Abstract
Objective
Although the role of microRNA-17 (miR-17) has been identified as a tumour biomarker in various studies, its prognostic value in cancers remains unclear. Therefore, we performed a systematic review and meta-analysis to analyse and summarise the relationship between the miR-17 status and clinical outcome in a variety of human cancers.
Design
Systematic review and meta-analysis.
Data sources
PubMed, Web of Science and Embase from the first year of records to 15 May 2017.
Outcomes
The patients’ survival results were pooled, and pooled HRs with 95% CIs were calculated and used for measuring the strength of association between miR-17 and the prognosis of cancers, including hepatocellular carcinoma, lung cancer, osteosarcoma, glioma, T-cell lymphoblastic lymphoma and colon cancer. Heterogeneity, publication bias and subgroup analysis were also conducted.
Results
A total of 1096 patients were included in this meta-analysis from 12 articles. The results indicated that the increased expression of miR-17 played an unfavourable role in overall survival in various human carcinomas with the HR of 1.342 taking into account the publication bias. In subgroup analysis, HR of ethnicity (Caucasian HR=1.48 and Asian HR=1.40), disease (digestive system HR=1.36 and blood system cancer (HR=2.38), detection method (quantitative real-time PCR HR=1.40 and in situ hybridisation, HR=2.59) and detection sample (tissue HR=1.45 and serum HR=1.32) were significant with p<0.05. For the analysis of disease-free survival and recurrence-free survival, the increased expression of miR-17 was associated with unfavourable prognosis (HR=1.40).
Conclusions
miR-17 may be a useful biomarker in predicting the clinical outcome of human cancers, but due to the limitations of the current studies, further verification of the role of miR-17 in human malignancies is urgently needed.
PROSPERO registration number
CRD42017065749
Keywords: microrna-17, cancer, outcome, prognosis, meta-analysis
Strengths and limitations of this study.
This is the first meta-analysis that summarised and reported the microRNA-17 as a novel potential cancer prognostic biomarker in the clinical field.
We used strict, broad search strategy of the internet databases to minimise any potential publication bias.
We conducted the subgroup analysis and found that the upregulated expression of microRNA-17 may imply poor clinical outcome in digestive system cancers.
The major limitation of our meta-analysis is the inclusion of a limited number of studies carried out on Western populations decreasing the applicability of our results among other ethnicities. MicroRNA-17 detection is not routine clinical practice, and the prognostic value of microRNA-17 remains controversial. In the future, additional clinical trials are needed to verify the prognostic significance of microRNA-17.
Introduction
Despite significant advances in clinical research over the past few decades, cancer is still a key health burden and a leading cause of death worldwide. In the year 2017, it is estimated that 1 688 780 patients were diagnosed with cancers with 600 920 cancer deaths in the USA.1 Due to the advanced screening methods and adjuvant systemic therapies for newly diagnosed cases, the mortality rate for cancers is declining in the developed countries,2 whereas the clinical outcome of cancers in the low/middle-income countries is still poor.3 4
There are several independent factors for identifying and evaluating the clinical outcome of human cancers, including tumour size, histological grade, age of the patients and metastasis to lymph nodes.5–8 Tissue-based and serum-based tumour biomarkers are widely used to predict the prognosis of neoplasms. However, these techniques are far from satisfactory due to the low specificity and sensitivity.9–11 Thus, a less-invasive and more accurate biomarker would be of great value for the prognosis of human tumours.
The discovery of microRNAs (miRNAs) provided an innovative method for the prognosis of cancers by a less-invasive detection method.12 miRNAs, a class of endogenous non-coding single-stranded RNAs with the length of 18–25 nucleotides, act as regulators of gene expression by pairing with the complementary nucleotides in the 3’-untranslated regions (3’-UTR) of their target mRNAs. miRNAs may act as regulators of cell growth, proliferation, differentiation and apoptosis.13 Because of these fundamental activities, numerous studies have shown that miRNAs function as tumour suppressors or oncogenes. It has also been reported that some miRNAs are differentially expressed between tumour and non-tumour tissues, and the abnormal expression of tumour-associated miRNAs can be detected in patient’s blood, cancerous tissue and faecal samples.14 15 Recent studies have demonstrated that aberrantly expressed miRNAs, especially those acting as tumour suppressors or oncogenes, are related to cancer development, progression and patients’ response to therapy.16–18 Therefore, miRNAs can be considered as useful prognostic biomarkers for various human cancers.
