IL1RAP is an emerging target for AML therapy. Studying its cell-intrinsic function revealed that IL1RAP interacts with and amplifies signaling through c-KIT and FLT3 in AML cells. This novel promiscuous role of IL1RAP in AML has implications for therapeutic targeting.
Abstract
The surface molecule interleukin-1 receptor accessory protein (IL1RAP) is consistently overexpressed across multiple genetic subtypes of acute myeloid leukemia (AML) and other myeloid malignancies, including at the stem cell level, and is emerging as a novel therapeutic target. However, the cell-intrinsic functions of IL1RAP in AML cells are largely unknown. Here, we show that targeting of IL1RAP via RNA interference, genetic deletion, or antibodies inhibits AML pathogenesis in vitro and in vivo, without perturbing healthy hematopoietic function or viability. Furthermore, we found that the role of IL1RAP is not restricted to the IL-1 receptor pathway, but that IL1RAP physically interacts with and mediates signaling and pro-proliferative effects through FLT3 and c-KIT, two receptor tyrosine kinases with known key roles in AML pathogenesis. Our study provides a new mechanistic basis for the efficacy of IL1RAP targeting in AML and reveals a novel role for this protein in the pathogenesis of the disease.
Graphical Abstract
Introduction
In acute myeloid leukemia (AML), there is a current need for molecular understanding of pathways relevant in disease initiation and for targeted therapies that selectively and directly inhibit these pathways. We and others previously identified the surface molecule IL-1 receptor accessory protein (IL1RAP) as consistently overexpressed in AML hematopoietic stem and progenitor cells (HSPC) across multiple genetic subtypes of AML (Barreyro et al., 2012; Askmyr et al., 2013; Ho et al., 2016; Sadovnik et al., 2017), as well as in high-risk myelodysplastic syndromes (MDS), hematologic malignancies that often progress to AML. As a result of low IL1RAP expression on normal HSPCs (Barreyro et al., 2012; Ho et al., 2016) and apparent dispensability of IL1RAP for the viability of mammalian organisms (Cullinan et al., 1998), IL1RAP has emerged as a promising target for leukemic stem cell (LSC)-directed immunotherapeutic approaches in myeloid malignancies (Järås et al., 2010; Askmyr et al., 2013; Herrmann et al., 2014; Ågerstam et al., 2015; Jiang et al., 2016; Landberg et al., 2016; Warfvinge et al., 2017); however, little is known about whether IL1RAP has a cell-intrinsic role in AML. Current IL1RAP-targeting strategies rely on immune effector cell recruitment, despite most AML patients having compromised immune systems. Here, we used antibody targeting, RNA-interference, and genetic deletion to study the functional role of IL1RAP in oncogenic signaling and leukemic transformation. We show that targeting IL1RAP delays AML pathogenesis in the absence of immune effector cells and without perturbing healthy hematopoiesis. In exploring the molecular basis for these effects, we unexpectedly found that IL1RAP is a more promiscuous coreceptor than previously appreciated, and its role is not restricted to the IL-1 receptor (IL-1RI) pathway. Specifically, IL1RAP physically interacts with and mediates signaling through FLT3 and c-KIT, two receptor tyrosine kinases with significant roles in AML pathogenesis (Ikeda et al., 1991; Lisovsky et al., 1996; Scheijen and Griffin, 2002; Stirewalt and Radich, 2003). Our study reveals novel functional and mechanistic roles of IL1RAP in AML pathogenesis and provides a rationale for the further exploration of therapeutic strategies directly targeting IL1RAP and its functions.
Results
IL1RAP-directed antibodies inhibit AML growth cell-intrinsically through induction of differentiation and apoptosis
We tested various antibodies that target the extracellular portion of the IL1RAP protein for effects on growth of the AML cell line THP-1, which expresses high IL1RAP levels (Barreyro et al., 2012; Fig. S1 A). We identified several antibodies with growth inhibitory effects, including a polyclonal anti-IL1RAP antibody (referred to as IL1RAP pAb), as well as two monoclonal antibodies (referred to as IL1RAP mAb 1 and mAb 2). IL1RAP antibodies showed a cytostatic effect on the growth of THP-1 cells (Figs. 1, A and B; and Fig. S1 B). Antibodies directed against another highly expressed surface protein on THP-1 cells, CD13, did not affect their growth (Fig. S1 H). As a further test for specificity, we tested the effect of IL1RAP antibodies on an AML cell line with low IL1RAP expression. Although most AML cell lines tested expressed high levels of IL1RAP, we identified one cell line, KG-1a, that had low levels of surface IL1RAP by flow cytometry. Treatment of KG-1a cells with IL1RAP pAb did not lead to growth inhibition (Fig. S1 I). Together, these experiments support an IL1RAP-specific effect.
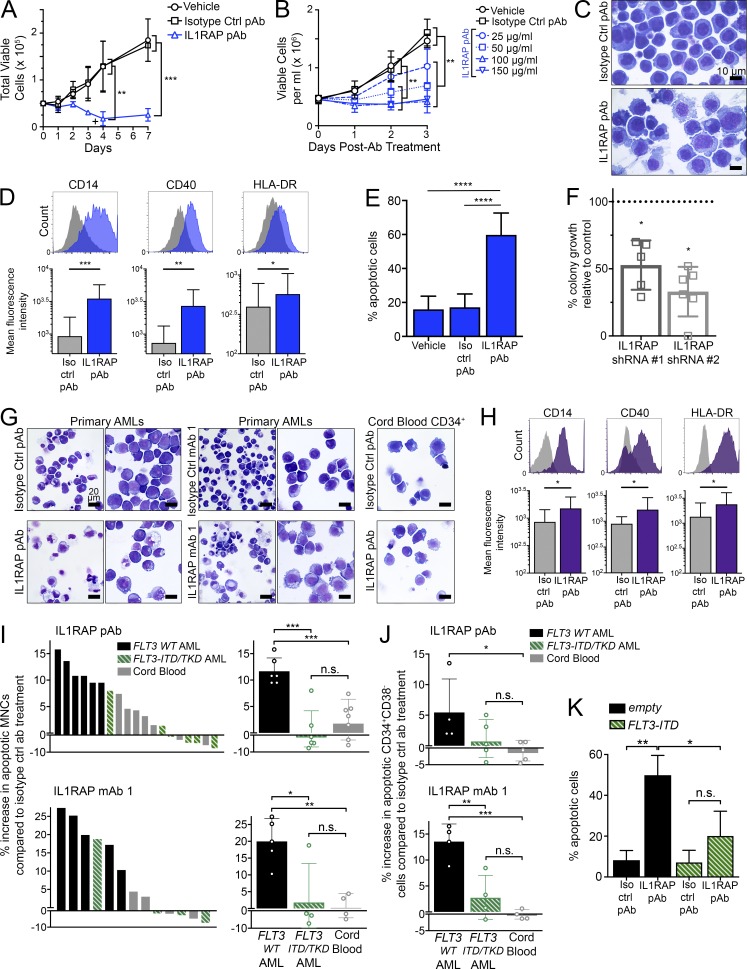
Figure 1.
Targeting of IL1RAP reduces growth of human AML cells by inducing differentiation and apoptosis, without affecting healthy hematopoietic cells. (A) Cell proliferation of THP-1 AML cells with replenishment of IL1RAP polyclonal antibody (pAb). 100 µg/ml of each antibody was added at day 0 and where indicated by the symbol +. Data represent the mean ± SD of two independent experiments. P-values were calculated using unpaired two-tailed t tests, and multiple comparisons were corrected for using the Holm-Sidak method. (B) Cell proliferation of THP-1 cells treated with different doses of IL1RAP pAb. Data represent the mean ± SD of three independent experiments. P-values were calculated using unpaired two-tailed t tests and multiple comparisons were corrected for using the Holm-Sidak method. (C) Morphology of THP-1 cells after treatment with 150 µg/ml IL1RAP pAb or isotype control pAb for 24 h.(D) Expression of macrophage differentiation markers in THP-1 cells by flow cytometry 24 h after addition of 150 µg/ml IL1RAP pAb. Representative histograms are shown (top). Bar graphs (bottom) represent the mean ± SD of five independent experiments. P-values were calculated using ratio paired two-tailed t tests comparing raw MFI values of isotype control pAb– versus IL1RAP pAb–treated cells. (E) Percentage of apoptotic (annexinV+) THP-1 cells after treatment with 150 µg/ml IL1RAP pAb for 72 h. Data represent the mean ± SD of five independent experiments. P-values were calculated using unpaired two-tailed t tests. (F) Colony formation of primary AML MNCs in semisolid media after knockdown of IL1RAP by two shRNAs. Fold change relative to a control shRNA (dotted line) is shown. Each point represents an individual patient: MDS01, MDS02, AML03, AML07, AML13, and AML15 (#2 only). Mean ± SD is shown. P-values were calculated using ratio paired two-tailed t tests comparing raw colony numbers of control shRNA– versus IL1RAP shRNA–treated cells. (G) Morphology of primary AML MNCs or cord blood CD34+ cells after treatment with 300 µg/ml IL1RAP antibodies for 48 h. AML samples shown are AML05 (left) and AML17 (right) for each antibody. 50×, 0.95 NA. (H) Expression macrophage differentiation markers in primary AML samples by flow cytometry 24 h after addition of 300 µg/ml IL1RAP pAb. Histograms from sample AML18 are shown (top). Bar graphs (bottom) represent the mean ± SD of six different primary AML patient samples (AML01, AML05, AML07, AML16, AML18, and AML019). P-values were calculated using Wilcoxon test comparing raw MFI values of isotype control pAb– versus IL1RAP pAb–treated cells. (I) Increase in apoptotic (annexinV+) primary AML or primary cord blood MNCs after treatment with 300 µg/ml IL1RAP pAb (top graphs) or IL1RAP mAb 1 (bottom graphs) for 48 h. Percent increase in apoptosis relative to isotype control pAb treatment is represented two different ways. Left, each bar represents an individual patient. Solid black bars indicate AML patients without FLT3 mutations, striped green bars indicate AML patients harboring FLT3 activating mutations (ITD or TKD), and gray bars represent healthy cord blood samples. Right, patient samples are grouped by type: FLT3 WT AML, FLT3-ITD or -TKD AML, or cord blood. Each point (open circles) represents an individual patient. Mean ± SD is shown. P-values were calculated using unpaired two-tailed t tests. (J) Increase in apoptotic (annexinV+) primary AML or primary cord blood CD34+CD38− cells after treatment with 300 µg/ml IL1RAP pAb (top graph) or IL1RAP mAb 1 (bottom graph) for 48 h. Patient samples are grouped by type: FLT3 WT AML, FLT3-ITD or -TKD AML, or cord blood. Each point (open circles) represents an individual patient. Mean ± SD is shown. P-values were calculated using unpaired two-tailed t tests. (K) Percentage of apoptotic (annexinV+) THP-1 cells transduced with MSCV-GFP empty control or MSCV-FLT3-ITD-GFP and treated with 150 µg/ml IL1RAP pAb for 72 h. Data represent the mean ± SD of three independent experiments. P-values were calculated using unpaired two-tailed t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We also noted that IL1RAP antibody treatment caused THP-1 cells, which are immature monoblastic leukemia suspension cells, to adhere to the culture dish, accumulate cytoplasmic vacuoles, and acquire a smaller nuclear to cytoplasmic ratio, features characteristic of macrophage differentiation (Fig. 1 C and Fig. S1, C and D). In line with these morphological observations, IL1RAP antibody treatment led to a rapid induction of expression of the monocyte/macrophage maturation markers CD14, CD40, and HLA-DR (Fig. 1 D and Fig. S1 E). In addition, IL1RAP antibody treatment significantly increased cell death of THP-1 cells (Fig. 1 E and Fig. S1 F). Costaining for differentiation and apoptosis markers after treatment with IL1RAP antibodies showed that differentiation markers were up-regulated as early as 12 h, including in the preapoptotic population, indicating that differentiation preceded apoptosis (unpublished data). Of note, treatment of THP-1 cells with the physiological IL-1 receptor antagonist, IL-1Ra, did not phenocopy IL1RAP antibody treatment. We treated THP-1 cells with two doses of IL-1Ra, 600 nM (twice the dose shown to be inhibitory to CML CD34+CD38− cells [Zhang et al., 2016]), and a dose 10 times higher, 6 µM. The modest induction of apoptosis by IL-1Ra at the higher dose (P = 0.08) was significantly less than that induced by a 10-fold lower dose of IL1RAP antibody (Fig. S1 G). This result suggests that the effects of IL1RAP inhibition in AML cells are not restricted to inhibition of the IL-1 signaling pathway.
Collectively, these data show that targeting IL1RAP leads to reduced cell growth and induction of differentiation and apoptosis in a cell-intrinsic manner, in the absence of immune effector cells, and thus provide evidence for the functional relevance of IL1RAP in human AML cells.
Targeting of IL1RAP inhibits primary human AML cells, but does not affect healthy hematopoietic cells
Next, we tested IL1RAP inhibition in primary human AML and high-risk MDS cells. We knocked down IL1RAP in mononuclear cells (MNCs) from primary MDS/AML specimens using IL1RAP shRNAs and isolated transduced cells by FACS using the GFP marker encoded by the shRNA lentiviral constructs (Fig. S2 A). shRNA-mediated down-regulation of IL1RAP significantly reduced colony-forming capacity of primary MDS/AML cells (Fig. 1 F; Fig. S2 B; and Table S1). In line with our observations in AML cell lines, IL1RAP antibodies led to a more differentiated morphology in primary AML MNCs, but did not lead to differentiation of normal cord blood CD34+ cells (Fig. 1 G). This was expected, because cord blood CD34+ cells have low IL1RAP expression (Fig. S2 C). Differentiation was confirmed by flow cytometric analysis of six primary AML samples, which showed significantly increased expression of the monocytic differentiation markers CD14, CD40, and HLA-DR after treatment with IL1RAP antibody (Fig. 1 H).
