Schmidt et al. propose a new concept for colon cancer therapy that advocates specific and simultaneous targeting of different tumor cell subpopulations with high NOTCH- and MAPK-signaling pathway activity. This overcomes treatment resistance by tumor cell plasticity and strongly improves therapy response.
Abstract
In colorectal cancer, signaling pathways driving tumor progression are promising targets for systemic therapy. Besides WNT and MAPK signaling, activation of NOTCH signaling is found in most tumors. Here, we demonstrate that high NOTCH activity marks a distinct colon cancer cell subpopulation with low levels of WNT and MAPK activity and with a pronounced epithelial phenotype. Therapeutic targeting of MAPK signaling had limited effects on tumor growth and caused expansion of tumor cells with high NOTCH activity, whereas upon targeting NOTCH signaling, tumor cells with high MAPK activity prevailed. Lineage-tracing experiments indicated high plasticity between both tumor cell subpopulations as a mechanism for treatment resistance. Combined targeting of NOTCH and MAPK had superior therapeutic effects on colon cancer growth in vivo. These data demonstrate that tumor cells may evade systemic therapy through tumor cell plasticity and provide a new rationale for simultaneous targeting of different colon cancer cell subpopulations.
Introduction
Colorectal cancer is a major cause of cancer morbidity and mortality, ranking third in incidence among men and women (Jemal et al., 2010). Although complete surgical removal of the tumor may be curative, treatment of advanced disease relies on systemic therapy including the use of biologically active agents that target signaling pathways related to tumor progression (Heinemann et al., 2014). In this context, targeting MAPK signaling by blocking EGFR with therapeutic antibodies is a commonly used approach (Miyamoto et al., 2017), while more recently MEK inhibition also is being clinically evaluated (Bennouna et al., 2011). However, targeting of MAPK signaling has limited effects and on average prolongs patient survival by a few months only (Van Cutsem et al., 2009; Douillard et al., 2014), indicating the urgent need for radical improvements in targeted therapy for patients with colorectal cancer.
Besides WNT- and MAPK-signaling pathways that often are activated by mutations and contribute to tumor progression (Cancer Genome Atlas Network, 2012), active NOTCH signaling has been observed in colon cancer (Sonoshita et al., 2011). NOTCH is an evolutionary conserved signaling pathway involved in embryonic development, cell fate decisions, and tissue homeostasis (Bray, 2006). Signaling is activated by binding of NOTCH ligands to their receptors with sequential proteolytic processing, including an intracellular cleavage by γ-secretases, that generate active NOTCH intracellular domains (NICDs). NICDs then form DNA-binding complexes with other protein partners, such as RBPJκ, and activate the expression of typical NOTCH effectors, including HES1 (Sang et al., 2010). In colon cancer, high NOTCH activity has been linked to the cancer stem cell phenotype (Bu et al., 2013) and to epithelial-mesenchymal transition (EMT; Brabletz et al., 2011), both of which are drivers of tumor progression. Moreover, high NOTCH activity has been associated with poor survival (Yuan et al., 2015), suggesting that NOTCH contributes to tumor progression and that targeting NOTCH may be clinically effective. However, in conflict with this idea, others demonstrated repressive functions of NOTCH on WNT and MAPK signaling (Kim et al., 2012; Rampias et al., 2014), and treatment trials with γ-secretase inhibitors repressing NOTCH in colon cancer so far have been disappointing (Strosberg et al., 2012; Tolcher et al., 2012). The role of NOTCH signaling for colon cancer progression and its translational relevance for therapeutic targeting therefore still remain unclear.
Signaling pathways that are active in colon cancer can be strongly regulated within the tumor. Specifically, WNT and MAPK signaling typically are high in tumor cells at the infiltrative tumor margin or tumor edge only, where putative colon cancer stem cells reside and where tumor cells undergo EMT (Brabletz et al., 2001; Horst et al., 2012; Blaj et al., 2017). On the contrary, colon cancer cells that are located more centrally within the tumor have comparatively low activity for both pathways and assume more differentiated epithelial phenotypes (Vermeulen et al., 2010; Cernat et al., 2014; Blaj et al., 2017). However, the intratumoral distribution of NOTCH activity and associated tumor cell phenotypes have remained poorly characterized. Moreover, it is unknown to what extent intratumoral heterogeneity of signaling pathways contributes to resistance against targeted therapies of colon cancer. To shed light on these issues, we characterized NOTCH-signaling activity in colon cancer in detail, tested effects of targeted therapy on tumor cell subpopulations with differential pathway activity, and derived a more efficient treatment strategy by targeting different tumor cell subpopulations at the same time.
Results
High NOTCH activity indicates a distinct tumor cell subpopulation in colon cancer
To obtain insights into the role of the NOTCH pathway in colorectal cancer, we examined tissue specimens of a total of 328 adenocarcinomas for accumulation of NICD, which indicates activation of NOTCH signaling. Immunostaining revealed widespread nuclear accumulation of NICD in tumor cells of most cases (80.5%; Fig. 1 A). Interestingly, however, NICD was not evenly distributed within these tumors. Specifically, colon cancer cells that were located at the tumor edge were negative for NICD in 89.4% of these cases, whereas, in contrast, tumor cells located closer to the tumor center abruptly became NICD positive (Fig. 1 A). We then examined the NOTCH effector HES1 in a subset of 225 cases. Similar to the pattern of NICD, we also found expression in the center of colorectal cancers (66.2%), whereas its expression was diminished or absent in tumor cells at the tumor edge (Fig. S1 A). Collectively, these findings showed that NOTCH signaling is activated in the center of colorectal cancers but unexpectedly down-regulated at the infiltrative tumor edge.
Figure 1.
Distribution and phenotype of colorectal cancer cells with high NOTCH activity. (A) Representative immunostaining for NICD in primary colon cancer tissue. Right panel shows higher magnification of area boxed in the left panel. Arrowheads indicate tumor cells at the tumor edge, and arrows indicate tumor cells toward the tumor center; open arrowhead indicates an endothelial cell. (B, D, F, and H) Double immune fluorescence for indicated proteins in representative colon cancer tissues. Arrowheads indicate tumor cells at the tumor edge, and arrows indicate tumor cells toward the tumor center. Open arrow indicates endothelial cells at the tumor edge. (C, E, G, and I) Quantification of co–immune fluorescence signals. Relative fluorescence intensities (% RFI) for indicated proteins in tumor cells with high (upper quartile) and low (lower quartile) NICD staining intensity are shown. Data are derived from n ≥ 500 tumor cells in n = 10 different colorectal cancer cases. Error bars indicate mean ± SD. **, P < 0.01; ***, P < 0.001 by t test. Bars: 200 µm (A, left); 20 µm (A, right); 50 µm (B, D, F, and H).
Colon cancer cells at the tumor edge are known to activate MAPK and WNT signaling (Blaj et al., 2017). Therefore, we compared the activity of both pathways with the status of the NOTCH pathway. Using FRA1 and nuclear β-catenin as indicators for MAPK and WNT activity, respectively, we found that tumor cells with strong staining for these markers showed significantly decreased or absent staining for NICD (Fig. 1, B–E). On the contrary, colon cancer cells with high levels of NICD showed much lower expression of FRA1 and nuclear β-catenin. Moreover, tumor cells that were positive for NICD on average were more numerous and also showed higher proliferation rates than tumor cells with FRA1 expression (Fig. S1, B and C). High activities of NOTCH and MAPK/WNT therefore were mutually exclusive in colon cancer cells and marked distinct tumor cell subpopulations. Next, we tested for an association of NOTCH signaling and EMT. Double immune fluorescence staining showed that colon cancer cells with high LAMC2 expression levels, a marker indicating EMT in colon cancer (Sánchez-Tilló et al., 2011), were devoid of strong NICD accumulation, whereas, in contrast, colon cancer cells with high NICD levels showed low LAMC2 expression (Fig. 1, F and G). Moreover, colon cancer cells with high NICD levels had significantly higher expression of the epithelial cell adhesion molecule E-cadherin, when compared with colon cancer cells with low NICD levels (Fig. 1, H and I). These findings demonstrate that colon cancers are composed of distinct tumor cell subpopulations, including tumor cells at the tumor edge with high MAPK and WNT activity undergoing EMT and tumor cells with high NOTCH activity in the tumor center that have a more epithelial phenotype.
