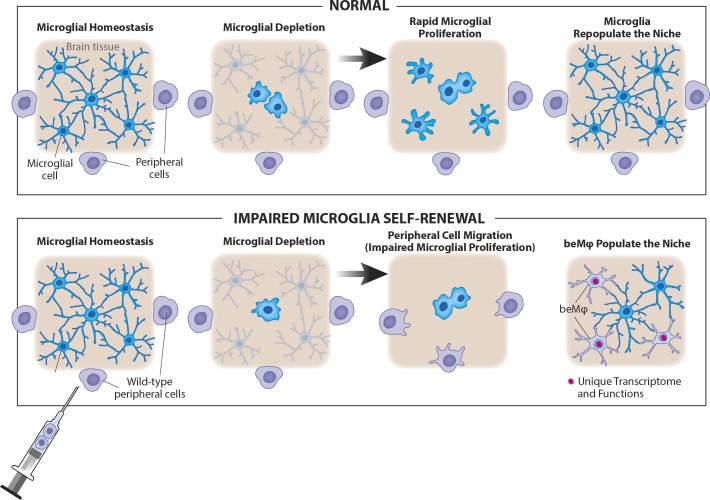
Peripherally derived macrophages can engraft the brain in the context of chronic microglia loss without brain irradiation and maintain a unique transcriptional signature throughout different experimental contexts.
Abstract
Peripherally derived macrophages infiltrate the brain after bone marrow transplantation and during central nervous system (CNS) inflammation. It was initially suggested that these engrafting cells were newly derived microglia and that irradiation was essential for engraftment to occur. However, it remains unclear whether brain-engrafting macrophages (beMφs) acquire a unique phenotype in the brain, whether long-term engraftment may occur without irradiation, and whether brain function is affected by the engrafted cells. In this study, we demonstrate that chronic, partial microglia depletion is sufficient for beMφs to populate the niche and that the presence of beMφs does not alter behavior. Furthermore, beMφs maintain a unique functional and transcriptional identity as compared with microglia. Overall, this study establishes beMφs as a unique CNS cell type and demonstrates that therapeutic engraftment of beMφs may be possible with irradiation-free conditioning regimens.
Graphical Abstract
Introduction
Although most tissue resident macrophage populations are initially populated by primordial yolk sac–derived macrophages (Alliot et al., 1999; Ginhoux and Merad, 2011), some are replaced with cells derived from fetal monocytes or hematopoietic stem cells, leading to either complete replacement of yolk sack–derived macrophages or mixed populations that are dominated by cells of the fetal monocyte or hematopoietic stem cell lineage (Epelman et al., 2014; Hoeffel et al., 2015; Sheng et al., 2015). Microglia, however, are a notable exception to this rule, and under homeostatic conditions, they self-renew from the original yolk sac lineage throughout the life of the animal (Ajami et al., 2007; Elmore et al., 2014; Epelman et al., 2014; Bruttger et al., 2015; Hoeffel et al., 2015; Sheng et al., 2015).
Monocytes do not enter the healthy brain but are seen within the brain parenchyma under certain pathological conditions, where their contribution to central nervous system (CNS) pathology is highly debated (Butovsky et al., 2012; Jung and Schwartz, 2012; Chiu et al., 2013; Prinz and Priller, 2014; Yamasaki et al., 2014; Jay et al., 2015; Thériault et al., 2015; Wang et al., 2016). Further, the conditions required for macrophage engraftment into the CNS parenchyma are not well understood. However, hematopoietic cells readily engraft the brain after lethal whole-body irradiation and bone marrow transplantation (BMT), often assumed to be secondary to blood–brain barrier (BBB) opening after irradiation (Priller et al., 2001; Mildner et al., 2007).
These peripherally derived brain-engrafting macrophages (beMφs) were initially noted to spatially replace microglia, tiling with resident microglia, and to develop ramifications similar to those of microglia (Priller et al., 2001; Mildner et al., 2007). These findings led to the hypothesis that hematopoietic-derived macrophages are capable of differentiating into true microglia upon engraftment. Indeed, several groups have found that the tissue environment directs macrophage differentiation, transcriptomes, and function (Lavin et al., 2014; Gibbings et al., 2015; Beattie et al., 2016; Scott et al., 2016; van de Laar et al., 2016), supporting the concept that the brain environment may be sufficient to drive differentiation of peripheral-derived microglia.
In the context of clinical implications, it has been shown that macrophage engraftment after BMT is beneficial in lysosomal storage disease (Walkley et al., 1994; Krivit et al., 1995, 1999; Platt and Lachmann, 2009), a mouse model of obsessive-compulsive disorder (Chen et al., 2010), and some mouse models of neurodevelopmental disorders (Derecki et al., 2012; Hsiao et al., 2012). However, the role of beMφs in other pathologies, such as CNS injury, Alzheimer’s disease, and amyotrophic lateral sclerosis, remains a topic of debate (Prinz and Priller, 2014).
Major questions about beMφs remain. What are the conditions necessary for engraftment into the brain, and is irradiation required for engraftment? Do beMφs alter brain function? Do beMφs become true microglia, or are they an independent class of resident brain macrophages that exists under defined conditions?
Results
Partial microglia depletion leads to beMφ engraftment independent of irradiation, without affecting behavior
We first set out to test the hypothesis that creation of a brain niche by chronic microglia depletion is sufficient to drive beMφ engraftment, independent of irradiation. To deplete microglia chronically, we used Cx3cr1CreER/+::Csf1rFlox/Flox mice fed tamoxifen chow to chronically excise Csf1r (the gene encoding colony-stimulating factor 1 receptor, which is critical for microglia survival) from cells expressing Cx3cr1, which in the brain is restricted to microglia (Li et al., 2006; Goldmann et al., 2013; Yona et al., 2013).
Before analyzing the brain, we assessed the peripheral immune system, which contains many cells that express Cx3cr1. Cx3cr1CreER/+::Csf1rFlox/Flox mice treated with tamoxifen demonstrated chronic deficiency in Cx3cr1+ lamina propria intestinal macrophages (Fig. S1, A–C). Although Cx3cr1lo/negLy6Chi monocytes did not experience significant changes upon tamoxifen treatment, Cx3cr1hiLy6Clo monocytes were severely depleted in these mice (Fig. S1, D–F), consistent with expression of Cre driven by the Cx3cr1 promoter.
We then moved on to analyze the brain. Although no differences were observed in microglia counts between Cx3cr1CreER/+::Csf1rFlox/Flox and Cx3cr1+/+::Csf1rFlox/Flox animals fed control chow (Fig. S2 A), 1 wk on a tamoxifen diet induced an ∼25% reduction of microglia throughout the brain, which was maintained for the duration of tamoxifen treatment (Fig. 1, A and B). When Csf1r levels were analyzed after 12 wk on a tamoxifen diet, both protein and gene expression were reduced, but not completely eliminated, as compared with Cre-negative controls (Fig. 1 C and Fig. S2, B and C), consistent with partial microglia depletion. To help explain why microglia were only partially depleted, we immunostained for Ki67 and assessed the incorporation of BrdU. We found that microglia were Ki67+ and BrdU+ in Cx3cr1CreER/+::Csf1rFlox/Flox mice, but not Cx3cr1+/+::Csf1rFlox/Flox mice, fed tamoxifen chow (Fig. S2, D and E). These data demonstrate that the microglia niche begins to proliferate in response to inducible Csf1r excision, suggesting that microglia loss is countered by proliferation of the remaining Csf1r-expressing microglia, or that signaling through another growth factor receptor is capable of driving microglia proliferation.
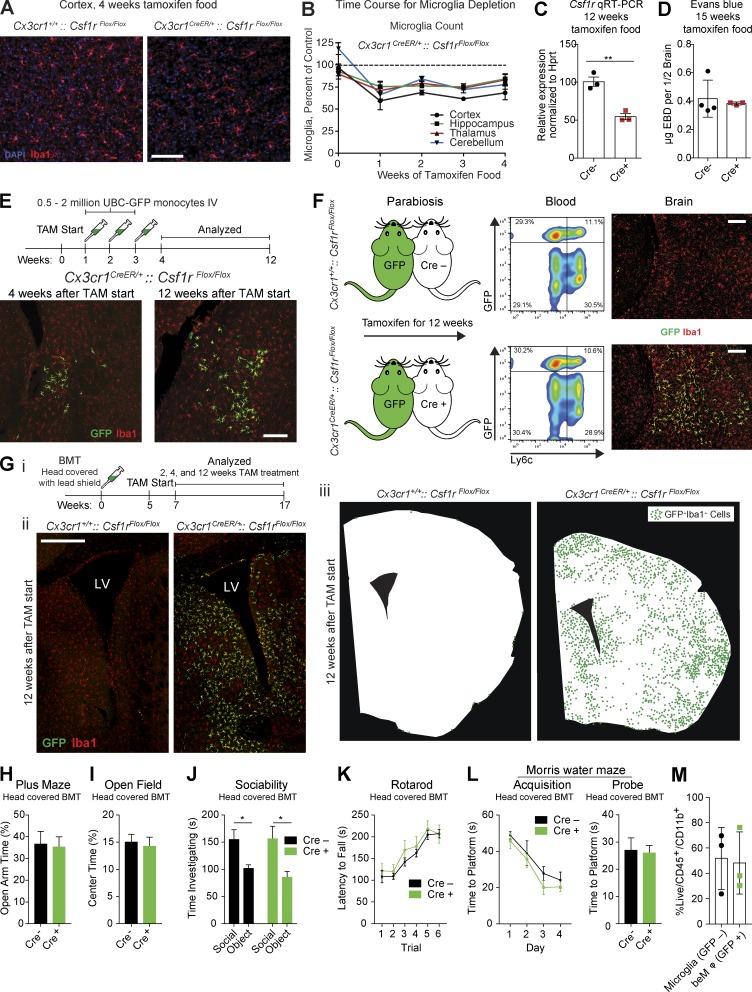
Figure 1.
Partial microglia depletion leads to beMφ engraftment independent of irradiation. (A) Representative images of Iba1+ microglia (red) after 4 wk of tamoxifen treatment in Cx3cr1CreER/+::Csf1rFlox/Flox mice and Cre-negative controls. Nuclei (DAPI) are shown in blue (n = 3 Cre− and 4 Cre+ mice). Bar, 100 µm. (B) Deleting Csf1r from microglia results in ∼25% chronic reduction of microglia throughout the brain (n = 3–4 mice per group for each time point; representative of two experiments). (C) Gene expression by quantitative RT-PCR of CD115/Csf1r on sorted microglia (n = 3 per group; two-tailed Student’s t test, **, P < 0.01; performed once). (D) Quantification of Evans blue dye in brains of Cx3cr1+/+::Csf1rflox/flox and Cx3cr1CreER/+::Csf1rflox/flox mice fed tamoxifen diet for 15 wk (n = 4 and 3 mice per group, two-tailed Student’s t test, not significant; performed once). (E) Top: Injection strategy for GFP+ monocytes. Bottom: Representative images of GFP+ beMφs (green) infiltrating adjacent to a lateral ventricle 1 and 9 wk after the last monocyte injection. All brain macrophages (resident microglia and beMφs) are positive for Iba1 (red). Images are representative of n = 3 mice (representative of two experiments). Bar, 200 µm. (F) Left: Cartoon of parabiotic pairings. UBC-GFP mice were paired to Cx3cr1CreER/+::Csf1rFlox/Flox mice and Cre-negative controls. After 12 wk of tamoxifen treatment, blood was analyzed by flow cytometry (middle), and brains were analyzed by immunohistochemistry (right). Although the percentage of GFP+ cells in the blood was similar between Cx3cr1CreER/+::Csf1rFlox/Flox mice and Cre-negative controls, GFP+ beMφs (green) were only found in Cre-positive mice. All brain macrophages, including microglia, were Iba1 positive (red). Images/data are representative of n = 3 mice per group. Bars, 200 µm. (G) Strategy to assess engraftment of beMφs after BMT using lead to shield the head (i). Representative images of beMφs (green) after 12 wk on tamoxifen (ii). All macrophages, including microglia, are Iba1+ (red), whereas beMφs are also GFP+ (green). Bar, 500 µm. LV, lateral ventricle. Illustrations of beMφ engraftment after 12 wk on tamoxifen (iii). Silhouettes of brain sections were generated on actual brain slices and beMφs locations were marked with a green dot. Each dot represents a single GFP+Iba1+ beMφ. Images are representative of n = 3–6 mice per group (representative of two independent experiments). (H–L) No differences in behavior were observed in mice containing beMφs (Cre+). Cx3cr1CreER/+::Csf1rFlox/Flox mice and Cre-negative controls underwent BMT with head shielding. After recovery, mice were treated with tamoxifen for 12 wk and then placed back on a regular diet for 4 wk before behavioral testing. Mice were tested on the plus maze (H; not significant, two-tailed Student’s t test; n = 24, 23; pooled data from two independent cohorts), open field (I; not significant, two-tailed Student’s t test; n = 24, 23; pooled data from two independent cohorts), three-chamber social assay (J; not significant for genotype and *, P < 0.05 for social variable, two-way repeated measures ANOVA with Sidak’s post hoc; n = 15; pooled data from two independent experiments), rotarod (K; not significant for genotype, two-way repeated measures ANOVA; n = 9, 8; experiment performed once), and water maze (L; not significant for acquisition, two-way repeated measures ANOVA and not significant for probe trial, two-tailed Student’s t test; n = 9, 8; performed once). (M) Quantification of brain macrophages from mice in behavior assays by flow cytometry. Mice were analyzed after behavior assays were complete, at least 8 wk after they had been placed back on regular diet. Brains of Cre-positive mice contained 48.2% ± 14.2 SEM beMφs (GFP+) out of total CD45/CD11b+ cells (not significant, two-tailed Student’s t test; n = 3 samples per cell type with 3–4 mice pooled per sample; performed once). Error bars represent ±SEM.
