Voltage-gated proton channels open appropriately in myriad physiological situations because their gating is powerfully modulated by both pHo and pHi. Cherny et al. serendipitously identify a histidine at the inner end of the S3 helix that is required for the response to pHi.
Abstract
We recently identified a voltage-gated proton channel gene in the snail Helisoma trivolvis, HtHV1, and determined its electrophysiological properties. Consistent with early studies of proton currents in snail neurons, HtHV1 opens rapidly, but it unexpectedly exhibits uniquely defective sensitivity to intracellular pH (pHi). The H+ conductance (gH)-V relationship in the voltage-gated proton channel (HV1) from other species shifts 40 mV when either pHi or pHo (extracellular pH) is changed by 1 unit. This property, called ΔpH-dependent gating, is crucial to the functions of HV1 in many species and in numerous human tissues. The HtHV1 channel exhibits normal pHo dependence but anomalously weak pHi dependence. In this study, we show that a single point mutation in human hHV1—changing His168 to Gln168, the corresponding residue in HtHV1—compromises the pHi dependence of gating in the human channel so that it recapitulates the HtHV1 response. This location was previously identified as a contributor to the rapid gating kinetics of HV1 in Strongylocentrotus purpuratus. His168 mutation in human HV1 accelerates activation but accounts for only a fraction of the species difference. H168Q, H168S, or H168T mutants exhibit normal pHo dependence, but changing pHi shifts the gH-V relationship on average by <20 mV/unit. Thus, His168 is critical to pHi sensing in hHV1. His168, located at the inner end of the pore on the S3 transmembrane helix, is the first residue identified in HV1 that significantly impairs pH sensing when mutated. Because pHo dependence remains intact, the selective erosion of pHi dependence supports the idea that there are distinct internal and external pH sensors. Although His168 may itself be a pHi sensor, the converse mutation, Q229H, does not normalize the pHi sensitivity of the HtHV1 channel. We hypothesize that the imidazole group of His168 interacts with nearby Phe165 or other parts of hHV1 to transduce pHi into shifts of voltage-dependent gating.
Introduction
Voltage-gated proton channels, HV1, possess several distinctive or unique properties (DeCoursey, 2015). They are exquisitely proton selective with single-channel currents in the femtoampere range (Cherny et al., 2003). Perhaps because the conduction pathway is narrow and lacks a continuous water wire (Kulleperuma et al., 2013; Morgan et al., 2013; Chamberlin et al., 2014; Dudev et al., 2015), no high-affinity blockers exist that act by simple occlusion of the pore. The most potent inhibitor is Zn2+, which binds competitively with protons to two histidines in the external vestibule of human HV1 (hHV1) and interferes with channel opening (Cherny and DeCoursey, 1999; Takeshita et al., 2014). Although controversial (Bennett and Ramsey, 2017; DeCoursey, 2017), the conduction pathway includes an aspartate that appears to be obligatorily protonated and deprotonated during permeation (DeCoursey, 2003; Musset et al., 2011; Smith et al., 2011; Dudev et al., 2015; van Keulen et al., 2017). Consequently, H+ conduction has anomalously large temperature dependence (DeCoursey and Cherny, 1998; Kuno et al., 2009) and deuterium isotope effects (DeCoursey and Cherny, 1997) when compared with other channels. HV1 in many species are dimers (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008), but monomeric constructs also function with fairly subtle differences from the dimer (Koch et al., 2008; Tombola et al., 2008; Musset et al., 2010).
Perhaps the most unusual property of HV1 is ΔpH-dependent gating, a unique mechanism that is essential to all its functions. Although HV1 is a voltage-gated ion channel, the voltage at which all known HV1 open is strongly dependent on pH. Increasing pHo or decreasing pHi shifts the H+ conductance (gH)-V relationship negatively by ∼40 mV/unit. At symmetrical pH (pHo = pHi), the threshold for activation in most species is 10–20 mV (Cherny et al., 1995; DeCoursey, 2013). The combination of these properties ensures that under almost all conditions encountered by living cells, HV1 opens only when the electrochemical gradient for H+ is outward, so that opening HV1 channels results in acid extrusion from cells (DeCoursey, 2003). Functions of HV1 beyond acid extrusion, such as compensating electrically for the electrogenic activity of NADPH oxidase (Henderson et al., 1987; DeCoursey et al., 2003; DeCoursey, 2010), also require ΔpH dependent gating to enable HV1 to open appropriately (Murphy and DeCoursey, 2006).
The phenomenon of ΔpH-dependent gating seems to be remarkably robust. Despite at least 171 different HV1 mutants having been characterized electrophysiologically (DeCoursey et al., 2016), mutation at only one position, Trp207 (in hHV1 numbering), significantly altered ΔpH-dependent gating (Cherny et al., 2015). Mutation to this Trp resulted in premature saturation of the response to pHo but did not affect pHi responses. Recently identified HV1 in a few species (Cherny et al., 2015; Chaves et al., 2016), including the voltage-gated proton channel gene in the snail Helisoma trivolvis (HtHV1; Thomas et al., in this issue), have an exaggerated pHo response, shifting more than 40 mV/unit. Evaluation of three dozen hHV1 mutants revealed shifts of the absolute voltage dependence over a 270-mV range, but every mutant still exhibited an ∼40-mV/pH unit shift from whatever its starting voltage was (Ramsey et al., 2010). We were therefore surprised to find that despite a robust pHo response, the WT HtHV1 channel has anomalously weak pHi dependence, with the gH-V relationship shifting just 20 mV/unit or less (Thomas et al., 2018). While testing several possible explanations for the rapid gating of snail HtHV1, we serendipitously discovered that replacement of a single amino acid, His168, in the human channel nearly abolishes its pHi dependence. We conclude that His168 is a crucial component of pHi sensing in hHV1.
Materials and methods
Electrophysiological and mutagenesis methods for HtHV1 are described in the accompanying article (Thomas et al., 2018); mutagenesis of hHV1 was described previously (Musset et al., 2010). A triple mutant of hHV1 (H167N/H168V/K168N) was supplied by I. Scott Ramsey and David E. Clapham (Harvard Medical School, Boston, MA). Statistical comparison of groups was done by using Student’s t test.
Sequence analysis
46 full-length HV1 sequences, including 11 electrophysiologically confirmed and 13 high-confidence sequences from protists—previously shown to be more diverse than animal HV1 (Smith et al., 2011)—were aligned with MAFFT (Katoh and Standley, 2013). Sequences were trimmed to the voltage-sensing domain as described previously (Smith et al., 2011). Sequences that were not confirmed electrophysiologically were excluded, and empty columns removed from the alignment. The activation (opening) kinetics of electrophysiologically confirmed sequences were characterized as fast (H. trivolvis, HtHV1; Lymnaea stagnalis, LsHV1; and Strongylocentrotus purpuratus, SpHV1) or slow (Homo sapiens, hHV1; Mus musculus, mHV1; Oryctolagus cuniculus, OcHV1; Rattus norvegicus, RnHV1; and Cricetulus griseus, CgHV1); sequences with intermediate or unknown kinetics were removed. To identify sequence sites that distinguish sequence subfamilies, the alignment was submitted to the SPEER server (Chakraborty et al., 2012) with either user-defined (two subfamilies) or automated subgrouping and the following parameters: single-submission eight sequences, Relative Entropy term, PC Property Distance term, and Type. The same alignment but in randomized sequence order was also submitted with the automated subgrouping parameter and all other parameters as above. Sites corresponding to His167 and His168 in hHV1 were identified as appearing in the top 10 sites identified in all analyses and having a P value of <0.05 in at least one analysis.
Results
Is His168 responsible for the slow gating kinetics of hHV1?
