STIM1 activates store-operated Ca2+ entry (SOCE) by translocating to endoplasmic reticulum–plasma membrane junctions. Chang et al. reveal that STIM1 localization and SOCE are regulated by a dynamic trapping mechanism mediated by STIM1 binding to EB1 at growing microtubule ends.
Abstract
The endoplasmic reticulum (ER) Ca2+ sensor STIM1 forms oligomers and translocates to ER–plasma membrane (PM) junctions to activate store-operated Ca2+ entry (SOCE) after ER Ca2+ depletion. STIM1 also interacts with EB1 and dynamically tracks microtubule (MT) plus ends. Nevertheless, the role of STIM1–EB1 interaction in regulating SOCE remains unresolved. Using live-cell imaging combined with a synthetic construct approach, we found that EB1 binding constitutes a trapping mechanism restricting STIM1 targeting to ER–PM junctions. We further showed that STIM1 oligomers retain EB1 binding ability in ER Ca2+-depleted cells. By trapping STIM1 molecules at dynamic contacts between the ER and MT plus ends, EB1 binding delayed STIM1 translocation to ER–PM junctions during ER Ca2+ depletion and prevented excess SOCE and ER Ca2+ overload. Our study suggests that STIM1–EB1 interaction shapes the kinetics and amplitude of local SOCE in cellular regions with growing MTs and contributes to spatiotemporal regulation of Ca2+ signaling crucial for cellular functions and homeostasis.
Introduction
Ca2+ is a universal second messenger that governs many important cellular functions such as secretion, cell migration, differentiation, and apoptosis (Berridge et al., 2000; Dupont et al., 2011; Lewis, 2011). Elevation of cytosolic Ca2+ via inositol 1,4,5-triphosphate–induced Ca2+ release from the ER store after cell surface receptor activation is the key to Ca2+ signaling. Animal cells have evolved a feedback mechanism, namely store-operated Ca2+ entry (SOCE), that links ER Ca2+ store depletion to a Ca2+ influx across the plasma membrane (PM) from the extracellular space to support sustained Ca2+ signaling and ER Ca2+ store refill (Feske and Prakriya, 2013; Prakriya and Lewis, 2015). The importance of SOCE is demonstrated by patients with mutations in SOCE components manifesting the symptoms of immunodeficiency, autoimmunity, skeletal myopathy, and ectodermal dysplasia with anhidrosis (Feske, 2011).
SOCE is mediated by the ER Ca2+ sensor STIM1 and the PM Ca2+ channel Orai1 (Prakriya and Lewis, 2015). The activation of SOCE is a dynamic process involving changes in STIM1 subcellular localization. STIM1 is an ER transmembrane (TM) protein with an N-terminal Ca2+-sensing EF hand-sterile α motif domain in the ER lumen (Fig. 1 A). The cytosolic portion of STIM1 contains coiled-coil domains (CC1–3), a serine/proline region, and a C-terminal region (CT; amino acids 633–685; Fig. 1 A) with a polybasic motif (PB). In the resting state, STIM1 binds to Ca2+ in the ER lumen and localizes diffusely throughout the ER (Liou et al., 2005). After ER Ca2+ store depletion, Ca2+-free STIM1 rapidly oligomerizes, leading to a conformational extension of the PB and the Orai1 activation domain, namely CAD, SOAR, or CCb9 that roughly corresponds with the CC2–3 domains (Kawasaki et al., 2009; Park et al., 2009; Yuan et al., 2009; Prakriya and Lewis, 2015). The oligomerized/exposed PB binds to phosphatidylinositol 4,5-bisphosphate and other phospholipids at the PM (Liou et al., 2007; Ercan et al., 2009; Korzeniowski et al., 2009; Walsh et al., 2009; Chen et al., 2017). STIM1–phospholipid interaction traps STIM1 at ER–PM junctions, where the ER and the PM form close appositions, allowing STIM1 at the ER to activate Orai1 at the PM, resulting in SOCE (Liou et al., 2007; Prakriya and Lewis, 2015). STIM1 targeting to ER–PM junctions is a rate-limiting step in the activation of SOCE. Although STIM1 oligomerization occurs within 5 s after ER Ca2+ store depletion, it takes >40 s for STIM1 to translocate to ER–PM junctions (Liou et al., 2007). The mechanism underlying the time discrepancy between STIM1 oligomerization and translocation is not clear.
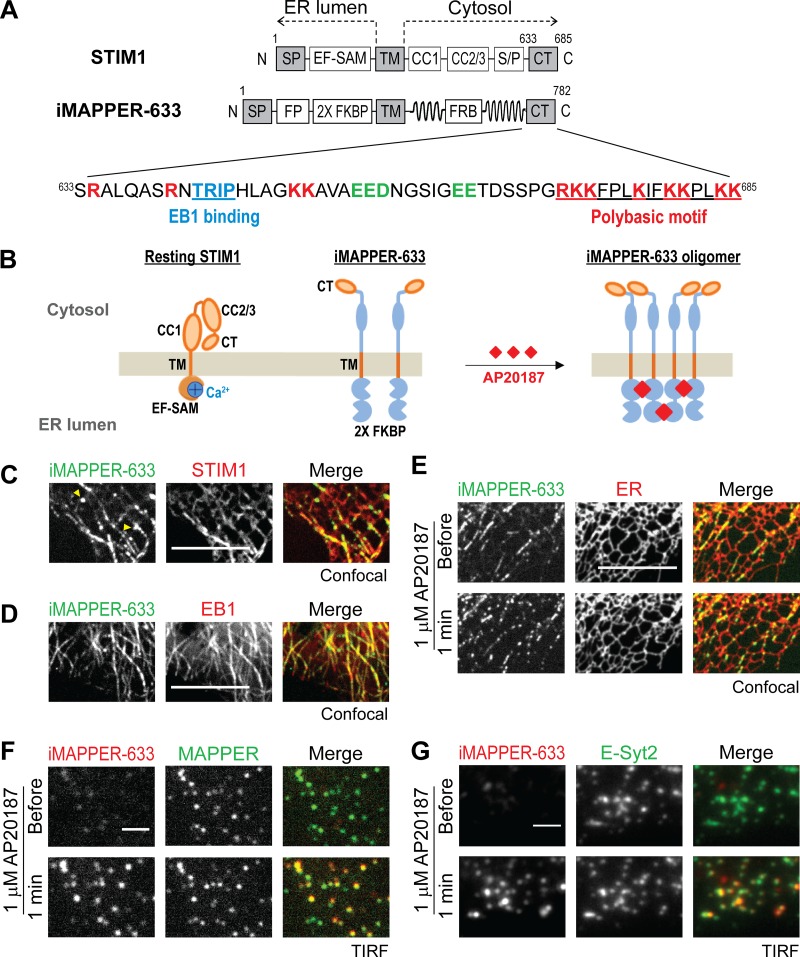
Figure 1.
iMAPPER-633: A synthetic construct for dissecting targeting mechanisms of STIM1. (A) Diagrams of STIM1 and iMAPPER-633. Amino acid number and domains are indicated. EF-SAM, EF hand and sterile α motif; FRB, FKBP–rapamycin binding domain. Identical domains between STIM1 and iMAPPER-633 are in gray. The amino acid sequences of STIM1 CT are displayed. Core EB1 binding motifs are labeled in blue, positively charged residue are in red, and negatively charged residues are in green. (B) Schematic diagram depicting resting STIM1 and iMAPPER-633 and oligomerized iMAPPER-633 after AP20187 treatment. Domains are indicated as in A. (C) Localization of YFP–iMAPPER-633 in HeLa cells coexpressing mCherry-STIM1, monitored by confocal microscopy. Yellow arrowheads indicate iMAPPER-633 puncta without STIM1 colocalization, possibly formed because of loss of EB1 binding during MT catastrophe. (D) Localization of YFP–iMAPPER-633 in HeLa cells coexpressing EB1-mCherry, monitored by confocal microscopy. (E) YFP–iMAPPER-633 displays punctate localization after 1 µM AP20187 treatment, monitored by confocal microscopy in HeLa cells cotransfected with mCherry-ER. (F) Translocation of mCherry–iMAPPER-633 to ER–PM junctions after 1 µM AP20187 treatment, monitored by TIRF microscopy in HeLa cells cotransfected with GFP-MAPPER. (G) Translocation of mCherry–iMAPPER-633 to ER–PM junctions after 1 µM AP20187 treatment, monitored by TIRF microscopy in HeLa cells cotransfected with GFP–E-Syt2. Bars: (C–E) 10 µm; (F and G) 2 µm.