We also measured apoptosis in primary AML MNCs after treatment with IL1RAP antibodies. We observed induction of apoptosis in many, but not all, primary AML samples compared with treatment with the respective isotype control antibodies. When we examined the molecular characteristics of the samples that did not undergo apoptosis in response to IL1RAP antibody treatment, we observed that they all had activating mutations in the FLT3 gene, either internal tandem duplications in the juxtamembrane domain-coding sequence (FLT3-ITD) or tyrosine kinase domain mutations (FLT3-TKD). Although one patient with FLT3-ITD showed increased apoptosis in response to treatment with IL1RAP antibodies, all other FLT3-ITD/TKD samples showed minimal or no response. Overall, IL1RAP antibodies selectively induced apoptosis in FLT3 WT AML MNCs compared with cord blood MNCs, but did not significantly affect viability of FLT3 mutant MNCs. Importantly, IL1RAP antibodies did not lead to apoptosis of healthy cord blood MNCs compared with isotype control antibody treatment (Fig. 1 I). We next examined the effect of IL1RAP antibodies on more immature, leukemic stem and progenitor cell–enriched populations in AML samples. IL1RAP antibody treatment led to significant apoptosis in CD34+CD38− AML cells from FLT3 WT patients, but not FLT3 mutant patients, compared with healthy CD34+CD38− cord blood cells (Fig. 1 J). These data suggest that in certain patient subsets, IL1RAP is functionally important not only in AML blasts, but also in immature AML cells.
To determine if activating FLT3 mutations are merely associated with or are causal of reduced effectiveness of IL1RAP inhibition, we introduced FLT3-ITD into the FLT3 WT cell line THP-1. We transduced THP-1 cells with an MSCV-FLT3-ITD-GFP vector (containing a 21-bp duplication in exon 14 of FLT3) or an MSCV-GFP empty control vector and treated transduced cells with IL1RAP antibodies. Strikingly, we found that IL1RAP antibody–induced apoptosis was abrogated upon expression of FLT3-ITD in THP-1 cells (Fig. 1 K and Fig. S2 D), indicating a causal relationship.
To further investigate the therapeutic window for targeting IL1RAP, we studied the effects of IL1RAP deletion on the hematopoietic system using Il1rap knockout (Il1rap−/−) mice (Cullinan et al., 1998). We monitored wt and Il1rap−/− mice up to 14 mo of age and found no difference in peripheral blood (PB) counts upon Il1rap knockout (Fig. S2 E). We also measured the proportions of mature white blood cell types in wt and Il1rap−/− mice by flow cytometry and found no significant differences (Fig. S2 F). Next, we analyzed the bone marrow (BM) of wt and Il1rap−/− mice by flow cytometry and found no alteration in the percentages of phenotypic hematopoietic stem and progenitor cells (Lineage−c-Kit+Sca-1+ or Lineage−c-Kit+Sca1− cells) in the absence of IL1RAP (Fig. S2 G). Together, these data indicate that IL1RAP is dispensable for the formation of hematopoietic stem and progenitor cells and that healthy, steady-state hematopoiesis is effective even in the absence of IL1RAP.
Lastly, to determine any defects in stem cell functional capacity as a result of Il1rap knockout, we performed competitive transplantation experiments with wt versus Il1rap−/− BM and monitored the engraftment of recipient mice for 20 wk. We did not observe any competitive disadvantage in the repopulation capacity of Il1rap−/− cells (Fig. S2 H), indicating IL1RAP loss does not significantly impair normal HSPC function.
Down-regulation, antibody targeting, or genetic deletion of IL1RAP inhibits AML pathogenesis in vivo
To assess the effect of IL1RAP down-regulation in vivo, we transduced two AML cell lines, THP-1 and HEL, with IL1RAP-directed or control shRNAs and transplanted these cells into immunodeficient recipient (NSG) mice (Fig. 2 A and Fig. S3, A and B). We compared leukemic cell chimerism in the BM at fixed endpoints (day 80 for THP-1 cells; day 30 for HEL cells) and found significantly reduced infiltration of AML cells after IL1RAP knockdown (Fig. 2 B). Of note, leukemic cells isolated from HEL transplanted mice at the endpoint no longer exhibited IL1RAP knockdown, indicating that the leukemia cells had silenced or otherwise compensated for expression of the shRNA transgene and outcompeted cells with IL1RAP knockdown in vivo (Fig. S3 C).
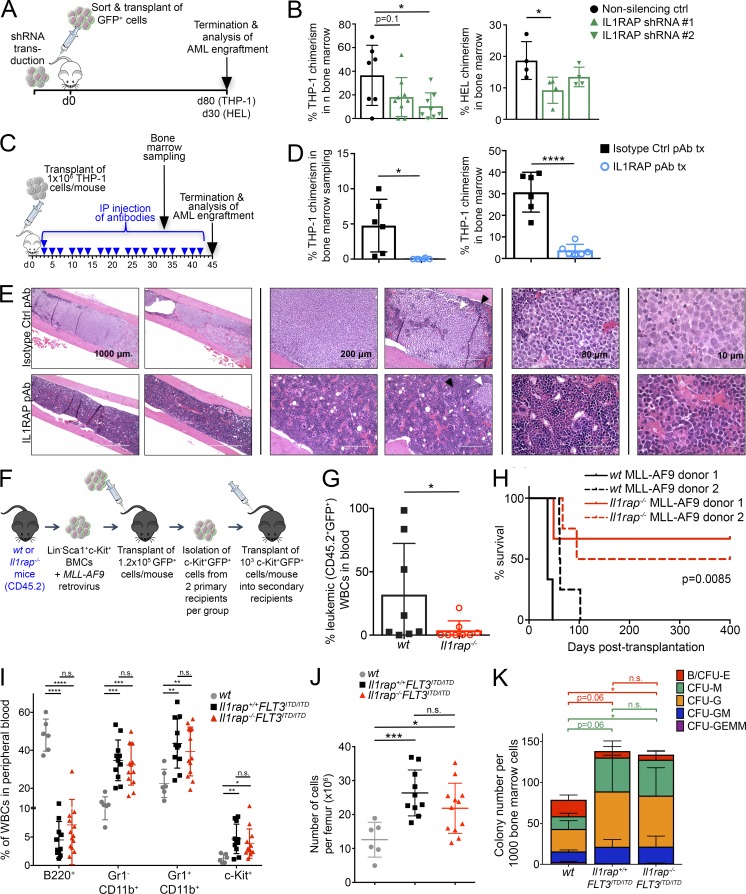
Figure 2.
Down-regulation, antibody targeting, or genetic deletion of IL1RAP inhibits AML pathogenesis in vivo. (A) Experimental scheme for B. THP-1 AML cells or HEL AML cells were transduced with lentiviral control or IL1RAP-directed shRNAs, isolated by FACS using the GFP marker, and 2.5 × 105 (THP-1) or 2.8 × 105 (HEL) transduced cells were transplanted in each NSG mouse. (B) Chimerism of shRNA-transduced human THP-1 cells or HEL cells in the BM of mice at termination. Each symbol represents an individual mouse. Mean ± SD is shown. P-values were calculated using unpaired two-tailed t tests. (C) Experimental scheme for D–E. THP-1 cells were transplanted into NSG mice. Mice were dosed three times per week with 10 mg/kg IL1RAP pAb or isotype control pAb for 6 wk. Blue arrowhead indicates one dose. (D) Chimerism of THP-1 cells in BM of treated mice 33 d (left) and 45 d (right) after transplant. Each symbol represents an individual mouse. Mean ± SD is shown. P-values were calculated using unpaired two-tailed t tests. tx, treatment. (E) Sectioned and H&E–stained femurs of transplanted mice showing THP-1 cell infiltration (white arrowheads) and residual normal mouse BM (black arrowheads). Four magnifications of femurs of multiple mice are shown. (F) Experimental scheme for G–H. MLL-AF9 transduced Il1rap−/− or wt BM LSK cells were expanded ex vivo and transplanted into primary (1.2 × 105 cells/mouse) then secondary recipients (103 cells/mouse). (G) Leukemic cell chimerism in the blood of MLL-AF9 secondary recipient mice 4 wk after transplant. P-value was calculated using an unpaired two-tailed t test. (H) Kaplan-Meier plot showing survival of secondary recipient mice transplanted with wt versus Il1rap−/− MLL-AF9 cells. Recipient mice of cells from each donor are shown as individual curves. P-value was calculated using log-rank (Mantel-Cox) test. (I) Flow cytometric analysis of PB populations in wt, homozygous FLT3-ITD knock-in mice (Il1rap+/+FLT3ITD/ITD), and Il1rap−/−FLT3ITD/ITD mice at 12 mo of age. P-values were calculated using unpaired two-tailed t tests. (J) Cell number per femur in wt, Il1rap+/+FLT3ITD/ITD, and Il1rap−/−FLT3ITD/ITD mice at 12 mo of age. P-values were calculated using unpaired two-tailed t tests. (K) BM cells isolated from wt, Il1rap+/+FLT3ITD/ITD, and Il1rap−/−FLT3ITD/ITD mice at 12 mo of age were plated in methylcellulose medium and scored for colony formation after 14 d. Each colony type is indicated by a different color: B/CFU-E, burst-forming unit erythroid or colony forming unit erythroid; CFU-M, macrophage; CFU-G, granulocyte; CFU-GM, granulocyte/macrophage; CFU-GEMM, granulocyte, erythroid, macrophage, and megakaryocyte. Data represent the mean ± SD of three independent experiments. P-values were calculated using unpaired two-tailed t tests comparing counts for each type of colony. Statistical significance bar color refers to the colony type indicated in the legend. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. n.s, not significant.
Next, to test the efficacy of IL1RAP targeting using a more therapeutically relevant approach, NSG mice were transplanted with THP-1 cells and dosed with isotype control antibody or IL1RAP polyclonal antibody three times per week for 6 wk (Fig. 2 C). IL1RAP antibody treatment was extremely well tolerated by the mice, and no negative side effects or weight loss was observed as a result of the treatment (Fig. S3 D). IL1RAP antibody treatment resulted in a striking reduction in leukemic cells in the BM, both in analysis of BM aspirates taken 4 wk after the start of the treatment and in total BM analysis at the experimental endpoint (Fig. 2 D). The effectiveness of the IL1RAP antibody in eliminating leukemic cells from the BM was further illustrated in sectioned and H&E–stained femurs of treated mice (Fig. 2 E). Photos in the right panel of Fig. 2 E show high magnification images where human leukemic cells can be easily identified, as they are much larger than the residual healthy mouse cells and are a homogenous population of lighter-staining blasts with a large nuclear to cytoplasmic ratio. Femur sections shown at lower magnifications display the near-complete elimination of human leukemia cells after IL1RAP antibody treatment. Because THP-1 cells home not only to the BM but also to the liver in vivo, we monitored leukemic cell infiltration in the livers of treated mice. We observed a dramatic reduction of tumors in the liver (Fig. S3 E, left), and H&E–stained liver sections confirmed substantially reduced infiltration of human leukemia cells in mice treated with IL1RAP antibody (Fig. S3 E, right).
To address whether IL1RAP expression is functionally relevant for leukemia pathogenesis in a genetic model of AML, we transplanted wt or Il1rap−/− mouse BM HSPCs transduced with the MLL-AF9 fusion oncogene into sublethally irradiated congenic recipient mice (Fig. 2 F). MLL-AF9–transduced wt BM cells displayed overexpression of IL1RAP compared with residual normal mouse BM cells (Fig. S3 F). Although most primary recipient mice transplanted with Il1rap−/− MLL-AF9 cells developed leukemia at the same rate as the wt group (Fig. S3 G), all mice in the wt group succumbed to leukemia within 21 wk, but two out of nine mice transplanted with Il1rap−/− MLL-AF9 cells did not develop AML for the duration of the study (Fig. S3 H). c-Kit+ cells have been shown to contain the leukemia-initiating population in the MLL-AF9 retroviral model (Krivtsov et al., 2006). To test the effect of IL1RAP deletion on LSC function in this model, we performed secondary transplantations of c-Kit+ leukemic cells from a total of four primary leukemic mice (two per group). The donor mice for secondary transplant had initial disease latencies of 145 d (wt donor 1), 58 d (wt donor 2), 61 d (Il1rap−/− donor 1), and 58 d (Il1rap−/− donor 2). wt and Il1rap−/− secondary donor cells had similar immunophenotypes: c-Kit+GFP+ cells lacked expression of lymphoid and erythroid lineage markers, had similar expression of myeloid markers (Gr-1/CD11b), and lacked expression of Sca-1 (Fig. S3 I). In secondary transplants, we observed significantly delayed leukemic progression (Fig. 2 G) and increased overall survival in the Il1rap−/− group (P = 0.0085), with more than half of mice transplanted with Il1rap−/− MLL-AF9 primary leukemia cells still alive after 400 d (Fig. 2 H). Lack of differences in the immunophenotypes of transplanted cells indicates that the differences in disease are a result of functional defects rather than the result of alterations in the cell types transplanted. In line with our findings, a recent study found that pharmacologic inhibition of IRAK1, a downstream effector of IL-1RI/IL1RAP, inhibits MLL-AF9–driven leukemia (Liang et al., 2017). Our data provide genetic evidence that deletion of Il1rap impairs AML pathogenesis in vivo.
The effects of IL1RAP antibodies in primary AML patient samples indicated that IL1RAP inhibition is not as effective in the context of activating FLT3 mutations such as FLT3-ITD. To further test this, we assessed the effect of IL1RAP deletion in FLT3-ITD–driven myeloproliferative disease. We crossed Il1rap−/− mice with FLT3-ITD knock-in mice to generate Il1rap−/−FLT3ITD/ITD mice. As reported (Lee et al., 2007), FLT3ITD/ITD mice developed myeloproliferative disease characterized by increased BM cellularity and splenomegaly as a result of expanded granulocyte (Gr-1+/CD11b+), monocyte (Gr-1−CD11b+), and immature (c-Kit+) cell populations with an associated decreased lymphoid population (B220+) in the PB and spleen. This phenotype was not rescued by Il1rap deletion (Fig. 2, I and J; and Fig. S3 J). Furthermore, BM cells isolated from wt, Il1rap+/+FLT3ITD/ITD, and Il1rap−/−FLT3ITD/ITD mice at 12 mo of age were examined for their colony-forming potential in methylcellulose. Compared with wt, both Il1rap+/+FLT3ITD/ITD and Il1rap−/−FLT3ITD/ITD cells exhibited increased colony formation, with expanded output of granulocyte and monocyte/macrophage colonies and diminished output of erythroid colonies (Fig. 2 K). Overall, these data demonstrate that Il1rap is dispensable for FLT3-ITD–driven myeloproliferative disease, consistent with our patient data suggesting that IL1RAP inhibition is mostly ineffective in the setting of constitutive FLT3 activation.
