Abstract
Somatic rearrangements contribute to the mutagenized landscape of cancer genomes. Here, we systematically interrogated rearrangements in 560 breast cancers by using a piecewise constant fitting approach. We identified 33 hotspots of large (>100 kb) tandem duplications, a mutational signature associated with homologous-recombination-repair deficiency. Notably, these tandem-duplication hotspots were enriched in breast cancer germline susceptibility loci (odds ratio (OR) = 4.28) and breast-specific ‘super-enhancer’ regulatory elements (OR = 3.54). These hotspots may be sites of selective susceptibility to double-strand-break damage due to high transcriptional activity or, through incrementally increasing copy number, may be sites of secondary selective pressure. The transcriptomic consequences ranged from strong individual oncogene effects to weak but quantifiable multigene expression effects. We thus present a somatic-rearrangement mutational process affecting coding sequences and noncoding regulatory elements and contributing a continuum of driver consequences, from modest to strong effects, thereby supporting a polygenic model of cancer development.
Whole-genome sequencing (WGS) has permitted unrestricted access to the human cancer genome and has consequently led to a search for driver mutations that may confer selective advantage throughout the human genome. Recurrent somatic mutations in coding sequences are often interpreted as driver mutations, particularly when they are supported by transcriptomic changes or functional evidence. However, recurrent somatic mutations in noncoding sequences are less straightforward to interpret. TERT promoter mutations in malignant melanoma1,2 and NOTCH1 3′-region mutations in chronic lymphocytic leukemia3 have successfully been demonstrated to act as driver mutations. By contrast, multiple noncoding loci have been highlighted as being recurrently mutated, though evidence that these mutations are true drivers remains lacking. Indeed, in a recent exploration of 560 breast cancer whole genomes4, the largest cohort of WGS cancers to date, statistically significant recurrently mutated noncoding sites (by substitutions and insertions or deletions (indels)) have been identified, but alternative explanations for localized increases in mutability, such as a propensity to form secondary DNA structures, have been observed4.
Previous efforts have focused on recurrent substitutions and indels, and an analysis seeking sites that are recurrently mutated through rearrangements has not been formally performed. Such sites may be indicative of driver loci under selective pressure (such as amplification of ERBB2 and CCND1) or may represent highly mutable sites that are simply prone to double-strand break (DSB) damage. Sites that are under selective pressure generally have a high incidence in a particular tissue type; are highly complex; and reflect multiple classes of rearrangement, including deletions, inversions, tandem duplications and translocations. By contrast, sites that are simply breakable may show a low frequency of occurrence and a preponderance of a particular class of rearrangement, thus indicating susceptibility to specific mutational processes.
An anecdotal observation in the cohort of 560 breast cancers has indicated sites in the genome that appear to be rearranged recurrently, albeit at a low frequency, and by a very specific rearrangement class of tandem duplications. Rarely, tandem duplications recur at approximately the same locus in the same cancer and result in the appearance of nested tandem duplications. No explanation has been provided for this observation. Here, we used a new approach to systematically identify sites in the human cancer genome that are recurrently mutagenized by rearrangements, specifically tandem duplications, to fully characterize the prevalence and the effects of these sites of recurrent tandem duplications in this cohort of breast cancers.
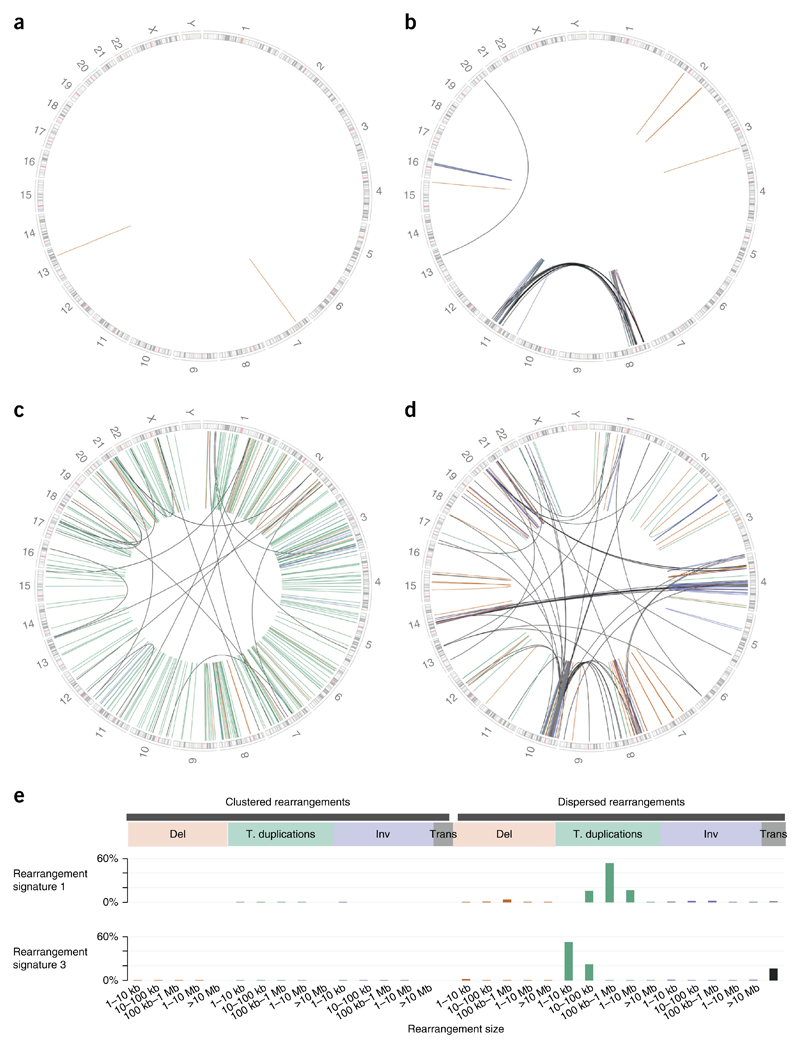
In total, 77,695 rearrangements, including 59,900 intrachromosomal (17,564 deletions, 18,463 inversions and 23,873 tandem duplications) and 17,795 interchromosomal translocations, have previously been identified in this cohort. The distribution of rearrangements within each cancer is complex (Fig. 1a–d); some cancers have few rearrangements without distinctive patterns, and others have collections of focally occurring rearrangements such as amplifications, whereas many have rearrangements distributed throughout the genome. This variability indicates very different sets of underlying mutational processes.
