Abstract
Animals respond to diurnal shifts in their environment with a combination of behavioral, physiological, and molecular changes to synchronize with regularly-timed external cues. Reproduction, movement, and metabolism in cnidarians have all been shown to be regulated by diurnal lighting, but the molecular mechanisms that may be responsible for these phenotypes remain largely unknown. The starlet sea anemone, Nematostella vectensis, has oscillating patterns of locomotion and respiration, as well as the molecular components of a putative circadian clock that may provide a mechanism for these light-induced responses. Here, we compare transcriptomic responses of N. vectensis when cultured under a diurnal lighting condition (12 hours light: 12 hours dark) with sea anemones cultured under constant darkness for 20 days. More than 3,000 genes (~13% of transcripts) had significant differences in expression between light and dark, with most genes having higher expression in the photoperiod. Following removal of the light cue 678 genes lost differential expression, suggesting that light-entrained gene expression by the circadian clock has temporal limits. Grouping of genes differentially expressed in light:dark conditions showed that cell cycle and transcription maintained diel expression in the absence of light, while many of the genes related to metabolism, antioxidants, immunity, and signal transduction lost differential expression without a light cue. Our data highlight the importance of diel light cycles on circadian mechanisms in this species, prompting new hypotheses for the role of photoreception in major biological processes, e.g., metabolism, immunity.
Keywords: Nematostella, diel, gene expression, microarray, light:dark, gene-ontology
1. Introduction
Nearly all organisms on Earth exhibit biological rhythms driven by environmental cues, or Zeitgebers, set by biotic and abiotic factors (e.g., temperature and food availability), with solar (day/night) irradiance as a prominent natural cue. The endogenous, self-sustained oscillations in eukaryotes are propelled by core molecular machinery conserved across distinct phylogenetic lineages composed of three essential parts: the proteins that make up the clock itself, input environmental signals and regulatory proteins, and output genes/proteins (clock controlled genes, CCGs; Allada et al., 1998; Dunlap, 1999; Hoadley et al., 2016; Reitzel et al., 2010; Reppert, 2000). These components work together to entrain the clock to environmental cues and coordinate timing of clock-driven gene expression (Dunlap, 1999). The genetic basis of circadian rhythmicity has been defined in several model animals, including insects (Allada and Chung, 2010; Yuan et al., 2007), mammals (Buhr and Takahashi, 2013; Dodd et al., 2005; Panda et al., 2002), and more recently in sponges (Jindrich et al., 2017) and cnidarians (Hoadley et al., 2011; Reitzel et al., 2010; Vize, 2009, see below). Each of these studies identified several feedback loops, transcription factors, promoter regions, or regulator proteins related to circadian rhythmicity that are likely conserved deep in animal evolution (Allada and Chung, 2010; Sebens and DeRiemer, 1977; Yuan et al., 2007; Zhu et al., 2008).
Cnidarians (corals, jellyfish, sea anemones, hydroids) are an early diverging animal lineage that, as an outgroup to Bilateria, comprise an insightful system to study circadian biology from a comparative and functional context. The starlet sea anemone, Nematostella vectensis, has been developed as a model cnidarian due to its ease of laboratory culture and sequenced genome (Hand and Uhlinger, 1992; Hand and Uhlinger, 1994; Putnam et al., 2007), becoming a focal species for cnidarian circadian research (Hendricks et al., 2012; Oren et al., 2015; Peres et al., 2014; Reitzel et al., 2010; Reitzel et al., 2013b). In addition to its informative phylogenetic position, N. vectensis inhabits a highly variable estuarine environment, with seasonal and daily fluctuations in abiotic factors (Reitzel et al., 2013a; Sheader et al., 1997) known to influence behavior and physiology; e.g., gametogenesis, reproductive cycles, oxygen consumption and locomotion (Fritzenwanker and Technau, 2002; Hand and Uhlinger, 1994; Hendricks et al., 2012; Maas et al., 2016).
Our understanding of what molecular components are involved in the cnidarian clock is fairly limited (see reviews by Hoadley et al., 2016; Reitzel et al., 2013b). Initial studies have provided evidence that cnidarians possess light-sensing proteins responsible for photoreception and genes orthologous to core transcription factors, e.g., Clock and Cycle (Hoadley et al., 2011; Levy et al., 2007; Reitzel et al., 2010); they however lack the prototypical bilaterian clock repressors PER and TIM. Reitzel et al. (2010) described three cryptochromes in N. vectensis and determined that expression of two (NvCry1a and NvCry1b) are diel and likely play a role in the circadian regulatory pathway, whereas the potential involvement of other light-sensing proteins like opsins is yet to be determined (Reitzel et al., 2010; Suga et al., 2008). Although circadian cycling as it relates to light-dependent cues is responsible for numerous physiological, behavioral, and molecular processes, our understanding of which genes respond to daily signals is poor. Potential cnidarian CCGs have been studied in a few species of scleractinian corals: Acropora cervicornis (Hemond and Vollmer, 2015), Acropora millepora (Brady et al., 2011; Hoadley et al., 2011; Levy et al., 2007; Vize, 2009), and Favia fragum (Hoadley et al., 2011) as well as the sea anemone N. vectensis (Oren et al., 2015; Reitzel et al., 2010). Oren et al. (2015) characterized changes in gene expression in N. vectensis over a light:dark period and showed that approximately 180 genes had cyclic expression. Under light:dark conditions, potential CCGs that exhibit diel patterns of expression are from diverse gene families and can be categorized into particular functional subgroups; e.g., metabolism, cell cycle, immunity, sensory processes, and essential circadian CCGs. Previous studies that have compared gene expression in light:dark and constant dark conditions have suggested a mixed response in cnidarians. Reitzel et al. (2010) showed that long periods of darkness resulted in loss of cyclic gene expression for putative circadian clock genes in N. vectensis. Peres et al. (2014) showed that this loss of cyclic expression occurred over a few days, in a gene-dependent manner. In corals, three days of constant darkness resulted in loss of oscillating gene expression for F. fragum (Hoadley et al., 2011). Similarly, Brady et al. (2011) showed that brief periods of total darkness (one day) resulted in loss of cyclical gene expression for circadian clock genes in A. millepora. It is presently unclear how the prolonged absence of light impacts transcriptome-wide diel gene expression in cnidarians. In this study, we aim to explore genes influenced by the photoperiod that putatively govern daily cycles and circadian regulation in sea anemones.
Here, we compare transcriptome-wide differences in gene expression by N. vectensis cultured in light:dark and long-term constant dark conditions to differentiate molecular responses that may be due to diel lighting and circadian regulation, respectively. By comparing individuals in the middle of the day and night, we show that diel lighting strongly influences gene expression and the absence of light results in a decrease in the number of differentially expressed genes (DEG). Grouping of these genes into functional categories further supports the hypothesis that many key molecular processes are regulated by consistent diel lighting conditions.
