Abstract
Objectives
Many studies have attempted to discriminate patients with schizophrenia from healthy controls by machine learning using structural or functional MRI. We included both structural and diffusion MRI (dMRI) and performed random forest (RF) and support vector machine (SVM) in this study.
Methods
We evaluated the performance of classifying schizophrenia using RF method and SVM with 504 features (volume and/or fractional anisotropy and trace) from 184 brain regions. We enrolled 47 patients and 23 age- and sex-matched healthy controls and resampled our data into a balanced dataset using a Synthetic Minority Oversampling Technique method. We randomly permuted the classification of all participants as a patient or healthy control 100 times and ran the RF and SVM with leave one out cross validation for each permutation. We then compared the sensitivity and specificity of the original dataset and the permuted dataset.
Results
Classification using RF with 504 features showed a significantly higher rate of performance compared to classification by chance: sensitivity (87.6% vs. 47.0%) and specificity (95.9 vs. 48.4%) performed by RF, sensitivity (89.5% vs. 48.0%) and specificity (94.5% vs. 47.1%) performed by SVM.
Conclusions
Machine learning using RF and SVM with both volume and diffusion measures can discriminate patients with schizophrenia with a high degree of performance. Further replications are required.
Keywords: Classification, Diffusion MRI, Random forest, Support vector machine, Schizophrenia
Highlights
-
•
Schizophrenia was classified with dMRI and volume data by random forest and support vector machine.
-
•
Both machine learning methods show high classification performance.
-
•
There was no significant difference of classification between two methods.
1. Introduction
Patients diagnosed with schizophrenia demonstrate a wide variety of clinical symptoms including hallucinations, delusions, formal thought disorder, and cognitive dysfunctions (Sadock and Sadock, 2007; Weinberger and Harrison, 2011). Currently, diagnosing schizophrenia or evaluating the severity of the illness is determined based on clinical symptoms and an interview, without using objective biomarkers (American Psychiatric Association, 2013). Biomarker refers to objective indications of medical state observed from outside the patient, which can be measured accurately and reproducibly (Strimbu and Tavel, 2010). Also, it can be used to diagnose the disease or to predict the severity of the illness. Establishing diagnosis without using objective information may sometimes lead to misdiagnosis, which is affected by some factors such as the race and sex of the patients, and even the experience of the clinician (Green et al., 2012; Neighbors et al., 1989). In addition, psychotic symptoms cannot be easily determined based on an interview with the patients (Fanous et al., 2012). More objective biomarkers would be beneficial to help psychiatrists to diagnose and evaluate the illness. Many studies have tried to identify core pathological structural or functional changes in patients diagnosed with schizophrenia in order to establish biomarkers of the disease (Kubicki et al., 2007; Shenton et al., 2001).
While thousands of imaging studies have reported various structural as well as functional abnormalities in schizophrenia, these cannot be used as biomarkers at this time for several reasons: 1) some abnormalities are only present in some patients (Shenton et al., 2001), 2) there is not a clear separation between patients and healthy controls because the range of values in all measures, when compared separately, are both wide and overlapping, and, 3) patients in each study show very heterogenous clinical characteristics.
According to several theories, schizophrenia should be considered a brain disease where subtle changes in various brain locations coexist (van Erp et al., 2016). Traditional univariate methods, which focus on gross differences in one structure at a group level, cannot detect widely distributed or subtle changes in the brain (Orru et al., 2012). Because multiple measures have been introduced and reported to be abnormal in subsets of patients, and each explains only a small percentage of the variance, a multivariate approach should be considered for diagnosing schizophrenia using MRI. Furthermore, given the enormous number of values extracted from MRI images, machine learning approaches are needed to discriminate patients from healthy controls. Several such methods can be used to create classification models by means of pattern recognition of input data, i.e., linear discriminant analysis (LDA), support vector machine (SVM), and random forest (RF).
Some studies have previously been carried out to examine psychiatric diseases using T1, PET (positron emission tomography), or dMRI with SVM, LDA, or RF (Orru et al., 2012; Bansal et al., 2012). The performance rate for predicting schizophrenia using imaging data varies, however, depending on the statistical method and the population of patients used. For example, studies using structural MRI report a performance rate that ranges from 54 to 91% (Castellani et al., 2012; Davatzikos et al., 2005; Greenstein et al., 2012; Karageorgiou et al., 2011; Kasparek et al., 2011; Kawasaki et al., 2007; Mandl et al., 2013; Mourao-Miranda et al., 2012; Nieuwenhuis et al., 2012; Schnack et al., 2014; Sun et al., 2009; Takayanagi et al., 2011; Zanetti et al., 2013). Moreover, a recent meta-analysis revealed that, when compared to structural MRI, resting state functional MRI (fMRI) shows higher sensitivity (84% vs. 76%) and lower specificity (77% vs. 79%) (Kambeitz et al., 2015). Fewer studies using dMRI alone, or in conjunction with structural MRI, have been performed than have studies using structural or functional MRI (Arbabshirani et al., 2017).
Of note, Ardekani et al., (2011) reported a high rate of performance (96% sensitivity and 92% specificity for Fractional Anisotropy (FA) images, 96% sensitivity and 100% specificity for Mean Diffusivity (MD) images) when LDA was used. Using FA (Caan et al., 2006), reported a classification error of 25% in a cohort of 34 patients with schizophrenia and 24 controls, estimated using five-fold cross validation. On the other hand (Caprihan et al., 2008), achieved a classification error of 20% using a leave-one-out cross validation approach in a sample of 45 patients and 45 healthy volunteers. Other studies have reported a 70–91% performance rate including sensitivity and/or specificity (Ingalhalikar et al., 2010; Pettersson-Yeo et al., 2013; Rathi et al., 2010). While some studies with a small sample size have shown very high performance rates (≥95% sensitivity) (Ardekani et al., 2011; Fekete et al., 2013; Tang et al., 2012), a recent meta-analysis and review article reported the overall rate of accuracy for the classification of schizophrenia using imaging data ranges between 80.3% and 82% (Kambeitz et al., 2015; Arbabshirani et al., 2017).