Together, these data demonstrated that inducible deletion of Csf1r in microglia leads to chronic, partial microglia loss, leaving a partially unfilled niche. We could therefore use this model to test whether the presence of a niche (formed without irradiation) that cannot be filled by microglia, could allow beMφ engraftment. Importantly, we did not detect increased BBB permeability in tamoxifen-treated Cre-positive as compared with Cre-negative mice (Fig. 1 D). We first injected ∼95% pure Ly6ChiCD115+ sorted bone marrow monocytes expressing GFP (Fig. S2 F) intravenously into Cx3cr1CreER/+::Csf1rFlox/Flox mice on a tamoxifen diet. Starting 1 wk after tamoxifen, mice were given three weekly intravenous injections of GFP+ monocytes. Mice were analyzed 1 or 9 wk after the final monocyte transfer (4 or 12 wk after tamoxifen initiation). The brains of Cre-positive animals contained ramified GFP+ beMφs (Fig. 1 E), whereas none were observed in brains of Cre-negative mice (Fig. S2 G). These results supported the hypothesis that in the context of chronic microglia deficiency (and without CNS irradiation, infection, or BBB compromise), circulating monocytes can engraft into the CNS and persistently fill the available niche created by depleted microglia, as cells were still present 9 wk after the final cell transfer.
It is possible that sorted bone marrow monocytes possess a unique ability to engraft the CNS that is not possessed by circulating blood monocytes. To confirm that circulating cells are capable of engrafting the CNS, we used parabiotic mice. Cre-negative or Cre-positive mice were parabiotically joined to UBC-GFP mice and placed on a tamoxifen diet for 12 wk. As expected, Cre-negative mice had no detectable GFP+ cells in their brains, whereas Cre-positive mice contained GFP+ ramified beMφs in the parenchyma (Fig. 1 F). Because the bone marrow niche remains undisturbed in parabiosis, these results provide evidence that circulating monocytes (or, at a minimum, circulating leukocytes) indeed possess the ability to engraft the brain and become beMφs when the microglia-vacant niche is present.
Although monocyte transfer and parabiosis demonstrated that beMφs can and will engraft the microglia-depleted brain without irradiation, these experiments did not reveal the potential extent of beMφ engraftment. To understand the full extent of peripheral-derived engraftment into the CNS in tamoxifen-treated Cx3cr1CreER/+::Csf1rFlox/Flox mice, we decided to completely replace the peripheral immune system with GFP-expressing cells. To this end, we performed BMT with lead shielding of the head, a well-established technique to prevent beMφ engraftment after BMT (Butovsky et al., 2006, 2007; Rolls et al., 2008; Shechter et al., 2009; Derecki et al., 2012). 5 wk after BMT, nearly every circulating immune cell (except T cells, which is a known phenomenon; Bosco et al., 2010) was GFP+, including nearly 100% of monocytes (Fig. S2, H–M).
We then assessed beMφ engraftment at 2, 4, or 12 wk after tamoxifen. We could only detect rare GFP+Iba1+ ramified beMφs after 2 wk, primarily in circumventricular regions (Fig. S3, A–C), but after 4 wk, the circumventricular regions of these mice had considerable engraftment of beMφs (Fig. S3, A–D). Interestingly, we observed that although GFP+ cells could be found in the choroid plexus and ventricle walls of all mice, ramified GFP+Iba1+ beMφs were only present in the brain parenchyma in Cx3cr1CreER/+::Csf1rFlox/Flox mice (Fig. S3 D). After 12 wk, we found GFP+Iba1+ macrophage engraftment throughout the brains of Cx3cr1CreER/+::Csf1rFlox/Flox animals, as opposed to their Cre-negative counterparts, in which no ramified GFP+ cells were observed anywhere in the brain parenchyma (Figs. 1 G and S3 C). Furthermore, if we irradiated the entire mouse (including the head) beMφ engraftment into Cx3cr1CreER/+::Csf1rFlox/Flox mice was greatly accelerated compared with Cre-negative controls (Fig. S3, E and F). This suggests that although CNS irradiation induces beMφ engraftment, this is substantially enhanced by impairment of microglia self-renewal by deletion of Csf1r.
We next set out to interrogate whether beMφ affect brain function using a battery of behavioral assays. Head-covered BMT was performed in Cx3cr1CreER/+::Csf1rFlox/Flox and Cx3cr1+/+::Csf1rFlox/Flox mice as described above, and mice were fed a tamoxifen diet for 12 wk, followed by 4 wk of control chow to eliminate the potential effects of tamoxifen on behavior. We found no difference in any behavioral assays tested, such as elevated plus maze (Fig. 1 H), open field (Fig. 1 I), sociability (Fig. 1 J), rotarod (Fig. 1 K), or Morris water maze (Fig. 1 L). Importantly, we found that Cre-positive mice had a mean of 48% GFP+ beMφs out of all CNS macrophages/microglia at the conclusion of behavioral testing, at which point mice had been on control chow for a minimum of 8 wk (Fig. 1 M). Together, these results suggest that even with significant microglia replacement by beMφs, overall brain function is not affected.
beMφs are transcriptionally distinct from microglia
We next set out to test the hypothesis that beMφs are an independent class of brain macrophage that maintains a unique functional and transcriptomic profile. To this end, we decided to perform RNA sequencing on beMφ and microglia in three unique models of beMφ engraftment. First, we sorted live CD45+CD11b+ and either GFP-negative (microglia) or GFP-positive (beMφ) cells from the same brains of tamoxifen-treated Cx3cr1CreER/+::Csf1rFlox/Flox mice. Microglia and beMφ clustered strongly by cell type, with cell type accounting for 96% of the transcriptional variance (Fig. 2 A). Compared with microglia, beMφs had 1,512 differentially up-regulated and 1,598 differentially down-regulated genes (Fig. 2 B and Table S1).
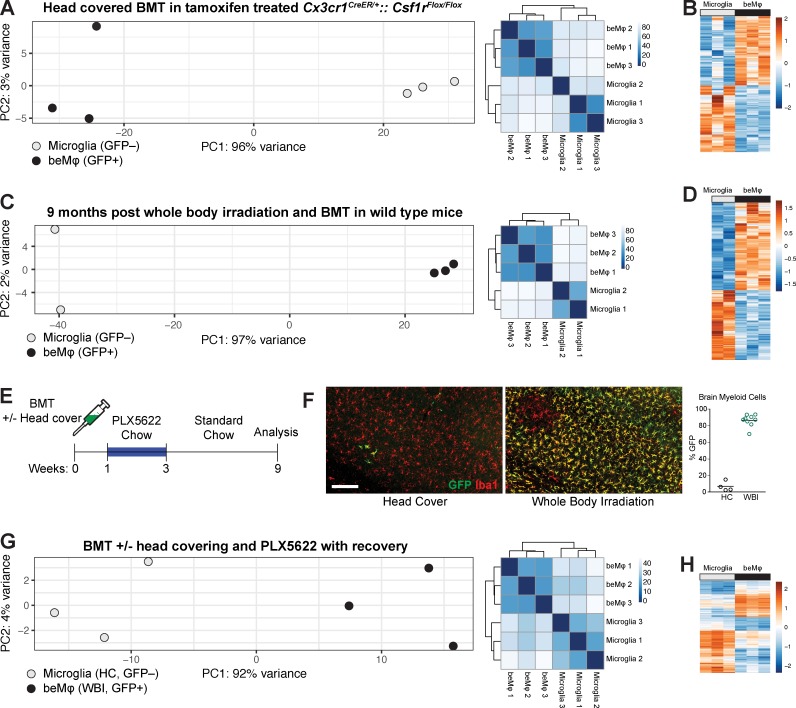
Figure 2.
beMφs are a transcriptionally distinct cell type. (A) PCA plot and heatmap of distance between samples for beMφs and microglia in the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment. Mice were treated with tamoxifen for 12 wk, followed by a minimum of 8 wk on control chow (each dot represents a pooled sample from at least three mice). (B) Differentially expressed genes (adjusted P < 0.05) between beMφs and microglia in the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment. The values are standardized rlog-transformed values across samples. (C) PCA plot and heatmap of distance between samples for the beMφs and microglia when using traditional BMT (each dot represents a pooled sample from at least three mice). (D) Differentially expressed genes (adjusted P < 0.05) between beMφs and microglia when using traditional BMT. The values are standardized rlog-transformed values across samples. (E) Strategy for achieving beMφ engraftment using BMT with or without head covering and PLX5622 treatment. (F) Representative images and quantification of GFP+ beMφs in mice treated as in E. Bar, 200 µm. HC, head covered; WBI, whole-body irradiation. (G) PCA plot and heatmap of distance between samples for beMφs and microglia when using BMT/PLX5622 (each dot represents a pooled sample from at least three mice). (H) Differentially expressed genes (adjusted P < 0.05) between beMφs and microglia when using BMT/PLX5622. The values are standardized rlog-transformed values across samples.
In our second model of beMφ engraftment, we replicated classic beMφ engraftment models by performing whole-body irradiation and BMT with UBC-GFP bone marrow. We then allowed cells to engraft and remain for 9 mo and sorted beMφs and microglia from the same brains. Again, we found that cell type accounted for the large majority of variance (97%; Fig. 2 C), and when compared with microglia, beMφs had a large number of differentially up-regulated (2,008) and differentially down-regulated genes (1,596; Fig. 2 D and Table S1).
