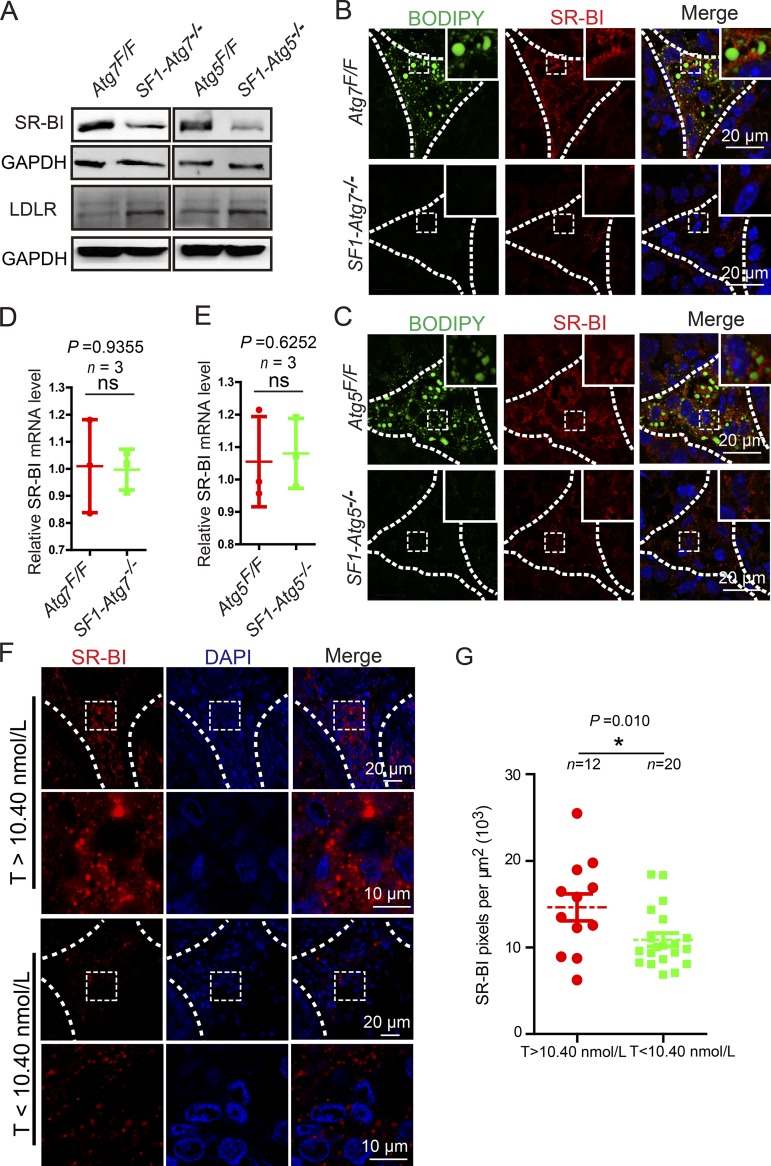
Because autophagy deficiency in Leydig cells is associated with reduced levels of serum testosterone in both rats and mice (Midzak et al., 2009; Bassil and Morley, 2010; Bassil, 2011; Li et al., 2011; Yoshii et al., 2016), we speculated that low levels of serum testosterone in patients might be correlated with autophagy deficiency in some hypogonadism patients. To test this hypothesis, we recruited 20 patients diagnosed as having azoospermia or oligospermia with low-serum testosterone levels (testosterone < 10.40 nmol/L, 22–35 yr old; Table S2) and 12 patients with normal serum testosterone levels (testosterone > 10.40 nmol/L, 22–39 yr old; Table S1) for open biopsy of the testis. We then examined the expression of the microtubule-associated protein light chain 3 (LC3), an autophagic marker (Klionsky et al., 2016), using immunofluorescence staining of the Leydig cells obtained from their testes. The results showed that LC3 expression and puncta number per square micrometer were significantly decreased in the Leydig cells from the patients with low testosterone levels compared with those of the control group (Fig. 1, A–C), suggesting that autophagy deficiency might be correlated with the decline of serum testosterone in some patients with azoospermia or oligospermia.
Figure 1.
The serum testosterone level is closely related to autophagy. (A) The expression level of LC3 was decreased in Leydig cells of the low-serum testosterone (T) level azoospermia patients. Immunofluorescence staining of LC3 (green) in the testes of azoospermia patients. (B) Quantification of the fluorescence intensity per μm2 of LC3 in A. (C) Quantification of the puncta number per μm2 of LC3 in A. (D) The expression level of LC3 was increased in Leydig cells during development. Immunofluorescence staining of LC3 (green) in the testes of mice at d7, d14, d21, and d56. *, P < 0.05; **, P < 0.01. (E) The frozen tissue sections of mouse testes at d7, d14, d21, and d56 were stained with ORO (red). (F) The localization of LC3 and LDs was detected in adult mouse testes by LC3 (green) immunofluorescence analysis and BODIPY (red) staining. Arrowheads indicate LC3 colocalized with LDs, and arrows indicate LC3 punctate signals out of LDs. Insets are marked by boxes and are 7.7 µM wide. Nuclei were stained with DAPI (blue).
Autophagy is active in Leydig cells
To further explore the correlation between testosterone production and autophagy in Leydig cells, we characterized the in vivo autophagy activity in mouse Leydig cells at various developmental stages. Four distinct types of Leydig cells have been identified and characterized: stem Leydig cells (postnatal days 1–14; d1–14), progenitor Leydig cells (d14–28), immature Leydig cells (d28–56), and adult Leydig cells (>d56; Chamindrani Mendis-Handagama and Ariyaratne, 2001; O’Shaughnessy et al., 2002a,b; Chen et al., 2009). Immunofluorescence staining of LC3 was performed in the testes at various developmental stages, and we found that LC3 expression was initiated in progenitor Leydig cells (d14; Fig. 1 D, d14 panel) and peaked in adult Leydig cells (d56; Fig. 1 D, d56 panel). The expression of LC3 was closely correlated with the accumulation of lipid droplets (LDs) as revealed by oil red O (ORO) staining (Fig. 1 E; Bilińska, 1994). Thus, consistent with previous studies and hypotheses, autophagy is extremely active in adult mouse Leydig cells.
As autophagy might participate in the utilization of LDs to provide cholesterol for steroidogenesis in Leydig cells, such as testosterone synthesis (Chen et al., 2009), we speculated that autophagy might be involved in testosterone synthesis via lipid metabolism in Leydig cells. To further test this hypothesis, we examined the precise localization of LC3 and LDs in adult mouse testes (Fig. 1 F). Consistent with previous findings in hepatocytes and macrophage foam cells (Singh et al., 2009), the LC3 signal colocalized with LDs (Fig. 1 F, arrowheads) as revealed by 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a, 4a-diaza-s-indacene (BODIPY) staining (Gocze and Freeman, 1994). In addition, many LC3 puncta unassociated with LDs could be also detected in Leydig cells (Fig. 1 F, arrows), indicating that in addition to participating in lipid metabolism, autophagy might also play other unknown roles in Leydig cells. Based on these studies, we concluded that autophagy is extremely active in adult mouse Leydig cells and might regulate steroidogenesis via lipid metabolism.
Impaired autophagy affects sexual behavior
To further investigate whether autophagy is involved in steroidogenesis in Leydig cells, we knocked out Atg7 or Atg5 in Leydig cells by crossing mice with floxed Atg7 or Atg5 alleles to SF1-Cre mice, which could express Cre recombinase in Leydig cells and other steroidogenic cells (Komatsu et al., 2005; Dhillon et al., 2006; Hara et al., 2006). These steroidogenic cell–specific Atg7 and Atg5 knockout mice were named SF1-Atg7−/− and SF1-Atg5−/−, respectively.
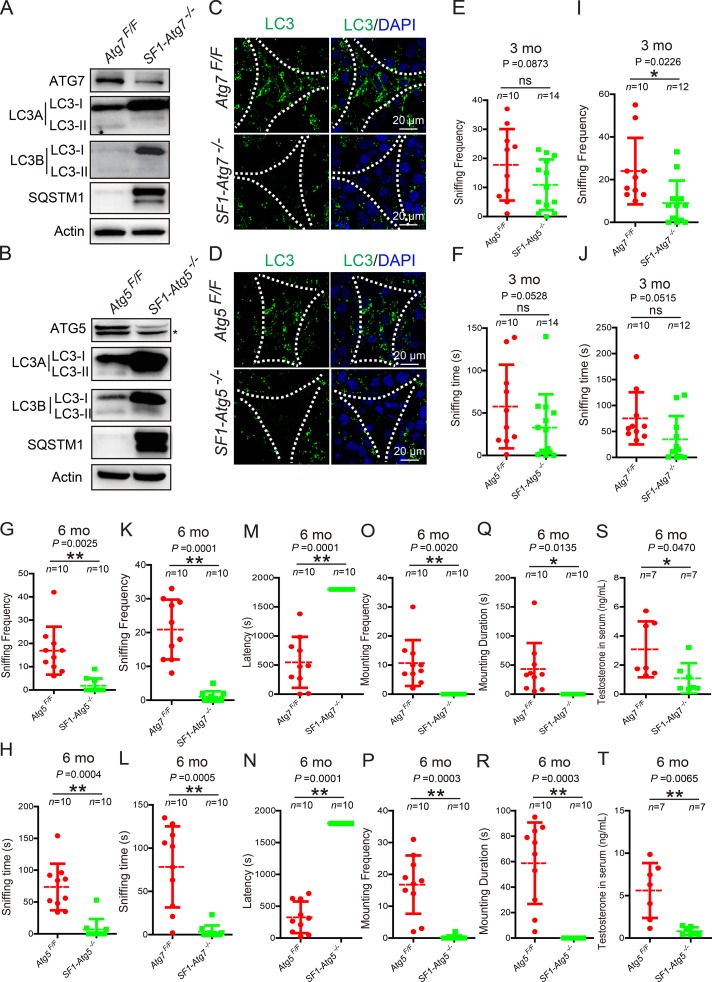
To examine the knockout efficiency, we isolated Leydig cells from SF1-Atg7−/−, SF1-Atg5−/−, Atg7Flox/Flox, and Atg5Flox/Flox mouse testes and subsequently extracted total protein. Immunoblotting data showed that the protein levels of either ATG7 or ATG5 were dramatically reduced in knockout Leydig cells compared with their control groups (Fig. 2, A and B). Consistent with a role for ATG7 and ATG5 in autophagy, the cytosolic form LC3-I but not the membrane-associated form LC3-II along with the autophagic substrate SQSTM1/p62 accumulated in either Atg7- or Atg5-deficient Leydig cells (Fig. 2, A and B). The immunofluorescence analysis of LC3 also showed the disappearance of punctate structures (representing autophagosomes) in both SF1-Atg7−/− and SF1-Atg5−/− mouse Leydig cells (Fig. 2, C and D), suggesting that the autophagic flux may be disrupted (Klionsky et al., 2016).
Figure 2.