MAPK and NOTCH activity are associated with colon cancer progression
Next, we investigated the clinical relevance of tumor cell subpopulations with high MAPK and NOTCH activity by scoring FRA1 and NICD in our collection of colorectal cancer cases (Fig. 2 A). 225 of these cases were UICC stage II colorectal cancers with recorded clinical follow-up data (Table S1). Kaplan-Meier statistics revealed that FRA1-negative cases were associated with a tendency toward better cancer-specific and disease-free survival when compared with FRA1-positive cases (Fig. 2 B). We then tested for associations with NICD staining and found that colorectal cancers with <10% NICD-positive tumor cells (NICD low) also showed significantly better cancer-specific and disease-free survival than cases with higher frequencies of NICD-positive tumor cells (NICD high; Fig. 2 B). Furthermore, when testing a combined evaluation of FRA1 and NICD, we found that patients whose tumors were both FRA1 negative and NICD low almost perfectly survived the follow-up period, with no event of cancer-specific death and only one event of tumor progression (Fig. 2 B). In contrast, patients whose tumors were either FRA1 positive, NICD high, or both showed significantly poorer cancer-specific and disease-free survival. Testing for associations with other clinical and pathological variables revealed that FRA1 positivity and high NICD levels were more frequent in low than in high-grade colon cancers, whereas the other core clinical variables T-category, age, and sex, as well as KRAS mutation status were not linked to FRA1 or NICD (Table S1). We then included these variables into proportional hazards regression analyses and found that combined absence of FRA1 and NICD was an independent predictor of favorable outcome for disease-free survival (Table S2).
Figure 2.
Prognostic relevance of MAPK and NOTCH in colorectal cancer. (A) Assessment of FRA1 and NICD in primary human colorectal cancers. For FRA1, tumors were categorized as negative or positive, based on absence or presence of detectable immunostaining in tumor cells. For NICD, cases were categorized as low or high, based on less or more than 10% tumor cells with strong NICD staining, respectively. (B) Survival associations of FRA1, NICD, and their combination in 225 UICC stage II colorectal cancers. Kaplan-Meier plots for cancer-specific survival and disease free survival are shown. P-values are log-rank test results. Ratios on curves indicate the number of events over the number of patients per group. (C) Association of FRA1, NICD, and their combination with liver metastasis in a matched case-control collection of 92 colon cancers. *, P < 0.05; **, P < 0.01 by χ2 test. (D) Representative staining for FRA1 and NICD in a primary colon cancer and corresponding liver metastasis. Arrowheads indicate tumor cells at the tumor edge, and arrows indicate tumor cells toward the tumor center; open arrowheads indicate endothelial cells. Bars, 50 µm.
Furthermore, we evaluated 92 colon cancers of a case-control collection of matched tumor pairs, with and without synchronous liver metastasis (Table S3), applying the same scoring method as for the UICC II collection. We observed that tumors with liver metastases were significantly more often FRA1 positive, NICD high, or both, whereas on the contrary, none of the few tumors that were FRA1 negative and NICD low had metastasized (Fig. 2 C; Table S3). We then also examined another 11 colorectal cancers of which we obtained paired tissues of primary tumors and corresponding metastases to the liver (n = 6), the peritoneum (n = 3), or the lung (n = 2). Interestingly, nine metastases had recapitulated the patterns of FRA1- and NICD-positive tumor cells of their primary tumors (Fig. 2 D), whereas only two, in contrast to their primary tumors, showed absent or lower FRA1 or NICD staining, respectively. Collectively, these findings suggested that tumor cell subpopulations with MAPK and NOTCH activity are both important for colon cancer progression in early- and late-stage disease, frequently show similar presence in primary tumors and corresponding metastases, and that best clinical outcome may be expected if the activity of both pathways is low.
Colon cancers evade MAPK- or NOTCH-targeted therapy by shifting their phenotype
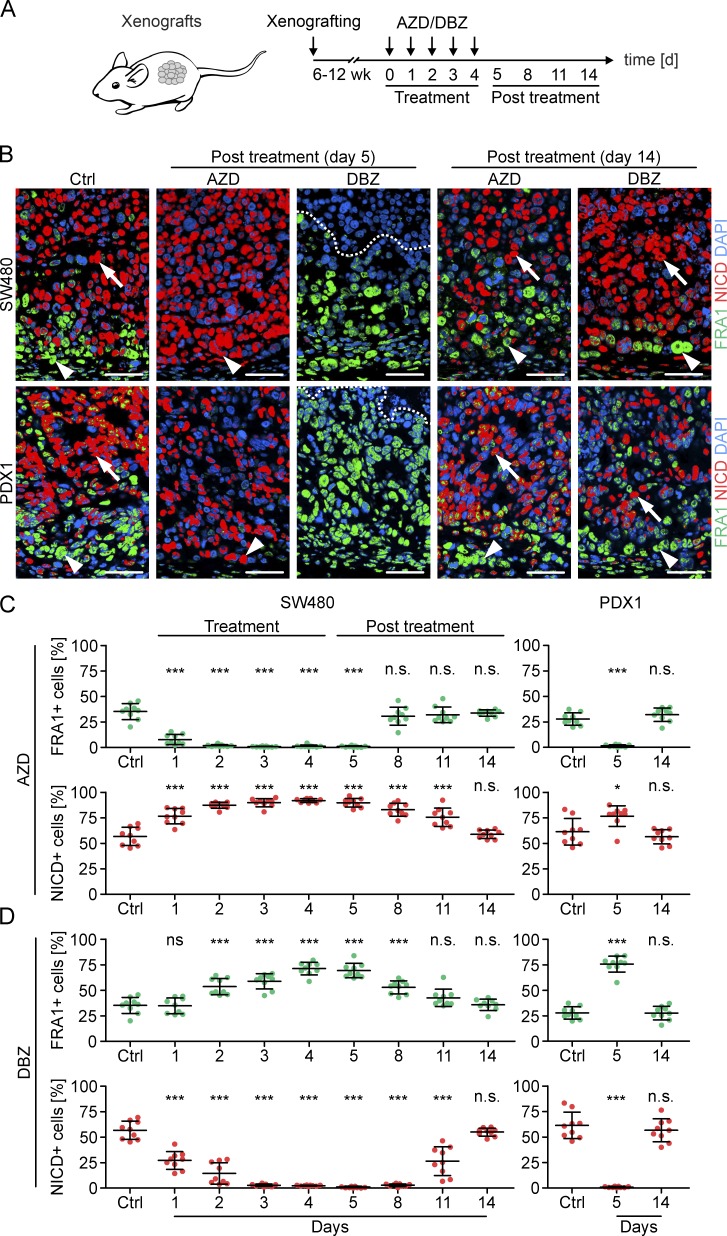
MAPK and NOTCH signaling can be repressed with the MEK inhibitor selumetinib (AZD) and the γ-secretase inhibitor dibenzazepine (DBZ), respectively. To evaluate the effects of either treatment, we used mouse models of cell line–derived SW480 or patient-derived PDX1 colon cancer xenografts (Fig. 3 A). Xenograft tumors were composed of tumor cell subpopulations with strong expression of FRA1 at the tumor edge, and accumulation of NICD toward the tumor center and thus adequately modeled the intratumoral composition and distribution of MAPK and NOTCH activity in primary colon cancers (Fig. 3 B). We then treated mice bearing these xenografts with AZD for 5 d and observed that FRA1 expression was completely lost in these tumors (Fig. 3, B and C). Time course analysis showed that in SW480 xenografts this already occurred after 2 d of treatment (Fig. 3 C), which indicated strong and rapid repressive effects of AZD on MAPK signaling, as expected. Surprisingly, however, the frequency of NICD-positive tumor cells significantly expanded under AZD treatment in SW480 and PDX1 xenografts, and these cells then directly reached the tumor edge, suggesting an expansion of intratumoral NOTCH activity under MAPK repression (Fig. 3, B and C). In addition, AZD treatment reduced the number of cleaved caspase-3–labeled tumor cells, indicating that the loss of FRA1-positive tumor cells was not a result of increased apoptosis (Fig. S2). These findings suggest that colon cancer cell subpopulations switched from MAPK to NOTCH activity upon MAPK repression.