2. Methods
2.1 Animal care and conditions
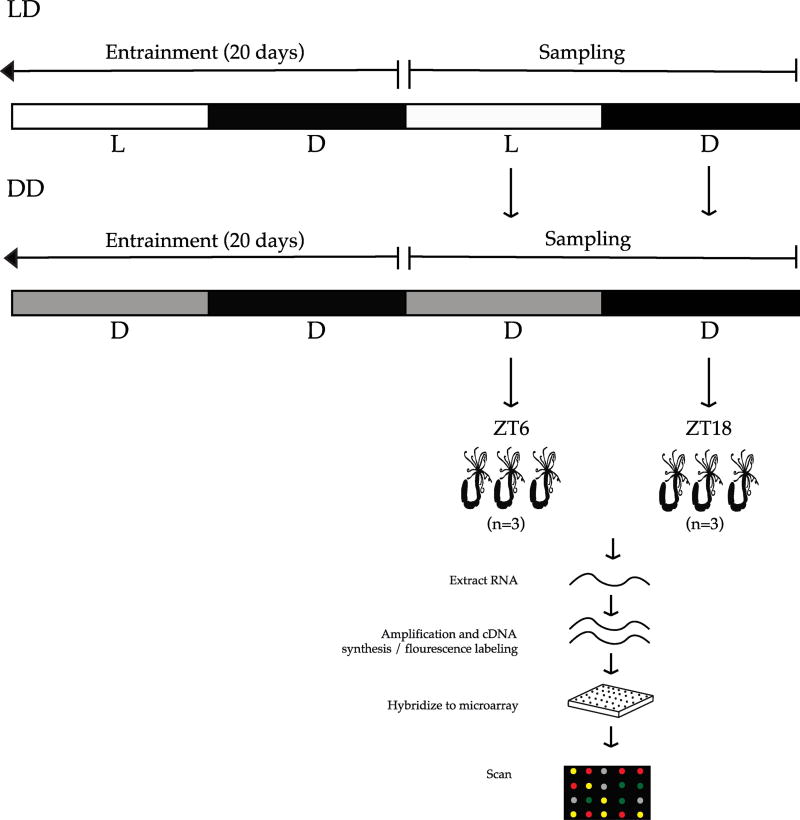
N. vectensis were cultured as described in (Peres et al., 2014). Briefly, adult sea anemones were kept under a 12:12 hour light:dark (LD) cycle – lights on at 7:00 hour (Hawaii standard time), Zeitgeber Time ZT0 – lights off at 19:00 hour; Zeitgeber Time ZT12 under full spectrum lights for 20 days. Animals were fed twice weekly and water was changed every day at ZT0, with no observable effect on the animals. A second group of individuals were kept in constant darkness (DD) for 20 days under the same conditions. Following exposure to respective light treatments, animals were sampled from LD and DD at two time points over a 24-hour cycle, once during subjective day at ZT6 and once during subjective night at ZT18 for gene expression analysis (Figure 1). Culturing, sampling, and animal transfer into RNAlater was done in the dark with minimal background lighting to eliminate the unknown effects of red light on the experiment.
Fig. 1. Experimental design for transcriptome wide profiling of diel expressed genes.
Nematostella vectensis were exposed to 12:12 hour light:dark cycle (LD) or constant dark conditions (DD) for 20 days prior to sampling, after which animals were preserved in RNAlater at two time points over a 24-hour period. RNA was extracted from whole animals, cDNA synthesized and labeled with fluorescent dye, and then prepared for hybridization to the custom microarray.
2.2 RNA extraction and cDNA synthesis
Three replicates per treatment were sampled at ZT6 and ZT18 and preserved in RNAlater. Total RNA was extracted from whole sea anemones using TriPure Reagent (Roche Inc., USA) according to the manufacturer’s specifications (Figure 1). Briefly, adult animals were lysed in 0.5 ml TriPure reagent and incubated for 5 minutes at room temperature. Two hundred µl of 1-bromo-3-chloropropane (BCP) was added to the tubes and centrifuged at 12,000×g for 15 minutes. The aqueous phase was transferred to a fresh tube, treated with DNase (Ambion, Life Technologies, Grand Island, NY, USA), and total RNA was pelleted by precipitation with isopropyl alcohol and centrifugation at 12,000×g for 10 minutes. The RNA pellet was washed with 75% ethanol and pelleted at 7,500×g for 5 minutes then air-dried at room temperature. RNA pellets were reconstituted in RNase-free water. RNA was quantified spectrophotometrically at 260 nm with 260/280 ratios between 1.8 and 2.0. RNA quality was also checked by 1.4% agarose gel electrophoresis stained with 5 µlml-1 ethidium bromide. Complementary DNA (cDNA) was synthesized with a Superscript III double-stranded cDNA synthesis kit (Invitrogen, USA), followed by Cy3 labeling (NimbleGen One-Color DNA Labeling Kit). Microarray hybridization was performed according to manufacturer's recommendations and scanned with a GenePix Personal 4100a scanner.
2.3 Microarray data processing
We used microarray analysis to discover differentially expressed genes during LD and DD light treatments. The 4-plex NimbleGen microarray chip contained 72,000 features providing complete coverage of the N. vectensis annotated transcriptome. The oligonucleotide-based chip (Roche) was custom designed as previously described (DuBuc et al., 2014; Layden et al., 2016) with three replicate probes per gene. The high-density oligonucleotide array dataset can be accessed from ArrayExpress under the accession number E-MTAB-6198. Our array results were analyzed in CLC Genomics Workbench 10.1.0 following a log-transformation (Log2) and quantile-normalization of raw expression values. Quality control was then performed on the normalized expression values by performing hierarchical clustering of features using Euclidean distance, and by clustering the criteria set to the average linkage. Statistical analysis of the clustered features was done using empirical analysis of differential gene expression to estimate tagwise dispersions with a total count filter cutoff of 5.0. The exact test parameters compared all pairs and p-values were False Discovery Rate (FDR) corrected.
2.4 Analysis of differentially expressed genes
In order to identify differentially expressed genes (DEGs) of interest we used a combination of a minimal fold change and significance values during LD and compared these with DD. DEGs that occur over a 12:12 LD cycle (p-value < 0.05 and absolute value of fold change greater than two) and not in constant darkness (p-value > 0.05 and absolute value of fold change less than two) were considered to follow a diel cycle. Genes that were differentially expressed when comparing LD and DD (p-value < 0.05 and absolute value of fold change greater than two) were also identified for the day (ZT6) and night (ZT18) time points (LD6 vs. DD6 and LD18 vs. DD18) respectively. Each subset of genes recovered in our microarray analysis were BLASTed against species with well characterized proteomes (Supplemental Table 1) to identify genes of interest related to diel cycling that have not been characterized in N. vectensis.
2.5 Gene annotation and functional analysis of differentially expressed genes
Information for DEGs of interest were retrieved and assigned putative function based on the N. vectensis genome project (Putnam et al., 2007). All of the genes were classified using the publicly available protein databases: Gene Ontology (GO) and Eukaryotic Clusters of Orthologous Groups of proteins (KOG). Genes were also screened via tBLASTn searches for potential circadian related genes that have been characterized in other taxa (Supplemental Table 1).