Steroidogenic cell–specific knockout of Atg7- or Atg5-affected testosterone synthesis. (A and B) The autophagic flux was disrupted in autophagy-deficient Leydig cells. Immunoblotting analyses of ATG7, SQSTM1/p62, and LC3 were performed in both Atg7Flox/Flox (left) and SF1-Atg7−/− Leydig cells (right; A). The ATG7 protein level was dramatically reduced, and actin served as a loading control. Similar results were observed in Atg5Flox/Flox mice (left) and SF1-Atg5−/− mice (right) as shown in B. The asterisk indicates nonspecific signals. (C and D) The LC3 punctate structures disappeared in Leydig cells of autophagy-deficient mice. Immunofluorescence analysis using LC3 (green) in both Atg7Flox/Flox (top) and SF1-Atg7−/− Leydig cells (bottom; C). Similar results were obtained in Atg5Flox/Flox mice (top) and SF1-Atg5−/− mice (bottom) as shown in D. Nuclei were stained with DAPI (blue). (E–L) Aging autophagy-deficient male mice spent less time sniffing female targets than the control male mice. 3-mo-old Atg5-deficient mice showed no significant difference in sniffing frequency and time (E and F), whereas 6-mo-old autophagy-deficient mice spent less time sniffing female genital odor than the control groups (G, H, K, and L). (M and R) Aging autophagy-deficient male mice showed a failure of mounting. 6-mo-old autophagy-deficient male mice mounted female mice with a longer latency (M and N), less frequency (O and P), and shorter duration (Q and R) than the control groups. (S and T) Testosterone production was significantly reduced in autophagy-deficient mice. *, P < 0.05; **, P < 0.01.
After the steroidogenic cell–specific knockout of Atg7 or Atg5, the autophagy-deficient male mice appeared more docile compared with the control groups. Mouse sexual behavior analysis confirmed our observations and revealed that the frequency and total time of sniffing in Atg5-deficent male mice were significantly decreased compared with the control groups at 6 mo (Fig. 2, G and H). Conversely, at 3 mo, some indexes showed no significant difference (Fig. 2, E and F). A significant decrease in the frequency and total time of sniffing could also be observed in 6-mo-old SF1-Atg7−/− male mice (Fig. 2, K and L). Further analysis of 6-mo-old autophagy-deficient male mice showed they mounted female targets with a longer latency, less frequency, and shorter duration than the control groups (Fig. 2, M–R), indicating that the steroidogenic cell–specific knockout of Atg7 or Atg5 might affect sexual behavior in aged male mice, similar to the symptoms of LOH (Bassil and Morley, 2010; Bassil, 2011).
Reproductive steroid hormones such as testosterone are essential for normal reproductive function, and the deficiency in serum testosterone is characteristic of LOH (Bassil and Morley, 2010; Bassil, 2011). Next, we examined whether the abnormal sexual behavior in aged autophagy-deficient male mice reflected the reduction of serum testosterone. The serum testosterone concentrations in aged autophagy-deficient mice were measured, showing a significant decrease compared with the control groups (Fig. 2, S and T). Collectively, these results provide evidence that the disruption of autophagy might affect the sexual behavior of male mice as a result of the sharp reduction in testosterone in the serum, similar to the symptoms of LOH. Thus, these autophagy-deficient mice might serve as a novel LOH animal model.
Cholesterol decreases in autophagy-deficient Leydig cells
Because the different developmental stages of Leydig cells show distinct steroidogenic capacities, abnormal Leydig cell proliferation or differentiation might influence testosterone levels (Chen et al., 2009). To test whether the disruption of autophagy affected the proliferation/differentiation of Leydig cells, we first assessed the fetal–adult Leydig cell transition in SF1-Atg7−/− mouse testes via immunofluorescence staining of Thrombospondin2 (TSP2 or Thbs2), which is expressed solely in fetal Leydig cells (O’Shaughnessy et al., 2002b). In SF1-Atg7−/− mouse testes, the TSP2 signal was detected in d4 mouse Leydig cells but disappeared in adult Leydig cells (Fig. S1 A), similar to TSP2 signals in Atg7Flox/Flox mice, indicating that a disruption of autophagy might not influence Leydig cell differentiation. Subsequently, Leydig cells were isolated from autophagy-deficient mouse testes. Total cell counts showed that the number of Leydig cells in autophagy-deficient mouse testes was not reduced compared with the control groups (Fig. S1, B and C). These results suggest that the disruption of autophagy in Leydig cells has no obvious influence on the proliferation or differentiation of Leydig cells.
A decline in testosterone in the serum commonly reflects extrinsic factors, intrinsic factors, or both (Chen et al., 2009; Midzak et al., 2009). In regard to extrinsic factors, the steroidogenic function of Leydig cells is strictly controlled by the hypothalamus–pituitary–gonad (HPG) axis and the pituitary gonadotropin (Gn)-luteinizing hormone (LH) released from the anterior pituitary gland (Ellis et al., 1983). In autophagy-deficient mice, the LH and follicle-stimulating hormone (FSH) concentrations in serum were similar to those in the control groups (Fig. S1, D–G). Thus, although the HPG axis was affected to some extent, the decline in testosterone in autophagy-deficient mice was probably not caused by either decreased LH or FSH.
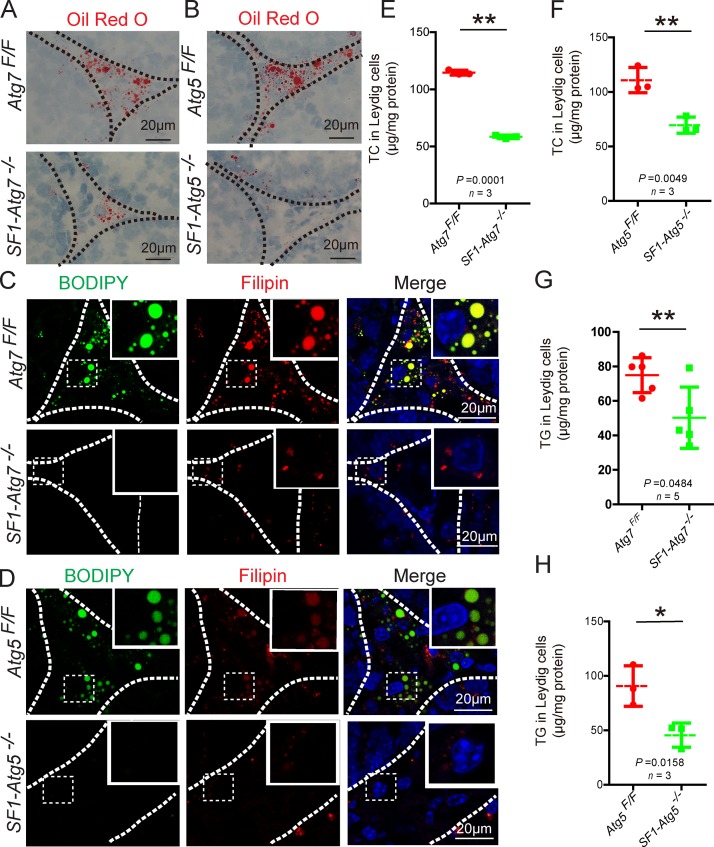
Next, we turned to intrinsic factors. Cholesterol serves as the initial raw material for testosterone synthesis in Leydig cells (Azhar et al., 2003), and autophagy has been implicated in lipid metabolism to provide the cell with cholesterol (Singh et al., 2009; Singh, 2010). If autophagy is involved in testosterone synthesis mainly by promoting lipid metabolism in Leydig cells, then cholesterol should accumulate in autophagy-deficient Leydig cells. To test this hypothesis, we performed ORO staining to detect lipid storage in Leydig cells as cholesterol is commonly stored in cytoplasmic LDs in the form of cholesterol esters (Haider, 2004). Surprisingly, LDs clearly decreased in autophagy-deficient testes compared with control groups (Fig. 3, A and B), in complete contrast with our working hypothesis and previous results obtained from hepatocytes (Singh et al., 2009). Similar to the ORO staining results, the BODIPY signals were also extremely reduced in autophagy-deficient testes (Fig. 3, C and D). Because ∼10% of testosterone is synthesized in the adrenal gland and SF1-Cre is also expressed in this gland, we also studied the effect of Atg5 and Atg7 knockout on the adrenal gland. We determined that once Atg5 or Atg7 was knocked out, corticosterone decreased in the serum and LDs clearly decreased in the adrenal cortex (Fig. S2). These results suggest that lipid metabolism in the adrenal gland is also affected from steroidogenic cell–specific autophagy disruption mice. Because the LDs in the adrenal cortex were similar to those of the Leydig cells, we used Leydig cells to address the underlying mechanism. To help elucidate the underlying mechanism, we characterized the concentration of total cholesterol (TC) in Leydig cells and showed that TC was significantly reduced in autophagy-deficient Leydig cells but not in either total testis or serum (Fig. 3, E and F; and Fig. S3, A–D), indicating that the disruption of autophagy affected cholesterol levels in Leydig cells. Next, we measured TG concentrations in Leydig cells, which are also stored in LDs (Martin and Parton, 2006). TGs were significantly reduced in autophagy-deficient Leydig cells but not in total testis and serum (Fig. 3, G and H; and Fig. S3, E–H). Next, we directly detected cholesterol levels in Leydig cells using Filipin staining. In autophagy-deficient testes, Filipin signals were notably decreased compared with control groups (Fig. 3, C and D). These results suggest that an insufficient supply of cholesterol in autophagy-deficient Leydig cells might be the primary reason for the decline in the serum testosterone concentration in autophagy-deficient mice. Although the decreased cholesterol levels in autophagy-deficient Leydig cells were not expected, the LC3 puncta unassociated with LDs (Fig. 1 F) suggest that in addition to a role in lipid metabolism, autophagy might play some novel unknown role in cholesterol regulation in Leydig cells.
Figure 3.
Dramatically reduced LDs and cholesterol in autophagy-deficient Leydig cells. (A and B) LDs dramatically decreased in autophagy-deficient Leydig cells. ORO (red) staining in both Atg7Flox/Flox (top) and SF1-Atg7−/− Leydig cells (bottom; A). The ORO (red) staining results in Atg5Flox/Flox mice (top) and SF1-Atg5−/− mice (bottom) are shown in B. (C and D) Lipid and cholesterol storage were extremely reduced in autophagy-deficient Leydig cells. BODIPY (green) and Filipin (red) staining were performed in both Atg7Flox/Flox (top) and SF1-Atg7−/− Leydig cells (bottom; C). BODIPY (green) and Filipin (red) staining results in Atg5Flox/Flox mice (top) and SF1-Atg5−/− mice (bottom) are shown in D. Insets are marked by boxes and are 15 µM wide. Nuclei were stained with DAPI (blue). (E and F) The concentration of TC in autophagy-deficient Leydig cells was significantly reduced compared with the control groups. (G and H) The concentration of TG in autophagy-deficient Leydig cells was significantly reduced compared with the control groups. *, P < 0.05; **, P < 0.01. See also Figs. S1, S2, and S3.
Impaired autophagy perturbs cholesterol uptake
Next, we determined the factors contributing to the decreased LD and cholesterol levels in autophagy-deficient Leydig cells. Decreasing levels might be caused by either increased consumption or decreased supply. The decrease in LDs and cholesterol was somewhat similar to that observed in autophagy-deficient adipocytes, accompanied by increased copy numbers of mitochondria (Zhang et al., 2009). Thus, the decrease in LDs and cholesterol might reflect a superactive metabolism resulting from increased numbers of mitochondria. To test this hypothesis, autophagy-deficient Leydig cells were isolated and stained with MitoTracker red. Results showed that the amount of mitochondria was not increased; instead, a clear decrease was observed compared with the control groups (Fig. S1, H and I). To further confirm these results, we crossed SF1-Atg5Flox/− mice with mitochondrial-DsRed transgenic mice (CAG/su9-DsRed2 mice; Hasuwa et al., 2010), and progeny showed that the number of mitochondria was dramatically reduced as characterized with fluorescence microscopy (Fig. S1 J). These results suggest that the decrease in LDs and cholesterol is not likely caused by increased consumption in autophagy-deficient Leydig cells.