In addition to its ability to bind phospholipids and Orai1 at the PM, STIM1 can directly interact with the microtubule (MT) plus end–binding proteins EB1 and, to a lesser extent, EB3 (Grigoriev et al., 2008). These interactions are mediated by an EB1-binding motif that resides in the CT (Honnappa et al., 2009). STIM1 interaction with EB1 at MT plus ends can be visualized using live-cell imaging as fluorescent protein (FP)-tagged STIM1 adopts MT-like organization and displays comet-like movement (Baba et al., 2006; Mercer et al., 2006; Grigoriev et al., 2008; Honnappa et al., 2009). This observed movement represents a traveling wave of STIM1 concentration at transient contacts between MT plus ends and the ER (Grigoriev et al., 2008). Mutation of the core EB1-binding TRIP residues to TRNN disrupts STIM1–EB1 interaction, resulting in the disappearance of MT plus end tracking by STIM1 (Honnappa et al., 2009). Nevertheless, the significance of STIM1–EB1 interaction in regulating STIM1 translocation to ER–PM junctions and SOCE remains unclear.
To dissect the contribution of EB1 binding to STIM1 localization and function, we generated inducible membrane-attached peripheral ER (MAPPER; iMAPPER)-633, a synthetic construct that contains STIM1 ER membrane targeting motifs and the STIM1 CT, consisting of both the EB1-binding motif and the phospholipid-binding PB (Fig. 1 A). iMAPPER-633 with an exposed PB tracks MT plus ends rather than localizing to ER–PM junctions. Oligomerization or inhibition of EB1 binding results in iMAPPER-633 translocation to ER–PM junctions, indicating that oligomerization promotes binding to PM phospholipids, whereas EB1 binding impedes PM targeting. Similarly, EB1 binding limits full-length STIM1 access to ER–PM junctions in the resting state and during ER Ca2+ store depletion. Disruption of EB1 binding facilitates Orai1 recruitment and SOCE activation, resulting in Ca2+ overload. Together, our findings indicate that EB1 binding provides a trapping mechanism regulating STIM1 localization and SOCE and suggest that STIM1-mediated Ca2+ signaling may be locally regulated by binding to EB1 on growing MT ends.
Results
iMAPPER-633: A synthetic construct for dissecting targeting mechanisms of STIM1
The localization of STIM1 is regulated by multiple factors including ER Ca2+ levels, protein conformation, oligomerization, phosphorylation, binding to Orai1 and phospholipids at the PM, and binding to EB1 at MT plus ends. Previous studies have shown that ER Ca2+ depletion–induced conformational change, oligomerization, and subsequent binding to phospholipids at the PM via a C-terminal PB are the critical steps for STIM1 targeting to ER–PM junctions (Liou et al., 2007; Prakriya and Lewis, 2015). Nevertheless, the role of EB1 binding in regulating STIM1-mediated Ca2+ signaling at ER–PM junctions is not well understood.
To dissect the contribution of EB1 binding in the key steps for STIM1 targeting to ER–PM junctions, we engineered a synthetic construct that contains the minimal targeting motifs of STIM1. It includes the signal peptide (SP) and TM domain of STIM1 for ER membrane localization as well as the CT of STIM1, enabling its binding to EB1 at MT plus ends and phospholipids at the PM (Fig. 1 A). A tandem FK506-binding protein (FKBP) motif (2× FKBP) was inserted into the ER luminal region following an FP to enable oligomerization upon treatment with small-molecule AP20187 and optical imaging, respectively (Fig. 1 B). We further included the cytosolic linker region of the synthetic ER–PM junctional marker MAPPER (Chang et al., 2013), which has been shown to provide the proper length spanning the gap at ER–PM junctions. We named this synthetic construct iMAPPER-633 because its design resembles MAPPER and features inducible translocation to ER–PM junctions via the CT of STIM1 starting at residue 633. An intermediate construct containing the SP, an FP, 2× FKBP, and the TM displayed ER localization when expressed in HeLa cells, indicating successful ER targeting (Fig. S1 A). Unlike the CT in full-length STIM1, which is partially buried in the resting state (Zhou et al., 2013), the CT in iMAPPER-633 is expected to be fully exposed, facilitating assessment of the contribution of EB1-binding and phospholipid-binding motifs to STIM1 localization (Fig. 1 B).
When examined using confocal microscopy, iMAPPER-633 appeared to track MT plus ends (Video 1) and colocalize with STIM1 in the resting state (Fig. 1 C). Consistent with the expectation that the CT of iMAPPER-633 is exposed and more accessible to EB1 binding than that of STIM1, iMAPPER-633 appeared to be more concentrated in MT-like structures than in the ER compared with STIM1 (Fig. 1 C). The localization of iMAPPER-633 at MT-like structures was accompanied by a few highly concentrated iMAPPER-633 clusters with apparent movement toward the nucleus, possibly formed because of loss of EB1 binding during MT catastrophe (Fig. 1 C, yellow arrowheads; and Video 1). In addition, iMAPPER-633 colocalized with EB1 (Fig. 1 D) and an ER luminal marker (Fig. 1 E, top), supporting that iMAPPER-633 is an ER protein with concentration at transient contacts between MT plus ends and the ER.
When AP20187 was applied to induce oligomerization, iMAPPER-633 rapidly translocated into puncta, whereas the bulk ER structure was unaffected (Fig. 1 E, bottom). Colocalization of iMAPPER-633 with MAPPER, monitored by total internal reflection fluorescence (TIRF) microscopy, indicates that iMAPPER-633 puncta formation corresponds with its translocation to ER–PM junctions (Fig. 1 F). Consistently, AP20187 induced colocalization of iMAPPER-633 with E-Syt2 and E-Syt3, two proteins that constitutively localize at ER–PM junctions (Giordano et al., 2013) as monitored using TIRF microscopy (Figs. 1 G and S1 B). These results indicate that in spite of an exposed PB, iMAPPER-633 predominantly binds EB1 at MT plus ends. Oligomerization of iMAPPER-633 likely potentiates its ability to bind phospholipids at the PM, resulting in translocation to ER–PM junctions.
EB1-binding prevents the PB from mediating PM targeting
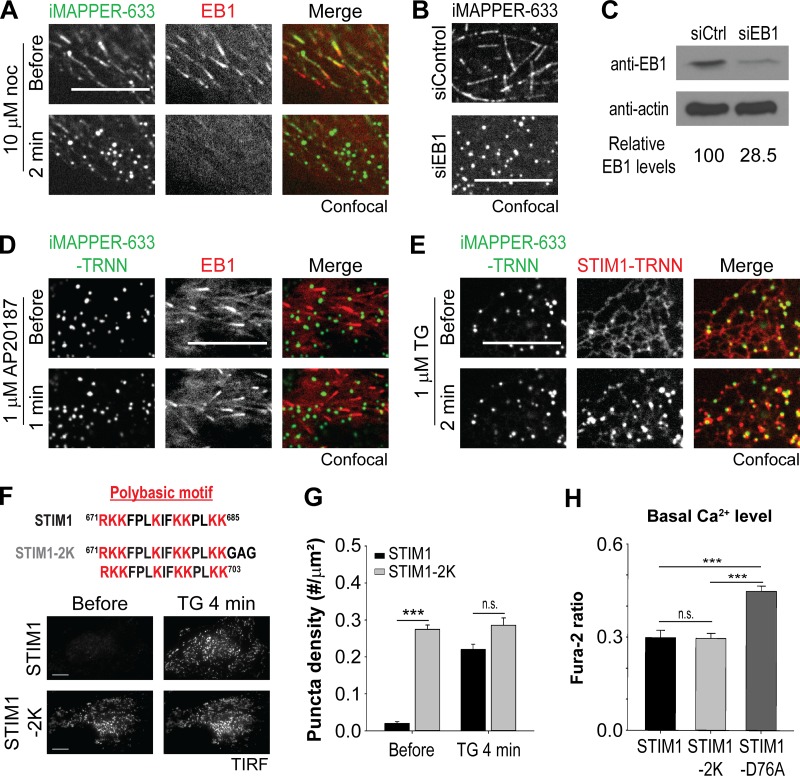
It is plausible that tracking MT plus ends via EB1 binding prevents the PB of iMAPPER-633 from mediating PM targeting. To test this possibility, we applied nocodazole, an inhibitor of MT polymerization, to disrupt EB1–MT association. We found that iMAPPER-633 rapidly translocated to ER–PM junctions, accompanying nocodazole-induced EB1 dissociation from MT plus ends (Fig. 2 A). Consistently, iMAPPER-633 readily localized at ER–PM junctions in cells with EB1 knockdown by siRNA against EB1 (siEB1; Fig. 2, B and C). We further generated the iMAPPER-633–TRNN mutant, disrupting its EB1-binding ability, and found that iMAPPER-633–TRNN prelocalized to ER–PM junctions (Fig. 2 D, top). The intensity of iMAPPER-633–TRNN at ER–PM junctions remained similar after AP20187 treatment, suggesting that the majority of iMAPPER-633–TRNN was prelocalized at ER–PM junctions as a result of its inability to bind EB1 (Fig. 2 D, bottom). These data indicate that the PB is sufficient for targeting iMAPPER-633 to ER–PM junctions in the absence of EB1 binding and that EB1 binding traps iMAPPER-633 at contacts between the ER and MT plus ends, preventing its localization at ER–PM junctions.