Down-regulation, antibody targeting, or genetic deletion of IL1RAP inhibits FLT3 ligand and stem cell factor (SCF)-mediated signaling and growth
We and others have previously shown that IL1RAP is up-regulated in phenotypically defined LSCs in human AML, and here we have shown that IL1RAP is functionally important for growth and survival of AML cells. However, the molecular pathways governed by IL1RAP in AML cells are unknown. We sought to investigate this by examining molecular effects upon IL1RAP down-regulation, specifically the effect on key signaling pathways in AML. Given the difference in effectiveness of IL1RAP inhibition depending on FLT3 mutational status, we first measured the effect of IL1RAP targeting on proteins and phosphoproteins in the FLT3 signaling pathway. We used shRNAs to knock down IL1RAP in THP-1 cells, which express the WT FLT3 receptor (Fig. S4, A and K), and stimulated the cells with FLT3 ligand (FLT3L). As expected, in cells expressing a control shRNA, FLT3L stimulation led to activation of FLT3 as well as AKT and ERK, downstream mediators of receptor tyrosine kinase signaling. However, knockdown of IL1RAP nearly abolished this FLT3L-induced phosphoprotein activation (Fig. 3 A and Fig. S4 D). This was a striking result, because IL1RAP has not been previously implicated in FLT3 signaling, one of the key oncogenic pathways in AML (Lisovsky et al., 1996; Scheijen and Griffin, 2002; Stirewalt and Radich, 2003; Brandts et al., 2005; Choudhary et al., 2005; Kiyoi et al., 2005; Leung et al., 2013). Next, we tested whether antibody-mediated interference of IL1RAP could inhibit signaling through the FLT3 pathway in AML cell lines and primary AML patients’ samples. We found that even in the absence of exogenous FLT3L, treatment with IL1RAP antibodies reduced FLT3 phosphorylation and total FLT3 levels, indicating a strong inhibition of the FLT3 pathway (Fig. S4 E). We then treated THP-1 AML cells or primary AML MNCs lacking FLT3-activating mutations with IL1RAP or isotype control antibody and stimulated the cells with FLT3L. As expected, FLT3L stimulation led to increased phosphorylation of FLT3 in the presence of the control antibody. Strikingly, IL1RAP antibody treatment led to a significant decrease in FLT3L-induced FLT3 phosphorylation (Fig. 3 B, C, and E; and Fig. S4 F). Given that IL1RAP antibodies did not lead to apoptosis in primary AML patient samples containing FLT3 activating mutations, we assessed if this was associated with a decreased effect on FLT3 signaling. We found that in primary AML MNCs from patients with FLT3 activating mutations, whereas FLT3L stimulation led to slightly increased phosphorylation of FLT3 in combination with the control antibody, a phenomenon previously reported in FLT3-ITD AML cells (Zheng et al., 2011), IL1RAP antibody treatment did not lead to a significant decrease in FLT3 phosphorylation (Fig. 3 D and E; and Fig. S4 I). Finally, because reduction or inhibition of IL1RAP reduced WT FLT3 signaling, we tested whether overexpression of IL1RAP could drive FLT3 signaling. We transduced 293T cells with an IL1RAP expression plasmid and examined protein lysates for phospho-FLT3 by Western blot. We saw a drastic induction of p-FLT3, demonstrating that IL1RAP overexpression can increase signaling through the FLT3 pathway (Fig. S4 G).
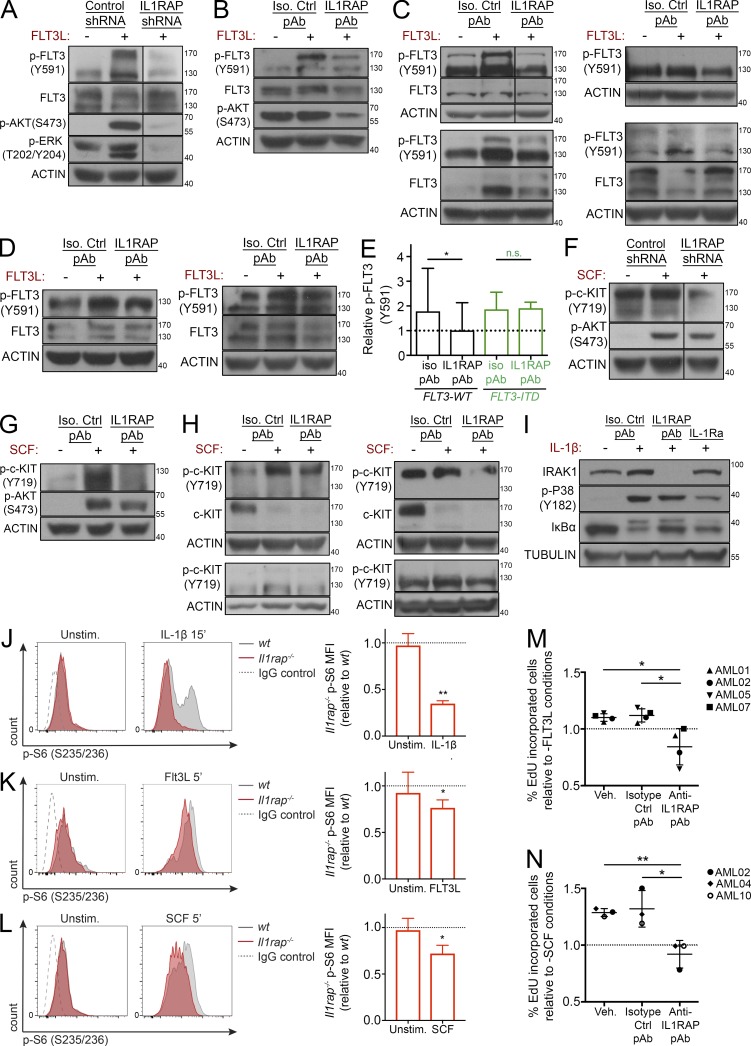
Figure 3.
Down-regulation, antibody targeting, or genetic deletion of IL1RAP inhibits FLT3 ligand and SCF-mediated signaling and growth. (A) IL1RAP or control shRNA–transduced THP-1 cells were stimulated with 50 ng/ml rhFLT3L for 5 min. Protein lysates were examined for p-FLT3 (Y591), total FLT3, p-AKT (S473), and p-ERK (T202/Y204) by immunoblotting. Actin served as a loading control for all immunoblotting experiments unless otherwise stated. (B) THP-1 cells were treated with IL1RAP pAb (100 µg/ml) for 24 h and stimulated with 50 ng/ml rhFLT3L for 5 min. Protein lysates were examined for p-FLT3 (Y591), total FLT3, and p-AKT (S473) by immunoblotting. (C) FLT3 WT primary AML MNCs were treated with IL1RAP pAb (100 µg/ml) and stimulated with 50 ng/ml rhFLT3L for 24 h. Protein lysates were examined for p-FLT3 (Y591) and total FLT3 by immunoblotting. Samples AML02 (top left), AML01 (bottom left), AML05 (top right), and AML16 (bottom right) are shown. (D) FLT3-ITD primary AML MNCs were treated with IL1RAP pAb (100 µg/ml) and stimulated with 50 ng/ml rhFLT3L for 24 h. Protein lysates were examined for p-FLT3 (Y591) and total FLT3 by immunoblotting. Samples AML10 (left) and AML04 (right) are shown. (E) Quantification of p-FLT3 (Y591) from Western blots of FLT3 WT (n = 6) and FLT3-ITD (n = 3) primary AML MNCs treated with isotype control pAb or IL1RAP pAb (100 µg/ml) and stimulated with 50 ng/ml rhFLT3L for 24 h. Dotted line indicates unstimulated, isotype control antibody treated cells. Protein levels relative to actin are shown. P-values were calculated using Wilcoxon test. (F) IL1RAP or control shRNA-transduced TF-1 cells were stimulated with 50 ng/ml rhSCF for 5 min. Protein lysates were examined for p-c-KIT (Y719) and p-AKT (S473) by immunoblotting. (G) HEL cells were treated with IL1RAP pAb (200 µg/ml) for 24 h and stimulated with 50 ng/ml rhSCF for 5 min. Protein lysates were examined for p-c-KIT (Y719) and p-AKT (S473) by immunoblotting. (H) Primary AML MNCs were treated with IL1RAP pAb (100 µg/ml) and stimulated with 50 ng/ml rhSCF for 24 h. Protein lysates were examined for p-c-KIT (Y719) and total c-KIT by immunoblotting. Samples AML06 (top left), AML07 (bottom left), AML04 (top right), and AML10 (bottom right) are shown. (I) THP-1 cells were treated with IL1RAP pAb (100 µg/ml; 670 nM) or IL-1Ra (75 µg/ml; 4.4 µM) for 24 h and stimulated with 20 ng/ml rhIL-1β for 15 min. Protein lysates were examined for IRAK, p-P38 (Y182), and IκBα levels by immunoblotting. β-Tubulin served as loading control. (J) Left, representative p-S6 (S235/236) flow cytometry histograms of Lineage−c-Kit+ BM cells from wt or Il1rap−/− mice, unstimulated, or stimulated for 15 min with 100 ng/ml rmIL-1β. Right, geometric MFI of p-S6 in Il1rap−/− cells relative to wt (n = 3). Data in J–L are expressed as mean ± SD. P-values were calculated using ratio paired two-tailed t tests comparing raw MFI values. (K) Left, representative p-S6 (S235/236) flow cytometry histograms of Lineage−c-Kit+Flt3+ BM cells from wt or Il1rap−/− mice, unstimulated or stimulated for 5 min with 100 ng/ml rmFlt3L. Right, MFI of p-S6 in Il1rap−/− cells relative to wt (n = 3). (L) Left, representative p-S6 (S235/236) flow cytometry histograms of Lineage−c-Kit+ BM cells from wt or Il1rap−/− mice, unstimulated or stimulated for 5 min with 2.5 ng/ml rmSCF. Right, MFI of p-S6 in Il1rap−/− cells relative to wt (n = 3). (M) Primary AML MNCs were treated with IL1RAP pAb (100 µg/ml) and 50 ng/ml rhFLT3L in the presence of EdU for 24 h. EdU incorporation was measured at 24 h by flow cytometry. Fold change relative to conditions with antibody, but without FLT3L (dotted line), is shown. Each symbol represents an individual patient (indicated in legend); data represent the mean ± SD of four different patient samples. P-values were calculated using unpaired two-tailed t tests. (N) Primary AML MNCs were treated with IL1RAP pAb (100 µg/ml) and 50 ng/ml rhSCF in the presence of EdU. EdU incorporation was measured at 24 h by flow cytometry. Fold change relative to conditions with antibody, but without SCF (dotted line), is shown. Each point represents an individual patient (indicated in legend); data represent the mean ± SD of three different patient samples. P-values were calculated using unpaired two-tailed t tests. *, P < 0.05; **, P < 0.01. n.s., not significant.
These findings led us to investigate the ability of IL1RAP antibodies to block signaling through other AML-relevant pathways. Like FLT3, the receptor c-KIT is a class III receptor tyrosine kinase that plays a pivotal role in AML (Ikeda et al., 1991; Reilly, 2002; Scheijen and Griffin, 2002). Because THP-1 cells do not express high levels of c-KIT, we knocked down IL1RAP in TF-1 cells, which express both IL1RAP and c-KIT at high levels (Fig. S4 B), and stimulated the cells with KIT ligand (SCF). SCF stimulation led to increases in p-c-KIT as well as p-AKT in control cells, and knockdown of IL1RAP attenuated this phosphoprotein activation (Fig. 3 F and Fig. S4 D). We used another AML cell line that highly expresses IL1RAP and c-KIT, HEL (Fig. S4 C), to test the effect of IL1RAP antibodies on this pathway. Again, we found increased phosphorylation of c-KIT and AKT upon stimulation with SCF; this effect was reversed after IL1RAP antibody treatment (Fig. 3 G and Fig. S4 H). These findings were validated in several primary AML samples (Fig. 3 H and Fig. S4 H).
Because IL1RAP is known to be critical for IL-1 signaling (Greenfeder et al., 1995; Dunne and O’Neill, 2003), we also examined the effect of IL1RAP targeting on IL-1 downstream targets in AML cells, using a natural antagonist of the pathway, IL-1 receptor antagonist (IL-1Ra), as a positive control. We treated THP-1 cells with IL1RAP antibody, isotype control antibody, or IL-1Ra, and stimulated the cells with IL-1β. We found that IL1RAP antibody antagonized IL-1β–mediated phosphorylation of P38 and IκBα degradation, indicating that the IL-1 pathway is strongly inhibited by IL1RAP interference in AML cells. IL1RAP antibody treatment abolished IRAK1 expression (Fig. 3 I and Fig. S4 J), corroborating a recent study showing that inhibition of IRAK1 reduces MDS and AML leukemic colony formation and prolongs survival in a xenograft model of MDS (Rhyasen et al., 2013).
As an additional approach to investigate the importance of IL1RAP in these two signaling pathways, we used the Il1rap−/− mouse model. We performed phospho–flow cytometry on HSPCs isolated from wt and Il1rap−/− mice; cells were stimulated with FLT3L, SCF, or IL-1β as a positive control, and intracellular phospho–S6 protein was measured as a readout of downstream signaling though these pathways (Kalaitzidis and Neel, 2008). We found that Il1rap−/− BM HSPCs had a significantly reduced response to stimulation with FLT3L, SCF, and IL-1β (Fig. 3, J–L). These data further validate the role of IL1RAP in signaling through the FLT3 and c-KIT pathways.
Finally, to determine whether IL1RAP-mediated inhibition of multiple signaling pathways translates to functional growth inhibition, we treated primary AML cells with IL1RAP or isotype control antibody, stimulated the cells with either FLT3L or SCF, and measured EdU incorporation by flow cytometry as a readout of proliferation. IL1RAP antibody treatment antagonized both FLT3L- and SCF-stimulated proliferation in multiple primary AML patients’ samples (Fig. 3, M and N).
Together, these data indicate that targeting of IL1RAP inhibits signaling and AML cell growth through the FLT3 and c-KIT pathways in addition to inhibition of the canonical IL-1 receptor signaling pathway.