Until now, most studies that have tried to predict schizophrenia have been performed with LDA or SVM using structural or functional MRI. In addition, studies with dMRI have mainly used diffusion measures in WM as features for classification. However, RF has shown a remarkable classifying ability regarding Alzheimer's disease with MRI data(Lebedev et al., 2014), possibly due to the following merits; 1) RF can estimate feature importance during training with little additional processes, providing better insight into the biological sense in the classification model; 2) RF is an ensemble of several decision trees and each tree is grown using a random subset of training sets and a random subset of features, which provides potential higher performance for the generalizations compared to decision tree; and, finally, 3) since RF produces non-linear decision boundaries due to the usage of decision tree, RF can outperform linear methods in capturing diverse patterns of structural or functional features that are distributed across the whole brain (Breiman, 2001; Venkataraman et al., 2010; Venkataraman et al., 2012). Despite of these merits of RF, its classification performance is not always better than other machine learning methods. It depends on the sample size, machine learning method, and features used in classification (Khondoker et al., 2016; Salvador et al., 2017; Katuwal et al., 2015).
DMRI is a method that is very sensitive to microstructural abnormalities (Beaulieu, 2002; Kanaan et al., 2005) including demyelination, axonal loss, edema, and inflammation (Assaf and Pasternak, 2008). Therefore, dMRI can detect subtle structural GM changes in intrinsic connections (Barbas and Pandya, 1989; Tardif and Clarke, 2001). In addition, some studies have reported that decreases in membranes, axon terminals, dendrites, and dendritic spines are among the causes of the reduced GM volume seen in schizophrenia (Bennett, 2011; Costa et al., 2001; Glantz and Lewis, 2000). In fact, some studies have reported abnormalities in the GM using dMRI (Lee et al., 2016; Lee et al., 2009; Moriya et al., 2010),
There are few studies using diffusion measures on GM as a feature for classification method. In this study, we evaluated the rate of performance for the classification of schizophrenia using the RF method and SVM method, with volume and dMRI measures in GM and WM as features, and we identified which structures were important for discriminating patients with schizophrenia from healthy controls.
2. Materials and methods
2.1. Participants and clinical variables
Subjects were enrolled from the Asan Medical Center, a university–affiliated hospital. Patients who were right-handed and were between the ages of 20 and 40 years old were eligible to participate in this study. Any patients with diseases that affect brain function were excluded. In addition, patients were excluded if they were unable to complete neuropsychological testing or MRI scanning sessions. Subjects within the patient group had a diagnosis of schizophrenia, which was determined by a psychiatrist according to the Diagnostic and Statistical Manual of Mental Disorders-IV-Text Revision (DSM-IV-TR) criteria. Moreover, they also displayed psychotic symptoms such as delusions or hallucinations for <5 years. In addition, subjects in the control group did not have any Axis I psychiatric diagnosis themselves or any first-degree relatives with an Axis I psychiatric diagnosis.
We enrolled 91 subjects in the study, but 11 cases were excluded due to poor image quality or incidental brain lesions. Ten patients were additionally excluded because their diagnoses changed to other psychotic disorders, such as bipolar disorder, when they were re-evaluated 1–6 months after the initial enrollment. The final dataset, consisting of 70 subjects (patients: N = 47; controls: N = 23), was used for the analysis.
Written informed consent was obtained from all subjects. Ethical approval for the study was obtained from the local Institutional Review Board, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
2.2. Assessment of symptoms and neurocognition
Assessment of symptoms, neurocognition, and social cognition was completed within one week of the date of the MRI examination. All subjects were evaluated using an age- and sex-adjusted short form of the Wechsler Adult Intelligence Scale-Third edition (WAIS-III), which consisted of 6 subtests including digit span, vocabulary, arithmetic, picture arrangement, block design, and digit symbol. The psychiatric symptoms of patients were evaluated by a psychiatrist using the Positive and Negative Syndrome Scale (PANSS).
2.3. MRI protocol and image processing
MRI scans were performed using an 8 channel SENSE head coil on a 3 Tesla scanner (Philips Achieva). DMRI images were acquired with an echo planar imaging (EPI) dMRI sequence. One baseline (b = 0) image and 32 diffusion gradient directions with b = 1000 s/mm2 were also acquired. The scan parameters were as follows: field of view (FOV): 224 ∗ 224 ∗ 135 mm, voxel size: 2 ∗ 2 ∗ 3 mm3, echo time (TE): 70 ms, flip angle: 90°, repetition time (TR): 5422 ms. Structural T1 MRI images with turbo field echo were acquired and the scan parameters were as follows: FOV: 240 ∗ 240 ∗ 170, voxel size 1 ∗ 1 ∗ 1 mm3, TE: 4.6 ms, TR: 9 ms, flip angle: 8°.
DMRI images were upsampled to 1 ∗ 1 ∗ 1 mm3 voxel size using Slicer V. 4.4 (Fedorov et al., 2012). Subsequently, diffusion tensor imaging (DTI) images were calculated with weighted least squares with an added procedure to correct tensors with negative eigenvalues from dMRI. To exclude the meninges or cerebral spinal fluid (CSF), we eroded the boundary voxels of each DTI image. Using Slicer V. 4.4 and an in-house program, we estimated FA and Trace (TR, equal to 3*MD) images from DTI images.
T1 images were parcellated into discrete anatomical regions using the Desikan-Killiany atlas of FreeSurfer V. 5.3 (Fischl et al., 2002). This was followed by non-linear registration of T1 images with a b = 0 baseline images of dMRI by Advanced Neuroimaging Tools (Avants et al., 2010; Avants et al., 2011). The same registration transformation was then applied to FreeSurfer parcellated labels. We then extracted the volume and/or mean diffusion measures (FA and TR) of 184 ROIs and used these values as features for classification. The volume of each ROI was corrected using individual total cranial volume. We have described all 504 features in Supplementary Table 1.
2.4. Resampling, classification with random forest and support vector machine
Since our dataset was unbalanced, with the patient group about twice as large as that of the healthy control group, and classification with RF and SVM results may be biased by an unbalanced distribution of the sample, we resampled our data into a balanced dataset (46 patients and 46 controls) using the Synthetic Minority Oversampling Technique (SMOTE) (Chawla et al., 2002). The SMOTE is one of the most popular methods for addressing the issue of unbalance and the general idea is to artificially generate cases of the minority class using K-nearest neighbors algorithm of these cases. In addition, to make a more balanced dataset, the majority class cases are also under-sampled (Chawla et al., 2002).
