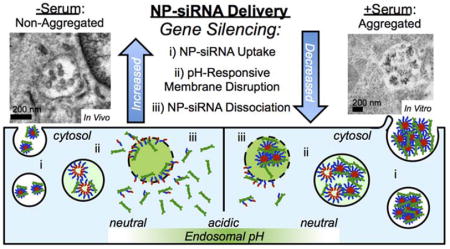
Abstract
Nanoparticles (NP) interact with complex protein milieus in biological fluids, and these interactions have profound effects on NP physicochemical properties and function. Surprisingly, most studies neglect the impact of these interactions, especially with respect to NP-mediated siRNA delivery. Here, the effects of serum on colloidal stability and siRNA delivery of a pH-responsive micellar NP delivery system were characterized. Results show cationic NP-siRNA complexes aggregate in ≥ 2% serum in buffer, but are stable in serum-free media. Furthermore, non-aggregated NP-siRNA delivered in serum-free media result in 4-fold greater siRNA uptake in vitro, compared to aggregated NP-siRNA. Interestingly, pH-responsive membrane lysis behavior, which is required for endosomal escape, and NP-siRNA dissociation, necessary for mRNA knockdown, are significantly reduced in serum. Consistent with these data, non-aggregated NP-siRNA in serum-free conditions result in highly efficient gene silencing, even at doses as low as 5 nM siRNA. NP-siRNA diameter was measured at albumin and IgG levels mimicking biological fluids. Neither albumin nor IgG alone induces NP-siRNA aggregation, implicating other serum proteins in NP colloidal instability. Finally, as a proof-of-principle that stability is maintained in established in vivo models, transmission electron microscopy reveals NP-siRNA are taken up by ductal epithelial cells in a non-aggregated state when injected retroductally into mouse salivary glands in vivo. Overall, this study shows serum-induced NP-siRNA aggregation significantly diminishes efficiency of siRNA delivery by reducing uptake, pH-responsive membrane lysis activity, and NP-siRNA dissociation. Moreover, these results highlight the importance of local NP-mediated drug delivery, and are broadly applicable to other drug delivery systems.
Keywords: nanoparticle, aggregation, siRNA, micelle, serum, mesenchymal stem cell, salivary gland
Graphical Table of Contents
Illustration shows nanoparticle (NP)-siRNA complexes aggregate in the presence of serum. Non-aggregated NP-siRNA result in greater gene silencing due to increased siRNA uptake, pH-responsive membrane disruption, and NP-siRNA dissociation, compared to aggregated NP-siRNA.
Small interfering RNA (siRNA) has extensive therapeutic versatility, as it can specifically silence any gene of interest.1,2 As of 2015, over 30 siRNA candidates have reached various stages of clinical trials for indications including cancer, ophthalmic conditions, genetic disorders, and infectious diseases.3 However, hurdles have stalled universal adoption of siRNA pharmacologically. Only recently has the number of viable siRNA drug candidates grown due to the development of drug delivery systems, including dendrimers, liposomes, and nanoparticles (NPs).4,5 These drug delivery systems provide multifunctionality required to overcome barriers to siRNA delivery, which include tissue targeting, nuclease-mediated degradation, cellular uptake, and endosomal escape.6,7 The majority of success has been limited to studies employing systemically-delivered lipid NPs for liver-related indications due to nonspecific size and opsonization-mediated NP accumulation in the liver after intravenous injections.2,8,9 However, localized delivery to confined organs with easy access, such as the eye, has also realized clinical success and highlights the advantages of localized siRNA delivery.10
Regardless of route of administration, the physicochemical properties of NPs change as a function of their biological milieu. Specifically, a protein corona forms around NPs due to the adsorption of biomolecules in vitro or in vivo.11,12 This NP-protein corona results in surface properties that are vastly different from naïve NPs, which are typically characterized in protein-free solutions. For example, exposure to human plasma results in negative surface charges, even for cationic NPs, with as many as 300 different proteins identified from the NP surface after a 30-second incubation.13 Many NP parameters influence the protein-corona composition and quantity, including size, charge, and surface functionalization.13–15 NP surface properties can also alter the structure of adsorbed proteins, further changing NP characteristics.16,17 In addition to affecting how NPs interact with cells, the protein corona has been shown to mask targeting moieties at the NP surface, resulting in non-specific cell uptake in transferrin-functionalized NPs.18 Furthermore, it is widely known that NP-protein interactions significantly alter NP colloidal stability, resulting in aggregation or stabilization depending on NP properties and proteins present in the biological media.19
As these NP-protein interactions are complex and critical for therapeutic development, fundamental understanding of these phenomena is imperative. Our lab has successfully developed diblock copolymers that self-assembles into spherical NPs, in which the corona is cationic to complex anionic siRNA and the core is hydrophobic and pH-responsive to facilitate self-assembly and endosomal escape of the siRNA payload.20 This NP-siRNA delivery system has been used to modulate gene expression in mesenchymal stem cells in vitro,21,22 to protect salivary glands from radiation-induced cell damage via delivery of protective siRNA,23 and to expedite fracture healing by localizing regenerative NP-siRNA to fracture sites using degradable hydrogel depots.24 Others have modified similar NPs via PEGylation for systemic delivery applications.25 While initial studies showed this class of NPs protects siRNA from serum-mediated degradation,20 it remains unknown if protein adsorption occurs, and how absorbed proteins affect NP-siRNA delivery.
To determine how protein adsorption affects NP function, NP-mediated siRNA delivery was directly compared in serum-containing and serum-free conditions. Changes in NP diameter due to protein adsorption or aggregation were measured using dynamic light scattering (DLS) at serum levels relevant to in vitro cell culture. Due to our in vivo success using local NP-siRNA delivery23,24, changes in NP diameter were also measured in buffer containing albumin and IgG levels consistent with those reported in saliva and interstitial fluids. Additionally, in vitro NP-siRNA delivery outcomes, such as uptake and gene silencing were assessed in serum-containing and serum-free conditions to determine the effect of serum on NP-siRNA delivery efficacy. Finally, the effects of serum on pH-responsive NP membrane lytic behavior and NP-siRNA dissociation were characterized, as these mechanisms are critical for successful cytosolic delivery of siRNA.
Results and Discussion
Understanding the influence of NP interactions with biological proteins on drug delivery is critical, as these interactions have been shown to drastically change NP physicochemical properties, colloidal stability, and NP-cell interactions.13,16,26 In this study, NP-siRNA complexes were characterized for physicochemical changes in various biological milieus. Results show NP-siRNA complexes significantly increase in diameter and polydispersity index (PDI) in serum, suggesting NP-siRNA aggregation that was confirmed using TEM. To determine the in vitro effects of serum-mediated aggregation on NP-siRNA delivery, NP-siRNA were delivered to hMSCs in serum-containing and serum-free media. Results show that non-aggregated NPs are less cytocompatible but mediate more efficient siRNA uptake than aggregated NP-siRNA. Additionally, aggregated complexes also exhibited less pH-dependent membrane lysis and siRNA displacement from NPs. Furthermore, gene silencing is more efficient in non-aggregated NP-siRNA, and could achieve significantly greater gene knockdown at 3-fold lower NP-siRNA doses compared to aggregated NP-siRNA. Interestingly, NP-siRNA incubated with only BSA at concentrations relevant for several biological milieus increased NP-siRNA diameter, albeit less than serum, but did not cause aggregation. Finally, NP-siRNA complexes were directly injected in mouse salivary glands and TEM reveals uptake of non-aggregated NP-siRNA by ductal epithelial cells in vivo, emphasizing the importance of localized NP-siRNA delivery.