Figure 1.
Spectrum of distribution of rearrangements in human breast cancers. Circos plots depicting somatic rearrangements, with chromosomal ideograms on the outermost right and lines representing rearrangements (green, tandem duplications (t. duplications); orange, deletions (del); blue, inversions (inv); gray, interchromosomal events (trans)). (a) Quiescent tumor. (b) Tumor with focal ‘clustered’ rearrangements. (c) Tumor with mainly tandem duplications distributed throughout the genome (‘dispersed’ rearrangements). (d) Tumor with a mixed pattern of dispersed rearrangements and clustered rearrangements. (e) Rearrangement signatures 1 and 3 primarily comprise tandem duplications but are predominantly characterized by tandem duplications of different lengths (>100 kb and <10 kb, respectively).
Thus, large focal collections of ‘clustered’ rearrangements were first separated from rearrangements that were widely distributed or ‘dispersed’ in each cancer, then distinguished on the basis of class (inversion, deletion, tandem duplication or translocation) and size (1–10 kb, 10–100 kb, 0.1–1 Mb, 1–10 Mb and >10 Mb)4, before a mathematical method for extracting mutational signatures was applied5. Six rearrangement signatures (RS1–RS6) were extracted, representing discrete rearrangement mutational processes in breast cancer4. Two distinctive mutational processes in particular were associated with dispersed tandem duplications. RS1 and RS3 were primarily characterized by large (>100 kb) and small (<10 kb) tandem duplications, respectively (Fig. 1e). Although both signatures are associated with tumors deficient in homologous recombination (HR) repair4,6–9, RS3 is specifically associated with inactivation of BRCA1. Therefore, because each signature represents distinct biological defects in human cells, we proceeded with a systematic analysis of sites of recurrent mutagenesis by regarding these two mutational signatures as independent processes.
Previously, tumors have been described as having a large degree of genomic instability10,11 and even a tandem-duplicator phenotype12–14, but earlier studies have lacked sufficient resolution to distinguish among different tandem-duplication signatures. Here, we show the importance of using a mutational signature approach, highlighting differences in behavior between short (<10 kb) and long (>100 kb) tandem duplications.
We identified an unexpectedly high number of rearrangement hotspots dominated by the RS1 mutational process characterized by long (>100 kb) tandem duplications4. Intuitively, a hotspot of mutagenesis that is enriched in a particular mutational signature suggests a propensity for DNA DSB damage and specific recombination-based-repair mutational mechanisms that may explain these tandem-duplication hotspots. However, we found additional intriguing features associated with these hotspots.
Results
Identification of rearrangement hotspots
To systematically identify hotspots of tandem duplications throughout the genome, we first considered the background distribution of rearrangements, which is known to be nonuniform. We performed a regression analysis to detect and quantify the associations between the distribution of rearrangements and a variety of genomic landmarks, including replication-time domains, gene-rich regions, background copy number, chromatin state and repetitive sequences (Online Methods and Supplementary Fig. 1). The associations learned were considered in creating an adjusted background model and were also applied during simulations; these steps were critical to the following phase of hotspot detection. Adjusted background models and simulated distributions were calculated for RS1 and RS3 tandem-duplication signatures separately because the vastly differing numbers of rearrangements in each signature (5,944 and 13,498, respectively) might have biased the detection of hotspots for the different signatures.
We next used the principle of intermutation distance15 (IMD)—the distance from one breakpoint to the one immediately preceding it in the reference genome—and used a piecewise constant fitting (PCF) approach16,17, a method of segmentation of sequential data that is frequently used in analyses of copy-number data. We applied PCF to the IMDs of RS1 and RS3 separately, seeking segments of the breast cancer genomes where groups of rearrangements exhibited short IMDs, which indicate hotspots that are more frequently rearranged than the adjusted background model (Fig. 2 and Supplementary Note). The parameters used for the PCF algorithm were optimized against simulated data (Online Methods and Supplementary Fig. 2). We sought to detect a conservative number of hotspots while minimizing the number of false-positive hotspots. Notably, all highly clustered rearrangements, such as those causing driver amplicons, had previously been identified in each sample and removed, and thus did not contribute to these hotspots. However, to ensure that a hotspot did not comprise only a few samples with multiple breakpoints each, a minimum of eight samples was required to contribute to each hotspot. This method obviated the use of genomic bins and permitted detection of hotspots of varying genomic size.
Figure 2.
Identifying hotspots of rearrangements. (a) Schematic of dispersed rearrangements in the genomes of five hypothetical patients, with regions identified as hotspots by the PCF algorithm highlighted in beige. Notably, differing sizes of each putative hotspot are permitted through this method that negates the use of bins. (b) Workflow of PCF application to rearrangement signatures. (c) Rainfall plots of chromosome 8 rearrangements for tandem-duplication signatures RS1 (>100 kb) (top) and RS3 (<10 kb) (bottom). Inter-rearrangement distance is plotted on a logarithmic scale on the y axis. Black lines indicate PCF-defined hotspots. RS1 is 20 times more likely to form hotspots than RS3, and these hotspots are visible as punctuated collections of breakpoints in these plots.
We applied the PCF method to RS1 and RS3 rearrangements separately, seeking loci with a rearrangement density exceeding twice the local adjusted background density for each signature and involving a minimum of eight samples. Interestingly, 0.5% of 13,498 short RS3 tandem duplications contributed to four RS3 hotspots. By contrast, 10% of 5,944 long RS1 tandem duplications formed 33 hotspots, thus demonstrating that long RS1 tandem duplications were 20 times more likely than short RS3 tandem duplications to form a rearrangement hotspot. Indeed, these hotspots were visible as punctuated collections of rearrangements in genome-wide plots of rearrangement breakpoints (Fig. 2c and Supplementary Table 1).