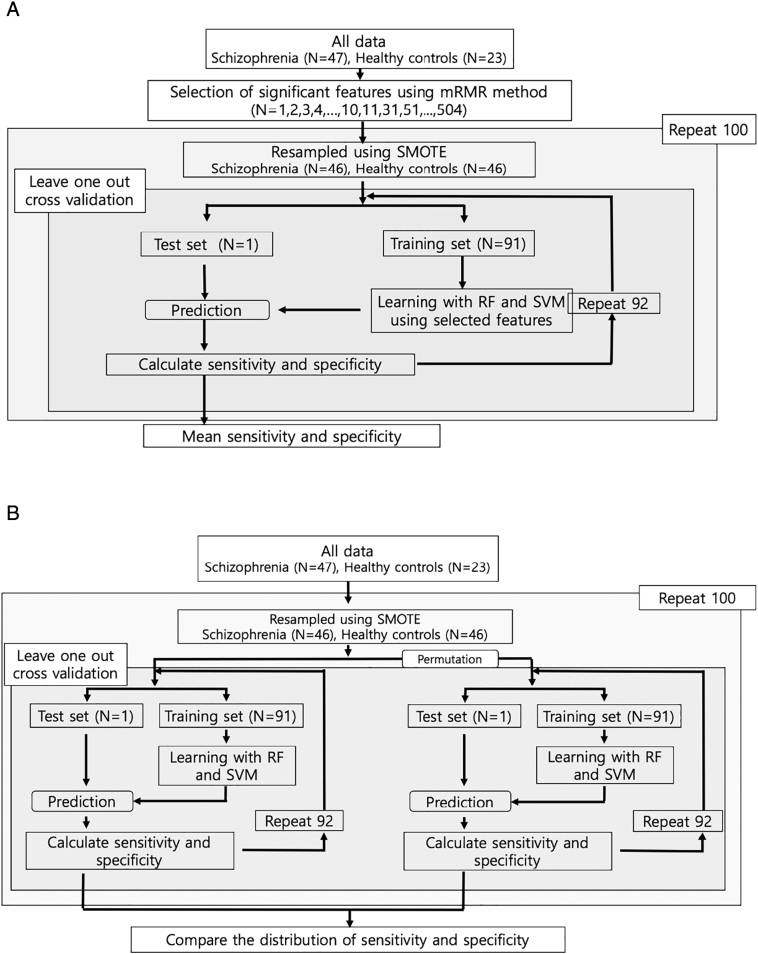
Because there might be a chance of correlation among the features, we performed maximum relevance minimum redundancy (mRMR, feature selection method proposed by Peng et al., (2005)) method to select the number of important features for classification from all features (N = 504). Then we gradually increased the number of features (N = 1, 2, 3, …, 9, 10, 11, 31, 51, …, 491, 504) from the total number of features. After that, we performed learning and classification using these selected features and investigated the performance of later classification. (Fig. 1a).
Fig. 1.
a. Flow chart of classifications of RF and SVM using selected features by mRMR method
Abbreviations: RF (Random forest), SVM (Support vector machine), mRMR (maximum relevance minimum redundancy)
b. Flow chart of comparisons of classification performances of RF and SVM between original class and randomly permuted class
Abbreviations: RF (Random forest), SVM (Support vector machine).
In addition, to test the statistical significance of the classification using RF and SVM, we randomly permuted the classification of all participants as a patient or healthy control 100 times and ran the RF and SVM with leave-one-out cross-validation (LOOCV, special case of k-cross validation when k is 92) for each permutation. (Fig. 1b) We then compared the sensitivity, and specificity of the original data set and the permuted data set.
The parameters of RF used during the learning phase were as follows: number of trees (5000), number of features randomly selected at each node (square root of the number of features, 22 in our study), and size of the node (Sadock and Sadock, 2007). Size of the node in random forest is the minimal number of observations allowed in the terminal nodes of each tree, which indirectly limits the tree size in the RF method. The test sets were predicted to be a patient or a control using the model from the learning phase of the RF. By comparing the real classification of participants (patient or healthy control) with the classification predicted by the RF and SVM, we calculated measures of performance such as sensitivity and specificity using the following formulas: sensitivity = (number of true positives)/(number of true positives + false negatives), specificity = (number of true negatives)/(number of true negatives + false positives). Since the RF generates the Out of Bag (OOB) error, we calculated the OOB error estimate.
2.5. Comparisons of volume, FA, and TR in important predictors between groups
As described above, we sorted the importance of features using mRMR method. We summarized the list of 20 important features and compared their values between schizophrenia patients and healthy controls to verify that these features make biological sense.
2.6. Statistical analysis
All statistical analyses were performed using R (ver. 3.4.1) (R Core Team, 2017), DMwR (ver. 0.4.1) for the SMOTE method (Torgo, 2010), random forest packages (ver. 4.6–12) (Liaw and Wiener, 2002), and e1071 for SVM (ver. 1.6–8) (e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R package version 1.6–8, 2017).
3. Results
3.1. Summary of demographic and clinical characteristics
We enrolled 70 participants (23 healthy controls and 47 patients) in this study. There was no significant difference between groups for males (controls: 34.8%, patients: 38.3%, X2 = 0.082, p = 0.775) and for age (controls: 29.70 ± 5.15, patients: 28.68 ± 6.23, t = 0.676, p = 0.501). Patients had a decreased adjusted IQ score compared to healthy controls (controls: 120.39 ± 9.32; patients: 97.91 ± 15.84; t = 7.44, p < 0.0001, Welch's t-test). The total PANSS score in patients was 61.11 ± 14.92 (positive syndrome score: 15.91 ± 6.51; negative syndrome score: 16.77 ± 7.08; general score: 28.43 ± 6.82). The olanzapine equivalent dose of atypical antipsychotics at the time of the MRI scan was 15.52 ± 8.60 mg/day (N = 45), and the olanzapine equivalent dose of typical antipsychotics at the time of the MRI scan was 19.33 ± 26.58 mg/day (N = 3). The duration of illness in the patients was 1.02 ± 1.58 years (Table 1).
Table 1.