One such example is of miR-17 that is aberrantly expressed in patients with cancer.19–21 The miR-17 family, which includes six members, is one of the most extensively studied miRNA clusters.22 These miRNAs are located within an 800 base-pair region of human chromosome 13, play an essential role in the development of the heart, lung and human immune system.23 Recent studies have found that miR-17 may play a critical role in the development of human cancers.24 25 Increased expression of miR-17 promotes the metastasis of lung and pancreatic cancers, suggesting its role as an oncogene.26 27 However, other studies have reported that miR-17 inhibits tumour cell invasion and metastasis in breast cancer.28 In all, the role of miR-17 in cancer development and the exact mechanism are not yet clearly described. According to the miRBase (http://www.mirbase.org), miR-17 includes two members, miR-17–5p and miR-17–3p which are located in the sequence of miR-17 with a stem-loop structure. As a result, the detection of miR-17–5p, miR-17–3p has the same effect as detecting miR-17.29–33
Several published results indicate that the higher expression of the miR-17 is indicative of poor prognosis in patients with cancer.26 27 34–43 However, several confounding factors, including race, detection method and tumour site, may affect the observations making the relationship between aberrant expression of miR-17 and the clinical outcome of patients with cancer inconsistent. We, therefore, conducted a meta-analysis of available studies to evaluate the clinical utility of miR-17 as a novel cancer prognostic indicator.
Material and methods
Data source and search strategy
The following online electronic databases were used for the literature search: PubMed, Web of Science and Embase. The search period was up to 15 May 2017. Key search words used were: (1) prognosis OR prognostic OR survival OR outcome OR mortality; (2) cancer OR tumur OR tumour OR carcinoma OR neoplasm; (3) miR-17 OR microRNA-17 OR hsa-mir-17. Details are listed in the online supplementary table 1. Additionally, we also searched the references and relevant published articles via Google Scholar.
bmjopen-2017-018070supp001.pdf (79.2KB, pdf)
Inclusion and exclusion criteria
The inclusion criteria of the articles were: (1) the cancers were diagnosed by the histological examination or any other accepted standard, (2) miR-17 was studied in human cancers, (3) the expression of miR-17 and the clinical outcome of patients were included in the research and (4) reports with survival outcome and the data analysed HR with 95% CI and HR with a p value.
The exclusion criteria were: (1) duplicate publications; (2) articles focused on other genes; (3) case reports, reviews, letters and animal trails; (4) unqualified or insufficient data; (5) HR, 95% CI and p value were not provided or could not be calculated and (6) articles concentrated on the polymorphisms or methylation patterns of miRNAs.
Questions of suitability of articles to be included were examined and discussed by the authors after reviewing the abstract and full-text manuscript. The final decision was made by the academic committee.
Data extraction and quality assessment
All included studies were decided by the two investigators (CH and XY) independently based on titles and abstracts. Full text of the articles was required if the articles were potentially suitable for the meta-analysis. Furthermore, the literature search was performed again in the excluded articles to avoid missing any article potentially relevant for the study. The original authors of the articles were contacted if any supplementary data were needed. Any disagreement was resolved by the two authors (CH and XY). The extracted details of the articles were as follows: (1) publication information: the name of the authors, publication area and publication year; (2) patient’s characteristics: diseases, stage of the disease, RNA detection method, type of tissue sample and follow-up years; (3) the measurement of miR-17 measurement and its cut-off value and (4) HR of miR-17 for overall survival (OS), disease-free survival (DFS) and recurrence-free survival (RFS), as well as their 95% CI and p values. The HRs and their 95% CI were extracted from the original articles or via emails from the authors. If not, we calculated HR and 95% CI using the data of observed deaths, cancer recurrences or the original data provided by the authors. All calculations mentioned above were based on the methods provided by Parmar et al.44 The quality of the included articles was assessed based on a systematic review checklist of the Dutch Cochrane Centre proposed by Meta-analysis Of Observational Studies in Epidemiology.45
Statistical analysis
The test of heterogeneity of pooled HRs was carried out by using Cochran’s Q test and Higgins I2 statistic. A p value of <0.05 or I2 >50% was considered as statistically significant. The 95% CI of I2 was calculated by the method introduced by Hedges et al.46 If heterogeneity existed, the random-effects model was performed among the included studies; otherwise, the fixed-effects model was selected. I2 value ranged from 0% to 100%. All p values were two sided.
HR >1 presents of upregulated expression of miR-17 indicated poor prognosis in patients, and HR <1 suggested a better prognosis. Publication bias was evaluated by the Begg’s test and Egger’s test.47 48 If the publication bias did exist, the trim and fill method introduced by Duval and the Tweedit’s was used to adjust the results.49 The STATA software V.14.0 (StataCorp) was used in all of the statistical analyses.
Patients and public involvement statement
The patients or public were not involved in the study.