IL1RAP physically interacts with FLT3 and c-KIT in human AML cells
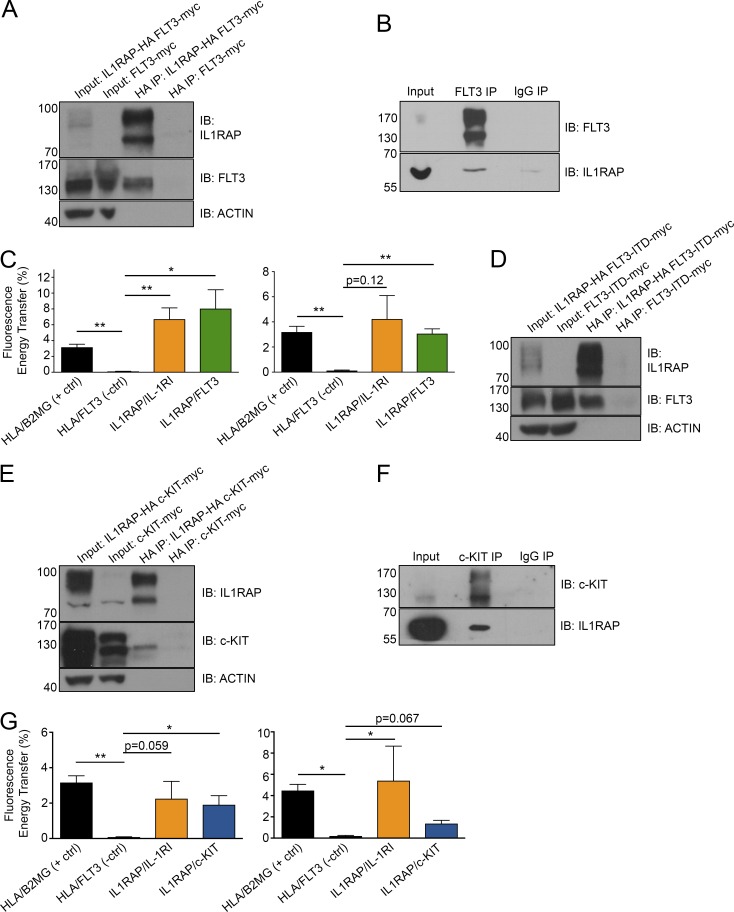
Given that inhibition of IL1RAP interfered with FLT3 and c-KIT signaling in AML cells, we hypothesized that IL1RAP may physically interact with these receptors. We generated constructs encoding HA-tagged IL1RAP and myc-tagged FLT3 and c-KIT and coexpressed these fusion proteins in 293T cells. Whole cell lysates were prepared and subjected to immunoprecipitation with anti-HA magnetic beads. FLT3 and c-KIT were detected in the eluted proteins from HA (IL1RAP) pulldown (Fig. 4, A and E, respectively), indicating they each interact with IL1RAP. We next sought to measure these interactions in AML cells. We used FLT3 and c-KIT antibodies for coimmunoprecipitation in THP-1 whole cell lysates and detected IL1RAP by immunoblot of proteins eluted from either FLT3 or c-KIT pulldown (Fig. 4, B and F; and Fig. S4 L), demonstrating IL1RAP–FLT3 and IL1RAP–c-KIT interactions under endogenous, nonoverexpression conditions in AML cells. To confirm these interactions in AML cells through an independent method, we used a flow cytometric Förster resonance energy transfer (FRET) assay on AML cell lines with high expression of IL1RAP and FLT3 or IL1RAP and c-KIT (Fig. S4 K). We stimulated cells with IL-1β, FLT3L, or SCF before fixation and antibody labeling. Fluorescent-labeled antibodies for IL1RAP and IL-1RI, FLT3, or c-KIT served as FRET donor and acceptor molecules. We used flow cytometry to measure the overlap of the emission spectrum of the fluorescent donor molecule (phycoerythrin [PE]) with the excitation spectrum of the acceptor molecule (allophycocyanin [APC]), known as FRET (Förster, 1948). This energy transfer occurs at physical distances between 10 and 100 Å, indicating a physical interaction is occurring. Strikingly, we observed an energy transfer between IL1RAP–FLT3 and IL1RAP–c-KIT that was comparable to the well-documented protein interactions between MHC complex members (HLA–β2MG), as well as the canonical IL1RAP–IL-1RI complex. HLA and FLT3, which are not documented or expected to interact, served as a negative control (Fig. 4, C and G).
Figure 4.
IL1RAP physically interacts with cytokine receptors FLT3 and c-KIT in human AML cells. (A) Coimmunoprecipitation of HA-tagged IL1RAP in protein lysates from 293T cells transfected with IL1RAP-HA and FLT3-myc fusion vectors. Immunoblot (IB) was performed for IL1RAP and FLT3. (B) Coimmunoprecipitation of endogenous FLT3 in protein lysates from THP-1 cells. Immunoblot was performed for FLT3 and IL1RAP. (C) Fluorescence energy transfer (FRET) between IL1RAP and FLT3. THP-1 (left) or MOLM-13 (right) cells were stimulated with 200 ng/ml rhIL-1β or 100 ng/ml rhFLT3L. For C and G, cells were stained with fluorescent antibodies against the indicated proteins for flow cytometry analysis and FRET was determined for cells expressing both receptors. Data represent the mean ± SD of three independent experiments. (D) Coimmunoprecipitation of HA-tagged IL1RAP in protein lysates from 293T cells transfected with IL1RAP-HA and FLT3-ITD-myc fusion vectors. Immunoblot was performed for IL1RAP and FLT3. (E) Coimmunoprecipitation of HA-tagged IL1RAP in protein lysates from 293T cells transfected with IL1RAP-HA and c-KIT-myc fusion vectors. Immunoblot was performed for IL1RAP and c-KIT. (F) Coimmunoprecipitation of endogenous c-KIT in protein lysates from THP-1 cells. Immunoblot was performed for c-KIT and IL1RAP. (G) FRET between IL1RAP and c-KIT. THP-1 (left) or HEL (right) cells were stimulated with 200 ng/ml IL-1β or 100 ng/ml SCF. Data represent the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01.
Interestingly, we saw interactions between IL1RAP and FLT3 in a cell line heterozygous for FLT3-ITD, MOLM13, indicating either that we were detecting interactions with the remaining WT FLT3 in these cells or that IL1RAP also interacts with the mutant FLT3 receptor. To clarify this result and considering the difference in sensitivity of FLT3 WT and FLT3 mutant AMLs to IL1RAP inhibition, we assessed if IL1RAP can physically interact with mutant FLT3. We generated a construct encoding myc-tagged FLT3 with an internal tandem duplication (21 bp in exon 14), coexpressed it with HA-IL1RAP in 293T cells and performed immunoprecipitation with anti-HA magnetic beads as described above. FLT3 was detected in the eluted proteins from HA (IL1RAP) pulldown (Fig. 4 D), indicating IL1RAP can still interact with the mutant FLT3 receptor. Given our data showing that IL1RAP targeting does not abrogate FLT3-ITD signaling (Fig. 3, D and E), we conclude that the presence of FLT3-ITD does not abolish physical interaction with IL1RAP, but instead leads to constitutively active FLT3 signaling that is no longer dependent on IL1RAP in AML cells.
Collectively, our data demonstrate that endogenous interactions between IL1RAP–FLT3 and IL1RAP–c-KIT are occurring in AML cells, and show for the first time that IL1RAP can cooperate with and amplify multiple key pathways in AML.
Discussion
Over the past 25 years, several lines of evidence have shown that AML arises from primitive hematopoietic cells that gain initial mutations, giving rise to preleukemic stem cells (pre-LSC) and, upon progressive acquisition of further aberrations, become fully transformed LSCs capable of initiating and maintaining the disease (Baum et al., 1992; Lapidot et al., 1994; Bonnet and Dick, 1997; Passegué et al., 2003; Sarry et al., 2011; Jan et al., 2012; Pandolfi et al., 2013; Shlush et al., 2014; Yasuda et al., 2014). Although traditional cytotoxic chemotherapies often reduce leukemic burden in patients to undetectable levels, pre-LSC and LSC are generally chemoresistant and thus are not eliminated. Through resistance to this initial treatment or acquisition of additional mutations via therapy-related DNA damage, these cell populations drive relapse and death in the majority of AML patients. Thus, it is vital that we gain a molecular understanding of these cells and derive therapies that eliminate pre-LSC and LSC to prevent relapse and ultimately cure AML (Saito et al., 2010; Pandolfi et al., 2013; Corces-Zimmerman et al., 2014).
Heterogeneity in the cellular compartment of origin as well as the mutational spectrum of AML have challenged the identification of commonly dysregulated pathways that are relevant in the initiation and maintenance of the disease (Sarry et al., 2011; Jan and Majeti, 2013; Papaemmanuil et al., 2016). It has been proposed that age-associated chronic inflammation can facilitate clonal hematopoiesis and progression to hematopoietic malignancies including AML. These inflammatory signaling pathways lead to cell survival, cell cycling, and DNA damage, promoting cell transformation (Culver-Cochran and Starczynowski, 2018). In addition, chronic IL-1 signaling has recently been linked to the establishment of a myeloid differentiation bias and impaired blood homeostasis (Pietras et al., 2016), conditions that may enable pre-leukemic clones to expand and eventually transform (Pietras, 2017). Targeting of inflammatory pathways in myeloid malignancies is a focus of ongoing efforts including clinical trials (Warr et al., 2011; Rhyasen et al., 2013; Gañán-Gómez et al., 2015; Hemmati et al., 2017). Our group has previously reported that IL1RAP is one of the few genes commonly up-regulated in pre-LSC and LSC-containing immunophenotypic populations from BM samples of patients with AML (Barreyro et al., 2012). Of note, IL1RAP is directly upstream of the MyD88/IRAK1/IRAK4/TRAF6 innate immunity signaling axis which has been implicated in MDS and AML pathogenesis (Gañán-Gómez et al., 2015; Rhyasen and Starczynowski, 2015; Fang et al., 2017). Although expression of IL1RAP is minimal in healthy HSPCs (Barreyro et al., 2012), IL1RAP has been shown to be an important inflammatory regulator in its role in the IL-1 pathway (O’Neill, 2008), and its overexpression in myeloid malignancies may be linked to this proinflammatory state. Studies in Il1rap−/− mice show that in response to IL-1 injection, IL1RAP is required in a variety of cell types for IL-6 secretion and E-selectin expression, two molecules involved in recruitment of immune cells to infection sites (Cullinan et al., 1998), and we have independently reproduced these findings (unpublished data). In addition, Il1rap, but not Il-1r1, was shown to be necessary for both IL-33 and IL-36–driven skin inflammatory phenotypes consisting of immune cell infiltration and increased cytokine and chemokine expression (Blumberg et al., 2007; Rankin et al., 2010; Liang et al., 2011), indicating IL1RAP’s contribution to a proinflammatory state. In future studies, it will be interesting to assess whether possible proinflammatory effects of IL1RAP overexpression are functionally relevant and contribute to disease pathogenesis in MDS/AML.
Because of the low levels of IL1RAP on normal HSPC and its apparent nonessentiality for mammalian organisms given that Il1rap−/− mice have no major steady-state phenotype (Cullinan et al., 1998), IL1RAP is currently emerging as a promising candidate surface target in various myeloid malignancies, including AML, and approaches to target IL1RAP are underway at various preclinical and clinical stages with encouraging initial results (Ågerstam et al., 2015, 2016; Jiang et al., 2016). However, most current strategies rely on recruitment of the immune system and very little is known about possible cell-intrinsic functions of IL1RAP in AML cells. This could be of particular importance for several reasons: (1) most patients with AML have significantly impaired immune systems, which may make the activation of immune cells challenging; and (2) if the target is not functionally relevant, development of therapy resistance is more likely as cells could down-regulate IL1RAP without disturbing their function.
In this study, we show that disruption of IL1RAP function via RNA interference, genetic deletion, or through direct antibody targeting significantly delays AML pathogenesis both in vitro and in vivo in the absence of immune effector cells and without perturbing healthy hematopoiesis. Specifically, our data show that IL1RAP antibodies preferentially lead to apoptosis in AML MNCs and AML CD34+CD38− cells, compared with these populations in healthy samples. Accordingly, we did not observe hematopoietic dysfunction in Il1rap−/− mice, suggesting that there is a therapeutic window for targeting IL1RAP. Our study reveals a rationale for the further exploration of therapeutic strategies directly targeting IL1RAP; ones that exploit functional dependence of AML cells on IL1RAP.
IL1RAP is commonly known to be the coreceptor for the IL-1 receptor and it is required to transduce IL-1 signaling, leading to proinflammatory gene expression and cell survival (O’Neill, 2000; Weber et al., 2010). Our findings demonstrate a much broader role for IL1RAP in AML than previously appreciated. In exploring the molecular basis for the inhibitory effects of IL1RAP targeting, we found that IL1RAP amplifies signaling through known, important oncogenic AML pathways and physically interacts with the receptor tyrosine kinases FLT3 and c-KIT in AML cells. Of note, findings in other cell types are also in support of the notion of a cell type–dependent promiscuity of IL1RAP; e.g., it was reported that IL1RAP can form a complex with the IL-33 receptor and c-Kit in mast cells and with the IL-36 receptor in T cells and is essential for signaling through these pathways (Towne et al., 2004; Ali et al., 2007; Lingel et al., 2009; Drube et al., 2010). These novel roles of IL1RAP in AML uncover an adaptor molecule-dependent mechanism used by AML cells to amplify signaling that is not a result of a direct activating mutation in signaling receptors. In line with this finding, our experiments reveal that in the setting of constitutive activation of the FLT3 receptor, signal amplification by IL1RAP may become less relevant, as IL1RAP targeting in this setting did not significantly alter FLT3 signaling. The FLT3 and c-KIT receptors are both class III receptor tyrosine kinases that share a similar structure, containing an extracellular ligand-binding domain with five immunoglobulin-like domains, a single-pass transmembrane domain, juxtamembrane domain, and intracellular cytoplasmic domains with tyrosine kinase activity (Reilly, 2002). Because c-KIT–activating mutations are relatively rare in AML, we were not able to study whether these mutations would alter the effectiveness of IL1RAP inhibition. However, we would speculate that constitutive activation of c-KIT may abrogate the inhibition of SCF-driven signaling and growth mediated by targeting of IL1RAP. Given our previous findings that IL1RAP overexpression occurs on early LSCs (Barreyro et al., 2012) and considering that FLT3 and c-KIT activating mutations are a relatively late event in leukemic stem cell transformation (Paschka et al., 2013; Corces-Zimmerman et al., 2014), we posit that even in FLT3 or c-KIT mutated leukemias there may be early pre-LSC or LSC subclones where IL1RAP plays a role in sustaining cell survival and expansion, thus still may be a therapeutically relevant target in these mutational subsets of the disease.