Figure 3.
Effects of MAPK and NOTCH repression in colon cancer xenografts. (A) Schema and experimental schedule for xenografting, inhibitor treatment, and tumor analysis. (B) Immunofluorescence for FRA1 and NICD in SW480 and patient-derived (PDX1) xenografts. Vehicle-treated tumors (Ctrl) and AZD- or DBZ-treated tumors at indicated time points were analyzed. Arrowheads indicate tumor cells at the tumor edge, and arrows indicate tumor cells toward the tumor center. Areas above dotted lines are tumor necrosis. Bars, 50 µm. (C and D) Quantification of FRA1- and NICD-positive tumor cells in SW480 and PDX1 xenografts. Vehicle-treated tumors (Ctrl) and tumors at indicated time points during and after AZD (C) or DBZ (D) treatment were analyzed. Error bars are mean ± SD. *, P < 0.05; ***, P < 0.001 by t test; n.s., not significant; compared with Ctrl. n ≥ 3 independent biological replicates.
Next, to repress NOTCH signaling, we treated SW480 and PDX1 xenografts with DBZ and found complete depletion of NICD accumulation in these tumors that in time course analysis in SW480 xenografts was fully effective after 3 d of treatment (Fig. 3, B and D). However, under DBZ treatment, the frequency of FRA1-positive tumor cells significantly increased, indicating expanded MAPK signaling upon repression of NOTCH activity (Fig. 3, B and D). Furthermore, in contrast to MAPK inhibition, DBZ treatment significantly increased the number of cleaved caspase-3–labeled tumor cells (Fig. S2), suggesting that colon cancer cells with high NOTCH activity were at least in part lost from the tumor through apoptosis.
With these findings in mind, we then analyzed xenograft tumors of mice that had been treated with AZD or DBZ and subsequently had been taken off treatment for up to 10 d before analysis. Astonishingly, in these tumors the original distributions and frequencies of colon cancer cells with FRA1 expression at the tumor edge and NICD accumulation in more centrally located tumor cells were readily restored (Fig. 3, B and D). Time course analysis in SW480 xenografts further demonstrated a quicker recovery of FRA1-positive tumor cells than of NICD-positive tumor cells from respective treatments (Fig. 3, C and D). Collectively, these data indicated that colon cancers may evade targeted treatment against MAPK or NOTCH signaling by a reversible shift in predominating pathway activity.
MAPK and NOTCH have opposite effects on epithelial differentiation in colon cancer
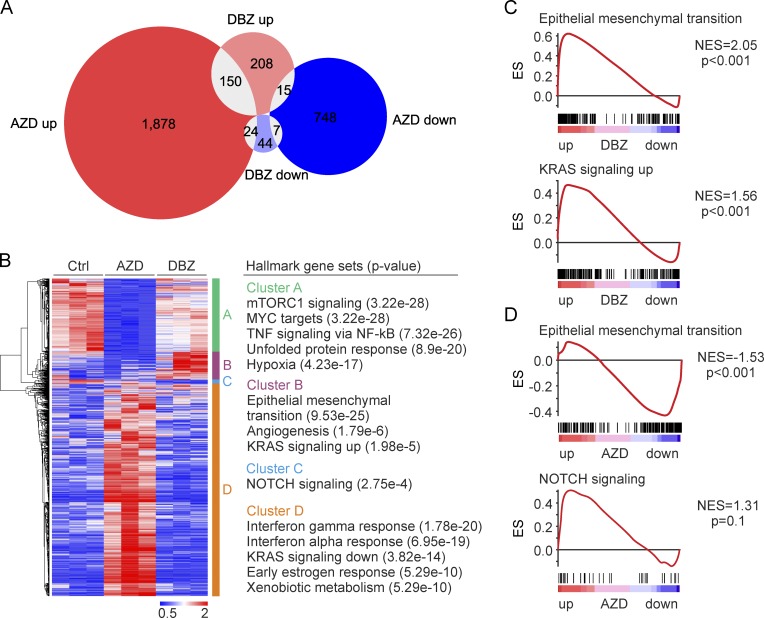
To shed more light on the effects of therapeutic targeting of MAPK and NOTCH signaling in colon cancer, we analyzed gene expression in SW480 xenografts after AZD or DBZ treatment by RNA-Seq. Considering genes with at least twofold change in expression, we found that AZD treatment affected 12.1% (2,822 genes) of the detected transcriptome, whereas DBZ treatment only deregulated 1.9% (448 genes). Differentially expressed genes only partially overlapped, indicating discriminative effects of both treatments (Fig. 4 A). Unsupervised hierarchical clustering of gene expression then revealed four major clusters that were characterized by repression (cluster A) or up-regulation (cluster D) upon AZD treatment or by repression (cluster C) or up-regulation (cluster B) upon DBZ treatment, respectively (Fig. 4 B). Searching for functional associations, we found that genes, which were repressed by AZD treatment (cluster A), were enriched for hallmark gene sets known to be related to MAPK activity, such as mTORC1 signaling or MYC target genes. Surprisingly however, when characterizing genes that were up-regulated by DBZ treatment (cluster B), we found strong enrichment for hallmark gene sets linked to tumor progression and, most significantly, to EMT (Fig. 4 B). GSEA analyses on unfiltered RNA-Seq data of DBZ-treated xenograft tumors confirmed a highly significantly enriched expression of EMT hallmark genes (Fig. 4 C). On the contrary, when analyzing data from AZD-treated tumors, we found that EMT-related genes were strongly repressed (Fig. 4 D). At the same time, NOTCH repression by DBZ caused overexpression of genes related to KRAS signaling (Fig. 4 C), whereas MAPK repression with AZD marginally up-regulated genes of NOTCH signaling (Fig. 4 D).
Figure 4.
Impact of MAPK and NOTCH repression on gene expression in colon cancer in vivo. (A) Venn diagram of genes with significantly (P < 0.05) differential expression and two or more fold change in SW480 xenografts that were treated for five consecutive days with AZD or DBZ compared with vehicle treatment (Ctrl). (B) Heat map and unsupervised hierarchical clustering of genes with significantly (P < 0.05) differential expression and two or more fold change (left). Rows represent genes and columns represent biological replicates. Four main clusters are indicated. Hallmark gene sets most enriched in each cluster as determined by GSEA (right). (C and D) GSEA for indicated gene sets using unfiltered gene expression data of DBZ- (C) and AZD-treated (D) tumors compared with controls. ES, enrichment score; NES, normalized enrichment score. (A–D) n = 3 independent biological replicates.