Effect of serum on NP-siRNA Diameter
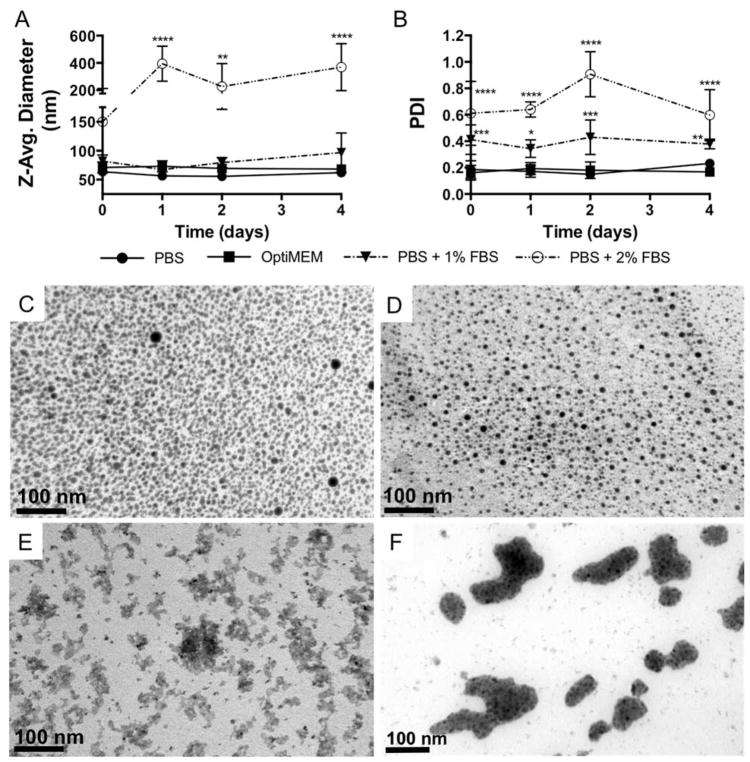
NPs often encounter various biological environments containing a diverse array of proteins and ions that vary widely across different biological fluids. However, NP physicochemical properties are often characterized under physiological salt concentrations and then introduced to serum-containing cell culture media or other biological fluids, and NP-protein interactions that can drastically alter NP colloidal stability and physicochemical properties are surprisingly ignored.19 Here, siRNA was complexed to NP at a charge ratio (+/−) = 4 and DLS was used to measure NP-siRNA diameter in the presence of serum–containing buffer and in serum-free OptiMEM media. Measurements in the presence of 10% FBS, mimicking traditional cell culture media, were unsuccessful due to abrupt NP-siRNA aggregation and sedimentation, rendering samples unfit for DLS. Therefore, DLS measurements of NP-siRNA diameter were made in the presence of 1% FBS and the serum level was increased until NP-siRNA began to aggregate. Figure 1A shows that NP-siRNA Z-average diameter in OptiMEM and PBS + 1% FBS was not significantly different than NP-siRNA in PBS alone, and were stable over 4 days. NP-siRNA diameter was significantly increased to ~150 nm immediately in PBS + 2% FBS and further increased to ~400 nm after one day, after which size was stable over the 4 day experiment. Although there was no change in NP-siRNA diameter in PBS + 1% FBS, the PDI was significantly increased, as it was also for NP-siRNA in PBS + 2% FBS (Figure 1B). PDI of NP-siRNA in OptiMEM media remained unchanged. Multiple studies have shown serum protein-induced aggregation in a variety of NP systems, but these studies are limited to rigid, solid NPs typically composed of gold or poly(styrene).27–31 The effect of serum on the NP-siRNA delivery system used herein has been previously limited to demonstration of siRNA protection from serum-mediated degradation.20 In fact, the majority of literature reporting micellar NP serum stability only consider serum-mediated NP disassembly and not aggregation.25,32,33 To evaluate the effects of cell culture serum levels on NP-siRNA diameter, which was not possible via DLS, NP-siRNA were embedded in PEG hydrogels to facilitate thin sectioning for electron microscopy. Figure 1C and D shows that control (no serum) and 1% FBS exposed NP-siRNA are singular and evenly distributed. However, Figure 1E and F shows that, when incubated in PBS + 2% or 10% FBS prior to encapsulation, NP-siRNA form large irregularly shaped aggregates, consistent with previous reports.34 ImageJ was used to quantify NP-siRNA diameter in the TEM images (Table S1). Results show diameter of NP-siRNA in the absence of serum is 7.3 ± 1.7 nm and 6.2 ± 1.7 nm in 1% FBS. This corroborates data from DLS results that NP-siRNA do not aggregate in 1% FBS. It should be noted that significantly smaller NP diameters in TEM analysis is due to artifacts of drying associated with TEM processing. Similarly, NP-siRNA diameter is increased nearly tenfold in 10% FBS (74.5 ± 38.6 nm). NP-siRNA diameter could not be quantified in 2% FBS samples due to interconnected aggregates that could not be resolved in image analysis. These data are critical, as size and shape are known to affect NP-cell interactions and subsequent uptake mechanisms.35,36 For example, NP diameters of 50 nm, which are similar to the sizes of non-aggregated NP-siRNA here, are optimum for efficient uptake of multiple NP types across multiple cell lines35. Additionally, NPs with diameters similar to those used in this study are taken up via mechanisms that include clathrin- and caveolae-mediated endoctytosis and non-specific endocytosis37,38. In contrast, intracellular accumulation of aggregated NPs, with sizes of 200–400 nm, is observed with our studies as well, albeit at much lower frequencies. Through TEM analysis, multiple uptake mechanisms of the aggregated NPs are implicated including macropinocytosis (Figure S1) which is consistent with uptake of cationic NPs with diameters > 150 nm, including PEI-DNA complexes39 and polymeric micellar NPs40.
Figure 1.
NP-siRNA complexes aggregate in the presence of serum, but are stable in serum-free OptiMEM media. NPs were complexed to siRNA at a charge ratio (+/−) = 4 and incubated in various serum-free and serum-containing buffers. A) DLS reveals NP-siRNA diameter is stable over four days in PBS, OptiMEM media, and at 1% fetal bovine serum (FBS), but significantly increases in 2% FBS. B) Polydispersity Indices (PDI) from DLS measurements show size distribution significantly increases at 1% and 2% FBS, but remains stable in OptiMEM culture media. n = 6; *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001 compared to PBS group at the same time point using two-way ANOVA with Dunnett’s test for multiple comparisons; Error bars represent the standard deviation. NP-siRNA complexes were incubated in PBS (C) or PBS + 1% FBS (D), 2% FBS (E), and 10% FBS overnight and embedded in hydrogels to allow sectioning for TEM. TEM micrographs confirm serum-mediated NP-siRNA aggregation.
Effect of serum on NP-siRNA cytocompatibility
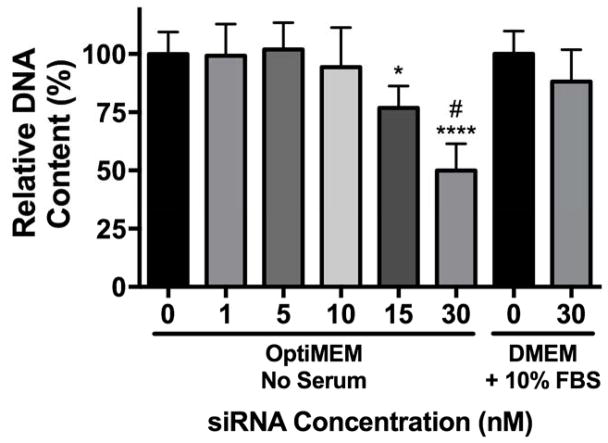
In general, positively charged NPs exhibit lower levels of cytocompatibility compared to negative or neutral NPs41. To determine the effect of serum on NP-siRNA cytocompatibility, hMSCs were treated in vitro with NPs complexed to a non-targeting negative control siRNA at varying concentrations in serum-free OptiMEM media, and DNA content was analyzed as a measure of cytocompatibility (Figure 2). DNA content was quantified because it is linearly correlated with cell number, it can be assayed using a standard curve for reference, and it exhibits greater sensitivity versus metabolic assays traditionally used to assess cell viability.42 siRNA treatments at 30 nM in DMEM + 10% FBS do not reduce cytocompatibility and were used as a benchmark for comparison based on our previous work.22 Cytocompatibility was maintained in serum-free OptiMEM up to 10 nM siRNA. However, treatments at 15 and 30 nM siRNA significantly reduced DNA content to 80% and 50% relative to untreated cells, indicative of cytotoxicity. This is most likely due to retained cationic charge on the NP-siRNA complex. Additionally, several reports have shown reduced cytotoxicity in the presence of serum, attributed to a reduction in uptake.41,43,44 However, it remains unknown if reduced uptake in serum is due to changes in NP size, colloidal stability, or reduced membrane interactions.
Figure 2.
NP-siRNA are more cytotoxic in serum-free culture media. hMSCs were incubated for 24 hours with NPs complexed to a non-targeting negative control siRNA at varying concentrations at charge ratio (+/−) = 4 in DMEM + 10% FBS or serum-free OptiMEM media. At 30 nM siRNA, NP-siRNA complexes are more cytotoxic in OptiMEM compared to DMEM + 10% FBS. Cytotoxicity is observed in OptiMEM at concentrations > 10 nM siRNA. n=6; *p<0.05, ***p<0.001 compared to no treatment (NT); #p<0.0001 compared to 30 nM siRNA in DMEM + 10% FBS. Significance determined using two-way ANOVA with Dunnett’s test relative to NT control and Bonferroni’s test for 30 nM pairwise comparison. Error bars represent the standard deviation.