3. Results and Discussion
3.1 Differential gene expression
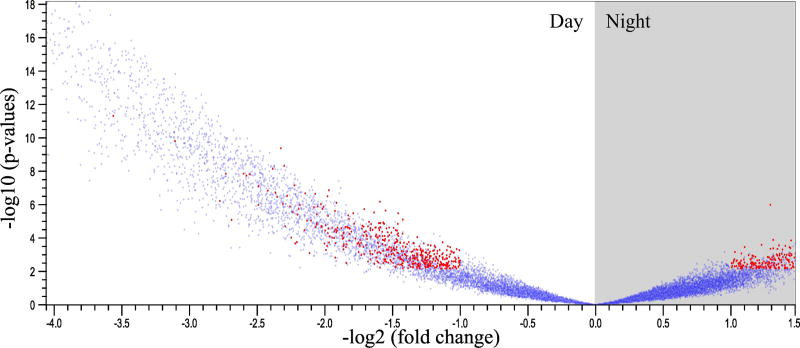
Using a transcriptome-wide microarray, we compared global changes in gene expression between N. vectensis exposed to diurnal light conditions and animals kept in constant darkness. Comparisons of gene expression at ZT6 and ZT18 in both conditions was separated into two analyses: time-dependent differences in gene expression in LD vs. DD (comparisons at mid-day and mid-night; LD6 vs. LD18) and differences in gene expression in a "light" period for each treatment (comparisons during mid-day; LD6 vs. DD6). Time-dependent diel expression of N. vectensis revealed a total of 3,138 DEG in a LD cycle (p-value <0.05, Figure 2; Supplemental Table 2). Comparisons of gene expression at ZT6 and ZT18 in each light treatment identified 253 and 13 DEGs, respectively (p-value <0.05, Supplemental Table 3-4).
Fig. 2. Volcano plot of differentially expressed genes in N. vectensis under oscillating light conditions.
The volcano plot shows transcriptome-wide oscillations in gene expression (blue dots) of approximately 24,000 genes, where ~13% were differently expressed in the light or dark when cultured under LD conditions (red dots).
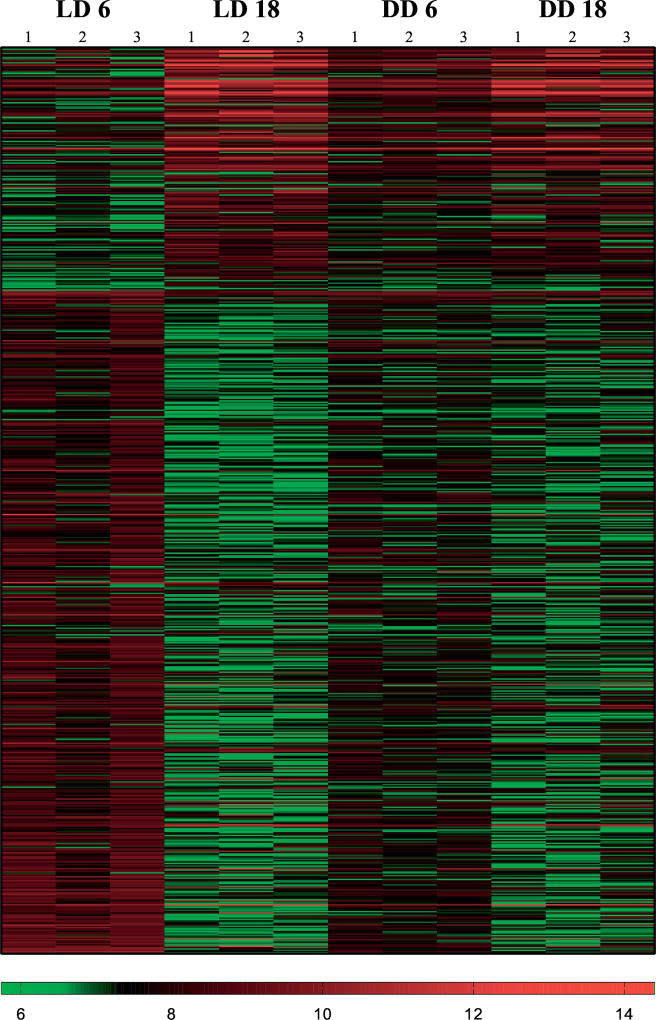
Functional categorization of DEGs focuses on differences in expression for genes in LD, but not in DD over our two sampling points. This subset of 678 genes lost diel expression in the absence of the light cue at these time points (LD6-18 vs. DD6-18; p-value <0.05). Of these, 467 were significantly upregulated during ZT6 (p-value <0.05, Figure 3; Supplemental Table 5). Due to the small number of genes assigned at ZT18, they were not considered in further analyses. Functional annotation and pathway analyses of DEGs between both LD and DD light treatment identified biological and molecular processes that provide insight to the daily cycles sea anemones exhibit (Table 1). Each transcript recovered at least one BLAST hit at NCBI (Supplemental Table 6) and was assigned to pathways involving signaling, metabolism, cell cycle, immunity, and others in broadly defined categories (Table 2). Next, we used a combination of analyses to connect the large differences in gene expression between these lighting treatments with potential functions and molecular pathways.
Fig. 3. Heat map of DEG in the absence of light.
Heat map of differentially expressed genes under 12:12 hour light:dark (LD) conditions or constant dark conditions (DD). Red bars show upregulated genes. Green bars show downregulated genes. Each column represents replicates in each light condition (LD, DD) at each time point (ZT6, ZT18).
Table 1.
Top Differentially Expressed Genes under LDvsDD
| Top | JGID | BLASTX | Fold Change⊥ | FDR p |
|---|---|---|---|---|
| 1 | 241170 | F-box B protein | −11.804 ↑ | 3.52E-10 |
| 2 | 83142 | Succinate dehydrogenase assembly factor 1 | −8.607 ↑ | 7.69E-09 |
| 3 | 241916 | Basic leucine zipper/WD-containing protein | −6.844 ↑ | 1.33E-05 |
| 4 | 215456 | XK-related protein | −6.638 ↑ | 4.30E-07 |
| 5 | 122916 | G-protein coupled receptor | −6.445 ↑ | 1.40E-04 |
| 6 | 247149 | Mitochondrial inner membrane protease | −6.061 ↑ | 4.30E-07 |
| 7 | 185016 | Histone 2A | −5.972 ↑ | 5.44E-07 |
| 8 | 96929 | Riboflavin-binding protein | −5.869 ↑ | 4.55E-07 |
| 9 | 185016 | Transcription elongation factor SPT4 | −5.620 ↑ | 2.10E-05 |
| 10 | 247038 | Alpha-CaTuliN (Catenin/vinculin related) | −5.615 ↑ | 2.02E-06 |
|
| ||||
| 1 | 110258 | Thrombospondin | 4.789 ↓ | 1.58E-06 |
| 2 | 200639 | Mucin-2 precursor | 4.711 ↓ | 4.56E-02 |
| 3 | 98287 | Extensin precursor (cell wall glycoprotein) | 3.975 ↓ | 3.63E-02 |
| 4 | 52111 | SH3 and multiple Ankyrin repeat domain | 3.887 ↓ | 2.91E-03 |
| 5 | 239622 | Neuropilin | 3.731 ↓ | 3.80E-03 |
| 6 | 201293 | Lysyl oxidase-like 3 | 3.535 ↓ | 4.23E-03 |
| 7 | 222647 | Solute carrier family 12 member 1 | 3.495 ↓ | 3.84E-02 |
| 8 | 115225 | Neurocalcin | 3.485 ↓ | 3.85E-02 |
| 9 | 222687 | Terminal uridylytransferase 4 | 3.458 ↓ | 5.04E-03 |
| 10 | 213978 | Outer defense fiber protein 3 | 3.339 ↓ | 1.30E-02 |
The presented fold-change value represents mean expression at ZT18 minus ZT6 for each gene. Thus, a negative fold-change indicates higher expression during the day time period of the 24-hour cycle, denoted by ↑.