Our third model was designed to take advantage of the synergistic effect on engraftment we had observed with the combination of CNS irradiation and Csf1r deletion in microglia (Fig. S3, E and F). Instead of Csf1r deletion using our genetic model, we used the Csf1r inhibitor PLX5622 in chow to eliminate the irradiation-damaged, yet still viable, microglia. Mice were given BMT with UBC-GFP bone marrow with or without head covering during irradiation, followed by 1 wk of recovery, 2 wk of chow containing PLX5622, and finally 6 wk of recovery on standard chow (Fig. 2 E). In this model, mice with head covering had minimal GFP+ beMφ engraftment and had effectively repopulated the CNS with endogenous microglia with minimal beMφ engraftment (Fig. 2 F), consistent with previously published data (Elmore et al., 2014). However, mice that had received whole-body irradiation demonstrated robust engraftment of GFP+ beMφ (Fig. 2 F), mimicking the synergistic effects of inducible deletion of Csf1r and whole-body irradiation with BMT in our previous experiments. Because of the robust level of engraftment in this model, we sorted GFP+ beMφs from whole-body–irradiated mice and GFP− microglia from head-covered mice for RNA sequencing. We again found that cell type accounted for the large majority of transcriptional variance (92%; Fig. 2 G) and that when compared with microglia, beMφs had 829 differentially up-regulated and 1,275 differentially down-regulated genes (Fig. 2 H and Table S1).
beMφs are functionally distinct from microglia
To assess beMφ function in each of the three engraftment models, we used gene set variation analysis (GSVA; Hänzelmann et al., 2013) to identify commonly enriched Gene Ontology Biological Process (GOBP) terms in beMφ versus microglia, and vice versa. There were 117 GOBP terms commonly enriched in all three beMφ datasets and 47 GOBP terms commonly enriched in all three microglia datasets (Table S2). Within these commonly enriched functional terms, several notable functions and themes were apparent. Consistently, beMφs were enriched for “regulation of wound healing, spreading of epidermal cells,” as well as multiple functions for extracellular uptake, growth factor signaling and production, extracellular matrix interaction and migration, regulation of vasculogenesis, interactions with neurons and glia, lipid metabolism, and a large number of immunological processes (Fig. 3 A and Table S2). Microglia also demonstrated common functions for interaction with neurons and glia and lipid metabolism, but also notably had multiple functions related to neurotransmitters and steroids (Fig. 3 B and Table S2). These results suggest that although beMφs and microglia may have some similar functional themes, they are not functionally interchangeable cell types, supporting the hypothesis that these are two distinct populations of CNS macrophages.
Figure 3.
beMφs and microglia maintain unique predicted functions in multiple experimental models. (A) Selected beMφ Gene Ontology biological functions that are identified as enriched by GSVA in the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment, traditional BMT, and BMT/PLX5622 when compared with the microglia in the same experiment. The boxplots show the distribution of the −log10(FDR-adjusted p-value) of the corresponding functional term calculated for each of the three experiments. A complete list of functions commonly up-regulated in beMφs versus microglia in all datasets can be found in Table S2. (B) Selected microglia Gene Ontology biological functions that are identified as enriched by GSVA in the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment, traditional BMT, and BMT/PLX5622 when compared with beMφs in the same experiment. Boxplots show the distribution with mean of the −log10(FDR-adjusted p-value) of the corresponding functional term calculated for each of the three experiments. A complete list of functions commonly up-regulated in microglia versus beMφs in all datasets can be found in Table S2.
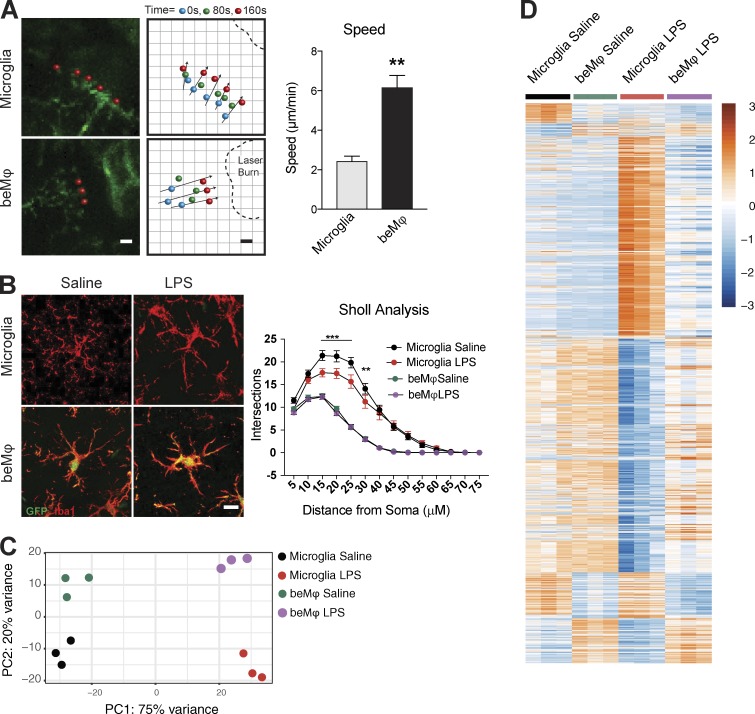
Although our RNA-sequencing analysis suggested that microglia and beMφs are unique cell types, we decided to experimentally confirm differential response to stimuli. Using our system of BMT and PLX5622 treatment for beMφ engraftment, which allows substantial repopulation of microglia by beMφs, we first tested response to laser burn injury in live multiphoton imaging. Our results demonstrate that beMφs moved toward the laser burn faster than microglia (Fig. 4 A). We then assessed response to in vivo LPS injection. Mice were given i.p. LPS or saline injection, and the brains were collected for analysis 6 h later. Sholl analysis of Iba1 staining revealed that beMφs were less ramified than microglia and demonstrated no change in ramification after LPS, whereas microglia were significantly more ramified and demonstrated significant reduction in ramification after LPS (Fig. 4 B). Principal component analysis (PCA) of the corresponding RNA-sequencing dataset demonstrated distinct clustering of samples by cell type and treatment (Fig. 4 C). Differential expression (DE) analysis revealed a very large number of differentially expressed genes after LPS treatment (3,627 up-regulated and 4,910 down-regulated genes in beMφs as compared with microglia; Fig. 4 D and Table S1).
Figure 4.
beMφs have distinct morphology and response to stimuli compared with microglia. (A) Representative images from two-photon in vivo imaging of microglia and beMφs responding to laser injury. Bar, 5 µm. Dots represent movement of processes over time. beMφ processes move more rapidly toward the injury site (Student’s t test, **, P < 0.01; n = 3 mice; representative of two independent experiments). (B) Representative images of microglia and beMφs in response to LPS. All brain macrophages are Iba1+. Bar, 10 µm. Sholl analysis of microglia and beMφs 6 h after LPS injection (i.p.). beMφs are less complex than microglia and do not change complexity after LPS (two-way ANOVA P < 0.0001 for an interaction between type of macrophages and branching over distance; ***, P < 0.0001; **, P < 0.001; n = 60 microglia from three different mice per group; performed once). Error bars represent ±SEM. (C) PCA plots of RNA-sequencing transcriptional data from saline or LPS treated microglia and beMφs. (D) Differentially expressed genes (adjusted P < 0.05) between saline- or LPS-treated microglia and beMφs. DE comparisons were made between saline- and LPS-treated samples separately to determine two lists of DE genes, which were combined and displayed for all samples in the heatmap. The values are standardized rlog-transformed values across samples.
beMφs have a predictable genetic signature
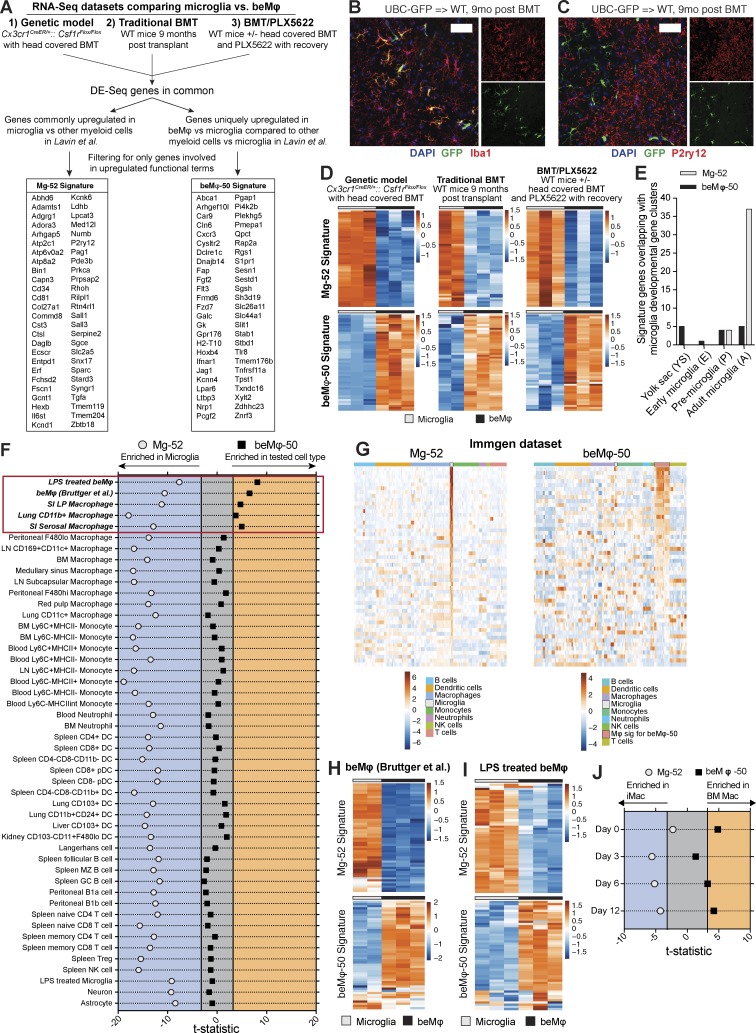
In our RNA-sequencing analysis of saline- or LPS-treated beMφs and microglia, we noted gene clusters that remained consistently differentially expressed between beMφs and microglia regardless of treatment (Fig. 4 D), suggesting that there are core genetic signatures that may be used to define and identify beMφs versus microglia. To this end, we generated genetic signatures for beMφs and microglia by first taking the commonly up-regulated genes among all three of the models of beMφ engraftment. For the beMφ signature, we then refined it by eliminating genes that were differentially expressed between peripheral myeloid cells and microglia in an RNA-sequencing study performed by Lavin et al. (2014); the goal of this refinement was to include only core genes that define beMφs versus microglia and not other myeloid cells versus microglia. For the microglia signature, we refined the signature using the study by Lavin et al. (2014) by only including genes that were commonly unique to microglia in their study and to the microglia in our datasets. Finally, both signatures were refined by eliminating all genes that were not included in functionally enriched terms, thereby creating “functional” genetic signatures. This process created a 52-gene microglia signature (Mg-52; Fig. 5 A and Table 1) and a 50-gene beMφ signature (beMφ-50; Fig. 5 A and Table 2).
Figure 5.
beMφs have a predictable genetic signature distinct from microglia. (A) Schematic showing how the Mg-52 and beMφ-50 signatures were generated by intersecting differentially expressed genes between beMφs and microglia (fold change > 1.5 and adjusted P < 0.05) from the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment, traditional BMT, and BMT/PLX5622 RNA-sequencing datasets in this study and the myeloid cells in Lavin et al. (2014). (B and C) Representative images of brains from mice 9 mo after BMT (n = 4 mice per group; performed once). Bars, 50 µm. Donor bone marrow was from a transgenic mouse that expresses GFP under a UBC promoter. beMφs (green, GFP+) express Iba1 (B), but not P2ry12 (C). (D) Heatmap of the Mg-52 and beMφ-50 in the Cx3cr1CreER/+::Csf1rFlox/Flox model with head-covered BMT and tamoxifen treatment, traditional BMT, and BMT/PLX5622 RNA-sequencing datasets. The values are standardized rlog-transformed values across samples. (E) Overlap of the Mg-52 and beMφ-50 in the microglia developmental stages from Matcovitch-Natan et al. (2016). (F) Enrichment of the Mg-52 and beMφ-50 signatures in various cell types. Axes represent the value of the CAMERA test statistic for each signature (beMφs over microglia), which is used to calculate the corrected p-value (FDR corrected). Samples falling outside of the gray area indicate a statistically significant (FDR <0.001) enrichment of the signature. Samples falling to the left of the gray area demonstrate enrichment for the signature in microglia as compared with the tested cell type. Samples falling to the right of the gray area demonstrate enrichment for the signature in the tested cell type as compared with microglia. Statistics can be found in Table 4. (G) Heatmap of the Mg-52 and beMφ-50 signatures in the Immgen microarray dataset. Values are standardized rlog-transformed values across samples. (H and I) Heatmaps of the Mg-52 and beMφ-50 signatures in beMφs and microglia in Bruttger et al. (2015) (H) and BMT/PLX5622 cells in mice treated with LPS (I); neither dataset was used in the generation of the signatures. The values are standardized rlog-transformed values across samples. (J) Enrichment of the Mg-52 and beMφ-50 signatures in induced pluripotent stem cell–derived microglia-like cells and BM Macs co-cultured with neurons from Takata et al., 2017. The enrichment method is the same as described in F. Statistics can be found in Table 4.