Effect of serum on nanoparticle-mediated siRNA uptake in hMSCs in vitro
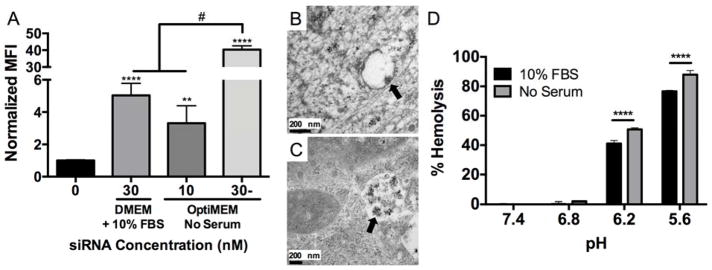
To determine the effect of serum on NP-mediated siRNA uptake, NPs were complexed to fluorescent siRNA and used to treat hMSCs. Figure 3A shows that there was an 8-fold increase in hMSC median fluorescence intensity (MFI) when 30 nM NP-siRNA was delivered in serum-free OptiMEM media compared to DMEM + 10% FBS.22 This increase in NP-siRNA uptake could account for greater cytotoxicity due to the dramatic increase of intracellular NP payload. Moreover, these results are consistent with previous findings that aggregation of gold NPs reduced uptake in multiple cell types by up to 25%.45 Due to increased NP-siRNA cytotoxicity in OptiMEM using a 30 nM dose, hMSC were also treated with 10 nM NP-siRNA in OptiMEM. MFI levels were not significantly different than those achieved with a 3-fold higher dose (30 nM) in DMEM + 10% FBS, indicating non-aggregated NP-siRNA result in more efficient siRNA uptake. NP-siRNA complexes began to sediment upon introduction in PBS + 10% FBS, and this sedimentation was observed via TEM when delivered to hMSCs in serum-containing media (Figure S2). Sedimentation of the NP-siRNA, which reduces the amount of NP-siRNA available to cells, in concert with increased aggregate size, leads to less efficient NP-siRNA uptake. Transmission electron microscopy (TEM) was used to obtain qualitative visual evidence of NP-siRNA uptake. Figure 3B shows nano-sized structures within intracellular vesicles with diameters consistent with stable, non-aggregated NP-siRNA from DLS measurements (Figure 1) when delivered in serum-free OptiMEM media for six hours. hMSCs treated in OptiMEM were also investigated at 12 and 24 hours post-treatment, but no NP-siRNA were observed at these later time points. When treated in DMEM + 10% FBS, larger nanostructures with diameters consistent with aggregated NP-siRNA from DLS analysis (Figure 1) had accumulated in intracellular vesicles 24 hours post-treatment (Figure 3C).
Figure 3. NP-siRNA delivered using serum-free media, which prevents NP-siRNA aggregation, results in significantly greater uptake and pH-responsive membrane lysis.
Fluorescent siRNA was complexed to NPs at a charge ratio (+/−) = 4 and delivered to hMSCs in vitro in DMEM + 10% FBS or serum-free OptiMEM media and uptake was detected via flow cytometry. A) NP-siRNA delivered in serum-free OptiMEM media results in significantly increased median fluorescence intensity (MFI) 24 hours post-treatment compared to DMEM + 10% FBS at 30 nM siRNA. A 3-fold lower NP-siRNA dose (10 nM) delivered in OptiMEM achieves similar uptake levels as 30 nM NP-siRNA dose in DMEM + 10% FBS. n=6, **p<0.01, ****p<0.0001 compared to untreated, #p<0.0001. B) TEM micrograph reveals NP structures (arrow) with diameters similar to values obtained by DLS are visible in hMSC membrane bound vesicles 6 hours post-treatment in serum-free OptiMEM media in vitro, and were no longer detectable at later time points. C) TEM micrograph shows NP structures (arrow) with diameters consistent with aggregated NPs from DLS experiments that accumulate in intracellular vesicles 24 hours post-treatment when delivered in DMEM + 10% FBS. D) Hemolysis data shows pH-responsive membrane lysis activity is significantly lower in serum-mediated NP-siRNA aggregated state at endosomal pH 6.2 and 5.6. Representative data from a single experiment using four sample replicates, ***p<0.001; Significance determined using two-way ANOVA with Dunnett’s test to compare to no treatment (NT) control and Tukey’s test for multiple pairwise comparisons. Error bars represent standard deviation.
These data suggest that serum-induced aggregation reduces the pH-responsive behavior necessary for endosomal escape. Therefore, a hemolysis assay was performed at physiological and endosomal pH (7.4–5.6) to determine if NP pH-dependent membrane disruption is affected by serum mediated NP-siRNA aggregation. Reduction of NP pH-response could result in NP accumulation in the endolysosomal compartment after uptake due to decreased endosomolysis, thus reducing siRNA delivery. Results show 10% FBS-induced NP-siRNA aggregation significantly reduces NP-siRNA membrane lysis ability at late endosomal pH 6.2 and 5.6 (Figure 3D). Previous studies have shown that the NP-protein corona is retained within endosomes.16,46 This could explain the decrease in pH-responsive behavior, as the acidic endosomal environment is less accessible to NP cores within aggregates. The stability of these aggregates within acidic vesicles reduces diblock exchange, leading to lower levels of NP destabilization that is required for endosomal escape. This is especially important for cytosolically active drugs, such as siRNA.
Effect of serum on siRNA displacement from NP via poly(anion) competition
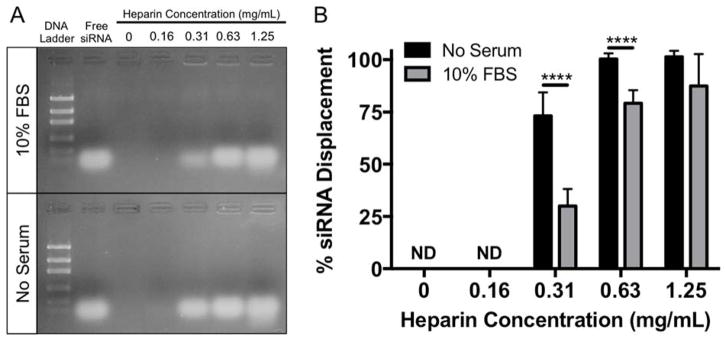
In addition to uptake and endosomal escape, siRNA dissociation from the carrier is a critical barrier to siRNA delivery, as siRNA must dissociate from NP in the cytoplasm after endosomal escape for siRNA-mediated knockdown to occur. Therefore, to determine if serum-induced NP-siRNA aggregation affects the ability of siRNA to be displaced from NPs by other poly(anions), NP-siRNA complexes were incubated overnight with 10% FBS to induce aggregation. After 24 hours, varying concentrations of heparin, a poly(anion), was added to NP-siRNA complexes, and free (displaced) siRNA was detected via gel electrophoresis and compared to unaggregated controls (Figure 4A). Quantification shows that siRNA was not displaced at 0 and 0.16 mg/mL heparin (Figure 4B). Approximately 75% of siRNA was displaced from non-aggregated NP-siRNA complexes in serum-free conditions at 0.31 mg/mL heparin. Displacement of siRNA from aggregated NP-siRNA complexes at the same heparin concentration is significantly reduced to ~25%. Significantly less siRNA displacement was also observed in aggregated NP-siRNA at 0.63 mg/L compared to unaggregated NPs. 1.25 mg/mL heparin displaced nearly 100% of the siRNA in both groups. In summary, our data show serum-mediated NP aggregation can reduce both pH-responsiveness and siRNA dissociation, making siRNA delivery less efficient, regardless of uptake levels.
Figure 4.
siRNA displacement from NPs is significantly reduced in the presence of serum. siRNA was complexed to NPs at a charge ratio (+/−) = 4 and incubated overnight with or without 10% FBS. Heparin, a poly(anion), was added to NP-siRNA complexes at varying concentrations to displace siRNA. Displaced (Free) siRNA was detected via gel electrophoresis (A), and quantified using ImageJ (B). n=3 gels for each condition; ****p<0.0001; Significance determined using two-way ANOVA with Bonferroni’s test to correct for multiple comparisons. Error bars represent the standard deviation.