Results
Literature selection
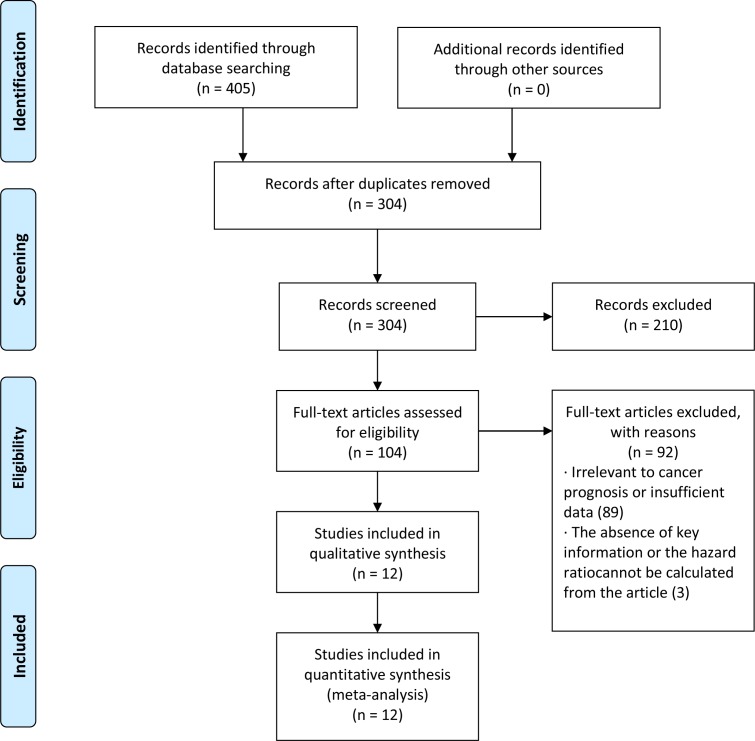
We started with 405 articles associated with miR-17 and cancer prognosis was identified from online database searches. After removing the replicate records, 304 miR-17-related articles were left. The first screening based on the species, article type and language eliminated 210 citations from the analysis. Subsequently, the remaining 104 studies were carefully assessed by reviewing the abstract and full text of each article. After that, 89 articles were excluded from the study because they were unrelated to miR-17 expression levels or because of the lack of survival statistics such as HRs, 95% CI or p value. Finally, 15 studies, which investigated the potential relationship between miR-17 expression and prognosis of human cancers, remained for further detailed screening and data extraction. Three of the studies that explained the relationship between miR-17 expression and the clinical outcome of cancer had to be removed because the authors did not provide the exact HR value, or the value cannot be calculated from the data. Thus, 12 articles (12 studies)26 27 34–43 were included in this meta-analysis (figure 1).
Figure 1.
Flow diagram of the studies selection phase.
Characteristics of selected studies
All 12 studies included in the meta-analysis were retrospective studies published between 2010 and 2016.26 27 34–43 Patient’s OS was reported in all 12 studies, and 3 studies also examined the DFS or RFS. The type of the cancers included gastrointestinal cancers (colorectal cancer, gastric cancer), lung cancer, pancreatic cancer, hepatocellular cancer, osteosarcoma, glioma, T-cell lymphoblastic lymphoma and oesophageal squamous cell carcinoma. A total of 1096 patients with various types of cancers were from People’s Republic of China, Japan, Spain and Brazil. Quantitative real-time PCR (qRT-PCR) was used to assess the expression of miR-17 in 12 studies, and 1 study used the in situ hybridisation (ISH). All studies used tissue and serum samples as the source of the miR-17. The majority (10 of 12) of the HRs reported in the present analysis were included in the multivariate analysis. The remaining two HRs could be estimated by Kaplan-Meier analysis and relative risk values. Most of the studies have the follow-up research for at least 38 months. The clinical characteristics of the studies included in this article are summarised in table 1.
Table 1.
