Abstract
CD44, an abundantly expressed adhesion molecule, and its alternative splice variants have been associated with tumorigenesis and metastasis. In the context of gastric cancer (GC), de novo expression of CD44 variant 6 (CD44v6) is found in more than 60% of GCs, but its role in the pathogenesis and progression of this type of cancer remains unclear. Using a combination of media conditioning experiments and decellularized extracellular matrices (ECMs), this study investigates the hypothesis that CD44v6 overexpression enhances tumor cell malignant behavior by modulating stromal cell-mediated ECM remodeling. Our findings indicate that soluble factors secreted by CD44v6 expressing GC cells particularly increase proliferation and myofibroblastic differentiation of adipose stromal cells (ASCs). These changes in ASC phenotype mediate the deposition of fibrotic/desmoplastic ECM that, in turn, stimulates GC proliferation and inhibits GC clustering. Pharmacological inhibition of matrix metalloproteinase (MMP) activity in tumor cells abrogated matrix-induced changes in tumor cell malignant behavior. Additionally, studies in mice confirmed the pathological relevance of CD44v6 expression and consequential changes in ECM remodeling to gastric tumorigenesis in vivo. Collectively, these results indicate a direct link between CD44v6, ECM remodeling, and GC malignant behavior opening new insights into potential CD44v6-targeted therapies.
Introduction
Gastric cancer (GC) remains the third leading cause of cancer-related mortality worldwide,1 with high incidence and low survival rates mostly due to its detection in advanced stages of the disease.2 While very few GC-specific, de novo expressed molecules have been identified, CD44 has garnered significant interest as a potential therapeutic molecular target.3 Nevertheless, its role in tumorigenesis still remains controversial, as it can function as both an oncogene and a tumor suppressor.4,5 Moreover, limited insights exist regarding the correlation between CD44 protein expression and clinicopathological features of GC.6
CD44 is a ubiquitously expressed cell surface molecule that binds to the extracellular matrix (ECM), primarily to hyaluronic acid. CD44 has been shown to regulate several cell functions, ranging from cell–cell and cell–matrix interactions, cell invasion and migration, to tumor progression and metastasis.5,7 Due to alternative splicing, the CD44 locus gives rise to multiple transcripts and corresponding protein isoforms, which have been detected in a variety of other human tumors such as lung,8 breast,9 ovarian,10 and colorectal cancer.11 Among CD44 isoforms, CD44 variant 6 (CD44v6) has been shown to play a major role in cancer progression due in part to its ability to directly bind to major cytokines produced in the tumor micro-environment.3,12 Furthermore, we have previously demonstrated that CD44v6 is expressed de novo in pre-malignant and malignant lesions of the stomach but not in normal gastric glands,13 bringing into question its role in the pathogenesis and progression of GC.
It is widely accepted that malignant behavior and cancer progression is dependent on the evolving crosstalk between tumor cells and their surrounding microenvironment, which is regulated not only by altered cell–cell interactions and soluble factor signaling, but also by the highly dynamic nature of ECM.14–17 Nevertheless, the functional contribution of the ECM to GC malignant behavior is still poorly understood due in part to a lack of appropriate model systems. Tumor-associated changes in ECM homeostasis occur because of an imbalance between new ECM deposition and proteolytic remodeling by enzymes such as matrix metalloproteinases (MMPs).18–20 Under pathophysiological conditions this imbalance leads to changes in ECM composition, structure, and mechanics, which in turn can modulate tumor cell behavior via integrin-dependent signaling pathways.21–23
Fibronectin and type I collagen are the most common and abundant fibrillar ECM proteins found in cancer-associated ECM. Their increase is a result of excessive fibrotic remodeling, also referred to as desmoplasia, which is largely mediated by alpha smooth muscle actin (α-SMA)-expressing myofibroblasts.24,25 Fibroblasts and bone marrow-derived mesenchymal stem cells are generally considered as the main source of myofibroblasts.26 However, adipose stromal cells (ASCs) are also prone to undergo myofibroblast differentiation when exposed to aberrant ECM biophysical properties and tumor-secreted soluble factors.27–30 While some studies have reported the interaction between these other stromal cells and GC cells,31–34 the tumor-promoting role of ASC-mediated ECM remodeling in GC remains largely elusive. Yet this understanding is critical as adipose tissue, which harbors ASCs in its stromal vascular fraction, is a common component of the microenvironment that GC invades during the process of metastasis.35
To gain an improved mechanistical understanding of how microenvironmental features regulate GC malignant behavior, biomaterials-based, three-dimensional (3D) culture models are increasingly utilized. Such systems aim to recapitulate key features of the complex organization of the stromal ECM using a range of naturally derived or synthetic polymers in combination with appropriate fabrication techniques.36–40 Additionally, decellularized matrices provide invaluable tools to elucidate the effect of cell–matrix interactions on various aspects of tumor behavior. These cell-derived matrices represent key biophysical and biochemical features of native ECM (including 3D fibrillar architecture, complex composition, physiologically relevant mechanical properties) more accurately as compared to chemically defined biomaterials and thus, allow studying cellular interactions with their ECM niche under relevant conditions.41–43
In the present work, we investigated the hypothesis that CD44v6 overexpression in tumor cells modulates ASC-mediated ECM remodeling and desmoplasia, which in turn provides a positive feedback mechanism that is advantageous for tumor cell proliferation. We addressed this hypothesis by first treating human ASCs with tumor conditioned media (TCM) from isogenic human GC cell lines, either overexpressing or lacking isoform CD44v6, to study their differentiation into myofibroblasts and respective changes in ECM deposition. Afterwards, ASC-derived matrices were decellularized and used as a native human ECM source to access their influence on GC cell behavior in vitro. The contribution of ECM remodeling to tumor cell behavior was evaluated by seeding GC cells on decellularized matrices in the presence or absence of a broad-spectrum MMP inhibitor. Finally, mouse studies confirmed the pathological relevance of CD44v6 expression and consequential changes in ECM remodeling to gastric tumorigenesis in vivo providing insights that may inform future therapeutic approaches.