Effect of serum on NP-siRNA mediated gene silencing
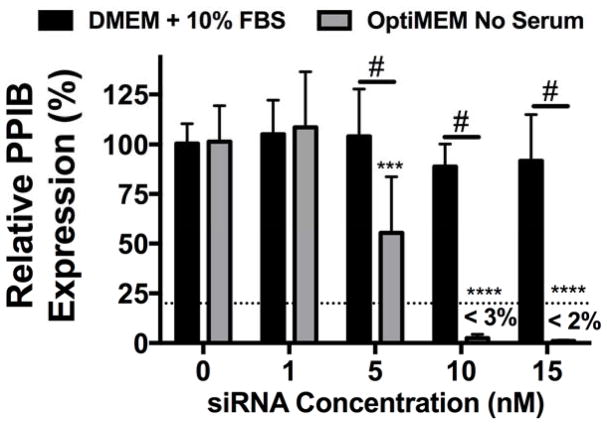
As both pH-responsiveness and siRNA dissociation are required for siRNA to interact with the RNAi machinery for successful gene silencing6,47 and are compromised after NP aggregation, knockdown was investigated. Specifically, hMSC were treated with NP complexed to siRNA targeting peptidylprolyl isomerase B (PPIB), a non-essential housekeeping gene in humans, at varying siRNA concentrations and PPIB expression was quantified using RT-PCR (Figure 5). No gene silencing was evident for groups treated with aggregated NP-siRNA complexes in DMEM + 10% FBS. However, treatment with non-aggregated NP-siRNA complexes at 5 nM siRNA resulted in ~50% knockdown. Furthermore, over 97% and 98% gene knockdown was observed at 10 and 15 nM NP-siRNA treatments, respectively. Even when delivered at levels of similar uptake (30 nM in DMEM + 10% FBS22 and 10 nM in serum-free OptiMEM), NP-siRNA in serum-free conditions (e.g., non-aggregated) result in significantly increased gene silencing, which can be attributed to increased pH-responsive membrane disruption (Figure 3D) and NP-siRNA displacement (Figure 4). Many studies investigating siRNA delivery systems show successful gene silencing in the presence of serum; however, very few perform direct comparisons to assess the effect of serum on the delivery system. Those that do use high concentrations of siRNA (50–100 nM) to achieve even modest gene silencing levels (20–30%) relative to those achieved herein.48,49
Figure 5.
NP-siRNA delivered in serum free media results in significantly more efficient gene silencing compared to NP-siRNA delivered in serum containing media. siRNA targeting human peptidylprolyl isomerase B (PPIB) gene was complexed to NPs at a charge ratio (+/−) = 4 and delivered to hMSCs in vitro in DMEM + 10% FBS or serum free OptiMEM media at varying concentrations and PPIB expression was detected via RT-PCR. PPIB expression is relative to no treatment (NT) groups and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Dashed line represents gene silencing previously achieved using 30 nM NP-siRNA in DMEM + 10% FBS.22 n=6; ***p<0.001, ****p<0.0001 compared to no treatment (NT) control within a treatment group; #p<0.001; Significance determined using two-way ANOVA with Dunnett’s test to compare to no treatment (NT) control and Tukey’s test for multiple pairwise comparisons. Error bars represent the standard deviation.
NP-siRNA stability in physiologically relevant serum albumin levels and in mouse salivary glands in vivo
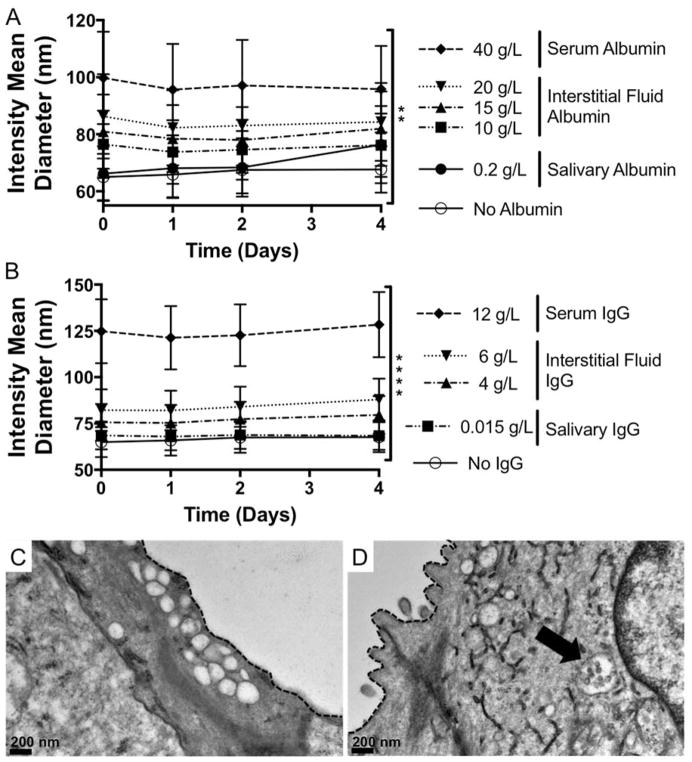
Despite serum-mediated reductions in delivery efficacy, this NP-siRNA delivery system demonstrates therapeutic efficacy in multiple in vivo environments. Retroductal injection of NP-siRNA results in protection from radiation-induced tissue damage in mouse salivary glands23. Furthermore, NPs complexed to osteogenic siRNA and delivered locally to a fracture site via degradable hydrogel depots significantly enhanced bone healing.24 In both contexts, NP-siRNA were delivered locally, and thus were not introduced to the complex protein-rich serum milieu encountered during systemic administration. Nevertheless, interstitial fluids and saliva contain albumin, which is anionic and also the most abundant serum protein.50 Studies report interstitial albumin concentrations ranging from ~10–20 g/L in musculoskeletal tissues50–52, and 0.2 g/L in saliva.53 Additionally, IgG is another protein shown to interact with NPs13,54 found abundantly in serum (11.2 g/L),55 interstitial fluid (4 – 6 g/L),56,57 and saliva.53 Therefore, NP-siRNA diameter was measured in the presence of albumin or IgG at concentrations mimicking these biological fluids. Figure 6A reveals albumin alone significantly increases NP-siRNA diameter which is constant over four days, but diameters are smaller than aggregated NPs (Figure 1). IgG had a similar effect, as shown in Figure 6B. This suggests that albumin and IgG adsorbs to the NP-siRNA surface but remains colloidally stable, which has been shown in other NP systems58,59. Infact, However, this is not consistent with a recent study that identified IgG as a serum component that causes NP aggregation.60 This discrepancy could be due to differences in NP physicochemical properties that dictate underlying NP-protein interactions. To determine the colloidal state of locally delivered NP-siRNA in vivo, NP-siRNA complexes were injected retroductally into mouse salivary glands, which were excised, fixed one hour post-treatment, and processed for TEM imaging. Images demonstrate the presence of intracellular vesicles in salivary gland ductal epithelial cells of both control and NP-treated glands (Figure 6B, C). However, NP structures with diameters consistent with DLS results in Figure 4. 1A are only observed in treated mouse salivary glands (Figure 6C), and appear to be colloidally stable. This provides qualitative, yet powerful evidence that NP-siRNA, which may be unfit for systemic administration due to serum-mediated aggregation, are colloidally stable and readily taken up in vivo when delivered locally.
Figure 6. NP-siRNA are colloidally stable in the presence of serum albumin and IgG and when delivered locally to mouse salivary glands in vivo.
DLS shows presence of bovine serum albumin (BSA) (A) or IgG (B) alone at concentrations matching biological fluids does not cause NP aggregation, but does increase NP diameter with increasing BSA and IgG concentration and is stable over time. n = 6 from two independent experiments each containing triplicate measurements (A). n = 9 from three independent experiments containing triplicate measurements. **p < 0.05, ****p < 0.0001 using two-way ANOVA to determine if protein concentration affects NP diameter. Error bars represent the standard error of the mean. C, D) Representative TEM micorographs show intracellular membrane-bound vesicles, which are empty in untreated mice (C) and contain stable NPs in treated mice (D, arrow). Dashed line indicates cell membrane separating intracellular ductal compartment and the ductal lumen.
Conclusion
Overall, this study demonstrates the importance of characterizing the effects of serum on NP-mediated siRNA delivery. Cationic NP-siRNA complexes aggregated in as little as 2% serum, and aggregation significantly impeded the efficiency of siRNA delivery. In addition to decreasing NP-siRNA uptake, this study shows serum-mediated NP aggregation decreases the pH-responsive membrane destabilizing behavior required for endosomal escape and NP-siRNA decomplexation, both of which contribute to decreased cytoplasmic delivery of the siRNA payload. Furthermore, physiological concentrations of either albumin or IgG alone do not cause NP-siRNA aggregation and NP-siRNA are colloidally stable in vivo when delivered locally to the murine submandibular gland. The impact of this study is multifold: 1) It stresses the importance of characterizing the effect of serum on NP-siRNA properties and siRNA delivery efficiency, 2) It reveals reduction of polymer pH-responsive behavior and siRNA dissociation as mechanisms by which aggregation can decrease siRNA delivery, and 3) It exemplifies the importance of localized delivery, such that NP that are unfit for systemic administration are effective when delivered locally. This information is crucial for future characterization of NP-siRNA delivery systems and will guide the next generation of functional siRNA delivery systems.