Table 2.
Differentially Expressed Genes Involved in Major Pathways
| JGID | BLASTx | Fold Change⊥ | FDR p |
|---|---|---|---|
|
Antioxidant
| |||
| 103340 | Catalase (XP_001633745.1) | −3.57 ↑ | 2.10E-03 |
| 236349 | Superoxide dismutase (XP_001641079.1) | −7.68 ↑ | 3.95E-09 |
| 89807 | Thioredoxin (XP_001638442.1) | −12.72 ↑ | 5.42E-13 |
| 81388 | Glutathione peroxidase (XP_001641220.1) | −9.75 ↑ | 1.21E-12 |
| 195315 | HSP70 (XP_001622423.1)* | −3.81 ↑ | 1.32E-03 |
| 178640 | HSP90 (X P_001640689.1) | −6.16 ↑ | 1.59E-06 |
|
Cell Cycle and Regulation
| |||
| 110405 | PDZ binding protein (XP_001631518.1)* | 2.24 ↓ | 0.05 |
| 208897 | Dynein light chain Tctex-type 1 (XP_001619966.1)* | −3.08 ↑ | 3.55E-04 |
| 185341 | E3 ubiquitin-protein ligase MYLIP (XP_001633631.1)* | −2.55 ↑ | 6.75E-03 |
| 234384 | Adenylate cyclase (XP_001637931.1)* | −3.24 ↑ | 3.27E-03 |
| 110258 | Thrombospondin-1 (XP_001631080)* | 4.79 ↓ | 1.58E-06 |
| 98238 | Homeobox protein (XP_001635465.1)* | −2.63 ↑ | 2.81E-03 |
| 86719 | Astacin-like (XP_001639324.1)* | 2.02 ↓ | 0.04 |
| 114959 | Wnt signaling protein (XP_001630030.1)* | 2.04 ↓ | 0.04 |
| 2028 | Low-density lipoprotein receptor (XP_001623138.1)* | 2.53 ↓ | 0.03 |
|
Immunity
| |||
| 204961 | RIPL1-like protein (XP_001634829.1)* | −2.5 ↑ | 0.04 |
| 170435 | LBP (XP_001629157.1) | −3.43 ↑ | 6.33E-04 |
| 82163 | MyD88 (XP_001641005.1) | −8.64 ↑ | 2.77E-09 |
| 1816 | TIR (XP_001637587.1) | −6.78 ↑ | 5.44E-07 |
| 18 | Complement (C3) precursor (BAH22723.1) | −3.72 ↑ | 7.22E-05 |
| 196263 | MAC/PF (XP_001642092.1) | −2.84 ↑ | 3.57E-03 |
| 112390 | TRAF6 (XP_001630936.1) | 2.42 ↓ | 0.01 |
|
Metabolism
| |||
| 248519 | Malate dehydrogenase (XP_001622276.1) | −23.45 ↑ | 3.46E-14 |
| 186479 | Carbonic anhydrase (XP_001632619.1) | −3.48 ↑ | 2.37E-03 |
| 244812 | Fructose-bisphosphate aldolase (XP_001629735.1) | −2.93 ↑ | 6.27E-03 |
| 237552 | Ferritin H (XP_001624357.1) | −3.77 ↑ | 0.02 |
| 173595 | Glutamate transporter (XP_001625720.1) | −3.44 ↑ | 7.53E-04 |
|
Signal Transduction
| |||
| 50930 | Melanopsin (XP_001631416.1)* | 2.7 ↓ | 8.63E-03 |
| 101020 | HIOMT/melatonin (XP_001634470.1) | −5.49 ↑ | 5.18E-07 |
| 105524 | RAS GTPase (XP_001633090.1)* | −2.33 ↑ | 6.93E-03 |
| 40487 | Voltage-gated potassium channel (XP_001640129.1)* | 2.69 ↓ | 0.01 |
| 205216 | Rhodopsin-like receptor (XP_001634610.1)* | 2.24 ↓ | 0.03 |
| 118227 | Latrophilin receptor (XP_001620953.1) | −2.78 ↑ | 0.02 |
| 246569 | Voltage-gated calcium channel (XP_001626501.1)* | −2.13 ↑ | 3.66E-03 |
| 88314 | HIF1 (XP_001638872.1) | −16 ↑ | 5.83E-13 |
| 88486 | CK1δ/ε (XP_001638852.1)* | −2.25 ↑ | 0.01 |
| 248021 | bZIP CREB (XP_001623517.1) | −7.83 ↑ | 3.20E-08 |
| 242269 | bZIP DNA Binding Site (XP_001634376.1) | −20.78 ↑ | 1.71E-15 |
| 150375 | bZIP HLF (XP_001619763.1)* | −4.71 ↑ | 5.31E-05 |
|
Other
| |||
| 120351 | Frataxin (XP_001628857.1)* | −2.19 ↑ | 2.27E-03 |
| 238195 | TIMP (XP_001641336.1)* | −3.34 ↑ | 3.58E-03 |
Indicates maintenance of DE in the absence of a light cue.
The presented fold-change value represents mean expression at ZT18 minus ZT6 for each gene. Thus, a negative fold-change indicates higher expression during the daytime period of the 24-hour cycle, denoted by ↑.