Materials and methods
Transfection of MKN74 parental cells to generate CD44v6 cells
The human stomach adenocarcinoma epithelial cell line MKN74 was kindly provided by C. Oliveira, a co-author from the Institute of Molecular Pathology and Immunology of University of Porto (IPATIMUP, Portugal). De novo expression of v6 containing isoform CD44v6 within this cell line (henceforth referred to as CD44v6 cells) was achieved by transfection of the variant CD44-04 – ENST00000415148 (OriGene) cloned into a pIRES-EGFP2 expression vector (C. Oliveira, unpublished data). Transfection of the vector was performed with Lipofectamine 2000 (Invitrogen). Upon transfection, CD44v6 expressing cells were cultured in selection medium containing 1 mg mL−1 of geneticin and subjected to three consecutive rounds of bead sorting with CELLection™ Pan Mouse IgG magnetic beads (Thermo Scientific) and using mouse anti-CD44V6 antibody – clone MA54 (Thermo Scientific). Transfection and selection efficiency was assessed by immunocytochemistry and flow cytometry.
Cell culture
MKN74 cells (passages 49–62), CD44v6 cells (passages 7–20) and 3T3-L1 (ATCC) preadipocytes (up to passage 12) were routinely cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Atlanta Biologicals) and 1% penicillin/streptomycin (Pen/Strep, Gibco), and supplemented with 1% geneticin (Gibco) in the case of CD44v6 cells. Human ASCs (Lonza) were cultured in their corresponding growth media (ADSC-GM, Lonza) and utilized up to passage 6. Cell cultures were maintained at 37 °C under a 5% CO2 humidified atmosphere.
Preparation of tumor-conditioned media (TCM)
MKN74 and CD44v6 cells were maintained in their growth media until 90% confluence, at which time they were switched to low serum DMEM (1% FBS and 1% Pen/Strep). After 24 h, conditioned media was collected, normalized to cell number (counted by hemocytometer), concentrated 10-fold in an Amicon centrifugal filter tube (Millipore, MWCO 3 kDa) and subsequently reconstituted with media containing 1% FBS and 1% Pen/Strep specifically 1: 1 DMEM/F12 for ASCs and DMEM for 3T3-L1 cells. Control media was incubated for 24 h in the absence of cells and then treated similarly. Conditioned and Control media were analyzed for transforming growth factor-β (TGF-β) levels by the human Quantikine ELISA (R&D Systems) in accordance with the manufacturers’ instructions.
Analysis of ASCs growth and proliferation
After exposing ASCs and 3T3-L1s to TCM or Control media for 3 days, cells were counted manually using a hemocytometer. To assess proliferation, 10 μM bromodeoxyuridine (BrdU, Sigma-Aldrich) was added to cells for 20 h (ASCs) and 12 h (3T3-L1s) prior to fixation in 4% paraformaldehyde (PFA). Afterwards, samples were pretreated with ice cold 1 N HCL, followed by 37 °C 2 N HCL, and then 0.1 M borate buffer at room temperature prior to adding biotinylated mouse anti-BrdU (Thermo Scientific). Finally, BrdU and nuclei were stained with streptavidin conjugated Alexa Fluor 555 (Thermo Scientific) and DAPI (Thermo Scientific), respectively. Imaging was performed on a Zeiss Observer Z.1 microscope and an AxioCam MRm camera. The percentage of BrdU-positive cells was determined by manually counting ten representative images per sample with four samples per condition using ImageJ software.