Methods
Diblock copolymer synthesis and characterization
The synthesis and characterization of a pH-responsive diblock copolymer has been previously reported22. Briefly, reversible addition-fragmentation chain transfer polymerization was used to synthesize diblock copolymers. The cationic 1st block was synthesized by mixing dimethylaminoethyl methacrylate (DMAEMA), 4-cyano-4-[(ethylsulfanylthiocarbonyl)-sulfanyl]pentanoic acid (ECT) as the chain transfer agent (CTA), and the radical initiator 2,2′-Azobis(2-methylpropionitrile) (AIBN) such that the [monomer]/[CTA]/[Initiator] = 100/10/1 in dimethylformamide (DMF) at 25 wt%. The reaction was purged with nitrogen for 40 minutes and the reaction proceeded for 6 hours at 60 °C. The resulting poly(DMAMEA)-macroCTA was isolated via precipitation and washing with centrifugation using 80:20 pentane:diethyl ether and dried overnight under vacuum. This macroCTA used in the 2nd block reaction in which DMAEMA, 2-propylacrylic acid (PAA), and butyl methacrylate (BMA) monomers at 1:1:2 molar ratio were mixed with macroCTA and AIBN such that [monomer]/[macroCTA]/[intiatior] = 400/10/1 in 25 wt% of solids in DMF. The reaction proceeded for 24 hours at 60 °C under nitrogen. The diblock polymer was isolated via precipitation and centrifugation in 80:20 pentane: diethyl ether and dried under vacuum. Absolute molecular weight and polydispersity (PDI, Mw/Mn) of pDMAEMA-macroCTA 1st block and the diblock copolymer were obtained via gel permeation chromatography (GPC) (Shimadzu Technologies) using a TSKgel Guard SuperH-H guard column (Tosoh Biosciences) and a TSKgel Super HM-N for separation (Tosoh Biosciences) using a column oven at 60 °C. The system includes a miniDAWN TREOS multi-angle light scattering detector (Wyatt Technologies), and an Optilab T-rEX differential refractometer (Wyatt Technologies) to determine absolute molecular weights using reported dn/dc values for p(DMAEMA) (0.06 mL/g).61–63 GPC analysis reveals 1st block Mn = 11.1 kg/mol with PDI = 1.03, and diblock Mn = 30.4 kg/mol with PDI = 1.2.
NP-self-assembly and complexation with siRNA
NP self-assembly and complexation with siRNA have been previously described22. Briefly, raw polymer was dissolved in ethanol (EtOH) at 4 mg/mL, diluted in an equal volume of 1× PBS, and dialyzed using 3500 Da molecular weight dialysis tubing against distilled, deionized H2O for at least 24 hours. NP solutions were filtered with 0.2 μm syringe filters. NP concentration was verified by lyophilizing 1 mL aliquots and measuring the dry polymer mass.
Gel retardation assays were used to determine the critical charge ratio, which is defined as the ratio between the positively charged protonated DMAEMA residues of the pDMAEMA block, where 50% of the residues are protonated at physiological pH, and the negative charges from the siRNA at which there is no free siRNA. This was empirically determined by complexing a dose of siRNA with varying amounts of NP in a total volume of 16 μL. BlueJuice Gel Loading Buffer (4 μL) (Invitrogen) was added to each sample and thoroughly mixed. 20 μL of each sample was loaded into the wells of a 2% agarose gel in 1× tris-acetate-EDTA (TAE) buffer. Free siRNA was separated via gel electrophoresis run at 80 V for 45 minutes. The gel was stained with SybrGold Nucleic Acid Gel Stain (Invitrogen, 1:100 in 1× TAE) for 45 minutes with gentle rocking and protected from light. Stained gels were placed on a UV illumination table and images of illuminated siRNA bands were captured using a digital camera. ImageJ gel analysis tool was used to quantify siRNA band intensities and were normalized to free siRNA. This critical charge ratio was used to determine NP volume necessary to achieve a charge ratio (CR, +/−) = 4 for NP-siRNA treatments. To form NP-siRNA complexes, the appropriate amount of siRNA was added to a 1.5 mL tube, diluted in 1× PBS, and then a volume of NP were added to achieve CR=4 and NP-siRNA solution was incubated for 25 minutes to allow complexation.
NP-siRNA diameter measurements
NP-siRNA complexes were diluted in 1× Dulbecco’s phosphate buffered saline (PBS, Gibco) or 1× PBS with a range of fetal bovine serum (FBS), bovine serum albumin (BSA), Immunoglobulin G (IgG), or OptiMEM I media without phenol red (Gibco). Note: all protein solutions in PBS were filtered (0.2 μm) prior to introduction of NP-siRNA. NP-siRNA diameters were measured at 25 °C in disposable cuvettes using a Malvern Zetasizer Nano ZS with auto attenuation and a 173 ° scattering angle. Cumulants analysis was used to report Z-average diameter and polydispersity for measurements made in FBS due to the presence of multiple peaks that change throughout the duration of the measurements due to dynamic NP-protein interactions and aggregation. To ensure signal collected was not from FBS proteins, blank samples of PBS and FBS with no NP-siRNA were run at matching attenuator positions of samples with NP-siRNA. Blank samples did not produce enough signal for analysis. For NP-siRNA in BSA measurements, distribution analysis was used and intensity mean diameter of NP-siRNA peak is reported, as the NP-siRNA peak is well-defined and separated from the characteristic BSA peak at ~7 nm.
NP-siRNA hydrogel embedding for transmission electron microscopy
Serum in NP solutions poses problems for EM analysis, including the formation of a thick protein layer on the grid that can obscure NP structure, and contamination of the EM interior with protein. Therefore, poly(ethylene glycol) (PEG) hydrogels were used as tissue phantoms to embed NP-siRNA and allow for thin sectioning, similar to tissue. PEG (4-arm, 10 kDa) was functionalized with norbornene (PEGN) and characterized via NMR as previously described64. Hydrogels were formed via thiol-ene photopolymerization as previously described65,66. Briefly, PEGN was dissolved with 3.5 kDa PEG-dithiol crosslinker at a 1:1 thiol:ene ratio in PBS at 10 wt% PEGN and 0.05 wt% lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, synthesized as previously described67) to form a precursor solution. NP-siRNA complexes were formed with FBS to induce aggregation and then added to hydrogel precursor solutions. 40 μL of hydrogel-NP-siRNA precursor solution was added to a cylindrical mold and polymerized under 5 mW/cm2 365 nm UV light for 3 minutes. Gels formed with NP-siRNA complexes only in PBS were used as controls. Hydrogels were fixed in 2.5% gluteraldehyde and submitted to the University of Rochester Medical Center’s Electron Microscopy core for standard processing. Briefly, fixed gels were post-fixed in osmium tetroxide (OsO4), processed through a series of graded alcohols, infiltrated with epoxy resin, and embedded. Embedded gels were polymerized overnight at 70 °C. Semi-thin 1–2 μm sections are cut to select an appropriate area for ultra-thin sectioning with a diamond knife. Sections are placed on grids and stained with uranyl acetate and lead citrate. Grids were imaged using a Hitachi 7650 transmission electron microscope (TEM). NP diameter was quantified using in ImageJ using the Analyze Particles tool.
hMSC cell culture
Human bone marrow aspirate was obtained from Lonza and MSCs were isolated as previously described68. hMSCs growth media (GM) is composed of low glucose Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) with 10% FBS (Atlanta Biologics), and 1% antibiotic-antimycotic (Gibco). For expansion, GM was supplemented with 1 ng/mL recombinant human basic fibroblast growth factor-2 (Corning) at every media change, and was omitted when hMSCs were plated for in vitro experiments. hMSCs were cultured in a humidified incubator at 37 °C with 5% CO2, and passaged using 0.25% trypsin-EDTA (Gibco). For all in vitro experiments, hMSCs at passages 2–4 were seeded on multiwell plates at 8,000 cells/cm2 and allowed to adhere overnight.
Preparation and administration of NP-siRNA treatments
NP-siRNA were prepared at 10× the final concentration using siRNA complexed to NP in 1× PBS at a charge ratio (CR, +/−) = 4 as previously described22. Concentrated treatments were added directly to cell culture media and untreated samples received an equal treatment volume of 1× PBS.