A summary table of the meta-analysis
| Study | Year | Country | Diseases | Case no | Stage | Sample | Assay | Cut-off value | HR | Follow-up (months) | Type of miR-17 detection |
| Chen et al37 | 2012 | China | HCC | 120 | I–IV | Tissue | qRT-PCR | Median | RR | 46 | miR-17–5p |
| Qun et al27 | 2013 | China | Lung cancer | 221 | I–IV | Tissue | qRT-PCR | Median | Given | 50 | miR-17 |
| Li et al41 | 2014 | China | Osteosarcoma | 117 | I–III | Tissue | qRT-PCR | Median | Given | 44 | miR-17 |
| Lu et al35 | 2012 | China | Glioma | 108 | I–IV | Tissue | qRT-PCR | Mean | RR | 60 | miR-17 |
| Xi et al42 | 2015 | China | T-cell lymphoblastic lymphoma | 57 | III, IV | Tissue | qRT-PCR | Median | Given | Up to 13 years | miR-17 |
| Yu et al40 | 2012 | China | Colon cancer | 48 | I–IV | Tissue | qRT-PCR | Median | Given | 5–66 | miR-17 |
| Manuel et al39 | 2011 | Spain | Gastrointestinal cancer | 38 | I–IV | Tissue | qRT-PCR | Mean | Given | 38 | miR-17 |
| Robaina et al38 | 2016 | Brazil | Burkitt lymphoma | 41 | I–IV | Tissue | ISH | Median | Given | 69 | miR-17 |
| Xu et al36 | 2014 | China | Oesophageal squamous cell carcinoma | 105 | I–IV | Tissue | qRT-PCR | Mean | Given | 52 | miR-17 |
| Jun et al26 | 2010 | Japan | Pancreatic cancer | 80 | I–IV | Tissue | qRT-PCR | Median | Given | 60 | miR-17–5p |
| Wang et al43 | 2011 | China | Gastric cancer | 65 | I–IV | Serum | qRT-PCR | Median | Given | 36 | miR-17–5p |
| Zheng et al34 | 2013 | China | HCC | 96 | I–IV | Serum | qRT-PCR | Median | Given | NG | miR-17–5p |
HCC, hepatocellular carcinoma; ISH, in situ hybridisation; miR-17, microRNA-17; NG, not given; OS, overall survival; qRT-PCR, quantitative real-time PCR; RR, risk ratio.
Association between miR-17 and OS
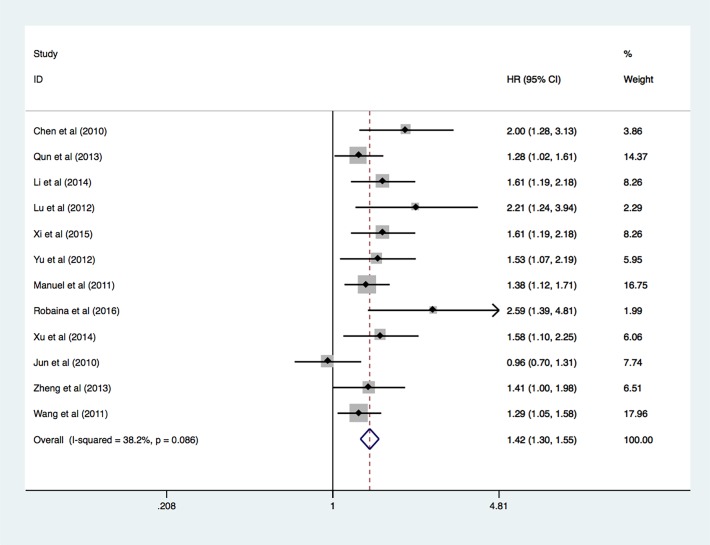
Due to low heterogeneity, fixed-effects model was used to calculate and analyse the pooled HR value. High expression level of miR-17 was associated with the poor OS in patients with diverse cancers. The statistical power of Q test is low when there are limited studies included in the meta-analysis. We, therefore, conducted random-effect analysis on the OS (HR 1.45, 95% CI 1.29 to 1.63, p<0.001), which was not significantly different compared with the analysis of fixed-effect model. Details of the meta-analysis are systematically summarised in the figure 2.
Figure 2.
Forest plot of meta-analysis of overall survival in association with miR-17 expression.
To demonstrate the predictive role of miR-17, subgroups analysis was conducted based on patients’ ethnicity, cancer type, methods identifying miRNAs and type of tissue samples. Clinical association between miR-17 and OS was found in the Asian and Caucasian patients (figure 3A). The association was also significant in other subgroups, including digestive system cancers and blood cancers (figure 3B), qRT-PCR detection method (figure 3C), and tissue and serum samples (figure 3D). miR-17 includes two members, miR-17–5p and miR-17–3p which are located in the sequence of miR-17 with a stem-loop structure. Therefore, analysis of miR-17–5p or miR-17–3p afforded the same effect (or result) as miR-17. To clarify the heterogeneity, we conducted a subgroup analysis concerning the detection method of miR-17 and found that the clinical value was also significant in miR-17 group and miR-17–5p group. There was no significant difference between the two groups (figure 3E), implying that same effect existed when detecting miR-17 and miR-17–5p. Details of the subgroup analysis are listed in the table 2.
Figure 3.
Forest plots of subgroup meta-analysis of OS in association with miR-17 expression. (A) Forest plots of the merged analyses of OS in different ethnic groups. Squares and lines correspond to the study-specific HRs and 95% CIs, respectively. The area of the squares represents the weight, and the diamonds represent the summary of HRs and 95% CIs. (B) Forest plots of the merged analyses of OS in different diseases groups. (C) Forest plots of the merged analyses of OS in different RNA detection methods groups. (D) Forest plots of the merged analyses of OS in different sample groups. (E) Forest plots of the merged analyses of OS in the detection method of miR-17. ISH, in situ hybridisation; miR-17, microRNA-17; OS, overall survival; qRT-PCR, quantitative real-time PCR.