Analysis of myofibroblast differentiation and ECM deposition
ASCs and 3T3-L1s were plated on glass coverslips, cultured for 7 days, fixed with 4% PFA, and then analyzed for myofibroblast differentiation and ECM deposition via immunofluorescence image analysis. For immunofluorescence of α-SMA, fibronectin (Fn) and collagen type I, fixed cells were permeabilized with 0.05% Triton-X (Thermo Scientific) in PBS (PBS-X), blocked with 1% bovine serum albumin (BSA, Thermo Scientific) in PBS, and then incubated with primary antibodies directed against α-SMA (Abcam), Fn (Sigma-Aldrich), and collagen I (Millipore) overnight at 4 °C. Following three washes in PBS, samples were labeled with anti-mouse Alexa Fluor 488-conjugated and anti-rabbit Alexa Fluor 568-conjugated secondary antibodies (Thermo Scientific), while cell nuclei were labeled with DAPI. Imaging was performed on a Zeiss Observer Z.1 microscope and an AxioCam MRm camera. The levels of α-SMA, Fn, and collagen I were determined by quantifying the staining intensity via the RGB measure function in ImageJ and normalizing to cell number in each field of view based on the number of DAPI stained nuclei.
Western blot analysis
ASCs were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 1× protease and phosphatase inhibitor cocktail (all from Thermo Scientific), and 1 mM PMSF (Calbiochem). Protein concentrations were measured using a bicinchoninic acid protein assay kit (Thermo Scientific) and equal amounts of protein samples were loaded on gels (Bio-Rad), separated by reducing SDS-PAGE, and transferred to PVDF membranes (Bio-Rad). After blocking with 5% milk powder in TBS-T, membranes were incubated overnight at 4 °C with primary antibodies raised against α-SMA and fibronectin, as well as the housekeeping protein HSP90 (Santa Cruz). Following incubation with species-specific HRP-conjugated secondary antibodies (Novus Bio) chemiluminescence detection was performed using an ECL kit (Thermo Scientific). Densitometric analysis was performed with Image Lab software (Bio-Rad).
Decellularization
ASCs were seeded on plastic coverslips (Thermanox, Thermo Scientific) coated with human plasma Fn (Thermo Scientific) at a concentration of 30 μg mL−1 in PBS to facilitate cell adhesion. After preconditioning cells in Control media or TCM for 7 days, cultures were decellularized using a modified protocol44 that included extra wash steps. Briefly, ECMs were incubated with extraction buffer containing 0.5% PBS-X and 20 mM NH4OH at 37 °C for 2–3 min, then washed with PBS and deionized water. Afterwards, samples were blocked for non-specific binding with PBS containing 1% BSA at 37 °C for 30 min and washed multiple times. For immunofluorescence studies, matrices were fixed for 1 h at 4 °C and stained for Fn using a primary mouse antibody and an anti-mouse Alexa Fluor 488-conjugated secondary antibody, as described above. DAPI was used as a nuclear counterstain and Alexa Fluor 568 phalloidin enabled detection of the F-actin cytoskeleton. Matrix thickness was determined by analysis of confocal microscopy Z-stack images captured with a Zeiss LSM 710 confocal microscope.
Analysis of tumor cell responses to ASC-derived ECMs
To analyze whether GC cell-mediated variations of ASC-derived ECMs feedback into altered tumor cell behavior, MKN74 and CD44v6 cells were cultured on either the different decellularized matrices or tissue culture plastic (TCPS) in low serum media (DMEM, 1% FBS, 1% Pen/Strep) for 3 days. Cell growth was determined by counting trypsinized cells using a hemocytometer, while proliferation was assessed by immunofluorescence image analysis of BrdU incorporation, as described above. To quantify cell clustering, decellularized ECMs were analyzed by immunofluorescence image analysis of Fn, F-actin cytoskeleton, and DAPI. Cell nuclei from five representative images for each sample for a total of four samples per condition were manually counted and the results expressed as the percentage of individual cells and clusters. For inhibition studies, media was supplemented with 20 μM broad-spectrum MMP inhibitor BB-94 (Batimastat, Tocris).
In vivo studies
MKN74 and CD44v6 cells were injected individually or in combination with ASCs (maximum number of cells = 9.5 × 105 in 100 μL of DMEM, 10% FBS, 1% Pen/Strep) into the subcutaneous space of five-week-old, female NOD.CB17-Prkdcscid/J (SCID) mice (4 mice, 2 tumors per mouse) per condition in accordance with protocol 2009-0117 as approved by Cornell’s University Institutional Animal Care and Use Committee (IACUC). Explants were harvested eight weeks after implantation, imaged, and formalin fixed for subsequent paraffin-embedding and histological analysis. Five-μm sections were stained with Hematoxylin and Eosin (H&E) or Masson’s Trichrome. To assess the degree of desmoplasia, H&E sections were scored by a pathologist (NLS) in a blinded manner using the following rubric:
Grade 1: delicate indistinct fibrovascular stroma diffusely throughout sheets of neoplastic cells
Grade 2: thin distinct bands of fibrovascular stroma throughout sheets of neoplastic cells
Grade 3: thin distinct fibrovascular bands separating neoplastic cells into packeted arrangements
Grade 4: streaming thick fibrovascular bands separating neoplastic cells into packeted arrangements.
