Abstract
Cytochrome P450 (P450) enzymes are responsible for metabolizing drugs. Expression of P450s can directly affect drug metabolism, resulting in various outcomes in therapeutic efficacy and adverse effects. Several nuclear receptors are transcription factors that can regulate expression of P450s at both basal and drug-induced levels. Some long noncoding RNAs (lncRNAs) near a transcription factor are found to participate in the regulatory functions of the transcription factors. The aim of this study is to determine whether there is a transcriptional regulatory network containing nuclear receptors and lncRNAs controlling both basal and drug-induced expression of P450s in HepaRG cells. Small interfering RNAs or small hairpin RNAs were applied to knock down four nuclear receptors [hepatocyte nuclear factor 1α (HNF1α), hepatocyte nuclear factor 4α (HNF4α), pregnane X receptor (PXR), and constitutive androstane receptor (CAR)] as well as two lncRNAs [HNF1α antisense RNA 1 (HNF1α-AS1) and HNF4α antisense RNA 1 (HNF4α-AS1)] in HepaRG cells with or without treatment of phenobarbital or rifampicin. Expression of eight P450 enzymes was examined in both basal and drug-induced levels. CAR and PXR mainly regulated expression of specific P450s. HNF1α and HNF4α affected expression of a wide range of P450s as well as other transcription factors. HNF1α and HNF4α controlled the expression of their neighborhood lncRNAs, HNF1α-AS1 and HNF4α-AS1, respectively. HNF1α-AS1 and HNF4α-AS1 was also involved in the regulation of P450s and transcription factors in diverse manners. Altogether, our study concludes that a transcription regulatory network containing the nuclear receptors and lncRNAs controls both basal and drug-induced expression of P450s in HepaRG cells.
Introduction
Enzymes belonging to the cytochrome P450 (P450) superfamily are responsible for primarily metabolizing 70%–80% of prescription drugs (Evans and Relling, 1999). Interindividual variability in P450-mediated drug metabolism exists in the general population that leads to variability in therapeutic efficacy and adverse drug reactions (Zanger and Schwab, 2013). In addition to genetic polymorphisms in the P450 genes, the variability in P450 expression is a major cause of interindividual differences in P450-mediated drug metabolism (Nebert and Vesell, 2004). Therefore, it is critical to understand how P450 expression is regulated, particularly at the transcriptional level.
Transcription of P450 expression is regulated under two different conditions: normal physiologic condition (basal level) and challenged conditions when exposed to drugs or xenobiotics (induced level). It has been accepted that the basal expression of P450s is regulated by two key transcription factors: hepatocyte nuclear factor 1α (HNF1α) and hepatocyte nuclear factor 4α (HNF4α). HNF1α and HNF4α have been proven to be key transcriptional regulators in the control of expression of a wide range of hepatic genes, including P450s, under normal physiologic conditions across different species (Chung and Bresnick, 1997; Cheung et al., 2003; Bell and Michalopoulos, 2006; Kamiyama et al., 2007; Wortham et al., 2007; Matsunaga et al., 2008; Rana et al., 2010; Chiang et al., 2014; Dong et al., 2015). HNF1α and HNF4α mainly control the transcription of their target genes by directly binding to the promoter regions (Waxman, 1999; Honkakoski and Negishi, 2000). The existence of binding sites for liver-enriched transcription factors in numerous liver-specific genes further supports the important roles of HNF1α and HNF4α in hepatic gene regulation (Cereghini, 1996; Schrem et al., 2002).
The induced levels of P450s are controlled by two xenobiotic sensor nuclear receptors: constitutive androstane receptor (CAR) and pregnane X receptor (PXR). Activation of these nuclear receptors by drugs can result in induced expression of target P450 genes (Jana and Paliwal, 2007; Timsit and Negishi, 2007; Tompkins and Wallace, 2007). CAR mainly regulates induction of the CYP2B subfamily and can be activated by phenobarbital (PB) (Sueyoshi et al., 1999). PXR mainly regulates induction of the CYP3A subfamily and can be activated by rifampicin (RIF) (Lehmann et al., 1998).
Studies have shown the complexity of the network interactions between nuclear receptors in the regulation of P450s. HNF1α deficiency in mice has been shown to alter expression of several nuclear receptors (Cheung et al., 2003). HNF4α has been shown to regulate PXR and CAR, which affect the induction of downstream P450s (Tirona et al., 2003). Furthermore, HNF4α was reported to positively regulate HNF1α (Kuo et al., 1992). However, no research has studied the network among PXR, CAR, HNF1α, and HNF4α in a single system.
Long noncoding RNAs (lncRNAs) refer to RNA transcripts longer than 200 nucleotides that have no protein coding function (Guttman et al., 2009; Cabili et al., 2011). Recent studies have shown that lncRNAs are involved in the regulation of their neighborhood genes (Ørom et al., 2010; Kim et al., 2012; Batista and Chang, 2013; Villegas and Zaphiropoulos, 2015; Engreitz et al., 2016). The involvement of lncRNAs has been demonstrated in multiple physiologic processes, including metabolism and disease pathogenesis (Kornfeld and Brüning, 2014; Kwok and Tay, 2017). The gene encoding lncRNA HNF1α antisense RNA 1 (HNF1α-AS1) is located on human chromosome 12, next to the HNF1α gene. HNF1α-AS1 was first identified to regulate cell proliferation and migration of human esophageal adenocarcinoma cells (Yang et al., 2014). Following studies showed that HNF1α-AS1 also is involved in tumorigenesis and metastatic progression of several other cancer types (Dang et al., 2015; Wang et al., 2017; Zhang et al., 2017, 2018). The gene encoding lncRNA HNF4α antisense RNA 1 (HNF4α-AS1) is located on human chromosome 20, which overlaps with the HNF4α gene. Based on previous data on lncRNAs, we hypothesized that HNF1α-AS1 and HNF4α-AS1 are involved in a transcription network with other transcription factors to regulate the expression of P450s.