3.2 Gene ontology analysis
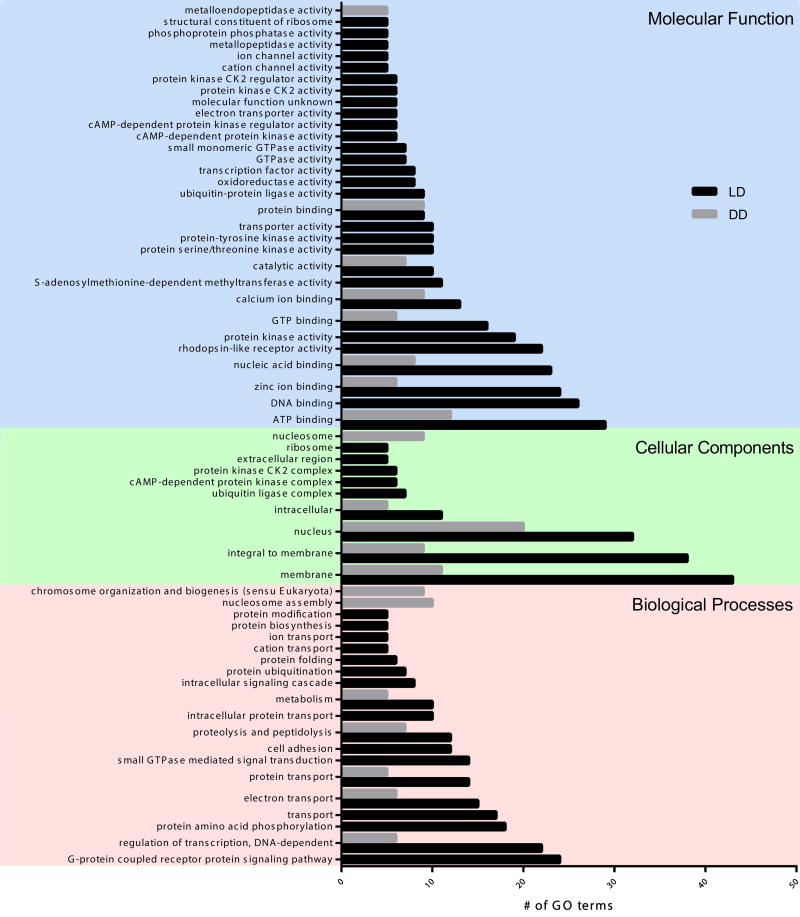
We used Gene-Ontology (GO) analysis to partition DEGs into broad functional categories in the biological processes, cellular components, and molecular function ontologies for both lighting treatments. Enrichment analysis showed significant diverse functional groups for each condition, with the primary difference being the number of genes expressed in each treatment (Figure 4). We considered only the subset of 678 genes that were differentially expressed (DE) between LD6 and LD18, and not in DD. Approximately half (340) of these DEGs were associated with 1,333 different GO groups. Two hundred fifty-three of these genes had significant differences in expression between LD6 and DD6 (Supplemental Table 3), of which 144 were associated with 625 GO groups. The difference in the number of GO terms assigned to each light treatment is shown in Figure 4. In the biological processes domain, DEG resulting from diel light conditions recovered predominantly genes related to regulation of transcription (GO:0006355; n=22), G-protein coupled receptors (GO:0007186; n=24), protein amino acid phosphorylation (GO:0006468; n=18), and transport (GO:0006810; n=17). In the molecular function domain, we recovered genes related to rhodopsin-like receptor activity (GO:0001584; n=22), ATP binding (GO:0005524; n=29), catalytic activity (GO:0003824; n=10), GTP binding and activity (GO:0005525; n=16), and zinc ion binding (GO:0008270; n=24). Genes related to other ion channels were significantly enriched under LD conditions, including calcium ion binding (GO:0005509; n=13), cation channel activity (GO:0005261; n=5), and ion channel activity (GO:0005216; n=5). In the cellular component domain of the same treatment, we recovered genes related to cAMP-dependent protein (GO:0005952; n=6), ubiquitin ligase complex (GO:0000151; n=7), and casein kinase activity (GO:0004680, GO:0004681; n=6). Furthermore, proteolysis and peptidolysis (GO:0006508; n=7) of the biological processes domain, nucleus (GO:0005634; n=20) of the cellular component domain, and ATP binding (GO:0005524; n=12) of molecular function domain remained enriched in DD, although not to the level of those under diel lighting (Figure 4). Genes related to G-protein coupled receptors (GO:0007186; n=0), rhodopsin-like receptor activity (GO:0001584; n=0), casein kinase activity (GO:0004680, GO:0004681; n=0) and the ion channel groups (GO:0005509, GO:0005261, GO:0005216; n=0) were not enriched in the absence of light. Genes from these pathways are discussed in greater detail later in the discussion section.
Fig. 4. Enriched Gene Ontology (GO) Category Distribution of DEG.
Classification of DEGs in LD (black bars) and DD (grey bars) treatment by Gene Ontology (GO) annotations and categorized into three main categories: biological processes, cellular component, and molecular function. A total of 340 genes were assigned GO terms in LD and 144 were assigned GO terms in DD. GO accession numbers are listed in Supplementary Figure 7. *There were no genes in LD18 vs. DD18 included for these comparisons (see Results).
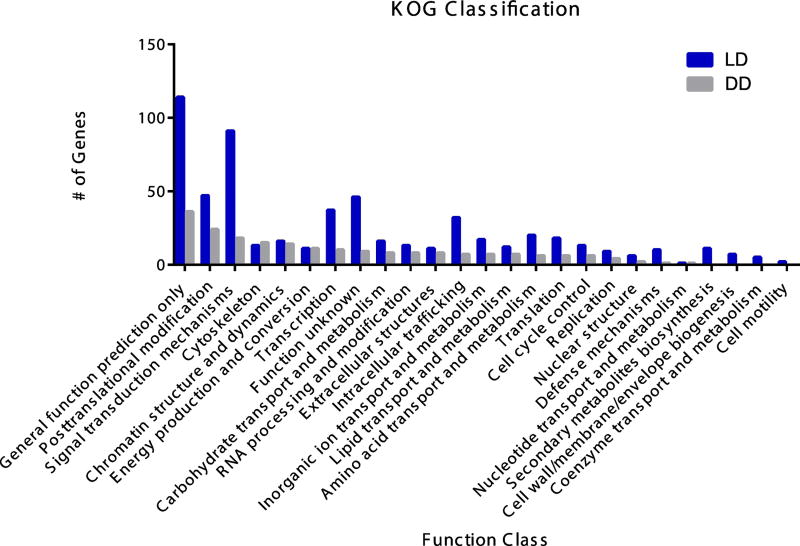
3.3 KOG classification
In order to address functionality, we used the Eukaryotic Clusters of Orthologous Groups database to assign DEGs to 25 categories under four broader groups (poorly characterized, metabolism, cellular processes, information storage and processing; Supplemental Table 7). In the subset of 678 genes that lost DE in the absence of light, 578 were identified with the three largest groups being: 1. general function prediction only (n=114), 2. signal transduction mechanisms (n=91), and 3. posttranslational modification/protein turnover/chaperones (n=114). Interestingly, only a few genes were assigned to nucleotide transport and metabolism (n=1) and cell motility (n=2). Between LD vs. DD at ZT6, 208 genes were assigned to 21 categories with the most genes falling under: 1. general function prediction only (n=36), 2. posttranslational modification/protein turnover/chaperones (n=24), and 3. signal transduction mechanisms (n=18). Defense mechanisms (n=1) and nucleotide transport and metabolism (n=1) had the fewest number of genes assigned. The principal difference between treatments is the number of genes assigned to signal transduction mechanisms, intracellular trafficking, and transcription (Figure 5).
Fig. 5. Eukaryotic Clusters of Orthologous Groups (KOG) Classification.
A total of 792 differentially expressed genes were assigned to 25 functional categories and are shown in functional categories of differentially expressed genes with LD (blue bars) and DD (grey bars) treated animals.