Collectively, our findings indicate that IL1RAP represents a potent therapeutic target by permitting the simultaneous targeting of multiple oncogenic pathways in AML (Fig. 5). Importantly, as IL1RAP is also overexpressed in HSPC of chronic myeloid leukemia (Järås et al., 2010; Gerber et al., 2013; Herrmann et al., 2014; Zhao et al., 2014; Sadovnik et al., 2017; Warfvinge et al., 2017) and high-risk MDS (Barreyro et al., 2012; Shastri et al., 2017), and a recent paper has uncovered that IL1RAP has an active super enhancer in AML (McKeown et al., 2017), there is significant therapeutic potential in further developing IL1RAP-directed targeting strategies.
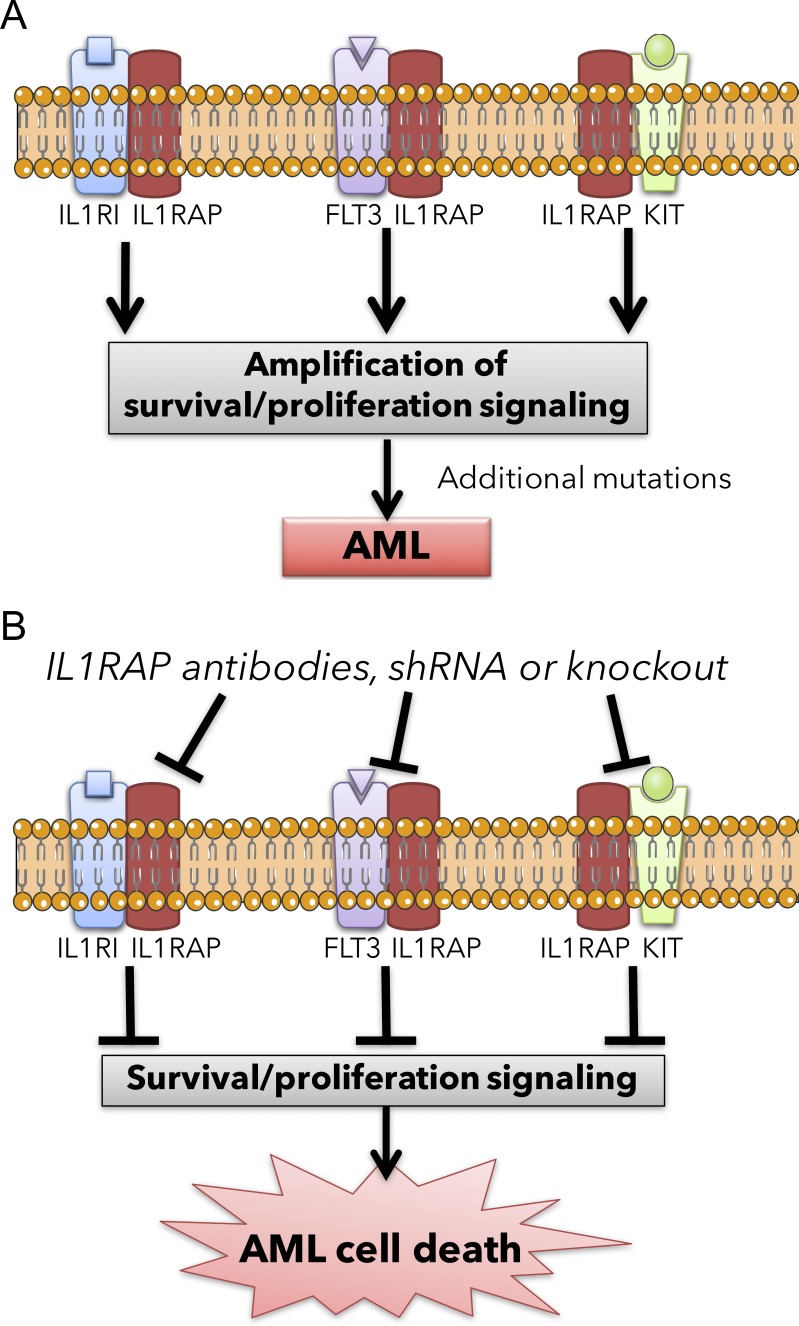
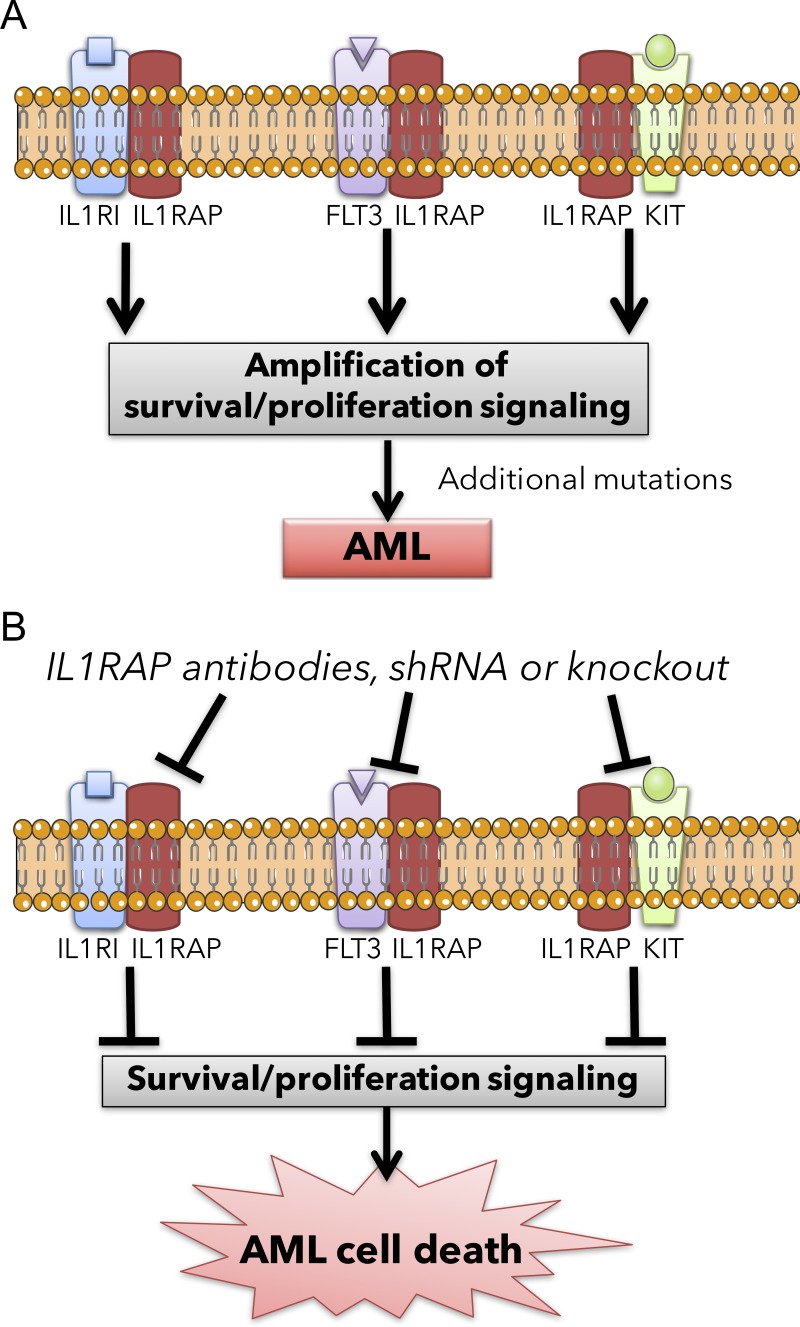
Figure 5.
Proposed model of IL1RAP targeting in acute myeloid leukemia leading to the simultaneous inhibition of multiple oncogenic pathways. (A) Schematics of IL1RAP-driven amplification of multiple signaling pathways in AML stem and progenitor cells. (B) Schematics of the same signaling pathways in AML stem and progenitor cells upon IL1RAP inhibition.
Materials and methods
Reagents and cell lines
THP-1, KG1-a, HEL, and TF-1 cells were obtained from the American Type Culture Collection (ATCC). MOLM-13 cells were obtained from DSMZ. 293T cells were purchased from Open Biosystems. THP-1 cells were cultured in RPMI 1640 medium containing 2% heat-inactivated (hi) FBS and 1% penicillin/streptomycin (P/S). KG-1a cells were cultured in IMDM medium containing 2% hiFBS and 1% P/S. TF-1 cells were cultured in RPMI 1640 medium containing 10% hiFBS, 1% P/S and 2 ng/ml rhGM-CSF. HEL and MOLM-13 cells were cultured in RPMI 1640 medium containing 10% hiFBS and 1% P/S. 293T cells were cultured in DMEM medium containing 10% hiFBS and 1% P/S. Cells were incubated at 37°C with 5% CO2.
The polyclonal anti-IL1RAP antibody was purchased from Santa Cruz (sc-47056 LS). The monoclonal anti-IL1RAP antibodies were purchased from Abnova (H00003556-M03, clone 3D8; and H00003556-M06, clone 2A3). IL-1 receptor antagonist (IL-1Ra) was purchased from Sigma-Aldrich (SRP3084). Normal goat IgG was purchased from Santa Cruz (sc-3887 LS). The monoclonal isotype control antibodies (mouse IgG2a and IgG2b) were purchased from Abcam (ab18449 and ab18457). The polyclonal anti-CD13 antibody was purchased from Santa Cruz (sc-6995 LS).
For flow cytometry, cells were preincubated with human or mouse TruStain FcX Fc Receptor Blocking Solution (422302 and 101320; Biolegend). IL1RAP expression was measured using anti–human IL-1RAcP/IL-1R3 PE antibody (FAB676P; R&D) or APC antibody (FAB676A; R&D) with mouse IgG1 PE (IC002P; R&D) or APC (17-4714; eBioscience) as controls. CD13 expression was measured using anti–human CD13 APC/Cy7 antibody (301710; Biolegend). Recombinant human FLT3 ligand (rhFLT3L), rhSCF, rhTPO, rhIL-3, and rhIL-6 were purchased from Gemini (300-118, 300-185, 300-188, 300-151, and 300-906). rhIL-1β was purchased from R&D (201-LB). rhGM-CSF was purchased from Gemini (300-124). Recombinant mouse FLT3L (rmFLT3L), rmSCF, and rmTPO were purchased from Miltenyi Biotec (300-306, 300-348, and 300-351). rmIL-3 and rmIL-6 were purchased from Gemini (300-324 and 300-327). rmIL-1β was purchased from R&D (401-ML).
Primary samples
Umbilical cord blood, adult AML/MDS BM, and PB samples were obtained after written informed consent and following Albert Einstein College of Medicine institutional review board approval (CCI 2008-842 and 2006-536). For functional studies, MNCs were isolated by Ficoll-Paque density gradient centrifugation (Ficoll Hypaque was purchased from GE Healthcare). Primary MDS/AML cells were cultured in IMDM medium containing 2% hiFBS, 1% P/S, 50 ng/ml rhFLT3L, 50 ng/ml rhSCF, 50 ng/ml rhTPO, 10 ng/ml rhIL-3, and 25 ng/ml rhIL-6. Characteristics of the primary MDS/AML samples used for experiments are found in Table 1.
Table 1. Characteristics of AML specimens.
| Sample number | Age/ sex | Diagnosis | Sample type | Molecular | Chromosomal abnormalities | Fold change in IL1RAP MFI (relative to isotype) |
|---|---|---|---|---|---|---|
| MDS01 | 60/M | High risk MDS | PB | N/A | Monosomy 7 | 5.9 |
| MDS02 | N/A | High risk MDS | PB | N/A | Monosomy 7 | 7.1 |
| AML01 | 76/M | AML M4, myelomonocytic | BM | U2AF1 S34F; TET2 frameshift & C1378Y & D1587G; ASXL1 L1304V; NRAS G13D | None detected | 10.4 |
| AML02 | 68/M | Residual AML after induction therapy with monocytic differentiation; history of MPD, DLBCL | PB | JAK2; SRSF2 P95H; TET2 R550X; TP53 R175H | Complex cytogenetics & t(4:11) | 6.1 |
| AML03 | M | High risk MDS/AML | PB | N/A | del(13)(q12q22) and der(18)t(13;18)(q22;q23) | 11.8 |
| AML04 | 25/M | AML M2 | BM | FLT3-ITD | Trisomy 8 | 4.4 |
| AML05 | 62/F | AML/MPN | PB | JAK2 V617F; TP53 R248W; IDH2 R140Q | None detected | 5.9 |
| AML06 | 49/F | Relapsed AML | PB | FLT3-ITD | 46,XX | 29 |
| AML07 | 73/M | AML | PB | N/A | 46,XY,t(1;4)(p21;q21)[3]/ 46,XY[7] | 4.1 |
| AML08 | 70/M | AML (monocytic) | PB | ASXL1 L823* (nonsense); BCOR R1183*; RUNX1 frameshift; SRSF2 P95H; TET2 insertion & R1516* | None detected | 10.3 |
| AML09 | 54/F | AML (monocytic) | PB | FLT3-TKD; DNMT3A R882H; IDH1 R132H; MLL-PTD | None detected | 22 |
| AML10 | 61/F | AML M5 with hyperleucocytosis | BM | FLT3-ITD; DNMT3A R882H; NPM1 frameshift | None detected | 12 |
| AML11 | 48/F | AML (monocytic) | PB | CEBPA frameshift; CSF3R R698H; NF1 SNP | None detected | 2.5 |
| AML12 | 37/F | AML | PB | FLT3-ITD | 47,XX,+8[15]/ 46,XX[5] 21–24% trisomy 8 (D8Z2,RUNX1T1)x3[42-48/200] | 1.9 |
| AML13 | 67/M | AML | PB | MLL-PTD | Trisomy 8 | 13.2 |
| AML14 | 59/F | AML | PB | FLT3-ITD; NPM1; WT1 | None detected | 3.1 |
| AML15 | 38/M | AML | PB | NRAS | 46,XY t(9;11)(p22;q23) | 3.6 |
| AML16 | 54/M | AML | PB | DNMT3A, NPM1 | None detected | 12.7 |
| AML17 | 38/M | AML | PB | N/A | N/A | 8.3 |
| AML18 | 29/M | AML | BM | None detected | t(8;21)(q22;q22) 45,X,-Y,t(8;21)(q22;q22)[20] | 19.4 |
| AML19 | 61/M | MDS/MPN/AML | PB | JAK2 | 46,XY,der(8) ?t(8;21)(q22;q22), del(9)(q13q22), dup(12)(q14q24.1), -22,+1-2mar[8]/46,xy(3) | N/A |
Age, sex, diagnosis, sample type, molecular abnormalities, chromosomal abnormalities (fluorescence in situ hybridization, karyotyping), and IL1RAP expression (fold change in MFI relative to isotype control antibody stain) are shown where data were available. * indicates stop codon.