Table 1. List of genes in the Mg-52 signature.
| Symbol | Ensembl | Entrez | Name |
|---|---|---|---|
| Abhd6 | ENSMUSG00000025277 | 66082 | Abhydrolase domain containing 6 |
| Adamts1 | ENSMUSG00000022893 | 11504 | A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
| Adgrg1 | ENSMUSG00000031785 | 14766 | Adhesion G protein–coupled receptor G1 |
| Adora3 | ENSMUSG00000000562 | 11542 | Adenosine A3 receptor |
| Arhgap5 | ENSMUSG00000035133 | 11855 | Rho GTPase-activating protein 5 |
| Atp2c1 | ENSMUSG00000032570 | 235574 | ATPase, Ca2+ sequestering |
| Atp6v0a2 | ENSMUSG00000038023 | 21871 | ATPase, H+ transporting, lysosomal V0 subunit A2 |
| Atp8a2 | ENSMUSG00000021983 | 50769 | ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| Bin1 | ENSMUSG00000024381 | 30948 | Bridging integrator 1 |
| Capn3 | ENSMUSG00000079110 | 12335 | Calpain 3 |
| Cd34 | ENSMUSG00000016494 | 12490 | CD34 antigen |
| Cd81 | ENSMUSG00000037706 | 12520 | CD81 antigen |
| Col27a1 | ENSMUSG00000045672 | 373864 | Collagen, type XXVII, alpha 1 |
| Commd8 | ENSMUSG00000029213 | 27784 | COMM domain containing 8 |
| Cst3 | ENSMUSG00000027447 | 13010 | Cystatin C |
| Ctsl | ENSMUSG00000021477 | 13039 | Cathepsin L |
| Daglb | ENSMUSG00000039206 | 231871 | Diacylglycerol lipase, beta |
| Ecscr | ENSMUSG00000073599 | 68545 | Endothelial cell surface–expressed chemotaxis and apoptosis regulator |
| Entpd1 | ENSMUSG00000048120 | 12495 | Ectonucleoside triphosphate diphosphohydrolase 1 |
| Erf | ENSMUSG00000040857 | 13875 | Ets2 repressor factor |
| Fchsd2 | ENSMUSG00000030691 | 207278 | FCH and double SH3 domains 2 |
| Fscn1 | ENSMUSG00000029581 | 14086 | Fascin actin-bundling protein 1 |
| Gcnt1 | ENSMUSG00000038843 | 14537 | Glucosaminyl (N-acetyl) transferase 1, core 2 |
| Hexb | ENSMUSG00000021665 | 15212 | Hexosaminidase B |
| Il6st | ENSMUSG00000021756 | 16195 | Interleukin 6 signal transducer |
| Kcnd1 | ENSMUSG00000009731 | 16506 | Potassium voltage-gated channel, Shal-related family, member 1 |
| Kcnk6 | ENSMUSG00000046410 | 52150 | Potassium inwardly rectifying channel, subfamily K, member 6 |
| Ldhb | ENSMUSG00000030246 | 16832 | Lactate dehydrogenase B |
| Lpcat3 | ENSMUSG00000004270 | 14792 | Lysophosphatidylcholine acyltransferase 3 |
| Med12l | ENSMUSG00000056476 | 329650 | Mediator complex subunit 12-like |
| Numb | ENSMUSG00000021224 | 18222 | Numb homologue (Drosophila) |
| P2ry12 | ENSMUSG00000036353 | 70839 | Purinergic receptor P2Y, G-protein–coupled 12 |
| Pag1 | ENSMUSG00000027508 | 94212 | Phosphoprotein associated with glycosphingolipid microdomains 1 |
| Pde3b | ENSMUSG00000030671 | 18576 | Phosphodiesterase 3B, cGMP inhibited |
| Prkca | ENSMUSG00000050965 | 18750 | Protein kinase C, alpha |
| Prpsap2 | ENSMUSG00000020528 | 212627 | Phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| Rhoh | ENSMUSG00000029204 | 74734 | Ras homologue family member H |
| Rilpl1 | ENSMUSG00000029392 | 75695 | Rab interacting lysosomal protein-like 1 |
| Rtn4rl1 | ENSMUSG00000045287 | 237847 | Reticulon 4 receptor-like 1 |
| Sall1 | ENSMUSG00000031665 | 58198 | Sal-like 1 (Drosophila) |
| Sall3 | ENSMUSG00000024565 | 20689 | Sal-like 3 (Drosophila) |
| Serpine2 | ENSMUSG00000026249 | 20720 | Serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| Sgce | ENSMUSG00000004631 | 20392 | Sarcoglycan, epsilon |
| Slc2a5 | ENSMUSG00000028976 | 56485 | Solute carrier family 2 (facilitated glucose transporter), member 5 |
| Snx17 | ENSMUSG00000029146 | 266781 | Sorting nexin 17 |
| Sparc | ENSMUSG00000018593 | 20692 | Secreted acidic cysteine rich glycoprotein |
| Stard3 | ENSMUSG00000018167 | 59045 | START domain containing 3 |
| Syngr1 | ENSMUSG00000022415 | 20972 | Synaptogyrin 1 |
| Tgfa | ENSMUSG00000029999 | 21802 | Transforming growth factor alpha |
| Tmem119 | ENSMUSG00000054675 | 231633 | Transmembrane protein 119 |
| Tmem204 | ENSMUSG00000024168 | 407831 | Transmembrane protein 204 |
| Zbtb18 | ENSMUSG00000063659 | 30928 | Zinc-finger and BTB domain containing 18 |
List of the 52 up-regulated genes identified to distinguish microglia from beMφ via the approach outlined in Fig. 5 A.
Table 2. List of genes in the beMφ-50 signature.
| Symbol | Ensembl | Entrez | Name |
|---|---|---|---|
| Abca1 | ENSMUSG00000015243 | 11303 | ATP-binding cassette, subfamily A (ABC1), member 1 |
| Arhgef10l | ENSMUSG00000040964 | 72754 | Rho guanine nucleotide exchange factor 10-like |
| Car9 | ENSMUSG00000028463 | 230099 | Carbonic anhydrase 9 |
| Cln6 | ENSMUSG00000032245 | 76524 | Ceroid-lipofuscinosis, neuronal 6 |
| Cxcr3 | ENSMUSG00000050232 | 12766 | Chemokine (C-X-C motif) receptor 3 |
| Cysltr2 | ENSMUSG00000033470 | 70086 | Cysteinyl leukotriene receptor 2 |
| Dclre1c | ENSMUSG00000026648 | 227525 | DNA cross-link repair 1C |
| Dnajb14 | ENSMUSG00000074212 | 70604 | DnaJ heat shock protein family (Hsp40) member B14 |
| Fap | ENSMUSG00000000392 | 14089 | Fibroblast activation protein |
| Fgf2 | ENSMUSG00000037225 | 14173 | Fibroblast growth factor 2 |
| Flt3 | ENSMUSG00000042817 | 14255 | FMS-like tyrosine kinase 3 |
| Frmd6 | ENSMUSG00000048285 | 319710 | FERM domain containing 6 |
| Fzd7 | ENSMUSG00000041075 | 14369 | Frizzled class receptor 7 |
| Galc | ENSMUSG00000021003 | 14420 | Galactosylceramidase |
| Gk | ENSMUSG00000025059 | 14933 | Glycerol kinase |
| Gpr176 | ENSMUSG00000040133 | 381413 | G protein–coupled receptor 176 |
| H2-T10 | ENSMUSG00000079491 | 15024 | Histocompatibility 2, T region locus 10 |
| Hoxb4 | ENSMUSG00000038692 | 15412 | Homeobox B4 |
| Ifnar1 | ENSMUSG00000022967 | 15975 | Interferon (alpha and beta) receptor 1 |
| Jag1 | ENSMUSG00000027276 | 16449 | Jagged 1 |
| Kcnn4 | ENSMUSG00000054342 | 16534 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| Lpar6 | ENSMUSG00000033446 | 67168 | Lysophosphatidic acid receptor 6 |
| Ltbp3 | ENSMUSG00000024940 | 16998 | Latent transforming growth factor beta binding protein 3 |
| Nrp1 | ENSMUSG00000025810 | 18186 | Neuropilin 1 |
| Pcgf2 | ENSMUSG00000018537 | 22658 | Polycomb group ring finger 2 |
| Pgap1 | ENSMUSG00000073678 | 241062 | Post-GPI attachment to proteins 1 |
| Pi4k2b | ENSMUSG00000029186 | 67073 | Phosphatidylinositol 4-kinase type 2 beta |
| Plekhg5 | ENSMUSG00000039713 | 269608 | Pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| Pmepa1 | ENSMUSG00000038400 | 65112 | Prostate transmembrane protein, androgen induced 1 |
| Qpct | ENSMUSG00000024084 | 70536 | Glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| Rap2a | ENSMUSG00000051615 | 76108 | RAS related protein 2a |
| Rgs1 | ENSMUSG00000026358 | 50778 | Regulator of G-protein signaling 1 |
| S1pr1 | ENSMUSG00000045092 | 13609 | Sphingosine-1-phosphate receptor 1 |
| Sesn1 | ENSMUSG00000038332 | 140742 | Sestrin 1 |
| Sestd1 | ENSMUSG00000042272 | 228071 | SEC14 and spectrin domains 1 |
| Sgsh | ENSMUSG00000005043 | 27029 | N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| Sh3d19 | ENSMUSG00000028082 | 27059 | SH3 domain protein D19 |
| Slc26a11 | ENSMUSG00000039908 | 268512 | Solute carrier family 26, member 11 |
| Slc44a1 | ENSMUSG00000028412 | 100434 | Solute carrier family 44, member 1 |
| Slit1 | ENSMUSG00000025020 | 20562 | Slit homologue 1 (Drosophila) |
| Stab1 | ENSMUSG00000042286 | 192187 | Stabilin 1 |
| Stbd1 | ENSMUSG00000047963 | 52331 | Starch binding domain 1 |
| Tlr8 | ENSMUSG00000040522 | 170744 | Toll-like receptor 8 |
| Tmem176b | ENSMUSG00000029810 | 65963 | Transmembrane protein 176B |
| Tnfrsf11a | ENSMUSG00000026321 | 21934 | Tumor necrosis factor receptor superfamily, member 11a, NF-κB activator |
| Tpst1 | ENSMUSG00000034118 | 22021 | Protein-tyrosine sulfotransferase 1 |
| Txndc16 | ENSMUSG00000021830 | 70561 | Thioredoxin domain containing 16 |
| Xylt2 | ENSMUSG00000020868 | 217119 | Xylosyltransferase II |
| Zdhhc23 | ENSMUSG00000036304 | 332175 | Zinc-finger, DHHC domain containing 23 |
| Znrf3 | ENSMUSG00000041961 | 407821 | Zinc and ring finger 3 |
List of the 50 up-regulated genes identified to distinguish beMφ from microglia via the approach outlined in Fig. 5 A.
Of note, several previously identified microglia-specific genes were included in the Mg-52 signature, such as Cst3, Hexb, P2ry12, and Sall1, and Tmem119. Based on this, we tested P2ry12 as a marker of microglia versus beMφs. Indeed, although both beMφs and microglia stained for Iba1 (Fig. 5 B), P2ry12 was a unique marker for microglia (Fig. 5 C).
We next evaluated the reliability of our signatures within the datasets used to generate them. As expected, all three datasets universally demonstrated enrichment of Mg-52 in microglia and enrichment of the beMφ-50 in beMφ (Fig. 5 D). In a study by Matcovitch-Natan et al. (2016), gene clusters involved in stages of microglia development were identified. We assessed the overlap of our signatures with these microglia developmental gene clusters and found that the only substantial overlap occurred with the Mg-52 signature and the “Adult Microglia” developmental gene cluster (37/52 genes in Mg-52; Fig. 5 E). These results supported the idea that the Mg-52 signature is specific to adult microglia and that the beMφ-50 signature does not correlate to any microglia developmental programs.