HepaRG cells were selected as the experimental model due to a previous study showing that HepaRG cells express comparable levels of P450s and transcription factors as primary human hepatocytes (Hart et al., 2010). Furthermore, HepaRG cells also respond to different P450 inducers and inhibitors (Aninat et al., 2006; Andersson et al., 2012; Gerets et al., 2012). To study the control of expression of P450s by a regulatory network containing nuclear receptors and lncRNAs, we used small interfering RNA (siRNA) and small hairpin RNA (shRNA) to perform gene knockdown in HepaRG cells and investigated the effects on expression of selected nuclear receptors, lncRNAs, and P450s.
Materials and Methods
Chemicals and Reagents.
HepaRG cells were kindly provided by Biopredic International (Rennes, France). ADD710 growth additives and ADD720 differentiation additives were purchased from Biopredic International. William’s E Medium, collagen I coated T-25 flasks, collagen I coated 12-well plates, Glutamax Supplement, and Opti-MEM medium were obtained from Thermo Fisher Scientific (Carlsbad, CA). siRNAs, including a negative control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), CAR (siRNA ID: s19369), PXR (siRNA ID: 138582), HNF4α (siRNA ID: s6698), HNF1α (siRNA ID: s13868), HNF1α-AS1 (siRNA ID: n265372), and HNF4α-AS1 (siRNA ID: n356309), Lipofectamine RNAiMAX transfection reagent, and Lipofectamine stem transfection reagent were provided by Thermo Fisher Scientific. shRNA negative control and shRNAs targeting HNF1α-AS1 were obtained from GeneCopoeia (Rockville, MD). Phenobarbital sodium salt and rifampicin were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies against GAPDH, CAR, and PXR were obtained from Abcam (Cambridge, MA). Anti-HNF1α antibody and TRIzol reagent were purchased from Invitrogen (Carlsbad, CA). Anti-HNF4α antibody and anti-rabbit IgG antibody were obtained from Cell Signaling Technology (Danvers, MA).
Cell Culture.
HepaRG cells were cultured following a protocol from Biopredic International. Briefly, a three-step protocol was used to generate fully differentiated HepaRG cells. Cells were first thawed and cultured in a HepaRG growth medium (William’s E Medium supplied with Glutamax and ADD710 growth additives) for 2 weeks to reach full confluence. Cells were next cultured in a 1:1 mixture of the HepaRG growth medium and a HepaRG differentiation medium (William’s E Medium supplied with Glutamax and ADD720 growth additives) for 1 week. In the last step, cells were cultured in the HepaRG differentiation medium for another week to reach a full differentiation status. During the whole process, cells were incubated at 37°C and 5% CO2 with the medium changed every 3 days. Fully differentiated cells were either used directly in T-25 flash or trypsinized and seeded into collagen I coated 12-well plates for further treatment.
siRNA Transfection.
For siRNA transfection, fully differentiated HepaRG cells were transfected with different siRNAs (negative control, HNF1α, HNF1α-AS1, HNF4α, HNF4α-AS1, CAR, and PXR) using the Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer’s protocol. Sixteen hours after transfection, the siRNA-containing medium was replaced with a normal HepaRG differentiation medium. RNAs and proteins harvested at 48 hours after siRNA transfection were used for knockdown efficiency and analysis of basal expression of P450s.
shRNA Transfection.
For shRNA transfection, undifferentiated HepaRG cells were stably transfected with four different shRNAs targeting HNF1α-AS1 or a negative control shRNA using the Lipofectamine stem transfection reagent. Positive transfected cells were selected by puromycin treatment (3 μg/ml). After antibiotic selection, cells were cultured to fully differentiated status and harvested for RNA isolation.
Drug Treatment.
For drug treatment, siRNA transfected HepaRG cells were treated with 1 mM phenobarbital sodium salt, 10 µM rifampicin, or phosphate-buffered saline (vehicle) for 24 hours. After drug treatment, RNAs were isolated for analysis of P450 induction.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (RT-PCR).
Total RNAs were isolated from HepaRG cells using a TRIzol reagent according to the manufacturer’s protocol. RNA concentrations were measured by a Nano Drop spectrophotometer (Nano Drop Technologies, Wilmington, DE) at 260 nm and RNA integrity was evaluated using an Agilent 2200 Tape Station (Agilent Technologies, Santa Clara, CA). One microgram of total RNAs was subjected to cDNA synthesis using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). RT-PCR was performed using a CFX96 Real-Time System (Bio-Rad Laboratories) with the primer sequences shown in Supplemental Table 1. Expression of GAPDH, HNF1α, HNF4α, CAR, PXR, aryl hydrocarbon receptor (AHR), HNF1α-AS1, CYP1A2, 2B6, 2C8, 2C9, 2C19, 2E1, and 3A4 were measured using an iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) and expression of HNF4α-AS1 was measured using a TaqMan Gene expression assay (Life Technologies, Carlsbad, CA). Relative mRNA expression levels were determined by normalizing examined gene expression against GAPDH expression using a ΔΔCt method.
Protein Sample Preparation and Quantification by Western Blot.