Molecular diagnostics
Conventional cytogenetics and fluorescence in situ hybridization were performed on primary patient BM aspirates or PB at the Albert Einstein College of Medicine Molecular Cytogenetic Core. Next-generation sequencing was performed on genomic DNA to detect mutations in the following genes: ASXL1, CBL, DNMT3A, ETV6, EZH2, IDH1, IDH2, JAK2, KIT, MPL, NPM1, NRAS, PHF6, RUNX1, TET2, SETBP1, SF3B1, SRSF2, TP53, U2AF1, ZRSR2, and MLL-PTD according to standard methods (AML molecular panel, Genoptix). FLT3-ITD mutations were detected by PCR as previously described (Kiyoi et al., 1999).
Flow cytometry
FACS was done using a Becton Dickinson (BD) FACS Aria, five-laser BD FACSAria II Special Order System, or MoFlo Astrios (Beckman Coulter). Flow cytometry analysis was performed on a five-laser BD FACSAria II Special Order System or BD LSRII containing a yellow laser (BD LSR II-Y). MFI refers to geometric mean fluorescence intensity. Log10 scales are used for plots showing flow cytometry fluorescence intensity data.
Cell proliferation, differentiation, and apoptosis assays
For liquid culture assays, cell lines and primary cells were plated at 500,000 cells/ml. For liquid culture growth assays longer than 72 h, fresh media and antibody were added every 72 h. For proliferation assays, cells were manually counted by trypan blue exclusion using a hemocytometer. Expression of macrophage markers was measured by flow cytometry using following antibodies: CD14 (MHCD1406; Invitrogen), HLA-DR (560943; BD PharMingen), and CD40 (313004 or 313005; Biolegend). Effects of IL1RAP antibodies and IL-1Ra on cell death were measured with DAPI and fluorescently labeled annexinV by flow cytometry as previously described (Barreyro et al., 2012). For primary AML and cord blood samples, cells were treated for 48 h with IL1RAP or control antibodies and then costained with antibodies against CD34 (343512; Biolegend), CD38 (25-0389; eBioscience), annexinV (556420; BD PharMingen), and DAPI to determine apoptotic proportions in bulk or specific cell subsets. Cord blood CD34+ cell enrichment was performed before treatment with IL1RAP or control antibodies using the human CD34 MicroBead kit (130-046-702; Miltenyi Biotec) according to the manufacturer’s instructions.
For FLT3-ITD transduction, THP-1 cells were placed in virus-coated plates (see below) with 8 µg/ml polybrene to facilitate transduction and spun at 1,000 rcf for 1 h at 32°C. Transduced (GFP+) THP-1 cells were gated on for flow cytometric apoptosis analysis after IL1RAP antibody treatment. The transgene was detected in THP-1 cells by isolating genomic DNA and amplifying the FLT3-ITD–containing exons using Amplitaq gold (4311806; Applied Biosciences) as previously described (Kiyoi et al., 1999).
Knockdown of IL1RAP and colony formation assays
For knockdown of IL1RAP, we cloned shRNA template oligonucleotides into the pSIH1-H1-copGFP shRNA vector (System Biosciences) and produced lentiviral particles as previously described (Barreyro et al., 2012). THP-1, HEL, or TF-1 cells were transduced with a nonsilencing control (luciferase) shRNA- or IL1RAP-specific shRNAs (MOI = 1), and transduced (GFP+) cells were FACS sorted before transplantation. For all shRNA transduction experiments, 8 µg/ml polybrene was added to cells before incubation with virus, and spin infection was performed after addition of virus (1,000 rcf for 1 h at 32°C) to facilitate transduction. Knockdown efficiency was measured by flow cytometry using anti–human IL-1RAcP/IL-1R3 APC or PE (FAB676A and FAB676P; R&D) and mouse IgG1 APC (17-4714; eBioscience) or PE (IC002P; R&D) as controls.
Primary human MDS and AML MNCs were transduced with IL1RAP shRNA lentiviruses (MOI = 1). 24 h after shRNA infection, GFP+ cells were FACS sorted and plated between 1.5 × 105 and 2.5 × 105 cells/ml in human complete methylcellulose medium (HSC003; R&D) supplemented with 40 µg/ml low-density lipoproteins (L7914; Sigma-Aldrich). Colonies were scored after 10–14 d.
Mouse transplantation experiments
For xenograft experiments, THP-1 or HEL cells were intravenously injected into sublethally irradiated (200 rads total body irradiation) NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; stock 005557; Jackson Laboratory). For antibody treatment of mice, mice were injected intraperitoneally with 10 mg/kg IL1RAP pAb three times per week, with a first dose of 20 mg/kg 3 d after transplantation. Mice were sacrificed at a fixed endpoint or at signs of disease. For endpoint analysis of xenografted mice, one femur and/or a piece of the liver was saved for histology. Cells from the BM were isolated by crushing of tibiae, femurs, and pelvic bones of mice in PBS containing 2 mM EDTA (PBS EDTA). Isolation of cells from spleens and livers was performed by manual crushing in PBS EDTA. Erythrocytes in each organ were lysed using ammonium chloride lysing (ACK) solution, pH 7.4. Chimerism analysis was performed on cells aspirated from the hind femurs or on total BM cells. BM cells were stained with antibodies against human CD15 (MHCD1528; Invitrogen) or human CD45 (560975; BD PharMingen) and mouse CD45.1 (25-04553-82 or 17-0453-81; eBioscience) and analyzed by flow cytometry. IL1RAP expression was measured in mCD45.1−GFP+ cells using anti–human IL-1RAcP/IL-1R3 PE antibody (FAB676P; R&D).
MSCV-MLL-AF9-IRES-GFP, MSCV-FLT3-ITD-IRES-GFP, and MSCV-IRES-GFP retroviral vectors and respective packaging vectors were transfected into 293T cells to produce viral supernatants. To facilitate virus transduction, virus was spun (MOI = 0.5; 1,000 rcf for 1 h at 32°C) in 12-well plates coated with 24 µg retronectin (T100A; Takara) per well. Viral supernatant was then removed and cells were added to plates for spin infection (see below).
Il1rap knockout (Il1rap−/−) mice were obtained from Jackson Laboratory (B6;129S1-Il1raptm1Roml/J; stock 003284) and backcrossed for over 10 generations to a C57BL/6J background. wt littermates were used as controls. For MLL-AF9 experiments, BM was isolated from Il1rap−/− and wt C57BL/6J mice (CD45.2). Lin−c-Kit+Sca-1+ (LSK) cells were isolated by FACS and cultured overnight in Stemspan media (MACS 130-091-680; Miltenyi Biotec) containing 1% l-glutamine, 2% P/S, 50 ng/ml rmTPO, rmSCF and rmFLT3L, and 10 ng/ml rmIL-3 and rmIL-6. LSK cells were transferred to virus-coated plates at 5 × 105 cells/ml in the same medium as above. Polybrene (5 µg/ml) was added to the media before spin infection at 1,000 rcf for 1 h at 32°C. LSK cells were subsequently plated at 20,000 cells/ml in mouse complete methylcellulose medium (HSC007; R&D) for 5 d. GFP+ cells were then isolated by FACS, and 120,000 GFP+ cells/mouse were retroorbitally transplanted into sublethally irradiated (650 rads) wt CD45.1 mice (B6.SJL-PtprcaPepcb/BoyJ; stock 002014; Jackson Laboratory). For secondary transplantation, GFP+c-Kit+ cells were isolated by FACS from BM of leukemic primary recipient mice, and 1,000 cells/mouse were transplanted into secondary recipient mice as described above. Chimerism analysis was performed on cells aspirated from the hind femurs of mice or on blood obtained from cheek bleeds using CD45.2 (47-0454-82; eBioscience) and CD45.1 (12-0453-82; eBioscience) antibodies after erythrocyte lysis.
IL1RAP expression in cells from MLL-AF9 mice was measured using an anti–mouse IL-1RAcP antibody provided by Amgen (M215), followed by a fluorescently conjugated anti–rat secondary antibody (eBioscience 17–4812). Immunophenotypes of MLL-AF9 cells used for secondary transplant were measured using the following antibodies: CD4 (13-0041-82; eBioscience), CD8a (13-0081-82; eBioscience), B220 (13-0452-82; eBioscience), CD19 (13-0193-82; eBioscience), Ter119 (13-5921-82; eBioscience), streptavidin-PE (12-4317-87; eBioscience), c-Kit (17-1178-42; eBioscience), Gr-1 (25-5931-85; eBioscience), CD11b (25-0112-82; eBioscience), and Sca-1 (108127; Biolegend).
For competitive transplants, BM was isolated from wt or Il1rap−/− CD45.2+ mice and wt CD45.1+ competitor mice, lysed with ACK buffer, and mixed in a 1:1 ratio. 2 × 106 cells were transplanted into each lethally irradiated (950 rads) congenic recipient mouse (CD45.1+). We monitored wt versus Il1rap−/− donor cell (CD45.2+) chimerism by sampling PB every 4 wk for 20 wk with antibodies against CD45.2 (EB: 47-0454-82; eBioscience) and CD45.1 (560580; BD PharMingen).
Mice were housed in Albert Einstein College of Medicine animal facilities. All experiments were performed in compliance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine (protocols 2013-1202 and 2016-1003).
Cytomorphology and histopathology
To assess cell morphology after antibody treatment, cells were spun onto glass slides (5 min at 600 rpm) using a StatSpin CytoFuge 2 (Beckman Coulter) and May-Grünwald Giemsa stained or stained with a modified Giemsa stain (Shandon Kwik-Diff Stains; Thermo Scientific). Femurs and liver sections from mice were fixed in 10% neutral buffered formalin, decalcified (for femurs), and paraffin embedded. Femurs and livers were sectioned and H&E–stained for histological analysis. Images were captured at room temperature on an EVOS FL Auto Imaging System with a color high-sensitivity CMOS, 1/2” 2,048 × 1,536, 3.1 megapixel camera (Life Technologies), exported as TIFF files, and images were processed in Adobe Photoshop.
Analysis of Il1rap−/− mice and Il1rap−/−FLT3ITD/ITD mice
PB flow cytometry analysis on Il1rap−/− mice was done using the following antibodies: CD3 (48-0031-82; eBioscience), B220 (11-0452-82; eBioscience), CD11b (17-0112-81; eBioscience), and Gr-1 (12-5931-82; eBioscience). The lineage cocktail for flow cytometry was made up of the following antibodies: CD4 (13-0041-82; eBioscience), CD8a (13-0081-82; eBioscience), B220 (13-0452-82; eBioscience), CD19 (13-0193-82; eBioscience), Ter119 (13-5921-82; eBioscience), Gr-1 (13-5931-85; eBioscience), and CD11b (13-0112-82; eBioscience). For flow cytometry analysis of HSPC, streptavidin-PE (12-4317-87; eBioscience), c-Kit (17-1178-42; eBioscience), and Sca-1 (108127; Biolegend) were used.
FLT3-ITD knock-in mice (B6.129-Flt3tm1Dgg/J; stock 011112) were obtained from Jackson Laboratory and backcrossed for over 10 generations to a C57BL/6J background. Mice were bred to Il1rap−/− mice to obtain Il1rap−/−FLT3ITD/ITD and Il1rap+/+FLT3ITD/ITD mice. Blood was isolated by cardiac puncture of euthanized mice, erythrocytes were lysed with ACK buffer, and cells were analyzed by flow cytometry using the following antibodies: B220 (47-0452-82; eBioscience), Gr-1 (12-5931-82; eBioscience), CD11b (17-0112-81; eBioscience), and c-Kit (11-1178; eBioscience). Isolation of cells from BM and spleens was performed as described above. For colony assays, total BM cells were plated at 103 cells/ml in mouse complete methylcellulose medium (HSC007; R&D), and colonies were scored after 14 d.
Western blotting and EdU incorporation experiments
For Western blotting, THP-1, TF-1, and HEL cells were plated at 106 cells/ml in RPMI 1640 serum-free medium with 1% P/S for antibody treatment and stimulations. shRNA-transduced cells were plated in RPMI 1640 serum-free medium for 2 h before stimulation with cytokines. For antibody treatment Western blots, cells were incubated with IL1RAP pAb and normal goat IgG or IL-1Ra for 24 h. After 24 h, 50 ng rhFLT3L or rhSCF was added and incubated with cells for 5 min at 37°C or 20 ng/ml rhIL-1β was added and incubated with cells for 15 min at 37°C.
For Western blotting and EdU incorporation experiments, primary AML MNCs were plated at 5 × 105 cells per ml in IMDM medium containing 2% FBS, 1% P/S, 50 ng/ml rhTPO, 50 ng/ml rhSCF (excluded for ± SCF stimulation), 50 ng/ml rhFLT3L (excluded for ± FLT3L stimulation), 10 ng/ml rhIL-3, and 25 ng/ml of rhIL-6. Cells were incubated with IL1RAP pAb or normal goat IgG and 50 ng/ml rhFLT3L or 50 ng/ml rhSCF for 24 h where indicated. EdU incorporation was measured after incubation of cells with 2 mM EdU for 24 h using the Click-IT EdU Flow Cytometry Assay kit according to the manufacturer’s instructions (C10424; Invitrogen).