To test the specificity of Mg-52 and beMφ-50 for their respective cell types, we used a competitive gene set test (CAMERA; Wu and Smyth, 2012). Publically available transcriptomic datasets of immune cells (Heng et al., 2008), neurons (Srinivasan et al., 2016), astrocytes (Srinivasan et al., 2016), beMφs (Bruttger et al., 2015; Table 3), and our LPS-treated beMφ data (which were not used in the generation of signatures) were used to validate the signatures. All of these datasets contain microglia samples that were used as a common reference cell in our CAMERA analysis. As expected, neither neurons nor astrocytes were enriched for beMφ-50, but microglia were enriched for the Mg-52 signature as compared with neurons and astrocytes (Fig. 5 F and Table 4). Similarly, LPS-treated microglia were not enriched for beMφ-50 as compared with control microglia, whereas control microglia were enriched for Mg-52 (Fig. 5 F and Table 4).
Table 3. List of publically available datasets used in this study.
| GEO dataset | First author | Conditions | Study type | GEO link | Publication link |
|---|---|---|---|---|---|
| GSE68376 | Bruttger | Microglia versus CNS engrafting bone marrow–derived macrophages (beMφs) | RNA sequencing | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68376 | http://www.ncbi.nlm.nih.gov/pubmed/26163371 |
| GSE15907 | ImmGen | Microglia versus peripheral immune cells for signature validation | Affymetrix Array | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15907 | Multiple, see citations on GEO page |
| GSE63340 | Lavin | Comparison of microglia to peripheral macrophages/monocytes for generation of signatures | RNA sequencing | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63340 | http://www.ncbi.nlm.nih.gov/pubmed/25480296 |
| GSE75246 | Srinivasan | LPS-treated and control neurons/astrocytes/microglia | RNA sequencing | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75246 | https://www.ncbi.nlm.nih.gov/pubmed/27097852 |
| GSE99078 | Takata | iMac versus BM Mac | RNA sequencing | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99078 | https://www.ncbi.nlm.nih.gov/pubmed/28723550 |
Table 4. CAMERA statistics used in the validation of the Mg-52 and beMφ-50 genetic signatures.
| GEO dataset | Control | Experimental | Mg-52 | beMφ-50 | ||
|---|---|---|---|---|---|---|
| T statistic | FDR | T statistic | FDR | |||
| GSE75246 | Microglia, vehicle treated | Microglia, LPS treated | −9.18997599139088 | 8.73894827192082E-20a | −0.960428754944643 | 0.336853025380644 |
| GSE75246 | Microglia, vehicle treated | Neuron, vehicle treated | −9.28373097503757 | 3.65539900363888E-20a | −1.6069073647106 | 0.108093118311235 |
| GSE75246 | Microglia, vehicle treated | Astrocyte, vehicle treated | −8.43449685504562 | 7.17579519282282E-17a | −0.982701094445303 | 0.325768448416933 |
| GSE75246 | Microglia, LPS treated | Neuron LPS treated | −5.703178515505 | 2.3906543497579E-08a | −1.23253311708603 | 0.217766916347967 |
| GSE75246 | Microglia, LPS treated | Astrocyte, LPS treated | −3.82020514805529 | 0.000267622477204a | −0.76323460910686 | 0.445334000895242 |
| GSE75246 | Neuron, vehicle treated | Neuron, LPS treated | −2.48230960173674 | 0.026125771365367 | 1.03261170555403 | 0.30180022362708 |
| GSE75246 | Neuron, vehicle treated | Astrocyte, vehicle treated | 2.39665178928464 | 0.033112577014953 | 0.940103366793486 | 0.347177795693465 |
| GSE75246 | Astrocyte, vehicle treated | Astrocyte, LPS treated | −0.355557598200872 | 0.722176280064764 | −0.55668524588016 | 0.722176280064764 |
| GSE15907 | Microglia | SI LP macrophage | −11.2065064200678 | 9.18045352544835E-29a | 4.71522334166825 | 2.43012363462998E-06a |
| GSE15907 | Microglia | Lung CD11b+ macrophage | −17.8986346561823 | 8.2656485448573E-71a | 3.75505071435236 | 0.000173778763046a |
| GSE15907 | Microglia | SI serosal macrophage | −12.8781234055138 | 1.66788609720075E-37a | 4.98232404004903 | 6.33293438301929E-07a |
| GSE15907 | Microglia | Peritoneal F4/80lo macrophage | −13.8346052104306 | 4.90645664168948E-43a | 1.33690258052763 | 0.181269070989656 |
| GSE15907 | Microglia | LN CD169+CD11c+ macrophage | −16.7938329111185 | 1.40723697488701E-62a | 0.323384063567144 | 0.746407650677293 |
| GSE15907 | Microglia | BM macrophage | −14.1038712769937 | 1.1592597363317E-44a | −0.91021289075231 | 0.362720802630218 |
| GSE15907 | Microglia | Medullary sinus macrophage | −16.9449218942956 | 1.12872637834688E-63a | 0.394208861959158 | 0.693430887937153 |
| GSE15907 | Microglia | LN subcapsular macrophage | −16.8108255068152 | 1.06072069454246E-62a | −0.569435569747971 | 0.569066737637983 |
| GSE15907 | Microglia | Peritoneal F4/80hi macrophage | −13.2907475407257 | 7.64121015322315E-40a | 1.77706541396221 | 0.075572099377594 |
| GSE15907 | Microglia | Red pulp macrophage | −13.8968419518646 | 2.07736897631509E-43a | 0.800634377157631 | 0.423352494668722 |
| GSE15907 | Microglia | Lung CD11c+ macrophage | −12.449942293778 | 3.74528948133128E-35a | −1.84381236333192 | 0.065224715729658 |
| GSE15907 | Microglia | BM Ly6c+MHCII− monocyte | −15.9185653482659 | 2.03921322346964E-56a | −0.818538144350527 | 0.413059355130746 |
| GSE15907 | Microglia | BM Ly6c−MHCII− monocyte | −16.995529069284 | 4.82435079029921E-64a | −0.4980128606501 | 0.618480246889288 |
| GSE15907 | Microglia | Blood Ly6c+MHCII+ monocyte | −16.4580514220967 | 3.54539257686525E-60a | 0.935598972668471 | 0.349490573927721 |
| GSE15907 | Microglia | Blood Ly6c+MHCII− monocyte | −13.4521196565583 | 8.89007050093597E-41a | 0.93218558886987 | 0.351251449984407 |
| GSE15907 | Microglia | LN Ly6c+MHCII− monocyte | −16.8225925914844 | 8.72001502349316E-63a | 1.24084625878423 | 0.214676554440318 |
| GSE15907 | Microglia | Blood Ly6c−MHCII+ monocyte | −18.884692403881 | 1.39532275643974E-78a | 0.270993599010383 | 0.786398646520675 |
| GSE15907 | Microglia | Blood Ly6c−MHCII− monocyte | −16.5767118715121 | 5.08670106411122E-61a | −0.514869830796944 | 0.60664945933478 |
| GSE15907 | Microglia | Blood Ly6c−MHCIIint monocyte | −13.856797569651 | 3.61293182305638E-43a | 0.232110933264168 | 0.816454138490933 |
| GSE15907 | Microglia | Blood neutrophil | −12.9632371395544 | 5.56623874137407E-38a | −1.79279030852971 | 0.073020996049815 |
| GSE15907 | Microglia | BM neutrophil | −11.4003953949682 | 1.02230860670541E-29a | −1.72096439038072 | 0.08527216840678 |
| GSE15907 | Microglia | Spleen CD4+ DC | −13.9384560198774 | 1.16689659747184E-43a | −0.245307707479572 | 0.806220606319135 |
| GSE15907 | Microglia | Spleen CD8+ DC | −13.7057882876266 | 2.87179757732365E-42a | 0.398151064886531 | 0.690522924468198 |
| GSE15907 | Microglia | Spleen CD4−CD8−CD11b− DC | −15.1108056316066 | 5.1513600603475E-51a | −0.371877852772889 | 0.709987586561984 |
| GSE15907 | Microglia | Spleen CD8+ pDC | −11.9560451648539 | 1.54734296499501E-32a | −0.5330479854046 | 0.594006063116689 |
| GSE15907 | Microglia | Spleen CD8− pDC | −12.0888471854728 | 3.13529730173023E-33a | −0.686519292990229 | 0.492393361276459 |
| GSE15907 | Microglia | Spleen CD4−CD8−CD11b+ DC | −16.7499579333542 | 2.91605319710899E-62a | −0.773651602002031 | 0.439145647090047 |
| GSE15907 | Microglia | Lung CD103+ DC | −12.9476715882146 | 6.80692723941832E-38a | 1.56459587282508 | 0.117692921772633 |
| GSE15907 | Microglia | Lung CD11b+CD24+ DC | −14.2347565935749 | 1.83032966037038E-45a | 1.79831123076597 | 0.072142206176879 |
| GSE15907 | Microglia | Liver CD103+ DC | −14.59556587753 | 1.0361250320237E-47a | 0.882871388829489 | 0.377315974859225 |
| GSE15907 | Microglia | Kidney CD103−CD11b+F4/80lo DC | −13.3863273878228 | 2.14353221609188E-40a | 1.96342185139998 | 0.049610489223897 |
| GSE15907 | Microglia | Langerhans cell | −13.5648053990461 | 1.94992734843495E-41a | −0.368430279415978 | 0.71255617554319 |
| GSE15907 | Microglia | Spleen follicular B cell | −11.8221570653282 | 7.60363847464607E-32a | −2.06435720774193 | 0.038996239349848 |
| GSE15907 | Microglia | Spleen MZ B cell | −12.8680272621344 | 1.89888380743561E-37a | −2.21791907042995 | 0.02657112051891 |
| GSE15907 | Microglia | Spleen GC B cell | −11.6270160082023 | 7.5026540207794E-31a | −2.63836018619294 | 0.008337012761183 |
| GSE15907 | Microglia | Peritoneal B1a cell | −12.8368524481395 | 2.83249315683069E-37a | −2.25999214810576 | 0.023832043427742 |
| GSE15907 | Microglia | Peritoneal B1b cell | −12.4704022163508 | 2.90329306882604E-35a | −2.03073615939647 | 0.042294469723543 |
| GSE15907 | Microglia | Spleen naive CD4+ T cell | −12.0514689371187 | 4.92231507926634E-33a | −1.32085874766837 | 0.186563009465559 |
| GSE15907 | Microglia | Spleen naive CD8+ T cell | −15.632336222605 | 1.79884156914574E-54a | −1.88247758455911 | 0.059785157409316 |
| GSE15907 | Microglia | Spleen memory CD4+ T cell | −12.7721168022034 | 6.47878151163567E-37a | −0.442027244316272 | 0.658474090360188 |
| GSE15907 | Microglia | Spleen memory CD8+ T cell | −13.5419211345285 | 2.65618103458737E-41a | −1.33469326750991 | 0.181991378524863 |
| GSE15907 | Microglia | Spleen T regulatory cell | −15.4254434586749 | 4.36485159999069E-53a | −1.26238816488991 | 0.206823310056597 |
| GSE15907 | Microglia | Spleen natural killer cell | −15.8677797159762 | 4.54101007200768E-56a | −1.28565291148151 | 0.198578545975841 |
| GSE68376 | Microglia | beMφ | −10.6110235228595 | 6.8924083456567E-26a | 6.56936921593136 | 5.25863738290986E-11a |
| GSE108575 | Microglia, LPS treated | beMφ, LPS treated | −7.64393242343623 | 2.23315767992962E-14a | 8.126057045216 | 9.55133453377967E-16a |
| GSE99078 | iMac, day 0 | BM Mac, day 0 | −2.27409571243689 | 0.022973398251917 | 4.85289376959958 | 2.45636395979254E-06a |
| GSE99078 | iMac, day 3 | BM Mac, day 3 | −5.60231923708091 | 4.29964257929522E-08a | 1.34724285505885 | 0.177921083688115 |
| GSE99078 | iMac, day 6 | BM Mac, day 6 | −5.17686685222183 | 4.56742989186354E-07a | 3.23262604923775 | 0.001229061186423 |
| GSE99078 | iMac, day 12 | BM Mac, day 12 | −4.26858897016626 | 2.48966525049818E-05a | 4.21696975941865 | 2.48966525049818E-05a |
Publically available transcriptomic datasets and transcriptomic data generated in the current study were used to assess for enrichment of Mg-52 and beMφ-50 gene signatures. CAMERA analysis was performed as described in Materials and methods.
Significance threshold FDR <0.001.