Contrasting RS3 hotspots and RS1 hotspots
RS3 hotspots had different characteristics from those of RS1 hotspots (Fig. 3). The four RS3 hotspots were highly focused, occurred in small genomic windows and exhibited very high rearrangement densities (61.8–658.3 breakpoints per megabase; Fig. 3b). In contrast, the 33 RS1 hotspots had densities between 7.6 and 83.2 breakpoints per megabase and demonstrated other striking characteristics (Fig. 3a). In several RS1 hotspots, duplicated segments showed genomic overlap among patients, even when most patients had only one tandem duplication, as depicted in a cumulative plot of duplicated segments for samples contributing rearrangements to a hotspot (Fig. 3c and Supplementary Fig. 3). Interestingly, the nested tandem duplications incidentally observed previously4 were a particular characteristic of the RS1 hotspots. The hotspots of RS1 and RS3 were distinct from one another, apart from one locus where two long noncoding RNAs, NEAT1 and MALAT1, reside (Supplementary Note).
Figure 3.
Hotspots of dispersed rearrangements. A large (>100 kb) tandem-duplication mutational process shows distinctive genomic overlap among patients and coincides with germline susceptibility loci and super-enhancer regulatory elements. (a) Summary of 33 hotspots of long tandem duplications (RS1). (b) Four hotspots of short tandem duplications (RS3). Top plot, density of rearrangement breakpoints within hotspots, and their positions on chromosomes. The black horizontal lines denote the expected breakpoint density according to the background model. Bottom plot, frequency of each hotspot in the cohort of 560 patients. Hotspots containing breast cancer–susceptibility SNPs are marked with blue circles, and breast-specific super-enhancers are marked with red triangles. Genes that may be relevant are highlighted, although their true relevance is uncertain. Chr, chromosome. (c) Two different hotspots of RS1. Left plots (chromosome 12: 11.8–12.8 Mb) coincide with two breast-tissue-specific super-enhancers, and right plots (chromosome 8: 116.6–117.7 Mb) coincide with a breast cancer germline susceptibility locus. Nearby cancer genes are annotated, although the relevance of these genes is uncertain. The top six panels at left and right depict genomic rearrangements for six individual patients at each locus. Copy number (y axis) is depicted as black dots (10-kb bins). Green lines indicate tandem-duplication breakpoints. Precise genomic overlap is apparent among patients. Bottom plots show cumulative numbers of samples with a rearrangement involving this genomic region, emphasizing at its peak the region of critical genomic overlap between samples. Thick red lines represent breast-tissue-specific super enhancers. The blue vertical line represents the position of the breast cancer germline susceptibility locus. The relevant reference SNP cluster ID is provided.
In assessing the potential genomic consequences of RS1 and RS3 tandem duplications on functional components of the genome12, we observed that RS1 rearrangements duplicated important driver genes and regulatory elements, whereas RS3 rearrangements mainly transected them (Fig. 4, Supplementary Table 2 and Online Methods). This result is likely to be related to the size of tandem duplications in these signatures. Short (<10 kb) RS3 tandem duplications were more likely to duplicate very small regions, with an effect equivalent to disruption of genes or regulatory elements. In contrast, RS1 tandem duplications were long (>100 kb) and were more likely to duplicate whole genes or regulatory elements.
Figure 4.
Genomic consequences of the tandem-duplication signatures. Tandem duplications can transect or duplicate genomic features such as regulatory elements or genes. (a) Tandem duplications attributed to rearrangement signature RS1 often duplicate genomic regions containing breast cancer–predisposition SNPs, breast-tissue super-enhancers and oncogenes. RS1 rearrangements in hotspots show a particular enrichment in comparison with RS1 rearrangements that occur in other regions and with simulated rearrangements. There are 524 RS1 duplications in hotspots and 4,916 duplications outside of hotspots. (b) Tandem duplications attributed to RS3 in hotspots are enriched in transecting cancer genes more than in the rest of genome, or in simulated data. There are 57 RS3 duplications in hotspots and 10,967 RS3 duplications outside of hotspots. Asterisks indicate statistically significant enrichment of any particular genomic feature within hotspots compared with outside hotspots, as calculated by two-sided Fisher’s exact test. ****P ≤ 0.0001; **P ≤ 0.01; *P ≤ 0.05. Error bars show the s.d. across ten different simulated data sets.
Strikingly, the effects were stronger for tandem duplications that contributed to hotspots of RS1 and RS3 than they were for tandem duplications that were not in hotspots or that were simulated. Thus, although the likelihood of transection or duplication may be governed by the size of tandem duplications, the particular enrichment in hotspots appears to carry important biological implications.
The enrichment in disruption of tumor suppressor genes in RS3 hotspots (OR = 167; P = 9.4 × 10−41 by Fisher’s exact test) is relatively simple to understand, because these hotspots are likely to be under selective pressure. Accordingly, two of the four RS3 hotspots occurred within well-known tumor suppressors, PTEN and RB1. Other rearrangement classes were also enriched in these genes, in agreement with their being driver events (Online Methods and Supplementary Table 3). Furthermore, these sites have been identified as putative driver loci in an independent analysis seeking driver rearrangements through gene-based methods4.