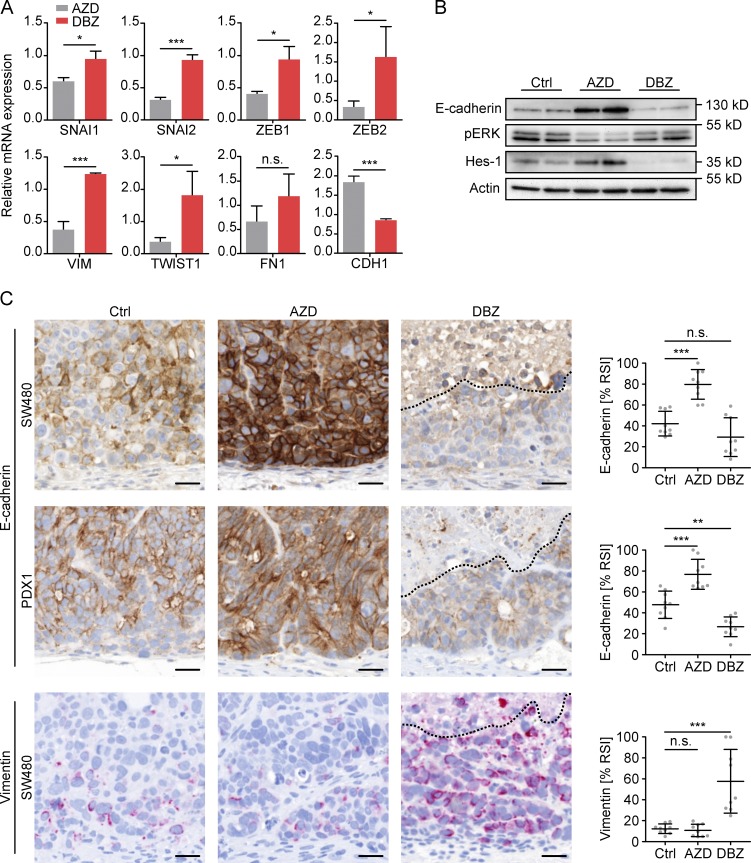
Because these data suggested opposing effects of MAPK and NOTCH repression on EMT, we next looked at individual factors that were linked to EMT in colon cancer. ZEB1/2, SNAI1/2, and TWIST, which encode for well-known key EMT regulators, but also VIM, which indicates an EMT phenotype, showed significantly higher expression levels in DBZ than in AZD treated xenografts (Fig. 5 A). In addition, CDH1, which encodes for E-cadherin and indicates epithelial differentiation, was repressed by DBZ and up-regulated by AZD. By immunoblotting, we confirmed overexpression of E-cadherin upon AZD treatment on the protein level, although it was reduced in DBZ-treated tumors (Fig. 5 B). Also, immunostaining showed strongly increased and expanded E-cadherin expression in tumor cells of SW480 and PDX1 xenografts after AZD treatment, whereas, on the contrary, DBZ treatment reduced E-cadherin levels in these tumors (Fig. 5 C). Furthermore, tumor cells of SW480 xenografts became strongly positive for Vimentin after DBZ treatment (Fig. 5 C), whereas PDX1 tumors did not express detectable Vimentin levels. Collectively, these data demonstrated that MAPK and NOTCH repression had opposing effects on epithelial differentiation in colon cancer, with NOTCH repression causing an overall shift toward an EMT phenotype.
Figure 5.
Effects of MAPK and NOTCH repression on EMT in colon cancer xenografts. (A) Relative expression levels of selected EMT-related genes in SW480 xenografts after 5 d of treatment with AZD or DBZ. Data are mean and error bars indicate SD. n = 3 independent biological replicates. (B) Immunoblotting for indicated proteins on tumor lysates of SW480 xenografts after 5 d of vehicle (Ctrl), AZD, or DBZ treatment. n ≥ 3 independent biological replicates, 2 of which are shown. (C) Representative immunostainings (left) and quantification of relative staining intensities (% RSI, right) for E-cadherin and Vimentin in SW480 and/or PDX1 xenografts after 5 d of vehicle (Ctrl), AZD, or DBZ treatment. Areas above dotted lines are tumor necrosis. Bars, 25 µm. Error bars are mean ± SD. **, P < 0.01; ***, P < 0.001 by t test; n.s., not significant. n ≥ 3 independent biological replicates.
Plasticity of MAPK and NOTCH signaling in colon cancer cells
To further learn about the dynamics of tumor cell subpopulations with active MAPK and NOTCH signaling, we developed a lentiviral Cre recombinase–sensitive system for lineage tracing in colon cancer xenografts (Fig. 6 A). This system consists of three lentiviral vectors, two of which mediate doxycycline-inducible expression of an estrogen receptor-Cre fusion protein (pLenti rtTA3G and pLenti TetO-CreERT2), and a third vector that upon Cre recombination irreversibly switches from expression of RFP to YFP (pLenti Trace). We transduced all three vectors into SW480 colon cancer cells and xenografted them into immune-compromised NOD/SCID mice. Tumor-bearing mice then were treated with AZD or DBZ, causing loss of FRA1- or NICD-positive tumor cell subpopulations, respectively. Vehicle-treated tumors were included as controls. During treatment and in nontreated controls, recombination was then induced with doxycycline and tamoxifen (Fig. 6 B). 2 d after recombination, we observed that individual or small clusters of tumor cells had been labeled by YFP in all xenograft tumors (Fig. 6 C and Fig. S3). Importantly, in AZD-treated tumors, the frequency of NICD–/YFP–double positive tumor cells was significantly higher than in nontreated controls, whereas FRA1-positive tumor cells were completely absent (Fig. 6 D). However, DBZ-treated xenografts had higher frequencies of FRA1–/YFP–double positive tumor cells than control tumors, but contained no NICD-positive tumor cells (Fig. 6 D). However, at 15 d after recombination, and after mice had been taken off treatment, we found that clonal patches of YFP-positive tumor cells then had formed that in all xenografts included both FRA1- and NICD-positive tumor cell subpopulations (Fig. 6 C and Fig. S3). Importantly, the frequencies of FRA1–/YFP– and NICD–/YFP–double positives were then similar to those in nontreated control tumors (Fig. 6 E).
Figure 6.
Lineage tracing of colon cancer cells after MAPK or NOTCH inhibition. (A) Lentiviral vectors for expression of rtTA (pLenti rtTA3G), doxycycline dependent CreERT2 (pLenti TetO-CreERT2), and the Cre-responsive color transgene (pLenti Trace). Upon Cre-recombination, the RFP transgene element flanked by loxN will be removed, causing an irreversible switch from expression of RFP to YFP fluorescence. BlastR/PuroR, blasticid and puromycin resistance genes; LTR, long terminal repeat; PRE posttranscriptional regulatory element; TRE, tetracycline response element. (B) Triple transduced colon cancer cells were xenografted into NOD/SCID mice. Experimental schedule for Cre-recombination by doxycycline (DOX) and tamoxifen (TAM) in AZD- or DBZ-treated xenografts. (C) Representative double immune fluorescence images for YFP, FRA1, and NICD at 2 and 15 d after recombination in AZD- and DBZ-treated SW480 xenografts, as indicated. Narrow panels are higher magnifications of areas boxed in squared panels. Arrowheads point to FRA1- and NICD-positive tumor cells within single YFP-positive clones at 15 d after recombination. Bars, 25 µm. (D and E) Quantification of FRA1–/YFP– and NICD–/YFP–double positive tumor cells in vehicle- (Ctrl), AZD-, and DBZ-treated SW480 xenografts at 2 d (D) and 15 d (E) after recombination. Error bars are mean ± SD. *, P < 0.05; ***, P < 0.001 by t test; n.s., not significant. n ≥ 3 independent biological replicates.
We also treated PDX1 xenograft tumors with AZD or DBZ and then labeled remaining NICD- or FRA1-positive tumor cells, respectively, with BrdU (Fig. S4 A). Analyzing tumors 7 d after AZD or DBZ treatment revealed that the label then had expanded to reappeared FRA1- or NICD-positive tumor cells which at the time of labeling were absent from the tumor (Fig. S4 B). Furthermore, the frequencies of NICD–/BrdU– and FRA1–/BrdU–double positive tumor cells in AZD- and DBZ-treated tumors, respectively, were higher at 6 h after labeling than at 7 d, where they showed similar frequencies, irrespective of the precedent treatment (Fig. S4 C). Collectively, these findings demonstrate that tumor cell subsets with high MAPK or NOTCH activity can be restored from remaining tumor cells during recovery from AZD or DBZ treatment and provide evidence for plasticity of signaling pathway activity in colon cancer cells.