Likewise, the Mg-52 signature was enriched in microglia compared with all immune cell types tested from the publically available Immgen microarray dataset (Heng et al., 2008), again supporting the fidelity of this signature for the detection of microglia versus nonmicroglia (Fig. 5, F and G; and Table 4). However, three peripheral immune cell types were significantly enriched for beMφ-50: small intestine serosal macrophages, lung CD11b+ macrophages, and small intestine lamina propria macrophages (Fig. 5, F and G; and Table 4). Interestingly, all three of these are monocyte-derived macrophages, providing further evidence of a monocyte origin for beMφ.
Importantly, there was strong enrichment for beMφ-50 in beMφ transcriptomes generated in another laboratory (Bruttger et al., 2015; Fig. 5, F and H; and Table 4). We also observed strong enrichment for beMφ-50 in LPS-treated beMφ (Fig. 5, F and I; and Table 4), confirming our initial observation that beMφs and microglia have core genes that can be used to define each cell type regardless of a strong stimulus.
Finally, we used our signatures to interrogate recently published data that purports to have generated induced pluripotent stem cell–derived microglia-like cells (iMacs; Takata et al., 2017). When CAMERA analysis was applied to RNA-sequencing data from iMacs versus bone marrow–derived macrophages (BM Macs) over the course of an in vitro co-culture with neurons, we found that Mg-52 became significantly enriched in iMac by day 3 of co-culture, and remained enriched through day 12 (Fig. 5 J and Table 4). By comparison, beMφ-50 was enriched in BM Mac on days 0 and 12 (Fig. 5 J and Table 4). These results supported the conclusions of Takata et al. (2017) that they had generated microglia-like cells in vitro.
Overall, these data confirmed that Mg-52 and beMφ-50 represent core signatures that may be used to define and identify microglia and beMφs in a direct comparison. In addition, the fact that each cell type has a distinct core genetic program lends strong evidence that beMφ indeed represent an independent type of macrophage that could have a long-lived presence and unique functional implications in the CNS.
Discussion
The role of peripherally derived engrafting macrophages in the CNS has long been a subject of scientific interest and debate (Prinz and Priller, 2014; Larochelle et al., 2016). In this work, we investigated several outstanding questions regarding these enigmatic cells. First, using a genetic model of chronic partial microglia deficiency, we showed that persistent loss of microglia and their inability to repopulate the niche is sufficient to induce beMφ engraftment into the CNS (i.e., in the absence of irradiation). Second, we demonstrated that beMφs do not alter brain function as measured by a wide array of behavioral tests. Third, we found that beMφs maintain a unique transcriptional and functional identity in three different models of beMφ engraftment. Fourth, we generated functional genetic signatures capable of detecting either beMφs (beMφ-50) or microglia (Mg-52) in a direct comparison. Finally, we validated their ability to specifically detect microglia (Mg-52) or beMφs and other monocyte-derived macrophages (beMφ-50) in the datasets that we generated and in other, publically available transcriptomic datasets.
It was previously unclear as to whether or not beMφ would eventually differentiate into bona fide microglia, and thereby take on the exact physiological roles of microglia in the CNS. Here, we have provided evidence to suggest that beMφs are in fact a unique cell type and, although capable of taking up long-term residence in the CNS, maintain a unique transcriptional and functional identity. This may help to explain the reported therapeutic roles for beMφs (Walkley et al., 1994; Krivit et al., 1995, 1999; Platt and Lachmann, 2009; Chen et al., 2010; Derecki et al., 2012; Hsiao et al., 2012) and suggests that beMφs may not only “replace” dysfunctional microglia but also in fact provide unique therapeutic benefits based on their unique identity and functional profile. Further, the fact that chronic microglia depletion (along with the inability of the remaining microglia to proliferate and refill the niche) is sufficient to drive beMφ engraftment into the brain gives hope that in the future, it may be possible to use radiation-free conditioning regimens to achieve substantial beMφ engraftment in patients. Such an approach would focus on specifically targeting microglia proliferation while leaving the circulating monocyte pool intact. This may be a challenging task, because both microglia and other myeloid cells primarily use Csf1 signaling for proliferation and survival; alternative targets for specific microglia depletion would likely be necessary in the clinical context. It is also encouraging that we did not find any substantial behavioral abnormalities upon beMφ engraftment, as this could be a significant concern if such high levels of beMφ engraftment were achieved in patients.
Our models demonstrate that beMφ could replace microglia only when microglia are impaired in their ability to repopulate the niche, without the need for irradiation, inflammation, or BBB disruption. It is important to note that previously published data demonstrating the ability of microglia to self-repopulate are not refuted by our findings (Elmore et al., 2014; Bruttger et al., 2015; Huang et al., 2018). On the contrary, we support the concept that under most physiological circumstances microglia will repopulate the niche through self-renewal. It is conceivable, however, that during the long lifespan of a human, beMφs may engraft into a brain under certain pathological conditions (in which microglia self-renewal is affected), changing the myeloid niche of the brain. Our data also suggest that impairment of microglia self-renewal is likely one of the primary effects of irradiation-induced repopulation of the brain by beMφs.
Together, our findings demonstrate that peripherally derived CNS macrophages are a unique cell type capable of replacing microglia in the context of microglia deficiency without irradiation. These findings reframe the identity of beMφs and firmly place them as an independent class of brain macrophage. Our findings may be therapeutically exploited in the future to achieve beMφ engraftment without irradiation.
Materials and methods
Animal experiment approval
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Virginia.
Mice
Mice were initially purchased from The Jackson Laboratory and subsequently maintained and bred in-house under standard housing conditions (12 h light/dark cycle and fed ad libitum). All mice were on a C57BL/6J background, and both males and females were used unless stated otherwise. Strains used were C57BL/6J, C57BL/6-Tg(UBC-GFP)30Scha/J, B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J, and B6.Cg-Csf1rtm1.2Jwp/J. Cx3cr1CreER/+::Csf1rFlox/Flox mice were generated by breeding Cx3cr1CreER/+::Csf1rFlox/Flox mice with Cx3cr1+/+::Csf1rFlox/Flox mice. For experiments using tamoxifen, mice were fed TD.130856 at 250 mg/kg diet (purchased from Harlan) starting at 4 wk after birth or as otherwise specified in figures. Experimental groups were blinded before beginning the experiment and remained blinded until the end.
Irradiation and BMT
Mice were γ irradiated with a lethal dose of 1,000 rad. 4 h after irradiation, mice were intravenously injected with 5 × 106 bone marrow cells. Mice were given water supplemented with trimethoprim-sulfamethoxazole for 2 wk and monitored daily for the first 4 d. A lead shield was placed over the head during irradiation in some experiments, as indicated.
Tissue collection
Mice were euthanized with Euthasol and transcardially perfused with 0.01 M PBS containing 5 U/ml heparin. Blood was collected from the retinal artery after removing the eye and placed in heparinized tubes. For immunohistochemistry, brains were carefully removed and dropped fixed in 4% paraformaldehyde for 48 h. Brains were then washed with PBS and stored at 4°C until further processing. For isolating cells, brains were harvested and immediately processed as described below. Intestinal cell suspensions were prepared as previously described (Cronk et al., 2015). In brief, the entire length of the small intestine was excised and opened longitudinally. Luminal contents were washed in Ca/Mg-free PBS, chopped in 2.5-cm pieces, and placed in conical tubes containing 30 ml HBSS, 5% FBS, and 2 mM EDTA. The tubes were shaken at 37°C and 250 rpm for 20 min, after which the intestines were strained through a nylon mesh and washed again under the same conditions. After the last wash, the intestines were transferred into 20 ml HBSS containing 900 U/ml Collagenase VIII (Sigma) and 40 U/ml DNase-I (Sigma) and then shaken for 15 min at 200 rpm to digest. After the incubation, the tubes were vortexed thoroughly and the resulting cell suspension passed through 70-µm cell strainers into clean tubes. The cells were washed twice with cold HBSS, 5% FBS, and 2 mM EDTA and centrifuged at 4°C, 425 relative centrifugal force (RCF), for 5 min. After the last wash, the supernatant was decanted and the pellet was resuspended in FACS buffer containing 0.01 M PBS, 1% BSA, 2 mM EDTA, and 0.1% sodium azide and prepared for FACS analysis.
Flow cytometry
After tissue collection (as described above), cells were incubated with flow cytometry antibodies at 4°C for 30 min in a total volume of 200 µl of flow cytometry buffer (PBS containing 0.1% sodium azide and 1% BSA), washed with 5 ml flow cytometry buffer, pelleted at 300 RCF, decanted, and analyzed on a Gallios flow cytometer (Beckman Coulter). Antibodies were purchased from BD PharMingen except for CD115 (BioLegend).
Immunohistochemistry
Brains were collected as described above and cryoprotected in 30% sucrose. After freezing in optimum cutting temperature compound (Sakura Finetek), 40-µM sections were cut on a cryostat (Leica). Floating sections were stored in PBS containing Azide (0.02%) until further processing. For immunohistochemistry, brain sections were permeabilized with PBS containing 0.5% Triton X-100 and then blocked with 10% chicken serum in PBS containing 0.05% Triton X-100 for 1 h. Sections were then incubated overnight with primary antibody in PBS containing 2% chicken serum. Primary antibodies included rabbit anti-GFP (1:1,000; Abcam), goat anti-Iba1 (1:300; Abcam), rabbit anti-Iba1 (1:300; Wako), rabbit-anti Ki67 (1:500; Abcam), and rabbit anti-P2ry12 (1:10,000; gift from O. Butovsky, Ann Romney Center for Neurologic Diseases, Brigham and Women's Hospital, Harvard Medical School, Boston, MA). After incubating primary antibody, sections were washed three times and then incubated with fluorescently conjugated secondary antibodies (1:1,000) in PBS containing 0.05% Triton X-100 and 2% chicken serum for 1 h. Sections were then washed three times with PBS containing 0.05% Triton X-100. DAPI was added to the second wash to stain DNA containing nuclei. Sections were then mounted on microscope slides with Aquamount. All images were collected on a Leica SP8 confocal microscope or Leica wide-field microscope.
BrdU proliferation assay
BrdU injection and analysis were performed as previously described (Lu et al., 2011). In brief, mice were injected twice (i.p.) with 50 mg/mg BrdU, with 2 h between each injection. Brains were analyzed by immunohistochemistry 24 h after the last BrdU injection.
Cell sorting
To FACS sort microglia and beMφ, brains were collected after euthanization and systemic perfusion with PBS, and then meninges were removed. Brains were physically minced and incubated in a 15-ml tube in 5 ml HBSS containing Mg and Ca, 2 mg/ml papain, 50 U/ml DNASE-I (Sigma), and Glutamax (Invitrogen) at 37°C for 15 min. After gentle trituration, the brains were incubated at 37°C for an additional 15 min, triturated, incubated at 37°C for an additional 15 min, and triturated a third time. After this, the tubes were filled with DMEM/F12 containing 10% FBS and filtered through a 70-µM cell strainer. Cells were pelleted at 300 RCF and sorted as follows. Cells were incubated with CD11b+ microglia magnetic selection beads according to manufacturer’s protocol (Miltenyi). Cells were then positively selected by AutoMACS and used for downstream applications or FACS sorted by gating on live cells by DAPI exclusion (DAPI-negative cells, high side scatter exclusion, singlet events, CD45Lo, CD11b+, and either GFP− [microglia] or GFP+ [beMφs]). Monocytes were isolated from bone marrow using mouse bone marrow monocyte selection beads and sorted on an LS magnetic columns according to the manufacturer’s protocol (Miltenyi). Sorted monocytes were at least 95% pure monocytes (CD11b+Ly6CHiCD115+) by flow cytometry.
RNA sequencing and quantitative RT-PCR
RNA was isolated from FACS-sorted microglia using an RNeasy mini kit (QIAGEN). Each sample used for RNA-sequencing was pooled from three or four total mice per sample. For quantitative RT-PCR, cDNA was generated using High Capacity cDNA kit (Applied Biosystems) and Csf1r gene expression was analyzed with the Mm00432691_m1 TaqMan Gene Expression assay. For RNA sequencing, all postprocessing (including linear RNA amplification and cDNA library generation) and sequencing was performed by Hudson Alpha Genomic Services Laboratory.