For protein isolation, cells were washed out of medium with PBS containing 1 mM sodium orthovanadate (Na3VO4). Lysates were prepared using modified radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (50 mM Tris-HCl, pH 7.4, 1% NP-40 [vol/vol], 0.25% Na-deoxycholate [wt/vol], 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 ug/ml each aprotinin, leupeptin, pepstatin [Roche], 1 mM Na3VO4, 1 mM NaF, and 20 mM β-glycerophosphate). Cells were lysed for 30 min at 4°C and sonicated for 5 min (Diagenode Bioruptor). Protein concentration was measured on a spectrophotometer (read at 595 nm) using Bradford reagent (500-0006; Biorad). SDS-PAGE was performed and cell lysates were resolved on 8% polyacrylamide gels. The following antibodies were used for immunoblotting: Phospho-FLT3 (Y591; 3461; Cell Signaling Technology [CST]), FLT3 (C-20; sc-479; Santa Cruz), phospho–c-KIT (Y719; 3391; CST), phospho-AKT (S473; D9E XP; 4060; CST), phospho-ERK1/2 (T202/Y204; 9101; CST), IRAK1 (4359; CST), phospho-P38 (Y182; ab47363; Abcam), IκBα (9242; CST), β-tubulin (T8328; Sigma-Aldrich) and actin (A2066; Sigma-Aldrich). Primary antibodies were incubated overnight at 4°C in blocking solution (TBS containing 0.1% [vol/vol] Tween-20 with 5% [wt/vol] BSA). Secondary antibody anti–rabbit IgG HRP conjugated (sc-2004; Santa Cruz) or anti–mouse IgG HRP conjugated (sc-2005; Santa Cruz) was used for detection of primary antibodies.
Lineage depletion, cytokine stimulation, and phosphoflow cytometry of mouse BM cells
BM cells were labeled with lineage antibodies, and lineage-positive BM was depleted using magnetic anti–rat dynabeads (11035; Invitrogen) following the manufacturer’s instructions. The lineage cocktail for magnetic depletion was made up of the following antibodies: CD4 (13-0041-82; eBioscience), CD8a (13-0081-82; eBioscience), B220 (13-0452-82; eBioscience), CD19 (13-0193-82; eBioscience), Ter119 (13-5921-82; eBioscience), Gr-1 (13-5931-85; eBioscience), and CD11b (13-0112-82; eBioscience). The lineage-negative (Lineage−) BM was incubated for 1 h in serum free IMDM at 37°C before stimulation with recombinant mouse IL-1β (rmIL-1β; 100 ng/ml for 15 min), Flt3 ligand (rmFlt3L; 100 ng/ml for 5 min) or stem cell factor (rmSCF; 2.5 ng/ml for 5 min). After fixation and permeabilization with Cytofix solution (BD) and ice-cold methanol, intracellular staining for phospho-S6 ribosomal protein (Ser235/236; 4851; CST), and staining for c-Kit (11-1178-42; eBioscience) and Flt3 (12-1357-42; eBioscience) was performed for flow cytometry analysis.
Overexpression of IL1RAP, FLT3, and c-KIT
Human IL1RAP and c-KIT were PCR-amplified from cDNA from human AML cell lines (THP-1 and HEL, respectively) and cloned into pCMV-HA-N and pCMV-myc-N vectors (631604 and 635689; Clontech). Human FLT3 was digested from a MSCV-FLT3-neo vector and ligated into the pCMV-myc-N vector. Human FLT3-ITD was digested from an MSCV-FLT3-ITD-GFP vector and ligated into the pCMV-myc-N vector. 293T cells were transfected with FLT3, FLT3-ITD, c-KIT, and IL1RAP plasmids using FuGENE HD Transfection Reagent (E2311; Promega) according to the manufacturer’s guidelines.
Coimmunoprecipitation
Lysates from 293T cells or THP-1 cells were prepared using modified RIPA buffer containing protease and phosphatase inhibitors and ATP (50 mM Tris-HCl, pH 7.4, 1% NP-40 [vol/vol], 150 mM NaCl, 0.1% SDS [wt/vol], 1 mM PMSF, 1 ug/ml each aprotinin, leupeptin, pepstatin [Roche], 1 mM Na3VO4, 1 mM NaF, 20 mM β-glycerophosphate, and 10 mM ATP). Lysates were homogenized by passing through a 27 G needle. 1 mg of lysate was precleared for 1 h with 30 µl of sheep anti–mouse M-280 Dynabeads (11201D; Life Technologies) or sheep anti–rabbit IgG Dynabeads M-280 (11203D; Life Technologies). For endogenous coimmunoprecipitations, 50 µl of dynabeads were coupled to 5 µg of FLT3 rabbit IgG antibody (C-20; sc-479; Santa Cruz), c-KIT Rabbit IgG antibody (C-19; sc-168; Santa Cruz), c-KIT Mouse IgG (3308; CST), Normal Rabbit IgG antibody (sc-3888; Santa Cruz), or Mouse (G3A1) mAb IgG1 Isotype Control (5415; CST) for 1 h. 25 ul (precoupled) anti-HA magnetic beads (88836; Pierce) were used for HA coimmunoprecipitations. Antibody-bead complexes were washed in lysis buffer without SDS or ATP, added to 500 µg protein lysate, and rotated at 4°C overnight. Bound proteins were isolated by boiling antibody–bead complexes in SDS sample buffer. Immunoblotting was performed as described above using the following antibodies: human IL-1RAcP/IL-1R3 biotinylated antibody (BAF676; R&D), FLT3 rabbit IgG antibody (C-20; sc-479; Santa Cruz), c-KIT rabbit IgG antibody (C-19; sc-168; Santa Cruz) or c-KIT mouse IgG antibody (3308; CST). Secondary antibodies Pierce High Sensitivity Streptavidin-HRP (21134; Thermo Scientific), TrueBlot ULTRA anti–rabbit Ig HRP (18-8816-31; Rockland), and TrueBlot ULTRA anti–mouse Ig HRP (18-8817-30; Rockland) were used for detection of primary antibodies using autoradiography film.
Cytokine receptor expression and FRET
Fluorescent antibodies against surface receptors were incubated with cells for 30 min: anti–human HLA-A,B,C APC or PE conjugate (311410 and 311406; Biolegend), anti–human β2-microglobulin APC or PE (316312 and 316306; Biolegend), anti–human IL-1RAcP/IL-1R3 APC or PE (FAB676A and FAB676P; R&D), anti–human IL1RI APC or PE (FAB269A and FAB269P), anti–human CD135 (FLT3) APC or PE (17-1357 and 12-1357; eBioscience), and human CD117 (c-KIT) APC or PE (17-1178 and 12-1178; eBioscience). Mouse IgG1 APC (17-4714; eBioscience) or PE (IC002P; R&D) was used to set gates. For FRET experiments, cells were left unstimulated or stimulated with either 200 ng/ml rhIL-1β, 100 ng/ml rhFLT3L, or 100 ng/ml rhSCF for 3 min before fixation with PBS containing 0.1% formaldehyde. Cells were labeled with PE and APC antibodies at saturating concentration and analyzed with a BD FACSAria II Special Order System. Analysis of FRET was done on cells expressing both proteins of interest.
The following samples were used for the determination of FRET: Sample 1, unlabeled cells; Sample 2, cells with epitope A labeled with PE; Sample 3, cells with epitope A labeled with APC; Sample 4, cells with epitope B labeled with PE; Sample 5, cells with epitope B labeled with APC; Sample 6, cells with epitope A labeled with PE and epitope B labeled with APC; Sample 7, cells with epitope B labeled with PE and epitope A labeled with APC.
The MHC class I molecules comprising the heavy chain labeled with APC and the light chain (β2 microglobulin) labeled with PE were used as a positive control. The dye/protein molar ratio (r) of IL1RAP-PE, IL1RAP-APC, FLT3-PE, FLT3-APC, KIT-APC, and KIT-PE is 1:1. The molar absorption coefficient (ε) used for PE is 1,200,000 M-1cm-1 and APC is 5,000 M-1cm-1. Cytokine-stimulated or unstimulated cells were washed twice with ice-cold PBS. One million cells per sample were labeled with 5–50 µg/ml final concentration of antibodies in 50 µl total volume of PBS containing 0.1% BSA for 30 min on ice. Cells were washed twice with ice-cold PBS and fixed with 500 µl PBS containing 1% formaldehyde. Samples were stored at 4°C until measurement.
For flow cytometric determination of FRET the BD FACSAria was set according to the parameters in Table S2 and flow cytometric measurements were performed following published instructions (György Vereb and Szöllősi, 2011). FRET was calculated as described previously (Batard et al., 2002). In brief, the correction α factor was calculated with the following formula:
Fd,a(x,y) represents the MFI obtained in the absence (d = 0) or presence (d = 1) of a donor, in the absence (a = 0) or presence (a = 1) of an acceptor after excitation at x nm and detection at y nm. F0,0(488,575) represents the autofluorescence of cells (no donor and no acceptor, d = 0 and a = 0) when excited with the argon laser (488 nm) and detected at 575 nm. F1,0(488,575) represents the specific and optimal signal of cells labeled only with an antibody conjugated with PE (presence of donor and absence of acceptor, d = 1 and a = 0) when excited at 488 nm and detected at 575 nm. The α factor was obtained by analyzing three samples: cells alone, cells saturated with an antibody conjugated with PE, and cells saturated with the same antibody conjugated with APC.
The sensitized emission of APC was detected with excitation at 488 nm and emission at 675 nm. The sensitized emission was corrected for the direct contribution of PE. The S1 correction factor was calculated by using the following formula:
The sensitized emission was corrected for the direct contribution of APC. The S2 correction factor was calculated by using the following formula:
The APC emission was corrected for the direct contribution of PE. The S3 correction factor was calculated by using the following formula:
An intermediate value, A, was calculated by using the following formula:
FRET efficiency was obtained as follows:
Statistical analysis
Flow cytometry data were analyzed using FACSDiva software (v7; BD) or FlowJo (v10.0; Tree Star, Inc.). Western blot quantification was performed using ImageJ (v1.44o, National Institutes of Health; Schneider et al., 2012). P-values were calculated using unpaired or paired two-tailed t tests or log-rank tests for survival analysis. GraphPad Prism (version 7.0c for Mac; GraphPad Software) was used for statistical tests. Results represent the mean ± standard deviations of independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****; P < 0.0001.
Online supplemental material
Fig. S1 shows that additional IL1RAP antibodies reduce growth of AML cells by inducing differentiation and apoptosis. Fig. S2 shows the effects of shRNA targeting of IL1RAP in primary human AML cells, expression of IL1RAP in healthy cord blood HSPC, and the effects of IL1RAP deletion on the mouse hematopoietic system. Fig. S3 shows additional data from the in vivo experiments shown in Fig. 2. Fig. S4 shows that down-regulation and antibody targeting of IL1RAP inhibit FLT3L, SCF, and IL-1–mediated signaling. Table S1 shows absolute colony number output for IL1RAP shRNA-transduced primary AML samples in Fig. 1 F. Table S2 shows the flow cytometer settings used for determination of FRET in Figs. 4 (C and G).
Supplementary Material
Acknowledgments
We would like to thank all past and current members of the Steidl laboratory for their helpful suggestions and discussions and I. Kaur, R. Stanley, A. Pandolfi, I. Schulze, J. Wheat, D. Walter, M. Ferreira, T. Yatsenko, V. Thiruthuvanathan, and L. Benard for their guidance and assistance with experiments. We thank F. Aodengtuya, Y. Zhang, and J. Zhang from the AECOM Flow Cytometry Core Facility, D. Sun from the AECOM Stem Cell Isolation and Xenotransplantation Facility (funded through New York Stem Cell Science grant no. C029154), and P. Schultes from the AECOM Department of Cell Biology. We thank J. Ruthberg for help in preparing this manuscript and graphic design expertise.
This work was supported by National Institutes of Health grants R01CA166429 (to U. Steidl), R01CA196973 (to K. Gritsman), and U10CA180827 (to E. Paietta), as well as research grants of the Feldstein Medical Foundation and the Leukemia & Lymphoma Society to U. Steidl. K. Mitchell was partially supported by the Training Program in Cellular and Molecular Biology and Genetics (National Institutes of Health, T32 GM007491), and S.J. Taylor was supported by the Einstein Training Program in Stem Cell Research from the Empire State Stem Cell Fund (NYSTEM; New York State Department of Health, contract C30292GG). U. Steidl is the Diane and Arthur B. Belfer Scholar in Cancer Research of the Albert Einstein College of Medicine and is a Research Scholar of the Leukemia & Lymphoma Society.
The authors declare no competing financial interests.
Author contributions: K. Mitchell, L. Barreyro, S.J. Taylor, I. Antony-Debré, and U. Steidl designed the study and experiments; K. Mitchell, L. Barreyro, T.I. Todorova, S.J. Taylor, I. Antony-Debré, and S.-R. Narayanagari performed experiments; L.A. Carvajal and K. Gritsman provided guidance with experiments and study design; I. Mantzaris, G. Pendurti, E. Paietta, and A. Verma provided primary patient specimens and diagnostic information; J. Leite and Z. Piperdi provided technical assistance; K. Mitchell, L. Barreyro, S.J. Taylor, and I. Antony-Debré analyzed and interpreted data; K. Mitchell and U. Steidl wrote the manuscript.