Table 2.
Subgroup analysis
| Subgroup | No of studies | Heterogeneity | Pooled HR (95% CI) | P values | |
| I2 (95% CI) | P values | ||||
| Total | 12 | 38.2% (0% to 68.7%) | 0.086 | 1.42 (1.30 to 1.55) | <0.001 |
| Ethnic subtotal | |||||
| Caucasian | 2 | 71.6% (0% to 93.6%) | 0.06 | 1.48 (1.21 to 1.81) | <0.001 |
| Asian | 10 | 36.1% (0% to 69.5%) | 0.12 | 1.40 (1.27 to 1.55) | <0.001 |
| Disease subtotal | |||||
| Digestive system | 7 | 34.8% (0% to 72.4%) | 0.163 | 1.36 (1.22 to 1.51) | <0.001 |
| Respiratory system | 1 | NA | NA | 1.28 (1.02 to 1.61) | 0.036 |
| Blood system | 2 | 0 | 0.713 | 2.38 (1.56 to 3.63) | <0.001 |
| Glioma | 1 | NA | NA | 1.61 (1.19 to 2.18) | 0.002 |
| Osteosarcoma | 1 | NA | NA | 1.61 (1.19 to 2.18) | <0.001 |
| Detected method subtotal | |||||
| qRT-PCR | 11 | 29.0% (0% to 65.0%) | 0.169 | 1.40 (1.28 to 1.53) | <0.001 |
| ISH | 1 | NA | NA | 2.59 (1.39 to 4.81) | 0.003 |
| Detected sample subtotal | |||||
| Tissue | 10 | 46.2% (0% to 74.1%) | 0.053 | 1.45 (1.31 to 1.61) | <0.001 |
| Serum | 2 | 0 | 0.662 | 1.32 (1.10 to 1.57) | 0.002 |
| Detection of miR-17 subtotal | |||||
| miR-17 | 8 | 60.1% (13.2% to 81.7%) | 0.057 | 1.29 (1.11 to 1.49) | <0.001 |
| miR-17–5p | 4 | 7.5% (0% to 43.4%) | 0.372 | 1.50 (1.34 to 1.67) | 0.001 |
ISH, in situ hybridisation; miR-17, microRNA-17; miR-17-5p, microRNA-17-5p; NA, not available; qRT-PCR, quantitative real-time PCR.
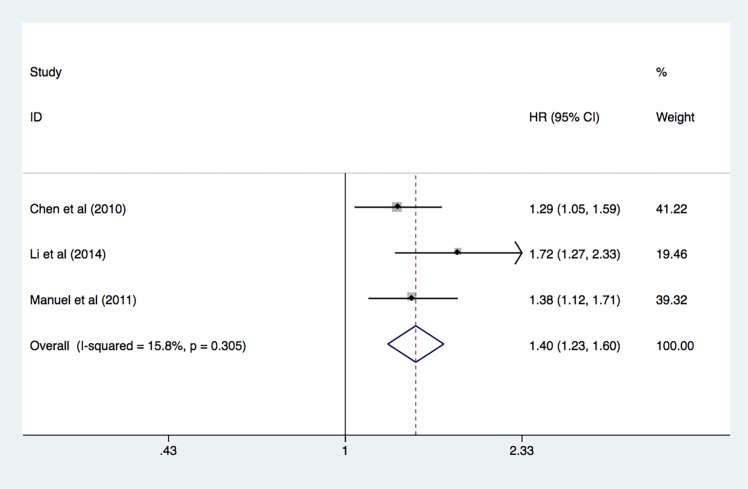
Correlation between miR-17 and DFS and RFS
A total of three studies37 38 41 were included in the analysis of DFS and RFS. The analyses revealed a predictive role of increased expression of miR-17 for the prognosis of patients with cancer (pooled HR 1.40, 95% CI 1.23 to 1.60, p<0.001) as determined by the fixed-effect model (I2=15.8%, p=0.305) (figure 4).
Figure 4.
Forest plot of disease-free survival and recurrence-free survival in association with miR-17 expression. miR-17, microRNA-17.