Cell lysates were prepared from HeapRG cells cultured in collagen I coated T-25 flasks with a radioimmunoprecipitation assay buffer (supplied with protease inhibitor cocktail). Protein concentrations were determined using a Qubit 2.0 Fluorometer (Invitrogen). Eighty micrograms of proteins were loaded and run on a polyacrylamide gel using a Mini-PROTEAN Tetra System (Bio-Rad Laboratories). Proteins were then transferred onto a polyvinylidene difluoride membrane and blocked in 5% bovine serum albumin for 1 hour. After blocking, the membrane was incubated with an antibody diluted in 2.5% bovine serum albumin (anti-GAPDH, 1:4000; anti-HNF1α, 1:1000; anti-HNF4α, 1:1000; anti-CAR, 1:500; and anti-PXR, 1:1000) overnight. Then, the membrane was incubated in an anti-rabbit IgG antibody (1:2000) diluted in 2.5% bovine serum albumin. Protein bands were visualized using a ChemiDoc MP Imaging System (Bio-Rad Laboratories). Semi quantification of proteins was calculated as the relative expression against GAPDH using an ImageJ program (National Institutes of Health, Bethesda, MD).
Statistical Analysis.
The data are shown as mean ± S.D. The significance of differences between mean values was determined using a two-tailed unpaired Student’s t test. Each t test was done independently with no correction for multiple comparisons. Statistical analyses were performed using Prism7, version 7.01 from GraphPad Software, Inc. (La Jolla, CA). Differences were regarded as statistically significant if P < 0.05.
Results
Impact on Regulation of P450s by Knockdown of CAR.
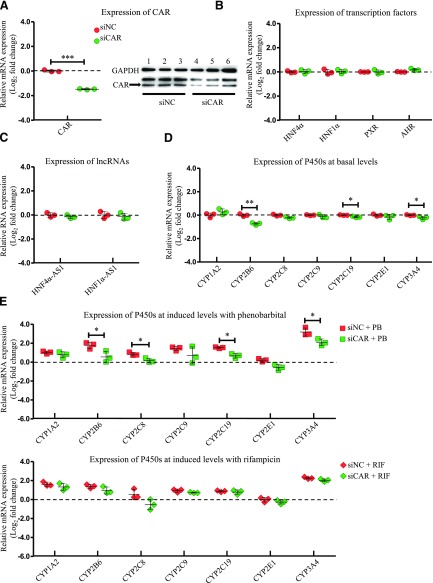
To evaluate the role of CAR in affecting the expression of the selected transcription factors, lncRNAs, and P450s, the HepaRG cells were transfected with a siRNA targeting CAR, and then the mRNA expression of the selected genes was evaluated by RT-PCR. Knockdown efficiency of CAR after siRNA transfection into HepaRG cells was confirmed with a decrease of 76% in mRNA level determined by RT-PCR and a decrease of 65% in protein level detected by western blot (Fig. 1A). Next, the mRNA expression of other examined transcription factors involved in the P450 regulation was analyzed. As shown in Fig. 1B, mRNA expression of HNF4α, HNF1α, PXR, and AHR was not affected by CAR knockdown. The expression of lncRNAs, HNF4α-AS1, and HNF1α-AS1, were also not affected by CAR knockdown (Fig. 1C). Among the selected drug-metabolizing P450s, basal mRNA expression of CYP2B6, 2C19, and 3A4, which were known CAR-regulated P450s, showed statistically significant decreases (Fig. 1D). In contrast, mRNA expression of CYP1A2 was increased in HepaRG cells with CAR knockdown (Fig. 1D). To measure the effect of CAR knockdown on drug-induced P450 expression, HepaRG cells were treated with PB or RIF after CAR knockdown by siRNA. As shown in Fig. 1E, the induction levels were lower for CYP2B6 (3.4- to 1.5-fold change in the induction level after CAR knockdown by PB treatment), CYP2C19 (2.9- to 1.6-fold change in the induction level after CAR knockdown by PB treatment), and CYP3A4 (9.3- to 4.3-fold change in the induction level after CAR knockdown by PB treatment).
Fig. 1.
Changes of gene expression by CAR knockdown. (A) Suppression of CAR expression in both mRNA (left) and protein (right) levels by siRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after CAR knockdown. (C) Expression of HNF4α-AS1 or HNF1α-AS1 after CAR knockdown. (D) Expression of the selected P450s at basal levels after CAR knockdown. (E) Expression of the selected P450s at induced levels with treatment of either PB or RIF after CAR knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
Impact on Regulation of P450s by Knockdown of PXR.
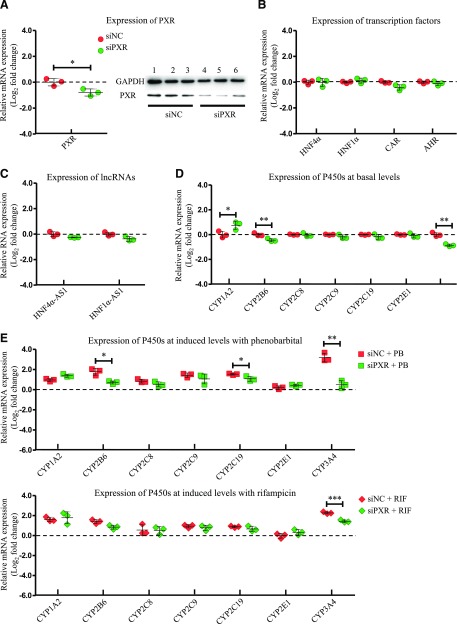
The role of PXR in expression of the selected transcription factors, lncRNAs, and P450s was studied with a similar experimental design as CAR. Knockdown efficiency of PXR was evaluated by RT-PCR and western blot after siRNA transfection into HepaRG cells. As shown in Fig. 2A, expression of PXR mRNA and protein was decreased to approximately 43% and 40% of the control, respectively. Expression of HNF4α, HNF1α, and AHR was not affected by PXR knockdown, while expression of CAR decreased after PXR knockdown (Fig. 2B). For lncRNAs, there were slight decreases in expression of HNF4α-AS1 and HNF1α-AS1 with no statistical significance (Fig. 2C). For basal P450 expression, only the PXR-regulated P450s CYP2B6 and 3A4 were decreased, whereas CYP1A2 was increased (Fig. 2D). Furthermore, induction of the PXR-regulated P450s was significantly repressed by PXR knockdown compared with the control group, as seen in CYP2B6 (3.4- to 1.6-fold change in the induction level after PXR knockdown by PB treatment) and CYP3A4 (9.3- to 1.4-fold change in the induction level after PXR knockdown by PB treatment and 4.7- to 1.6-fold change in the induction level after PXR knockdown by RIF treatment) with statistical significance.