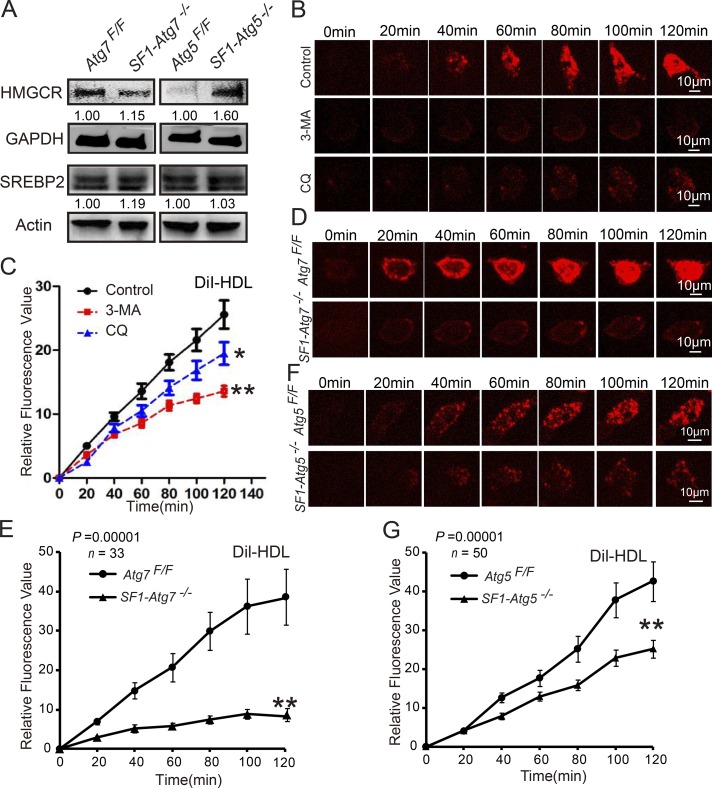
Next, we examined the effect of autophagy disruption on the supply of LDs and cholesterol in Leydig cells. There are two major sources for cholesterol within steroidogenic cells: de novo synthesis from acetate and transport of lipoproteins through plasma membrane–bound receptors (Hou et al., 1990; Azhar and Reaven, 2002). To investigate which aspects were influenced in autophagy-deficient Leydig cells, we first examined protein levels of the cholesterol biosynthetic enzymes 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMGCR) and the cholesterol biosynthesis–related transcription factor SREBP-2 (Eberlé et al., 2004) using immunoblotting in autophagy-deficient Leydig cells and control groups. Results showed that the protein levels of HMGCR and SREBP-2 were not decreased in autophagy-deficient Leydig cells compared with control groups (Fig. 4 A), likely reflecting a compensatory response to the cholesterol-insufficient status of the Leydig cells. Thus, the disruption of autophagy might not inhibit cholesterol synthesis in Leydig cells.
Figure 4.
Impaired autophagy perturbs cholesterol uptake. (A) The levels of cholesterol synthesis–related proteins did not decrease in autophagy-deficient Leydig cells. Immunoblotting analysis of HMGCR and SREBP2 in Atg7Flox/Flox, SF1-Atg7−/−, Atg5Flox/Flox, and SF1-Atg5−/− Leydig cells. GAPDH and actin served as loading controls. (B) The isolated Leydig cells were treated with physiological saline (control), 3-MA, or CQ. The in vitro cholesterol absorption experiment was subsequently performed in Leydig cells using the fluorescence-labeled cholesterol-rich lipoprotein DiI-HDL and were examined using live-cell imaging. (C) The absorption curves of DiI-HDL in 3-MA– or CQ-treated Leydig cells and control groups. (D and F) The steroidogenic cell–specific knockout Atg7 or Atg5 affected cholesterol uptake. Absorption of DiI-HDL (red) was examined using live-cell imaging. (E and G) The absorption curve of DiI-HDL in autophagy-deficient Leydig cells and control groups. Error bars show means ± SD. *, P < 0.05; **, P < 0.01.
Next, we examined whether autophagy was involved in cholesterol uptake in Leydig cells. High-density lipoprotein (HDL) cholesterol is a major source of cholesterol for testosterone production in Leydig cells (Landschulz et al., 1996). To examine the potential role of autophagy in the uptake of cholesterol from HDL cholesterol, in vitro cholesterol uptake experiments were performed in isolated Leydig cells using the fluorescence-labeled cholesterol-rich lipoprotein 1,1’-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI)-HDL (Landschulz et al., 1996). Two autophagy/lysosome inhibitors, 3-methyladenine (3-MA) and chloroquine (CQ; Wang et al., 2014), were added to inhibit the autophagy–lysosome pathway. After treatment with these inhibitors, the rates and amounts of DiI-HDL absorbed by Leydig cells were significantly reduced compared with the control group (Fig. 4, B and C), indicating that autophagy might be involved in cholesterol uptake. To further confirm this result, autophagy-deficient Leydig cells were isolated and subsequently tested using a cholesterol absorption experiment. Similar to these results, the autophagy-deficient Leydig cells showed a defect in cholesterol uptake compared with the control groups (Fig. 4, D–G). Collectively, these results suggest that autophagy plays a role in cholesterol uptake in Leydig cells.
Leydig cell cholesterol uptake requires the SR-BI and the low-density lipoprotein (LDL) receptor (LDLR), two major lipoprotein receptors that mediate the high-capacity selective uptake of cholesterol esters from the core of HDL, LDL, or very-low-density lipoprotein (DeAngelis et al., 2014). To further investigate the mechanism of autophagy in cholesterol uptake, we determined the protein levels of SR-BI and LDLR in autophagy-deficient Leydig cells using immunoblotting. Compared with the control groups, the protein levels of SR-BI in autophagy-deficient Leydig cells were dramatically reduced, whereas the LDLR protein levels were not affected (Fig. 5 A). These results suggest that autophagy might regulate cholesterol uptake by up-regulating SR-BI. To further confirm this result, an immunofluorescence analysis of SR-BI was performed in autophagy-deficient testes. Similar to the immunoblotting results, SR-BI signals in autophagy-deficient testes were also reduced compared with the control groups (Fig. 5, B and C). These results suggest that the decreased cholesterol pool in autophagy-deficient Leydig cells results from impaired cholesterol uptake likely induced by the down-regulation of the lipoprotein receptor SR-BI. We next determined the expression of SR-BI in the Leydig cells of patients with low-serum testosterone–level azoospermia or oligospermia and found that the SR-BI signals were also significantly decreased (Fig. 5, F and G), suggesting that autophagy deficiency in some of the patients with low-serum testosterone might also influence serum testosterone levels through the down-regulation of SR-BI.
Figure 5.
The disruption of autophagy in Leydig cells inhibits cholesterol uptake by down-regulating SR-BI. (A) The protein levels of SR-BI were extremely reduced in autophagy-deficient Leydig cells. Immunoblotting analyses of SR-BI and LDLR were performed in Atg7Flox/Flox, SF1-Atg7−/−, Atg5Flox/Flox, and SF1-Atg5−/− Leydig cells. GAPDH served as a loading control. (B and C) SR-BI was decreased in the autophagy-deficient Leydig cells. (B) Immunofluorescence analysis of SR-BI (red) and BODIPY (green) staining in Atg7Flox/Flox (top) and SF1-Atg7−/− (bottom) Leydig cells. (C) Immunofluorescence analysis of Atg5Flox/Flox (top) and SF1-Atg5−/− mice (bottom). Insets are marked by boxes and are 8 µM wide. (D and E) Relative mRNA level of SR-BI in autophagy-deficient Leydig cells. The mRNA levels of SR-BI showed no changes in autophagy-deficient Leydig cells compared with the control groups. (F) SR-BI immunofluorescence analysis (red) in Leydig cells of the low-serum testosterone (T) level azoospermia or oligospermia patients and control groups. The nuclei were stained with DAPI (blue). (G) Quantification of the fluorescence intensity per μm2 of SR-BI in F. *, P < 0.05.
Autophagy regulates SR-BI by eliminating NHERF2
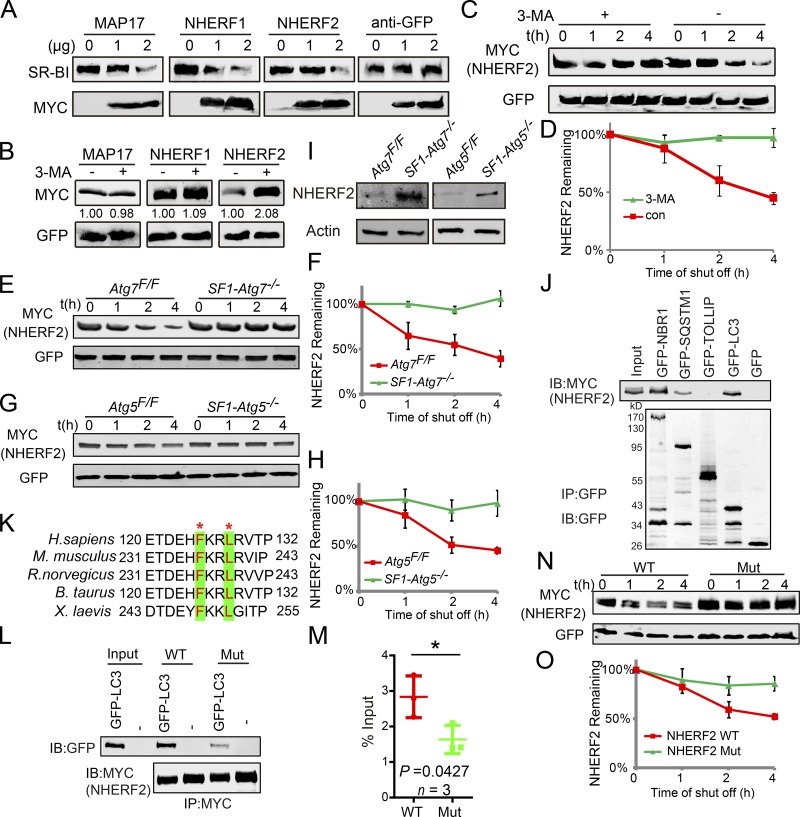
Next, we determined how autophagy down-regulates SR-BI in Leydig cells. First, we detected the mRNA expression level of SR-BI in autophagy-deficient Leydig cells and found no significant differences between autophagy-deficient and control groups (Fig. 5, D and E), indicating that autophagy might mediate the degradation of some negative regulators of SR-BI to maintain proper protein levels of SR-BI at the posttranscriptional level. Three major SR-BI–negative regulators have been reported: MAP17, NHERF1, and NHERF2, whose accumulation down-regulates SR-BI via posttranscriptional regulation (Silver et al., 2003; Hu et al., 2013). We cooverexpressed these genes with constant amounts of SR-BI in HEK293T cells and detected that increased expression of these proteins was accompanied by decreased expressions of SR-BI protein (Fig. 6 A). To determine whether these negative regulators of SR-BI might be regulated by autophagy, 3-MA was added to the media of HEK293T cells transfected with the three genes. After treatment with the autophagy inhibitor, NHERF2 protein, but not the other two proteins, accumulated in the cells (Fig. 6 B). Furthermore, in autophagy-deficient mouse testes, immunofluorescence analyses of these three proteins showed that the signal for NHERF2 but not the other two proteins clearly increased compared with control groups (Fig. S4), suggesting that NHERF2 might be eliminated by the autophagy–lysosome pathway. To further confirm this result, cycloheximide (CHX) chase assays were performed, showing that NHERF2 degradation was strongly delayed after 3-MA treatment (Fig. 6, C and D). Next, we characterized NHERF2 degradation in autophagy-deficient Leydig cells. Results showed that the disruption of autophagy strongly delayed NHERF2 degradation compared with control groups (Fig. 6, E–H). In addition, NHERF2 protein levels increased in autophagy-deficient Leydig cells compared with control cells (Fig. 6 I). Thus, the degradation of NHERF2 is dependent on the autophagy–lysosome pathway. Furthermore, coimmunoprecipitation experiments in HEK293T cells showed that NHERF2 could physically interact with LC3 and the autophagic adapter proteins NBR1 and SQSTM1 but not Toll-interacting protein (TOLLIP; Fig. 6 J; Lu et al., 2014). We also observed that NHERF2 partially colocalized with LC3, NBR1, SQSTM1, and LAMP2 under starvation conditions in Leydig cells. LAMP2 is a lysosome membrane–integrated protein (Fig. S5; Saftig and Klumperman, 2009), suggesting the presence of lysosomes or autolysosomes. Usually, selective autophagy substrates have one or more LC3-interacting region (LIR) motifs that are essential for the protein’s interaction with LC3 and further degradation (Rogov et al., 2014). After protein sequence analysis, we identified a potential LIR motif in NHERF2 (Fig. 6 K), which is highly conserved in different species. We then mutated this LIR motif to F236A/L239A and found that once this LIR motif was disrupted, the interaction between NHERF2 and LC3 was impaired (Fig. 6, L and M), and its degradation was dramatically delayed (Fig. 6 N and O). Collectively, these results suggest that NHERF2 is degraded via the autophagy–lysosome pathway.