In our companion article (Thomas et al., 2018), we reported the identification of an HV1 in a snail H. trivolvis (HtHV1) that exhibited rapid activation kinetics as reported for native proton currents in other snails (Byerly et al., 1984). We performed an analysis of subfamily determining sites to distinguish HV1 sequences with fast versus slow gating kinetics. Species categorized as having fast gating included snails, HtHV1 (Thomas et al., 2018) and L. stagnalis, LsHV1 (Byerly et al., 1984); and sea urchin, S. purpuratus, SpHV1 (Sakata et al., 2016) and are shaded green in Fig. 1. Species with slow gating are human, hHV1 (Bernheim et al., 1993; DeCoursey and Cherny, 1993; Demaurex et al., 1993); mouse, mHV1 (Kapus et al., 1993); rabbit, O. cuniculus, OcHV1 (Nordström et al., 1995); rat, R. norvegicus, RnHV1 (DeCoursey, 1991); and Chinese hamster, C. griseus, CgHV1 (Cherny et al., 1997) and are shaded pink in Fig. 1. This analysis pointed to two residues corresponding to His167 and His168 in hHV1 numbering; these residues are in the intracellular S2/S3 loop, extending into the S3 transmembrane helix. Sakata et al. (2016) found that substituting either Ser or Thr for His164 in mHV1, which corresponds to His168 in hHV1 (Fig. 1, asterisk), accelerated activation kinetics by an order of magnitude, raising the possibility that His in this position governs activation kinetics, acting as a “throttle.” The same substitutions at the neighboring His163 had no effect. For these reasons we focused our experiments on the S2/S3 loop area.
Figure 1.
Alignment of HV1 S2–S3 linker and nearby regions (E153–D185 in hHV1). HV1 from several species were characterized as activating rapidly (green background: HtHV1, LsHV1, and SpHV1) or slowly (pink background: hHV1, mHV1, OcHV1, RrHV1, and CgHV1) based on available electrophysiological data. Unshaded species exhibit intermediate kinetics. In hHV1 numbering, the asterisk (*) indicates the position corresponding to the throttle histidine H168; the caret (^) indicates the position corresponding to F165. The S2–S3 linker in hHV1 encompasses K157–F165 (Li et al., 2015).
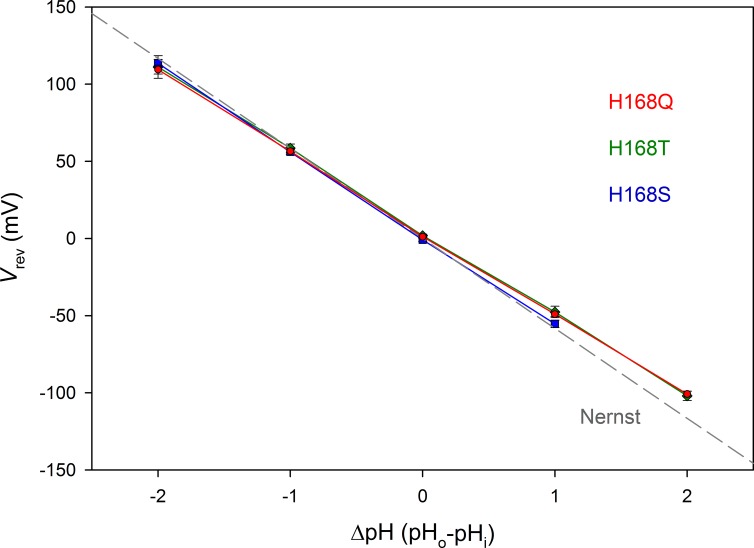
Similar to Sakata et al. (2016), we engineered mutations of His168, replacing it with Ser (H168S) or Thr (H168T). Because activation of snail HtHV1 is distinctly faster than that of sea urchin SpHV1, we reasoned that replacing His168 with Gln, which is at the corresponding position in HtHV1 (H168Q), should cause even faster kinetics. As a control, we replaced the neighboring His167 in hHV1 with Lys (H167K), which occupies this position in HtHV1 (Fig. 1). Finally, we tested a triple mutant, in which three consecutive titratable amino acids (His168 and its immediate neighbors) were all replaced by neutral residues (H167N/H168V/K169N). Robust proton selective currents were observed in all mutants. Results from H168S, H168T, and H168Q mutants were indistinguishable, and we thus refer to all such mutants as H168X. Fig. 2 shows that the reversal potential (Vrev) in the three H168X mutants was close to the Nernst potential, confirming proton selectivity.
Figure 2.
His168 mutants are proton selective. Mean ± SEM values for the reversal potential (Vrev) are plotted for 12 H168Q, 9 H168T, and 6 H168S cells or patches. The linear regression slope for each mutant is −52.6, −53.2, and −56.3 mV/unit change in ΔpH, respectively. For comparison, the gray dashed line shows the Nernst potential, EH, expected for perfect H+ selectivity.
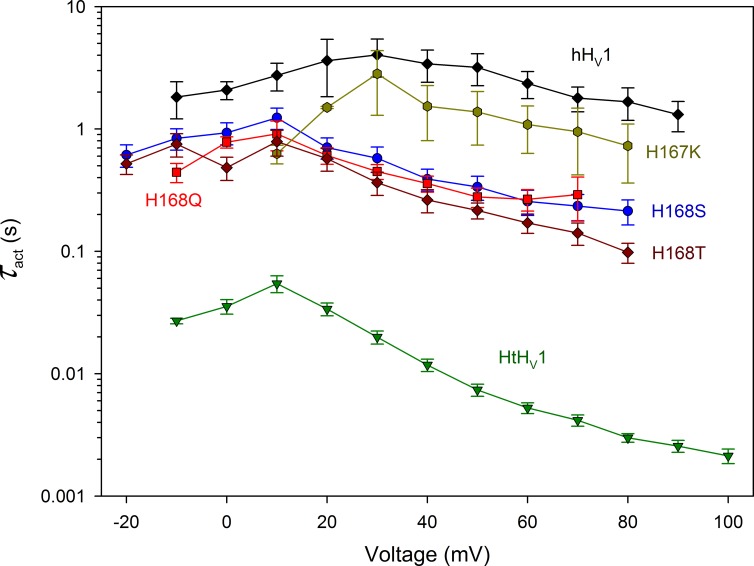
Fig. 3 illustrates the effects of these mutations on channel-opening kinetics. Replacing His168 with any of the three amino acids (Ser, Thr, or Gln) produced a 10-fold speeding of the activation (opening) time constant (τact) in the positive voltage range, much like the analogous effect in mHV1 (Sakata et al., 2016). Mutating the neighboring His167 had no clear effect. That the effects of the three substituents (Ser, Thr, or Gln) were identical suggests that the speeding of activation is caused by removing the imidazole group of His and is not caused by introducing other side chains at this position. Thus, the hydroxyl group of Thr or Ser was no more or less effective than the amide group of Gln. Presumably, His168 interacts specifically with other parts of the hHV1 channel in a manner that results in slower activation. Nevertheless, despite the distinct acceleration caused by His168 removal, τact remained much slower than in the WT snail channel HtHV1.
Figure 3.
The throttle histidine explains only a fraction of the rapid activation of HtHV1. Activation time constants (τact) were determined by single exponential fits to currents in WT HtHV1, WT hHV1, and in four hHV1 mutants, as indicated, all at pHo 7 and pHi 7. Mean ± SEM is plotted for 3–12 WT hHV1, 2–3 H167K, 3–6 H168S, 3–9 H168Q, 3–6 H168T, and 7–17 WT HtHV1. At all voltages from −10 to +80 mV, the WT hHV1 value is significantly larger than that of H168T, H168S, and H168Q combined and larger than HtHV1 (P < 0.01). The WT hHV1 data were taken from Cherny et al. (2015) and the HtHV1 data from Thomas et al. (2018).