Fig. 2.
Changes of gene expression by PXR knockdown. (A) Suppression of PXR expression in both mRNA (left) and protein (right) levels by siRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after PXR knockdown. (C) Expression of HNF4α-AS1 or HNF1α-AS1 after PXR knockdown. (D) Expression of the selected P450s at basal levels after PXR knockdown. (E) Expression of the selected P450s at induced levels with treatment of either PB or RIF after PXR knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
Impact on Regulation of P450s by Knockdown of HNF1α.
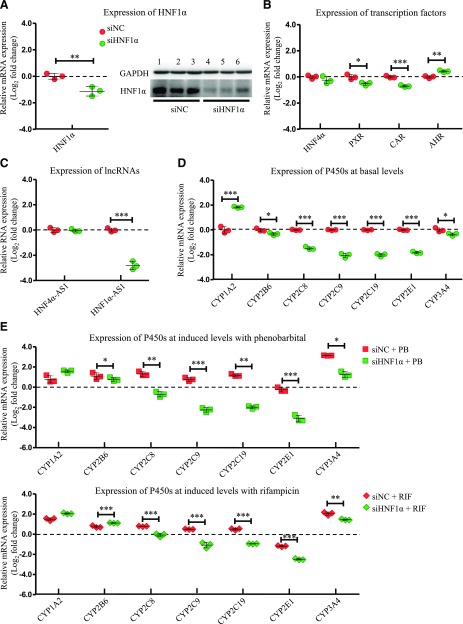
Next, the role of HNF1α was studied in an experiment with the same design as CAR and PXR. Transfection of small interfering HNF1α resulted in a decrease of 54% in HNF1α mRNA level and a decrease of 59% in HNF1α protein level (Fig. 3A). HNF1α knockdown also repressed mRNA expression of PXR and CAR, while mRNA expression of AHR was increased. The expression of HNF4α was not affected (Fig. 3B). For lncRNAs, the expression of HNF4α-AS1 was not affected (Fig. 3C). However, HNF1α knockdown caused a decrease of 85% of HNF1α-AS1 expression as shown in Fig. 3C. HNF1α knockdown also affected the expression of multiple P450s in both basal and induced levels. Basal expression of CYP2B6, 2C8, 2C9, 2C19, 2E1, and 3A4 showed decreases with statistical significance (Fig. 3D). The expression of CYP2B6 and 3A4 was still induced by treatment with either PB or RIF, but at a much lower fold compared with the control group. Furthermore, the induction of CYP2C8, 2C9, and 2C19 was completely abolished by HNF1α knockdown (Fig. 3E).
Fig. 3.
Changes of gene expression by HNF1α knockdown. (A) Suppression of HNF1α expression in both mRNA (left) and protein (right) levels by siRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after HNF1α knockdown. (C) Expression of HNF4α-AS1 or HNF1α-AS1 after HNF1α knockdown. (D) Expression of the selected P450s at basal levels after HNF1α knockdown. (E) Expression of the selected P450s at induced levels with treatment of either PB or RIF after HNF1α knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
Impact on Regulation of P450s by Knockdown of HNF4α.
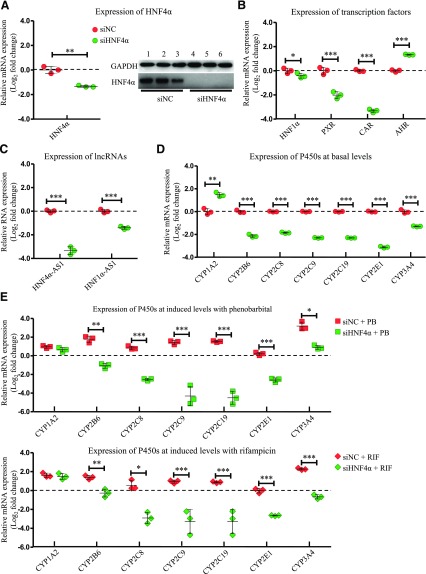
Cells transfected with small interfering HNF4α showed 60% less expression of HNF4α mRNA and almost depleted expression of HNF4α protein (Fig. 4A). In the analysis of the selected transcription factors, expression levels of HNF1α, PXR, and CAR all decreased, while AHR expression increased with HNF4α knockdown (Fig. 4B). Expression of both lncRNAs was almost depleted with approximately a 90% decrease in HNF4α-AS1 expression and a decrease of 63% in HNF1α-AS1 expression (Fig. 4C). HNF4α knockdown also decreased basal expression of CYP2B6, 2C8, 2C9, 2C19, 2E1, and 3A4, but to a much greater extent than HNF1α knockdown (Fig. 4D). P450 induction by treatment with PB or RIF was also largely diminished by HNF4α knockdown (Fig. 4E).
Fig. 4.