Relative Quantification of hMSC DNA content
NP-siRNA complexes containing varying concentrations of ON-TARGETplus Non-targeting Control Pool siRNA (Dharmacon) were delivered to hMSCs for 24 hours. After NP-siRNA incubation, cells were washed 2× with PBS and lysed using 1× Luciferase Cell Culture Lysis Reagent (Promega) in PBS, and lysates were quickly sonicated using a probe sonicator to homogenize samples. 10 μL of lysate was diluted in 90 μL of 1× TE buffer and Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) was used according to manufacturer’s protocol using λDNA standard curve to quantify DNA content. Treatment groups were normalized to untreated hMSCs.
Quantification of NP-mediated siRNA uptake via flow cytometry
NP-siRNA complexes containing varying concentrations of Silencer FAM-labeled Negative Control No. 1 siRNA (Ambion) were delivered to hMSCs for 24 hours. After incubation, cells were washed 2× with PBS, trypsinized, transferred to 1.5 mL tubes, and collected via centrifugation. Collected cells were washed once more and resuspended in 90 μL of flow buffer (PBS containing of 0.5 w/v% BSA and 0.01 v/v% trypan blue to quench extracellular fluorescence69). 10 μL of diluted propidium iodide (1:500 in flow buffer, Molecular Probes) was added to each sample just prior to analysis to discriminate dead cells using an Accuri C6 flow cytometer (BD Biosciences). 5,000 cells were gated for analysis using single stained controls for gating and compensation. Data analysis was performed using FlowJo software.
TEM assessment of NP-siRNA stability in vitro
Circular glass coverslips (VWR) were autoclaved and placed in multiwell tissue culture plates. hMSCs were seeded on coverslips as described for other in vitro experiments herein. Cells were treated with 30 nM NP-siRNA for serum-containing treatments and 10 nM NP-siRNA for serum-free treatments. Media was removed and cells were immediately fixed in 2.5% gluteraldehyde after continuous treatment with NP-siRNA for 6, 12, and 24 hours. Fixed coverslips post-fixed with osmium tetroxide (OsO4) were processed for transmission electron microscopy (TEM) imaging using the “Pop-off” technique70.
Hemolysis assay
NP-siRNA treatments were made at 20× the final treatment concentration of 30 nM siRNA complexed to NP at CR = 4. Either FBS or PBS was added to NP-siRNA complexes at 10% the final volume and incubated overnight prior the hemolysis assay. The day of the assay, 25 mL of human blood was retrieved from consenting donors by a trained phlebotomist in compliance with the University of Rochester’s Institutional Review Board. Blood was drawn directly into K2-EDTA coated vacutainers to prevent coagulation. Erythrocytes were isolated and washed in 150 mM NaCl via centrifugation and used for a hemolysis assay as previously described71. Briefly, washed erythrocytes were split evenly into 4 tubes washed once with phosphate buffer (PB) at each pH to be tested. 1 mL of erythrocytes at each pH was diluted into 49 mL of PB at the corresponding pH. 10 μL of 20× NP-siRNA samples were pipetted in to U-bottom 96 well plates in quadruplicate. At each pH, 10 μL of 20% Triton X-100 was used for positive controls to set 100% hemolysis, and 10 μL PBS with or without 10% FBS was used as negative controls to subtract any baseline hemolysis at each pH. 190 μL of prepared erythrocytes were added to each well using a multi-channel pipette. Plates were sealed with adhesive covers and incubated at 37 °C for 1 hour. Plates were centrifuged to pellet intact erythrocytes and 100 μL of supernatant was transferred to a clear 96 well plate. Absorbance of the released hemoglobin in supernatant due to hemolysis was measured at 541 nm. At each pH, baseline signal from PBS negative controls were subtracted and experimental samples were normalized signal obtained from Triton X-100 samples. Results from a single experiment are shown in Figure 3D. A duplicate experiment was performed and results are shown in Figure S3. Donor-to-donor variability in collected human blood leads to variability across separate experiments. However, both experiments show similar trends, but saturation of hemolysis was observed in one experiment.
NP-siRNA displacement assay
siRNA (4 μM) was complexed to NP at CR = 4 in a 16 μL volume of PBS. FBS or PBS was added to 10% of the final volume. Complexes were incubated overnight to allow aggregation in 10% FBS samples. The next day, 4 μL of varying concentrations of heparin sodium salt (Sigma) was added to NP-siRNA samples and incubated for 30 minutes. At this time, free siRNA controls were made with the same amount of siRNA contained in NP-siRNA samples. In the FBS group, a volume of FBS was added to free siRNA control when heparin was added to NP-siRNA to account for any change in signal due to potential FBS-mediated degradation of released siRNA. BlueJuice Gel Loading Buffer (Invitrogen) (5 μL) was added to each sample and thoroughly mixed. 20 μL of each sample was loaded into the wells of a 2% agarose gel in 1× tris-acetate-EDTA (TAE) buffer. Displaced siRNA was separated via gel electrophoresis as described previously for gel retardation assays in “NP self-assembly and complexation with siRNA” section.
Assessing NP-siRNA mediated gene knockdown
hMSC were treated with NP-siRNA at varying concentrations in either hMSC GM or OptiMEM I media for 24 hours. Media was removed and replaced with fresh GM for an additional 24 hours to allow siRNA-meditated knockdown to occur. Total RNA was isolated and reverse transcribed into cDNA used for RT-PCR as previously described22 with RNA normalized to 5 ng/μL prior to RT and 5 μL of cDNA was used in each RT-PCR reaction. Primer efficiencies were calculated from each well as previously described72 using 3% and 6% of the maximum amplification to set two thresholds. Relative PPIB expression was calculated using the Pfaffl equation relative to untreated samples and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression73.
Assessment of NP-siRNA uptake in mouse salivary glands via TEM
Adult female C57/BL6 mice 6–10 weeks of age were used for NP-siRNA injections in accordance with the University Committee on Animal Resources at the University of Rochester, Rochester, NY. Retroductal injections into the Wharton’s Duct of the submandibular gland (SMG) were administered through the oral cavity according to previously described protocols23,74. Briefly, mice were anesthetized using 100 mg/kg ketamine and 10 mg/kg xylazine, and body temperature was maintained using heating pads throughout the procedures. The sublingual orifice of the Wharton’s duct was cannulated using a mouse intracranial catheter (#CS-32) to allow NP-siRNA injection directly into the SMG. Atropine was administered intraperitoneally (1 μg/g body weight) to prevent salivary backflow. Sterile NP-siRNA were prepared the same day prior to injections using NPs complexed to 4 μg of siRNA in 1× PBS in a 50 μL volume. A single mouse was given NP-siRNA injection in each gland using a 27-gauge Hamilton microliter syringe (#7637-01) and kept immobilized for 15 minutes post-injection, after which the catheter was removed. After 1 hour the treated mouse and an untreated control mouse were euthanized and SMG were immediately surgically dissected and placed in 2.5% gluteraldehyde + 4% paraformaldehyde for fixation overnight. Fixed glands were submitted to the University of Rochester Medical Center’s Electron Microscopy core and were processed identically to NP-siRNA hydrogel samples previously described.
Statistical analysis
Unless otherwise noted, all experiments were performed in triplicate in at least two independent experiments. One- or two-way ANOVA was used with appropriate post-hoc testing to correct for multiple comparisons, as indicated, to assess significant differences between means (α = 0.05). Statistical analyses were performed using Prism6.0. For all plots, the mean is represented with standard deviation shown as error bars.
Supplementary Material
Table S1: Quantification of NP-siRNA diameter after embedding in hydrogels for TEM imaging.
Figure S1: hMSC take up NP-siRNA aggregates through various mechanisms.
Figure S2: NP-siRNA aggregates sediment in serum containing media.
Figure S3: Results from NP-siRNA hemolysis assay repeat shows similar serum-mediated reduction in NP-siRNA pH-dependent membrane lysis behavior.
Acknowledgments
The Authors would like to thank Karen de Mesy Bentley, M.S., and Gayle Schneider at the University of Rochester Medical Center’s (URMC) Electron Microscopy Shared Resource Laboratory for EM sample preparation and imaging. Additionally, the authors would like to thank the URMC’s Flow Cytometry Core for valuable training and guidance, and the University of Rochester’s McGrath Lab for use of facilities. Funding for this work was provided by the National Science Foundation (DMR-1206219), the New York State Stem Cell Science Program (NYSTEM IDEA-N11G-035), and the National Institutes of Health (R01 AR064200, R01 AR056696, R01 DE018023, P30 AR069655 (Sub-Project 5278), and R56 DE025098). JJV is supported NIH/National Cancer Institute (NCI) Ruth L. Kirschstein National Research Service Award F30 CA206296. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, National Institutes of Health, or NYSTEM.