By contrast, the enrichment in oncogene duplication in RS1 hotspots (OR = 1.49; P = 4.1 × 10−3 by Fisher’s exact test) was apparent12 although not as strong as the enrichment in transections of cancer genes in RS3 hotspots. More notably, enrichment in other putative regulatory features was also observed. Indeed, susceptibility loci associated with breast cancer18,19 were 4.28 times more frequent in RS1 hotspots than in the rest of the tandem-duplicated genome (P = 3.4 × 10−4 by Poisson test; Supplementary Figs. 4a, 5 and 6). Additionally, 18 of 33 (54.5%) RS1 tandem-duplication hotspots contained at least one breast super-enhancer. The density of breast super-enhancers was 3.54 times higher in hotspots compared with the rest of the tandem-duplicated genome (P = 7.0 × 10−16 by Poisson test; Supplementary Figs. 4b, 5 and 6). This effect was much stronger than that for non-breast-tissue super-enhancers (OR = 1.62) or enhancers in general (OR = 1.02; Supplementary Table 4). This gradient highlights the tissue-specific relationship between tandem-duplication hotspots and regulatory elements classified as super-enhancers.
The reasons underlying these observations in RS1 hotspots, however, are less clear. Single or nested tandem duplications in RS1 hotspots effectively increase the number of copies of a genomic region but do so only incrementally. The enrichment in breast cancer–susceptibility loci, super-enhancers and oncogenes at hotspots of a very particular mutational signature may reflect an increased likelihood of damage and thus susceptibility to a passenger mutational signature that occurs because of the high transcriptional activity associated with such regions. However, it is also intriguing to consider that the resulting copy-number increase might confer a more modest selective advantage and contribute to the driver landscape. To investigate the latter possibility, we explored the effects of RS1 tandem duplications on gene expression.
Effects of RS1 hotspots on expression
Several RS1 hotspots involve validated breast cancer genes12 (for example, ESR1 and ZNF217; Supplementary Figs. 7 and 8) and may conceivably contribute to the driver landscape through increasing the number of copies of a gene, even if by only a single copy.
ESR1 is an example of a breast cancer gene that is a target of an RS1 hotspot. A breast-specific super-enhancer and a breast cancer–susceptibility locus are present in the vicinity of ESR1. We found that 14 samples contributed to this hotspot, of which ten had only one tandem duplication or simple nested tandem duplications of this site. Six samples had expression data, and all showed significantly elevated levels of ESR1 despite a modest copy-number increase (Supplementary Fig. 7a). Four samples had a small number of rearrangements (<30) yet had a highly specific tandem duplication of ESR1, thus suggesting selection (Supplementary Fig. 9). Most other samples with rearrangements in the other 32 hotspots were triple-negative tumors. By contrast, samples with rearrangements in the ESR1 hotspot showed a different preponderance: 11 of 14 were estrogen-receptor-positive tumors. Thus, we propose that the duplications in the ESR1 hotspot are putative drivers that would not have been detected through previous customary copy-number approaches but are likely to be important to identify because of the associated risk of developing resistance to antiestrogen chemotherapeutics20,21.
MYC encodes a transcription factor that coordinates a diverse set of cellular programs and is deregulated in many different cancer types22,23. Thirty patients contributed to the RS1 hotspot at the MYC locus, with modest copy-number gains. A spectrum of genomic outcomes was observed, including single or nested tandem duplications, flanking (16 samples) or wholly duplicating the gene body of MYC (14 samples) (Fig. 5a). Notably, a breast-tissue super-enhancer and two germline susceptibility loci are present in the vicinity of MYC19,24 (Fig. 5b). Because we had a larger number of samples with corresponding RNA-seq data, we modeled the expression levels of MYC, taking into account the breast cancer subtype and background copy number (whole-chromosome-arm gain is common for chromosome 8), and sought to determine whether tandem duplication was associated with increased transcription. We found that tandem duplications in the RS1 hotspot were associated with a doubling of the expression level of MYC (0.99 ± 0.28 (mean ± s.e.) log2 fragments per kilobase of transcript per million mapped reads (FPKM), P = 4.4 × 10−4 by t-test) (Supplementary Table 5 and Supplementary Fig. 10).
Figure 5.
From selective susceptibility to selective pressure. (a) The spectrum of genomic structural variation at MYC. Copy number (y axis) is depicted as black dots (10-kb bins). Lines represent rearrangement breakpoints (green, tandem duplications; pink, deletions; blue, inversions; purple, interchromosomal events). Genes other than MYC are marked as black lines at the top of the panel. (b) Cumulative number of samples with dispersed rearrangements within the MYC-related tandem-duplication hotspot. A peak is present very close to MYC but also flanking MYC, where two germline susceptibility loci are present. A large super-enhancer is also present upstream of MYC. (c) Proposed model of the cascade of events underlying the RS1-enriched hotspots in breast cancer. Sites enriched in super-enhancers (SENH) may be more highly transcribed and thus exposed to damage including DSB damage. Long tandem duplications, in contrast to other rearrangement classes, are particularly at risk of copying whole genes. Thus, although other rearrangement classes may be found (in low numbers in the same region), an enrichment in long tandem duplications is observed because of a small degree of selection in action.
The expression-related consequences of tandem duplications of putative regulatory elements, however, are more difficult to assess because of the uncertainty of the downstream targets of these regulatory elements. We therefore used a global gene-expression approach and applied a mixed effects model to understand the contribution of tandem duplications of these elements, while controlling for breast cancer subtype and background copy number. We found that tandem duplications involving a super-enhancer or breast cancer–susceptibility locus were associated with an increase in levels of global gene expression, even when the gene itself was not duplicated. The effect was strongest on oncogenes (0.30 ± 0.20 log2 FPKM; P = 0.12 by likelihood-ratio test) than for other genes (0.16 ± 0.04 log2 FPKM; P = 1.8 × 10−4) within RS1 hotspots or for genes in the rest of the genome (Supplementary Table 5).
Thus, tandem duplications of cancer genes demonstrated strong expression effects on individual genes (for example, ESR1 and MYC), whereas tandem duplications of putative regulatory elements demonstrated modest but quantifiable global gene-expression effects. The spectrum of functional consequences at these loci may hence range from insignificance, through mild enhancement, to strong selective advantage—all of which are consequences of the same somatic-rearrangement mutational process.