Figure 2.
EB1 binding prevents the PB from mediating PM targeting. (A) Translocation of YFP–iMAPPER-633 to ER–PM junctions after 10 µM nocodazole (noc) treatment, monitored by confocal microscopy in HeLa cells cotransfected with EB1-mCherry. (B) Subcellular localizations of YFP–iMAPPER-633, monitored by confocal microscopy in HeLa cells transfected with siControl or siEB1. (C) EB1 protein levels detected by Western blotting using anti-EB1 antibody in HeLa cells transfected with siControl (siCtrl) or siEB1. The intensity of bands was measured by ImageJ. Relative EB1 levels are indicated. (D) YFP–iMAPPER-633–TRNN distributes to ER–PM junctions in the absence or presence of AP20187 in HeLa cells, monitored by confocal microscopy. (E) Translocation of mCherry-STIM1-TRNN to ER–PM junctions labeled by YFP–iMAPPER-633–TRNN after 1 µM TG treatment in HeLa cells, monitored by confocal microscopy. (F) Localization of YFP-STIM1 and YFP-STIM1-2K with two PB in tandem in the CT in the absence or presence of 1 µM TG in HeLa cells, monitored by TIRF microscopy. Bars, 10 µm. (G) Quantification of the puncta density of YFP-STIM1 and YFP-STIM1-2K as described in F. Means ± SEM are shown (9–13 cells from two independent experiments). (H) Basal cytosolic Ca2+ levels, monitored by Fura-2 ratio in HeLa cells transfected with mCherry-STIM1, mCherry-STIM1-2K, or mCherry-STIM1-D76A. Means ± SD are shown (three independent experiments). ***, P < 0.001.
Unlike iMAPPER-633–TRNN, the STIM1-TRNN mutant showed a diffused distribution throughout the ER, with minimal prelocalization at ER–PM junctions (Fig. 2 E). ER Ca2+ store depletion by thapsigargin (TG) was required to trigger STIM1-TRNN translocation to ER–PM junctions and colocalization with iMAPPER-633–TRNN. These results are consistent with a previous finding that the CT in full-length STIM1 is partially buried and is not sufficient for PM binding until STIM1 activation after ER Ca2+ store depletion (Zhou et al., 2013). It is likely that the conformational change and oligomerization induced by dissociation from Ca2+ enable the PB to mediate PM binding for full-length STIM1.
We further generated a STIM1-2K construct by adding an extra PB to the very C terminus of STIM1 to enhance its ability to bind phospholipids at the PM (Fig. 2 F, top). In contrast with STIM1, this STIM1-2K construct with two PB in tandem in the CT prelocalized to ER–PM junctions without ER Ca2+ store depletion (Fig. 2 F, bottom). Consistently, quantitative analysis revealed elevated puncta density in cells transfected with STIM1-2K versus those transfected with STIM1 in the resting state (Fig. 2 G). Similar prelocalization at ER–PM junctions has been observed with the STIM1-D76A mutant that contains a point mutation that disrupts its ability to bind ER Ca2+ and exhibits an active conformation (Liou et al., 2005). Unlike STIM1-D76A, expression of STIM1-2K did not increase basal Ca2+ levels (Fig. 2 H), suggesting that STIM1-2K is not in an active conformation. These results indicate that a strong PB can mediate STIM1 localization at ER–PM junctions without inhibition of EB1 binding.
EB1 binding constitutes a trapping mechanism limiting STIM1 localization at ER–PM junctions
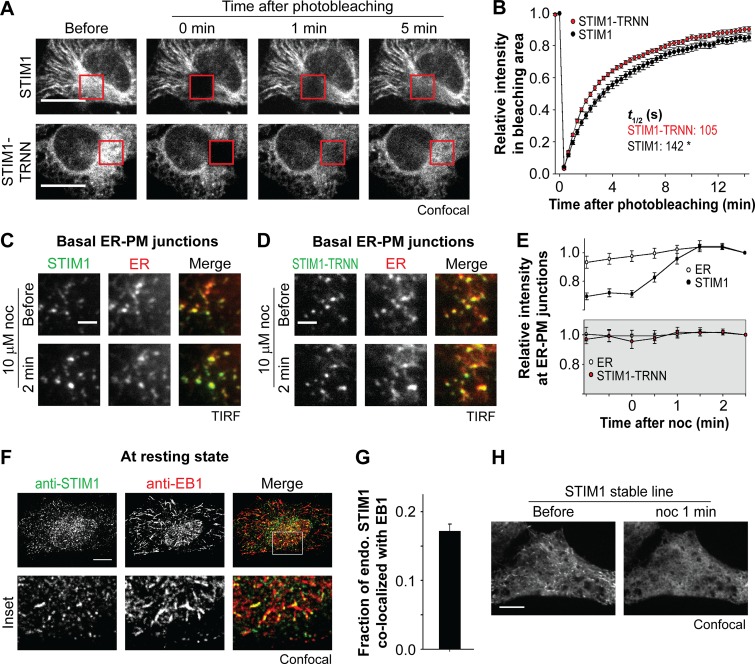
To determine how EB1 binding affects dynamic localization of STIM1, we performed FRAP experiments using cells transfected with STIM1 or STIM1-TRNN. We found that STIM1-TRNN fluorescence recovered faster than that of STIM1 in the bleached regions, with a significant difference in the time required to reach half recovery (t1/2; Fig. 3, A and B). It is likely that tracking MT plus ends by binding to EB1 limits the amount of STIM1 molecules accessing other ER regions, including ER–PM junctions.
Figure 3.
EB1 binding constitutes a trapping mechanism limiting STIM1 localization at ER–PM junctions. (A) Fluorescence recovery of YFP-STIM1 and YFP-STIM1-TRNN after photobleaching (red square boxes) in HeLa cells, monitored by confocal microscopy. (B) Relative intensity of YFP-STIM1 and YFP-STIM1-TRNN in the bleached areas as described in A. 19–20 cells from three independent experiments. Mean times to the half recovery (t1/2) are indicated. *, P < 0.05. (C and D) Changes in intensity of YFP-STIM1 (C) and YFP-STIM1-TRNN (D) at ER–PM junctions after 10 µM nocodazole (noc) treatment, monitored by TIRF microscopy in HeLa cells cotransfected with mCherry-ER. (E) Relative changes in intensity of STIM1 subtypes and mCherry-ER at ER–PM junctions derived from relative single puncta intensity as described in C and D. Intensity at the end time point is defined as 1. 13–14 cells from three to four independent experiments. (F) Localization of endogenous STIM1 and EB1 at resting state, visualized by immunostaining using confocal microscopy. (G) Fraction of STIM1 overlapping with EB1 calculated from quantification of endogenous (endo.) STIM1 and EB1 colocalization by immunostaining as described in F. Means ± SEM are shown. 29 cells from two independent experiments. (H) Changes in localization of YFP-STIM1 after 10 µM nocodazole treatment, monitored by confocal microscopy in HeLa cells stably expressing YFP-STIM1 at a low level. Bars: (A, F, and H) 10 µm; (C and D) 2 µm.
To test this hypothesis, we monitored ER–PM junctions in cells cotransfected with an mCherry-tagged ER luminal marker and YFP-tagged STIM1 or STIM1-TRNN using TIRF microscopy. After nocodazole treatment, an ∼25% increase in the intensity of STIM1 at ER–PM junctions was observed compared with the ER marker, whereas the intensity of STIM1-TRNN at ER–PM junctions remained unchanged (Fig. 3, C–E). These findings indicate that nocodazole treatment released STIM1 molecules trapped at the contacts between the ER and MT plus ends by EB1 binding, resulting in an increase of STIM1 molecules at ER–PM junctions in resting cells.
We further analyzed the interaction between endogenous STIM1 and EB1 in nontransfected HeLa cells. We used an antibody that specifically recognizes STIM1 as demonstrated by Western blotting using cells transfected with siRNA against STIM1 or a control (Fig. S2 A) and by immunostaining that detects peripheral STIM1 puncta after TG-induced ER Ca2+ depletion (Fig. S2 B). In addition, we used an anti-EB1 antibody that is capable of detecting subcellular localization of the endogenous EB1 protein by immunostaining, as demonstrated by the disappearance of MT plus end–like structures after nocodazole treatment (Fig. S2 C). Immunostaining of both STIM1 and EB1 in resting nontransfected HeLa cells revealed a subpopulation of STIM1 colocalized with EB1 (Fig. 3 F), which was estimated to be 17% of STIM1 (Fig. 3 G). These results indicate that the numerous transient contacts between growing MT and the ER network dynamically trap ∼17% of endogenous STIM1 by EB1 binding at contacts between the ER and MT plus ends.
Our findings are consistent with a previous study showing interaction between endogenous STIM1 and EB1 using immunoprecipitation (IP; Grigoriev et al., 2008). We further observed nocodazole-sensitive MT plus end–like structures of STIM1 in HeLa cells stably expressing YFP-STIM1 at the low level of twofold as compared with endogenous STIM1 (Figs. 3 H and S2 D). These results demonstrate that STIM1 trapping at MT plus ends via interaction with EB1 occurs at a low expression level.