3.4 Genes of interest
Genes involved in signal transduction
We have identified several candidate genes that show differential expression under LD conditions that are likely related to light-entrained functions. G-protein coupled receptors (GPCRs) respond to extracellular molecules or light to begin signal transduction pathways that elicit biological responses. Once activated, G-proteins relay messages in the cell by modulating targets, such as ion channels, that initiate second messenger cascades (Panda et al., 2005). Presence of GPCRs in cnidarians (Plachetzki et al., 2007; Suga et al., 2008) suggests this pathway as a potentiator for environmental light information, however their role in circadian entrainment, if any, is unknown. Our analysis showed genes associated with GPCR signaling pathways that were DE in LD and DD conditions (Table 2), including latrophilin receptor (XP_001620953.1), several rhodopsin-like receptors (XP_001634610.1, Figure 4) and GABA neuropeptide receptors (XP_001631044.1, XP_001632665.1) that bind members of the GPCR family.
Our study showed differential expression of the GPCR melanopsin under LD conditions, and like many other signal transduction genes, retained this pattern of expression in the absence of light (Table 2). Melanopsin is a nonvisual light sensitive photopigment that uses calcium as a second messenger in the eyes of vertebrates to signal the SCN to entrain circadian clocks by stimulating release of melatonin from the pineal gland (Berson et al., 2002; Panda et al., 2005). The presence of the G-protein coupled opsin, melanopsin in N. vectensis has been established (Churcher and Taylor, 2010; Hendricks et al., 2012; Peres et al., 2014), though the functional role of this protein has not been characterized in cnidarians. Testing the role of melanopsins in cnidarians would be particularly informative for understanding how light is used to entrain circadian clocks in these organisms.
Genes involved in the GPCR phototransduction pathway were also identified by our microarray analysis to be DE in response to LD and DD conditions. Ion channels are activated by G-proteins to initiate second messenger cascades and may serve as regulators of circadian input/output pathways that influence an organism’s behavior reviewed by (Ko et al., 2009). For example, calcium is used as the second messenger for the GPCR melanopsin (discussed previously) and can entrain circadian clocks (Love et al., 2004; Panda et al., 2005). Several voltage-gated ion channels, including voltage-gated calcium channels, voltage-gated potassium channels, and transient receptor cation channels are DE in N. vectensis under diel conditions and in constant darkness (Table 2). N. vectensis’ nervous system has an extensive voltage-gated ion channel repertoire with high functional diversity, including paralogs of many neuronal signaling components found in complex metazoans (Jegla et al., 2012; Li et al., 2015). Circadian regulation by ion channels in cnidarians has not been explored, however studies in invertebrate models like Drosophila indicate that they are important in maintaining daily behavioral rhythms and their loss inhibits free running oscillations (Nitabach et al., 2002). This implies that in addition to transcription factors and other regulatory proteins needed for sufficient clock function, electrical activity may be an essential element to animal circadian clocks. Many of the ion channels DE in this analysis were not disrupted by the absence of light, while other signaling molecules failed to be DE in these conditions. This indicates both light-driven and circadian regulation may play a role in ion channel function and signaling in cnidarians, as is seen in vertebrates (Ko et al., 2009).
The hormone melatonin, known for regulation of sleep-wake cycles, utilizes GPCR melatonin receptors to mediate diverse cascade events including opening and closing ion channels, activating protein kinases, and driving changes in gene expression (Morgan et al., 1994). Melatonin has been identified in cnidarians (Anctil, 2009) and previous research with N. vectensis has suggested that melatonin is differentially expressed under LD conditions (Peres et al., 2014; Roopin and Levy, 2012). Our microarray analysis recovered genes involved in the melatonin synthesis pathway: hydroxyl-O-methyltransferase (HIOMT, XP_001634470.1) and members of the tryptophan hydroxylase and phenylalanine hydroxylase group (TPH/PaH, XM_001623795.1 and XP_001623845.1) that bookend the classical melatonin synthesis pathway and have been previously described in N. vectensis (Anctil, 2009; Peres et al., 2014; Roopin and Levy, 2012). Consistent with these earlier studies, HIOMT was significantly DE and downregulated during the scotophase of LD, while TPH/PaH had no change in expression over the 24-cycle. Roopin and Levy (2012) showed peak melatonin expression 5 hours into the scotophase while Peres et al. (2014) showed peak melatonin expression at ZT12 with a peak in HOIMT at ZT8 consistent with our results (Table 2, Supplemental Table 8). The authors additionally observed a measurable increase in PaH expression at the light:dark transition, but because our study had limited sampling points, we were not able to discern the peak of expression for this gene. As the precursor to melatonin, it is not surprising that HOIMT expression is high in advance of peak melatonin expression. Collectively, these studies support that melatonin is expressed in a light-dependent manner and likely plays a role in cnidarian diel gene expression; however, the extent of melatonin involvement in endogenous circadian control is not clear.
Other genes involved in signaling pathways that were DE in response to LD conditions in this study include enzymes responsible for phosphorylation/dephosphorylation: casein kinase (CK1δ/ε, XP_001638852.1) and protein phosphatase (PP1, XP_001632147.1). Members of the casein kinase (CK) family have been shown to phosphorylate key targets in circadian pathways in bilaterian model organisms (Lee et al., 1998), driving molecular oscillations by phosphorylating clock genes and promoting their translocation back to the nucleus. Oren et al. (2015) reported at least six members of the CK family in N. vectensis and showed that inhibition of a CK1δ/ε homolog disrupted diel behavior in this sea anemone. Our results showed a significant upregulation of CK1δ/ε during the day but this transcript continued to be DE in the absence of light (Table 2). The circadian circuitry of cnidarians has not been resolved, however the current hypothesis proposes cryptochromes as negative repressors of the clock (Hoadley et al., 2016; Reitzel et al., 2013b). It is reasonable to hypothesize that a member of the CK family works to regulate the negative feedback loop through phosphorylation of proteins like cryptochromes in cnidarians. Together, these studies further support a hypothesized role of protein kinases in circadian networks in cnidarians.
Genes involved in transcription
Transcription factors regulate gene expression, often in response to environmental stimuli, including light. The cnidarian bZIP family of transcription factors have conserved features with vertebrate and insect homologs and their role in gene regulatory networks is well characterized (Amoutzias et al., 2007; Cyran et al., 2003; Jindrich and Degnan, 2016). A subset of bZIPs in the PAR family are hypothesized to have roles in cnidarian circadian clocks (Brady et al., 2011; Reitzel et al., 2013b). Our analysis revealed DE of transcription factors in this family including CREB (XP_001623517.1), DNA binding site protein (XP_001634376.1), and HLF (XP_001619763.1), all with upregulated expression during the day (Table 2). Of these, CREB and DNA binding site protein did not retain a differential signal in the absence of light, indicating an important role of diel light cues for this family of transcription factors as was previously shown for PAR-bZIPs (Reitzel et al., 2013b).