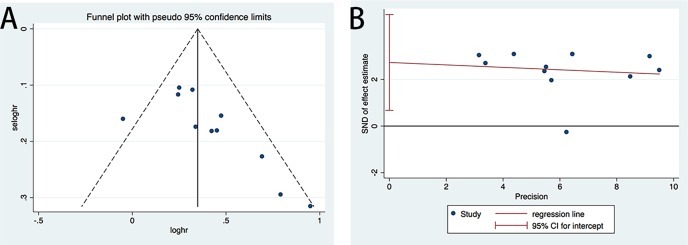
Publication bias
We used Begg’s funnel plot and Egger’s test to assess the possible publication bias of the included studies.47 48 In the analysis of relationship between miR-17 and the OS, the p values of Egger’s test and Begg’s test were 0.014 and 0.011, respectively. The funnel plot and Egger’s plot are displayed in figure 5A,B. Both Begg’s test and Egger’s test implied a publication bias, thus, the trim and fill method was performed to make pooled HR more reliable.49 The altered HR was 1.34, 95% CI 1.24 to 1.46, p<0.001, which was not significantly different from the pooled HR (online supplementary figure 1).
Figure 5.
(A) Funnel plot of merged analysis of OS comparing high or low expression of miR-17. (B) Egger’s test plot of merged analysis of OS comparing high or low expression of miR-17. miR-17, microRNA-17; OS, overall survival; SND, standard normal deviate.
bmjopen-2017-018070supp002.pdf (243.1KB, pdf)
Discussion
Previous studies have shown that miRNAs have a distinct expression profile in cancerous tissues which can be detected by qRT-PCR in frozen, formalin-fixed and paraffin-embedded tissues and in serum samples. Recently, miRNAs, serving as tumour suppressors or oncogenes, have been shown to play important roles in the evolution and progression of cancers. miRNAs are involved in a variety of crucial cellular pathways such as angiogenesis, innate and adaptive immune responses, cellular proliferation, invasion and metastasis.12 16 Several studies have reported the potential use of miRNAs as tumour biomarkers for detecting tumour occurrence, development and prognosis. Unfortunately, effective diagnosis techniques and prognosis indicators of cancer have not been found. Developing a novel less-invasive detection method with higher accuracy for cancer prognosis is of great significance in evaluating cancer progression as well as monitoring patients’ therapeutic response.
Over the last couple of decades, numerous studies have uncovered the involvement of miRNAs in the pathogenesis of cancer. Since miRNAs can be obtained non-invasively from the serum, urine and faecal samples, their utility as diagnostic and prognostic biomarkers in cancer and other diseases has been extensively explored. It has been reported that miRNA could be detected with higher accuracy than traditional cancer biomarkers in predicting the clinical outcome of the human colon cancers.50 However, adequate evidence is still lacking for the utility of miRNAs as cancer biomarkers in clinical practice.
miR-17, a widely studied miRNA, is aberrantly expressed in different kinds of cancers, such as glioma,35 oesophageal and oral squamous cell carcinomas,36 51 pancreatic cancer,26 gastrointestinal cancers,39 osteosarcoma52 and Burkitt lymphoma,38 and is significantly related to the clinical outcome of cancers. Our meta-analysis indicated that the elevated miR-17 expression is significantly associated with poor OS (HR=1.42) in patients with various types of carcinomas. The analysis using the Cochran’s Q test and Higgins I2 test implied low heterogeneity. As limited number of studies were included in the meta-analysis, the Q test had inadequate statistical power. We, therefore, applied the fixed-effects model to calculate and analyse the pooled HR value. We also conducted random-effect analysis on the OS, which was not significantly different when compared with analysis of fixed-effect model (figure 2). In the subgroup analysis, we found that the potential heterogeneity may have originated from the Caucasian group in the study conducted by Robaina et al.38 Unlike the commonly used RT-PCR, ISH technique was used to detect miR-17. Other factors contributing to the heterogeneity may include the limited number of patients (n=41) recruited in the study. However, both studies from Spain and Brazil recruited population of Caucasians decreasing the heterogeneity.
As the Begg’s test and the Egger’s test implied publication bias, we used the trim and fill method to obtain a more reliable pooled HR. We found that the adjusted HR was not significantly different from the pooled HR. In subgroup analysis, based on the characteristics of the individual studies, significant HR was found in the Caucasian and Asian groups, the qRT-PCR group and the tissue and serum sample groups. Furthermore, the increased expression of miR-17 indicated poor DFS and RFS in hepatocellular carcinoma (HCC) and gastrointestinal cancers. Several investigators have explored the functional roles of miR-17 and its involvement in human cancers. Yang et al found that the miRNA-17 was overexpressed in the HCC tissue, and promoted the phosphorylation of heat shock protein 27 (HSP27). The phosphorylated HSP27 then enhanced the migration of the HCC cells implying a significant role of miRNA-17 in the progression of HCC.53 Wang et al reported that the upregulated expression of miRNA-17–5p promoted cancer cells proliferation and inhibited apoptosis by post-transcriptional modulation of mRNA-p21 and tumour protein p53-induced nuclear protein 1.54 In the study by Ma et al, overexpression of miRNA-17 promoted cancer cells progression by targeting P130.55 Yan et al found overexpression of the miR-17–5p in pancreatic cancer. The miR-17–5p inhibitor promoted the expression of Bim protein by targeting the 3’-UTR of its mRNA and negatively regulating at the post-transcriptional level. Therefore, the authors suggested that the miR-17–5p inhibitor may be a novel therapeutic approach for pancreatic cancer.56 Together with our meta-analysis, these findings suggest that the detection of tissue or serum miR-17 expression may be a useful prognostic biomarker in patients with HCC, pancreatic cancer and gastrointestinal cancers.