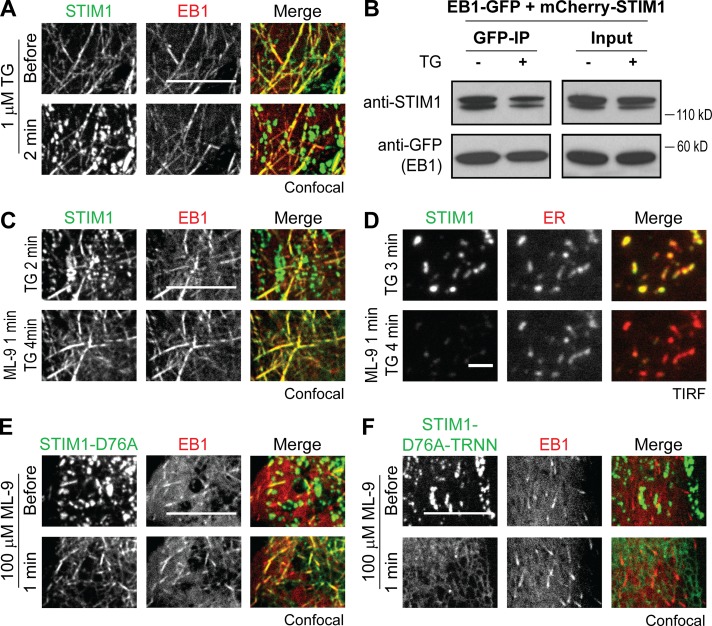
Activated STIM1 retains EB1 binding ability in ER Ca2+-depleted cells
It has been proposed that STIM1–EB1 interaction is disrupted by phosphorylation during ER Ca2+ store depletion (Pozo-Guisado et al., 2013). Nevertheless, we observed partial colocalization of STIM1 and EB1 after TG treatment (Fig. 4 A). Consistently, IP experiments showed that a portion of mCherry-STIM1 remained bound to EB1-GFP after TG treatment, whereas mCherry-STIM1-TRNN showed minimal interaction with EB1-GFP (Figs. 4 B and S3 A). We further applied 100 µM ML-9, which has been shown to rapidly trigger STIM1 dissociation from ER–PM junctions and reversion of puncta formation (Smyth et al., 2008), to cells cotransfected with STIM1 and EB1 during ER Ca2+ store depletion. After ML-9 treatment, TG-induced STIM1 puncta rapidly disappeared, and EB1-mediated trapping of STIM1 became apparent without refilling the ER Ca2+ store (Fig. 4 C). The disappearance of STIM1 puncta induced by ML-9 treatment was not caused by the disruption of ER–PM junctions, as monitored by an ER luminal marker using TIRF microscopy (Fig. 4 D). It is possible that ML-9 abolishes STIM1 binding to phospholipids at the PM because of STIM1 trapping at ER–PM junctions by Orai1 overexpression, which provides a PM targeting mechanism alternative to phospholipid binding (Park et al., 2009), was not affected by ML-9 treatment (Fig. S3 B). Furthermore, we found that STIM1-D76A, a constitutively active mutant that prelocalizes at ER–PM junctions without ER Ca2+ store depletion, was rapidly trapped by EB1 after ML-9 treatment (Fig. 4 E and Video 2). In contrast, STIM1-D76A-TRNN displayed ER localization after ML-9–induced dissociation from ER–PM junctions (Fig. 4 F). Together, these results demonstrate that STIM1 is capable of binding to EB1 at MT plus ends regardless of its activation state and the level of ER Ca2+ store. These findings indicate that activated STIM1 retains EB1-binding ability and is subjected to regulation by EB1 binding in ER Ca2+-depleted cells.
Figure 4.
Activated STIM1 retains EB1 binding ability in ER Ca2+-depleted cells. (A) Localization of YFP-STIM1 and EB1-mCherry in HeLa cells during the resting state and after 1 µM TG treatment, monitored by confocal microscopy. (B) IP of EB1-GFP with mCherry-STIM1 after 1 µM TG treatment in HeLa cells. Protein levels of EB1-GFP and mCherry-STIM1 in total cell lysates (Input) and IP were assessed by Western blotting using antibodies against GFP and STIM1. (C) Colocalization of YFP-STIM1 and EB1-mCherry in HeLa cells after 100 µM ML-9 treatment during ER Ca2+ depletion by 1 µM TG, monitored by confocal microscopy. (D) Disruption of TG-induced YFP-STIM1 accumulation at ER–PM junctions labeled by mCherry-ER in HeLa cells after 100 µM ML-9 treatment, monitored by TIRF microscopy. (E) Colocalization of YFP-STIM1-D76A and EB1-mCherry in HeLa cells after 100 µM ML-9 treatment, monitored by confocal microscopy. (F) YFP-STIM1-D76A-TRNN displayed ER localization without colocalizing with EB1-mCherry in HeLa cells after 100 µM ML-9 treatment, monitored by confocal microscopy. Bars: (A, C, E, and F) 10 µm; (D) 2 µm.
EB1 binding impedes STIM1 translocation to ER–PM junctions and Orai1 recruitment during ER Ca2+ depletion
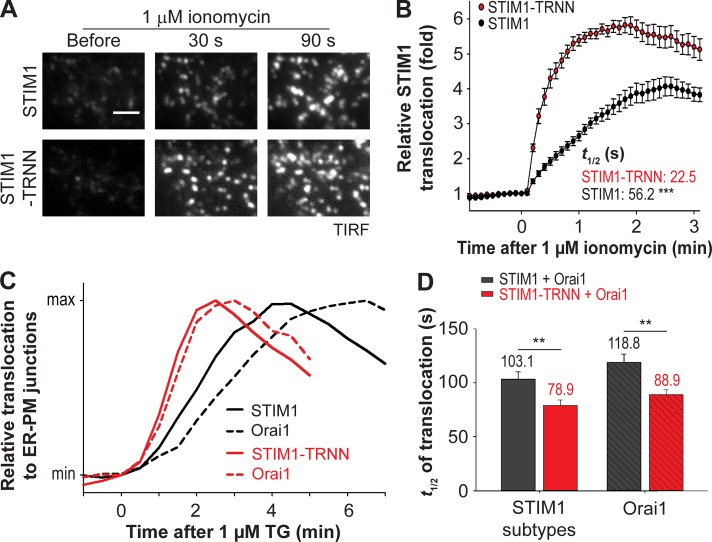
It is likely that EB1 binding impedes STIM1 translocation to ER–PM junctions after ER Ca2+ store depletion. Consistent with this notion, we observed nearly complete translocation of YFP-STIM1-TRNN 30 s after 1 µM ionomycin treatment, whereas YFP-STIM1 only began to accumulate at ER–PM junctions (Fig. 5 A). The t1/2 of STIM1 translocation to ER–PM junctions was 56.2 s (Fig. 5 B), which is comparable to a previous study (Liou et al., 2007). In contrast, STIM1-TRNN showed a significantly faster translocation than STIM1, with a t1/2 of 22.5 s. In addition, a higher amplitude of STIM1-TRNN translocation to ER–PM junctions than that of STIM1 was observed, indicating enhanced accumulation of STIM1-TRNN compared with STIM1 at ER–PM junctions (Fig. 5 B). The kinetic differences in translocation to ER–PM junctions between STIM1 and STIM1-TRNN were also detected in cells treated with TG (Figs. S4, A and B). Consistently, a significant increase in the rate of STIM1 translocation after ionomycin treatment was detected in siEB1-treated cells compared with those treated with siControl (Figs. S4, C and D). Notably, disruption of STIM1–EB1 interaction led to an accelerated STIM1 translocation by 23–34 s regardless of the rate of ER Ca2+ store depletion. These results indicate that trapping by EB1 at MT plus ends delays the translocation of STIM1 oligomers to ER–PM junctions during ER Ca2+ store depletion.
Figure 5.
EB1 binding impedes STIM1 translocation to ER–PM junctions and Orai1 recruitment during ER Ca2+ depletion. (A) Translocation of YFP-STIM1 and YFP-STIM1-TRNN to ER–PM junctions after 1 µM ionomycin treatment in HeLa cells, monitored by TIRF microscopy. Bar, 2 µm. (B) Relative translocation to ER–PM junctions of YFP-STIM1 and YFP-STIM1-TRNN as described in A. 14–15 cells from three independent experiments. Mean times to half-maximal translocation (t1/2) are indicated. (C) Relative translocation to ER–PM junctions of YFP-STIM1 subtypes and corresponding Orai1-mCherry after 1 µM TG treatment in HeLa cells, monitored by TIRF microscopy. Black, YFP-STIM1 coexpressed with Orai1-mCherry; red, STIM1-TRNN coexpressed with Orai1-mCherry. Mean traces are shown (15–23 cells from three to four independent experiments). (D) Time to the half-maximal translocation (t1/2) of YFP-STIM1 subtypes and Orai1-mCherry as described in C. Means ± SEM are shown. **, P < 0.01; ***, P < 0.001.