Long tandem-duplication hotspots are present and distinct in other cancers
We additionally explored other cancer cohorts for which sequence files were available. Pancreatic and ovarian cancers are two cancer types known to exhibit tandem duplications. We parsed raw sequence files through our mutation-calling algorithms and extracted rearrangement signatures as for breast cancers. We separately generated adjusted background models and simulations for these new data sets. The total numbers of available tumor samples (73 ovarian and 96 pancreatic)10,11 were much smaller than those for the breast cancer cohort, which is currently the largest cohort of WGS cancers of a single cancer type in the world. Thus, the power for detecting hotspots was substantially decreased, particularly for pancreatic cancer (Supplementary Fig. 11 for power calculation). Nevertheless, in ovarian tumors, we found 2,923 RS1 rearrangements and identified seven RS1 hotspots (Supplementary Table 6 and Supplementary Fig. 12), of which six were distinct from breast cancer RS1 hotspots. A marked enrichment in ovarian cancer–specific super-enhancers (11 super-enhancers over 20.2 Mb; OR = 2.9; P = 1.9 × 10−3 by Poisson test) was also noted for these hotspots. MUC1, a validated oncogene in ovarian cancer, was the focus of one of the hotspots. Thus, although larger cohorts of WGS cancers will be required to further examine this phenomenon, we propose that different cancer types may have different RS1 hotspots that are focused at highly transcribed sites specific to different tissues.
Discussion
Rearrangement signatures may, in principle, be mere passenger readouts of the stochastic mayhem in cancer cells. However, mutational signatures recurring at specific genomic sites, which also coincide with distinct genomic features, suggest a more directed nature—a sign of either selective susceptibility or selective pressure.
Perhaps as an attribute of being more highly active or transcribed (for example, super-enhancers), or some other as-yet-unknown quality (for example, germline single-nucleotide polymorphism (SNP) sites and other hotspots with no discerning features), these hotspots exemplify loci that are rendered more available for DSB damage and more dependent on repair that generates large tandem duplications6,25–27. These hotspots signify genomic sites that are inherently more susceptible to the HR-deficient tandem-duplication mutational process—sites of selective susceptibility.
An alternative argument may also hold true: the likelihood of damage and repair relating to this mutational process may be similar throughout the genome. However, by incrementally increasing the number of copies of genes driving tissue proliferation, survival and invasion (ESR1 and ZNF217) or noncoding regions with minor or intermediate modifying effects in cancer, such as germline susceptibility loci or super-enhancer elements, long tandem duplications may specifically enhance the overall likelihood of carcinogenesis, thus suggesting that these loci are subject to a degree of selective pressure and that this HR-deficient tandem-duplication mutational process is in fact a new mechanism of generating secondary somatic drivers.
Functional activity related to being a super-enhancer or a germline susceptibility locus may underlie primary susceptibility to mutagenesis of a given locus, but it requires a repair process that generates large tandem duplications to confer a selective advantage (Fig. 5c). Tandem-duplication mutagenesis is associated with DSB repair in the context of HR deficiency and is a potentially important mutagenic mechanism driving genetic diversity in evolving cancers by increasing the copy numbers of portions of the coding and noncoding genome. Tandem-duplication mutagenesis may directly increase the number of copies of an oncogene or alter noncoding sites where super-enhancers and risk loci28 are situated. Therefore, such mutagenesis may produce a spectrum of driver consequences29,30 ranging from strong effects in coding sequences to weaker effects in the coding and noncoding genome, thus supporting a polygenic model of cancer development.
In conclusion, structural mutability in the genome is not uniform. It is influenced by forces of selection and by mutational mechanisms, and recombination-based repair plays a critical role in specific genomic regions. Mutational processes may, however, not simply be passive contrivances, and some may be more harmful than others. We suggest that mutation signatures that confer a high degree of genome-wide variability are potentially more deleterious for somatic cells and thus are more clinically relevant. Translational efforts should be focused on identifying and managing these adverse mutational processes in human cancer.
Online Methods
Data set
The primary data set was obtained from a previous publication4. Briefly, 560 matched tumor and normal DNAs were sequenced with Illumina sequencing technology and aligned to the reference genome, and mutations were called with a suite of somatic-mutation-calling algorithms, as defined previously. In particular, somatic rearrangements were called via BRASS (Breakpoint AnalySiS) (https://github.com/cancerit/BRASS/) through discordant mapping of paired-end reads in the discovery phase. Clipped reads were not used to inform discovery. Primary-discovery somatic rearrangements were filtered against the germline copy-number variants (CNVs) in the matched normal samples as well as a panel of 50 normal samples from unrelated samples, to reduce the likelihood of calling germline CNVs and false positives.
In silico and/or PCR-based validation were performed in a subset of samples4. Primers were custom designed, and potential rearrangements were PCR amplified and identified as putatively somatic if a band observed in gel electrophoresis was seen in the tumor and not in the normal sample, in duplicate. Putative somatic rearrangements were then verified through capillary sequencing. Amplicons that were successfully sequenced were aligned back to the reference genome with Blat, to identify breakpoints at base-pair resolution. Alternatively, an in silico analysis was performed through local reassembly. Discordantly mapping read pairs that were likely to span breakpoints, as well as a selection of nearby properly paired reads, were grouped for each region of interest. With the Velvet de novo assembler31, reads were locally assembled within each of these regions, thus producing a contiguous consensus sequence of each region. Rearrangements, represented by reads from the rearranged derivative as well as the corresponding nonrearranged allele were instantly recognizable on the basis of a particular pattern of five vertices in the de Bruijn graph (a mathematical method used in de novo assembly of (short) read sequences) in Velvet. Exact coordinates and features of junction sequences (for example, microhomology or nontemplated sequences) were derived from this process, after alignment to the reference genome, as though they were split reads.
Only rearrangements that passed the validation stage were used in these analyses. Furthermore, additional post hoc filters were included to remove library-related artifacts (creating an excess of inversions in affected samples).
Rearrangement signatures
Previously, we had classified rearrangements as mutational signatures, as extracted with the non-negative matrix factorization framework.