Demographic and clinical information.
| Healthy control | Schizophrenia | Healthy control vs. Schizophrenia | |
|---|---|---|---|
| Number of subjects | 23 | 47 | |
| Number of Males (%) | 8 (34.8%) | 18(38.3%) | X2 = 0.082, p = 0.775 |
| Age (years) | 29.70 ± 5.15 | 28.68 ± 6.23 | t = 0.676, p = 0.501 |
| IQ | 120.39 ± 9.32 | 97.91 ± 15.84 | t = 7.44, p < 0.0001a |
| Duration of illness (years) | 1.02 ± 1.58 | ||
| PANSS | |||
| Total score | 61.11 ± 14.92 | ||
| Positive score | 15.91 ± 6.51 | ||
| Negative score | 16.77 ± 7.08 | ||
| General score | 28.43 ± 6.82 | ||
| Olanzapine equivalent dose of antipsychotics at time of MRI scan (mg/day) | |||
| Atypical antipsychotics (N = 45) | 15.52 ± 8.60 | ||
| Typical antipsychotics (N = 3) | 19.33 ± 26.58 | ||
Analyzed by Welch's t-test.
3.2. Classification results of RF and SVM by the number of used features
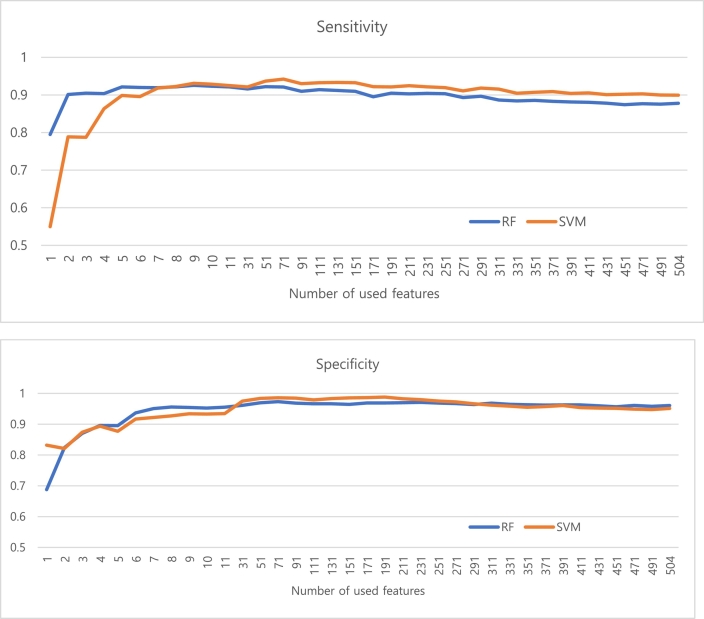
Generally, RF and SVM showed the highest sensitivity and specificity when used approximately 71 selected features (Fig. 2).
Fig. 2.
Performance of classification (upper: sensitivity, lower: specificity) based on the number of used features
Abbreviations: RF (Random forest), SVM (Support vector machine).
3.3. Permutation results
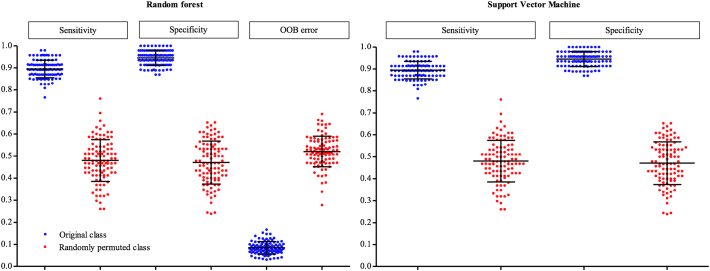
When the performance rate of the classification of the original group and the randomly permuted group were compared, the performance rate for the original group was significantly higher than the randomly permuted group (Table 2 and Fig. 3).
Table 2.
Comparisons of the performance rate of the classification of the original group and the randomly permuted group.
| Random forest (%, Mean ± SD (95% CI)) |
Support vector machine (%, Mean ± SD (95% CI)) |
|||
|---|---|---|---|---|
| Original class | Random class | Original class | Random class | |
| Sensitivity | 87.6 ± 4.2 (86.8–88.4) | 47.0 ± 8.3 (45.3–48.6) | 89.5 ± 3.9 (88.7–90.3) | 48.0 ± 9.4 (46.1–49.9) |
| Specificity | 95.9 ± 2.8 (95.3–96.4) | 48.4 ± 8.2 (46.8–50.1) | 94.5 ± 3.4 (93.8–95.2) | 47.1 ± 9.7 (45.2–49.0) |
| OOB error | 8.5 ± 2.8 (7.9–9.0) | 52.1 ± 6.9 (50.7–53.5) | ||
Abbreviations: OOB (Out-of-Bag), SD (standard deviation), CI (confidence interval).
Fig. 3.
Comparison of the distribution of performance between the original group and the randomly permuted group. The black lines indicate the mean and standard deviation of performance.
Abbreviations: OOB (Out-of-Bag).
3.4. Comparison values for significant predictors between the two groups
mRMR method showed the level of importance of all predictors in order to discriminate patients with schizophrenia from healthy controls. The values of the top 20 most important ROIs were compared using the Wilcoxon rank sum test between the two groups (Table 3).
Table 3.
Comparison of the values of the most important ROIs (N = 22) which most significantly contributed to the classification.