Activated STIM1 trapped at ER–PM junctions by binding to PM phospholipids can bind and recruit PM Ca2+ channel Orai1 to ER–PM junctions, resulting in SOCE. We further monitored Orai1-mCherry translocation to ER–PM junctions in cells coexpressing YFP-STIM1 or YFP-STIM1-TRNN. After TG treatment, translocation of STIM1 and STIM1-TRNN preceded Orai1 recruitment to ER–PM junctions, with a difference in t1/2 of 15 s (103.1 s for STIM1 and 118.8 s for Orai1) and 10 s (78.9 s for STIM1-TRNN and 88.9 s for Orai1), respectively (Fig. 5, C and D; and Fig. S4, E and F). Notably, a significant acceleration of Orai1 accumulation at ER–PM junctions by 30 s (88.9 s vs. 118.8 s) was observed in STIM1-TRNN–overexpressing cells compared with STIM1-overexpressing ones (Fig. 5 D). These data indicate that STIM1 trapping by EB1 at MT plus ends delays the binding and recruitment of Orai1 by STIM1 at ER–PM junctions during ER Ca2+ store depletion.
Disruption of STIM1–EB1 interaction facilitated SOCE and resulted in ER Ca2+ store overload
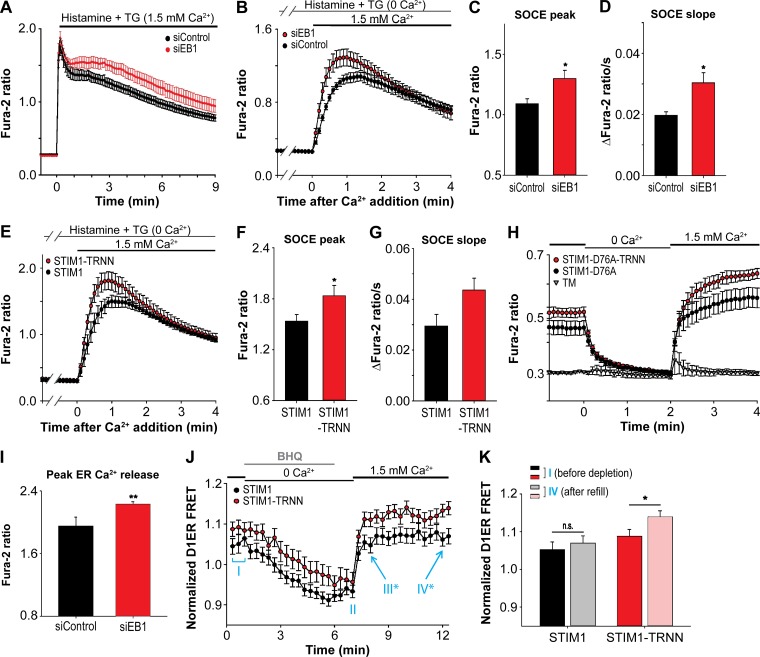
STIM1 interaction with Orai1 at ER–PM junctions initiates SOCE. Thus, regulation of STIM1 localization by EB1 binding in the resting state and during ER Ca2+ store depletion likely shapes the dynamics and extent of SOCE. Consistent with this notion, the sustained phase of cytosolic Ca2+ levels after ER Ca2+ depletion induced by histamine and TG treatment in siEB1-treated cells was higher than that in siControl-transfected cells, suggesting enhanced SOCE (Fig. 6 A). Next, we selectively monitored Ca2+ entry from the extracellular space after ER Ca2+ store depletion as a specific readout for SOCE and found that knockdown of EB1 resulted in a significant increase in the peak and slope of SOCE (Fig. 6, B–D). Consistently, STIM1-TRNN overexpression also led to a significant increase in the peak and a marked enhancement of the slope of SOCE compared with that mediated by STIM1 overexpression (Fig. 6, E–G). The effect of EB1-mediated STIM1 trapping on Ca2+ signaling was further exemplified in experiments using the constitutively active STIM1-D76A and STIM1-D76A-TRNN constructs. Overexpression of STIM1-D76A led to a marked increase in basal cytosolic Ca2+ levels compared with control (STIM1-D76A vs. TM; Fig. 6 H). This increase results from a constitutive SOCE because removal of extracellular Ca2+ rapidly decreased the cytosolic Ca2+ to the control level, and readdition of extracellular Ca2+ rapidly restored the elevated cytosolic Ca2+ level in STIM1-D76A–overexpressing cells. Overexpression of STIM1-D76A-TRNN further potentiates the elevated cytosolic Ca2+ level at basal and after readdition of extracellular Ca2+ (Fig. 6 H). Together, these results indicate that EB1 binding limits STIM1 localization at ER–PM junctions, dampening the amplitudes of STIM1-mediated SOCE.
Figure 6.
Disruption of STIM1–EB1 interaction facilitated SOCE and resulted in ER Ca2+ store overload. (A) Relative changes in cytosolic Ca2+ concentration after 100 µM histamine and 1 µM TG treatment, monitored by Fura-2 ratio in HeLa cells transfected with siControl or siEB1. Four independent experiments. (B) SOCE triggered by 100 µM histamine and 1 µM TG treatment, monitored by Fura-2 ratio in HeLa cells transfected with siControl or siEB1. Three independent experiments. (C) Peak of SOCE in HeLa cells treated with siControl or siEB1 as described in B. Three independent experiments. (D) Slope of SOCE in HeLa cells treated with siControl or siEB1 as described in B. Three independent experiments. (E) SOCE triggered by 100 µM histamine and 1 µM TG treatment, monitored by Fura-2 ratio in HeLa cells transfected with YFP-STIM1 or YFP-STIM1-TRNN. Three independent experiments. (F) Peak of SOCE in HeLa cells transfected with YFP-STIM1 and YFP-STIM1-TRNN as described in E. Three independent experiments. (G) Slope of SOCE in HeLa cells transfected with YFP-STIM1 and YFP-STIM1-TRNN as described in E. Three independent experiments. (H) Relative changes in cytosolic Ca2+ concentration in response to depletion and readdition of extracellular Ca2+, monitored by Fura-2 ratio in HeLa cells transfected with YFP-TM, YFP-STIM1-D76A, or YFP-STIM1-D76A-TRNN. Three to four independent experiments. (I) Peak ER Ca2+ release by 1 µM ionomycin treatment in the absence of extracellular Ca2+, monitored by Fura-2 ratio in HeLa cells transfected with siControl or siEB1. Three independent experiments. (J) Relative ER Ca2+ levels in the resting state (phase I), after 5 µM BHQ treatment (phase II), and after BHQ washout (phase III and IV), monitored by D1ER in HeLa cells transfected with mCherry-STIM1 or mCherry-STIM1-TRNN. 16–26 cells from three independent experiments. *, P < 0.05 between STIM1 and STIM1-TRNN. (K) Relative ER Ca2+ levels in phase I and IV as described in J, monitored by D1ER in HeLa cells transfected with mCherry-STIM1 or mCherry-STIM1-TRNN. Means ± SEM are shown. *, P < 0.05; **, P < 0.01.
In addition to maintaining cytosolic Ca2+ levels, SOCE is important to refill the ER Ca2+ store after depletion. We further tested whether enhanced SOCE caused by the absence of STIM1–EB1 interaction results in ER Ca2+ store overload. In resting cells, we observed a significantly elevated ER Ca2+ store in siEB1-treated cells as monitored by the release of ER Ca2+ by ionomycin treatment in the absence of extracellular Ca2+ (Fig. 6 I). We further tracked the dynamic changes in ER Ca2+ levels during store depletion and refill using an ER Ca2+ sensor, D1ER (Palmer et al., 2004), and a reversible sarco/ER Ca2+-ATPase inhibitor, 2,5-di-t-butyl-1,4-benzohydroquinone (BHQ). We found that the level of ER Ca2+ was moderately elevated in STIM1-TRNN–transfected cells compared with that in STIM1-transfected cells before and after depletion by BHQ (Fig. 6 J, phases I and II). After BHQ washout and readdition of extracellular Ca2+, a significant elevation in the ER Ca2+ level was observed in STIM1-TRNN–overexpressing cells compared with that in STIM1-overexpressing cells (Fig. 6 J, phases III and IV). Notably, the ER Ca2+ level after refill was comparable to that before BHQ-induced depletion in STIM1-overexpressing cells (Fig. 6 K); however, the level of ER Ca2+ after refill was significantly higher than that before depletion in STIM1-TRNN–overexpressing cells. These results indicate that EB1 binding constitutes a mechanism that optimizes SOCE and prevents ER Ca2+ store overload.