There are potential limitations of this study. The literature searches using authentic and widely used data bases found studies performed predominantly on Asian populations not encompassing sufficient numbers of other populations such as Caucasians. Our results of miR-17 as a potential biomarker may, therefore, not be applicable to other populations. The pooled HR values were also not sufficiently strong. Furthermore, the relatively limited sample size of 1031 patients weakened the statistical significance of the prognostic potential of miR-17 expression levels.
Conclusions
In summary, our meta-analysis suggested that miR-17 is a potential biomarker in various types of cancers. However, further multicentre clinical trials with larger sample size and prospective studies including Caucasians and patients representing other ethnicities are needed to confirm the prognostic value of miR-17 and its subsequent application as a prognostic biomarker in the routine clinical guidance of cancers.
Supplementary Material
Acknowledgments
The authors thank the grants from Guangdong Science and Technology Department, China.
Footnotes
Contributors: CH and MY conceived the study. CH and XY performed the data extraction and analysed the data. CH and MY wrote the paper. All authors had full access to all of the data and approved the final version of the manuscript.
Funding: The article is funded by Guangdong Science and Technology Department, China. (No.2014A020212636).
Competing interests: None declared.
Patient consent: Not required.
Ethics approval: The study does not include human participants or animals.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1. Siegel RL, Miller KD, Jemal A, et al. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians 2017;67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4. Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220–41. 10.3322/caac.21149 [DOI] [PubMed] [Google Scholar]
- 5. Fisher B, Slack NH, Bross ID. Cancer of the breast: size of neoplasm and prognosis. Cancer 1969;24:1071–80. [DOI] [PubMed] [Google Scholar]
- 6. Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 1957;11:359–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kroman N, Jensen MB, Wohlfahrt J, et al. Factors influencing the effect of age on prognosis in breast cancer: population based study. BMJ 2000;320:474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fisher B, Bauer M, Wickerham DL, et al. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer. An NSABP update. Cancer 1983;52:1551–7. [DOI] [PubMed] [Google Scholar]
- 9. Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes. Gut 2015;64:1637–49. 10.1136/gutjnl-2014-309086 [DOI] [PubMed] [Google Scholar]
- 10. Nicholson BD, Shinkins B, Pathiraja I, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev 2015;12:CD011134 10.1002/14651858.CD011134.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun W, Liu Y, Shou D, et al. AFP (alpha fetoprotein): who are you in gastrology? Cancer Lett 2015;357:43–6. 10.1016/j.canlet.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 12. Tricoli JV, Jacobson JW. MicroRNA: Potential for Cancer Detection, Diagnosis, and Prognosis. Cancer Res 2007;67:4553–5. 10.1158/0008-5472.CAN-07-0563 [DOI] [PubMed] [Google Scholar]
- 13. Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006;20:515–24. 10.1101/gad.1399806 [DOI] [PubMed] [Google Scholar]
- 14. Rokkas T, Kothonas F, Rokka A, et al. The role of circulating microRNAs as novel biomarkers in diagnosing colorectal cancer: a meta-analysis. Eur J Gastroenterol Hepatol 2015;27:819–25. 10.1097/MEG.0000000000000363 [DOI] [PubMed] [Google Scholar]
- 15. Yang X, Zhong J, Ji Y, et al. The expression and clinical significance of microRNAs in colorectal cancer detecting. Tumour Biol 2015;36:2675–84. 10.1007/s13277-014-2890-0 [DOI] [PubMed] [Google Scholar]
- 16. Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, et al. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther 2014;15:1444–55. 10.4161/15384047.2014.955442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho WC. Circulating MicroRNAs as Minimally Invasive Biomarkers for Cancer Theragnosis and Prognosis. Front Genet 2011;2:7 10.3389/fgene.2011.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avery-Kiejda KA, Braye SG, Mathe A, et al. Decreased expression of key tumour suppressor microRNAs is associated with lymph node metastases in triple negative breast cancer. BMC Cancer 2014;14:51 10.1186/1471-2407-14-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang Y, Yang W, Zhu Y, et al. Prognostic role of microRNA-203 in various carcinomas: evidence from a meta-analysis involving 13 studies. Springerplus 2016;5:1538 10.1186/s40064-016-3225-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z, Cai Q, Jiang Z, et al. Prognostic role of microRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit 2014;20:1668-74 10.12659/MSM.892096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Hong F, Yu Z. Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res 2013;41:596–602. 10.1177/0300060513485856 [DOI] [PubMed] [Google Scholar]