His168 mutation alters ΔpH-dependent gating of hHV1
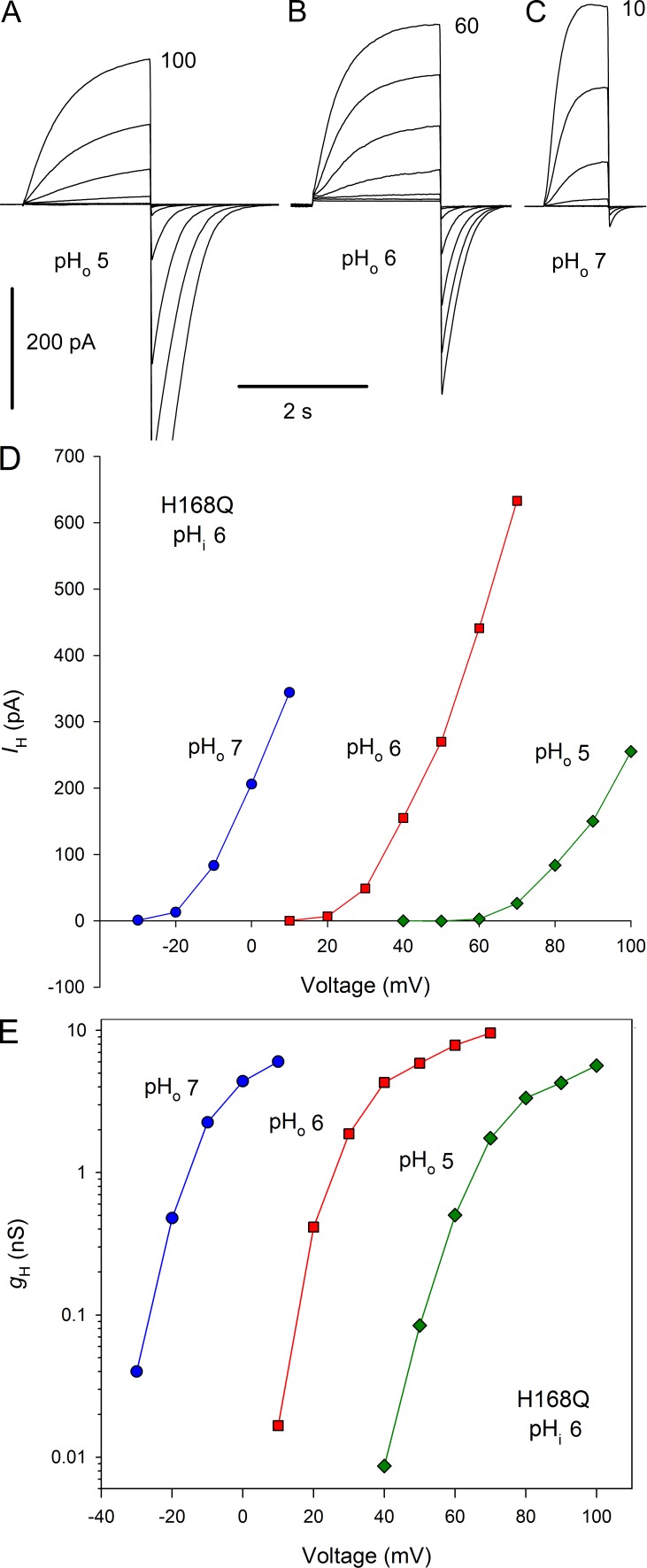
The His168 mutants exhibited normal responses to changes in pHo. Families of currents in a cell expressing the H168Q mutant are shown in Fig. 4, A–C. The currents appear qualitatively like WT hHV1 currents, although with somewhat faster activation. Changing pHo from 5 to 6 to 7 shifted the H+ current (IH)-V (Fig. 4 D) and gH-V relationships (Fig. 4 E) by ∼40 mV/unit. This behavior conforms to the classical “rule of forty” observed for HV1 in all species thus far examined (DeCoursey, 2013), except for HtHV1 as described in the companion article (Thomas et al., 2018).
Figure 4.
Gating of His168 mutants has normal pHo dependence. (A–C) Families of currents in a cell expressing H168Q with pHi 6 and pHo 5, 6, and 7, with pulses in 10-mV increments up to the voltage indicated. Holding potential, Vhold was −40 mV (A and B) or −60 mV (C). (D and E) Proton current–voltage curves (D) and gH-V relationships (E) from the families in A–C exhibit a normal 40-mV shift/unit change in pHo.
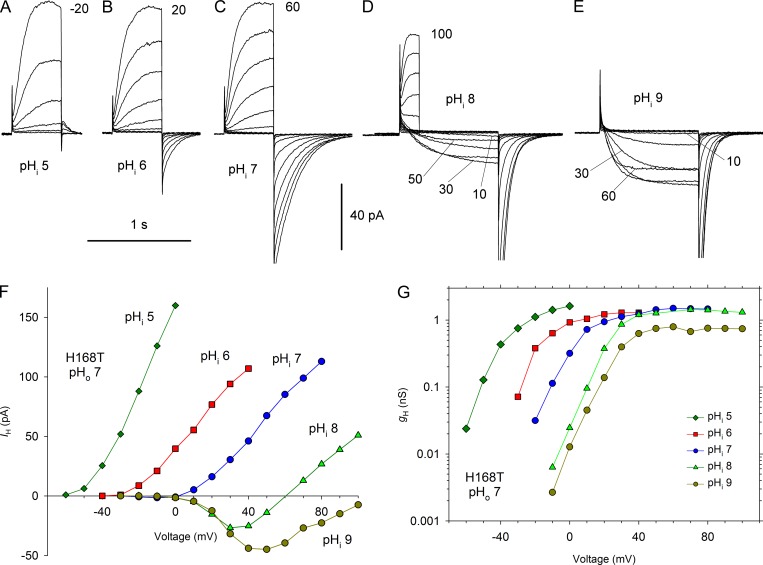
In contrast, mutation of His168 to Ser, Thr, or Gln greatly weakened the sensitivity of the mutant channels to pHi. Fig. 5, A–E shows currents in an inside-out patch with H168T at pHo 7 and pHi ranging from 5 to 9. Although the IH-V relationship does shift positively at higher pHi (Fig. 5 F), the shift is distinctly <40 mV, especially at high pHi, with the result that there are inward currents over a wide voltage range. Inward currents in WT hHV1 are rarely observed and occur mainly with large outward pH gradients (Musset et al., 2008). It is evident from the gH-V relationships (Fig. 5 G) that the shifts are ≤20 mV/unit over the entire pHi range studied.
Figure 5.
Mutation of His168 in hHV1 weakens the pHi dependence of gating. (A–E) Families of currents at several pHi values in an inside-out patch from a cell transfected with H168T, with pHo 7 in the pipette. Pulses were applied in 10-mV increments up to the voltage indicated from Vhold = −80 mV (A), −60 mV (B), or −40 mV (C–E). (D) Shorter pulses were applied to 60 mV and above, and the tail currents have been removed. (E) Pulses above 60 mV were omitted for clarity. (F) Current–voltage curves. (G) gH-V relationships from this experiment.
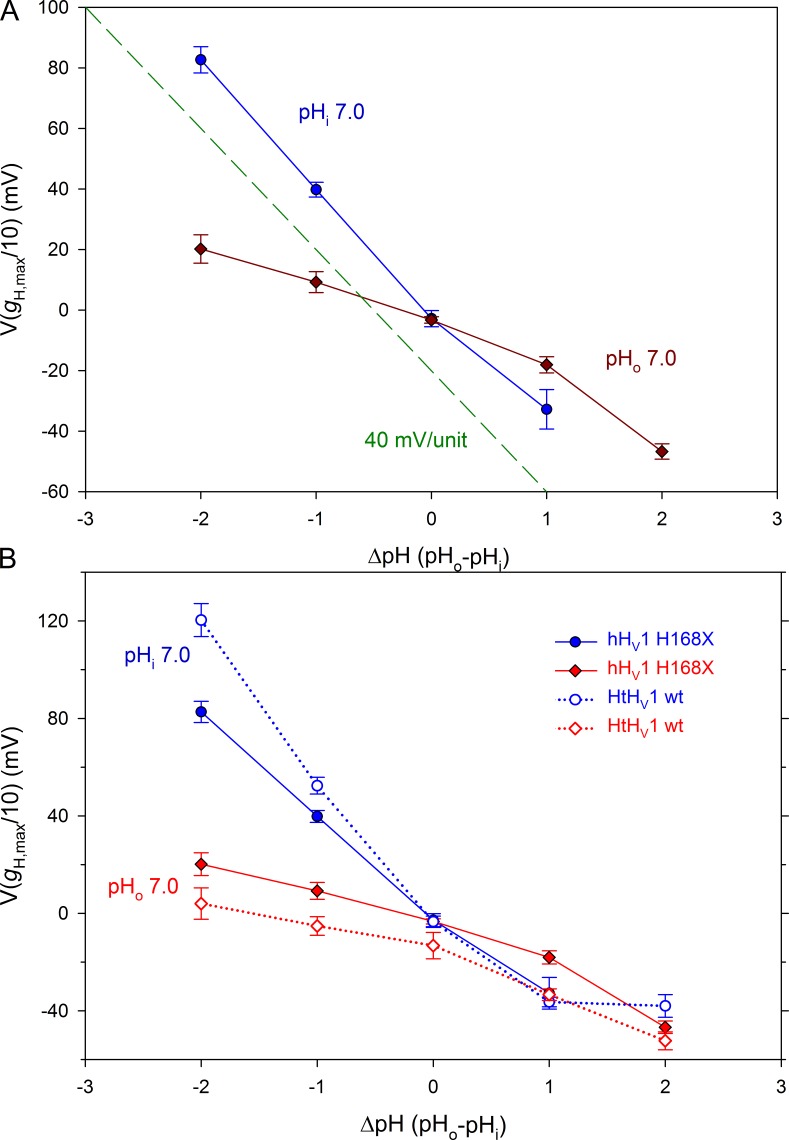
Measurements of the position of the gH-V relationship in several cells and patches are summarized in Fig. 6 A, quantified as V(gH,max/10), the voltage at which gH reached 10% of gH,max, its maximal value. Changes in pHo for the H168X mutants (Fig. 6 A, blue circles) produce a slope of 40 mV/unit up to pHo 8 (ΔpH = 1). This behavior is identical to the WT hHV1 response (Cherny et al., 2015). Data from inside-out patches (Fig. 6 A, dark red diamonds) reveal that the H168X mutants exhibit a greatly attenuated pHi response. The entire curve has a mean slope of only 16.1 mV/unit. However, the relationship appears steeper at high ΔpH. Omitting the point at ΔpH = 2 (pHo 7, pHi 5), the mean slope drops to 12.6 mV/unit.