| Nob | GM/ WM/ Sub |
Value | Side | Location | Mean ± SD (*10–3) |
Rank sum | p | |
|---|---|---|---|---|---|---|---|---|
| controls | patients | |||||||
| 1 | GM | TR | Rt | Middle temporal | 2.6 ± 0.09 | 2.8 ± 0.12 | 249 | 0.0003 |
| 2 | Sub | TR | Lt | Ventral DC | 3.0 ± 0.1 | 2.9 ± 0.1 | 817 | 0.0006 |
| 3 | GM | Vola | Lt | Parsopercularis | 5.0 ± 0.5 | 4.6 ± 0.5 | 774 | 0.0036 |
| 4 | WM | Vola | Rt | Inferior temporal | 6.4 ± 0.7 | 6.1 ± 0.4 | 730 | 0.0181 |
| 5 | Sub | Vola | Rt | Inf. Lat. Ventricle | 0.3 ± 0.1 | 0.5 ± 0.2 | 319 | 0.0057 |
| 6 | Sub | Vola | Lt | Hippocampus | 4.2 ± 0.4 | 3.9 ± 0.4 | 799 | 0.0013 |
| 7 | WM | FA | Lt | Transverse temporal | 327.6 ± 36.8 | 353.0 ± 41.3 | 349 | 0.0169 |
| 8 | Sub | Vola | Both | 4th Ventricle | 1.4 ± 0.4 | 1.8 ± 0.5 | 332 | 0.0093 |
| 9 | WM | FA | Lt | Precuneus | 396.6 ± 17.0 | 383.6 ± 22.2 | 709 | 0.0357 |
| 10 | WM | Vola | Lt | Lingual | 6.1 ± 0.7 | 5.6 ± 0.7 | 784 | 0.0024 |
| 11 | GM | TR | Lt | Caudal anterior cingulate | 2.7 ± 0.2 | 2.8 ± 0.2 | 291 | 0.0018 |
| 12 | GM | TR | Rt | Inf. Temporal | 2.5 ± 0.1 | 2.6 ± 0.1 | 280 | 0.0011 |
| 13 | WM | Vola | Lt | Inferior parietal | 11.2 ± 1.1 | 10.4 ± 1.0 | 748 | 0.0096 |
| 14 | Sub | Vola | Both | CC Anterior and middle | 0.6 ± 0.2 | 0.5 ± 0.1 | 666 | 0.1180 |
| 15 | GM | Vola | Rt | Parsopercularis | 4.3 ± 0.6 | 3.9 ± 0.5 | 758 | 0.0067 |
| 16 | GM | TR | Rt | Caudal anterior cingulate | 2.5 ± 0.1 | 2.6 ± 0.1 | 341.5 | 0.0131 |
| 17 | GM | FA | Rt | Middle temporal | 152.8 ± 7.2 | 146.4 ± 10.0 | 762 | 0.0057 |
| 18 | WM | Vola | Lt | Frontal pole | 0.2 ± 0.05 | 0.2 ± 0.04 | 735 | 0.0153 |
| 19 | Sub | Vola | Both | 3rd Ventricle | 0.8 ± 0.2 | 1.1 ± 0.4 | 290 | 0.0018 |
| 20 | WM | FA | Lt | Precentral | 393.7 ± 16.8 | 410.0 ± 25.8 | 350 | 0.0175 |
Analyzed by Wilcoxon rank sum test.
Abbreviations: GM, Gray matter; Sub, Subcortical structure; Whole, Whole brain; WM, White matter; FA, Fraction anisotropy; TR, Trace; Vol, Volume; Rt, Right; Lt, Left; Inf, Inferior; Lat, Lateral; Vent, Ventricle; Ventral DC, Ventral Diencephalon; CC, corpus callosum.
Corrected by estimated total intra-cranial volume.
No., order of importance of features to discriminate the patients from the healthy controls.
4. Discussion
This study shows that patients with schizophrenia can be classified with high sensitivity and specificity by two different machine learning methods (RF and SVM) when 504 features from volume, FA, and TR of the brain structure are used, including GM, WM, and subcortical structures. Our study showed a high rate of performance (sensitivity: 87.6%, 89.5%; specificity: 95.9%, 94.5%; RF, SVM respectively), which is higher than the results found by meta-analysis (Kambeitz et al., 2015). Most studies of recent onset schizophrenia or first-episode psychosis (FEP) reported 66–91.5% performance (sensitivity or specificity) (Karageorgiou et al., 2011; Kasparek et al., 2011; Mourao-Miranda et al., 2012; Sun et al., 2009; Takayanagi et al., 2011; Zanetti et al., 2013; Pettersson-Yeo et al., 2013; Rathi et al., 2010), which is lower than that with chronic schizophrenia (Kambeitz et al., 2015). Taking these previous studies into account, our results have more important implications. However, we should state that the application of our results to a general population of patients with schizophrenia should be carried out with caution since our small sample could not include all heterogeneous cases. Nonetheless studies that previously reported high performance (above 95% sensitivity) had even smaller sample sizes (Ardekani et al., 2011; Fekete et al., 2013; Tang et al., 2012) although a recent review paper has raised a concern about the issue of generalizability of studies with small sample size reported high performance (Arbabshirani et al., 2017). Larger study samples may include heterogeneous MRI information from both patients and healthy controls. Therefore, this increased heterogeneity can decrease the performance of the classification model.
Large training data sets, on the other hand, usually increase the performance of the classification system, and classification based on a small sample size is more unstable (Nieuwenhuis et al., 2012; Arbabshirani et al., 2017; Franke et al., 2010). We performed 100 repetitions of LOOCV to investigate the possibility of unstable results occurring due to a small sample size. One hundred repetitions of LOOCV also showed a very small range in the 95% confidence interval of sensitivity and specificity (RF: 86.8–88.4%, 95.3–96.4%, respectively; SVM:88.7–90.3%, 93.8–95.2%, respectively). This means that RF and SVM produced very stable classification. Of note, overfitting is one of important issues in machine-learning especially when trying to estimate parameters during the learning phase with small sample size and relatively large numbers of features as in our study (Arbabshirani et al., 2017). We could not find significant drop of overall performance rate when we repeatedly performed random forest with different numbers of features during the learning phase. We can assume that overfitting may therefore not significantly affect findings in our study.
We also tried to classify the cases using only MRI information. However, using MRI data alone may not be sufficient to classify cases in the early stages of the disease. Pina-Camacho et al. compared the rate of performance for predicting early onset FEP to develop into schizophrenia spectrum disorders (SSD) using different combinations of baseline clinical, neuropsychological, MRI, and biochemical information. Neuroimaging and biochemical information at baseline did not provide additional predictive value to the classification of developing SSD from FEP (Pina-Camacho et al., 2015). Besides early stages of disease, several studies have reported a more accurate rate of classifying schizophrenia when a combination of MRI and other information were used, such as genetic information (Greenstein et al., 2012) and neuropsychological results (Karageorgiou et al., 2011).