Changes of gene expression by HNF4α knockdown. (A) Suppression of HNF4α expression in both mRNA (left) and protein (right) levels by siRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after HNF4α knockdown. (C) Expression of HNF4α-AS1 or HNF1α-AS1 after HNF4α knockdown. (D) Expression of the selected P450s at basal levels after HNF4α knockdown. (E) Expression of the selected P450s at induced levels with treatment of either PB or RIF after HNF4α knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
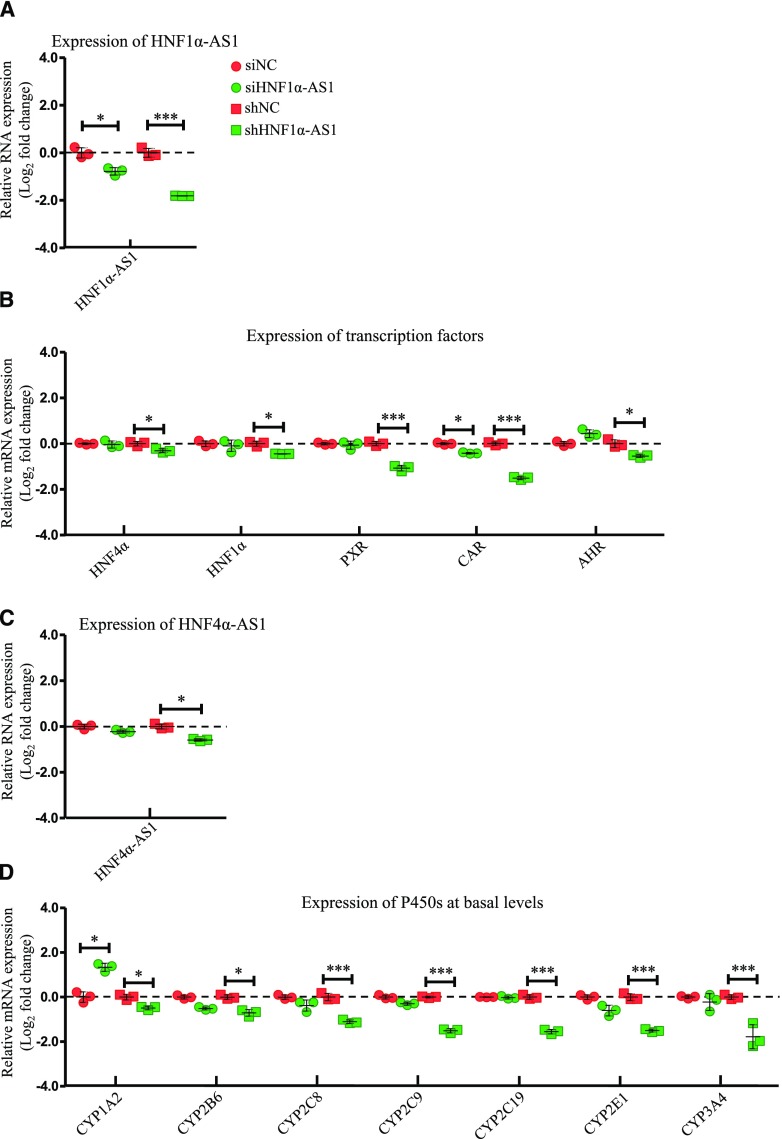
Impact on Regulation of P450s by Knockdown of HNF1α-AS1.
To study the role of the lncRNAs in the regulatory network in the control of expression of P450s, HepaRG cells were first transfected with siRNA or shRNA to knock down HNF1α-AS1. As shown in Fig. 5A, siRNA and shRNA transfection decreased HNF1α-AS1 expression by approximately 40% and 75%, respectively. In terms of transcription factors, HepaRG cells transfected with small interfering HNF1α-AS1 only showed decreased mRNA expression of CAR, while HNF1α, HNF4α, PXR, and AHR remained unchanged. However, HepaRG cells transfected with small hairpin HNF1α-AS1 showed decreased mRNA expression in all examined transcription factors, among which HNF4α, HNF1α, and AHR had only a minor decrease (15%, 28%, and 22%, respectively), while CAR and PXR had a major decrease (55% and 65%, respectively) (Fig. 5B). Expression of lncRNA HNF4α-AS1 was only affected by small hairpin HNF1α-AS1 transfection with a decrease of 33% (Fig. 5C). Basal expressions of several P450s were also affected by HNF1α-AS1 knockdown. HepaRG cells transfected with small interfering HNF1α-AS1 showed an increase in CYP1A2 expression and a decrease in CYP2E1 expression with statistical significance, accompanied by minor decreases in CYP2B6, 2C8, and 2C9 (Fig. 5D). HepaRG cells transfected with small hairpin HNF1α-AS1 showed extensively repressed effects on all examined P450s (Fig. 5D).
Fig. 5.
Changes of gene expression by HNF1α-AS1 knockdown. (A) Suppression of HNF1α-AS1 expression by siRNA or shRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after HNF1α-AS1 knockdown. (C) Expression of HNF4α-AS1 after HNF1α-AS1 knockdown. (D) Expression of the selected P450s at basal levels after HNF1α-AS1 knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
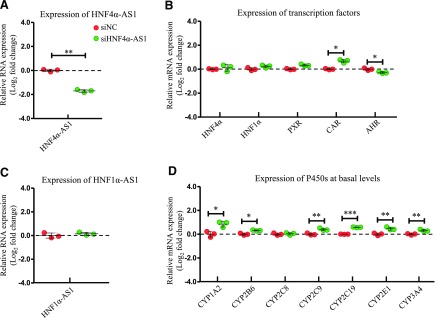
Impact on Regulation of P450s by Knockdown of HNF4α-AS1.
