Abstract
Massive strides have been made in technologies for collecting genome-scale data. However, tools for efficiently and flexibly assembling raw outputs into downstream analytical workflows are still nascent. aTRAM 1.0 was designed to assemble any locus from genome sequencing data but was neither optimized for efficiency nor able to serve as a single toolkit for all assembly needs. We have completely re-implemented aTRAM and redesigned its structure for faster read retrieval while adding a number of key features to improve flexibility and functionality. The software can now (1) assemble single- or paired-end data, (2) utilize both read directions in the database, (3) use an additional de novo assembly module, and (4) leverage new built-in pipelines to automate common workflows in phylogenomics. Owing to reimplementation of databasing strategies, we demonstrate that aTRAM 2.0 is much faster across all applications compared to the previous version.
Keywords: locus assembly, short-read sequencing, massively parallel sequencing, aTRAM, software
Introduction
Rapid, targeted locus assembly from massive sequencing runs is now commonly used across the biological sciences, with applications ranging from medicine to broad-scale phylogenomics. Sequencing methods vary from whole shotgun sequencing to reduced genome applications.1 In both reduced-genome and whole genome datasets, targeted assembly approaches can greatly reduce the computational scale of the assembly problem compared to full genome de novo assembly, and avoid errors introduced during whole genome assembly (Pop, 2009; Salzburg et al., 2005).2–5
aTRAM 1.0—automated Target Restricted Assembly Method—was developed to assemble any locus from next-generation sequencing (NGS) data.6,7 In this process, the locus of interest is targeted from the NGS reads and the assembly restricted to include only matching reads. It was written in Perl and designed to both work on individual computers and in high-performance computing contexts. The ability to parallelize assembly tasks at multiple scales and its implementation with a modular design philosophy crucially allows aTRAM to keep pace with development of new assembly approaches as they are published and requested by end users, all within a single, unified environment. Finally, aTRAM uses BLAST to find matching reads which allows for locus assemblies across very divergent taxa, those without a closely related sequenced genome. For example, protein-coding genes have been assembled across insects with > 150 million years of evolutionary distance.7
Here, we present aTRAM 2.0, a full reimplementation of aTRAM 1.0 in order to add new features, greatly increase control over assembly through more fine-tuning options, and reimplement previous methods for more efficient processing. The major target areas for improvement were as follows: (1) index both reads into BLAST databases to increase hits, (2) speed up sequence querying and retrieval through new databasing strategies, (3) reverse the filtering step such that assembled contigs BLAST against the target, (4) add an additional de novo assembler to enable further use cases, (5) add automated pipelines for rapid assembly of many loci from many taxa, and (6) enhance long-term sustainability and performance via a full code rewrite in Python.
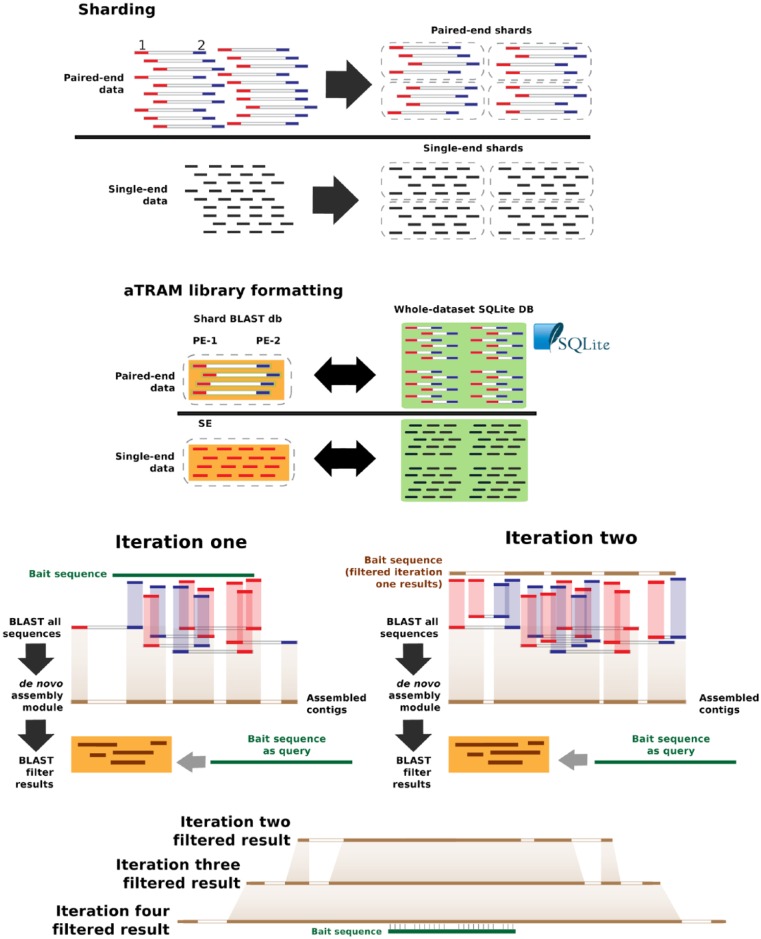
aTRAM 2.0 is currently a command-line python-based tool that works in two steps. First, users specify locus targets and a set of user options, and then an aTRAM-formatted library is built where the paired-end reads are separated into shards. Sharding is independent of sequence content, and is based on a database size threshold (250 Mb by default with the size and number of shards user-adjustable). The reads are grouped by mate-pairs and added to the shards in the order they appear from the original file until all reads have been included in a shard. This dataset division enables parallelized read queries and greatly improves performance even for serial queries. For each shard, a BLAST-formatted database is built for both mate-pairs. In the second step, the locus of interest is targeted