Treatment effects of MAPK and NOTCH repression in colon cancer
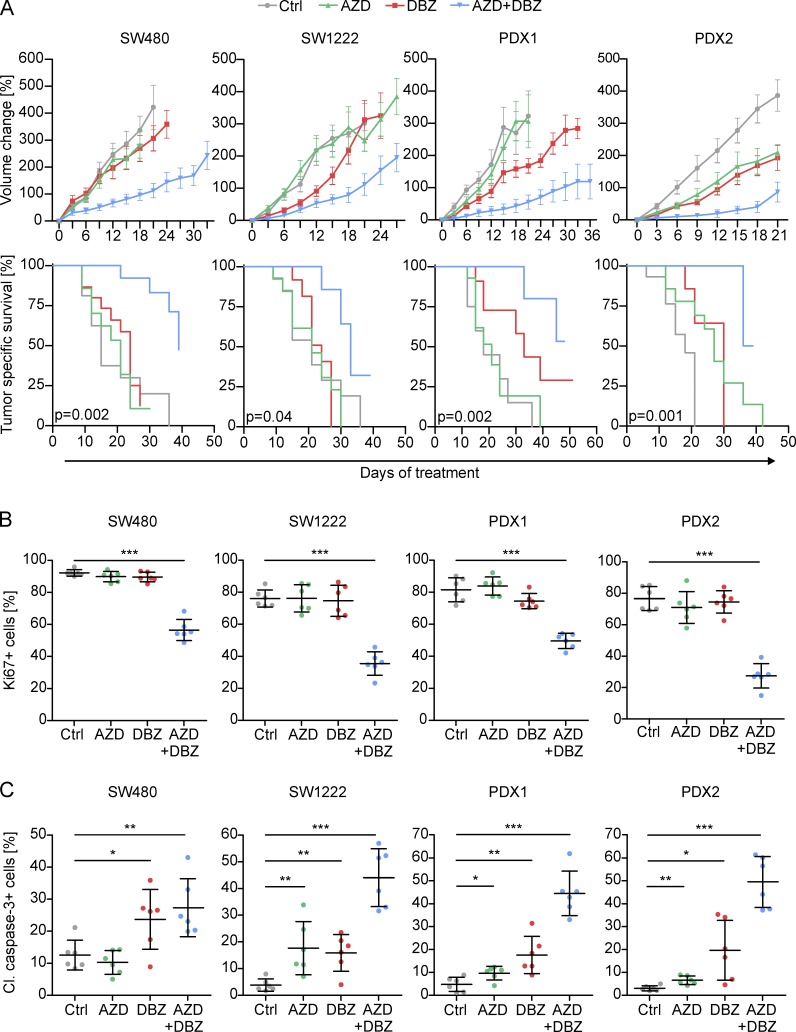
Finally, we evaluated the effects of targeting MAPK and NOTCH activity on tumor growth and survival in colon cancer xenografts. We applied AZD, DBZ, or a combination of both at treatment intervals of 3 d for several weeks. In addition to SW480 and PDX1, we included cell line–derived SW1222 and patient-derived PDX2 colon cancer xenografts, both of which also had the distribution of FRA1- and NICD-positive tumor cell subpopulations described above. We then evaluated tumor growth over time and observed that AZD treatment significantly slowed tumor growth of PDX2 tumors only, although it had no significant effects on growth of SW480, SW1222, and PDX1 xenografts (Fig. 7 A). Similarly, DBZ treatment slowed tumor growth in PDX2 tumors and also slightly in PDX1 tumors, whereas no overall effects on SW480 and SW1222 were observed. However, combined treatment with AZD and DBZ significantly slowed tumor growth and prolonged tumor-specific survival in all xenograft models, outweighing the effects of single agent treatments (Fig. 7 A). We then examined treated tumors and found that double treatments strongly reduced proliferation rates, as indicated by Ki67 staining, whereas single agent treatments had no significant effects on proliferation (Fig. 7 B and Fig. S5 A). Moreover, double treatment strongly increased apoptosis, as indicated by cleaved caspase-3, whereas single agent treatments again had lower or no significant effects (Fig. 7 C and Fig. S5 B). All tumors formed areas of tumor necrosis which variably increased upon treatment (Fig. 8, A and B). Of note, however, when analyzing double-treated xenograft tumors for FRA1 and NICD, we for both markers observed some remaining positive tumor cells, suggesting incomplete blockage of MAPK and NOTCH signaling with our treatment protocol (Fig. 8 C). Collectively, these data demonstrate superior therapeutic effects upon combined targeting of different tumor cell subpopulations with high MAPK and high NOTCH signaling in colon cancer.
Figure 7.
Therapeutic targeting of MAPK and NOTCH in colon cancer xenografts. (A) Long term treatment effects of AZD, DBZ, their combination, or vehicle (Ctrl) on SW480-, SW1222-, and patient-derived (PDX1 and PDX2) colon cancer xenografts, shown as growth curves (upper panels) and tumor specific survival in Kaplan-Meier plots (lower panels). Data are mean ± SE in growth curves. P-values are log-rank test results in Kaplan-Meier plots. n ≥ 10 independent biological replicates for each treatment group. (B and C) Quantification of immunostaining for Ki67 proliferation index (B) and cleaved (Cl.) caspase-3 (C) in treated xenografts. Error bars are mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test. n ≥ 3 independent biological replicates.
Figure 8.
Treatment effects of MAPK and NOTCH repression in colon cancer xenografts. (A) Quantification of necrosis in SW480, SW1222, PDX1, and PDX2 colon cancer xenografts after long-term treatment with AZD and/or DBZ or vehicle (Ctrl) as indicated. Data are mean ± SD *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test; n.s., not significant. n ≥ 3 independent biological replicates. (B) Representative overview micrographs of H&E-stained sections of xenograft tumors after indicated long term treatments. (C) Representative immunostainings for FRA1 and NICD in SW480 xenografts after long term treatment with vehicle (Ctrl), or AZD and DBZ. Arrowheads indicate remaining FRA1- and NICD-positive tumor cells. Bars: 1 mm (B); 25 µm (C).
Discussion
Here, we demonstrate that in colorectal cancer high NOTCH-signaling activity marks tumor cells with low levels of MAPK and WNT activity, and vice versa, indicating that high pathway activities for NOTCH and MAPK/WNT in colon cancer cells are mutually exclusive. Additionally, these pathway activities were linked to distinct tumor cell phenotypes. Although tumor cells with high MAPK activity resided at the tumor edge and underwent EMT, we found that tumor cells with high NOTCH activity had a pronounced epithelial phenotype and were located in the tumor center. On one hand, these findings can be explained when considering recent data that showed a repressive role of NOTCH on MAPK and WNT signaling (Kim et al., 2012; Rampias et al., 2014), both of which are strong inducers of EMT in colon cancer (Sánchez-Tilló et al., 2011; Blaj et al., 2017). On the other hand, our findings are unexpected in light of previous studies that suggested induction of EMT by NOTCH in various cancer types (Leong et al., 2007; Sahlgren et al., 2008; Yang et al., 2011; Fender et al., 2015). However, in contrast to these data that were mostly derived from cell culture experiments in vitro or from other tumor entities, such as lung or breast cancer, we here assessed the distribution of NOTCH activity in primary colon cancer tissues in situ. We therefore suggest that the emergence of colon cancer cell subpopulations with full NOTCH activation, their distribution within the tumor, and the associated epithelial phenotype depend on tumor entity and require the three-dimensional architecture of growing in vivo.
Therapeutic targeting of colon cancer cells with high MAPK or NOTCH activity by MEK or γ-secretase inhibitors caused a loss of respective tumor cell subpopulations in colon cancer xenografts. However, we demonstrate that tumor cells with high MAPK activity were unaffected or even expanded when targeting NOTCH, whereas the NOTCH-active tumor cell population expanded when targeting MAPK signaling. These shifts in predominating pathway activity were accompanied by changes in tumor cell phenotypes that on the gene expression and protein level indicated strongly increased EMT when repressing NOTCH, whereas MAPK repression had opposite effects. Because MAPK is a strong driver of EMT in colon cancer (Lemieux et al., 2009; Bakiri et al., 2015; Blaj et al., 2017), the varying contribution of tumor cell subsets with high MAPK activity may explain these findings. However, when further considering that EMT is a hallmark of cancer progression (Hanahan and Weinberg, 2011), we propose that solely targeting NOTCH may elicit limited or even adverse effects on the risk of tumor progression for patients with colon cancer. Our data therefore imply that single agent therapies that target specific signaling pathways require careful evaluation as a result of unexpected effects on overall tumor cell differentiation and may cause transitions into potentially aggressive tumor cell populations with intrinsically treatment-resistant phenotypes.