The role of HNF4α-AS1 in the regulatory network in the control of the expression of P450s was also studied in HepaRG cells transfected with siRNA targeting HNF4α-AS1. HNF4α-AS1 expression was decreased approximately 70% by siRNA transfection (Fig. 6A). Next, the expressions of the selected transcription factors were determined. Expression of HNF4α and HNF1α were increased slightly and expression of CAR was increased dramatically after HNF4α-AS1 knockdown, while expression of AHR was decreased with statistical significance (Fig. 6B). Knockdown of HNF4α-AS1 did not affect expression of HNF1α-AS1 (Fig. 6C). mRNA expressions of the selected P450s were measured. As shown in Fig. 6D, all selected P450s, except CYP2C8, had increased the expression after HNF4α-AS1 knockdown with statistical significance.
Fig. 6.
Changes of gene expression by HNF4α-AS1 knockdown. (A) Suppression of HNF4α-AS1 expression by siRNA transfection into HepaRG cells. (B) Expression of the selected transcription factors after HNF4α-AS1 knockdown. (C) Expression of HNF1α-AS1 after HNF4α-AS1 knockdown. (D) Expression of the selected P450s at basal levels after HNF4α-AS1 knockdown. The sample size for each group was n = 3. Data are presented as log2 fold change and shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 vs. control in two-tailed unpaired Student’s t tests.
Discussion
Transcription factors play a prominent role in the regulation of P450 expression (Honkakoski and Negishi, 2000; Schrem et al., 2002). In the present study, we studied the functions of CAR, PXR, HNF1α, and HNF4α. In addition, HNF1α-AS1 and HNF4α-AS1, which are the neighborhood lncRNAs of HNF1α and HNF4α, were also selected to test their roles in the regulation of P450 expression. Efficient knockdown of target genes was performed by siRNA or shRNA transfection into HepaRG cells. The effects on transcription of the selected transcription factors, lncRNAs, and drug-metabolizing P450 genes were measured by quantitative RT-PCR.
CAR and PXR have been reported to regulate expression of specific P450 subfamilies of CYP2Bs and CYP3As, respectively (Lehmann et al., 1998; Sueyoshi et al., 1999). In our experiment, knockdown of CAR or PXR only showed a minor effect on expression of other transcription factors or lncRNAs (Fig. 1, B and C; Fig. 2, B and C). Knockdown of CAR decreased basal and phenobarbital-induced expression of CYP2B6 (Fig. 1, D and E), whereas knockdown of PXR reduced basal and rifampicin-induced expression of CYP3A4 (Fig. 2, D and E) in HepaRG cells. In addition, CAR knockdown also showed a minor effect on phenobarbital-induced expression of CYP2C and 3A families (Fig. 1E), while PXR knockdown showed slightly diminished phenobarbital-induced expression of CYP2B and 2C families (Fig. 2E).
The role of HNF1α in the regulation of multiple hepatic expressions of P450s has previously been studied in an HNF1α-deficient mouse model (Cheung et al., 2003). Our data showed that HNF1α knockdown in HepaRG cells led to decreased expression of PXR and CAR, while it increased expression of AHR. Knockdown of HNF1α in HepaRG cells also decreased the basal mRNA levels of CYP2B6, 2C8, 2C9, 2C19, 2E1, and 3A4, while it increased the mRNA level of CYP1A2 (Fig. 3D). Compared with the HNF1α-deficient mice data, we observed a similar regulation pattern in the CYP2C and 2E subfamilies and an opposite regulation pattern in the CYP1A, 2B, and 3A subfamilies. Furthermore, the phenobarbital- and rifampicin-induced levels of the most examined P450s were also greatly inhibited by HNF1α knockdown, where the inhibition of CYP2B and 3A subfamilies might be partially due to decreased expression of CAR and PXR (Fig. 3E).
The role of HNF4α in the regulation of hepatic-specific P450 genes has been studied in several species. In HepaRG cells, we found that HNF4α knockdown decreased the expression of HNF1α, CAR, and PXR, while it increased the expression of AHR (Fig. 4B). Similar to HNF1α knockdown, HNF4α knockdown led to decreased expression of CYP2B6, 2C8, 2C9, 2C19, 2E1, and 3A4, while it increased the CYP1A2 expression (Fig. 4D). In addition, phenobarbital- and rifampicin-induced P450 expression was greatly inhibited, which might be partially contributed by the decreased expression of CAR and PXR (Fig. 4E).
In the current study, we found that knockdown of HNF1α and HNF4α was able to decrease the expression of PXR and CAR. This agrees with previous studies showing HNF4α crosstalks with PXR and CAR in the regulation of P450 expression (Li and Chiang, 2006; Miao et al., 2006). In addition, HNF4α has also been reported to directly regulate expression of PXR and CAR and subsequent induction of P450s through these two nuclear receptors (Kamiya et al., 2003; Tirona et al., 2003).
The interaction between HNF1α and HNF4α has been documented in several previous studies (Kuo et al., 1992; Bailly et al., 2001). A study showed that HNF4α positively regulates HNF1α in rat hepatoma cells, evidenced by the fact that deleting the HNF4α binding site in the promoter region of the HNF1α gene abolished the promoter activity (Kuo et al., 1992). Another study showed that HNF1α was able to negatively regulate HNF4α, evidenced by the identification of a binding site of HNF1α in the HNF4α promoter region (Bailly et al., 2001). However, in the study on HNF1α-deficient mice, the expression of HNF4α was not changed (Cheung et al., 2003). In our result, HNF1α knockdown only had a minor effect on mRNA expression of HNF4α (Fig. 3B), which agreed with the mouse data. HNF4α knockdown decreased mRNA expression of HNF1α with statistical significance (Fig. 4B), which agreed with the rat hepatoma cell data. Our data imply the existence of a transcription regulatory network containing transcription factors of HNF4α, HNF1α, and CAR/PXR in an order to control the basal and drug-induced expression of major P450s involved in drug metabolism.