Figure 6.
The His168 mutation recapitulates the anomalous ΔpH dependence of HtHV1. (A) The effect of pHo and pHi on the position of the gH-V relationship in the three H168x mutants combined (mean ± SEM) is plotted. The position of the gH-V relationship was defined in terms of V(gH,max/10). The dashed green line shows a slope of 40 mV/unit for reference. The dependence of these mutants on pHo is normal, whereas their pHi response is greatly attenuated. Linear regression on all points (ignoring the obvious nonlinearity) gives a slope of 38.9 mV/unit change in pHo and 16.1 mV/unit change in pHi. (B) The same data are replotted (shaded symbols) along with the analogous measurements for HtHV1 (open symbols) taken from Fig. 9 of the companion article (Thomas et al., 2018). In whole-cell measurements, pHo was varied with pHi 7 (blue symbols). When pHi was varied by using inside-out patches with pHo 7, there was very little shift of the gH-V relationship (red symbols). Numbers of cells for increasing ΔpH in H168X mutants for pHi 7 are 3, 10, 11, and 5 and for pHo 7 are 6, 10, 14, 11, and 4.
The behavior of the H168X mutants strikingly resembles that of WT HtHV1, as illustrated in Fig. 6 B. The data for HtHV1 from the companion article (Thomas et al., 2018) are replotted here (Fig. 6 B, open symbols and dotted lines) for comparison. Comparison of pHo responses reveals that HtHV1 has an exceptionally steep ΔpH dependence at pHo 7 or less, roughly 60 mV/unit. The response to pHo for H168X mutants (Fig. 6 B, shaded blue circles) is a 40-mV/unit shift, which is normal for human hHV1 (Cherny et al., 2015). The response of H168X mutants to pHi changes (Fig. 6 B, red diamonds) is notably weak and closely parallels the response of WT snail HtHV1. With respect to ΔpH-dependent gating, the mutation of a single amino acid, His168, effectively converts the human channel into a snail channel.
The immediate neighbors of His168 do not appear to contribute
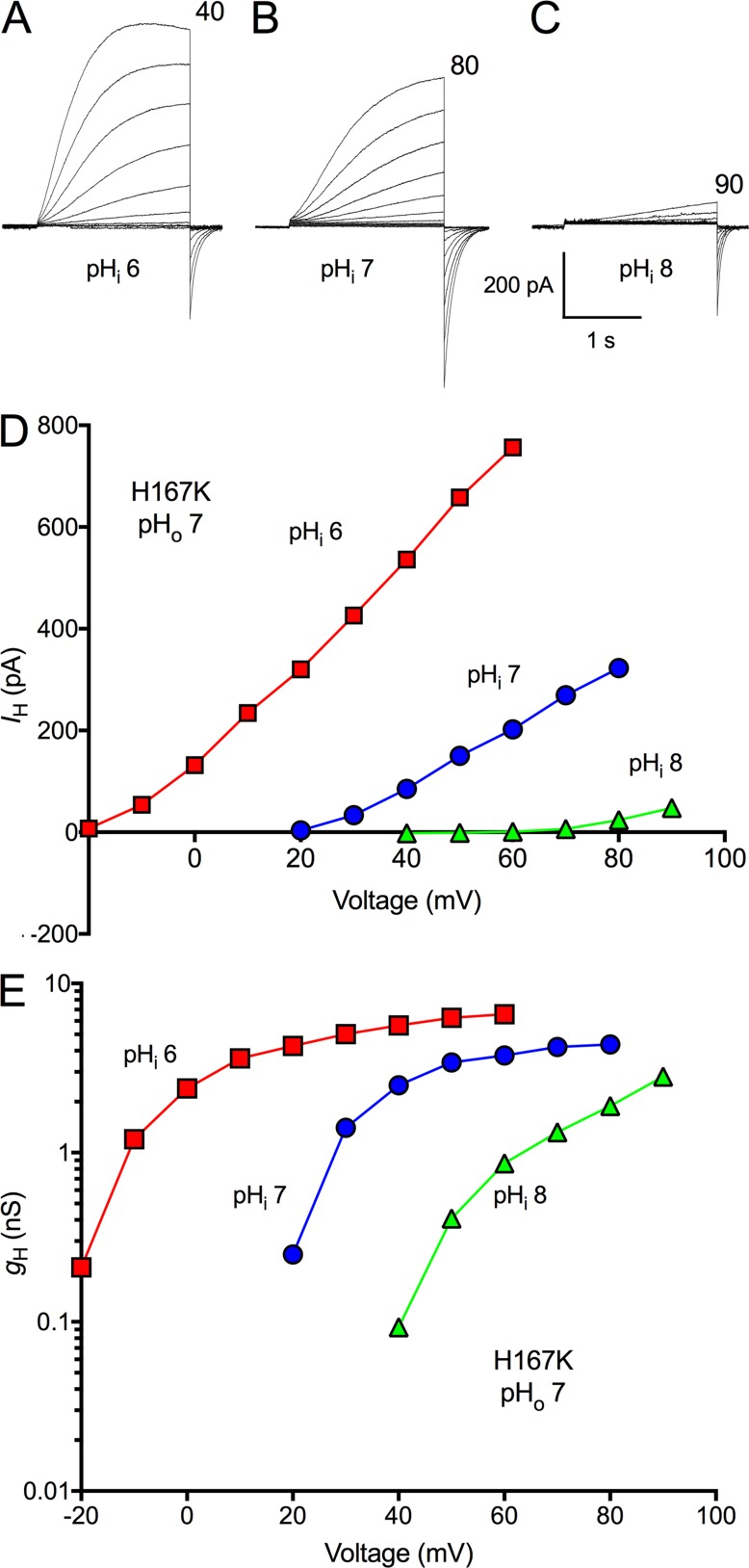
Located next the His168 that plays a critical role in pHi sensing is another His, His167. We wondered whether this nearby position might share the effect. However, the single mutation H167K did not impair pHi sensing. This specific replacement was selected because HtHV1 has Lys at the corresponding position (Fig. 1). Fig. 7 illustrates H+ currents in an inside-out patch expressing the H167K mutant at pHi 6, 7, and 8. The gH-V relationship shifted normally (Fig. 7 E). Mean data from three patches with H167K are plotted in Fig. 8, which shows that the pHi response of this mutant is indistinguishable from that of WT hHV1. Furthermore, Fig. 3 showed that the H167K mutation did not alter activation kinetics. Evidently, His168 but not His167 is located precisely where it can strongly influence gating in response to pHi.
Figure 7.