Several limitations should be noted in our study. First, classification may be carried out based on features of MRI that are related to brain abnormalities, such as those that can be indexed by IQ, and are not specific to schizophrenia. Previous studies have reported that patients with schizophrenia have a lower IQ compared to healthy controls (David et al., 1997) and IQ levels correlate with FA and MD (Fryer et al., 2008). Patients in our study also had a significantly lower IQ than healthy controls. Therefore, RF and SVM may be able to discriminate the cases with low IQ from the cases with high IQ, rather than discriminating subjects with schizophrenia from healthy controls. While it is not easy to control the effect of IQ on MRI, we regressed out IQ from all 504 features (volume, FA, and TR values). Then we ran 100 times LOOCV using RF with 504 features. Mean sensitivity and specificity was 82.8% and 92.7% respectively, which was slightly lower than those results (87.6% and 94.5%) of RF without adjustment of IQ. In addition to IQ, although there was no significant difference of age and gender between patients and healthy controls, these factors may be leading to overoptimistic estimates. The second limitation is that the structural differences that were used during the learning and classification phase may not be core pathological features of schizophrenia. Instead, these may be structural differences induced by several factors such as medications or life style (Dazzan et al., 2005; Jorgensen et al., 2015; Lieberman et al., 2005; Vita et al., 2015). Therefore, classification was carried out based on MRI differences that may not be related to core pathological changes. The last limitation is that, as previously mentioned, the sample size in our study was relatively small. The results in this study showed high prediction accuracy, while there was a chance that several confounding factors such as age, sex, medication or IQ might be leading to overoptimistic estimates. In addition, overfitting might happen during RF procedures due to relatively homogenous samples. Although we analyzed our sample data by cross-validation and permutation to overcome the problems, there is still a high risk of overfitting. It should be considered that even though the results showed high classification performance, their generalizability is limited. Our study used both volume and diffusion MRI data and performed RF method to discriminate patients with schizophrenia from healthy controls. The results of this study should be considered preliminary and require further replications.
5. Conclusion
We were able to accurately discriminate the patients with recent onset schizophrenia from healthy controls using volume, FA, and TR of GM and WM with an RF and SVM methods. In order to generalize our results further, more studies are needed with larger sample sizes that include patients with heterogeneous symptoms.
The following is the supplementary data related to this article.
List of features.
Funding sources
This study was supported by the National Research Foundation of Korea (NRF-2012R1A1A1006514 and NRF-2017R1D1A1B03032707 to JL), Veterans Affairs Merit Award (I01 CX000176-06 to MES), National Institute of Mental Health (U01MH109977 to MES, SB, MK and YR, R01MH097979 to YR, R01MH102377 to MK and K24MH110807 to MK) and Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD090641 to SB).
Conflict of interest
All authors declare that they have no conflict of financial interest.
Acknowledgement
None.
References
- American Psychiatric Association . 5th ed. American Psychiatric Association; Arlington, VA: 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage. 2017;145(Pt B):137–165. doi: 10.1016/j.neuroimage.2016.02.079. (Jan 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardekani B.A., Tabesh A., Sevy S., Robinson D.G., Bilder R.M., Szeszko P.R. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum. Brain Mapp. 2011;32(1):1–9. doi: 10.1002/hbm.20995. (Jan) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y., Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J. Mol. Neurosci. 2008;34(1):51–61. doi: 10.1007/s12031-007-0029-0. [DOI] [PubMed] [Google Scholar]
- Avants B.B., Yushkevich P., Pluta J., Minkoff D., Korczykowski M., Detre J., Gee J.C. The optimal template effect in hippocampus studies of diseased populations. NeuroImage. 2010;49(3):2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. (Feb 01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. (Feb 01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R., Staib L.H., Laine A.F., Hao X., Xu D., Liu J., Weissman M., Peterson B.S. Anatomical brain images alone can accurately diagnose chronic neuropsychiatric illnesses. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H., Pandya D.N. Architecture and intrinsic connections of the prefrontal cortex in the rhesus monkey. J. Comp. Neurol. 1989;286(3):353–375. doi: 10.1002/cne.902860306. (Aug 15) [DOI] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15(7–8):435–455. doi: 10.1002/nbm.782. (Nov–Dec) [DOI] [PubMed] [Google Scholar]
- Bennett M.R. Schizophrenia: susceptibility genes, dendritic-spine pathology and gray matter loss. Prog. Neurobiol. 2011;95(3):275–300. doi: 10.1016/j.pneurobio.2011.08.003. (Nov) [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. [Google Scholar]
- Caan M.W., Vermeer K.A., van Vliet L.J., Majoie C.B., Peters B.D., den Heeten G.J., Vos F.M. Shaving diffusion tensor images in discriminant analysis: a study into schizophrenia. Med. Image Anal. 2006;10(6):841–849. doi: 10.1016/j.media.2006.07.006. (Dec) [DOI] [PubMed] [Google Scholar]
- Caprihan A., Pearlson G.D., Calhoun V.D. Application of principal component analysis to distinguish patients with schizophrenia from healthy controls based on fractional anisotropy measurements. NeuroImage. 2008;42(2):675–682. doi: 10.1016/j.neuroimage.2008.04.255. (Aug 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani U., Rossato E., Murino V. Classification of schizophrenia using feature-based morphometry. J. Neural Transm. 2012;119(3):395–404. doi: 10.1007/s00702-011-0693-7. (Mar) [DOI] [PubMed] [Google Scholar]
- Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority oversampling technique. J. Artif. Intell. Res. 2002;16:321–357. [Google Scholar]
- Costa E., Davis J., Grayson D.R., Guidotti A., Pappas G.D., Pesold C. Dendritic spine hypoplasticity and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol. Dis. 2001;8(5):723–742. doi: 10.1006/nbdi.2001.0436. (Oct) [DOI] [PubMed] [Google Scholar]
- Davatzikos C., Shen D., Gur R.C. Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch. Gen. Psychiatry. 2005;62(11):1218–1227. doi: 10.1001/archpsyc.62.11.1218. (Nov) [DOI] [PubMed] [Google Scholar]
- David A.S., Malmberg A., Brandt L., Allebeck P., Lewis G. IQ and risk for schizophrenia: a population-based cohort study. Psychol. Med. 1997;27(6):1311–1323. doi: 10.1017/s0033291797005680. (Nov) [DOI] [PubMed] [Google Scholar]
- Dazzan P., Morgan K.D., Orr K. Different effects of typical and atypical antipsychotics on grey matter in first episode psychosis: the AESOP study. Neuropsychopharmacology. 2005;30(4):765–774. doi: 10.1038/sj.npp.1300603. (Apr) [DOI] [PubMed] [Google Scholar]
- e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version 1.6–8. https://cran.r-project.org/package=e1071 ([computer program]. Version); 2017.