References
- Ågerstam H., Karlsson C., Hansen N., Sandén C., Askmyr M., von Palffy S., Högberg C., Rissler M., Wunderlich M., Juliusson G., et al. 2015. Antibodies targeting human IL1RAP (IL1R3) show therapeutic effects in xenograft models of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 112:10786–10791. 10.1073/pnas.1422749112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ågerstam H., Hansen N., von Palffy S., Sandén C., Reckzeh K., Karlsson C., Lilljebjörn H., Landberg N., Askmyr M., Högberg C., et al. 2016. IL1RAP antibodies block IL-1-induced expansion of candidate CML stem cells and mediate cell killing in xenograft models. Blood. 128:2683–2693. 10.1182/blood-2015-11-679985 [DOI] [PubMed] [Google Scholar]
- Ali S., Huber M., Kollewe C., Bischoff S.C., Falk W., and Martin M.U.. 2007. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc. Natl. Acad. Sci. USA. 104:18660–18665. 10.1073/pnas.0705939104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askmyr M., Ågerstam H., Hansen N., Gordon S., Arvanitakis A., Rissler M., Juliusson G., Richter J., Järås M., and Fioretos T.. 2013. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 121:3709–3713. 10.1182/blood-2012-09-458935 [DOI] [PubMed] [Google Scholar]
- Barreyro L., Will B., Bartholdy B., Zhou L., Todorova T.I., Stanley R.F., Ben-Neriah S., Montagna C., Parekh S., Pellagatti A., et al. 2012. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 120:1290–1298. 10.1182/blood-2012-01-404699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batard P., Szollosi J., Luescher I., Cerottini J.C., MacDonald R., and Romero P.. 2002. Use of phycoerythrin and allophycocyanin for fluorescence resonance energy transfer analyzed by flow cytometry: advantages and limitations. Cytometry. 48:97–105. 10.1002/cyto.10106 [DOI] [PubMed] [Google Scholar]
- Baum C.M., Weissman I.L., Tsukamoto A.S., Buckle A.M., and Peault B.. 1992. Isolation of a candidate human hematopoietic stem-cell population. Proc. Natl. Acad. Sci. USA. 89:2804–2808. 10.1073/pnas.89.7.2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H., Dinh H., Trueblood E.S., Pretorius J., Kugler D., Weng N., Kanaly S.T., Towne J.E., Willis C.R., Kuechle M.K., et al. 2007. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 204:2603–2614. 10.1084/jem.20070157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., and Dick J.E.. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737. 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Brandts C.H., Sargin B., Rode M., Biermann C., Lindtner B., Schwäble J., Buerger H., Müller-Tidow C., Choudhary C., McMahon M., et al. 2005. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res. 65:9643–9650. 10.1158/0008-5472.CAN-05-0422 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Müller-Tidow C., Berdel W.E., and Serve H.. 2005. Signal transduction of oncogenic Flt3. Int. J. Hematol. 82:93–99. 10.1532/IJH97.05090 [DOI] [PubMed] [Google Scholar]
- Corces-Zimmerman M.R., Hong W.J., Weissman I.L., Medeiros B.C., and Majeti R.. 2014. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 111:2548–2553. 10.1073/pnas.1324297111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan E.B., Kwee L., Nunes P., Shuster D.J., Ju G., McIntyre K.W., Chizzonite R.A., and Labow M.A.. 1998. IL-1 receptor accessory protein is an essential component of the IL-1 receptor. J. Immunol. 161:5614–5620. [PubMed] [Google Scholar]
- Culver-Cochran A.E., and Starczynowski D.T.. 2018. Chronic innate immune signaling results in ubiquitination of splicing machinery. Cell Cycle. Apr 3:1–3. 10.1080/15384101.2018.1429082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drube S., Heink S., Walter S., Löhn T., Grusser M., Gerbaulet A., Berod L., Schons J., Dudeck A., Freitag J., et al. 2010. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 115:3899–3906. 10.1182/blood-2009-10-247411 [DOI] [PubMed] [Google Scholar]
- Dunne A., and O’Neill L.A.. 2003. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE. 2003:re3. [DOI] [PubMed] [Google Scholar]
- Fang J., Bolanos L.C., Choi K., Liu X., Christie S., Akunuru S., Kumar R., Wang D., Chen X., Greis K.D., et al. 2017. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 18:236–245. 10.1038/ni.3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster T. 1948. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 437:55–75. 10.1002/andp.19484370105 [DOI] [Google Scholar]
- Gañán-Gómez I., Wei Y., Starczynowski D.T., Colla S., Yang H., Cabrero-Calvo M., Bohannan Z.S., Verma A., Steidl U., and Garcia-Manero G.. 2015. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 29:1458–1469. 10.1038/leu.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J.M., Gucwa J.L., Esopi D., Gurel M., Haffner M.C., Vala M., Nelson W.G., Jones R.J., and Yegnasubramanian S.. 2013. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget. 4:715–728. 10.18632/oncotarget.990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S.A., Nunes P., Kwee L., Labow M., Chizzonite R.A., and Ju G.. 1995. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J. Biol. Chem. 270:13757–13765. 10.1074/jbc.270.23.13757 [DOI] [PubMed] [Google Scholar]
- György Vereb P.N., and Szöllősi J.. 2011. Flow Cytometry Protocols, Methods in Molecular Biology. In Flow Cytometric FRET Analysis of Protein Interaction. Hawley T., and Hawley R., editors. Humana Press, New York. 21 pp. [DOI] [PubMed] [Google Scholar]
- Hemmati S., Haque T., and Gritsman K.. 2017. Inflammatory Signaling Pathways in Preleukemic and Leukemic Stem Cells. Front. Oncol. 7:265 10.3389/fonc.2017.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H., Sadovnik I., Cerny-Reiterer S., Rülicke T., Stefanzl G., Willmann M., Hoermann G., Bilban M., Blatt K., Herndlhofer S., et al. 2014. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood. 123:3951–3962. 10.1182/blood-2013-10-536078 [DOI] [PubMed] [Google Scholar]
- Ho T.C., LaMere M., Stevens B.M., Ashton J.M., Myers J.R., O’Dwyer K.M., Liesveld J.L., Mendler J.H., Guzman M., Morrissette J.D., et al. 2016. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood. 128:1671–1678. 10.1182/blood-2016-02-695312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kanakura Y., Tamaki T., Kuriu A., Kitayama H., Ishikawa J., Kanayama Y., Yonezawa T., Tarui S., and Griffin J.D.. 1991. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 78:2962–2968. [PubMed] [Google Scholar]
- Jan M., and Majeti R.. 2013. Clonal evolution of acute leukemia genomes. Oncogene. 32:135–140. 10.1038/onc.2012.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Snyder T.M., Corces-Zimmerman M.R., Vyas P., Weissman I.L., Quake S.R., and Majeti R.. 2012. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4:149ra118 10.1126/scitranslmed.3004315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järås M., Johnels P., Hansen N., Agerstam H., Tsapogas P., Rissler M., Lassen C., Olofsson T., Bjerrum O.W., Richter J., and Fioretos T.. 2010. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc. Natl. Acad. Sci. USA. 107:16280–16285. 10.1073/pnas.1004408107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Liu B.Y., Huang J., Lu J., Sharmili R., Mishra M., Zhao X., Lin J., Hsi E.D., and Junutula J.R.. 2016. Proceedings of the 107th Annual Meeting of the American Association for Cancer Research. Abstr. 3337. [Google Scholar]
- Kalaitzidis D., and Neel B.G.. 2008. Flow-cytometric phosphoprotein analysis reveals agonist and temporal differences in responses of murine hematopoietic stem/progenitor cells. PLoS One. 3:e3776 10.1371/journal.pone.0003776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyoi H., Naoe T., Nakano Y., Yokota S., Minami S., Miyawaki S., Asou N., Kuriyama K., Jinnai I., Shimazaki C., et al. 1999. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 93:3074–3080. [PubMed] [Google Scholar]
- Kiyoi H., Yanada M., and Ozekia K.. 2005. Clinical significance of FLT3 in leukemia. Int. J. Hematol. 82:85–92. 10.1532/IJH97.05066 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822. 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Landberg N., Hansen N., Askmyr M., Ågerstam H., Lassen C., Rissler M., Hjorth-Hansen H., Mustjoki S., Järås M., Richter J., and Fioretos T.. 2016. IL1RAP expression as a measure of leukemic stem cell burden at diagnosis of chronic myeloid leukemia predicts therapy outcome. Leukemia. 30:255–258. 10.1038/leu.2015.135 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., and Dick J.E.. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648. 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Tothova Z., Levine R.L., Anderson K., Buza-Vidas N., Cullen D.E., McDowell E.P., Adelsperger J., Fröhling S., Huntly B.J., et al. 2007. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 12:367–380. 10.1016/j.ccr.2007.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.Y., Man C.H., and Kwong Y.L.. 2013. FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia. 27:260–268. 10.1038/leu.2012.195 [DOI] [PubMed] [Google Scholar]
- Liang K., Volk A.G., Haug J.S., Marshall S.A., Woodfin A.R., Bartom E.T., Gilmore J.M., Florens L., Washburn M.P., Sullivan K.D., et al. 2017. Therapeutic Targeting of MLL Degradation Pathways in MLL-Rearranged Leukemia. Cell. 168:59–72.e13. 10.1016/j.cell.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Seymour R.E., and Sundberg J.P.. 2011. Inhibition of NF-κB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J. Invest. Dermatol. 131:141–149. 10.1038/jid.2010.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A., Weiss T.M., Niebuhr M., Pan B., Appleton B.A., Wiesmann C., Bazan J.F., and Fairbrother W.J.. 2009. Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes. Structure. 17:1398–1410. 10.1016/j.str.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisovsky M., Estrov Z., Zhang X., Consoli U., Sanchez-Williams G., Snell V., Munker R., Goodacre A., Savchenko V., and Andreeff M.. 1996. Flt3 ligand stimulates proliferation and inhibits apoptosis of acute myeloid leukemia cells: regulation of Bcl-2 and Bax. Blood. 88:3987–3997. [PubMed] [Google Scholar]
- McKeown M.R., Corces M.R., Eaton M.L., Fiore C., Lee E., Lopez J.T., Chen M.W., Smith D., Chan S.M., Koenig J.L., et al. 2017. Superenhancer Analysis Defines Novel Epigenomic Subtypes of Non-APL AML, Including an RARα Dependency Targetable by SY-1425, a Potent and Selective RARα Agonist. Cancer Discov. 7:1136–1153. 10.1158/2159-8290.CD-17-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A. 2000. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE. 2000:re1 10.1126/stke.2000.44.re1 [DOI] [PubMed] [Google Scholar]
- O’Neill L.A. 2008. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 226:10–18. 10.1111/j.1600-065X.2008.00701.x [DOI] [PubMed] [Google Scholar]
- Pandolfi A., Barreyro L., and Steidl U.. 2013. Concise review: preleukemic stem cells: molecular biology and clinical implications of the precursors to leukemia stem cells. Stem Cells Transl. Med. 2:143–150. 10.5966/sctm.2012-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. 2016. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 374:2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P., Du J., Schlenk R.F., Gaidzik V.I., Bullinger L., Corbacioglu A., Späth D., Kayser S., Schlegelberger B., Krauter J., et al. 2013. Secondary genetic lesions in acute myeloid leukemia with inv(16) or t(16;16): a study of the German-Austrian AML Study Group (AMLSG). Blood. 121:170–177. 10.1182/blood-2012-05-431486 [DOI] [PubMed] [Google Scholar]
- Passegué E., Jamieson C.H., Ailles L.E., and Weissman I.L.. 2003. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Natl. Acad. Sci. USA. 100(Suppl 1):11842–11849. 10.1073/pnas.2034201100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin A.L., Mumm J.B., Murphy E., Turner S., Yu N., McClanahan T.K., Bourne P.A., Pierce R.H., Kastelein R., and Pflanz S.. 2010. IL-33 induces IL-13-dependent cutaneous fibrosis. J. Immunol. 184:1526–1535. 10.4049/jimmunol.0903306 [DOI] [PubMed] [Google Scholar]
- Reilly J.T. 2002. Class III receptor tyrosine kinases: role in leukaemogenesis. Br. J. Haematol. 116:744–757. 10.1046/j.0007-1048.2001.03294.x [DOI] [PubMed] [Google Scholar]
- Rhyasen G.W., and Starczynowski D.T.. 2015. IRAK signalling in cancer. Br. J. Cancer. 112:232–237. 10.1038/bjc.2014.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyasen G.W., Bolanos L., Fang J., Jerez A., Wunderlich M., Rigolino C., Mathews L., Ferrer M., Southall N., Guha R., et al. 2013. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 24:90–104. 10.1016/j.ccr.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovnik I., Herrmann H., Eisenwort G., Blatt K., Hoermann G., Mueller N., Sperr W.R., and Valent P.. 2017. Expression of CD25 on leukemic stem cells in BCR-ABL1+ CML: Potential diagnostic value and functional implications. Exp. Hematol. 51:17–24. 10.1016/j.exphem.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kitamura H., Hijikata A., Tomizawa-Murasawa M., Tanaka S., Takagi S., Uchida N., Suzuki N., Sone A., Najima Y., et al. 2010. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci. Transl. Med. 2:17ra9 10.1126/scitranslmed.3000349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarry J.E., Murphy K., Perry R., Sanchez P.V., Secreto A., Keefer C., Swider C.R., Strzelecki A.C., Cavelier C., Récher C., et al. 2011. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rγc-deficient mice. J. Clin. Invest. 121:384–395. 10.1172/JCI41495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheijen B., and Griffin J.D.. 2002. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene. 21:3314–3333. 10.1038/sj.onc.1205317 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri A., Will B., Steidl U., and Verma A.. 2017. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 129:1586–1594. 10.1182/blood-2016-10-696062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. HALT Pan-Leukemia Gene Panel Consortium . 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506:328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirewalt D.L., and Radich J.P.. 2003. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer. 3:650–665. 10.1038/nrc1169 [DOI] [PubMed] [Google Scholar]
- Towne J.E., Garka K.E., Renshaw B.R., Virca G.D., and Sims J.E.. 2004. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 279:13677–13688. 10.1074/jbc.M400117200 [DOI] [PubMed] [Google Scholar]
- Warfvinge R., Geironson L., Sommarin M.N.E., Lang S., Karlsson C., Roschupkina T., Stenke L., Stentoft J., Olsson-Strömberg U., Hjorth-Hansen H., et al. 2017. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 129:2384–2394. 10.1182/blood-2016-07-728873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr M.R., Pietras E.M., and Passegué E.. 2011. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip. Rev. Syst. Biol. Med. 3:681–701. 10.1002/wsbm.145 [DOI] [PubMed] [Google Scholar]
- Weber A., Wasiliew P., and Kracht M.. 2010. Interleukin-1 (IL-1) pathway. Sci. Signal. 3:cm1. [DOI] [PubMed] [Google Scholar]
- Yasuda T., Ueno T., Fukumura K., Yamato A., Ando M., Yamaguchi H., Soda M., Kawazu M., Sai E., Yamashita Y., et al. 2014. Leukemic evolution of donor-derived cells harboring IDH2 and DNMT3A mutations after allogeneic stem cell transplantation. Leukemia. 28:426–428. 10.1038/leu.2013.278 [DOI] [PubMed] [Google Scholar]
- Zhang B., Chu S., Agarwal P., Campbell V.L., Hopcroft L., Jørgensen H.G., Lin A., Gaal K., Holyoake T.L., and Bhatia R.. 2016. Inhibition of interleukin-1 signaling enhances elimination of tyrosine kinase inhibitor-treated CML stem cells. Blood. 128:2671–2682. 10.1182/blood-2015-11-679928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Yin L.L., Zhao D.M., Pan B., Chen W., Cao J., Cheng H., Li Z.Y., Li D.P., Sang W., et al. 2014. IL1RAP as a surface marker for leukemia stem cells is related to clinical phase of chronic myeloid leukemia patients. Int. J. Clin. Exp. Med. 7:4787–4798. [PMC free article] [PubMed] [Google Scholar]
- Zheng R., Bailey E., Nguyen B., Yang X., Piloto O., Levis M., and Small D.. 2011. Further activation of FLT3 mutants by FLT3 ligand. Oncogene. 30:4004–4014. 10.1038/onc.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.