The H167K mutant of hHV1 has normal pHi dependence. (A–C) Families of currents are shown in an inside-out patch with pHo 7 and pHi 6, 7, or 8, as labeled. Pulses were applied in 10-mV increments up to the voltage indicated, from Vhold = −60 mV (A) or -40 mV (B and C). (D and E) Current–voltage curves (D) and gH-V relationships from this patch are illustrated (E). At pHi 8, gH was estimated from tail current amplitudes and scaled according to gH calculated from the outward current at 90 mV.
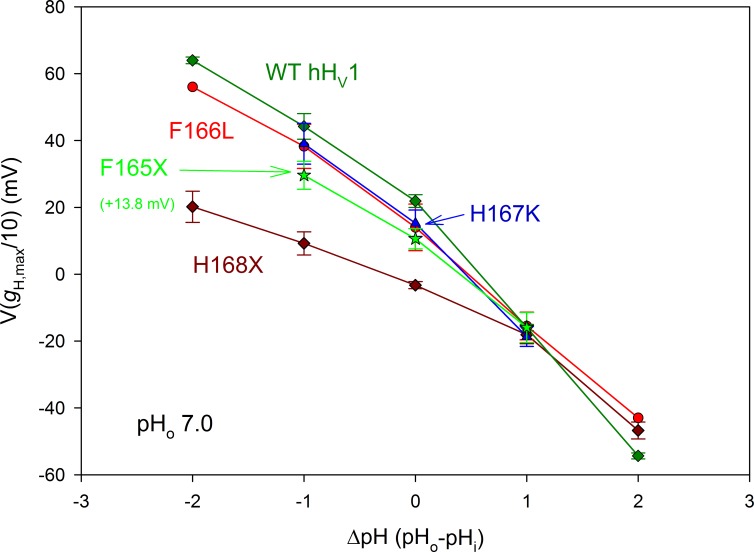
Figure 8.
The F166L and H167K mutations do not impair pHi sensing of hHV1. The pHi responses of the H168X mutants are replotted from Fig. 6. The pHi responses of WT hHV1, the F166L mutant of hHV1, and the H167K mutant of hHV1 are indistinguishable from each other. The F165X data are from three F165A and three F165H patches combined, and 13.8 mV has been added to all values to facilitate comparison with WT. All data (mean ± SEM) come from inside-out patches studied at pHo 7, with n = 2–9 (H168X), 1–4 (F166L), 3 (H167K), and 6 (F165X).
We also tested a triple mutant, in which three consecutive titratable amino acids at the inner end of the S3 helix were replaced by neutral residues (H167N/H168V/K169N). In inside-out patches with pHo 7.5, studied at pHi 5.5, 6.5, 7.5, and 8.5, the sensitivity to pHi was weak, like that of the H168X single mutants. The mean slope by linear regression of the relationship between ΔpH and V(gH,max/10) in seven patches was 20.1 mV/unit, similar to 16.1 mV/unit in H168X (Fig. 6), suggesting that the amino acids neighboring His168 were inactive. This triple mutant was examined previously, and normal ΔpH dependence was reported (Ramsey et al., 2010). However, in that study only pHo was varied, not pHi. The triple mutant, like all single His168 mutants, exhibits normal pHo dependence but abnormally weak pHi dependence.
Possible interaction between H168 and F165
Data on pHi sensitivity are somewhat less abundant than for pHo sensitivity, and much is derived from native tissues. Species with HV1 that have been shown to exhibit classical rule of forty behavior for changes in pHi include human (Demaurex et al., 1993; Gordienko et al., 1996; Schrenzel et al., 1996; Schilling et al., 2002), mouse (Kapus et al., 1993; Szteyn et al., 2012), Rana pipiens (Gu and Sackin, 1995), rat (Cherny et al., 1995; DeCoursey and Cherny, 1995), Karlodinium veneficum (Cherny et al., 2015), Emiliania huxleyi (Cherny et al., 2015), and Lingulodinium polyedrum (Rodriguez et al., 2017). The only violators of the rule thus far are the snails H. trivolvis (Thomas et al., 2018) and L. stagnalis (Byerly et al., 1984). Another intriguing exception is a short isoform of hHV1 expressed in sperm that lacks the first 68 amino acids of the N terminus and has weakened pHi sensing (Berger et al., 2017). Inspection of sequences (Fig. 1) for which pHi sensitivity has been measured directly (HtHV1, LsHV1, EhHV1, kHV1, LpHV1, hHV1, mHV1, and RnHV1) shows that Leu substitutes for Phe at the Phe165 position (human numbering) in two species with rapid gating and attenuated pHi responses, HtHV1 and LsHV1. Although in the available crystal structure of HV1 (Takeshita et al., 2014) this area did not have sufficient electron density to be visualized, molecular models of HV1 (Kulleperuma et al., 2013; Li et al., 2015) indicate that Phe165 could plausibly interact with the nearby His168 through π-stacking or cation-π interactions.
To test the idea that His168 interacts with Phe165, we individually mutated Phe165 and as a control, Phe166. F166L produced robust currents, and as shown in Fig. 8, pHi sensing of F166L was essentially normal and similar to that of WT hHV1. F165H and F165A (Fig. 8, F165X) produced currents that activated in a more negative voltage range than WT. To enable comparison of the slopes, 13.8 mV has been added to these data, so they superimpose on WT at ΔpH = 1. The pHi sensitivity of F165A and F165H was closer to WT than to that of the H168X mutants. Finally, the double mutant F165L/H168Q was also close to the single H168X mutant data (unpublished data). Although these results do not rule out the possibility of interaction between His168 and Phe165, it appears that His168 is distinctly more critical for pHi sensing.
The converse mutation Q229H fails to impart human characteristics to the H. trivolvis channel
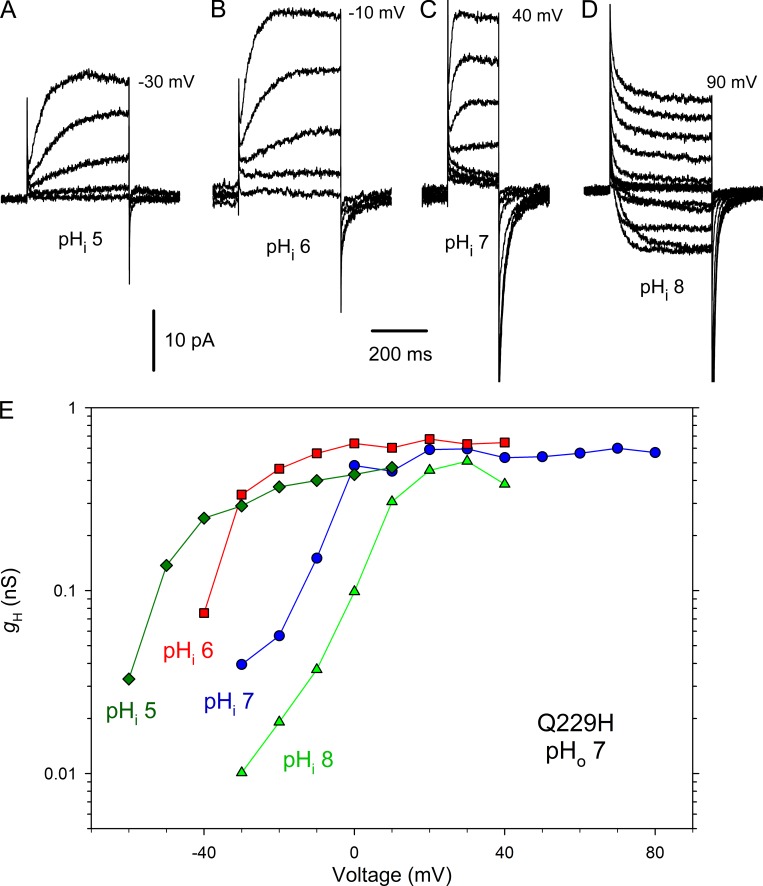
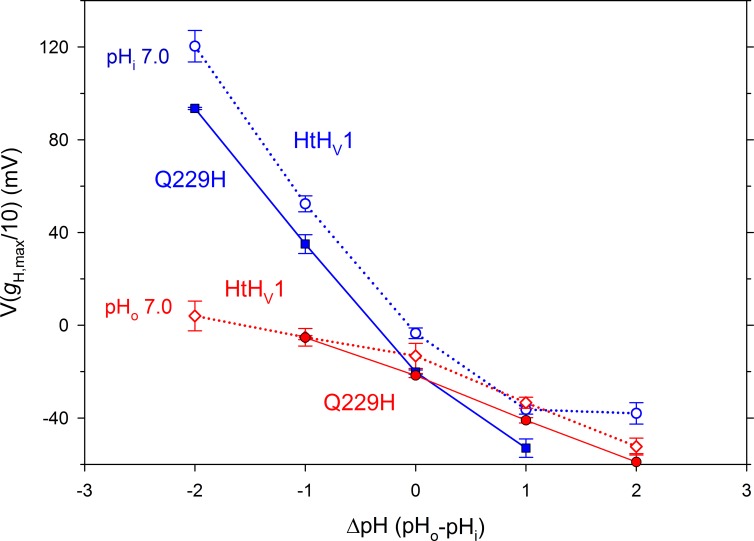
The single point mutation H168X (X = T, S, Q) profoundly alters the gating and pH sensing of the human channel hHV1, accelerating activation 10-fold and greatly attenuating the pHi response. We next considered whether the converse mutation (Q229H) would impart humanoid characteristics to HtHV1. In short, this mutation had no discernable effect on the gating kinetics or pH sensitivity of HtHV1 (Fig. 9). The Q229H mutation did not slow activation; at +40 mV at pHo 7, τact was 11.8 ± 1.4 ms (n = 17) in WT HtHV1 (Fig. 3) and 10.0 ± 2.0 ms (n = 8) in Q229H (P = 0.53). We assessed pHo sensitivity in whole-cell experiments. Like WT HtHV1, activation kinetics was extremely sensitive to pHo, whereas the deactivation or tail current (closing) time constant appeared to be pHo independent. Like WT, changes in pHo produced supernormal shifts in the position of the gH-V relationship, averaging 57 mV/unit between pHo 5 and 7 (Fig. 10), paralleling the WT response although the absolute voltages appear somewhat more negative. Similarly, the Q229H mutation did not discernibly change the weak response of HtHV1 to changes in pHi (Figs. 9 E and 10). In three inside-out patches studied at pHi 5, 6, 7, and 8, the slope of the mean dependence of V(gH,max/10) on ΔpH was 18 mV/unit by linear regression. It is clear that the extraordinary changes to the pHi response of the human channel resulting from His168 mutation reflect interactions that are not confined to this single amino acid and therefore are not easily transferred.
Figure 9.