In recent years, utilizing the Encyclopedia of DNA Elements projects, studies have revealed that over 70% of the human genome are capable of being transcribed into RNAs (Djebali et al., 2012). However, not all transcribed primary RNAs have protein coding ability. In human, only approximate 2% of the transcribed RNAs are translated into proteins (Szymanski and Barciszewski, 2002). Other than housekeeping noncoding RNAs (ribosomal RNAs, transfer RNAs, and small nuclear RNAs), whose biologic functions have been well defined, regulatory noncoding RNAs (siRNAs, microRNAs, and lncRNAs) have recently been shown to be actively involved in physiologic regulations (Djebali et al., 2012; Kornfeld and Brüning, 2014). Furthermore, lncRNAs have been shown to perform transcriptional regulation through interaction with transcription factors in different tissues (Clark and Blackshaw, 2014; Herriges et al., 2014).
We identified two lncRNAs as our target lncRNAs of interest, HNF1α-AS1 and HNF4α-AS1, which are located in the neighborhood regions of the HNF1α and HNF4α genes, respectively. Evidence has shown that antisense lncRNAs are likely to be involved in the regulatory functions of their neighborhood coding genes (Villegas and Zaphiropoulos, 2015). Hence, we examined the role of two selected lncRNAs in the regulation of P450 expression. We found that the expression of HNF1α-AS1 and HNF4α-AS1 is largely dependent on its neighborhood transcription factors of HNF1α and HNF4α, respectively. HNF1α knockdown depleted the expression of HNF1α-AS1 without alteration of expression of HNF4α-AS1 (Fig. 3C). The knockdown of HNF4α decreased expression of both HNF1α-AS1 and HNF4α-AS1 (Fig. 4C). We believed that HNF4α-AS1 was dependent on the expression of HNF4α and decreased expression of HNF1α was responsible for the decrease of HNF1α-AS1.
In the lncRNA knockdown experiments, HNF1α-AS1 knockdown by siRNA and shRNA showed very similar regulation trends on most examined genes (Fig. 5). Knockdown of HNF1α-AS1 by shRNA decreased mRNA expression of all examined transcription factors and basal levels of P450s as well as expression of HNF4α-AS1 (Fig. 5, B–D). The effect of HNF1α-AS1 knockdown was very similar to HNF1α knockdown, indicating that HNF1α-AS1 might work as a downstream cofactor of HNF1α in the regulation of other transcription factors and P450s. The knockdown of HNF4α-AS1 showed opposite regulatory effects on almost all examined genes compared with knockdown of HNF1α, HNF4α, and HNF1α-AS1 (Fig. 6, B–D), indicating the potential inhibitory role of HNF4α-AS1 in the regulation of P450s by the transcription factor network. These results confirmed our hypothesis that the lncRNAs of HNF1α-AS1 and HNF4α-AS1 participate in the regulation of drug-metabolizing P450 enzymes.
Even though we have observed involvement of the lncRNAs in the regulation of P450 expression, the underlying mechanisms are still unclear. Several mechanisms in regulation of gene expression by lncRNAs have been identified at different levels of gene expression, including transcriptional and post-transcriptional regulation levels (Engreitz et al., 2016; Schmitz et al., 2016). Generally, the regulatory roles of lncRNAs are dependent on the interactions with their partner molecules, such as proteins, RNAs, and DNAs (Villegas and Zaphiropoulos, 2015). Proteins are believed to be the major binding partners of lncRNAs due to the existence of distinct protein binding domains. By binding with protein molecules, lncRNAs can either work as scaffold molecules to promote the formation of protein complexes or work as decoy molecules to remove proteins from a specific location. Depending on the interacted protein functions, lncRNAs can have different impacts on expression of their regulated target genes (Ray et al., 2013). Interactions between lncRNAs and other RNAs can regulate targeted RNA properties, including RNA stability and translation efficiency. For example, an antisense lncRNA located next to mouse coding gene Uchl1 is able to regulate the translation efficiency of UCHL1 protein through activation of polysomes (Carrieri et al., 2012). Several specific examples have revealed the interactions between lncRNAs and DNAs, but whether these interactions are widespread in a genome is still unclear. For example, a noncoding RNA, promoter associated RNA, has been shown to mediate de novo methylation of its targeted DNAs by forming a DNA:RNA triplex, which can be recognized by DNA methyltransferases (Schmitz et al., 2010). This evidence suggests that lncRNAs are associated with a complexity of gene regulation processes. Based on current assays, in future experiments we will study how HNF1α-AS1 and HNF4α-AS1 regulate their targeted genes and their involvement in the regulation network in control of basal and drug-induced expression of P450s. In conclusion, the present study demonstrates the existence of a transcription regulatory network containing different nuclear receptors and lncRNAs in the regulation of both basal and drug-induced expression of P450s in HepaRG cells.
Acknowledgments
We thank Biopredic International (Rennes, France) for kindly providing HepaRG cells in this study.
Abbreviations
- AHR
aryl hydrocarbon receptor
- CAR
constitutive androstane receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HNF1α
hepatocyte nuclear factor 1α
- HNF1α-AS1
hepatocyte nuclear factor 1α antisense RNA 1
- HNF4α
hepatocyte nuclear factor 4α
- HNF4α-AS1
hepatocyte nuclear factor 4α antisense RNA 1
- lncRNA
long noncoding RNA
- P450
cytochrome P450
- PB
phenobarbital
- PXR
pregnane X receptor
- RIF
rifampicin
- RT-PCR
real-time polymerase chain reaction
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
Authorship Contributions
Participated in research design: Chen, Bao, Piekos, Zhang, Zhong.
Conducted experiments: Chen, Bao, Piekos.
Performed data analysis: Chen, Bao, Piekos, Zhong.