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest related to this work.
References
- 1.Wilson RC, Doudna JA. Molecular Mechanisms of Rna Interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wittrup A, Lieberman J. Knocking Down Disease: A Progress Report on Sirna Therapeutics. Nat Rev Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam JK, Chow MY, Zhang Y, Leung SW. Sirna Versus Mirna as Therapeutics for Gene Silencing. Mol Ther-- Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery Materials for Sirna Therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 5.Kozielski KL, Tzeng SY, Green JJ. Bioengineered Nanoparticles for Sirna Delivery. Wiley Interdiscip Rev: Nanomed Nanobiotechnol. 2013;5:449–468. doi: 10.1002/wnan.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Knocking Down Barriers: Advances in Sirna Delivery. Nat Rev Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williford JM, Wu J, Ren Y, Archang MM, Leong KW, Mao HQ. Recent Advances in Nanoparticle-Mediated Sirna Delivery. Annu Rev Biomed Eng. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 8.Patel HM, Moghimi SM. Serum-Mediated Recognition of Liposomes by Phagocytic Cells of the Reticuloendothelial System - the Concept of Tissue Specificity. Adv Drug Deliv Rev. 1998;32:45–60. doi: 10.1016/s0169-409x(97)00131-2. [DOI] [PubMed] [Google Scholar]
- 9.Blanco E, Shen H, Ferrari M. Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman-Aranguez A, Loma P, Pintor J. Small-Interfering Rnas (Sirnas) as a Promising Tool for Ocular Therapy. Brit J Pharmacol. 2013;170:730–747. doi: 10.1111/bph.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 13.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 14.Huhn D, Kantner K, Geidel C, Brandholt S, De Cock I, Soenen SJ, Rivera Gil P, Montenegro JM, Braeckmans K, Mullen K, Nienhaus GU, Klapper M, Parak WJ. Polymer-Coated Nanoparticles Interacting with Proteins and Cells: Focusing on the Sign of the Net Charge. ACS Nano. 2013;7:3253–3263. doi: 10.1021/nn3059295. [DOI] [PubMed] [Google Scholar]
- 15.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc Natl Acad Sci U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischer CC, Payne CK. Nanoparticle-Cell Interactions: Molecular Structure of the Protein Corona and Cellular Outcomes. Acc Chem Res. 2014;47:2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parveen R, Shamsi TN, Fatima S. Nanoparticles-Protein Interaction: Role in Protein Aggregation and Clinical Implications. Int J Biol Macromol. 2017;94:386–395. doi: 10.1016/j.ijbiomac.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Aberg C, Mahon E, Dawson KA. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 19.Moore TL, Rodriguez-Lorenzo L, Hirsch V, Balog S, Urban D, Jud C, Rothen-Rutishauser B, Lattuada M, Petri-Fink A. Nanoparticle Colloidal Stability in Cell Culture Media and Impact on Cellular Interactions. Chem Soc Rev. 2015;44:6287–6305. doi: 10.1039/c4cs00487f. [DOI] [PubMed] [Google Scholar]
- 20.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Development of a Novel Endosomolytic Diblock Copolymer for Sirna Delivery. J Controlled Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benoit DSW, Boutin ME. Controlling Mesenchymal Stem Cell Gene Expression Using Polymer-Mediated Delivery of Sirna. Biomacromolecules. 2012;13:3841–3849. doi: 10.1021/bm301294n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malcolm DW, Sorrells JE, Van Twisk D, Thakar J, Benoit DS. Evaluating Side Effects of Nanoparticle-Mediated Sirna Delivery to Mesenchymal Stem Cells Using Next Generation Sequencing and Enrichment Analysis. Bioeng Transl Med. 2016;1:193–206. doi: 10.1002/btm2.10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arany S, Benoit DS, Dewhurst S, Ovitt CE. Nanoparticle-Mediated Gene Silencing Confers Radioprotection to Salivary Glands in Vivo. Mol Ther. 2013;21:1182–1194. doi: 10.1038/mt.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Malcolm DW, Benoit DSW. Controlled and Sustained Delivery of Sirna/Nps from Hydrogels Expedites Bone Fracture Healing. Biomaterials. 2017;139:127–138. doi: 10.1016/j.biomaterials.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing Cationic and Hydrophobic Content of Pegylated Sirna Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen VH, Lee BJ. Protein Corona: A New Approach for Nanomedicine Design. Int J Nanomed. 2017;12:3137–3151. doi: 10.2147/IJN.S129300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rausch K, Reuter A, Fischer K, Schmidt M. Evaluation of Nanoparticle Aggregation in Human Blood Serum. Biomacromolecules. 2010;11:2836–2839. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- 28.Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J Am Chem Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 29.Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 30.Nuhn L, Gietzen S, Mohr K, Fischer K, Toh K, Miyata K, Matsumoto Y, Kataoka K, Schmidt M, Zentel R. Aggregation Behavior of Cationic Nanohydrogel Particles in Human Blood Serum. Biomacromolecules. 2014;15:1526–1533. doi: 10.1021/bm500199h. [DOI] [PubMed] [Google Scholar]
- 31.Lacerda SH, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF. Interaction of Gold Nanoparticles with Common Human Blood Proteins. ACS Nano. 2010;4:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Owen SC, Shoichet MS. Stability of Self-Assembled Polymeric Micelles in Serum. Macromolecules. 2011;44:6002–6008. doi: 10.1021/ma200675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diezi TA, Bae Y, Kwon GS. Enhanced Stability of Peg-Block-Poly(N-Hexyl Stearate L-Aspartamide) Micelles in the Presence of Serum Proteins. Mol Pharmaceutics. 2010;7:1355–1360. doi: 10.1021/mp100069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balog S, Rodriguez-Lorenzo L, Monnier CA, Obiols-Rabasa M, Rothen-Rutishauser B, Schurtenberger P, Petri-Fink A. Characterizing Nanoparticles in Complex Biological Media and Physiological Fluids with Depolarized Dynamic Light Scattering. Nanoscale. 2015;7:5991–5997. doi: 10.1039/c4nr06538g. [DOI] [PubMed] [Google Scholar]
- 35.Shang L, Nienhaus K, Nienhaus GU. Engineered Nanoparticles Interacting with Cells: Size Matters. J Nanobiotechnol. 2014;12:5. doi: 10.1186/1477-3155-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh N, Park JH. Endocytosis and Exocytosis of Nanoparticles in Mammalian Cells. Int J Nanomedicine. 2014;9(Suppl 1):51–63. doi: 10.2147/IJN.S26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC, Mahmoudi M. Cellular Uptake of Nanoparticles: Journey inside the Cell. Chem Soc Rev. 2017;46:4218–4244. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morteza Hasanzadeh K, Frances JH, Nicolas HV. Insights into Cellular Uptake of Nanoparticles. Current Drug Delivery. 2015;12:63–77. doi: 10.2174/1567201811666140821110631. [DOI] [PubMed] [Google Scholar]
- 39.Hufnagel H, Hakim P, Lima A, Hollfelder F. Fluid Phase Endocytosis Contributes to Transfection of DNA by Pei-25. Mol Ther. 2009;17:1411–1417. doi: 10.1038/mt.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki H, Bae YH. Evaluation of Drug Penetration with Cationic Micelles and Their Penetration Mechanism Using an in Vitro Tumor Model. Biomaterials. 2016;98:120–130. doi: 10.1016/j.biomaterials.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 41.Frohlich E. The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles. Int J Nanomed. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quent VM, Loessner D, Friis T, Reichert JC, Hutmacher DW. Discrepancies between Metabolic Activity and DNA Content as Tool to Assess Cell Proliferation in Cancer Research. J Cell Mol Med. 2010;14:1003–1013. doi: 10.1111/j.1582-4934.2010.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nafee N, Schneider M, Schaefer UF, Lehr CM. Relevance of the Colloidal Stability of Chitosan/Plga Nanoparticles on Their Cytotoxicity Profile. Int J Pharm. 2009;381:130–139. doi: 10.1016/j.ijpharm.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 44.Petri-Fink A, Steitz B, Finka A, Salaklang J, Hofmann H. Effect of Cell Media on Polymer Coated Superparamagnetic Iron Oxide Nanoparticles (Spions): Colloidal Stability, Cytotoxicity, and Cellular Uptake Studies. Eur J Pharm Biopharm. 2008;68:129–137. doi: 10.1016/j.ejpb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 45.Albanese A, Chan WC. Effect of Gold Nanoparticle Aggregation on Cell Uptake and Toxicity. ACS Nano. 2011;5:5478–5489. doi: 10.1021/nn2007496. [DOI] [PubMed] [Google Scholar]