The Q229H mutation does not restore human-like activation kinetics or pHi sensitivity to HtHV1. (A–D) Families of currents in an inside-out patch with pHo 7 in the pipette solution in 10-mV increments up to the voltage shown from Vhold = −80, −60, −40, and −40 mV, at pHi 5, 6, 7, and 8, respectively. (E) Currents from this patch converted to gH produce gH-V relationships that shift much less than 40 mV/unit. Corresponding measurements in WT HtHV1 are plotted in Fig. 5 G.
Figure 10.
The Q229H mutation does not restore human-like pHi sensitivity to HtHV1. The dependence of the position of the gH-V relationship on both pHo and pHi is similar in WT HtHV1 (open symbols and dotted lines) and in the Q229H mutant (shaded symbols and solid lines). The WT data are replotted from Fig. 6 B. Mean ± SEM are shown for n = 2–5 cells with pHi 7 and three inside-out patches with pHo 7.
Discussion
Is the human hHV1 channel slow because of the throttle residue, His168?
The sea urchin proton channel, SpHV1, exhibits rapid activation kinetics 20–60 times faster than the mouse mHV1. Sakata et al. (2016) investigated the structural basis for this property and, by creating progressively smaller chimerae with mHV1, finally identified a single amino acid (Ser) at the inner end of the S3 transmembrane segment that appeared largely responsible. Introducing Ser into mHV1 (H164S) accelerated mouse activation kinetics by more than an order of magnitude. In mHV1, H164T produced speeding similar to H164S, suggesting that the hydroxyl group is important (Sakata et al., 2016). Intriguingly, however, the converse mutation, and even substituting the entire S3 helix from mHV1 into SpHV1, failed to slow the kinetics of the latter.
We engineered mutations of human hHV1 at His168 (the position that corresponds to His164 in mHV1), replacing the His with Ser (H168S), Thr (H168T), or Gln (H168Q). In most respects, the results resembled those in mHV1: mutations at His168 accelerated activation by about one order of magnitude (Fig. 3). Because HtHV1 has Gln at the corresponding position and HtHV1 is distinctly faster than SpHV1, we expected that Gln substitution might produce faster activation; however, activation kinetics of H168Q were very similar to those of H168S and H168T, all of which remained much slower than that of WT HtHV1 (Fig. 3). Because the three mutants behaved indistinguishably, we conclude that the effects result from the removal of the imidazole side chain of His168 and not from the side chains that replace it.
The precise location of His168 is important. The His immediately adjacent to His168 does not share its impact on gating kinetics. Sakata et al. (2016) replaced His163 in mHV1 with the SpHV1 residue at the corresponding position (H163R); this mutation had little effect on kinetics of mHV1. We replaced His167 in hHV1 with Lys, which occupies the corresponding position in HtHV1. In neither species was activation kinetics appreciably affected (Fig. 3).
Also, similar to the results in mHV1, the converse mutation substituting the hHV1 residue for the HtHV1 throttle residue, Q229H, did not slow HtHV1 kinetics. Therefore, the mechanism by which His168 retards activation must require interaction with other parts of the protein that are absent in HV1 from species with rapid gating.
His168 in hHV1 is crucial for pHi sensing
In searching for a mechanistic explanation for the rapid gating kinetics of HtHV1, we serendipitously identified a histidine near the inner end of the channel that is crucial for pHi sensing. Transferring the Gln229 from the snail channel HtHV1 to replace His168 in the human channel hHV1 (Fig. 1) reproduced the impoverished pHi sensing of HtHV1 almost exactly (Fig. 6 B). Because three different substituents (Ser, Thr, and Gln) had indistinguishable effects (Figs. 3 and 6), it appears that a specific property of His in this position is involved. Because the converse mutation (Q229H) did not transfer human-like pHi sensing (or activation kinetics) to HtHV1 (Fig. 9), His168 must perform pHi sensing by interacting with its immediate environment in the human channel. This conclusion is reinforced by the fact that three unicellular species that lack His at this position (kHV1, LpHV1, and EhHV1; Fig. 1) exhibit normal pHi sensitivity (Cherny et al., 2015; Rodriguez et al., 2017). Replacing the neighboring His167 had no effect, and mutating both His167 and Lys169 (HHK→NVN) had no effect beyond that of His168 alone, indicating that the specific location at position 168 is critical.
When the ΔpH dependence of HV1 gating was discovered (Cherny et al., 1995), the simplest model that could reproduce the entire phenomenon postulated a single pH sensor with alternating access to both internal and external solutions. An expanded model with additional complexity includes distinct external and internal pH sensors. A “counter-charge” model has been suggested in which protonation disrupts charge-pair interactions, so that low pHi promotes opening and low pHo promotes closing (DeCoursey, 2018). A different kind of proposal for pH sensing does not invoke titratable pH sensors; instead, protonated waters in HV1 communicate information about local pH through electrostatic interactions with the voltage-sensing Arg residues in the S4 helix (Ramsey et al., 2010). Recently, the first mutation in HV1 documented to compromise pH sensing was reported (Cherny et al., 2015). In these Trp mutants, pHo but not pHi sensitivity was affected, suggesting that separate pHo and pHi sensors exist in HV1. Intriguingly, like the H168X mutants, these W207X mutants also dramatically accelerated hHV1 gating kinetics, and HtHV1 itself has extremely rapid gating.
That HtHV1 has normal pHo sensitivity but anemic pHi sensitivity suggests that its ΔpH dependence is accomplished by distinct external and internal pH sensors. That the H168x mutation in hHV1 selectively attenuates its pHi sensitivity without affecting its pHo sensitivity further supports the existence of distinct external and internal pH sensors.