Evans blue BBB permeability assay
To test for BBB permeability, mice were injected i.p. with Evans blue (13.3 µl/g of a 2% Evans blue solution dissolved in PBS) and samples were collected 1 h after injection. Mice were euthanized by Euthasol, perfused with ice cold PBS, and brains were removed. One half of each brain was homogenized in 1ml of PBS and mixed with one volume of 50% trichloroacetic acid, then incubated overnight at 4°C to precipitate out proteins and other particulates. Samples were centrifuged at 15,000 g for 30 min, 4°C. Supernatants were analyzed by 96-well plate reader at 620 nm (Multiskan FC; Fisher Scientific), and a standard curve was generated using known concentrations of Evans blue dye in PBS mixed 1:1 with 50% trichloroacetic acid.
Parabiosis
Surgery for parabiosis was performed as previously described (Radjavi et al., 2014). Female UBC-GFP mice were paired to Cx3cr1CreER/+::Csf1rFlox/Flox mice or Cre-negative controls matched for age, sex, and weight. After parabiosis surgery, mice were allowed 3 wk to recover before a 12-wk tamoxifen treatment.
Behavior
Behavioral testing was performed as previously described (Radjavi et al., 2014; Filiano et al., 2016). Before all behavior testing, mice were acclimated to the testing room for 1 h. Testing schedules were balanced for genotype and the observer was blinded to all conditions. Mazes were cleaned with 70% ethanol between trials and all behavioral assays were performed during the light hours avoiding 1 h after and before the lights turned on and off.
Plus maze
Mice were place into the center hub of the plus maze and were free to explore for 5 min. Movement was calculated with TopScan (CleverSys), and data were represented as percentage of time spent in the open arms during the 5-min trial.
Open field
Mice were placed into an open field (35 cm × 35 cm) and were free to explore for 15 min. Movement was monitored via TopScan, and data were represented as percentage of time spent in the center of the box (23 cm × 23 cm) during the 15-min trial.
Sociability
Social behavior was tested using the three-chamber assay. Mice were placed into the center chamber of a three-chamber social box and were free to explore all three rooms for 10 min per phase. For the habituation phase, empty wire cages (Spectrum Diversified Designs) were placed in the two outer rooms. After the initial habituation phase, the mice were returned to the center room for the social phase, where a novel mouse was placed under one cup (8–10-wk-old male habituated to the cup) and a novel object was placed under the other cup. Tracking was scored with TopScan, and time spent investigating around each cup was quantified.
Rotarod
Motor behavior was tested with an accelerated rotarod (MedAssociates). Mice were placed on an accelerating rotarod that accelerated from 4.0 to 40 rpm over 5 min. The latency of the mouse to fall off the rod was monitored via infrared beams. Mice were given six trials with a 4-h intertrial interval between trials 3 and 4.
Morris water maze
Cognitive function was tested with the water maze. For the acquisition phase, mice were placed in a 100-cm pool of opaque water with a hidden platform 1 cm under the surface. Mice were given three trials per day (maximum of 60 s) to find the platform with 30 min between each trial. This was repeated for 4 d. On the fifth day, for the probe trial, the platform was removed and mice were given 60 s to explore the maze. Video tracking of movement was performed with an EthoVision tracking system. For the acquisition phase, data were represented as latency to reach the platform. For the probe trial, data were represented as time spent in the quadrant that previously contained the platform.
Laser-burn injury
Brain macrophages were imaged by multiphoton microscopy. To target microglia, Cx3cr1CreER/+::Ai6 were irradiated with head shielding and given bone marrow from a wild-type donor mouse. To target beMφ, wild-type mice were irradiated and given bone marrow from a Cx3cr1GFP/+ donor mouse. After 4 wk of recovery, mice were fed tamoxifen for 4 wk to pulse-label macrophages and then placed back on normal chow. After 4 wk on normal chow, all mice were fed PLX5622 for 2 wk to clear out the brain macrophage pool and allowed to recover for 6 wk before imaging. For imaging, mice were anesthetized with ketamine/xylene (i.p.). Two-photon laser injury was performed by focusing a two-photon laser beam in the superficial layer of the cortex (∼50 µm deep) through a thinned intact skull as previously described (Davalos et al., 2005). In brief, a 780-nm two-photon laser (Chameleon Ultra II tunable Ti:Sapphire laser; Coherent) with a laser power of 60 to 80 mW was applied to a region of interest of 20 µm of diameter for ∼30 s (the efficiency of the injury was visualized by the bright autofluorescence sphere in the region of interest). The area was imaged for 30 min before the laser injury and 30 min after laser injury. Images were acquired using a ×25 water-immersion objective with 0.95 NA and external HyD nondescanned detectors (Leica). Four-dimensional imaging data were collected by obtaining images from the x, y, and z planes over time.
Image analysis
Automated image analysis algorithms were developed and implemented in the Virginia Image and Video Analysis laboratory (Acton). The software was written in the MATLAB (MathWorks) environment.
Process detection
Microglia and engrafted macrophage processes were detected from the images using area morphology implemented via connected filters (Acton and Mukherjee, 2000; Acton, 2001; Acton and Ray, 2006). Such connected filters have advantages over traditional filters in that they are edge preserving and do not depend on binarized versions of the image. The processes were detected that had a two-dimensional area (observed via maximum intensity projection) within a range of 15 to 75 µm2 and sufficient contrast with the background (>4% of intensity range). The positions of the processes were recorded by computing the region centroids.
Process tracking
Individual process locations were tracked temporally from frame to frame by finding a correspondence between detections (Scott Thomas Acton, 2006). The correspondence was determined by spatial proximity, consistency in direction of motion, and similarity in size. Group motion (and corresponding velocity) of the processes was computed using a dynamic Sholl analysis. The Sholl diagram was centered at the center of the burn site and consisted of five annuli of ring width of 17 µm. Groups of processes were tracked (in maximum intensity projection images) from the time of maximum population in the fifth annulus to the time of maximum population (of detected processes) in the third annulus (which was just outside the perimeter of the burn). This group tracking provided an overall speed computation that was robust to false positives and missed detections in the process detection task.
LPS challenge
To determine transcriptional and morphological changes of microglia and beMφs to an insult, mice were given a peripheral LPS challenge. Mice were first irradiated and given BMT with UBC-GFP bone marrow. To target microglia, head shielding was applied during irradiation in some mice. After 1 wk of recovery, mice were treated with PLX 5622 for 2 wk to deplete the brain macrophage niche and then placed on standard chow for 6 wk. Mice were injected with 50 µg LPS or saline (i.p.). 6 h later, brains were removed and macrophages sorted by MACS on CD11b beads as described above.
RNA-sequencing and functional analysis
The Gene Expression Omnibus (GEO) accession numbers for RNA-sequencing data generated for this publication are GSE84819, GSE108569, and GSE108575. All other previously published datasets used in this paper are listed with GEO under the accession numbers shown in Table 3 (GSE68376, GSE15907, GSE63340, GSE75246, and GSE99078). The raw sequencing reads (FASTQ files) went through two stages of preprocessing to remove low-quality reads and bases. First, they were chastity filtered, which removes any clusters that have a higher than expected intensity of the called base compared with other bases. They were then trimmed with Trimmomatic (Bolger et al., 2014) to remove low-quality bases (minimum read length after trimming, 36). After preprocessing, the quality of the reads was evaluated using FastQC (Andrews, 2010), and after passing quality control were aligned to the UCSC mm9 genome (Harrow et al., 2012) using the splice-aware read aligner STAR (Dobin et al., 2013). The quality of the alignments was next assessed by SAMStat (Lassmann et al., 2011), and any low-quality alignments were removed with samtools (Li et al., 2009; MAPQ <10). Next, the number of reads aligning to each gene was quantified with HTSeq (Anders et al., 2015), and then the Bioconductor package (Love et al., 2014). DESeq2 was used to normalize the raw counts, perform exploratory analysis (e.g., PCA), and DE analysis. Before DE analysis of the BMT/PLX dataset, surrogate variable analysis (Leek and Storey, 2007) was used to identify and adjust for latent sources of unwanted variation as implemented in the sva package (Leek et al., 2012). The p-values from the DE analysis were corrected for multiple hypothesis testing with the Benjamini–Hochberg false discovery rate (FDR) procedure. Heatmaps were generated with the R package pheatmap (Kolde, 2015), and UpSet plots (Lex et al., 2014) were created with the R package UpSetR (Gehlenborg, 2016). The functional terms enriched in beMφs and microglia for our RNA-sequencing datasets were determined with GSVA (Hänzelmann et al., 2013). The gene sets used for this analysis were from the GOBP (Ashburner et al., 2000; The Gene Ontology Consortium, 2017).
Microarray analysis
All of the microarrays were analyzed using a combination of the affy (Gautier et al., 2004), oligo (Carvalho and Irizarry, 2010), and limma (Ritchie et al., 2015) packages from Bioconductor. For the Affymetrix arrays, the expression values for each probe set were extracted using the robust multichip average (RMA) methodology (Irizarry et al., 2003). For the Agilent arrays, the probes were background corrected using a normal exponential convolution model (as implemented in limma; Silver et al., 2009) and quantile normalized. Replicate probes were summarized using the mean.
Signature creation
To create the Mg-52 signature, we first identified the differentially up-regulated genes in microglia versus beMφs in all three of the Kipnis datasets: the genetic model, traditional BMT, and BMT/PLX RNA sequencing. To be called significant, a gene needed to have a fold change >1.5 and a corrected p-value <0.05. These criteria for significance were used to identify all differentially expressed genes hereafter. Next, we identified the differentially up-regulated genes in microglia versus peripheral myeloid cell types (nine cell types in total) from the Lavin RNA-sequencing dataset (Lavin et al., 2014). The cell types in this dataset were peritoneal Mφ, small intestine Mφ, large intestine Mφ, monocytes, Kupffer Mφ, red pulp Mφ, alveolar Mφ, and neutrophils. The Mg-52 signature was defined as the 52 genes that were (a) up-regulated in microglia in all three of the Kipnis datasets, (b) up-regulated in eight out of nine microglia versus peripheral myeloid cell comparisons from the Lavin dataset, and (c) included in a gene set that was functionally enriched in microglia in at least one of the Kipnis datasets. To create the beMφ-50 signature, we first identified the differentially up-regulated genes in beMφ versus microglia in all three of the Kipnis datasets and the differentially up-regulated genes in the nine myeloid cell types versus microglia in the Lavin RNA-sequencing dataset. The beMφ-52 signature was defined as the 50 genes that were (a) up-regulated in beMφ in all three of the Kipnis datasets, (b) not up-regulated in the peripheral myeloid cells from the Lavin dataset, and (c) included in a gene set that was functionally enriched in beMφ in at least one of the Kipnis datasets.
Signature detection
The signatures were detected in the various transcriptomic datasets using CAMERA (Wu and Smyth, 2012; part of the limma package). The intergene correlation was set to 0.01. To use CAMERA with the RNA sequencing data, the raw counts needed to be transformed and normalized with the voom function (Law et al., 2014). This was not necessary for the microarray data. CAMERA returns both a test statistic and an FDR-corrected p-value. The test statistic was used as an enrichment score, with larger values of the statistic corresponding to a greater enrichment of the signature. To ensure the fidelity of the signature detection, we used a stringent corrected p-value threshold of 1E-3.
Online supplemental material
Fig. S1 shows that Cx3cr1-expressing resident myeloid cells in the periphery are depleted in Cx3cr1CreER/+::Csf1rflox/flox mice fed tamoxifen. Fig. S2 shows data supporting Fig. 1. Fig. S3 demonstrates engraftment of beMφs in Cx3cr1CreER/+::Csf1rflox/flox mice with or without head shielding and BMT. Table S1 shows DE analyses of RNA-sequencing data comparing beMφs and microglia. Table S2 shows functional analysis of beMφs and microglia.
Supplementary Material
Acknowledgments
We would like to thank all the members of the J. Kipnis laboratory for their valuable comments during multiple discussions of this work. We thank Anita Impagliazzo for the artwork (graphical abstract).
This work was supported by grants from the National Institutes of Health (AG034113 and NS081026 to J. Kipnis; T32-AI007496 to A.J. Filiano; and 5F30AI109984 to J.C. Cronk) and the Hartwell Foundation (to A.J. Filiano).
The authors declare no competing financial interests.