Figure 6.
Selective autophagy-dependent degradation of NHERF2. (A) The overexpression of NHERF1, NHERF2, and MAP17 down-regulated the protein levels of SR-BI. (B) Treatment with 3-MA resulted in the accumulation of NHERF2 but not the other two SR-BI negative regulators. (C) NHERF2 was stabilized in autophagic flux–disrupted cells. CHX chase assay of NHERF2 in HEK293T cells in the absence or presence of 3-MA. (D) Quantification of the relative NHERF2 levels in C. (E and G) NHERF2 was stabilized in autophagy-deficient Leydig cells. CHX chase assay of NHERF2 in Atg7Flox/Flox and SF1-Atg7−/− (E) as well as Atg5Flox/Flox and SF1-Atg5−/− (G) Leydig cells. (F and H) Quantification of the relative NHERF2 levels in E and G. (I) NHERF2 is accumulated in autophagy-deficient Leydig cells. The immunoblotting (IB) analysis of NHERF2 was performed in Atg7Flox/Flox and SF1-Atg7−/− as well as Atg5Flox/Flox and SF1-Atg5−/− Leydig cells. Actin served as a loading control. (J) NHERF2 interacts with LC3, NBR1, and SQSTM1. pCMV6-Myc-Nherf2 and pMXs-puro-GFP-SQSTM1, pMXs-IP-GFP-NBR1(38283), pEGFP-TOLLIP, or pEGFP-LC3 were cotransfected into HEK293T cells. 24 h after transfection, cells were collected for IP with anti-GFP antibody and analyzed with GFP antibodies, respectively. (K) Sequence alignment of NHERF2 LIR motifs from different species. Asterisks indicate conserved residues in LIR motifs. (L) The disruption of the LIR motif in NHERF2 influences its interaction with LC3. (M) Quantification of the relative binding affinity between NHERF2 (WT/mutant [F236A/L239A]) and GFP-LC3. Relative amounts of GFP-LC3 were quantified using Odyssey software and compared with the input. (N) The LIR motif of NHERF2 is essential for its degradation. CHX chase assay of NHERF2 (WT) and NHERF2 F236A/L239A (Mut) in HEK293T cells. (O) Quantification of the relative NHERF2 levels in N. The amounts of NHERF2 were quantified using Odyssey software. See also Figs. S4 and S5. Error bars show means ± SD. *, P < 0.05.
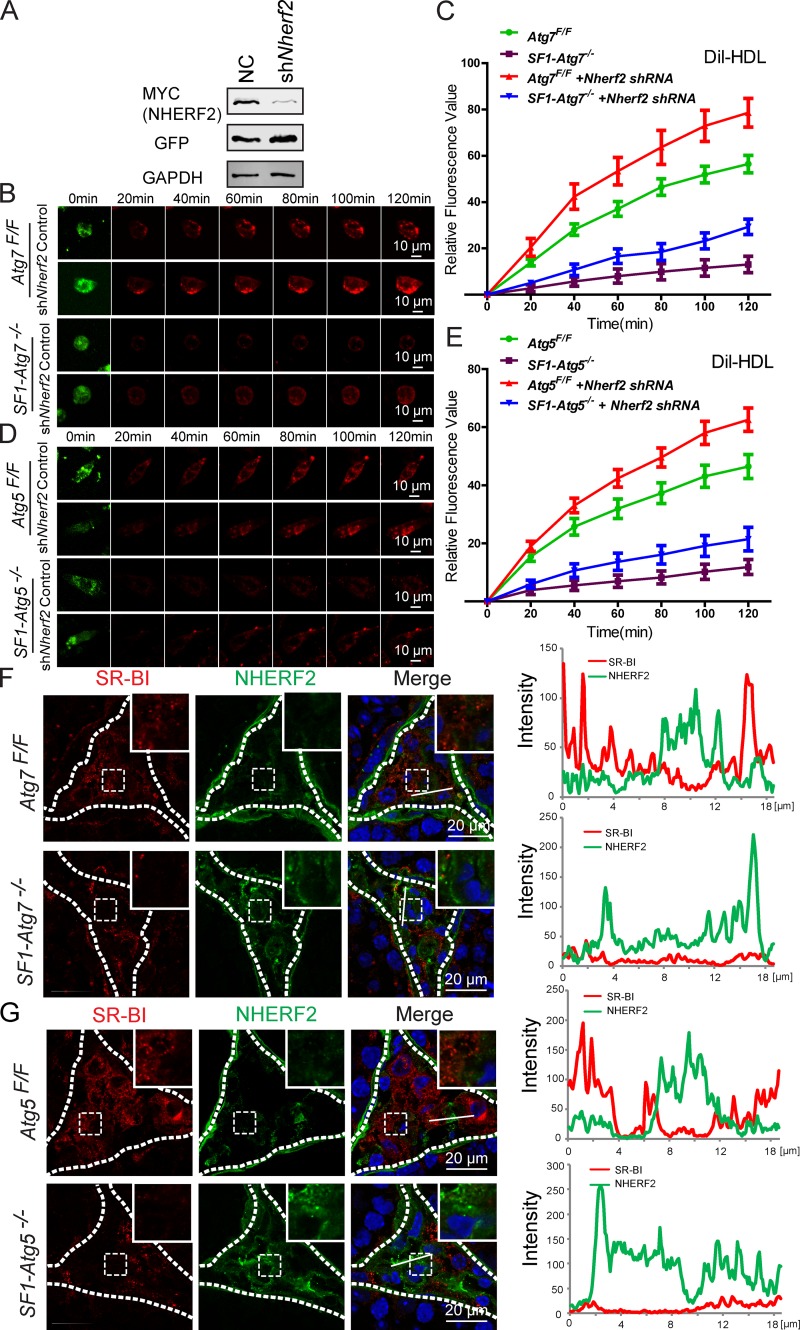
Next, we asked whether the accumulation of NHERF2 was the major cause of the cholesterol uptake defect in autophagy-deficient Leydig cells. If yes, then knockdown of Nherf2 in autophagy-deficient Leydig cells might at least partially rescue the impaired cholesterol uptake phenotype of the cells. In Nherf2-knockdown autophagy-deficient or WT Leydig cells, cholesterol uptake, both rates and amounts, was significantly increased compared with Nherf2-expressing cells, as indicated by DiI-HDL absorption (Fig. 7, A–E). Thus, NHERF2 appears to be the linker between autophagy and cholesterol uptake in Leydig cells.
Figure 7.
Autophagy regulates SR-BI by eliminating NHERF2 in the Leydig cells. (A) The knockdown of Nherf2 by shRNA could reduce NHERF2 expression effectively. pCMV6-Myc-Nherf2 was transfected into HEK293T cells, whereas the Nherf2 shRNA and control virus were added. 24 h after transfection, cells were collected for immunoblotting analysis with anti-Myc and anti-GFP antibodies, respectively. GAPDH served as a loading control. NC indicates the negative control group that was transfected with control virus. (B and D) The cholesterol uptake defects could be partially rescued by Nherf2 knockdown in autophagy-deficient and control Leydig cells. The Atg7Flox/Flox and SF1-Atg7−/− (B) as well as Atg5Flox/Flox and SF1-Atg5−/− (D) Leydig cells were infected by Nherf2 shRNA and control virus (green). The in vitro cholesterol absorption experiment was subsequently performed in the Leydig cells using the fluorescence-labeled cholesterol-rich lipoprotein DiI-HDL (red) and examined using live-cell imaging. (C and E) The cholesterol uptake defects could be partially rescued by Nherf2 knockdown in autophagy-deficient Leydig cells. Plots show absorption curves of DiI-HDL in autophagy-deficient and control Leydig cells infected by Nherf2 shRNA virus and control virus. Error bars show means ± SD. (F and G) The accumulation of NHERF2 in autophagy-deficient Leydig cells down-regulated SR-BI as well as SR-BI negatively correlated with NHERF2 in control Leydig cells. Immunofluorescence analysis was performed using SR-BI (red) and NHERF2 (green) in Atg7Flox/Flox (F, top), SF1-Atg7−/− (F, bottom), Atg5Flox/Flox (G, top), and SF1-Atg5−/− (G, bottom) Leydig cells, and the fluorescence intensity was quantified on the white lines. Insets are marked by boxes and are 10 µM wide. Nuclei were stained with DAPI (blue).
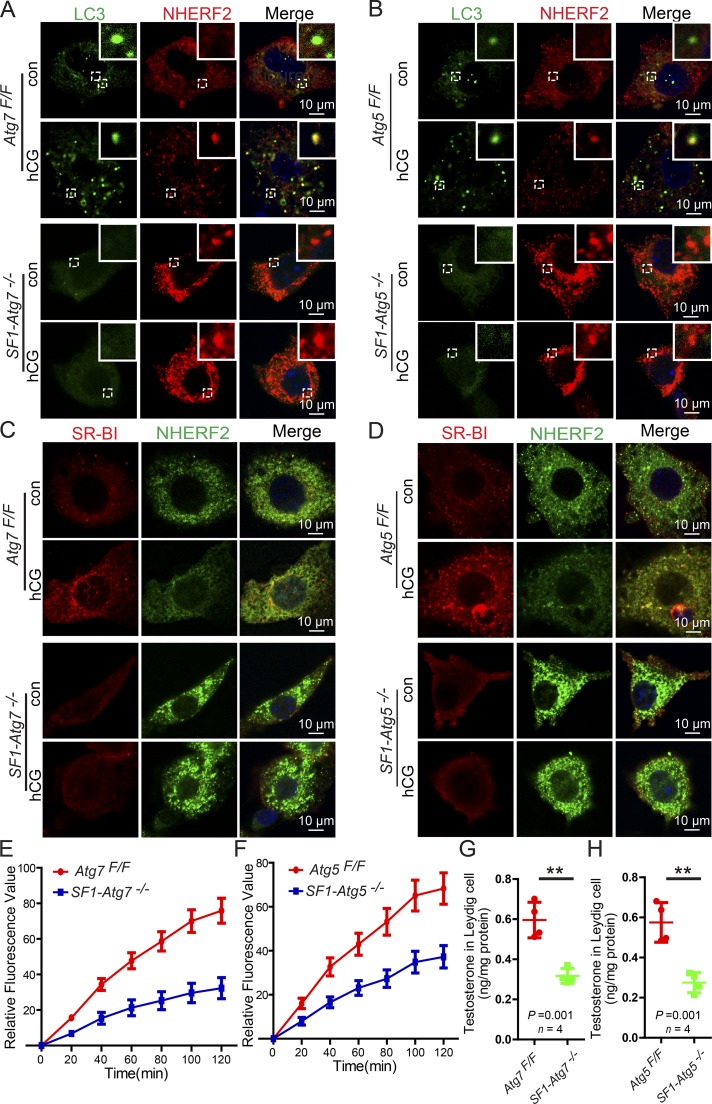
To further explore the relationship between NHERF2 accumulation and SR-BI protein levels, we assessed the expression and localization of NHERF2 and SR-BI in the testes of autophagy-deficient mice using immunofluorescence analysis. The NHERF2 and SR-BI signals showed a negative correlation in control mouse Leydig cells, and the accumulation of NHERF2 in autophagy-deficient mouse Leydig cells clearly showed reduced levels of SR-BI (Fig. 7, F and G). These results suggest that the disruption of autophagy in Leydig cells results in the abnormal accumulation of the SR-BI–negative regulator NHERF2, which in turn down-regulates SR-BI, leading to defective cholesterol uptake and a deficiency in testosterone synthesis.