Briefly, we first separated rearrangements that were focally clustered from widely dispersed rearrangements because we reasoned that the underlying biological processes generating these different rearrangement distributions are likely to be distinct. A PCF approach was applied to distinguish focally clustered rearrangements from dispersed ones. For each sample, both breakpoints of each rearrangement were considered separately, and all breakpoints were ordered on the basis of chromosomal position. The inter-rearrangement distance, defined as the number of base pairs from one rearrangement breakpoint to the one immediately preceding it in the reference genome, was calculated. Putative regions of clustered rearrangements were identified as having an average inter-rearrangement distance at least ten times less than the whole-genome average for the individual sample. PCF parameters used were γ = 25 and kmin = 10. The respective partner breakpoints of all breakpoints involved in a clustered region are likely to have arisen at the same mechanistic instant and therefore were considered as being involved in the cluster even if they were located at a distant chromosomal site.
In both classes of rearrangements (clustered and nonclustered), rearrangements were subclassified into deletions, inversions and tandem duplications, and then were further subclassified according to the size of the rearranged segment (1–10 kb, 10–100 kb, 0.1–1 Mb, 1–10 Mb and >10 Mb). The final category in both groups was interchromosomal translocations. The classification produces a matrix of 32 distinct categories of structural variants across 544 breast cancer genomes. This matrix was decomposed through the previously developed approach for deciphering mutational signatures by searching for the optimal number of mutational signatures that best explain the data5. A rearrangement was attributed to a signature if the posterior probability of the rearrangement being generated by the signature in a given sample exceeded 0.5 (ref. 8).
In total, six different rearrangement signatures were identified. RS1 and RS3 were two signatures that were mainly characterized by tandem duplications (Fig. 1e). RS1 was mainly characterized by large tandem duplications (>100 kb), whereas RS3 was mainly characterized by short tandem duplications. There is good reason to believe that these signatures are biologically distinct entities, because RS3 is very strongly associated with BRCA1 abrogation (germline or somatic mutation or promoter hypermethylation with concurrent loss of the wild-type allele). BRCA1 tumors also contain moderate numbers of RS1, but there are also samples with a larger excess of RS1 rearrangements that do not carry a specific genetic abnormality4.
To perform a systematic survey of tandem-duplication hotspots, we focused on these two rearrangement signatures. However, tandem duplications (and other rearrangements) are also not uniformly distributed through the genome. Thus, the following sections describe how we detected hotspots of tandem duplications of RS1 and RS3, after correcting for genomic biases.
Modeling the background distribution of rearrangements
Rearrangements are known to have an uneven distribution in the genome. There have been numerous descriptions linking genomic features such as replication timing with the nonuniform distribution of rearrangements. Thus, any analysis that seeks to detect regions of higher mutability than expected must take the genomic features that influence this nonuniform distribution into account in its background model. To formally detect and quantify associations between genomic features and somatic rearrangements in breast cancer, we conducted a multivariate genome-wide regression analysis (details in Supplementary Note).
Simulations of rearrangements
Simulations consisted of as many rearrangements as were observed for each sample in the data set, preserved the type of rearrangement (tandem duplication, inversion, deletion or translocation) and the length of each rearrangement (distance between partner breakpoints), and ensured that both breakpoints fell within mappable/callable regions in our pipeline. Simulations also took into account the genomic bias of rearrangements that were identified above (details in Supplementary Note).
Optimization of the PCF algorithm
The PCF algorithm is a method of segmentation of sequential data. We used PCF to find segments of the genome that had a rearrangement density much higher than that in the neighboring genomic regions and higher than expected according the background model. We demonstrated the significance of the identified hotspots by applying the same method to simulated data that followed the known genomic biases of rearrangements such as replication-time domains, transcription and background copy-number status.
Each rearrangement had two breakpoints, each of which was treated independently. Breakpoints were sorted according to reference-genome coordinates, and an intermutation distance (IMD) between two genome-sorted breakpoints was calculated for each breakpoint and then log-transformed to base 10. These log10 IMD values were fed into the PCF algorithm.
To call a segment of a genome with a higher rearrangement density a hotspot, a number of parameters had to be determined. The smoothness of segmentation was determined by the gamma (γ) parameter of the PCF analysis. A segment of genome was considered to be a peak only if it had a sufficient number of mutations, as specified by kmin. The average intermutation distance in the segment had to exceed an intermutation distance factor (i), which is the threshold used in comparing the breakpoint density in a segment with the genome-wide density of breakpoints:
where dseg is the density of breakpoints in a segment, defined as:
and dbg is the expected density of breakpoints in the segment, given the background model described above, which includes the genomic covariates of the segment. More specifically,
where bi is the expected number of breakpoints in the bins overlapping the segment, n is the number of overlapping bins, and s is the bin size (0.5 Mb).
The choice of parameters kmin, γ and i in the PCF algorithm was based on training on the observed data and on comparison of the outcomes with that of the simulated data.
Combinations of γ and i were explored to determine the optimal parameters for detection of hotspots. The sensitivity of detection of every hotspot in the observed data was balanced against the detection of false-positive hotspots in simulated data sets (Supplementary Fig. 2), as quantified according to the false discovery rate.
On the basis of the number of detected hotspots in observed and simulated data, we used γ = 8 and i = 2 in the final analyses, thus resulting in 33 hotspots of RS1 and 4 hotspots of RS3. In a further 1,000 simulated data sets, the same parameters resulted on average in 3.3 ± 1.9 (mean ± s.d.) and 0.1 ± 0.3) hotspots, respectively.
A data set that is not ‘clean’ and that contains many false-positive rearrangements may result in the identification of false-positive hotspots. Therefore, it is imperative to have a set of high-quality, highly curated rearrangement data (with better specificity than sensitivity), to avoid calling loci where algorithms tend to miscall rearrangements, as hotspots.
Workflow
Six rearrangement signatures were extracted from this data set of 560 breast tumors, as described above. Each rearrangement was probabilistically assigned to each rearrangement signature, given the six rearrangement signatures and the estimated contribution of each signature to each sample4.