To search for a structural basis for this species difference in pHi sensing, we compared the sequences of HV1 in species in which pHi responses have been documented to obey the rule of forty (human, mouse, rat, K. veneficum, E. huxleyi, and L. polyedrum; excepting R. pipiens for which we could find no sequence), to the sequence of HtHV1 with its anomalously weak sensitivity to pHi. We noticed a striking substitution in HtHV1, in which Phe165 (human numbering; Fig. 1) is replaced by Leu. This position is very highly conserved: in the alignment of 46 HV1 sequences that includes vertebrates, invertebrates, and protists, Phe is replaced by the conservative Tyr five times, by Leu twice, and by Met once. In this context, it is important to note that the protist HV1 sequences are more diverse than all animal sequences (Smith et al., 2011), highlighting the very high conservation of Phe at this position. The two snail species in which Leu replaces Phe (HtHV1 and LsHV1) both have rapid gating and weak pHi sensitivity (Fig. 1). In structural models of HV1 (Kulleperuma et al., 2013; Li et al., 2015), Phe165 is in a position in which it could plausibly interact with His168, through π-stacking or cation-π interactions. Reasoning that such an interaction might be part of the pHi-sensing mechanism, we replaced Phe165 in hHV1 with Ala or His to attempt to disrupt the interaction. Both F165A and F165H exhibited pHi sensing intermediate between WT and the H168x mutants but closer to WT hHV1. The neighboring F166L mutation did not affect pHi sensing (Fig. 8). For geometrical reasons, we consider it more likely that His168 could interact with Phe165 than with Phe166 because the latter would be constrained by proximity. Intrahelical i, i + 2 side-chain–side-chain contacts are statistically improbable (Walther and Argos, 1996). That Phe165 mutants were activated at more negative voltages than WT (Fig. 8) is consistent with F165-H168 interaction impeding hHV1 opening. That the insertion of His into the corresponding position in HtHV1 did not enhance pHi sensitivity indicates that the mere presence of His at this location is not sufficient, but that to affect pHi sensing requires additional interaction. Evidently, and perhaps surprisingly, the intracellular S2–S3 linker is of central importance in sensing pHi in human hHV1. Furthermore, the selective effect of mutations in this region on pHi sensing while leaving pHo sensing intact supports the idea that ΔpH-dependent gating of HV1 relies on independent internal and external pH sensors.
Acknowledgments
This work was supported by National Institutes of Health grants GM102336 (to T.E. DeCoursey and S.M.E. Smith) and GM121462 (to T.E. DeCoursey) and National Science Foundation grant MCB-1242985 (to T.E. DeCoursey and S.M.E. Smith).
The authors declare no competing financial interests.
Author contributions: S.M.E. Smith and T.E. DeCoursey conceptualized the study. T.E. DeCoursey curated the data. S. Thomas, S.M.E. Smith, V.V. Cherny, D. Morgan, and T.E. DeCoursey performed the formal analysis. T.E. DeCoursey and S.M.E. Smith acquired the funding. V.V. Cherny and D. Morgan performed the investigation. S.M.E. Smith and T.E. DeCoursey administered the project. S. Thomas and S.M.E. Smith provided the resources. S.M.E. Smith and T.E. DeCoursey visualized the study. T.E. DeCoursey wrote the original draft of the manuscript. S.M.E. Smith and T.E. DeCoursey reviewed and edited the manuscript.
Richard W. Aldrich served as editor.
References
- Bennett A.L., and Ramsey I.S.. 2017. CrossTalk opposing view: proton transfer in Hv1 utilizes a water wire, and does not require transient protonation of a conserved aspartate in the S1 transmembrane helix. J. Physiol. 595:6797–6799. 10.1113/JP274553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T.K., Fußhöller D.M., Goodwin N., Bönigk W., Müller A., Dokani Khesroshahi N., Brenker C., Wachten D., Krause E., Kaupp U.B., and Strünker T.. 2017. Post-translational cleavage of Hv1 in human sperm tunes pH- and voltage-dependent gating. J. Physiol. 595:1533–1546. 10.1113/JP273189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim L., Krause R.M., Baroffio A., Hamann M., Kaelin A., and Bader C.R.. 1993. A voltage-dependent proton current in cultured human skeletal muscle myotubes. J. Physiol. 470:313–333. 10.1113/jphysiol.1993.sp019860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L., Meech R., and Moody W. Jr. 1984. Rapidly activating hydrogen ion currents in perfused neurones of the snail, Lymnaea stagnalis. J. Physiol. 351:199–216. 10.1113/jphysiol.1984.sp015241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Mandloi S., Lanczycki C.J., Panchenko A.R., and Chakrabarti S.. 2012. SPEER-SERVER: a web server for prediction of protein specificity determining sites. Nucleic Acids Res. W242-W248 10.1093/nar/gks559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin A., Qiu F., Rebolledo S., Wang Y., Noskov S.Y., and Larsson H.P.. 2014. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 111:E273–E282. 10.1073/pnas.1318018111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves G., Derst C., Franzen A., Mashimo Y., Machida R., and Musset B.. 2016. Identification of an HV 1 voltage-gated proton channel in insects. FEBS J. 283:1453–1464. 10.1111/febs.13680 [DOI] [PubMed] [Google Scholar]
- Cherny V.V., and DeCoursey T.E.. 1999. pH-dependent inhibition of voltage-gated H+ currents in rat alveolar epithelial cells by Zn2+ and other divalent cations. J. Gen. Physiol. 114:819–838. 10.1085/jgp.114.6.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny V.V., Markin V.S., and DeCoursey T.E.. 1995. The voltage-activated hydrogen ion conductance in rat alveolar epithelial cells is determined by the pH gradient. J. Gen. Physiol. 105:861–896. 10.1085/jgp.105.6.861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny V.V., Henderson L.M., and DeCoursey T.E.. 1997. Proton and chloride currents in Chinese hamster ovary cells. Membr. Cell Biol. 11:337–347. [PubMed] [Google Scholar]
- Cherny V.V., Murphy R., Sokolov V., Levis R.A., and DeCoursey T.E.. 2003. Properties of single voltage-gated proton channels in human eosinophils estimated by noise analysis and by direct measurement. J. Gen. Physiol. 121:615–628. 10.1085/jgp.200308813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny V.V., Morgan D., Musset B., Chaves G., Smith S.M.E., and DeCoursey T.E.. 2015. Tryptophan 207 is crucial to the unique properties of the human voltage-gated proton channel, hHV1. J. Gen. Physiol. 146:343–356. 10.1085/jgp.201511456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 1991. Hydrogen ion currents in rat alveolar epithelial cells. Biophys. J. 60:1243–1253. 10.1016/S0006-3495(91)82158-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2003. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83:475–579. 10.1152/physrev.00028.2002 [DOI] [PubMed] [Google Scholar]
- DeCoursey T.E. 2010. Voltage-gated proton channels find their dream job managing the respiratory burst in phagocytes. Physiology (Bethesda). 25:27–40. 10.1152/physiol.00039.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2013. Voltage-gated proton channels: molecular biology, physiology, and pathophysiology of the HV family. Physiol. Rev. 93:599–652. 10.1152/physrev.00011.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2015. The voltage-gated proton channel: a riddle, wrapped in a mystery, inside an enigma. Biochemistry. 54:3250–3268. 10.1021/acs.biochem.5b00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2017. CrossTalk proposal: Proton permeation through HV1 requires transient protonation of a conserved aspartate in the S1 transmembrane helix. J. Physiol. 595:6793–6795. 10.1113/JP274495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2018. Voltage and pH sensing by the voltage-gated proton channel, HV1. J. R. Soc. Interface. 15:20180108 10.1098/rsif.2018.0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., and Cherny V.V.. 1993. Potential, pH, and arachidonate gate hydrogen ion currents in human neutrophils. Biophys. J. 65:1590–1598. 10.1016/S0006-3495(93)81198-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., and Cherny V.V.. 1995. Voltage-activated proton currents in membrane patches of rat alveolar epithelial cells. J. Physiol. 489:299–307. 10.1113/jphysiol.1995.sp021051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., and Cherny V.V.. 1997. Deuterium isotope effects on permeation and gating of proton channels in rat alveolar epithelium. J. Gen. Physiol. 109:415–434. 10.1085/jgp.109.4.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., and Cherny V.V.. 1998. Temperature dependence of voltage-gated H+ currents in human neutrophils, rat alveolar epithelial cells, and mammalian phagocytes. J. Gen. Physiol. 112:503–522. 10.1085/jgp.112.4.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E., Morgan D., and Cherny V.V.. 2003. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 422:531–534. 10.1038/nature01523 [DOI] [PubMed] [Google Scholar]