Autophagy is required for hormone-induced testosterone synthesis in Leydig cells
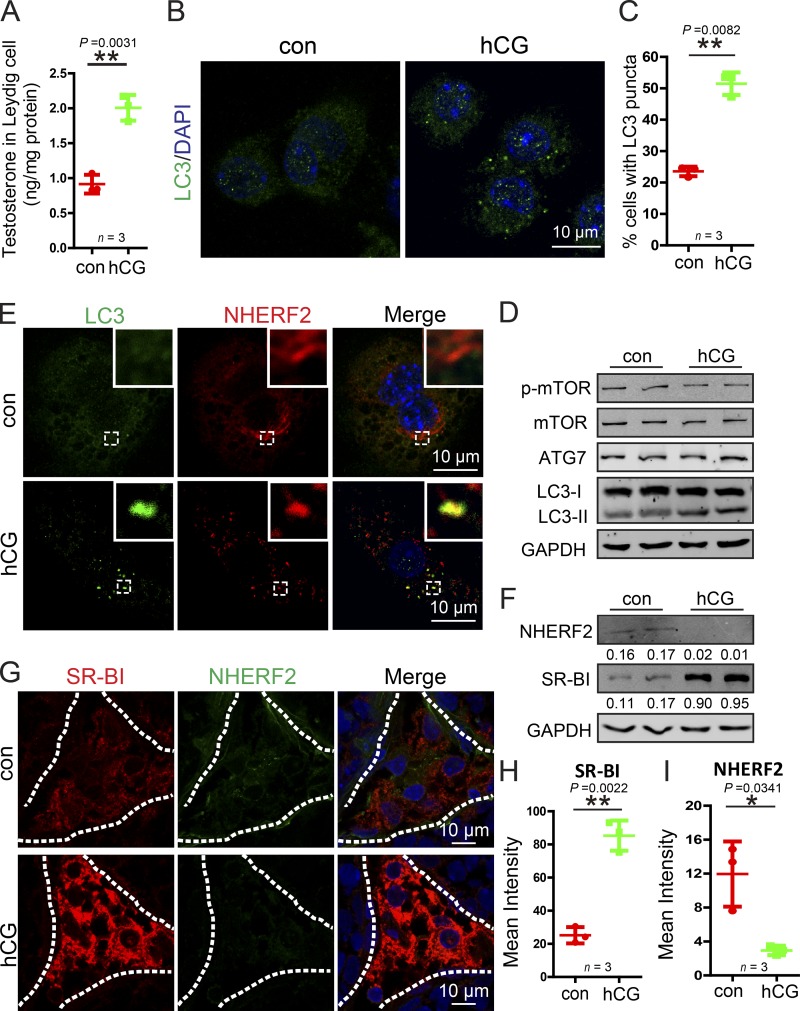
The testosterone synthesis in Leydig cells is controlled by the HPG axis and the pituitary Gn LH released from the anterior pituitary gland (Ellis et al., 1983); therefore, we speculated that autophagy might be involved in hormone-induced testosterone synthesis in Leydig cells. To test this possibility, isolated Leydig cells were directly treated with human chorionic Gn (hCG), the pituitary analogue of LH. Indeed, we found that testosterone synthesis was promoted by hCG treatment (Fig. 8 A). In addition, we characterized autophagy flux and autophagy-related proteins in hCG-treated Leydig cells. As shown in Fig. 8 D, although the levels of total mTOR, phosphor-mTOR, and ATG7 showed no significant difference between the hCG-treated and control groups, levels of the membrane-associated form of LC3-II (Fig. 8 D), the LC3 puncta, and the percentage of cells with LC3 puncta all increased (Fig. 8, B and C), suggesting that autophagy activity was induced after hCG treatment. Next, we examined whether the activated autophagy could facilitate NHERF2 degradation. In hCG-treated Leydig cells, NHERF2 clearly colocalized with LC3 as shown by immunofluorescence (Fig. 8 E). Immunoblotting analysis also showed that NHERF2 protein levels were significantly reduced, whereas SR-BI was obviously increased compared with control groups (Fig. 8 F). Finally, to examine the physiological condition in the testis, male mice were subcutaneously injected with 250 U/kg body weight hCG, whereas a control group received an equivalent volume of normal saline for 4 d. We then assessed the expression of NHERF2 and SR-BI by immunofluorescence. NHERF2 was significantly reduced, potentially allowing SR-BI to accumulate (Fig. 8, G–I). Thus, the hormone treatment in Leydig cells might activate autophagy to up-regulate SR-BI by facilitating the degradation of NHERF2.
Figure 8.
hCG treatment enhances autophagy to promote NHERF2 clearance and testosterone synthesis in Leydig cells. (A) Testosterone production was significantly increased in hCG-treated Leydig cells. (B) The LC3 puncta was significantly increased after hCG treatment in Leydig cells. Immunofluorescence analysis of LC3 (green) was performed in hCG-treated and control Leydig cells. (C) Quantification of the percentages of cells with LC3 puncta in B. (D) The membrane-associated form LC3-II was accumulated in hCG-treated Leydig cells. The immunoblotting analysis of phosphor-mTOR, mTOR, ATG7, and LC3 was performed in hCG-treated and control Leydig cells. GAPDH served as a loading control. (E) NHERF2 partially colocalized with LC3 in hCG-treated Leydig cells. Immunofluorescence analysis of NHERF2 (red) and LC3 (green) were performed in hCG-treated and control Leydig cells. Insets are marked by boxes and are 2.3 µM wide. (F) NHERF2 was reduced, and SR-BI accumulated in hCG-treated Leydig cells. The immunoblotting analysis of NHERF2 was performed in hCG-treated and control Leydig cells. GAPDH served as a loading control. Quantifications of Western blots for NHERF2 and SR-BI relative to GAPDH is shown below the corresponding bands. (G) A negative correlation between SR-BI and NHERF2 in hCG-treated and untreated Leydig cells. Immunofluorescence analysis of SR-BI (red) and NHERF2 (green) were performed in hCG-treated and control mice. Nuclei were stained with DAPI (blue). (H and I) Quantification of the fluorescence intensity of SR-BI and NHERF2 in G. *, P < 0.05; **, P < 0.01.
To test whether hCG-induced testosterone synthesis is dependent on autophagy-mediated NHERF2 degradation, we analyzed the expression and localization of LC3 and NHERF2 in autophagy-deficient Leydig cells after hCG treatment by immunofluorescence. We found that in hCG-treated autophagy-deficient Leydig cells, NHERF2 still accumulated, and hCG treatment did not induce LC3 puncta formation or promote colocalization of LC3 with NHERF2 (Fig. 9, A and B). In addition, hCG treatment did not lead to up-regulation of SR-BI in autophagy-deficient Leydig cells (Fig. 9, C and D). Consistent with these results, cholesterol uptake, which was stimulated by hCG treatment, was dramatically inhibited in autophagy-deficient Leydig cells (Fig. 9, E and F). Testosterone synthesis in hCG-treated autophagy-deficient Leydig cells was also significantly lower than in control groups (Fig. 9, G and H). These results suggested that autophagy might be activated in response to hormone stimulation, promotes SR-BI up-regulation through the degradation of NHERF2, enhances cholesterol uptake, and finally, stimulates testosterone synthesis in the Leydig cells.
Figure 9.
Autophagy-dependent NHERF2 clearance is required for hCG-induced testosterone synthesis in Leydig cells. (A and B) NHERF2 could not colocalize with LC3 in hCG-treated autophagy-deficient Leydig cells. Immunofluorescence analyses of NHERF2 (red) and LC3 (green) were performed in hCG-treated Atg7Flox/Flox and SF1-Atg7−/− as well as Atg5Flox/Flox and SF1-Atg5−/− Leydig cells. Insets are marked by boxes and are 2.5 µM wide. (C and D) Immunofluorescence analysis of SR-BI (red) and NHERF2 (green) were performed in hCG-treated Atg7Flox/Flox and SF1-Atg7−/− as well as Atg5Flox/Flox and SF1-Atg5−/− Leydig cells. Nuclei were stained with DAPI (blue). (E and F) The absorption curve of DiI-HDL in hCG-treated autophagy-deficient Leydig cells. Error bars show means ± SD. (G and H) Decreased testosterone production in hCG-treated autophagy-deficient Leydig cells. **, P < 0.01.
Discussion
Even before the molecular mechanism of the autophagic pathway was elucidated, autophagy has been observed to be extremely active in Leydig cells (Tang, 1988; Tang and Zhang, 1990; Yi and Tang, 1991, 1995, 1999; Tang et al., 1992). Recently, a steroidogenic decline has been associated with autophagic deficiency in rat and mouse Leydig cells (Li et al., 2011; Yoshii et al., 2016). However, the physiological role of autophagy in Leydig cells has not been well elucidated until now. We observe that autophagy indeed participates in testosterone production, but the underlying mechanism was far from our original hypothesis.
Cholesterol serves as the initial substrate for steroid hormone synthesis, and this compound is commonly stored in cytoplasmic LDs in the form of cholesterol esters in Leydig cells (Azhar et al., 2003; Haider, 2004; Martin and Parton, 2006). We observed that the expression of LC3 was highly consistent with the development of LDs at various developmental stages, and LC3 signals colocalized with LDs in adult mouse testes (Fig. 1, D–F). The results suggest that autophagy might also participate in lipid metabolism in Leydig cells, similar to research using other types of cells such as hepatocytes (Singh et al., 2009). Thus, we speculated that LDs might accumulate in autophagy-deficient Leydig cells, but surprisingly, the LDs in autophagy-deficient Leydig cells clearly decreased (Fig. 3). The decrease in LDs in autophagy-deficient Leydig cells is different from that in autophagy-deficient adipocytes, which is likely caused by a differentiation defect with accumulated mitochondria (Zhang et al., 2009). Further investigation uncovered a novel functional role for autophagy in cholesterol uptake, mediated by the lipoprotein receptor SR-BI (Figs. 4 and 5).
Autophagy regulates SR-BI by promoting the degradation of the SR-BI negative regulator NHERF2. NHERF2 (also called E3KARP) belongs to the NHERF family of PDZ scaffolding proteins, which regulate receptor and channel protein localization and function to scaffold intracellular signaling protein complexes (Seidler et al., 2009). NHERF2 binds to SR-BI and serves as a negative modulator that down-regulates SR-BI protein expression via the inhibition of de novo synthesis of this protein (Hu et al., 2013). In this study, we provide new evidence that shows that NHERF2 is degraded via the autophagy–lysosome pathway in Leydig cells to maintain proper protein levels of SR-BI to facilitate cholesterol uptake during testosterone synthesis, thereby providing a novel mechanism underlying the physiological role of autophagy in steroid hormone synthesis. Notably, the above-mentioned function might be just one of the physiological roles of autophagy, which may involve other substrates and participate in a manifold of processes such as lipolysis in Leydig cells.