Genes involved in metabolism and antioxidant activity
Although circadian clocks play significant roles in daily metabolism of bilaterians, little is known about the daily cycles in metabolism of cnidarians (Hoadley et al., 2016). Maas et al. (2016) showed that respiration rates in N. vectensis have a diel pattern when exposed to LD conditions, with peaks in oxygen consumption late during the light period, suggesting circadian control of respiration. The daily cycles of respiration were sustained over 6 days of constant conditions, indicating that N. vectensis is capable of maintaining a free-running metabolic rhythm (Maas et al., 2016). In contrast, our results showed that all genes related to metabolism failed to be DE in the absence of a light cue, but it is important to note that our study is not directly comparable due to differences in experimental design (20 days constant darkness) and sampling times (two time points). However, consistent with their physiological measurements, genes involved in metabolic pathways were DE in our analysis of the transcriptome when in LD. Genes involved in the glycolysis pathway and the citric acid cycle were identified to show diel expression with daytime peaks under LD conditions, but not in constant darkness. These were malate dehydrogenase (XP_001622276.1), carbonic anhydrase (XP_001632619.1), fructosebisphosphate aldolase (XP_001629735.1), and ferritin (XP_001624357.1, Table 2), which are responsible for the catalysis of malate to oxaloacetate in the citric acid cycle, conversion of carbon dioxide to bicarbonate to maintain pH, conversion of fructose-1,6-bisphophate to glyceraldehyde 3-phosphate in glycolysis, and iron storage and use, respectively. We also identified genes related to antioxidant activity DE in LD conditions but not in DD, including catalase (CAT, XP_001633745.1) and superoxide dismutase (SOD, XP_001641079.1). These genes scavenge reactive oxygen species that damage cells and were both upregulated during the day (Table 2), consistent with reports from Ruiz-Jones and Palumbi (2015) and Levy et al. (2007) in corals. Catalase and SODs were previously also shown to be differentially expressed in N. vectensis adults exposed to UV irradiation (Tarrant et al., 2014). Additional ‘defensome’ genes, thioredoxin (XP_001638442.1), glutathione peroxidase (XP_001641220.1), hypoxia inducible factor (HIF, XP_001638872.1), and heat shock proteins HSP70 (XP_001622423.1) and HSP90 (XP_001640689.1) were upregulated during the photoperiod (Goldstone, 2008; Reitzel et al., 2008). All genes related to antioxidant activity were only DE in LD conditions. Resolution of the involvement of light in these pathways would be informative in determining if N. vectensis exhibits endogenous circadian control or if light mediation is responsible for cycles of metabolism.
Genes involved in innate immunity
Cnidarians have a very limited repertoire of cells that are characterized as having immune function, namely circulating granular amoebocytes generally responsible for wound healing and ROS scavenging (Danielle and Smith, 1996). However, cnidarian genomes encode diverse genes associated with the innate immune pathway (Miller et al., 2007) including toll-like receptors or TLRs (Brennan et al., 2017). Our analysis showed some of these are differentially expressed in N. vectensis only under LD conditions. Candidates for the genes myeloid differentiation primary response 88 (MyD88, XP_001641005.1), lipid binding protein (LBP, XP_001629157.1) of the classic TLR pathway, and rab interacting lysosomal protein 1 (RILP-1, XP_001634829.1) were differentially expressed under LD conditions with an upregulation during the day but did not retain this expression pattern in the absence of light (Table 2). Additionally, we found members of the complement (C3 precursor, BAH22723.1) and membrane attack complex/perforin pathways (MAC/PF, XP_001642092.1) to have the same expression patterns. A gene matching TNF receptor associated factor (TRAF6, XP_001630936.1) was differentially expressed, however expression was highest during scotophase. Little research has been done to characterize the circadian regulation of innate immune pathways in invertebrates (Brennan et al., 2017; Stone et al., 2012), but our data indicates photoreception may be an important cue for cnidarian immune mechanisms, an area of future investigation.
‘Circadian’ Genes
Although we have identified several genes with differential expression in N. vectensis under LD conditions, canonical circadian genes (e.g., clock, cycle/bmal, cryptochromes, PARbZIP C) were not DE (Supplemental Table 8). Previous research involving these genes in cnidarians (reviewed in Hoadley et al., 2016) report oscillating expression in response to diel lighting conditions. It was not surprising that our study did not identify these genes as significantly differentially expressed, as they do not peak at ZT6 or ZT8. Due to limited sampling time points, we are unable to directly compare our results with other circadian experiments that sampled at multiple points during the photoperiod.
4. Conclusions
Our results describe a suite of molecular processes that are differentially regulated in response to the light environment. The decrease in differentially expressed genes when sea anemones are cultured in constant darkness indicates that consistent light cues are required to maintain oscillations for particular groups of genes. Previous research in N. vectensis and other cnidarians have shown that oscillations dampen or disappear for several hypothesized circadian clock genes when light is removed. Based on these combined data, light perception is a likely mediator in many molecular pathways that work synergistically to coordinate gene expression. We further support these observations by comparing across the transcriptome and have identified a subset of genes that did not retain significant differences in expression over a day period after individuals were in the dark for 20 days. While our study only samples at two time points, it is possible that some genes are not fully represented in our analysis due to simply not being at their peak or nadir at the time of sampling. A clear area of prospective future studies is to use a more frequent sampling protocol (as in Oren et al., 2015) in order to follow expression patterns of potential circadian genes that may peak before mid-day or mid-night. Further investigation of these candidate genes and the gene networks that connect them will provide new insights into the conservation of animal circadian clocks and its many functions. Moreover, additional research is needed to determine the role light plays in the entrainment of cnidarian circadian clocks and if cycles of expression in these candidate genes are mediated by environmental response to light or if they are endogenously generated.
Supplementary Material
Acknowledgments
WBL acknowledges and thanks the members of the Reitzel lab for support and Britney Phippen for assistance with analysis. Funding for this research was provided by NIH award R15GM114740 to AMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
All authors declare no competing interests.