Upon recovery from therapy, colon cancer cells with high MAPK or NOTCH activity, respectively, were quickly replenished. Using genetic and BrdU lineage tracing, we demonstrate clonal outgrowth of MAPK- and NOTCH-positive tumor cells from the remaining tumor cell population, irrespective of the pathway that was targeted, indicating phenotypic plasticity in signaling pathway activity as an underlying mechanism for treatment recovery. In line with these findings, a recent study demonstrated prompt reappearance of LGR5-positive colon cancer cells after their genetic ablation in tumor xenografts, indicating that tumor cell plasticity allowed reversion of differentiated tumor cells into colon cancer stem cells (Shimokawa et al., 2017). In this context, it remains to be determined whether colon cancer cells that express LGR5 or other putative cancer stem cell markers reside within MAPK- and/or NOTCH-positive tumor cell subpopulations. However, these data suggest that solely targeting colon cancer cell subpopulations with distinct phenotypes, such as EMT or enhanced stemness (Shibue and Weinberg, 2017), may clinically fail as a result of plasticity of phenotype and signaling pathway activity. Indeed, when we treated colon cancer xenografts for several weeks with MAPK or NOTCH inhibitors alone, effects on tumor growth either were nonsignificant or moderate only, which is in line with their limited effects in previous therapeutic trials (Bennouna et al., 2011; Strosberg et al., 2012). However, when combining both therapies, we found strong repressive effects on tumor cell proliferation and increased apoptosis, resulting in slowed tumor growth and prolonged tumor-specific survival. Given that these effects significantly outweighed those of single agent treatments, our findings denote that combined treatments mainly succeeded by restricting tumor cell plasticity. This lends support to a new concept for cancer therapy that advocates specific and simultaneous targeting of several different tumor cell subpopulations to strongly improve therapy response. Detailed analyses of targetable phenotypes and pathways found in different tumor cell subpopulations may thus pave the way for improved treatment options for patients with colorectal and other cancers.
The contribution of colon cancer cell subpopulations to tumor progression is not yet completely understood and our data on clinical relevance of MAPK and NOTCH activity shed useful light. Cancer progression requires invasion and dissemination of tumor cells, which are strongly driven by EMT (Massagué and Obenauf, 2016). However, it also requires seeding at metastatic sites which depends on the reverse process termed mesenchymal-epithelial transition (MET; Lamouille et al., 2014). Because we demonstrate that MAPK and NOTCH activity are linked to EMT and MET phenotypes, respectively, both pathways likely foster colon cancer progression in concert. This idea finds support in our observation that combined analyses of MAPK and NOTCH activity through FRA1 and NICD was most discriminatory in predicting patient outcome and tumor metastasis. Importantly, however, because patients whose tumors showed low activity for both pathways survived best and showed lowest tumor progression and metastasis rates, this further strengthened the rationale for combined targeted treatment against both pathways. Finally, because immunostainings for FRA1 and NICD readily indicated presence and extent of respective tumor cell subpopulations in colon cancer specimens and also often were consistent in primary colon cancers and their metastases, these may well be evaluated as predictive biomarkers for response to MAPK- and NOTCH-directed therapies.
In conclusion, we here provide evidence that combined targeting of MAPK and NOTCH signaling can improve therapeutic response in preclinical xenograft models of colorectal cancer. However, this study has certain limitations. Our data are derived from immune-compromised animals which partially lack the inflammatory microenvironment and tumor directed immune response, so that treatment effects in human patients with colorectal cancer may significantly differ. Moreover, because several substances for MAPK and NOTCH inhibition are clinically evaluated (Takebe et al., 2015; Miyamoto et al., 2017), most tolerable and effective drug combinations in human patients still remain to be determined. Toxic side effects, especially of combined MAPK and NOTCH inhibition also need to be thoroughly assessed. Finally, although repression of MAPK and NOTCH signaling significantly slowed tumor growth, blockage of both pathways was incomplete, and also, this treatment failed to regress established tumors. Further preclinical and clinical trials may therefore reveal if combined MAPK and NOTCH inhibition, in addition to established chemotherapeutic protocols, can improve therapy response in patients with colorectal cancer.
Materials and methods
Clinical samples
Colorectal cancer specimens from patients that underwent surgical resection at the University of Munich between 1994 and 2007 (LMU, Munich, Germany) were obtained from the archives of the Institute of Pathology. Follow-up data were recorded prospectively by the Munich Cancer Registry (data provided by J. Engel, LMU, Munich, Germany). Specimens were anonymized, and the study was approved by the institutional ethics committee of the Medical Faculty of the LMU. For the UICC stage II collection, inclusion criteria were colorectal adenocarcinomas with bowel wall infiltration (T3 and T4), but absence of nodal (N0) or distant metastasis (M0) at the time of diagnosis. The final collection consisted of 225 cases with 50 events of cancer-specific death and 71 events of tumor progression, either documented as tumor recurrence or metastasis. For the metastasis collection, a case control design was chosen that included tumor specimens of 92 patients. Half of the patients had colon cancers with synchronous liver metastasis (UICC stage IV), diagnosed by clinical imaging or liver biopsy. Controls consisted of colon cancer patients without distant metastasis at the time of diagnosis (UICC stages I–III) and with disease-free survival of at least 5 yr after primary surgical resection. Cases and controls were matched by tumor grade, T-category, and tumor location (all tumors were right-sided colon cancers), resulting in 46 matched pairs. Furthermore, 11 paired tissue samples of primary colorectal cancers and their metastases to different sites were collected. To establish PDX1 and PDX2 xenografts, vital tissue samples of two human primary colorectal adenocarcinomas were provided by the biobank under administration of the foundation Human Tissue and Cell Research (HTCR; Thasler et al., 2003).
For KRAS mutational testing, tumor tissue was scraped from deparaffinized tissue sections under microscopic control using sterile scalpel blades, and tumor DNA was extracted with QIAamp DNA Micro kits. KRAS exon 2 then was analyzed by pyrosequencing on a PyroMark Q24 Advanced instrument (Qiagen) with primers 5′-NNNGGCCTGCTGAAAATGACTGAA-3′ and 5′-Biotin-TTAGCTGTATCGTCAAGGCACTCT-3′ for amplification and 5′-TGTGGTAGTTGGAGCT-3′ for sequencing.
Lentiviral vectors
All template plasmids were obtained from Addgene. For the inducible pLenti TetO-CreERT2 expression vector, CreERT2 from pCAG-CreERT2 was amplified (a gift from C. Cepko, Harvard Medical School, Boston, MA) by PCR and inserted between BamHI and XbaI restriction sites of pLenti CMVTRE3G eGFP Puro (a gift from E. Campeau, Zenith Epigenetics Ltd., Calgary, Canada), replacing eGFP by CreERT2. For the Cre-sensitive recombination vector pLenti Trace, we inserted PCR-amplified mCherry-FLAG and EYFP-V5 into a plasmid with synthetic paired loxN sites. This cassette then was inserted between AgeI and SalI sites of pLenti PGK-GFP (a gift from D. Trono, École Polytechnique Fédérale de Lausanne, Switzerland), replacing GFP. Finally, the PGK promoter was replaced by an EF1-α promoter, yielding pLenti Trace. Modified vector elements were verified by restriction analysis and sequencing.