Desmoplasia scoring was confirmed with Masson’s Trichrome sections using digitally scanned images and Aperio ImageScope software. Tumor cell proliferation was assessed via manual counting of Ki67 (Dako, clone MIB-1) immunoreactive nuclei in 5 representative fields of view at 200-fold magnification per tumor using ImageJ software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6. Student’s t-tests were used to compare two data sets, whereas two-way analysis of variance (ANOVA) with Tukey’s post-hoc test was applied for multiple comparisons. Data are represented as mean ± standard error of the mean (SEM) and differences were considered statistically significant for p values < 0.05. In vitro results are shown for one representative study after similar results were replicated in three independent experiments.
Results
CD44v6 alters tumor growth and architecture in vivo
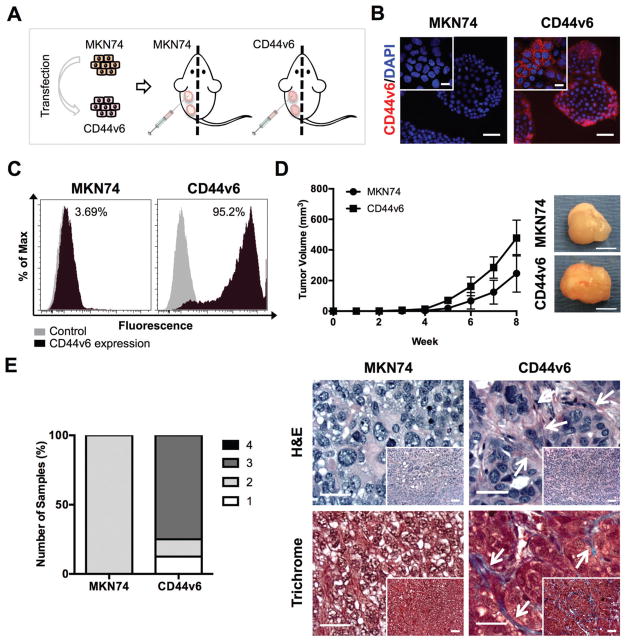
CD44v6 expression has been associated with poor prognosis in several human cancers, particularly those originating in the gastrointestinal tract.6,45 Nevertheless, the relationship between CD44v6 overexpression and the clinicopathological features of GC remains unknown. To unravel whether CD44v6 expression by tumor cells affects the desmoplastic reaction and tumor growth of GC, we used an isogenic human GC cell line over-expressing the CD44v6 isoform (CD44v6 cells), previously established by our group (unpublished data). This model was generated using the CD44-negative MKN74 cell line as a recipient to over-express the CD44v6 isoform (Fig. 1A). Transfection and selection efficiency was assessed by immunocytochemistry (Fig. 1B) and flow cytometry (Fig. 1C). To evaluate tumor formation and malignancy in vivo, parental MKN74 and CD44v6-expressing cells were injected individually into SCID mice (Fig. 1A). Analysis of tumor size after eight weeks suggested that tumors resulting from CD44v6 subcutaneous xenografts trended toward more rapid growth and increased tumor size when compared to MKN74 subcutaneous xenografts (Fig. 1D); albeit these differences were not statistically different. Enhanced growth was associated with increased desmoplastic (fibrotic) response as histopathological analysis revealed that CD44v6 tumors contained more fibrous stroma intersecting packets of malignant epithelial cells relative to MKN74 tumors (Fig. 1E). Furthermore, Masson’s Trichrome staining indicated greater content of collagen (blue) in tumors formed by CD44v6 cells as compared to tumors formed by MKN74 cells (Fig. 1E), confirming that overexpression of CD44v6 promotes tumor-associated desmoplasia.17
Fig. 1.
CD44v6 alters tumor growth and architecture in vivo. (A) Schematic showing the establishment of a gastric cancer (GC) cell line stably expressing CD44v6 and study design for subcutaneous xenograft injections of GC cells individually in SCID mice. (B) Immunofluorescence images showing CD44v6 isoform expression and nuclei (DAPI) on MKN74 and CD44v6 cells. Scale bars = 50 μm. Inset bars = 20 μm. (C) Representative histograms of MKN74 and CD44v6 analysis by flow cytometry quantifying CD44v6 expression. (D) Tumor volume from MKN74 (n = 3) and CD44v6 (n = 8) subcutaneous xenografts over eight weeks of growth and representative gross images of explanted tumors at week 8. Scale bars = 5 mm. (E) Histological grading and representative photomicrographs of the desmoplastic response of MKN74 and CD44v6 subcutaneous xenografts on Hematoxylin and Eosin (H&E) stained sections with confirmation of collagen deposition (blue) via Masson’s Trichrome-stained sections after 8 weeks of subcutaneous injection. Arrows indicate fibrotic tissue. Scale bars = 25 μm, inset bars = 50 μm.