Author’s contributions
WBL participated in data processing, interpretation, analysis and led drafting of the manuscript. JM and AMR contributed to data analysis and manuscript editing. RP participated in sample collection, experimentation, sample processing and data acquisition.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. An. Rev. of Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias G, Pichler E, Mian N, De Graaf D, Imsiridou A, Robinson-Rechavi M, Bornberg-Bauer E, Robertson D, Oliver S. A protein interaction atlas for the nuclear receptors: properties and quality of a hub-based dimerisation network. BMC Sys. Biol. 2007;1:34. doi: 10.1186/1752-0509-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: a genomic perspective. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2009;4:268–289. doi: 10.1016/j.cbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Sci. 2002;295:1070. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- Brady AK, Snyder KA, Vize PD. Circadian cycles of gene expression in the coral, Acropora millepora. PLoS ONE. 2011;6:e25072. doi: 10.1371/journal.pone.0025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD. Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-kappaB signal transduction, and development. Proc. Natl. Acad. Sci. 2017;114:E10122–e10131. doi: 10.1073/pnas.1711530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handb. of Exp. Pharm. 2013:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher AM, Taylor JS. The antiquity of chordate odorant receptors is revealed by the discovery of orthologs in the cnidarian Nematostella vectensis. Genome Biol. Evol. 2010;3:36–43. doi: 10.1093/gbe/evq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J. Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Danielle MCH, Smith VJ. Antibacterial properties of isolated amoebocytes from the sea anemone Actinia equina. Biol. Bull. 1996;191:441–451. doi: 10.2307/1543017. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Sci. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- DuBuc TQ, Traylor-Knowles N, Martindale MQ. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 2014;12:24. doi: 10.1186/1741-7007-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Fritzenwanker J, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa) Dev. Genes Evol. 2002;212:99–- 103. doi: 10.1007/s00427-002-0214-7. [DOI] [PubMed] [Google Scholar]
- Goldstone JV. Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis. Cell Biol. Toxi. 2008;24:483–502. doi: 10.1007/s10565-008-9107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand C, Uhlinger KR. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol. Bull. 1992;182:169–176. doi: 10.2307/1542110. [DOI] [PubMed] [Google Scholar]
- Hand C, Uhlinger KR. The unique, widely distributed, estuarine sea anemone, Nematostella vectensis Stephenson: A review, new facts, and questions. Estuaries. 1994;17:501. [Google Scholar]
- Hemond EM, Vollmer SV. Diurnal and nocturnal transcriptomic variation in the Caribbean staghorn coral, Acropora cervicornis. Mol. Ecol. 2015;24:4460–4473. doi: 10.1111/mec.13320. [DOI] [PubMed] [Google Scholar]
- Hendricks WD, Byrum CA, Meyer-Bernstein EL. Characterization of circadian behavior in the starlet sea anemone, Nematostella vectensis. PLoS One. 2012;7:e46843. doi: 10.1371/journal.pone.0046843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KD, Szmant AM, Pyott SJ. Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE. 2011;6:e19755. doi: 10.1371/journal.pone.0019755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KD, Vize PD, Pyott SJ. Current understanding of the circadian clock within cnidaria. In: Goffredo S, Dubinsky Z, editors. the cnidaria, past, present and future: the world of medusa and her sisters. Springer Intl. Pub.; 2016. pp. 511–520. [Google Scholar]
- Jegla T, Marlow HQ, Chen B, Simmons DK, Jacobo SM, Martindale MQ. Expanded functional diversity of shaker K+ channels in cnidarians is driven by gene expansion. PLOS ONE. 2012;7:e51366. doi: 10.1371/journal.pone.0051366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindrich K, Degnan BM. The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol. Biol. 2016;16:28. doi: 10.1186/s12862-016-0598-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindrich K, Roper KE, Lemon S, Degnan BM, Reitzel AM, Degnan SM. Origin of the animal circadian clock: diurnal and light-entrained gene expression in the sponge Amphimedon queenslandica. Front. Marine Sci. 2017:4. [Google Scholar]
- Ko GYP, Shi L, Ko ML. Circadian regulation of ion channels and their functions. J. Neurochem. 2009;110:1150. doi: 10.1111/j.1471-4159.2009.06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layden MJ, Johnston H, Amiel AR, Havrilak J, Steinworth B, Chock T, Röttinger E, Martindale MQ. MAPK signaling is necessary for neurogenesis in Nematostella vectensis. BMC Biol. 2016;14:61. doi: 10.1186/s12915-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, Hoegh-Guldberg O. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Sci. 2007;318:467–470. doi: 10.1126/science.1145432. [DOI] [PubMed] [Google Scholar]
- Li X, Martinson AS, Layden MJ, Diatta FH, Sberna AP, Simmons DK, Martindale MQ, Jegla TJ. Ether-à-go-go family voltage-gated K+; channels evolved in an ancestral metazoan and functionally diversified in a cnidarian–bilaterian ancestor. J. Expt. Biol. 2015;218:526. doi: 10.1242/jeb.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J, Dodd A, Webb A. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell. 2004;16:956–966. doi: 10.1105/tpc.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas AE, Jones IT, Reitzel AM, Tarrant AM. Daily cycle in oxygen consumption by the sea anemone Nematostella vectensis Stephenson. Biol. Open. 2016;5:161–164. doi: 10.1242/bio.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, Agata K, Bosch TC. The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PJ, Barrett P, Howell HE, Helliwell R. Melatonin receptors: localization, molecular pharmacology and physiological significance. Neurochem. Intl. 1994;24:101–146. doi: 10.1016/0197-0186(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Oren M, Tarrant AM, Alon S, Simon-Blecher N, Elbaz I, Appelbaum L, Levy O. Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci. Rep. 2015;5:11418. doi: 10.1038/srep11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nat. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Panda S, Nayak S, Campo B, Walker J, Hogenesch J, Jegla T. Illumination of the melanopsin signaling pathway. Sci. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- Peres R, Reitzel AM, Passamaneck Y, Afeche SC, Cipolla-Neto J, Marques AC, Martindale MQ. Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone Nematostella vectensis. Evodevo. 2014;5:26. doi: 10.1186/2041-9139-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Degnan BM, Oakley TH. The origins of novel protein interactions during animal opsin evolution. PLoS ONE. 2007;2:e1054. doi: 10.1371/journal.pone.0001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov V. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Sci. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS One. 2010;5:e12805. doi: 10.1371/journal.pone.0012805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Chu T, Edquist S, Genovese C, Church C, Tarrant AM, Finnerty JR. Physiological and developmental responses to temperature by the sea anemone Nematostella vectensis. Marine Ecol. Prog. Ser. 2013a;484:115–130. [Google Scholar]
- Reitzel AM, Sullivan JC, Traylor-knowles N, Finnerty JR. Genomic survey of candidate stress-response genes in the estuarine anemone Nematostella vectensis. Biol. Bull. 2008;214:233–254. doi: 10.2307/25470666. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM, Levy O. Circadian clocks in the cnidaria: environmental entrainment, molecular regulation, and organismal outputs. Integr. Comp. Biol. 2013b;53:118–130. doi: 10.1093/icb/ict024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopin M, Levy O. Temporal and histological evaluation of melatonin patterns in a 'basal' metazoan. J. Pineal Res. 2012;53:259–269. doi: 10.1111/j.1600-079X.2012.00994.x. [DOI] [PubMed] [Google Scholar]
- Sebens KP, DeRiemer K. Diel cycles of expansion and contraction in coral reef anthozoans. Marine Biol. 1977;43:247–256. [Google Scholar]
- Sheader M, Suwailem AM, Rowe GA. The anemone, Nematostella vectensis, in Britain: considerations for conservation management. Aquatic Cons. Marine and Freshwater Ecosy. 1997;7:13–25. [Google Scholar]
- Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila. PLoS Pathog. 2012;8:e1002445. doi: 10.1371/journal.ppat.1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. C. Biol. 2008;18:51–55. doi: 10.1016/j.cub.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Tarrant AM, Reitzel AM, Kwok CK, Jenny MJ. Activation of the cnidarian oxidative stress response by ultraviolet radiation, polycyclic aromatic hydrocarbons and crude oil. J. Exp. Biol. 2014;217:1444–1453. doi: 10.1242/jeb.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD. Transcriptome analysis of the circadian regulatory network in the coral Acropora millepora. Biol. Bull. 2009;216:131–137. doi: 10.1086/BBLv216n2p131. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, Reppert SM. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008;6:e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.