Wrote or contributed to the writing of the manuscript: Chen, Bao, Piekos, Zhu, Zhang, Zhong.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences of USA [Grant R01GM-118367] (to X.-b.Z.) and the National Natural Science Foundation of China [Grants 81173127 and 81773815] (to L.Z.). This study was also partially supported by the Institute for System Genomics at the University of Connecticut (to X.-b.Z.).
S.C.P. is an American Foundation for Pharmaceutical Education Pre-Doctoral Fellow.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Andersson TB, Kanebratt KP, Kenna JG. (2012) The HepaRG cell line: a unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin Drug Metab Toxicol 8:909–920. [DOI] [PubMed] [Google Scholar]
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C, Guillouzo A. (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34:75–83. [DOI] [PubMed] [Google Scholar]
- Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC. (2001) An enhancer element 6 kb upstream of the mouse HNF4α1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Res 29:3495–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AW, Michalopoulos GK. (2006) Phenobarbital regulates nuclear expression of HNF-4α in mouse and rat hepatocytes independent of CAR and PXR. Hepatology 44:186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491:454–457. [DOI] [PubMed] [Google Scholar]
- Cereghini S. (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J 10:267–282. [PubMed] [Google Scholar]
- Cheung C, Akiyama TE, Kudo G, Gonzalez FJ. (2003) Hepatic expression of cytochrome P450s in hepatocyte nuclear factor 1-alpha (HNF1α)-deficient mice. Biochem Pharmacol 66:2011–2020. [DOI] [PubMed] [Google Scholar]
- Chiang TS, Yang KC, Chiou LL, Huang GT, Lee HS. (2014) Enhancement of CYP3A4 activity in Hep G2 cells by lentiviral transfection of hepatocyte nuclear factor-1 alpha. PLoS One 9:e94885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Bresnick E. (1997) Identification of positive and negative regulatory elements of the human cytochrome P4501A2 (CYP1A2) gene. Arch Biochem Biophys 338:220–226. [DOI] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S. (2014) Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet 5:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu Y, Wang L, Wang Y, Huang Q. (2015) Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol 13:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. (2012) Landscape of transcription in human cells. Nature 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Chen Q, Liu X, Wen J, Jiang J, Deng Y. (2015) Role of specificity protein 1, hepatocyte nuclear factor 1α, and pregnane X receptor in the basal and rifampicin-induced transcriptional regulation of porcine cytochrome P450 3A46. Drug Metab Dispos 43:1458–1467. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV. (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286:487–491. [DOI] [PubMed] [Google Scholar]
- Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, Atienzar FA. (2012) Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol 28:69–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Li Y, Nakamoto K, Subileau EA, Steen D, Zhong XB. (2010) A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos 38:988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. (2014) Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28:1363–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. (2000) Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J 347:321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Paliwal J. (2007) Molecular mechanisms of cytochrome P450 induction: potential for drug-drug interactions. Curr Protein Pept Sci 8:619–628. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Inoue Y, Gonzalez FJ. (2003) Role of the hepatocyte nuclear factor 4α in control of the pregnane X receptor during fetal liver development. Hepatology 37:1375–1384. [DOI] [PubMed] [Google Scholar]
- Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. (2007) Role of human hepatocyte nuclear factor 4α in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab Pharmacokinet 22:287–298. [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. (2012) Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld JW, Brüning JC. (2014) Regulation of metabolism by long, non-coding RNAs. Front Genet 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Jr, Crabtree GR. (1992) A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355:457–461. [DOI] [PubMed] [Google Scholar]
- Kwok ZH, Tay Y. (2017) Long noncoding RNAs: lincs between human health and disease. Biochem Soc Trans 45:805–812. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiang JY. (2006) Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos 34:756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga N, Ikeda M, Takiguchi T, Koyanagi S, Ohdo S. (2008) The molecular mechanism regulating 24-hour rhythm of CYP2E1 expression in the mouse liver. Hepatology 48:240–251. [DOI] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. (2006) Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1α. J Biol Chem 281:14537–14546. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Vesell ES. (2004) Advances in pharmacogenomics and individualized drug therapy: exciting challenges that lie ahead. Eur J Pharmacol 500:267–280. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana R, Chen Y, Ferguson SS, Kissling GE, Surapureddi S, Goldstein JA. (2010) Hepatocyte nuclear factor 4α regulates rifampicin-mediated induction of CYP2C genes in primary cultures of human hepatocytes. Drug Metab Dispos 38:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Grote P, Herrmann BG. (2016) Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 73:2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J. (2002) Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev 54:129–158. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046. [DOI] [PubMed] [Google Scholar]
- Szymanski M, Barciszewski J. (2002) Beyond the proteome: non-coding regulatory RNAs. Genome Biol 3:reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. (2007) CAR and PXR: the xenobiotic-sensing receptors. Steroids 72:231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4α determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224. [DOI] [PubMed] [Google Scholar]
- Tompkins LM, Wallace AD. (2007) Mechanisms of cytochrome P450 induction. J Biochem Mol Toxicol 21:176–181. [DOI] [PubMed] [Google Scholar]
- Villegas VE, Zaphiropoulos PG. (2015) Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci 16:3251–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Mou L, Chai HX, Wang F, Yin YZ, Zhang XY. (2017) Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed Pharmacother 89:926–932. [DOI] [PubMed] [Google Scholar]
- Waxman DJ. (1999) P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys 369:11–23. [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4α, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710. [DOI] [PubMed] [Google Scholar]
- Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al. (2014) Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 63:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhang G, An X, Zhao H, Zhang Q, Zhao H. (2018) Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed Pharmacother 98:594–599. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. (2017) Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/β-catenin signaling pathway. Biomed Pharmacother 96:877–883. [DOI] [PubMed] [Google Scholar]