and assembled. To do this, a query sequence is BLASTed against the sharded database; matching reads are assembled with one of a set of de novo assembly modules. Assemblies are improved by an iterative approach: in the second iteration, the assembled contigs replace the original query, and are blasted against the short read database. Matching reads are then assembled de novo as in the first iteration. This process continues until the user-specified iteration limit is reached or no new contigs are assembled (Figure 1). Parallelization is possible at multiple levels because each gene is assembled independently many genes can be quickly assembled simultaneously in parallel. Furthermore, parallel processing is possible within each run as the database is split into shards, allowing each to be searched independently. Finally, parallelization control within assembly modules is also implemented. These processing steps are now highly customizable via user option settings. For example, it is possible to search only a fraction of the aTRAM library for high-coverage targets in large datasets (e.g. mitochondrial genomes); it is also possible to leverage many of the options in component de novo assembly modules.
Figure 1.
Overall aTRAM workflow, which includes a preparation process followed by assembly steps. The preparation process includes sharding raw data into an aTRAM library, including construction of a whole-dataset SQLite database from raw reads. The assembly process uses a bait sequence or set of sequences to perform an iterative assembly. The whole process generates assembled reads that are often longer than target baits as shown in later iterations below.
aTRAM 2.0 Implementation
Complete re-implementation in Python
We fully re-implemented the code from perl to Python3, redesigning the code with greater modularity. Our aim was to keep apace with the increase in bioinformatics development and availability of Python libraries. This reimplementation aligns with plans for broader development efforts and more sustainable, long-term growth of the toolkit.
SQLite database
The aTRAM database system was reimplemented as a SQLite database to store and greatly speed up retrieval of mate-pairs in each iteration.
Indexing and databasing of both mate-pairs
The aTRAM database now includes both mate pairs in the BLASTing and the indexing. Therefore, either read is allowed to match the target rather than the first read, resulting in a greater number of overall matches per iteration and typically fewer iterations required to reach an optimized assembly.
Reverse order of BLAST for faster speeds
aTRAM includes customizable filtering criteria for contigs, which is essential for controlling the quality of iterative assemblies. We have reversed the order of BLAST searches with each contig. Rather than large numbers of individual contig searches, after the de-novo assembly, a new BLAST database is built from assembled contigs in order to perform a single BLAST search against the target locus.
Multiplexed BLAST searches for many loci
We allow all query sequences to be in the same file, automatically piping data to individual locus assemblies.
Single- and paired-end data
aTRAM 2.0 now allows for single- as well as paired-end data, and the code base has also been modified with greater flexibility for multiple Illumina FASTQ and FASTA standards, enabling backwards compatibility with legacy datasets.
SPAdes assembler added
aTRAM 1.0 implemented Velvet,8 Abyss,9 and Trinity 10 as the de novo assemblers. aTRAM 2.0 additionally implements SPAdes,11 an assembler frequently used for target capture data in plants and especially beneficial for long targets.
Pipelines
We have implemented pipelines for common phylogenomics tasks. Users typically wish to assemble a set of loci from many libraries. Traditionally, tools of this type require users to develop custom loop scripts to perform these, which increase the learning curve for tools and can be inefficient if database queries are redundant. In aTRAM 2.0, when library and locus lists are provided, the software will automatically parallelize and distribute individual assembly tasks.