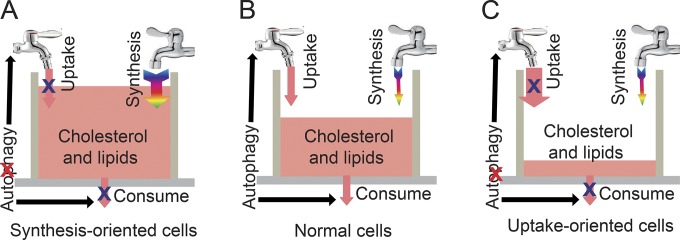
Thus, autophagy might simultaneously regulate the uptake and consumption of cholesterol in a single cell. In general, cholesterol homeostasis is maintained by supply and consumption through two sources for cholesterol: transport of lipoproteins through plasma membrane–bound receptors and de novo synthesis from acetate (Fig. 10 B; Hou et al., 1990; Azhar and Reaven, 2002). The major sources of cholesterol are distinct for different cell types. In synthesis-oriented cells such as hepatocytes, the disruption of autophagy primarily influences the utilization of cholesterol but not de novo cholesterol synthesis in the ER. Thus, cholesterol and lipids will accumulate in autophagy-deficient cells (Fig. 10 A). However, in uptake-oriented cells such as Leydig cells (Azhar et al., 2003), the disruption of autophagy blocks both the uptake and utilization of cholesterol. Without enough de novo synthesis, the cholesterol pool will be rapidly exhausted (Fig. 10 C). The third scenario might also apply to other types of uptake-oriented cells such as ovaries or adipocytes (Zhang et al., 2009; Gawriluk et al., 2014), which still requires further investigation.
Figure 10.
A proposed model for the role of autophagy in maintaining the cholesterol pool. (A–C) In normal cells (B), the cholesterol pool is regulated by the supply and consumption, and the cholesterol supply is derived from either uptake or de novo synthesis. Autophagy simultaneously regulates uptake and consumption to maintain the proper levels of cholesterol in a single cell. In synthesis-oriented cells (A) such as hepatocytes, the disruption of autophagy primarily influences the utilization but not the synthesis of cholesterol. Thus, cholesterol and lipids will accumulate in autophagy-deficient cells. In uptake-oriented cells (C) such as Leydig cells, the disruption of autophagy inhibits both the uptake and utilization of cholesterol. Without adequate synthesis, the cholesterol pool will rapidly be exhausted in autophagy-deficient cells.
The phenotypes of steroidogenic cell–specific autophagy-deficient mice are similar to those of LOH. Many aging Leydig cell–related steroidogenic symptoms could be identified in aged autophagy-deficient mice, including the reduction of serum testosterone and mitochondrial number (Figs. 2 and S1). Thus, steroidogenic cell–specific autophagy-deficient mice could serve as novel LOH animal models for the investigation of the pathology and potential treatment of LOH. Recent evidence suggests that men who take exogenous testosterone might face increased risks of stroke, heart attack, and prostate tumorigenesis (Corona et al., 2013). Moreover, administered testosterone might have suppressive effects on LH, resulting in lower Leydig cell testosterone production, reduced intratesticular testosterone concentrations, and reduced spermatogenesis. These effects make exogenous testosterone administration inappropriate for men wishing to father children. There are promising new approaches, such as Gn therapy, Gn-releasing hormone therapy, and stem Leydig cell replacement therapy (Jiang et al., 2014; Corona et al., 2015), for increasing serum testosterone by directly stimulating testosterone production in Leydig cells rather than by exogenous testosterone therapy, thus potentially avoiding some of the negative consequences of this hormone. Steroidogenic cell–specific autophagy-deficient mice provide an excellent platform with which to evaluate these new approaches, showing important theoretical and clinical significance for use in LOH studies. Our results also suggest an important potential for using autophagy stimulators to treat some LOH patient in the future.
Materials and methods
Patients and clinical samples
In this study, 32 patients diagnosed with azoospermia or oligospermia in the period from 2013 to 2016 were recruited to perform an open biopsy of the testis and serum testosterone detection. Before the testicular biopsy, the definite diagnosis of azoospermia or oligospermia was confirmed at least three times by semen analysis showing the absence of sperm in two consecutive ejaculates. Patients gave their written and informed consent to surgery, biopsy examination, and further analysis. This research have been approved by the Animal and Medical Research Ethics Committee of the Institute of Zoology and the Clinical Research Ethics Committee of First Hospital of Peking University (Reference number: 2016-078). Patients were subjected to an incision of 8–10 mm in length in the tunica albuginea of the testis, and then protruding testicular tissues were dissected using surgical microscissors. After dissection, the testicular pieces were immediately fixed with 4% PFA. Serum testosterone levels of all patients were measured using a radioimmunoassay (RIA) kit (Beijing Sino-UK Institute of Biological Technology). Based on the testosterone hormone analysis, azoospermia patients were further classified into two groups. Those patients with normal serum testosterone levels (testosterone > 10.40 nmol/L) formed a control group (n = 12) whose mean age was 30 yr old (range 22–39 yr). The rest of the patients (n = 20) displayed low-serum testosterone levels (testosterone < 10.40 nmol/L), and their mean age was 28 yr old (range 22–35 yr). All of the diagnoses were performed according to the guidelines of the World Health Organization.
Animals
The Atg5FloxFFlox mouse strain (RBRC02975; Hara et al., 2006) and Atg7FloxFFlox mouse strain (RBRC02759; Komatsu et al., 2005) were obtained from the Institute of Physical and Chemical Research BioResource Center with permission from N. Mizushima and M. Komatsu (Institute of Physical and Chemical Research, Ibaraki, Japan). The SF1-Cre mice (Dhillon et al., 2006) were purchased from The Jackson Laboratory. The Atg5Flox/Flox:SF1-Cre and Atg7Flox/Flox:SF1-Cre mice were bred from Atg5Flox/Flox mice, Atg7Flox/Flox mice, and SF1-Cre mice. The CAG/su9-DsRed2 mouse strain was obtained from O. Masaru and I. Masahito (Osaka University, Osaka, Japan; Hasuwa et al., 2010). Mice treated with hCG were injected subcutaneously with 250 U/kg body weight in 10 mg/ml mannitol and 10 mM sodium phosphate, pH 7.2, for 4 d, whereas control animals received an equivalent volume of 10 mg/ml mannitol and 10 mM sodium phosphate, pH 7.2, only. All animal studies were performed in accordance with the protocols of the Institutional Animal Care and Use Committee.
Antibodies
The rabbit anti-LC3 polyclonal antibody (ab58610) and anti-HMGCR monoclonal antibody (ab174830) were purchased from Abcam. The rabbit anti-ATG5 monoclonal antibody (12994), rabbit anti-ATG7 monoclonal antibody (8558), rabbit anti-SQSTM1/p62 polyclonal antibody (5114), and rabbit anti-LC3A/B (12741) anti-LC3A (4599) monoclonal antibody for immunoblotting were purchased from Cell Signaling Technology. The rabbit anti-TSP2 antibody (BA3616) was purchased from Boster. The mouse anti-SREBP2 antibody (557037) was purchased from BD. The rabbit anti–SR-BI antibody (NB400-104) was purchased from Novus Biologicals. The rabbit anti-LDLR (3839) was purchased from Biovision. The rabbit anti-NHERF1 (sc-134485), mouse anti-NHERF2 (sc-365388), and goat anti-MAP17 (sc-27375) antibodies were purchased from Santa Cruz Biotechnology, Inc. The mouse anti-MYC (M20002L) and antiactin (M20010L) antibodies were purchased from Abmart. The mouse anti-GAPDH antibody (ab1019t) was purchased from Bo Ao Rui Jing. The rabbit anti-LC3B polyclonal antibody (L7543) was purchased from Sigma-Aldrich. The rabbit anti-GFP polyclonal antibody was generated in rabbits using the corresponding recombinant proteins as antigens. The goat anti–rabbit FITC, goat anti–rabbit TRITC, rabbit anti–goat TRITC, goat anti–mouse FITC, and goat anti–mouse TRITC-conjugated secondary antibodies were purchased from Zhong Shan Jin Qiao. The Alexa Fluor 680–conjugated goat anti–mouse and Alexa Fluor 800–conjugated goat anti–rabbit secondary antibodies for immunoblotting were purchased from Invitrogen.
Other materials and reagents
DMEM–Ham’s nutrient mixture F-12 (F12/DMEM; SH30023.01B) was purchased from HyClone. HBSS (C141755001BT) without Ca2+ or Mg2+ and penicillin-streptomycin (15140–122) were purchased from Gibco. BSA (AP0027) was purchased from Amresco. Collagenase type IV (C5138), hCG (C1063), pregnant mare serum Gn (PMSG; G4877), 3-MA (M9281), CQ (C6628), CHX (R750107), β-NAD (N7004), trans-dehydroepiandrosterone (D4000), and nitroblue tetrazolium (N8876) were obtained from Sigma-Aldrich. Percoll (sc-296039) and Filipin III (sc-205323) were purchased from Santa Cruz Biotechnology, Inc. HDL labeled with DiI-HDL (H8910) was purchased from Solarbio. Radioimmunoprecipitation assay (RIPA) buffer (P0013C) and bicinchoninic acid kits (P0012) were purchased from Beyotime. TG (F001-1) and TC kits (F002-1) were purchased from Nanjing Jiancheng Bioengineering Institute, and BODIPY 493/503 (D3922) was purchase from Invitrogen.
Mating choice assay
The sexual behavior analysis was performed as described below. In brief, a sexually naive estrous 6-wk-old CD1 female mouse was introduced into the cage of each test male, which had not been changed for at least 4 d. Each assay lasted 30 min after the target mice were introduced. All activities were recorded using an infrared camera. The latency, frequency, and duration of mounting and sniffing of male targets were analyzed.
Immunofluorescence analyses
The testes were immediately embedded in an optimum cutting temperature compound (4583; Tissue-Tek) and cut into 8-µm sections using a microtome cryostat (CM1950; Leica Biosystems). Each section was fixed with 4% PFA (P1110; Solarbio) and rinsed three times in PBS, pH 7.4. After blocking with 5% BSA containing 0.1% Triton X-100, the primary antibody was added to the sections and incubated at 4°C overnight, followed by incubation with the secondary antibody. The nuclei were stained with propidium iodide (PI) or DAPI. The images were acquired using an LSM 780 microscope equipped with a Plan Apochromat 63× 1.4 NA oil-immersion differential interference contrast M27 objective lens (ZEISS) or a TCS SP8 microscope equipped with a high-contrast Plan Apochromat 63× 1.40 NA oil CS2 objective lens (Leica Microsystems). Zen software (2010; ZEISS) or Application Suite X software (2.0.0.14332; Leica Microsystems) was used to image acquisition. The images were adjusted for publication by Photoshop CS6 Extended (13.0.1; Adobe). For cellular immunofluorescence, the cells were plated onto a cover glass for 24 h, rinsed three times with PBS, fixed, and stained as detailed above. For BODIPY staining, the frozen sections were fixed in 4% PFA, rinsed with PBS, and subsequently incubated in 1 µg/ml BODIPY–PBS solution at RT for 10 min in the dark followed by immunofluorescence staining as detailed above. For Filipin staining, the frozen sections were fixed in 4% PFA, rinsed with PBS, and incubated in 20 mM NH4Cl at RT for 20 min. Subsequently, the sections were stained with Filipin working solution containing 50 µg/ml Filipin III for 1 h at RT in the dark followed by immunofluorescence staining as detailed above. The nuclei were stained with DAPI, and the images were immediately acquired using a LSM 780 microscope or a TCS SP8 microscope.