- 46.Doorley GW, Payne CK. Nanoparticles Act as Protein Carriers During Cellular Internalization. Chem Commun. 2012;48:2961–2963. doi: 10.1039/c2cc16937a. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of Sirna Therapeutics: Barriers and Carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar K, Maiti B, Kondaiah P, Bhattacharya S. Efficacious Gene Silencing in Serum and Significant Apoptotic Activity Induction by Survivin Downregulation Mediated by New Cationic Gemini Tocopheryl Lipids. Mol Pharmaceutics. 2015;12:351–361. doi: 10.1021/mp500620e. [DOI] [PubMed] [Google Scholar]
- 49.Nakanishi M, Patil R, Ren Y, Shyam R, Wong P, Mao HQ. Enhanced Stability and Knockdown Efficiency of Poly(Ethylene Glycol)-B-Polyphosphoramidate/Sirna Micellar Nanoparticles by Co-Condensation with Sodium Triphosphate. Pharm Res. 2011;28:1723–1732. doi: 10.1007/s11095-011-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellmerer M, Schaupp L, Brunner GA, Sendlhofer G, Wutte A, Wach P, Pieber TR. Measurement of Interstitial Albumin in Human Skeletal Muscle and Adipose Tissue by Open-Flow Microperfusion. Am J Physiol Endocrinol Metab. 2000;278:E352–356. doi: 10.1152/ajpendo.2000.278.2.E352. [DOI] [PubMed] [Google Scholar]
- 51.Fogh-Andersen N, Altura BM, Altura BT, Siggaard-Andersen O. Composition of Interstitial Fluid. Clin Chem. 1995;41:1522–1525. [PubMed] [Google Scholar]
- 52.Gilanyi M, Ikrenyi C, Fekete J, Ikrenyi K, Kovach AG. Ion Concentrations in Subcutaneous Interstitial Fluid: Measured Versus Expected Values. Am J Physiol. 1988;255:F513–519. doi: 10.1152/ajprenal.1988.255.3.F513. [DOI] [PubMed] [Google Scholar]
- 53.Rantonen PJF, Meurman JH. Correlations between Total Protein, Lysozyme, Immunoglobulins, Amylase, and Albumin in Stimulated Whole Saliva During Daytime. Acta Odontol Scand. 2000;58:160–165. doi: 10.1080/000163500429154. [DOI] [PubMed] [Google Scholar]
- 54.Minelli C, Garcia-Diez R, Sikora AE, Gollwitzer C, Krumrey M, Shard AG. Characterization of Igg-Protein-Coated Polymeric Nanoparticles Using Complementary Particle Sizing Techniques. Surf Interface Anal. 2014;46:663–667. [Google Scholar]
- 55.Gonzalez-Quintela A, Alende R, Gude F, Campos J, Rey J, Meijide LM, Fernandez-Merino C, Vidal C. Serum Levels of Immunoglobulins (Igg, Iga, Igm) in a General Adult Population and Their Relationship with Alcohol Consumption, Smoking and Common Metabolic Abnormalities. Clin Exp Immunol. 2008;151:42–50. doi: 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poulsen HL. Interstitial Fluid Concentrations of Albumin and Immunoglobulin G in Normal Men. Scand J Clin Lab Invest. 1974;34:119–122. [PubMed] [Google Scholar]
- 57.Rossing N, Worm AM. Interstitial Fluid: Exchange of Macromolecules between Plasma and Skin Interstitium. Clin Physiol. 1981;1:275–284. doi: 10.1111/j.1475-097x.1981.tb00896.x. [DOI] [PubMed] [Google Scholar]
- 58.Dominguez-Medina S, Blankenburg J, Olson J, Landes CF, Link S. Adsorption of a Protein Monolayer Via Hydrophobic Interactions Prevents Nanoparticle Aggregation under Harsh Environmental Conditions. ACS Sustain Chem Eng. 2013;1:833–842. doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baier G, Costa C, Zeller A, Baumann D, Sayer C, Araujo PH, Mailander V, Musyanovych A, Landfester K. Bsa Adsorption on Differently Charged Polystyrene Nanoparticles Using Isothermal Titration Calorimetry and the Influence on Cellular Uptake. Macromol Biosci. 2011;11:628–638. doi: 10.1002/mabi.201000395. [DOI] [PubMed] [Google Scholar]
- 60.Cukalevski R, Ferreira SA, Dunning CJ, Berggård T, Cedervall T. Igg and Fibrinogen Driven Nanoparticle Aggregation. Nano Res. 2015;8:2733–2743. [Google Scholar]
- 61.Horev B, Klein MI, Hwang G, Li Y, Kim D, Koo H, Benoit DSW. pH-Activated Nanoparticles for Controlled Topical Delivery of Farnesol to Disrupt Oral Biofilm Virulence. ACS Nano. 2015;9:2390–2404. doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gallow KC, Jhon YK, Genzer J, Loo YL. Influence of Gradient Strength and Composition Profile on the Onset of the Cloud Point Transition in Hydroxyethyl Methacrylate/Dimethylaminoethyl Methacrylate Gradient Copolymers. Polymer. 2012;53:1131–1137. [Google Scholar]
- 63.Kryuchkov MA, Detrembleur C, Jerome R, Prud’homme RE, Bazuin CG. Synthesis and Thermal Properties of Linear Amphiphilic Diblock Copolymers of L-Lactide and 2-Dimethylaminoethyl Methacrylate. Macromolecules. 2011;44:5209–5217. [Google Scholar]
- 64.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A Versatile Synthetic Extracellular Matrix Mimic Via Thiol-Norbornene Photopolymerization. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Hove AH, Beltejar MJ, Benoit DS. Development and in Vitro Assessment of Enzymatically-Responsive Poly(Ethylene Glycol) Hydrogels for the Delivery of Therapeutic Peptides. Biomaterials. 2014;35:9719–9730. doi: 10.1016/j.biomaterials.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shubin AD, Felong TJ, Graunke D, Ovitt CE, Benoit DS. Development of Poly(Ethylene Glycol) Hydrogels for Salivary Gland Tissue Engineering Applications. Tissue Eng Part A. 2015;21:1733–1751. doi: 10.1089/ten.tea.2014.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated Polymerization of Peg-Diacrylate with Lithium Phenyl-2,4,6-Trimethylbenzoylphosphinate: Polymerization Rate and Cytocompatibility. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pittenger MF. Mesenchymal Stem Cells from Adult Bone Marrow. Methods Mol Biol. 2008;449:27–44. doi: 10.1007/978-1-60327-169-1_2. [DOI] [PubMed] [Google Scholar]
- 69.Sahlin S, Hed J, Rundquist I. Differentiation between Attached and Ingested Immune Complexes by a Fluorescence Quenching Cytofluorometric Assay. J Immunol Methods. 1983;60:115–124. doi: 10.1016/0022-1759(83)90340-x. [DOI] [PubMed] [Google Scholar]
- 70.de Mesy Jensen KL. Tech Sample CY-1, Amer Soc Clin Pathol. 1987. “Pop-Off” Technique for Fna Smears for Diagnostic Electron Microscopy. [Google Scholar]
- 71.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of Ph-Responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. J Vis Exp. 2013:e50166. doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu W, Saint DA. A New Quantitative Method of Real Time Reverse Transcription Polymerase Chain Reaction Assay Based on Simulation of Polymerase Chain Reaction Kinetics. Anal Biochem. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 73.Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time Rt-Pcr. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng C, Voutetakis A, Kok MR, Goldsmith CM, Smith GB, Elmore S, Nyska A, Vallant M, Irwin RD, Baum BJ. Toxicity and Biodistribution of a First-Generation Recombinant Adenoviral Vector, in the Presence of Hydroxychloroquine, Following Retroductal Delivery to a Single Rat Submandibular Gland. Oral Dis. 2006;12:137–144. doi: 10.1111/j.1601-0825.2005.01170.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Quantification of NP-siRNA diameter after embedding in hydrogels for TEM imaging.
Figure S1: hMSC take up NP-siRNA aggregates through various mechanisms.
Figure S2: NP-siRNA aggregates sediment in serum containing media.
Figure S3: Results from NP-siRNA hemolysis assay repeat shows similar serum-mediated reduction in NP-siRNA pH-dependent membrane lysis behavior.