Test suite
We use a suite of pytests to ensure that current features work as expected. A set of regression tests also ensures that any code changes do not cause any unwanted effects.
Documentation
There is now a detailed manual with examples via a github README page (www.github.com/juliema/aTRAM/).
Performance Comparison with aTRAM 1.0
The end result of re-implementing aTRAM is a 3- to 10-fold increase in the combined speed of both the library preparation and locus assembly steps. Traditionally, aTRAM has been used to assemble protein coding genes from whole genome sequencing efforts. We compared the time it took to assemble five protein coding genes using the original test dataset of the chimpanzee louse, Pediculus schaeffi6 using aTRAM 1.0 and aTRAM 2.0 on the same Dell Precision server with Dual 20-core Xeon processors and 64 GB of RAM. We found that on average aTRAM 2.0 assembled the same gene 2.5 times faster with a range of (2.0× to 2.7×).
One of the key uses of aTRAM is to assemble targeted loci from enriched datasets. We compared assemblies of 5 ultraconserved elements (UCEs)12 from an unpublished mammal dataset (unpublished data, K. Bell). On average, aTRAM 2.0 assembled loci 10x faster than aTRAM 1.0 using the same test environment as above. On average, aTRAM 1.0 would assemble a UCE in ~100 seconds; aTRAM 2.0 now assembles equivalent or longer contigs in ~10 seconds. Furthermore, aTRAM 2.0 assembled, on average, 18% longer UCE loci than aTRAM 1.0. Due to the new data structure and greater use of mate-pair information in aTRAM 2.0, fewer iterations are needed to get to an equivalent or longer final assembly. For example, the number of reads found in iteration one is on average 1.8× the number of reads found in iteration 1 from aTRAM 1.0 (min = 1.18, max = 2.9×).
For reduced datasets that have a moderate target sequence length, the full locus is typically assembled in approximately half as many iterations. For runs constrained to the same number of iterations, aTRAM 2.0 is typically fivefold faster; hence, reimplementation of the code has resulted in improved runtimes both within and between iterations. Future work on this software will include comparisons with other targeted locus assemblers. These improvements set the stage for further rapid and cost-effective development of aTRAM as core software usable across the life sciences wherever workflows require assembly of protein coding genes, ultra-conserved elements, or any other type of targeted loci. As well, aTRAM 2.0 has particularly strong application and use for assembling genes from bacterial symbionts, mitochondrial genes and other associated genomes. aTRAM was designed as a multipurpose tool for researchers across the life sciences.
Acknowledgments
The authors wish to thank Kayce Bell, Bryan McLean, and Joe Cook for use of unpublished mammal data. Matt Gitzendanner is thanked for assistance in testing on the UF’s HiPerGator. Vijay Barve provided useful comments on this manuscript.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a pilot grant from the University of Florida Genetics Institute.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JMA, KPJ, and RPG designed the program; RAL wrote the code; JMA, RPG, RAF, and RF wrote documentation; JMA, RAF, RL, KPJ, and RPG wrote the manuscript.
Availability of Data: aTRAM 2.0 is available at http://www.github.com/juliema/aTRAM.
References
- 1. McCormack JE, Hird SM, Zellmer AJ. Applications of next-generation sequencing to phylogeography and phylogenetics. Molec Phylogen Evol. 2012;66:526–538. [DOI] [PubMed] [Google Scholar]
- 2. Alkan C, Saba S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson KP, Walden KKO, Robertson HM. Next-generation phylogenomics using a target restricted assembly method. Molec Phylogen Evol. 2013;66:417–422. [DOI] [PubMed] [Google Scholar]
- 4. Pop M. (2009). Genome assembly reborn: recent computational challenges. Brief Bioinformatics. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005;21:4320–4321. [DOI] [PubMed] [Google Scholar]
- 6. Allen JM, Huang DI, Cronk QC, Johnson KP. aTRAM—automated target restricted assembly method: a fast method for assembling loci across divergent taxa from next-generation sequencing data. BMC Bioinformatics. 2015;16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allen JM, Boyd B, Nguyen N, et al. Phylogenomics from whole genome sequences using aTRAM. Systemat Biol. 2017;66:786–798. doi: 10.1093/sysbio/syw105. [DOI] [PubMed] [Google Scholar]
- 8. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Research. 2009;19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bankevich A, Nurk S, Antipov D, et al. A new genome assembly algorithm and its applications to single cell sequencing. J Computat Biol. 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Systemat Biol. 2012;61:717–726. doi: 10.1093/sysbio/sys004. [DOI] [PubMed] [Google Scholar]