Discussion
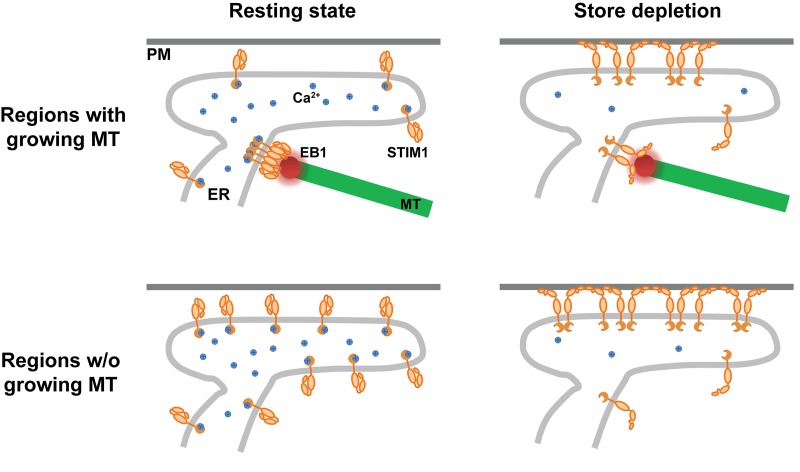
Based on our findings, we propose a model in which EB1-mediated trapping of STIM1 regulates SOCE at ER–PM junctions and prevents ER Ca2+ overload (Fig. 7). In the resting state, EB1 binding dynamically traps STIM1 at contacts between the ER and growing MT ends and restrains access to ER–PM junctions. After ER Ca2+ store depletion, EB1 binding continues to impede STIM1 localization to ER–PM junctions and activation of SOCE. In subcellular regions without growing MTs, STIM1 molecules in the ER can readily bind to PM to mediate SOCE at ER–PM junctions after ER Ca2+ store depletion. Without the dynamic trapping mechanism provided by EB1 binding, STIM1 in excess at ER–PM junctions results in elevated SOCE and ER Ca2+ store overload.
Figure 7.
Model. STIM1–EB1 interaction regulates STIM1 translocation to ER–PM junctions.
This model can be further applied to understand physiological functions in cells with polarized MT distribution. During directed cell migration, MT plus ends are oriented toward the front end of cells (Rodriguez et al., 2003). STIM1–EB1 interaction leads to polarized STIM1 distribution, accompanied by low cytosolic and ER Ca2+ levels, indicative of limited SOCE, at the front end of migrating cells (Tsai et al., 2014). Additionally, disrupting the polarized distribution by introducing STIM1-TRNN abolished cell migration, demonstrating the importance of STIM1–EB1 interaction in STIM1 distribution and in maintaining polarized Ca2+ signaling for cell migration.
STIM1 translocation to ER–PM junctions is a complex process involving in a series of signaling events, with multiple mechanisms contributing to STIM1 targeting to ER–PM junctions such as STIM1–phospholipid binding and STIM1–Orai1 interaction (Prakriya and Lewis, 2015). Moreover, several STIM1/Orai1-interacting proteins, including SARAF, (Palty et al., 2012), septins (Sharma et al., 2013), junctate (Srikanth et al., 2012), and CRACR2A (Srikanth et al., 2010), have been shown to regulate STIM1 translocation, STIM1–Orai1 interaction, or SOCE. These factors likely prevented previous studies from understanding of how STIM1–EB1 interaction contributes to SOCE and caused inconsistent results (Ribeiro et al., 1997; Bakowski et al., 2001; Baba et al., 2006; Smyth et al., 2007). By generating iMAPPER-633, a synthetic construct that contains the ER targeting motifs of STIM1, an FP, a chemically inducible oligomerization unit, cytosolic linkers, and the CT of STIM1, it was revealed that the PB in the exposed CT is sufficient for PM targeting because disrupting EB1 interaction led to a clear shift of iMAPPER-633 localization to ER–PM junctions without oligomerization. Nonetheless, the PB does not efficiently trap iMAPPER-633 at ER–PM junctions in the presence of EB1 interaction, resulting in concentration of iMAPPER-633 at contacts between the ER and MT plus ends. Oligomerization induced by AP20187 treatment increased the PM binding ability of iMAPPER-633, enabling a shift of concentration to ER–PM junctions. Thus, it is likely that STIM1 activation not only exposes the PB but also generates a strong PB by oligomerization, enabling STIM1 translocation to ER–PM junctions. Consistent with this notion, STIM1-2K with a strong PB distributes to ER–PM junctions before its activation.
Our analysis indicates that the numerous transient encounters between growing MTs and the ER network dynamically trap ∼17% of endogenous STIM1 molecules via EB1 binding at ER–MT plus end contacts at a given time in a HeLa cell. Because of the dynamic exchange between the EB1-free and EB1-bound pools of STIM1 molecules happening within minutes, the FRAP analysis used in this study can only reveal a slower recovery of STIM1 compared with STIM1-TRNN. It would be interesting to perform quantitative analysis of diffusion coefficients of EB1-free and EB1-bound STIM1 using single-molecule tracking. Notably, single-molecule tracking of wild-type STIM1 in resting cells revealed a broad range of diffusion coefficients (Wu et al., 2014), which may represent a mixed population of EB1-free and EB1-bound STIM1.
Consistent with a previous study, we found that ionomycin-induced STIM1 translocation to ER–PM junctions is a much slower process, with a t1/2 of ∼50 s compared with STIM1 oligomerization, which occurs almost instantly after ER Ca2+ depletion (Liou et al., 2007). STIM1-TRNN showed a significantly faster translocation, with an approximately twofold increase in t1/2. The enhanced STIM1-TRNN translocation to ER–PM junctions further led to an accelerated Orai1 accumulation at ER–PM junctions, indicating that EB1 binding regulates the kinetics of SOCE.
Moreover, we demonstrated that the trapping mechanism mediated by EB1 binding works continuously during SOCE because activated STIM1 became associated with EB1 after the disruption of PM binding by ML-9, an inhibitor of myosin light chain kinase (MLCK). The effects of ML-9 on STIM1 appeared to be independent of MLCK inhibition because knockdown of MLCK had no effect on SOCE (Smyth et al., 2008). Consistent with a previous observation that ML-9 was less efficient in inhibiting SOCE when both STIM1 and Orai1 were overexpressed (DeHaven et al., 2008), we found that the intensity of STIM1–Orai1 clusters remained similar after ML-9 treatment (Fig. S3 B). Thus, it is possible that ML-9 affects STIM1–phospholipid interaction at the PM. Multiple proteins localize at ER–PM junctions by binding to PM lipids to provide interorganelle signaling (Henne et al., 2015; Chang and Liou, 2016). Further work in defining the mechanisms underlying the actions of ML-9 may shed new light on STIM1 targeting as well as the function and regulation of ER–PM junctions.
A previous study demonstrated that STIM1 phosphorylation at residues S575, S608, and S621 by ERK1/2 is important for STIM1 dissociation from EB1 and translocation to ER–PM junctions during ER Ca2+ store depletion (Pozo-Guisado et al., 2013). Intriguingly, STIM1 phosphorylation was detected 2–5 min after TG treatment, arguing that STIM1 phosphorylation may occur after its translocation to ER–PM junctions. Nonetheless, STIM1 phosphorylation may provide a mechanism to disengage EB1 trapping for enhancing SOCE under certain conditions such as cell migration (Casas-Rua et al., 2015). Phosphorylation of STIM1 may also be relevant during cell division because dissociation of phosphorylated STIM1 from EB1 is required for exclusion of the ER from mitotic spindles (Smyth et al., 2012).
SOCE is one of the most important pathways for Ca2+ signaling and homeostasis. Thus, precise spatial-temporal regulation of SOCE is crucial for supporting cellular functions and health. In this study, we reveal an unexpected role of MT plus ends in optimizing STIM1 translocation and SOCE via an EB1 binding–mediated trapping mechanism and show that STIM1–EB1 interaction is important for preventing Ca2+ overload, which has been associated with apoptosis resulting from enhanced mitochondrial Ca2+ uptake and many pathological conditions including stroke, neurodegeneration, and cancer (Trump and Berezesky, 1995; Orrenius et al., 2003; Dong et al., 2006; Hajnóczky et al., 2006). Our study on the crosstalk between MT plus ends and STIM1-mediated SOCE may shed light on how cells dynamically coordinate MT growth to regulate Ca2+ signaling in physiological processes.
Materials and methods
Reagents
TG, pluronic F-127, and Fura-2–acetoxymethyl ester were purchased from Invitrogen. All chemicals for extracellular buffer (ECB; 125 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 20 mM Hepes, 10 mM glucose, and 1.5 mM CaCl2, pH 7.4), penicillin and streptomycin solution, ML-9, nocodazole, ionomycin, histamine, and EGTA were obtained from Sigma-Aldrich. AP20187 was purchased from Takara Bio Inc. BHQ was obtained from EMD Millipore. Anti-EB1 antibody (ab53358) and anti–β actin antibody (ab8227) were obtained from Abcam. Anti-GAPDH antibody (G-8795) was purchased from Sigma-Aldrich. Human cDNA library and siRNA used in this study were generated as described previously (Liou et al., 2005). Primers used for siRNA generation are listed in Table S1.