Cell culture and lentiviral transductions
HEK293 and SW480 cells were obtained from ATCC and SW1222 from the Ludwig Institute for Cancer Research. Cell lines were authenticated using short-tandem repeat profiling, tested negative for mycoplasma contamination, and cultured in DMEM containing 10% FBS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Biochrom). For transductions, lentivirus was produced in HEK293 cells by cotransfection with lentiviral vector, pCMV-dR8.91, and pMD2.G as previously described (Horst et al., 2012). Virus containing medium was passed through 0.45-µm filters (Millipore), mixed 1:1 with DMEM, and used to infect SW480 colon cancer cells in the presence of 8 mg/ml polybrene (Sigma-Aldrich). pLenti rtTA3G (gift from D. Esposito, National Cancer Institute, Rockville, MD), pLenti TetO-CreERT2, and pLenti Trace triple transduced cells then were single-cell sorted into 96-well plates on a FACSAria III instrument (BD Biosciences) and expanded. Recombination was tested in vitro by addition of 0.5 µg/ml doxycycline and 1 µM 4-hydroxytamoxifen (Sigma Aldrich) before xenotransplantation into mice.
Tumor xenografts and in vivo treatments
Mouse experiments were reviewed and approved by the Regierung von Oberbayern, and mice were housed in pathogen-free microisolator cages. Disaggregated primary colon cancers (PDX1 and PDX2), as well as SW1222 or SW480 colon cancer cells either native or carrying the lineage tracing constructs, were suspended in 100 µl of a 1:1 mixture of PBS and growth factor–depleted Matrigel (Corning) and injected subcutaneously into age- and gender-matched 6–8-wk-old NOD/SCID mice (NOD.CB17-Prkdcscid, The Jackson Laboratory) for xenograft formation. Mice were randomly assigned to control or treatment groups when tumor volumes reached 100 mm3. For short-term therapy and tracing experiments, mice were treated daily with 1.25 mg selumetinib (AZD6244; Selleckchem) p.o. or 0.35 mg DBZ (Axon Medchem) i.p. for 5 d. For lineage tracing, 2.5 µg doxycycline were given p.o. for 2 d starting on day 3, and recombination of pLenti Trace was induced by 7.5 ng tamoxifen i.p. (Sigma-Aldrich). For BrdU tracing, mice were injected with 1.25 mg BrdU (Sigma-Aldrich) 18 h after last inhibitor treatment. For long-term therapy, mice were treated with 1.25 mg AZD and 0.35 mg DBZ or vehicle as control every 3 d until tumors reached volumes of 1,000–1,300 mm3. Mice were sacrificed, tumors removed, and either formalin fixed and paraffin embedded for histology and immunostaining, or directly used for gene expression analyses and immunoblotting.
Immunoblotting
For immunoblotting, freshly harvested and snap-frozen tumor samples were ground in a liquid nitrogen–cooled mortar (Bel-Art) and lysed in radioimmunoprecipitation assay buffer (50 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP-40), supplemented with protease and phosphatase inhibitors (Roche). Samples then were sonicated for 20 s and centrifuged, and protein concentrations were measured with DC Protein Assays (Biorad). Laemmli sample buffer (Biorad) then was added to equal amounts of protein and heated for 5 min at 95°C. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes (Merck Millipore), and incubated with primary antibodies listed in Table S4. Bands were visualized using HRP-conjugated secondary mouse (Promega) or rabbit (Sigma) antibodies and chemiluminescent HRP Substrate (Millipore).
Immunohistochemistry and immune fluorescence
For immunohistochemistry, 5-µm tissue sections of colorectal cancer samples or xenografts were deparaffinized and stained on a Ventana Benchmark XT autostainer or manually by retrieving antigens in TRS6 (Dako Cytomation) for 20 min in a microwave oven. Primary antibodies used for incubation are listed in Table S4. Staining was visualized with ultraView or optiView DAB detection kits (Ventana Medical Systems) or by incubation with alkaline phosphatase coupled secondary antibodies and substrate kits (Vector). The intratumoral distribution of NICD and HES1 staining was determined by inspection of tumor edge and tumor center in each case. For survival analyses, NICD-positive tumor cells were scored in 10% steps by estimation. FRA1 expression was scored semiquantitatively, ranging from complete absence (score 0), weak (score 1), moderate (score 2), or strong expression (score 3). Cases then were classified as FRA1-negative (score 0) and FRA1-positive (scores 1–3).
For immune fluorescence, sections were deparaffinized, and antigens were retrieved in Target Retrieval Solution (Dako) or Epitope Retrieval Solution, pH 8.0 (Leica), for 20 min in a microwave oven. Sections then were incubated with primary antibodies listed in Table S4. NICD immunostaining was enhanced by tyramide signal amplification (PerkinElmer). For other epitopes, secondary Alexa Fluor 488–conjugated antibody (Invitrogen) was used for visualization, and nuclei were counterstained with DAPI (Vector Laboratories). Confocal fluorescence images then were taken on a LSM 700 laser-scanning microscope using the ZEN software (Zeiss). Contrast and brightness were adjusted in Photoshop (Adobe), and for NICD staining, a nuclear mask was applied. Colocalization of fluorescence signals was measured using Volocity 6.1.1 software (PerkinElmer) and plotted as percent values of maximum fluorescence intensity.
Gene expression analyses and Gene Set Enrichment Analysis (GSEA)
RNA was isolated from tumor xenografts using TRIzol (Invitrogen). Libraries were constructed using the mRNA Sense library preparation kit (Lexogen) according to manufacturer’s protocol. In brief, 500 ng of total RNA was captured on oligo dT beads, hybridized to random primers and stoppers for cDNA synthesis, and ligated. Single-stranded cDNAs with sequencing adapters then were amplified and barcoded, and libraries were purified with AMPure XP beads (Beckman Coulter), quantified, pooled at 10-nM concentration, and sequenced in multiplex on a HiSeq 1500 as 50-bp single reads. Data then were demultiplexed, adaptor sequences were removed, and the reads were mapped to the hg19 human reference genome. Sequence reads for annotated genes were counted with the HTseq count script from the DEseq2 package, and differentially expressed genes were identified with the edgeR package with a <1% false discovery rate. Heat maps and clustering were done with GENE-E (Broad Institute). Hallmark gene sets most enriched in each cluster were determined using GSEA tools. For enrichment curves GSEA analyses were run with 1,000 permutations. RNA-seq expression data are accessible through GEO (GSE98922).
Statistical analysis
Two-tailed Student’s t test was used to evaluate significant differences between two groups, and data indicate means ± SD, unless indicated otherwise. For patient outcome and mouse survival, the Kaplan-Meier method was used, and P-values were calculated by the log-rank test. Cox proportional hazards model was used for multivariate analysis. Differences were considered statistically significant when P < 0.05. Individual P-values are given within the figures. Statistics were calculated with Prism (GraphPad) or SPSS (IBM).
Online supplemental material
Fig. S1 shows the distribution of HES1 expression and characteristics of FRA1- and NICD-positive tumor cells in colorectal cancer. In Fig. S2, effects of short-term AZD and DBZ treatment on apoptosis in colon cancer xenografts are displayed. Fig. S3 demonstrates lineage tracing of tumor cells in vehicle-treated control colon cancer xenografts. Fig. S4 demonstrates BrdU tracing of colon cancer cells after MAPK and NOTCH inhibition. Fig. S5 shows the impact of long-term MAPK and NOTCH repression on proliferation and apoptosis in colon cancer xenografts. Table S1 contains information on clinical data of FRA1 and NICD expression in UICC stage II colorectal cancer. Table S2 shows results from multivariate analysis of disease-free survival. Table S3 contains information on clinical data of FRA1 and NICD expression in a case-control collection of colon cancers with and without distant metastasis. Table S4 lists primary antibodies used in this study.
Supplementary Material
Acknowledgments
We are grateful to Anja Heier, Jessica Kövi, Anne Küchler, Annegret Schäfer, Andrea Sendelhofert, and Karina Windhorst for experimental assistance.
This study was supported by grants from the Deutsche Krebshilfe (grant 11169 to D. Horst), the Rudolf Bartling Stiftung (to D. Horst and H. Hermeking), and the Curt-Bohnewand-Fonds (to C. Schaaf).
The authors declare no competing financial interests.