Tumor-secreted soluble factors from CD44v6 expressing tumor cells promote ASC myofibroblast differentiation in vitro and desmoplasia in vivo
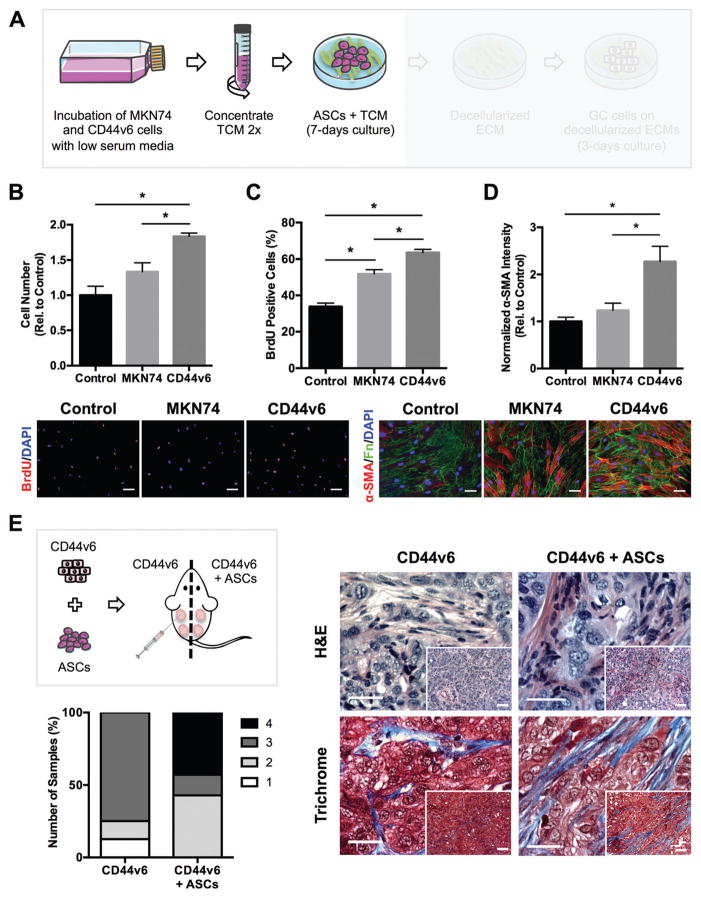
Based on our finding that tumors derived from CD44v6 over-expressing GC cells exhibit enhanced desmoplasia, we next sought to determine whether tumor cell-derived soluble factors mediate these differences by influencing ASC fate. Paracrine signaling between cancer cells and stromal cells supports the formation of a tumor-promoting microenvironment by inducing the differentiation of host stromal cells into myofibroblasts, which are key regulators of tumor desmoplasia.46 Nevertheless, the role that CD44v6 expression plays in this process is currently unknown. To evaluate the paracrine effects of MKN74 and CD44v6-secreted factors on ASC behavior, human ASCs were cultured in TCM from the two cell lines (Fig. 2A). Additionally, mouse 3T3-L1 preadipocytes were used to verify that results were not limited to primary ASCs, but are also relevant to other adipose stromal cells. Analysis of cell numbers (Fig. 2B and Fig. S1A, ESI†) and BrdU incorporation (Fig. 2C and Fig. S1B, ESI†) indicated that MKN74-conditioned media appeared to increase ASC proliferation; however, no significant difference in cell number was detected. In contrast, CD44v6 expression enabled GC cells to significantly increase ASC and 3T3-L1 proliferation via secretory factors in comparison with MKN74 cells that lack CD44v6 expression. Subsequently, ASC and 3T3-L1 differentiation into myofibroblasts was assessed by analysis of their expression of the myofibroblast marker α-SMA. Indeed, ASCs and 3T3-L1s exposed to CD44v6-TCM exhibited increased levels of α-SMA relative to control conditions, as evidenced by both immunofluorescence (Fig. 2D and Fig. S1C, ESI†) and Western blot analysis (Fig. S2, ESI†). Similarly, stromal cells have previously been shown to differentiate into myofibroblasts under the influence of tumor-derived factors.27,29,47,48 Among these factors, transforming growth factor-beta (TGF-β) plays a key role in myofibroblastic differentiation49 and is secreted at higher levels by more malignant tumor cells.27 Interestingly, comparison of Control media as well as TCM from MKN74 and CD44v6 cells via ELISA revealed that TGF-β is significantly enhanced in the TCM of CD44v6 expressing tumor cells (Fig. S3, ESI†), suggesting that CD44v6 expression up-regulates TGF-β secretion,50,51 which functions as an upstream mediator of ASC and 3T3-L1 differentiation into myofibroblasts27,52 (Fig. 2D and Fig. S3, ESI†). These results were further supported by tumor xenografts obtained after injecting a mixture of CD44v6 cells and ASCs in comparison to xenografts from CD44v6 cells alone. Histopathological evaluation of H&E-stained sections revealed increased heterogeneity of the desmoplastic response and a higher proportion of grade 4 desmoplasia when CD44v6 cells were implanted together with ASCs vs. alone (Fig. 2E). Additionally, increased fibrous tissue (desmoplasia) was apparent between nests of malignant epithelial cells in tumors derived from co-implantation of CD44v6 cells with ASCs vs. CD44v6 cells alone. Masson’s Trichrome staining confirmed this finding with greater collagen deposition (blue) in co-implanted tumors (Fig. 2E). Collectively, our data suggest that CD44v6 over-expression enhances hallmark features of myofibroblastic differentiation in ASCs, which contribute to a desmoplastic tumor microenvironment in GC.