To define hotspots of rearrangements in RS1 and RS3, the PCF algorithm was applied to the log10 IMD of RS1 or RS3 breakpoints separately, by using the following parameters: γ = 8, kmin = 8, and i = 2. Each locus was required to be represented by eight or more samples. The section below describes the hotspots that were identified by this method.
Identifying hotspots for individual rearrangement signatures
To explore hotspots associated with signatures of tandem duplications, we first separated rearrangements associated with the two signatures that were strongly characterized by tandem duplications (RS1 and RS3) (Fig. 2). PCF was performed on each of these two categories. We identified 33 hotspots of long RS1 tandem duplications and 4 hotspots of short RS3 tandem duplications, and they are listed and annotated in Supplementary Table 1.
RS3, characterized by short tandem duplications, also demonstrated four hotspots; two were likely drivers (PTEN and RB1), and the importance of the other two was less clear (CDK6 and NEAT1–MALAT1). The interpretation of duplications at the NEAT1–MALAT1 locus is provided in the Supplementary Note. Hotspots of remaining rearrangement signatures are also described in the Supplementary Note.
Genomic consequences of tandem duplications
We assessed the potential genomic consequences of the two rearrangement signatures associated with tandem duplications on gene function and on regulatory elements.
Rearrangements associated with the RS1 signature were usually long tandem duplications (>100 kb), which were more likely to duplicate whole genes and whole super-enhancer regulatory elements. In contrast, rearrangements associated with the RS3 signature were usually short tandem duplications (<10 kb) and therefore were more likely to duplicate smaller regions and had an effect equivalent to transecting genes or regulatory elements.
To formally assess the potential genomic consequences of RS1 and RS3 tandem duplications on gene function and on regulatory elements, we explored the following genomic elements: (i) breast cancer–susceptibility SNPs; (ii) breast-tissue-specific super-enhancer regulatory elements; (iii) oncogenes (if a duplication covered both a super-enhancer and an oncogene, it was counted in both categories); (iv) tumor suppressor genes; and (v) all genes.
An element was considered as being wholly duplicated by a tandem duplication if the element was completely between the two breakpoints. An element was considered as being transected by a tandem duplication if one or both breakpoints lay within the element. We did not consider the events in which only one breakpoint of duplication was within an element, because the effects of such events on genes and other elements are unclear.
We counted the number of times in which each of the five elements described above was duplicated or transected for RS1 and RS3, respectively, for: (i) RS1 or RS3 tandem duplications in hotspots (counted only once per sample, even if there were multiple tandem duplications affecting the same locus in the same person); (ii) RS1 or RS3 tandem duplications that were not within hotspots; (iii) RS1 and RS3 tandem duplications that had been simulated and corrected for all the characteristics described above.
Strikingly, RS1 hotspots were clearly enriched in duplication of whole oncogenes and whole super-enhancers, as compared with RS1 rearrangements not within hotspots and simulated RS1 rearrangements (Fig. 4 and Supplementary Table 2). This enrichment was not observed for RS3 hotspots. Furthermore, RS1 hotspot tandem duplications were scarcely found to transect genes or regulatory elements. In contrast, RS3 hotspots were strongly enriched in gene transections, in agreement with their being driver loci.
Thus, we provided evidence of different genomic consequences (whole gene/regulatory element duplications versus transections) of long or short tandem duplications in hotspots.
Germline loci for susceptibility to breast cancer
The list of breast cancer germline susceptibility alleles was derived from the literature18,32–40. Through this analysis, we sought to determine whether breast cancer–susceptibility SNP alleles are enriched in regions of recurrent rearrangement in breast cancer, to quantify this relationship and to provide a measure of statistical significance.
We performed an analysis comparing the density of SNPs in the genomic footprints of RS1 hotspots with the genomic footprints of other RS1 rearrangements in general (instead of simply comparing it with the rest of genome); this procedure controlled for unevenness in the distribution of tandem duplications. RS1 hotspots encompassed 58 Mb of the genome, whereas other segments of the genome covered by (at least) one tandem duplication encompassed 2,106 Mb.
The density of breast cancer–susceptibility SNPs outside of RS1 hotspots was 0.036 per megabase. Within RS1 hotspots, there were nine breast cancer–susceptibility SNPs, or 0.16 SNPs per megabase. Thus, the OR of finding a breast cancer–susceptibility SNP in RS1 hotspots compared with tandem-duplicated regions outside of RS1 hotspots was 4.28 (P = 3.4 × 10−4 by one-sided Poisson test).
Poisson tests were used to compare rates of events among genomic regions of different sizes and to account for uncertainty arising from the low number of events (nine SNPs) within the hotspots.
Enrichment in regulatory elements
The super-enhancer data set was obtained from the Super-Enhancer Archive (SEA)40. This archive uses publicly available H3K27ac ChIP-seq data sets and published super-enhancer lists to produce a comprehensive list of super-enhancers in multiple cell types and tissues. From this list (containing 2,282 unique super-enhancers for 15 human cell types or tissues), we extracted the super-enhancers active in breast cancer (755 elements) and the super-enhancers active in the other cell types or tissues (1,528 elements). Regulatory elements were verified to be mutually exclusive to each list, to ensure that each super-enhancer was analyzed in only one category, and a super-enhancer was placed in the breast cancer category, in which there was experimental evidence for multiple activations.
The list of general enhancers was obtained from the Ensembl Regulatory Build (GRCh37)41. We used the ‘Multicell’ list, containing 139,204 elements active in 17 different cell lines. From this list, we filtered out the enhancers that overlapped with super-enhancers, and we obtained a final list comprising 136,858 regulatory elements.
As described in the previous section, we divided the genome into RS1 hotspots (58 Mb) and other segments of the genome covered by a minimum of a single tandem duplication (2,106 Mb). We compared the densities of super-enhancers within RS1 hotspot segments and outside of the hotspots (Supplementary Fig. 4).