Cell culture and transfection
HeLa cells purchased from ATCC were cultured in MEM supplemented with 10% FBS (HyClone) and penicillin and streptomycin solution. DNA plasmids (25–50 ng) and siRNAs (25 nM) were transfected into HeLa cells with TransIT-LT1 reagent for 16–20 h and TransIT-TKO reagent for 48–72 h, respectively (Muris Bio).
DNA constructs
YFP-TM, YFP-STIM1, YFP-STIM1-D76A, MAPPER, mCherry-STIM1, mCherry-ER (KDEL), Orai1-mCherry, mCherry–K Ras tail, GFP-E-Syt2, and GFP-E-Syt3 were described previously (Liou et al., 2005, 2007; Chang et al., 2013; Giordano et al., 2013; Chen et al., 2017). mCherry-STIM1-D76A was constructed by replacing the YFP portion of YFP-STIM1-D76A with mCherry. iMAPPER-633 was generated by inserting PCR fragments of (a) 2× FKBP, (b) TM plus cytosolic regions of MAPPER (without PM-targeting motif), and (c) STIM1 CT containing amino acids 633–685 into the MAPPER (YFP or mCherry) construct digested with SpeI and BamHI. CT mutants of STIM1 and iMAPPER-633 were generated using a QuikChange site-direct mutagenesis kit (Agilent Technologies). STIM1-2K was generated by site-directed mutagenesis to insert a fragment encoding glycine, alanine, glycine, and amino acids 671–685 before the stop codon of STIM1. EB1-GFP and EB1-mCherry were cloned by inserting a PCR fragment containing EB1 into GFP-N1 and mCherry-N1 plasmids, respectively. All constructs listed in this section were verified by sequencing. All oligonucleotides used in this study are listed in Table S1.
IP
HeLa cells were cultured on six-well plates and transfected with EB1-GFP (300 ng/well) and mCherry-STIM1 subtypes (200 ng/well) overnight. Cells were then washed with warm PBS before lysis with 20 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 0.5% NP-40, and protease inhibitors on ice for 30 min. The lysates were subjected to centrifugation at 16,000 g for 15 min at 4°C, and the clear lysates (supernatants) were collected. The lysates were mixed with GFP-nAb agarose resin (Allele Biotechnology) and incubated with tumbling for 2 h at 4°C. The immunoprecipitated proteins were eluted with NuPAGE LDS sample buffer (Thermo Fisher Scientific) after washing the GFP-nAb agarose resin with 10 mM Tris buffer containing 150 mM NaCl and 0.5% NP-40 four times. The eluted proteins were analyzed by Western blotting using antibodies against GFP (ab290; Abcam) or STIM1 (4916; Cell Signaling Technology).
Live-cell confocal and TIRF microscopy and image analysis
HeLa cells were cultured and transfected on Lab-Tek chambered #1 coverglass (Nunc). Before imaging, cells were washed with ECB. Live-cell confocal and TIRF imaging experiments were performed at room temperature with CFI Apo 60× or 100× objectives (1.49 NA) and a confocal TIRF microscope custom built using an Eclipse Ti microscope (Nikon) with an HQ2 camera and an EM camera (c9100-13; Hamamatsu Photonics). The microscope was controlled by Micro-Manager software (Edelstein et al., 2010). FRAP was performed with a 60× objective and Andor spinning disk confocal and FRAPPA units on an Eclipse Ti microscope controlled by MetaMorph software (Molecular Devices). A 28-µm2 area of HeLa cells expressing YFP-STIM1 or YFP-STIM1-TRNN was subjected to photobleaching with three pulses of 515-nm laser for 300 µs at maximal intensity. The intensity traces of the photobleached areas were analyzed and normalized to a nonbleached area in the same cells. For the analyses of relative translocation of STIM1 subtypes to ER–PM junctions, 20–30 puncta in each cell from TIRF images were selected. The intensity traces of the selected puncta from the same cell were background subtracted, normalized to the value at time 0, and averaged.
Immunostaining
All procedures were performed at room temperature unless otherwise indicated, and all washing steps were done using PBS for 5 min. To detect the localization of endogenous STIM1 and EB1, nontransfected HeLa cells were rinsed with PBS and fixed with methanol at −20°C for 20 min. Fixed cells were washed three times and incubated with 0.3% Triton X-100 in PBS for 20 min for permeabilization. Permeabilized cells were then blocked with 5% normal donkey serum in PBS for 1 h followed by incubation with anti-STIM1 antibody (1:1,000 dilution) in PBS with 1% BSA at 4°C overnight. After three washes, the cells were incubated with fluorescent secondary antibody (1:2,000 in dilution) for 1 h. The cells were then washed three times and subjected to blocking and incubation with anti-EB1 antibody (1:1,000 dilution), followed by incubation with fluorescent secondary antibody (1:2,000 in dilution) as described above. After three washes, the immunostained samples were imaged with confocal microscopy. Endogenous STIM1 and EB1 colocalization by immunostaining was quantified by measuring the Manders coefficient of maximal-projected confocal images using the JACoP plugin in ImageJ (National Institutes of Health).
Cytosolic and ER Ca2+ level measurements
For measuring cytosolic Ca2+ levels, HeLa cells were loaded with 0.5 µM Fura-2–acetoxymethyl ester in ECB containing 0.05% pluronic F-127 and 0.1% BSA for 30 min at room temperature, avoiding light. Loaded cells were then washed with ECB containing 0.1% BSA and incubated in ECB for another 15–30 min before the experiments. Single-cell Ca2+ images were taken with a Plan Fluor 4× objective (0.15 NA) and an automated microscope custom built on an Eclipse Ti microscope with a camera (HQ2; Photometrics). The microscope was controlled by Micro-Manager software (Edelstein et al., 2010). Intracellular Ca2+ levels were indicated by ratio of 510-nm emission excited at 340 nm over those at 380 nm. The slope of SOCE represents the rate of increase in the Fura-2 ratio in the first five time points (30 s) after Ca2+ addition. To measure ER Ca2+ levels, HeLa cells were cotransfected with D1ER and mCherry-STIM1 or mCherry-STIM1-TRNN. Single-cell Ca2+ images were taken with a Plan Fluor 40× objective (1.30 NA) and a confocal TIRF microscope custom built using an Eclipse Ti microscope with an HQ2 camera and an EM camera (c9100-13). Dynamic changes in ER Ca2+ levels were indicated by the ratio of FRET (CFP excitation/YFP emission) signal to that of CFP.
Statistical analysis
Data were statistically analyzed by t test or one-way ANOVA using SigmaPlot software (Systat Software).
Online supplemental material
Fig. S1 shows successful ER targeting of the SP-FP–2× FKBP–TM construct and translocation of iMAPPER-633 to ER–PM junctions labeled by E-Syt3 after AP20187 treatment. Fig. S2 shows the specificity of the anti-STIM1 and anti-EB1 antibodies used in this study as demonstrated by Western blotting and immunostaining, and the low expression level of YFP-STIM1 in a stable HeLa cell line. Fig. S3 shows that STIM1-TRNN minimally interacts with EB1 as detected by IP and that STIM1 and Orai1 at ER–PM junctions remain stable after ML-9 treatment. Fig. S4 shows STIM1 and STIM1-TRNN translocation to ER–PM junctions after TG treatment, ionomycin-induced STIM1 translocation to ER–PM junctions in cells treated with siControl and siEB1, and recruitment of Orai1 to ER–PM junctions by STIM1 or STIM1-TRNN after TG treatment. Table S1 shows oligonucleotides used in this study. Video 1 is a time-lapse video demonstrating iMAPPER-633 binding to EB1 at MT plus ends in live HeLa cells. Video 2 is a time-lapse video demonstrating that STIM1 puncta rapidly disappeared and STIM1 colocalization with EB1 became apparent after ML-9 treatment during ER Ca2+ store depletion.
Supplementary Material
Acknowledgments
We thank the Liou laboratory members for valuable discussions and technical assistance. We also thank the University of Texas Southwestern Live-Cell Imaging Facility for assistance with FRAP experiments. We are grateful to Linda Patterson for administrative assistance.
This work was supported by National Institutes of Health grant GM113079, Welch Foundation grant I-1789, Howard Hughes Medical Institute graduate grant 56006776 (to the Mechanisms of Disease and Translational Science Ph.D. Track), Taiwan National Science Council grant 102-2917-I-564-019, and National Institutes of Health Cell and Molecular Biology training program grant T32 GM008203. J. Liou is a Sowell Family Scholar in Medical Research.
The authors declare no competing financial interests.
Author contributions: C.-L. Chang and J. Liou designed iMAPPER-633. C.-L. Chang, Y.-J. Chen, C.G. Quintanilla, and J. Liou performed experiments. C.-L. Chang, Y.-J. Chen, C.G. Quintanilla, T.-S. Hsieh, and J. Liou analyzed the results. J. Liou conceived and supervised the project. C.-L. Chang and J. Liou wrote the manuscript.