- DeCoursey T.E., Morgan D., Musset B., and Cherny V.V.. 2016. Insights into the structure and function of HV1 from a meta-analysis of mutation studies. J. Gen. Physiol. 148:97–118. 10.1085/jgp.201611619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N., Grinstein S., Jaconi M., Schlegel W., Lew D.P., and Krause K.H.. 1993. Proton currents in human granulocytes: regulation by membrane potential and intracellular pH. J. Physiol. 466:329–344. [PMC free article] [PubMed] [Google Scholar]
- Dudev T., Musset B., Morgan D., Cherny V.V., Smith S.M.E., Mazmanian K., DeCoursey T.E., and Lim C.. 2015. Selectivity mechanism of the voltage-gated proton channel, HV1. Sci. Rep. 5:10320 10.1038/srep10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordienko D.V., Tare M., Parveen S., Fenech C.J., Robinson C., and Bolton T.B.. 1996. Voltage-activated proton current in eosinophils from human blood. J. Physiol. 496:299–316. 10.1113/jphysiol.1996.sp021686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., and Sackin H.. 1995. Effect of pH on potassium and proton conductance in renal proximal tubule. Am. J. Physiol. 269:F289–F308. 10.1152/ajprenal.1995.269.3.F289 [DOI] [PubMed] [Google Scholar]
- Henderson L.M., Chappell J.B., and Jones O.T.G.. 1987. The superoxide-generating NADPH oxidase of human neutrophils is electrogenic and associated with an H+ channel. Biochem. J. 246:325–329. 10.1042/bj2460325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapus A., Romanek R., Qu A.Y., Rotstein O.D., and Grinstein S.. 1993. A pH-sensitive and voltage-dependent proton conductance in the plasma membrane of macrophages. J. Gen. Physiol. 102:729–760. 10.1085/jgp.102.4.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., and Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H.P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., and Larsson H.P.. 2008. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 105:9111–9116. 10.1073/pnas.0801553105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulleperuma K., Smith S.M.E., Morgan D., Musset B., Holyoake J., Chakrabarti N., Cherny V.V., DeCoursey T.E., and Pomès R.. 2013. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 141:445–465. 10.1085/jgp.201210856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno M., Ando H., Morihata H., Sakai H., Mori H., Sawada M., and Oiki S.. 2009. Temperature dependence of proton permeation through a voltage-gated proton channel. J. Gen. Physiol. 134:191–205. 10.1085/jgp.200910213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Letts J.A., and Mackinnon R.. 2008. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc. Natl. Acad. Sci. USA. 105:7692–7695. 10.1073/pnas.0803277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Shen R., Treger J.S., Wanderling S.S., Milewski W., Siwowska K., Bezanilla F., and Perozo E.. 2015. Resting state of the human proton channel dimer in a lipid bilayer. Proc. Natl. Acad. Sci. USA. 112:E5926–E5935. 10.1073/pnas.1515043112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D., Musset B., Kulleperuma K., Smith S.M.E., Rajan S., Cherny V.V., Pomès R., and DeCoursey T.E.. 2013. Peregrination of the selectivity filter delineates the pore of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 142:625–640. 10.1085/jgp.201311045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R., and DeCoursey T.E.. 2006. Charge compensation during the phagocyte respiratory burst. Biochim. Biophys. Acta. 1757:996–1011. 10.1016/j.bbabio.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Musset B., Cherny V.V., Morgan D., Okamura Y., Ramsey I.S., Clapham D.E., and DeCoursey T.E.. 2008. Detailed comparison of expressed and native voltage-gated proton channel currents. J. Physiol. 586:2477–2486. 10.1113/jphysiol.2007.149427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset B., Smith S.M.E., Rajan S., Cherny V.V., Sujai S., Morgan D., and DeCoursey T.E.. 2010. Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J. Physiol. 588:1435–1449. 10.1113/jphysiol.2010.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset B., Smith S.M.E., Rajan S., Morgan D., Cherny V.V., and DeCoursey T.E.. 2011. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277. 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström T., Rotstein O.D., Romanek R., Asotra S., Heersche J.N., Manolson M.F., Brisseau G.F., and Grinstein S.. 1995. Regulation of cytoplasmic pH in osteoclasts. Contribution of proton pumps and a proton-selective conductance. J. Biol. Chem. 270:2203–2212. 10.1074/jbc.270.5.2203 [DOI] [PubMed] [Google Scholar]
- Ramsey I.S., Mokrab Y., Carvacho I., Sands Z.A., Sansom M.S.P., and Clapham D.E.. 2010. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat. Struct. Mol. Biol. 17:869–875. 10.1038/nsmb.1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.D., Haq S., Bachvaroff T., Nowak K.F., Nowak S.J., Morgan D., Cherny V.V., Sapp M.M., Bernstein S., Bolt A., et al. . 2017. Identification of a vacuolar proton channel that triggers the bioluminescent flash in dinoflagellates. PLoS One. 12:e0171594 10.1371/journal.pone.0171594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata S., Miyawaki N., McCormack T.J., Arima H., Kawanabe A., Özkucur N., Kurokawa T., Jinno Y., Fujiwara Y., and Okamura Y.. 2016. Comparison between mouse and sea urchin orthologs of voltage-gated proton channel suggests role of S3 segment in activation gating. Biochim. Biophys. Acta. 1858:2972–2983. 10.1016/j.bbamem.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Schilling T., Gratopp A., DeCoursey T.E., and Eder C.. 2002. Voltage-activated proton currents in human lymphocytes. J. Physiol. 545:93–105. 10.1113/jphysiol.2002.028878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenzel J., Lew D.P., and Krause K.H.. 1996. Proton currents in human eosinophils. Am. J. Physiol. 271:C1861–C1871. 10.1152/ajpcell.1996.271.6.C1861 [DOI] [PubMed] [Google Scholar]
- Smith S.M.E., Morgan D., Musset B., Cherny V.V., Place A.R., Hastings J.W., and DeCoursey T.E.. 2011. Voltage-gated proton channel in a dinoflagellate. Proc. Natl. Acad. Sci. USA. 108:18162–18167. 10.1073/pnas.1115405108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szteyn K., Yang W., Schmid E., Lang F., and Shumilina E.. 2012. Lipopolysaccharide-sensitive H+ current in dendritic cells. Am. J. Physiol. Cell Physiol. 303:C204–C212. 10.1152/ajpcell.00059.2012 [DOI] [PubMed] [Google Scholar]
- Takeshita K., Sakata S., Yamashita E., Fujiwara Y., Kawanabe A., Kurokawa T., Okochi Y., Matsuda M., Narita H., Okamura Y., and Nakagawa A.. 2014. X-ray crystal structure of voltage-gated proton channel. Nat. Struct. Mol. Biol. 21:352–357. 10.1038/nsmb.2783 [DOI] [PubMed] [Google Scholar]
- Thomas S., Cherny V.V., Morgan D., Artinian L.R., Rehder V., Smith S.M.E., and DeCoursey T.E.. 2018. Exotic properties of a voltage-gated proton channel from the snail Helisoma trivolvis. J. Gen. Physiol. 10.1085/201711967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F., Ulbrich M.H., and Isacoff E.Y.. 2008. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 58:546–556. 10.1016/j.neuron.2008.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Keulen S.C., Gianti E., Carnevale V., Klein M.L., Rothlisberger U., and Delemotte L.. 2017. Does proton conduction in the voltage-gated H+ channel hHv1 involve Grotthuss-like hopping via acidic residues? J. Phys. Chem. B. 121:3340–3351. 10.1021/acs.jpcb.6b08339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D., and Argos P.. 1996. Intrahelical side chain-side chain contacts: the consequences of restricted rotameric states and implications for helix engineering and design. Protein Eng. 9:471–478. 10.1093/protein/9.6.471 [DOI] [PubMed] [Google Scholar]