Fig. 2.
Tumor-secreted soluble factors from CD44v6 expressing GC cells promote adipose-stromal cell (ASC) myofibroblast differentiation in vitro and desmoplasia in vivo. (A) Experimental setup showing collection of tumor-conditioned media (TCM) from GC cell lines and its incubation with ASCs. (B) Number of ASCs after culture in TCMs collected from Control, MKN74, and CD44v6 cells relative to Control (n = 3 samples per condition). (C) Immunofluorescence analysis of BrdU incorporation in ASCs treated with the different TCMs (n = 40 images per condition). Scale bars = 20 μm. (D) Immunofluorescence analysis of α-smooth muscle actin (α-SMA) of ASCs relative to Control (n = 20 images per condition). Scale bars = 20 μm. (E) Schematic of xenografting conditions. CD44v6 cells were injected into SCID mice either alone (n = 8) or in combination with ASCs (n = 7) and tumors were harvested after 8 weeks. Representative photomicrographs of H&E-stained cross-sections and confirmation of collagen deposition (blue) via Masson’s Trichrome stained sections. Corresponding histological scoring of desmoplasia based on H&E sections. Tumors were harvested 8 weeks after injection. Scale bars = 25 μm, Inset bars = 50 μm. * p < 0.05.
CD44v6 TCM-treated ASCs enhance fibrotic/desmoplastic ECM deposition and remodeling
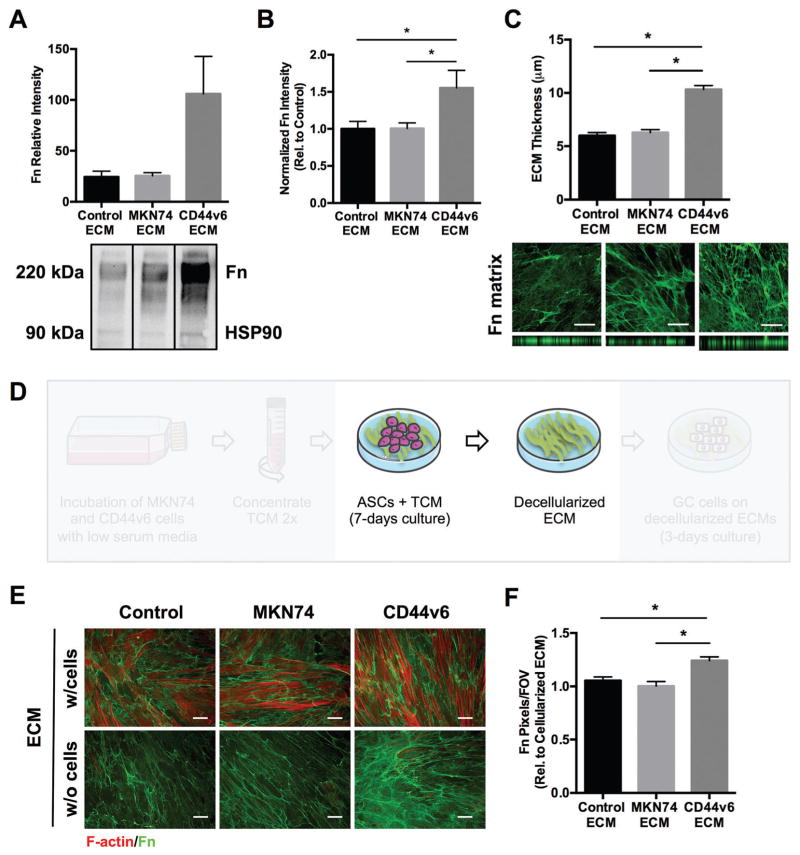
To evaluate whether the detected changes in ASC phenotype enhanced fibrotic/desmoplastic ECM deposition and remodeling, the levels of fibronectin, an ECM component primarily deposited by myofibroblasts and increased in GC,24,53 were analyzed by Western blot analysis (Fig. 3A). This analysis was further confirmed by fibronectin immunofluorescent staining of ASC and 3T3-L1-deposited ECMs (Fig. 3B and Fig. S1D, ESI†). Our results indicate that CD44v6-TCM-treated ASCs assembled denser fibronectin matrices with significantly increased thickness relative to control conditions (Fig. 3C), both parameters that were previously associated with increased ECM stiffness.47,52 In fact, increased levels of fibronectin had been previously detected in GC, mostly in the connective tissue at the invading edge of the tumors53 and stiffening of the stomach wall is a common feature of diffuse-type GC.54 Furthermore, consistent with our in vivo results, ASCs produced significantly more collagen I when treated with TCM from CD44v6 expressing cells rather than control conditions (Fig. S4, ESI†). This is a notable finding, as increased collagen I deposition contributes to increased tumor stiffness, motility/invasion and aggressiveness.22 Together, these results reveal that soluble factors from CD44v6 expressing GC cells promote the pro-fibrotic phenotype of ASCs, which enhance ECM deposition and remodeling.