ORO staining
ORO staining was performed as described below. The 7-µm frozen testis sections were fixed in 4% PFA for 15 min and rinsed three times with PBS. Subsequently, the sections were air-dried after incubation in 60% (vol/vol) isopropanol for 2 min. The sections were stained with ORO solution (O0625; Sigma-Aldrich; ORO saturated solution in 3:2 isopropanol/water) for 15 min as previously described (Bilińska, 1994). The sections were subsequently washed with 70% alcohol for 5 s to remove background staining. After rinsing with tap water, the sections were counterstained with Harris hematoxylin and mounted in 9:1 glycerol/PBS for further analysis.
Isolation and primary culture of Leydig cells
Leydig cells were isolated as described below. In brief, the 3-mo-old mice were euthanized using cervical dislocation, and the testes were removed and washed three times with PBS. Subsequently, the testes were decapsulated and incubated with 1 mg/ml collagenase IV in DMEM/F12 containing 5% FBS (10270; Gibco) in a shaking water bath (120 cycles/min) at 37°C for 15 min. After incubation, cold DMEM/F12 was added to stop the action of collagenase IV. Seminiferous tubules were separated from the interstitial cells by gravity sedimentation. The cells were collected by centrifugation (300 g for 6 min) and subsequently resuspended in 2 ml DMEM/F12 containing 5% FBS. To obtain purified Leydig cells, this suspension was loaded on top of a discontinuous Percoll gradient consisting of layers of 30%, 40%, 50%, and 60% Percoll dissolved in HBSS and centrifuged at 800 g at 4°C for 30 min. Leydig cells were primarily distributed from fraction 50% to 60%. Subsequently, the Leydig cells in the 50% to 60% fraction were carefully collected, washed with PBS, and centrifuged at 500 g for 5 min. The purity of Leydig cell preparations was >90% as determined using histochemical staining for 3β-hydroxysteroid dehydrogenase. These purified Leydig cells were cultured in DMEM/F12 supplemented with 15% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 34°C with 5% CO2.
Hormone measurement
Serum levels of mouse testosterone, LH, FSH, and corticosterone were specifically measured using a RIA kit. For hCG treatment, the Leydig cells were plated in 24-well culture plates and treated with 10 mU hCG for 2 h. Then, the Leydig cells were collected and washed once with ice-cold PBS. The cellular level of testosterone in Leydig cells was specifically measured using an RIA kit.
TG and TC measurement
The blood was withdrawn from the retroorbital vein of anesthetized mice; after standing for 40 min, blood was centrifuged at 5,000 rpm for 20 min, and the serum supernatants were stored at −80°C until measurement.
For TG and TC measurement in testes and Leydig cells, mice were euthanized, and tunica albuginea was removed from the testes. Then, testes or Leydig cell homogenates were prepared using a dounce homogenizer in cold RIPA-like buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with 1 mM phenylmethylsulfonyl fluoride and a protein inhibitor cocktail (04693132001; Roche). The homogenates were left standing for 15 min and centrifuged at 12,000 rpm for 10 min, and then the supernatant was collected. The protein concentrations were determined using a protein assay from Bio-Rad Laboratories. Total lipids were then extracted from the supernatant. In brief, the supernatant was mixed with 2:1 chloroform/methanol (vol/vol) at 37°C for 4 h and then centrifuged at 3,000 rpm for 10 min. The organic layer in the bottom were collected, evaporated with nitrogen gas to dryness for overnight, and resolubilized with double-distilled H2O, and then the TG and TC was measured by the TG kit (F001-1; Nanjing Jiancheng Bioengineering Institute) or TC kit (F002-1; Nanjing Jiancheng Bioengineering Institute) as described previously (Newman et al., 2007).
Cholesterol uptake and time-lapse imaging
Isolated Leydig cells were planted in gelatin-coated chambers and cultured as described above for 18 h. For knockdown experiments, the Nherf2 shRNA oligonucleotide (5′-GGGTTCAACCTGCATAGTGAC-3′) was cloned into the HpaI and XhoI site of the pSicoR-GFP lentiviral vector (11579; Addgene). The lentiviral plasmid was cotransfected into 293T cells with the packaging plasmids pMD2.G (12259) and psPAX2 (12260; Addgene), the virus was collected 48 h and 60 h after transfection, and then the cells were infected with 4 mg/ml polybrene. Approximately 10 µg/ml DiI-HDL replaced the culture medium before image acquisition. The cells were placed in a CO2 incubator chamber (5% CO2 at 37°C) fitted onto an UltraVIEW-VoX confocal laser microscope equipped with a 63× 1.40 NA oil-immersion objective lens (PerkinElmer). The images were automatically acquired every 5 min for 2 h. The fluorescence intensity of the cells was analyzed every 20 min by Volocity software (6.0; PerkinElmer). The images were prepared for publication by Photoshop.
Immunoblotting
Tissue or cell extracts were prepared using a dounce homogenizer in cold RIPA-like buffer supplemented with phenylmethylsulfonyl fluoride and cocktail. The homogenates were centrifuged at 12,000 rpm for 15 min, and the protein concentrations were determined using a protein assay from Bio-Rad Laboratories. The protein lysates (25 µg) were separated by SDS-PAGE and electrotransferred onto a nitrocellulose membrane. The membrane was scanned using the ODYSSEY Sa infrared imaging system (LI-COR Biosciences).
Real-time quantitative PCR (qPCR) analyses
Total RNA was extracted from the control and treatment groups using the RNeasy Mini kit (74104; QIAGEN) according to the manufacturer’s instructions. The RNA was pretreated with DNase (RNase-free DNase set; QIAGEN) according to the manufacturer’s instructions. The amount of total RNA was photometrically measured (BioPhotometer). cDNA was synthesized using the Prime Script reverse transcription reagent kit (RR037A; Takara Bio Inc.). Real-time PCR was performed using a Light Cycler 480II system (Roche) and the primers 5′-CCCTTCGTGCATTTTCTCAAC-3′ and 5′-CATCCCAACAAACAGGCCA-3′, and the results were analyzed using Light Cycle480 SW software, (1.5.1; Roche). In brief, the samples for qPCR were prepared using iQ SYBR green supermix (170-8882, Bio-Rad Laboratories), and the PCR cycles were run at 95°C for 10 s, 60–62°C for 45 s, 95°C for 60 s, and 55°C for 60 s, followed by a melting curve from 55–95°C in steps of 0.5°C and then subsequently held at 4°C (iCycler iQ; Bio-Rad Laboratories) after estimating the best reaction conditions using a temperature gradient. All values were normalized to GAPDH to balance potential irregularities in the RNA concentration. To control the effectivity of the process, a negative control (RT−) was consistently added to each qPCR assay. The 2ΔΔCt method was used to calculate the fold changes in gene expression.
Immunoprecipitation (IP)
Transfected cells were lysed in TAP lysis buffer (50 mM Hepes-KOH, pH 7.5, 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% NP-40, 10 mM NaF, 0.25 mM Na3VO4, and 50 mM β-glycerolphosphate) plus protease inhibitors (04693132001; Roche) for 30 min on ice and centrifuged at 13,000 g for 15 min. For IP, cell lysates were incubated with primary antibody overnight at 4°C and then incubated with protein A–Sepharose (17-1279-03; GE Healthcare) for 2 h at 4°C. Thereafter, the precipitants were washed two times with IP buffer (20 mM Tris, pH 7.4, 2 mM EGTA, and 1% NP-40), and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 55°C and analyzed by immunoblotting.
Plasmids
pCMV6-Myc-Nherf1, pCMV6-Myc-Nherf2, and pCMV6-Myc-MAP17 were purchased from Origene, and pMXs-puro-GFP-SQSTM1 (38277) and pMXs-IP-GFP-NBR1(38283) were purchased from Addgene. GFP-tagged TOLLIP was cloned into the EcoRI and BamHI site of pEGFP-N1 vector. pCMV6-Myc-Nherf2-F236A/L239A was generated by site-directed mutagenesis from pCMV6-Myc-Nherf2 vector. GFP-tagged LC3 was cloned into the XhoI and SacII sites of pEGFP-N1 vector. All plasmids were amplified in Escherichia coli.
Cell culture and transfection
HEK293T cells were maintained at 37°C and 5% CO2 in DMEM supplemented with penicillin, streptomycin, and 10% FBS. To generate cells with MAP17, NHERF1, and NHERF2 expression, HEK293T were transfected with the following plasmids: pCMV6-Myc-Nherf1, pCMV6-Myc-Nherf2, pCMV6-Myc-MAP17, and pEGFP-N1, respectively. 24 h after transfection, the expression of these proteins was confirmed by immunoblotting.
Inhibition assay
To identify intracellular protein degradation systems, the cells were treated with 25 mM NH4Cl, 10 mM 3-MA, and 25 mM CQ for 12 or 24 h to inhibit autophagy–lysosome pathways. Subsequently, NHERF2 levels were measured using immunoblotting.
Chase assay
CHX experiments were performed as described below. HEK293T cells or Leydig cells transfected with Nherf2 or its variant were treated with CHX (10 mg/ml) for the 1–4 h to inhibit de novo protein synthesis; subsequently, the degradation of NHERF2 was examined using immunoblotting.
Statistical analysis
All data are presented as means ± SD. The statistical significance of the differences between the mean values for the different genotypes was measured using Student’s t test. The data were considered significant when the p-value was <0.05 (*) or <0.01 (**).
Online supplemental materials
Fig. S1 shows that the disruption of autophagy did not affect the fetal–adult Leydig cell transition, proliferation, and Gn secretion. Fig. S2 shows that the lipids in the adrenal cortex and corticosterone in the serum decreased in the steroidogenic cell–specific autophagy disruption mice. Fig. S3 shows that the steroidogenic cell–specific disruption of autophagy did not affect the concentrations of TGs and TC in testes and serum. Fig. S4 shows that only NHERF2 was accumulated in autophagy-deficient Leydig cells. Fig. S5 shows that NHERF2 partially colocalized with LC3, NBR1, SQSTM1, and LAMP2 in starved Leydig cells. Table S1 shows characteristics of human patients in the normal serum testosterone level. Table S2 show characteristics of human patients in the low-serum testosterone level.
Supplementary Material
Acknowledgments
The authors would like to thank Noboru Mizushima, Masaaki Komatsu, Okabe Masaru, and Ikawa Masahito for providing the Atg5- and Atg7-floxed mice as well as the CAG/su9-DsRed2 mice, respectively. We thank Henry N. Ginsberg, Li Yu, Quan Chen, and Baas Tracey for critical reading of the manuscript.
This work was supported by the National Natural Science Foundation of China (grants 91649202), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA16020701), and the National Key R&D Program of China (grant 2016YFA0500901).
The authors declare no competing financial interests.
Author contributions: F. Gao, G. Li, C. Liu, H. Gao, and W. Liu performed most of the experiments and assisted in drafting the manuscript. H. Wang, M. Chen, Y. Shang, L. Wang, J. Shi, and W. Xia performed some of the experiments. W. Li, L. Chen, J. Li, F. Gao, and J. Jiao supervised the project. W. Li and L. Chen designed the experiments and drafted the manuscript.