References
- Baba Y., Hayashi K., Fujii Y., Mizushima A., Watarai H., Wakamori M., Numaga T., Mori Y., Iino M., Hikida M., and Kurosaki T.. 2006. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 103:16704–16709. 10.1073/pnas.0608358103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski D., Glitsch M.D., and Parekh A.B.. 2001. An examination of the secretion-like coupling model for the activation of the Ca2+ release-activated Ca2+ current I (CRAC) in RBL-1 cells. J. Physiol. 532:55–71. 10.1111/j.1469-7793.2001.0055g.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.J., Lipp P., and Bootman M.D.. 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1:11–21. 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- Casas-Rua V., Tomas-Martin P., Lopez-Guerrero A.M., Alvarez I.S., Pozo-Guisado E., and Martin-Romero F.J.. 2015. STIM1 phosphorylation triggered by epidermal growth factor mediates cell migration. Biochim. Biophys. Acta. 1853:233–243. 10.1016/j.bbamcr.2014.10.027 [DOI] [PubMed] [Google Scholar]
- Chang C.L., and Liou J.. 2016. Homeostatic regulation of the PI(4,5)P2-Ca(2+) signaling system at ER-PM junctions. Biochim. Biophys. Acta. 1861(8, 8 Pt B):862–873. 10.1016/j.bbalip.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.L., Hsieh T.S., Yang T.T., Rothberg K.G., Azizoglu D.B., Volk E., Liao J.C., and Liou J.. 2013. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Reports. 5:813–825. 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Chen Y.J., Chang C.L., Lee W.R., and Liou J.. 2017. RASSF4 controls SOCE and ER–PM junctions through regulation of PI(4,5)P2. J. Cell Biol. 216:2011–2025. 10.1083/jcb.201606047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHaven W.I., Smyth J.T., Boyles R.R., Bird G.S., and Putney J.W. Jr. 2008. Complex actions of 2-aminoethyldiphenyl borate on store-operated calcium entry. J. Biol. Chem. 283:19265–19273. 10.1074/jbc.M801535200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Saikumar P., Weinberg J.M., and Venkatachalam M.A.. 2006. Calcium in cell injury and death. Annu. Rev. Pathol. 1:405–434. 10.1146/annurev.pathol.1.110304.100218 [DOI] [PubMed] [Google Scholar]
- Dupont G., Combettes L., Bird G.S., and Putney J.W.. 2011. Calcium oscillations. Cold Spring Harb. Perspect. Biol. 3:a004226 10.1101/cshperspect.a004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14:Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan E., Momburg F., Engel U., Temmerman K., Nickel W., and Seedorf M.. 2009. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 10:1802–1818. 10.1111/j.1600-0854.2009.00995.x [DOI] [PubMed] [Google Scholar]
- Feske S. 2011. Immunodeficiency due to defects in store-operated calcium entry. Ann. N. Y. Acad. Sci. 1238:74–90. 10.1111/j.1749-6632.2011.06240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., and Prakriya M.. 2013. Conformational dynamics of STIM1 activation. Nat. Struct. Mol. Biol. 20:918–919. 10.1038/nsmb.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S.F., Pirruccello M., Milosevic I., Gracheva E.O., Bagriantsev S.N., Borgese N., and De Camilli P.. 2013. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 153:1494–1509. 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Gouveia S.M., van der Vaart B., Demmers J., Smyth J.T., Honnappa S., Splinter D., Steinmetz M.O., Putney J.W. Jr., Hoogenraad C.C., and Akhmanova A.. 2008. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 18:177–182. 10.1016/j.cub.2007.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnóczky G., Csordás G., Das S., Garcia-Perez C., Saotome M., Sinha Roy S., and Yi M.. 2006. Mitochondrial calcium signalling and cell death: Approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis. Cell Calcium. 40:553–560. 10.1016/j.ceca.2006.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Liou J., and Emr S.D.. 2015. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr. Opin. Cell Biol. 35:123–130. 10.1016/j.ceb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Lange I., and Feske S.. 2009. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem. Biophys. Res. Commun. 385:49–54. 10.1016/j.bbrc.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski M.K., Popovic M.A., Szentpetery Z., Varnai P., Stojilkovic S.S., and Balla T.. 2009. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J. Biol. Chem. 284:21027–21035. 10.1074/jbc.M109.012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.S. 2011. Store-operated calcium channels: New perspectives on mechanism and function. Cold Spring Harb. Perspect. Biol. 3:a003970 10.1101/cshperspect.a003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E. Jr., and Meyer T.. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Fivaz M., Inoue T., and Meyer T.. 2007. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. USA. 104:9301–9306. 10.1073/pnas.0702866104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J.C., Dehaven W.I., Smyth J.T., Wedel B., Boyles R.R., Bird G.S., and Putney J.W. Jr. 2006. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J. Biol. Chem. 281:24979–24990. 10.1074/jbc.M604589200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S., Zhivotovsky B., and Nicotera P.. 2003. Regulation of cell death: The calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 4:552–565. 10.1038/nrm1150 [DOI] [PubMed] [Google Scholar]
- Palmer A.E., Jin C., Reed J.C., and Tsien R.Y.. 2004. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc. Natl. Acad. Sci. USA. 101:17404–17409. 10.1073/pnas.0408030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R., Raveh A., Kaminsky I., Meller R., and Reuveny E.. 2012. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 149:425–438. 10.1016/j.cell.2012.01.055 [DOI] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo-Guisado E., Casas-Rua V., Tomas-Martin P., Lopez-Guerrero A.M., Alvarez-Barrientos A., and Martin-Romero F.J.. 2013. Phosphorylation of STIM1 at ERK1/2 target sites regulates interaction with the microtubule plus-end binding protein EB1. J. Cell Sci. 126:3170–3180. 10.1242/jcs.125054 [DOI] [PubMed] [Google Scholar]
- Prakriya M., and Lewis R.S.. 2015. Store-operated calcium channels. Physiol. Rev. 95:1383–1436. 10.1152/physrev.00020.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C.M., Reece J., and Putney J.W. Jr. 1997. Role of the cytoskeleton in calcium signaling in NIH 3T3 cells. An intact cytoskeleton is required for agonist-induced [Ca2+]i signaling, but not for capacitative calcium entry. J. Biol. Chem. 272:26555–26561. 10.1074/jbc.272.42.26555 [DOI] [PubMed] [Google Scholar]
- Rodriguez O.C., Schaefer A.W., Mandato C.A., Forscher P., Bement W.M., and Waterman-Storer C.M.. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609. 10.1038/ncb0703-599 [DOI] [PubMed] [Google Scholar]
- Sharma S., Quintana A., Findlay G.M., Mettlen M., Baust B., Jain M., Nilsson R., Rao A., and Hogan P.G.. 2013. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 499:238–242. 10.1038/nature12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J.T., DeHaven W.I., Bird G.S., and Putney J.W. Jr. 2007. Role of the microtubule cytoskeleton in the function of the store-operated Ca2+ channel activator STIM1. J. Cell Sci. 120:3762–3771. 10.1242/jcs.015735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J.T., Dehaven W.I., Bird G.S., and Putney J.W. Jr. 2008. Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J. Cell Sci. 121:762–772. 10.1242/jcs.023903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J.T., Beg A.M., Wu S., Putney J.W. Jr., and Rusan N.M.. 2012. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol. 22:1487–1493. 10.1016/j.cub.2012.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Jung H.J., Kim K.D., Souda P., Whitelegge J., and Gwack Y.. 2010. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12:436–446. 10.1038/ncb2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Jew M., Kim K.D., Yee M.K., Abramson J., and Gwack Y.. 2012. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA. 109:8682–8687. 10.1073/pnas.1200667109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trump B.F., and Berezesky I.K.. 1995. Calcium-mediated cell injury and cell death. FASEB J. 9:219–228. 10.1096/fasebj.9.2.7781924 [DOI] [PubMed] [Google Scholar]
- Tsai F.C., Seki A., Yang H.W., Hayer A., Carrasco S., Malmersjö S., and Meyer T.. 2014. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 16:133–144. 10.1038/ncb2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.M., Chvanov M., Haynes L.P., Petersen O.H., Tepikin A.V., and Burgoyne R.D.. 2009. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem. J. 425:159–168. 10.1042/BJ20090884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.M., Covington E.D., and Lewis R.S.. 2014. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol. Biol. Cell. 25:3672–3685. 10.1091/mbc.E14-06-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.P., Zeng W., Dorwart M.R., Choi Y.J., Worley P.F., and Muallem S.. 2009. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11:337–343. 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Srinivasan P., Razavi S., Seymour S., Meraner P., Gudlur A., Stathopulos P.B., Ikura M., Rao A., and Hogan P.G.. 2013. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 20:973–981. 10.1038/nsmb.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.