- Fanous A.H., Amdur R.L., O'Neill F.A., Walsh D., Kendler K.S. Concordance between chart review and structured interview assessments of schizophrenic symptoms. Compr. Psychiatry. 2012;53(3):275–279. doi: 10.1016/j.comppsych.2011.04.006. (Apr) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A., Beichel R., Kalpathy-Cramer J. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. (Nov) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete T., Wilf M., Rubin D., Edelman S., Malach R., Mujica-Parodi L.R. Combining classification with fMRI-derived complex network measures for potential neurodiagnostics. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. (Jan 31) [DOI] [PubMed] [Google Scholar]
- Franke K., Ziegler G., Kloppel S., Gaser C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. NeuroImage. 2010;50(3):883–892. doi: 10.1016/j.neuroimage.2010.01.005. (Apr 15) [DOI] [PubMed] [Google Scholar]
- Fryer S.L., Frank L.R., Spadoni A.D., Theilmann R.J., Nagel B.J., Schweinsburg A.D., Tapert S.F. Microstructural integrity of the corpus callosum linked with neuropsychological performance in adolescents. Brain Cogn. 2008;67(2):225–233. doi: 10.1016/j.bandc.2008.01.009. (Jul) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz L.A., Lewis D.A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. (Jan) [DOI] [PubMed] [Google Scholar]
- Green J.G., Gruber M.J., Kessler R.C., Lin J.Y., McLaughlin K.A., Sampson N.A., Zaslavsky A.M., Alegria M. Diagnostic validity across racial and ethnic groups in the assessment of adolescent DSM-IV disorders. Int. J. Methods Psychiatr. Res. 2012;21(4):311–320. doi: 10.1002/mpr.1371. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., Malley J.D., Weisinger B., Clasen L., Gogtay N. Using multivariate machine learning methods and structural MRI to classify childhood onset schizophrenia and healthy controls. Front. Psychol. 2012;3:53. doi: 10.3389/fpsyt.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M., Kanterakis S., Gur R., Roberts T.P., Verma R. DTI based diagnostic prediction of a disease via pattern classification. Med. Image Comput. Comput. Assist. Interv. 2010;13(Pt 1):558–565. doi: 10.1007/978-3-642-15705-9_68. [DOI] [PubMed] [Google Scholar]
- Jorgensen K.N., Skjaervo I., Morch-Johnsen L., Haukvik U.K., Lange E.H., Melle I., Andreassen O.A., Agartz I. Cigarette smoking is associated with thinner cingulate and insular cortices in patients with severe mental illness. J. Psychiatry Neurosci. 2015;40(4):241–249. doi: 10.1503/jpn.140163. (Jul) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambeitz J., Kambeitz-Ilankovic L., Leucht S., Wood S., Davatzikos C., Malchow B., Falkai P., Koutsouleris N. Detecting neuroimaging biomarkers for schizophrenia: a meta-analysis of multivariate pattern recognition studies. Neuropsychopharmacology. 2015;40(7):1742–1751. doi: 10.1038/npp.2015.22. (Jun) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan R.A., Kim J.S., Kaufmann W.E., Pearlson G.D., Barker G.J., McGuire P.K. Diffusion tensor imaging in schizophrenia. Biol. Psychiatry. 2005;58(12):921–929. doi: 10.1016/j.biopsych.2005.05.015. Dec 15. [DOI] [PubMed] [Google Scholar]
- Karageorgiou E., Schulz S.C., Gollub R.L. Neuropsychological testing and structural magnetic resonance imaging as diagnostic biomarkers early in the course of schizophrenia and related psychoses. Neuroinformatics. 2011;9(4):321–333. doi: 10.1007/s12021-010-9094-6. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasparek T., Thomaz C.E., Sato J.R. Maximum-uncertainty linear discrimination analysis of first-episode schizophrenia subjects. Psychiatry Res. 2011;191(3):174–181. doi: 10.1016/j.pscychresns.2010.09.016. (Mar 31) [DOI] [PubMed] [Google Scholar]
- Katuwal G.J., Cahill N.D., Baum S.A., Michael A.M. The predictive power of structural MRI in autism diagnosis. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2015;2015:4270–4273. doi: 10.1109/EMBC.2015.7319338. (Aug) [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Suzuki M., Kherif F. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. NeuroImage. 2007;34(1):235–242. doi: 10.1016/j.neuroimage.2006.08.018. (Jan 01) [DOI] [PubMed] [Google Scholar]
- Khondoker M., Dobson R., Skirrow C., Simmons A., Stahl D. A comparison of machine learning methods for classification using simulation with multiple real data examples from mental health studies. Stat. Methods Med. Res. 2016;25(5):1804–1823. doi: 10.1177/0962280213502437. (Oct) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki M., McCarley R., Westin C.F., Park H.J., Maier S., Kikinis R., Jolesz F.A., Shenton M.E. A review of diffusion tensor imaging studies in schizophrenia. J. Psychiatr. Res. 2007;41(1–2):15–30. doi: 10.1016/j.jpsychires.2005.05.005. (Jan–Feb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev A.V., Westman E., Van Westen G.J. Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness. Neuroimage Clin. 2014;6:115–125. doi: 10.1016/j.nicl.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Yoshida T., Kubicki M. Increased diffusivity in superior temporal gyrus in patients with schizophrenia: a diffusion tensor imaging study. Schizophr. Res. 2009;108(1–3):33–40. doi: 10.1016/j.schres.2008.11.024. (Mar) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Kim C.Y., Joo Y.H., Newell D., Bouix S., Shenton M.E., Kubicki M. Increased diffusivity in gray matter in recent onset schizophrenia is associated with clinical symptoms and social cognition. Schizophr. Res. 2016;176(2–3):144–150. doi: 10.1016/j.schres.2016.08.011. (Oct) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A., Wiener M. Classification and regression by randomforest. R News. 2002;(2/3):18–22. [Google Scholar]
- Lieberman J.A., Stroup T.S., McEvoy J.P. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N. Engl. J. Med. 2005;353(12):1209–1223. doi: 10.1056/NEJMoa051688. (Sep 22) [DOI] [PubMed] [Google Scholar]
- Mandl R.C., Brouwer R.M., Cahn W., Kahn R.S., Hulshoff Pol H.E. Family-wise automatic classification in schizophrenia. Schizophr. Res. 2013;149(1–3):108–111. doi: 10.1016/j.schres.2013.07.002. (Sep) [DOI] [PubMed] [Google Scholar]