Author contributions: J.C. Cronk designed and performed experiments and wrote the manuscript. A.J. Filiano designed and performed experiments and wrote the manuscript. A. Louveau helped with experimental design and performed experiments. I. Marin designed and performed experiments related to intestinal macrophages. R. Marsh assisted with experiments involving flow cytometry and image analysis. E. Ji assisted with experiments involving flow cytometry and image analysis. D.H. Goldman performed experiments related to live imaging. I. Smirnov performed most surgeries. N. Geraci designed and performed bioinformatic analysis. S. Acton designed analysis related to live imaging. C.C. Overall designed, performed, and oversaw bioinformatic analysis and wrote the manuscript. J. Kipnis oversaw the project, designed experiments, and wrote the manuscript.
References
- Acton S.T. 2001. Fast Algorithms for Area Morphology. Digit. Signal Process. 11:187–203. 10.1006/dspr.2001.0386 [DOI] [Google Scholar]
- Acton S.T., and Mukherjee D.P.. 2000. Area operators for edge detection. Pattern Recognit. Lett. 21:771–777. 10.1016/S0167-8655(00)00036-2 [DOI] [Google Scholar]
- Acton S.T., and Ray N.. 2006. Biomedical Image Analysis: Tracking. Morgan & Claypool Publishers. 152 pp. [Google Scholar]
- Ajami B., Bennett J.L., Krieger C., Tetzlaff W., and Rossi F.M.. 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10:1538–1543. 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Alliot F., Godin I., and Pessac B.. 1999. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain Res. 117:145–152. 10.1016/S0165-3806(99)00113-3 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed April 2016).
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium . 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie L., Sawtell A., Mann J., Frame T.C.M., Teal B., de Labastida Rivera F., Brown N., Walwyn-Brown K., Moore J.W.J., MacDonald S., et al. . 2016. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J. Hepatol. 65:758–768. 10.1016/j.jhep.2016.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco N., Swee L.K., Benard A., Ceredig R., and Rolink A.. 2010. Auto-reconstitution of the T-cell compartment by radioresistant hematopoietic cells following lethal irradiation and bone marrow transplantation. Exp. Hematol. 38:222–232. [DOI] [PubMed] [Google Scholar]
- Bruttger J., Karram K., Wörtge S., Regen T., Marini F., Hoppmann N., Klein M., Blank T., Yona S., Wolf Y., et al. . 2015. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity. 43:92–106. 10.1016/j.immuni.2015.06.012 [DOI] [PubMed] [Google Scholar]
- Butovsky O., Koronyo-Hamaoui M., Kunis G., Ophir E., Landa G., Cohen H., and Schwartz M.. 2006. Glatiramer acetate fights against Alzheimer’s disease by inducing dendritic-like microglia expressing insulin-like growth factor 1. Proc. Natl. Acad. Sci. USA. 103:11784–11789. 10.1073/pnas.0604681103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Kunis G., Koronyo-Hamaoui M., and Schwartz M.. 2007. Selective ablation of bone marrow-derived dendritic cells increases amyloid plaques in a mouse Alzheimer’s disease model. Eur. J. Neurosci. 26:413–416. 10.1111/j.1460-9568.2007.05652.x [DOI] [PubMed] [Google Scholar]
- Butovsky O., Siddiqui S., Gabriely G., Lanser A.J., Dake B., Murugaiyan G., Doykan C.E., Wu P.M., Gali R.R., Iyer L.K., et al. . 2012. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122:3063–3087. 10.1172/JCI62636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B.S., and Irizarry R.A.. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 26:2363–2367. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.K., Tvrdik P., Peden E., Cho S., Wu S., Spangrude G., and Capecchi M.R.. 2010. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 141:775–785. 10.1016/j.cell.2010.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., Morimoto E.T., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S., et al. . 2013. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 4:385–401. 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk J.C., Derecki N.C., Ji E., Xu Y., Lampano A.E., Smirnov I., Baker W., Norris G.T., Marin I., Coddington N., et al. . 2015. Methyl-CpG Binding Protein 2 Regulates Microglia and Macrophage Gene Expression in Response to Inflammatory Stimuli. Immunity. 42:679–691. 10.1016/j.immuni.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., and Gan W.B.. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8:752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- Derecki N.C., Cronk J.C., Lu Z., Xu E., Abbott S.B., Guyenet P.G., and Kipnis J.. 2012. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 484:105–109. 10.1038/nature10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S., Lavine K.J., Beaudin A.E., Sojka D.K., Carrero J.A., Calderon B., Brija T., Gautier E.L., Ivanov S., Satpathy A.T., et al. . 2014. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 40:91–104. 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano A.J., Xu Y., Tustison N.J., Marsh R.L., Baker W., Smirnov I., Overall C.C., Gadani S.P., Turner S.D., Weng Z., et al. . 2016. Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature. 535:425–429. 10.1038/nature18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B.M., and Irizarry R.A.. 2004. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 20:307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Gehlenborg N. 2016. UpSetR: A More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. R package version 1.2.2. Available at: https://rdrr.io/cran/UpSetR/ (accessed April 2016).
- Gibbings S.L., Goyal R., Desch A.N., Leach S.M., Prabagar M., Atif S.M., Bratton D.L., Janssen W., and Jakubzick C.V.. 2015. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood. 126:1357–1366. 10.1182/blood-2015-01-624809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., and Merad M.. 2011. [Microglia arise from extra-embryonic yolk sac primitive progenitors]. Med. Sci. (Paris). 27:719–724. 10.1051/medsci/2011278013 [DOI] [PubMed] [Google Scholar]
- Goldmann T., Wieghofer P., Müller P.F., Wolf Y., Varol D., Yona S., Brendecke S.M., Kierdorf K., Staszewski O., Datta M., et al. . 2013. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16:1618–1626. 10.1038/nn.3531 [DOI] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., and Guinney J.. 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 14:7 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. . 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22:1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., and Painter M.W.. Immunological Genome Project Consortium . 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., et al. . 2015. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 42:665–678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao E.Y., McBride S.W., Chow J., Mazmanian S.K., and Patterson P.H.. 2012. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc. Natl. Acad. Sci. USA. 109:12776–12781. 10.1073/pnas.1202556109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Xu Z., Xiong S., Sun F., Qin G., Hu G., Wang J., Zhao L., Liang Y.X., Wu T., et al. . 2018. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci. 10.1038/s41593-018-0090-8 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., and Speed T.P.. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L., et al. . 2015. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212:287–295. 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., and Schwartz M.. 2012. Non-identical twins - microglia and monocyte-derived macrophages in acute injury and autoimmune inflammation. Front. Immunol. 3:89 10.3389/fimmu.2012.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2015. Pretty Heatmaps. R package version 1.0.8. Available at: https://github.com/raivokolde/pheatmap (accessed April 2016).
- Krivit W., Sung J.H., Shapiro E.G., and Lockman L.A.. 1995. Microglia: the effector cell for reconstitution of the central nervous system following bone marrow transplantation for lysosomal and peroxisomal storage diseases. Cell Transplant. 4:385–392. 10.1177/096368979500400409 [DOI] [PubMed] [Google Scholar]
- Krivit W., Peters C., and Shapiro E.G.. 1999. Bone marrow transplantation as effective treatment of central nervous system disease in globoid cell leukodystrophy, metachromatic leukodystrophy, adrenoleukodystrophy, mannosidosis, fucosidosis, aspartylglucosaminuria, Hurler, Maroteaux-Lamy, and Sly syndromes, and Gaucher disease type III. Curr. Opin. Neurol. 12:167–176. 10.1097/00019052-199904000-00007 [DOI] [PubMed] [Google Scholar]
- Larochelle A., Bellavance M.A., Michaud J.P., and Rivest S.. 2016. Bone marrow-derived macrophages and the CNS: An update on the use of experimental chimeric mouse models and bone marrow transplantation in neurological disorders. Biochim. Biophys. Acta. 1862:310–322. 10.1016/j.bbadis.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Lassmann T., Hayashizaki Y., and Daub C.O.. 2011. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics. 27:130–131. 10.1093/bioinformatics/btq614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., and Smyth G.K.. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., and Storey J.D.. 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3:1724–1735. 10.1371/journal.pgen.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., and Storey J.D.. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 28:882–883. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A., Gehlenborg N., Strobelt H., Vuillemot R., and Pfister H.. 2014. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 20:1983–1992. 10.1109/TVCG.2014.2346248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen K., Zhu L., and Pollard J.W.. 2006. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis. 44:328–335. 10.1002/dvg.20219 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Elliott M.R., Chen Y., Walsh J.T., Klibanov A.L., Ravichandran K.S., and Kipnis J.. 2011. Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nat. Cell Biol. 13:1076–1083. 10.1038/ncb2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., et al. . 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 353:aad8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Mildner A., Schmidt H., Nitsche M., Merkler D., Hanisch U.K., Mack M., Heikenwalder M., Brück W., Priller J., and Prinz M.. 2007. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10:1544–1553. 10.1038/nn2015 [DOI] [PubMed] [Google Scholar]
- Platt F.M., and Lachmann R.H.. 2009. Treating lysosomal storage disorders: current practice and future prospects. Biochim. Biophys. Acta. 1793:737–745. 10.1016/j.bbamcr.2008.08.009 [DOI] [PubMed] [Google Scholar]
- Priller J., Flügel A., Wehner T., Boentert M., Haas C.A., Prinz M., Fernández-Klett F., Prass K., Bechmann I., de Boer B.A., et al. . 2001. Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat. Med. 7:1356–1361. 10.1038/nm1201-1356 [DOI] [PubMed] [Google Scholar]
- Prinz M., and Priller J.. 2014. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15:300–312. 10.1038/nrn3722 [DOI] [PubMed] [Google Scholar]
- Radjavi A., Smirnov I., Derecki N., and Kipnis J.. 2014. Dynamics of the meningeal CD4(+) T-cell repertoire are defined by the cervical lymph nodes and facilitate cognitive task performance in mice. Mol. Psychiatry. 19:531–533. 10.1038/mp.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls A., Shechter R., London A., Segev Y., Jacob-Hirsch J., Amariglio N., Rechavi G., and Schwartz M.. 2008. Two faces of chondroitin sulfate proteoglycan in spinal cord repair: a role in microglia/macrophage activation. PLoS Med. 5:e171 10.1371/journal.pmed.0050171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., Zheng F., De Baetselier P., Martens L., Saeys Y., De Prijck S., Lippens S., Abels C., Schoonooghe S., Raes G., et al. . 2016. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7:10321 10.1038/ncomms10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter R., London A., Varol C., Raposo C., Cusimano M., Yovel G., Rolls A., Mack M., Pluchino S., Martino G., et al. . 2009. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 6:e1000113 10.1371/journal.pmed.1000113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J., Ruedl C., and Karjalainen K.. 2015. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity. 43:382–393. 10.1016/j.immuni.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Silver J.D., Ritchie M.E., and Smyth G.K.. 2009. Microarray background correction: maximum likelihood estimation for the normal-exponential convolution. Biostatistics. 10:352–363. 10.1093/biostatistics/kxn042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F., et al. . 2016. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 7:11295 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K., Kozaki T., Lee C.Z.W., Thion M.S., Otsuka M., Lim S., Utami K.H., Fidan K., Park D.S., Malleret B., et al. . 2017. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity. 47:183–198. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45(D1):D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault P., ElAli A., and Rivest S.. 2015. The dynamics of monocytes and microglia in Alzheimer’s disease. Alzheimers Res. Ther. 7:41 10.1186/s13195-015-0125-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Laar L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B.N., and Guilliams M.. 2016. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity. 44:755–768. 10.1016/j.immuni.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Walkley S.U., Thrall M.A., Dobrenis K., Huang M., March P.A., Siegel D.A., and Wurzelmann S.. 1994. Bone marrow transplantation corrects the enzyme defect in neurons of the central nervous system in a lysosomal storage disease. Proc. Natl. Acad. Sci. USA. 91:2970–2974. 10.1073/pnas.91.8.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. . 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., and Smyth G.K.. 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40:e133 10.1093/nar/gks461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R., Lu H., Butovsky O., Ohno N., Rietsch A.M., Cialic R., Wu P.M., Doykan C.E., Lin J., Cotleur A.C., et al. . 2014. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med. 211:1533–1549. 10.1084/jem.20132477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. . 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.