Method 1. The OR of finding a super-enhancer active in breast tissue in RS1 hotspots, compared with regions of the genome rarely covered by RS1 duplications was 3.54 (P = 7.0 × 10−16 by one-sided Poisson test). The OR for observing a super-enhancer that was not associated with breast tissue was lower at 1.62, with P = 6.4 × 10−4. The OR for finding any enhancer in a RS1 hotspot was 1.02, with P = 0.12.
Method 2. The above analysis was based on the assumption that super-enhancers follow a Poisson distribution; however, this assumption might be violated by clusters of super-enhancer elements that exist in the genome. We therefore performed a set of simulations that did not depend on these assumptions.
To assess the likelihood of observing 59 super-enhancers within the regions of RS1 hotspots, the same number of regions of equivalent sizes was sampled from the genome. Similarly to the previous analysis, the random segments of the genome were drawn from genomic regions representative of nonhotspot tandem duplications (2,106 Mb). The procedure was repeated 10,000 times, and super-enhancers within the simulated segments were counted.
The observed overlap with 59 or more super-enhancers occurred zero times in 10,000 simulation rounds, by which we estimated the P value of the observation to be <10−4. The empirical distribution observed in the simulations is shown in Supplementary Figure 4c.
Analysis of gene expression
RNA expression levels of genes in the samples were obtained from RNA-seq data, as reported previously4. We set out to assess whether tandem duplications in the hotspots are associated with increased expression of affected genes. Statistical methods and results are presented in the Supplementary Note.
Hotspots of RS1 in other tumors
In addition to breast cancer, tumors of other tissue types sometimes show excess tandem duplications in their genomes. To investigate whether the rearrangements in other tumor types also accumulate in hotspots, we used previously published sequences of ovarian and pancreatic cancer genomes (details in Supplementary Note).
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments, because doing so was not relevant to the study.
Code availability
The code used to identify rearrangement hotspots can be found at https://github.com/DominikGlodzik/hotspots/tree/glodzik2016/.
Data availability
We used breast cancer data from a previous publication4. Our group had previously submitted raw data for breast cancer to the European Genome-phenome Archive under accession number EGAS00001001178. Somatic variants in breast cancer genomes have been deposited in the International Cancer Genome Consortium Data Portal (https://dcc.icgc.org/), and the data with computer code are provided in the Github repository above. Whole-genome data sets from pancreatic and ovarian cancers have been described previously10,11.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Acknowledgments
Data used in this analysis were funded through the ICGC Breast Cancer Working group by the Breast Cancer Somatic Genetics Study (BASIS), a European research project funded by the European Community’s Seventh Framework Programme (FP7/2010-2014) under grant agreement number 242006; the Triple Negative project, funded by the Wellcome Trust (grant reference 077012/Z/05/Z); and the HER2+ project, funded by Institut National du Cancer (INCa) in France (grant nos. 226-2009, 02-2011, 41-2012, 144-2008 and 06-2012). J.W.M.M. received funding for this project through an ERC Advanced grant (no. 322737). G.K. is supported by National Research Foundation of Korea grants (NRF 2015R1A2A1A10052578). The ICGC Asian Breast Cancer Project was funded through a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A111218-SC01). D.G. is supported by the EU-FP7-SUPPRESSTEM project. S.N.-Z. is funded by a Wellcome Trust Intermediate Fellowship (WT100183MA) and is supported as a Wellcome Beit Fellow.
Footnotes
Author Contributions
D.G. and S.N.-Z. designed the study, analyzed data and wrote the manuscript. M.R.S., P.J.C., D.E. and G.E. contributed to idea development. D.G. and S.M. performed all statistical analyses. H.D., S.M., J.D.-P., J.S., M.S. and X.Z. performed curation and contributed to analyses. M.S. contributed to curation and analysis of transcriptomic data. Y.L. and L.B.A. contributed to analysis. C.A.P., P.T.S., S.R.L., I.H.R. and H.R. contributed pathology assessment and/or samples and FISH analyses. K.R. contributed IT expertise. A.B.B., A.M.T., E.B., H.G.S., M.J.v.d.V., J.W.M.M., A.-L.B.-D., A.L.R., G.K. and A.V. contributed samples, clinical data collection and intellectual input to the project. All authors discussed the results and commented on the manuscript.
Competing Financial Interests
The authors declare competing financial interests: details are available in the online version of the paper.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 3.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morganella S, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383. doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patch AM, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 12.Menghi F, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci USA. 2016;113:E2373–E2382. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride DJ, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol. 2012;227:446–455. doi: 10.1002/path.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson B, Johansson M, Heyden A, Nelander S, Fioretos T. An improved method for detecting and delineating genomic regions with altered gene expression in cancer. Genome Biol. 2008;9:R13. doi: 10.1186/gb-2008-9-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsen G, et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398, e1–e2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis NA, Rass E, Scully R. Deciphering the code of the cancer genome: mechanisms of chromosome rearrangement. Trends Cancer. 2015;1:217–230. doi: 10.1016/j.trecan.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloan CA, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44:D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro-Giner F, Ratcliffe P, Tomlinson I. The mini-driver model of polygenic cancer evolution. Nat Rev Cancer. 2015;15:680–685. doi: 10.1038/nrc3999. [DOI] [PubMed] [Google Scholar]
- 30.Roy A, et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun. 2015;6:8891. doi: 10.1038/ncomms9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 33.Easton DF, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michailidou K, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 38.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, et al. SEA: a super-enhancer archive. Nucleic Acids Res. 2016;44:D172–D179. doi: 10.1093/nar/gkv1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used breast cancer data from a previous publication4. Our group had previously submitted raw data for breast cancer to the European Genome-phenome Archive under accession number EGAS00001001178. Somatic variants in breast cancer genomes have been deposited in the International Cancer Genome Consortium Data Portal (https://dcc.icgc.org/), and the data with computer code are provided in the Github repository above. Whole-genome data sets from pancreatic and ovarian cancers have been described previously10,11.