References
- Akishita M., and Yu J.. 2012. Hormonal effects on blood vessels. Hypertens. Res. 35:363–369. 10.1038/hr.2012.4 [DOI] [PubMed] [Google Scholar]
- Azhar S., and Reaven E.. 2002. Scavenger receptor class BI and selective cholesteryl ester uptake: partners in the regulation of steroidogenesis. Mol. Cell. Endocrinol. 195:1–26. 10.1016/S0303-7207(02)00222-8 [DOI] [PubMed] [Google Scholar]
- Azhar S., Leers-Sucheta S., and Reaven E.. 2003. Cholesterol uptake in adrenal and gonadal tissues: the SR-BI and ‘selective’ pathway connection. Front. Biosci. 8:998–1029. 10.2741/1165 [DOI] [PubMed] [Google Scholar]
- Bassil N. 2011. Late-onset hypogonadism. Med. Clin. North Am. 95:507–523. 10.1016/j.mcna.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Bassil N., and Morley J.E.. 2010. Late-life onset hypogonadism: a review. Clin. Geriatr. Med. 26:197–222. 10.1016/j.cger.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Bilińska B. 1994. Staining with ANS fluorescent dye reveals distribution of mitochondria and lipid droplets in cultured Leydig cells. Folia Histochem. Cytobiol. 32:21–24. [PubMed] [Google Scholar]
- Chamindrani Mendis-Handagama S.M., and Ariyaratne H.B.. 2001. Differentiation of the adult Leydig cell population in the postnatal testis. Biol. Reprod. 65:660–671. 10.1095/biolreprod65.3.660 [DOI] [PubMed] [Google Scholar]
- Chen H., Ge R.S., and Zirkin B.R.. 2009. Leydig cells: From stem cells to aging. Mol. Cell. Endocrinol. 306:9–16. 10.1016/j.mce.2009.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona G., Vignozzi L., Sforza A., and Maggi M.. 2013. Risks and benefits of late onset hypogonadism treatment: an expert opinion. World J. Mens Health. 31:103–125. 10.5534/wjmh.2013.31.2.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona G., Rastrelli G., and Maggi M.. 2015. The pharmacotherapy of male hypogonadism besides androgens. Expert Opin. Pharmacother. 16:369–387. [DOI] [PubMed] [Google Scholar]
- DeAngelis A.M., Roy-O’Reilly M., and Rodriguez A.. 2014. Genetic alterations affecting cholesterol metabolism and human fertility. Biol. Reprod. 91:117 10.1095/biolreprod.114.119883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon H., Zigman J.M., Ye C., Lee C.E., McGovern R.A., Tang V., Kenny C.D., Christiansen L.M., White R.D., Edelstein E.A., et al. . 2006. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 49:191–203. 10.1016/j.neuron.2005.12.021 [DOI] [PubMed] [Google Scholar]
- Eberlé D., Hegarty B., Bossard P., Ferré P., and Foufelle F.. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 86:839–848. 10.1016/j.biochi.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Ellis G.B., Desjardins C., and Fraser H.M.. 1983. Control of pulsatile LH release in male rats. Neuroendocrinology. 37:177–183. 10.1159/000123540 [DOI] [PubMed] [Google Scholar]
- Gawriluk T.R., Ko C., Hong X., Christenson L.K., and Rucker E.B. III. 2014. Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proc. Natl. Acad. Sci. USA. 111:E4194–E4203. 10.1073/pnas.1409323111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocze P.M., and Freeman D.A.. 1994. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 17:151–158. 10.1002/cyto.990170207 [DOI] [PubMed] [Google Scholar]
- Goginashvili A., Zhang Z., Erbs E., Spiegelhalter C., Kessler P., Mihlan M., Pasquier A., Krupina K., Schieber N., Cinque L., et al. . 2015. Insulin secretory granules control autophagy in pancreatic β cells. Science. 347:878–882. 10.1126/science.aaa2628 [DOI] [PubMed] [Google Scholar]
- Haider S.G. 2004. Cell biology of Leydig cells in the testis. Int. Rev. Cytol. 233:181–241. 10.1016/S0074-7696(04)33005-6 [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hasuwa H., Muro Y., Ikawa M., Kato N., Tsujimoto Y., and Okabe M.. 2010. Transgenic mouse sperm that have green acrosome and red mitochondria allow visualization of sperm and their acrosome reaction in vivo. Exp. Anim. 59:105–107. 10.1538/expanim.59.105 [DOI] [PubMed] [Google Scholar]
- Hou J.W., Collins D.C., and Schleicher R.L.. 1990. Sources of cholesterol for testosterone biosynthesis in murine Leydig cells. Endocrinology. 127:2047–2055. 10.1210/endo-127-5-2047 [DOI] [PubMed] [Google Scholar]
- Hu Z., Hu J., Zhang Z., Shen W.J., Yun C.C., Berlot C.H., Kraemer F.B., and Azhar S.. 2013. Regulation of expression and function of scavenger receptor class B, type I (SR-BI) by Na+/H+ exchanger regulatory factors (NHERFs). J. Biol. Chem. 288:11416–11435. 10.1074/jbc.M112.437368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidori A.M., Giannetta E., Gianfrilli D., Greco E.A., Bonifacio V., Aversa A., Isidori A., Fabbri A., and Lenzi A.. 2005. Effects of testosterone on sexual function in men: results of a meta-analysis. Clin. Endocrinol. (Oxf.). 63:381–394. 10.1111/j.1365-2265.2005.02350.x [DOI] [PubMed] [Google Scholar]
- Jiang M.H., Cai B., Tuo Y., Wang J., Zang Z.J., Tu X., Gao Y., Su Z., Li W., Li G., et al. . 2014. Characterization of Nestin-positive stem Leydig cells as a potential source for the treatment of testicular Leydig cell dysfunction. Cell Res. 24:1466–1485. 10.1038/cr.2014.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Arozena A.A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. . 2016. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 12:1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. . 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz K.T., Pathak R.K., Rigotti A., Krieger M., and Hobbs H.H.. 1996. Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J. Clin. Invest. 98:984–995. 10.1172/JCI118883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.R., Chen L., Chang Z.J., Xin H., Liu T., Zhang Y.Q., Li G.Y., Zhou F., Gong Y.Q., Gao Z.Z., and Xin Z.C.. 2011. Autophagic deficiency is related to steroidogenic decline in aged rat Leydig cells. Asian J. Androl. 13:881–888. 10.1038/aja.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Psakhye I., and Jentsch S.. 2014. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy. 10:2381–2382. 10.4161/15548627.2014.981919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., and Parton R.G.. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378. 10.1038/nrm1912 [DOI] [PubMed] [Google Scholar]
- Midzak A.S., Chen H., Papadopoulos V., and Zirkin B.R.. 2009. Leydig cell aging and the mechanisms of reduced testosterone synthesis. Mol. Cell. Endocrinol. 299:23–31. 10.1016/j.mce.2008.07.016 [DOI] [PubMed] [Google Scholar]
- Morales A., Bella A.J., Chun S., Lee J., Assimakopoulos P., Bebb R., Gottesman I., Alarie P., Dugré H., and Elliott S.. 2010. A practical guide to diagnosis, management and treatment of testosterone deficiency for Canadian physicians. Can. Urol. Assoc. J. 4:269–275. 10.5489/cuaj.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.W., Kaysen G.A., Hammock B.D., and Shearer G.C.. 2007. Proteinuria increases oxylipid concentrations in VLDL and HDL but not LDL particles in the rat. J. Lipid Res. 48:1792–1800. 10.1194/jlr.M700146-JLR200 [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy P.J., Johnston H., Willerton L., and Baker P.J.. 2002a Failure of normal adult Leydig cell development in androgen-receptor-deficient mice. J. Cell Sci. 115:3491–3496. [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy P.J., Willerton L., and Baker P.J.. 2002b Changes in Leydig cell gene expression during development in the mouse. Biol. Reprod. 66:966–975. 10.1095/biolreprod66.4.966 [DOI] [PubMed] [Google Scholar]
- Purvis K., Cusan L., and Hansson V.. 1981. Regulation of steroidogenesis and steroid action in Leydig cells. J. Steroid Biochem. 15:77–86. 10.1016/0022-4731(81)90261-2 [DOI] [PubMed] [Google Scholar]
- Rabinowitz J.D., and White E.. 2010. Autophagy and metabolism. Science. 330:1344–1348. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Saftig P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Seidler U., Singh A.K., Cinar A., Chen M., Hillesheim J., Hogema B., and Riederer B.. 2009. The role of the NHERF family of PDZ scaffolding proteins in the regulation of salt and water transport. Ann. N. Y. Acad. Sci. 1165:249–260. 10.1111/j.1749-6632.2009.04046.x [DOI] [PubMed] [Google Scholar]
- Silver D.L., Wang N., and Vogel S.. 2003. Identification of small PDZK1-associated protein, DD96/MAP17, as a regulator of PDZK1 and plasma high density lipoprotein levels. J. Biol. Chem. 278:28528–28532. 10.1074/jbc.M304109200 [DOI] [PubMed] [Google Scholar]
- Sinclair M., Grossmann M., Gow P.J., and Angus P.W.. 2015. Testosterone in men with advanced liver disease: abnormalities and implications. J. Gastroenterol. Hepatol. 30:244–251. 10.1111/jgh.12695 [DOI] [PubMed] [Google Scholar]
- Singh R. 2010. Autophagy and regulation of lipid metabolism. Results Probl. Cell Differ. 52:35–46. 10.1007/978-3-642-14426-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.M. 1988. The autophagic activity of Leydig cells in normal rat testes. Acta Biologiae Experimentalis Sinica. 21:119–129. [PubMed] [Google Scholar]
- Tang X.M., and Zhang H.X.. 1990. Cytochemical studies on the formation of multivesicular bodies in Leydig cells. Acta Biologiae Experimentalis Sinica. 23:453–463. PMID: [PubMed] [Google Scholar]
- Tang X.M., Zhang H.X., and Yi J.. 1992. Leydig cells–a normal cell model of cellular autophagy. Acta Biologiae Experimentalis Sinica. 25:39–47. PMID: [PubMed] [Google Scholar]
- Wang H., Wan H., Li X., Liu W., Chen Q., Wang Y., Yang L., Tang H., Zhang X., Duan E., et al. . 2014. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 24:852–869. 10.1038/cr.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Pan L., Zou Z., Wang D., Lu Y., Dong Z., and Zhu L.. 2017. Hypoxia reduces testosterone synthesis in mouse Leydig cells by inhibiting NRF1-activated StAR expression. Oncotarget. 8:16401–16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., and Tang X.M.. 1991. Lysosomes in the regulation of hormone secretion in endocrine cells. Acta Biologiae Experimentalis Sinica. 24:203–213. [PubMed] [Google Scholar]
- Yi J., and Tang X.M.. 1995. Functional implication of autophagy in steroid-secreting cells of the rat. Anat. Rec. 242:137–146. 10.1002/ar.1092420202 [DOI] [PubMed] [Google Scholar]
- Yi J., and Tang X.M.. 1999. The convergent point of the endocytic and autophagic pathways in leydig cells. Cell Res. 9:243–253. 10.1038/sj.cr.7290023 [DOI] [PubMed] [Google Scholar]
- Yoshii S.R., Kuma A., Akashi T., Hara T., Yamamoto A., Kurikawa Y., Itakura E., Tsukamoto S., Shitara H., Eishi Y., and Mizushima N.. 2016. Systemic Analysis of Atg5-Null Mice Rescued from Neonatal Lethality by Transgenic ATG5 Expression in Neurons. Dev. Cell. 39:116–130. 10.1016/j.devcel.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., and Jin S.. 2009. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA. 106:19860–19865. 10.1073/pnas.0906048106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.