- Moriya J., Kakeda S., Abe O. Gray and white matter volumetric and diffusion tensor imaging (DTI) analyses in the early stage of first-episode schizophrenia. Schizophr. Res. 2010;116(2–3):196–203. doi: 10.1016/j.schres.2009.10.002. (Feb) [DOI] [PubMed] [Google Scholar]
- Mourao-Miranda J., Reinders A.A., Rocha-Rego V. Individualized prediction of illness course at the first psychotic episode: a support vector machine MRI study. Psychol. Med. 2012;42(5):1037–1047. doi: 10.1017/S0033291711002005. (May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neighbors H.W., Jackson J.S., Campbell L., Williams D. The influence of racial factors on psychiatric diagnosis: a review and suggestions for research. Community Ment. Health J. 1989;25(4):301–311. doi: 10.1007/BF00755677. (Winter) [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis M., van Haren N.E., Hulshoff Pol H.E., Cahn W., Kahn R.S., Schnack H.G. Classification of schizophrenia patients and healthy controls from structural MRI scans in two large independent samples. NeuroImage. 2012;61(3):606–612. doi: 10.1016/j.neuroimage.2012.03.079. (Jul 02) [DOI] [PubMed] [Google Scholar]
- Orru G., Pettersson-Yeo W., Marquand A.F., Sartori G., Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci. Biobehav. Rev. 2012;36(4):1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. (Apr) [DOI] [PubMed] [Google Scholar]
- Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. (Aug) [DOI] [PubMed] [Google Scholar]
- Pettersson-Yeo W., Benetti S., Marquand A.F. Using genetic, cognitive and multi-modal neuroimaging data to identify ultra-high-risk and first-episode psychosis at the individual level. Psychol. Med. 2013;43(12):2547–2562. doi: 10.1017/S003329171300024X. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina-Camacho L., Garcia-Prieto J., Parellada M. Predictors of schizophrenia spectrum disorders in early-onset first episodes of psychosis: a support vector machine model. Eur. Child Adolesc. Psychiatry. 2015;24(4):427–440. doi: 10.1007/s00787-014-0593-0. (Apr) [DOI] [PubMed] [Google Scholar]
- R Core Team . Vienna; R Foundation: 2017. R: a Language and Environment for Statistical Computing. [Google Scholar]
- Rathi Y., Malcolm J., Michailovich O., Goldstein J., Seidman L., McCarley R.W., Westin C.F., Shenton M.E. Biomarkers for identifying first-episode schizophrenia patients using diffusion weighted imaging. Med. Image Comput. Comput. Assist. Interv. 2010;13(Pt 1):657–665. doi: 10.1007/978-3-642-15705-9_80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadock B.J., Sadock V.A. 10th ed. PA: LWW; Philadelphia: 2007. Synopsis of Psychiatry. [Google Scholar]
- Salvador R., Radua J., Canales-Rodriguez E.J. Evaluation of machine learning algorithms and structural features for optimal MRI-based diagnostic prediction in psychosis. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack H.G., Nieuwenhuis M., van Haren N.E., Abramovic L., Scheewe T.W., Brouwer R.M., Hulshoff Pol H.E., Kahn R.S. Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. NeuroImage. 2014;84:299–306. doi: 10.1016/j.neuroimage.2013.08.053. (Jan 01) [DOI] [PubMed] [Google Scholar]
- Shenton M.E., Dickey C.C., Frumin M., McCarley R.W. A review of MRI findings in schizophrenia. Schizophr. Res. 2001;49(1–2):1–52. doi: 10.1016/s0920-9964(01)00163-3. (Apr 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimbu K., Tavel J.A. What are biomarkers? Curr. Opin. HIV AIDS. 2010;5(6):463–466. doi: 10.1097/COH.0b013e32833ed177. (Nov) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., van Erp T.G., Thompson P.M. Elucidating a magnetic resonance imaging-based neuroanatomic biomarker for psychosis: classification analysis using probabilistic brain atlas and machine learning algorithms. Biol. Psychiatry. 2009;66(11):1055–1060. doi: 10.1016/j.biopsych.2009.07.019. (Dec 01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi Y., Takahashi T., Orikabe L. Classification of first-episode schizophrenia patients and healthy subjects by automated MRI measures of regional brain volume and cortical thickness. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Wang L., Cao F., Tan L. Identify schizophrenia using resting-state functional connectivity: an exploratory research and analysis. Biomed. Eng. Online. 2012;11:50. doi: 10.1186/1475-925X-11-50. (Aug 16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif E., Clarke S. Intrinsic connectivity of human auditory areas: a tracing study with DiI. Eur. J. Neurol. 2001;13(5):1045–1050. doi: 10.1046/j.0953-816x.2001.01456.x. (Mar) [DOI] [PubMed] [Google Scholar]
- Torgo L. CRC Press; Boca Raton: 2010. Data Mining with R: Learning with Case Studies. [Google Scholar]
- van Erp T.G., Hibar D.P., Rasmussen J.M. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry. 2016;21(4):585. doi: 10.1038/mp.2015.118. (Apr) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A., Kubicki M., Westin C.F., Golland P. Robust feature selection in resting-state fMRI connectivity based on population studies. Conf. Comput. Vis. Pattern Recognit. Workshops. 2010:63–70. doi: 10.1109/CVPRW.2010.5543446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A., Whitford T.J., Westin C.F., Golland P., Kubicki M. Whole brain resting state functional connectivity abnormalities in schizophrenia. Schizophr. Res. 2012;139(1–3):7–12. doi: 10.1016/j.schres.2012.04.021. (Aug) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita A., De Peri L., Deste G., Barlati S., Sacchetti E. The effect of antipsychotic treatment on cortical gray matter changes in schizophrenia: does the class matter? A meta-analysis and meta-regression of longitudinal magnetic resonance imaging studies. Biol. Psychiatry. 2015;78(6):403–412. doi: 10.1016/j.biopsych.2015.02.008. (Sep 15) [DOI] [PubMed] [Google Scholar]
- Weinberger D.R., Harrison P. 3rd ed. Wiley-Blackwell; Oxford: 2011. Schizophrenia. [Google Scholar]
- Zanetti M.V., Schaufelberger M.S., Doshi J. Neuroanatomical pattern classification in a population-based sample of first-episode schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2013;43:116–125. doi: 10.1016/j.pnpbp.2012.12.005. (Jun 03) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of features.