Author contributions: E.M. Schmidt and D. Horst developed methodology and performed experiments; E.M. Schmidt, S. Lamprecht, C. Blaj, S. Krebs, H. Blum, C. Schaaf, H. Hermeking, A. Jung, T. Kirchner, and D. Horst analyzed and interpreted data and wrote the manuscript; D. Horst supervised the study.
References
- Bakiri L., Macho-Maschler S., Custic I., Niemiec J., Guío-Carrión A., Hasenfuss S.C., Eger A., Müller M., Beug H., and Wagner E.F.. 2015. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFβ expression. Cell Death Differ. 22:336–350. 10.1038/cdd.2014.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennouna J., Lang I., Valladares-Ayerbes M., Boer K., Adenis A., Escudero P., Kim T.Y., Pover G.M., Morris C.D., and Douillard J.Y.. 2011. A Phase II, open-label, randomised study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Invest. New Drugs. 29:1021–1028. 10.1007/s10637-010-9392-8 [DOI] [PubMed] [Google Scholar]
- Blaj C., Schmidt E.M., Lamprecht S., Hermeking H., Jung A., Kirchner T., and Horst D.. 2017. Oncogenic Effects of High MAPK Activity in Colorectal Cancer Mark Progenitor Cells and Persist Irrespective of RAS Mutations. Cancer Res. 77:1763–1774. 10.1158/0008-5472.CAN-16-2821 [DOI] [PubMed] [Google Scholar]
- Brabletz S., Bajdak K., Meidhof S., Burk U., Niedermann G., Firat E., Wellner U., Dimmler A., Faller G., Schubert J., and Brabletz T.. 2011. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 30:770–782. 10.1038/emboj.2010.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L.A., Knuechel R., and Kirchner T.. 2001. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 98:10356–10361. 10.1073/pnas.171610498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689. 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Bu P., Chen K.Y., Chen J.H., Wang L., Walters J., Shin Y.J., Goerger J.P., Sun J., Witherspoon M., Rakhilin N., et al. . 2013. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 12:602–615. 10.1016/j.stem.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernat L., Blaj C., Jackstadt R., Brandl L., Engel J., Hermeking H., Jung A., Kirchner T., and Horst D.. 2014. Colorectal cancers mimic structural organization of normal colonic crypts. PLoS One. 9:e104284 10.1371/journal.pone.0104284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard J.Y., Siena S., Cassidy J., Tabernero J., Burkes R., Barugel M., Humblet Y., Bodoky G., Cunningham D., Jassem J., et al. . 2014. Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann. Oncol. 25:1346–1355. 10.1093/annonc/mdu141 [DOI] [PubMed] [Google Scholar]
- Fender A.W., Nutter J.M., Fitzgerald T.L., Bertrand F.E., and Sigounas G.. 2015. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J. Cell. Biochem. 116:2517–2527. 10.1002/jcb.25196 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Heinemann V., von Weikersthal L.F., Decker T., Kiani A., Vehling-Kaiser U., Al-Batran S.E., Heintges T., Lerchenmüller C., Kahl C., Seipelt G., et al. . 2014. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 15:1065–1075. 10.1016/S1470-2045(14)70330-4 [DOI] [PubMed] [Google Scholar]
- Horst D., Chen J., Morikawa T., Ogino S., Kirchner T., and Shivdasani R.A.. 2012. Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer Res. 72:1547–1556. 10.1158/0008-5472.CAN-11-3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Siegel R., Xu J., and Ward E.. 2010. Cancer statistics, 2010. CA Cancer J. Clin. 60:277–300. 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- Kim H.A., Koo B.K., Cho J.H., Kim Y.Y., Seong J., Chang H.J., Oh Y.M., Stange D.E., Park J.G., Hwang D., and Kong Y.Y.. 2012. Notch1 counteracts WNT/β-catenin signaling through chromatin modification in colorectal cancer. J. Clin. Invest. 122:3248–3259. 10.1172/JCI61216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J., and Derynck R.. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15:178–196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux E., Bergeron S., Durand V., Asselin C., Saucier C., and Rivard N.. 2009. Constitutively active MEK1 is sufficient to induce epithelial-to-mesenchymal transition in intestinal epithelial cells and to promote tumor invasion and metastasis. Int. J. Cancer. 125:1575–1586. 10.1002/ijc.24485 [DOI] [PubMed] [Google Scholar]
- Leong K.G., Niessen K., Kulic I., Raouf A., Eaves C., Pollet I., and Karsan A.. 2007. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 204:2935–2948. 10.1084/jem.20071082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., and Obenauf A.C.. 2016. Metastatic colonization by circulating tumour cells. Nature. 529:298–306. 10.1038/nature17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Suyama K., and Baba H.. 2017. Recent Advances in Targeting the EGFR Signaling Pathway for the Treatment of Metastatic Colorectal Cancer. Int. J. Mol. Sci. 18:752 10.3390/ijms18040752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias T., Vgenopoulou P., Avgeris M., Polyzos A., Stravodimos K., Valavanis C., Scorilas A., and Klinakis A.. 2014. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 20:1199–1205. 10.1038/nm.3678 [DOI] [PubMed] [Google Scholar]
- Sahlgren C., Gustafsson M.V., Jin S., Poellinger L., and Lendahl U.. 2008. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA. 105:6392–6397. 10.1073/pnas.0802047105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Tilló E., de Barrios O., Siles L., Cuatrecasas M., Castells A., and Postigo A.. 2011. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. USA. 108:19204–19209. 10.1073/pnas.1108977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L., Roberts J.M., and Coller H.A.. 2010. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol. Med. 16:17–26. 10.1016/j.molmed.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., and Weinberg R.A.. 2017. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14:611–629. 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., and Sato T.. 2017. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 545:187–192. 10.1038/nature22081 [DOI] [PubMed] [Google Scholar]
- Sonoshita M., Aoki M., Fuwa H., Aoki K., Hosogi H., Sakai Y., Hashida H., Takabayashi A., Sasaki M., Robine S., et al. . 2011. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 19:125–137. 10.1016/j.ccr.2010.11.008 [DOI] [PubMed] [Google Scholar]
- Strosberg J.R., Yeatman T., Weber J., Coppola D., Schell M.J., Han G., Almhanna K., Kim R., Valone T., Jump H., and Sullivan D.. 2012. A phase II study of RO4929097 in metastatic colorectal cancer. Eur. J. Cancer. 48:997–1003. 10.1016/j.ejca.2012.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P.J., Jeong W., Bando H., Kahn M., Yang S.X., and Ivy S.P.. 2015. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 12:445–464. 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thasler W.E., Weiss T.S., Schillhorn K., Stoll P.T., Irrgang B., and Jauch K.W.. 2003. Charitable State-Controlled Foundation Human Tissue and Cell Research: Ethic and Legal Aspects in the Supply of Surgically Removed Human Tissue For Research in the Academic and Commercial Sector in Germany. Cell Tissue Bank. 4:49–56. 10.1023/A:1026392429112 [DOI] [PubMed] [Google Scholar]
- Tolcher A.W., Messersmith W.A., Mikulski S.M., Papadopoulos K.P., Kwak E.L., Gibbon D.G., Patnaik A., Falchook G.S., Dasari A., Shapiro G.I., et al. . 2012. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. 30:2348–2353. 10.1200/JCO.2011.36.8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E., Köhne C.H., Hitre E., Zaluski J., Chang Chien C.R., Makhson A., D’Haens G., Pintér T., Lim R., Bodoky G., et al. . 2009. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med. 360:1408–1417. 10.1056/NEJMoa0805019 [DOI] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. . 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12:468–476. 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ahn Y.H., Gibbons D.L., Zang Y., Lin W., Thilaganathan N., Alvarez C.A., Moreira D.C., Creighton C.J., Gregory P.A., et al. . 2011. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J. Clin. Invest. 121:1373–1385. 10.1172/JCI42579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Ke J., Sun L., He Z., Zou Y., He X., Chen Y., Wu X., Cai Z., Wang L., et al. . 2015. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin. Exp. Metastasis. 32:169–179. 10.1007/s10585-015-9700-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.