- 22. Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 2013;20:1603–14. 10.1038/cdd.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008;133:217–22. 10.1016/j.cell.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q, Li YC, Wang J, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A 2008;105:2889–94. 10.1073/pnas.0800178105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ottman R, Levy J, Grizzle WE, et al. The other face of miR-17-92a cluster, exhibiting tumor suppressor effects in prostate cancer. Oncotarget 2016;7:73739 10.18632/oncotarget.12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu J, Ohuchida K, Mizumoto K, et al. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther 2010;10:748–57. 10.4161/cbt.10.8.13083 [DOI] [PubMed] [Google Scholar]
- 27. Chen Q, Si Q, Xiao S, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol 2013;30 10.1007/s12032-012-0353-2 [DOI] [PubMed] [Google Scholar]
- 28. Yu Z, Willmarth NE, Zhou J, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A 2010;107:8231–6. 10.1073/pnas.1002080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffiths‐Jones S. The microRNA Registry, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011;39:D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140–D144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng J, Dong P, Gao S, et al. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology 2013;60:549–52. 10.5754/hge12754 [DOI] [PubMed] [Google Scholar]
- 35. Lu S, Wang S, Geng S, et al. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol 2012;2012:1–6. 10.1155/2012/970761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu XL, Jiang YH, Feng JG, et al. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg 2014;97:1037–45. 10.1016/j.athoracsur.2013.10.042 [DOI] [PubMed] [Google Scholar]
- 37. Chen L, Jiang M, Yuan W, et al. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg 2012;25:156–61. 10.3109/08941939.2011.618523 [DOI] [PubMed] [Google Scholar]
- 38. Robaina MC, Faccion RS, Mazzoccoli L, et al. miR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosis. Ann Hematol 2016;95:881–91. 10.1007/s00277-016-2653-7 [DOI] [PubMed] [Google Scholar]
- 39. Valladares-Ayerbes M, Blanco M, Haz M, et al. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol 2011;39:1253 10.3892/ijo.2011.1112 [DOI] [PubMed] [Google Scholar]
- 40. Yu G, Tang JQ, Tian ML, et al. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol 2012;106:232–7. 10.1002/jso.22138 [DOI] [PubMed] [Google Scholar]
- 41. Li X, Yang H, Tian Q, et al. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma 2014;61:453–60. 10.4149/neo_2014_056 [DOI] [PubMed] [Google Scholar]
- 42. Xi Y, Li J, Zhang P, et al. Upregulation of miRNA-17 and miRNA-19 is associated with unfavorable prognosis in patients with T-cell lymphoblastic lymphoma. Exp Mol Pathol 2015;99:297–302. 10.1016/j.yexmp.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 43. Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep 2012;5:1514–20. 10.3892/mmr.2012.828 [DOI] [PubMed] [Google Scholar]
- 44. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- 45. Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008–12. [DOI] [PubMed] [Google Scholar]
- 46. Hedges LV, Pigott TD. The power of statistical tests in meta-analysis. Psychol Methods 2001;6:203–17. 10.1037/1082-989X.6.3.203 [DOI] [PubMed] [Google Scholar]
- 47. Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- 49. Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455–63. 10.1111/j.0006-341X.2000.00455.x [DOI] [PubMed] [Google Scholar]
- 50. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008;299:425–36. 10.1001/jama.299.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang CC, Yang YJ, Li YJ, et al. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncol 2013;49:923–31. 10.1016/j.oraloncology.2013.03.430 [DOI] [PubMed] [Google Scholar]
- 52. Li S, Gao Y, Wang Y, et al. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol Lett 2016;12:4905–10. 10.3892/ol.2016.5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang F, Yin Y, Wang F, et al. miR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology 2010;51:1614–23. 10.1002/hep.23566 [DOI] [PubMed] [Google Scholar]
- 54. Wang M, Gu H, Qian H, et al. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer 2013;49 10.1016/j.ejca.2012.12.017 [DOI] [PubMed] [Google Scholar]
- 55. Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun 2012;3:1291 10.1038/ncomms2276 [DOI] [PubMed] [Google Scholar]
- 56. Yan HJ, Liu WS, Sun WH, et al. miR-17-5p inhibitor enhances chemosensitivity to gemcitabine via upregulating Bim expression in pancreatic cancer cells. Dig Dis Sci 2012;57:3160–7. 10.1007/s10620-012-2400-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2017-018070supp001.pdf (79.2KB, pdf)
bmjopen-2017-018070supp002.pdf (243.1KB, pdf)