Fig. 3.
CD44v6 TCM-treated ASCs increase pro-fibrotic ECM deposition and remodeling. (A) Western blot quantification of fibronectin (Fn) deposited by TCM-treated ASCs relative to the corresponding HSP90 levels (n = 3 samples per condition). (B) Immunofluorescence image analysis of Fn deposited by TCM-treated ASCs relative to Control (n = 20 images per condition). Scale bars = 20 μm. (C) Quantification of deposited ECM thickness by confocal image analysis of Fn (n = 37 areas per condition). Scale bars = 20 μm. (D) Experimental setup illustrating in vitro ECM decellularization. (E and F) Immunofluorescence image analysis comparing Fn content post-decellularization relative to pre-decellularization (F-actin staining of ASCs) across experimental conditions (n = 40 images per condition). Scale bars = 20 μm. FOV, field of view. * p < 0.05.
As TCM similarly modulated ASC and 3T3-L1 behavior, subsequent studies were performed with the physiologically more relevant ASC primary cells.
Decellularized ASC-derived matrices retain native ECM composition and architecture
To obtain decellularized matrices for subsequent studies of GC cell behavior, ECMs deposited by ASCs in the presence of Control media or TCM were decellularized after 7 days of culture (Fig. 3D), leaving behind a cell-free fibrillar ECM (Fig. 3E). To evaluate decellularization efficiency and confirm that initial ECM characteristics were maintained, immunofluorescence analysis of fibronectin and F-actin was performed before and after decellularization. Image analysis confirmed the absence of cell remnants in decellularized matrices, and that the fibronectin network architecture was consistent before and after decellularization (Fig. 3E). Furthermore, upon decellularization, the levels of fibronectin remained significantly enhanced on CD44v6-TCM treated ASC-derived ECMs compared with MKN74 and Control ECMs (Fig. 3F). This confirmed our ability to generate decellularized matrices as a platform to interrogate the role of ASC-mediated ECM remodeling on GC cell behavior.
Decellularized matrices assembled by CD44v6-TCM treated ASCs promote GC phenotypes associated with malignancy
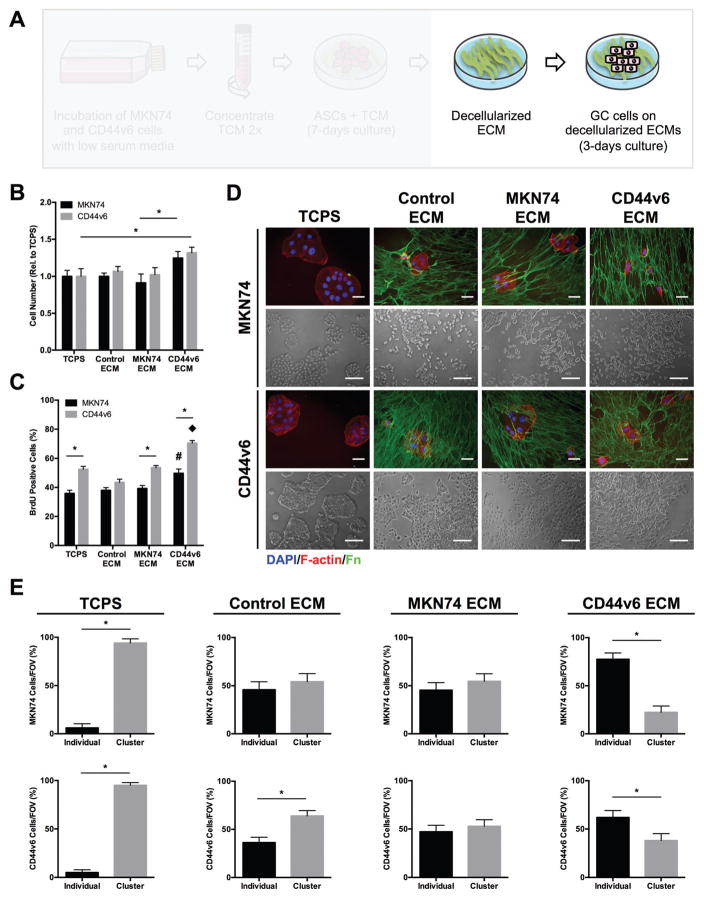
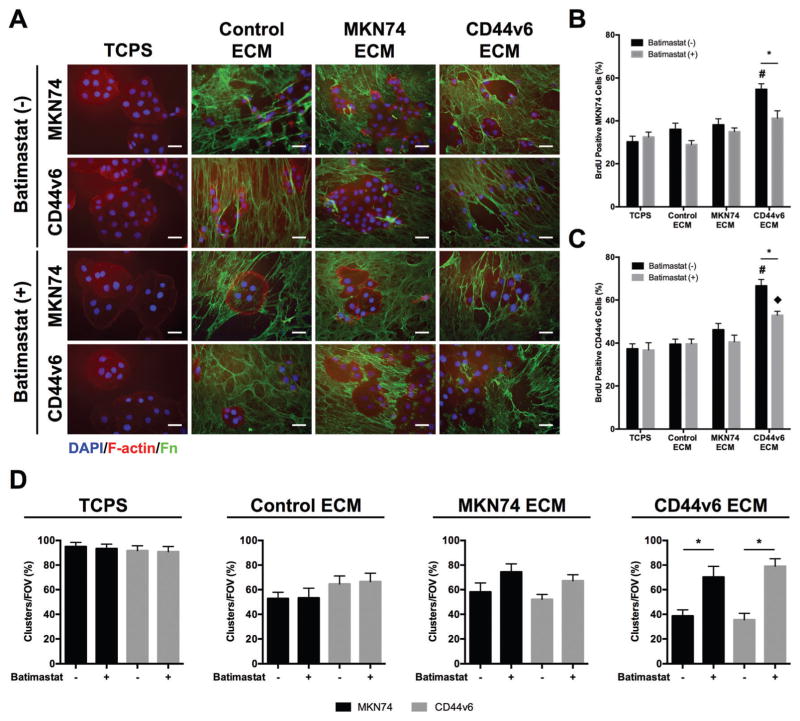
Given that ASC-mediated changes of ECM composition and structure can increase aggressiveness in other cancers,28,55 we next assessed whether GC cells also respond to varied ASC-dependent ECM properties. To this end, MKN74 and CD44v6 cells were seeded onto the different decellularized matrices described above (Fig. 4A). Cell counting indicated that MKN74 and CD44v6 cell growth was increased on decellularized matrices from CD44v6-conditioned ASCs relative to matrices from ASCs preconditioned with MKN74 TCM, Control media, or even on TCPS (Fig. 4B). BrdU incorporation confirmed increased proliferation of both GC cell lines on decellularized CD44v6- vs. MKN74-associated ECMs whereby a more pronounced effect was observed for CD44v6 proliferation (Fig. 4C). These observations imply that CD44v6 TCM-pretreatment enables ASCs to deposit matrix that enhances tumor cell proliferation via a positive feed-forward regulation. To determine whether CD44v6-associated ECM remodeling also regulates the level of direct cell–cell contact between GC cells, a parameter determining dissemination and thus, ultimately metastasis,56 the number of cell clusters and individual cells in all conditions was determined. Phase-contrast and immunofluorescence images were used to quantify the number of cell clusters and individual cells in all conditions (Fig. 4D). This analysis revealed that the different tumor cells primarily adhered in the form of multicellular clusters on TCPS, while culture on decellularized matrices increased the number of individual cells. Importantly, the number of individual cells further increased when both MKN74 and CD44v6 cells were cultured on CD44v6-TCM treated ASC-derived decellularized matrices relative to all other conditions (Fig. 4E). These observations suggest that ASC-mediated differences in ECM remodeling influence both the proliferation and intercellular adhesion characteristics of GC cells.
Fig. 4.
Decellularized ECMs assembled by CD44v6-TCM treated ASCs promote GC phenotypes associated with malignancy. (A) Experimental setup to analyze the effect of different ASC-assembled, decellularized ECMs on GC cells (MKN74 and CD44v6 cells). (B) Number of MKN74 and CD44v6 cells after 3 days of culture on decellularized ECMs relative to tissue culture plastic (TCPS) (n = 4) * p < 0.05. (C) BrdU incorporation by MKN74 and CD44v6 cells after 3 days of culture on decellularized ECMs and TCPS determined by immunofluorescence image analysis (n = 20 images per condition) * p < 0.05 from different cell lines within the same experimental condition, # and ◆ p < 0.05 between experimental conditions for the same cell line. (D) Representative immunofluorescence (Fn, F-actin, and nuclei [DAPI]) and phase contrast photomicrographs of MKN74 and CD44v6 cells on decellularized ECMs assembled under different TCM culture conditions. Scale bars = 20 μm. (E) Percentage of individual cells and clusters of MKN74 and CD44v6 cells after 3 days of culture on decellularized ECMs and TCPS as determined by image analysis (n = 25 images per condition) * p < 0.05.
Decellularized CD44v6-associated ECMs increase GC cell proliferation and decrease cell clustering in an MMP-dependent manner
Given our observations that ASC-dependent matrix properties influence GC cell behavior and because MMPs are widely accepted for their manifold contributions to matrix-induced changes of tumorigenesis,20 we next tested whether a functional link exists between GC cell responses to the varied matrices and MMP activity. To this end, GC cells were seeded onto the different decellularized matrices and their MMP activity was blocked pharmacologically using the broadband MMP inhibitor BB-94 (batimastat) (Fig. 5A). Image analysis of BrdU incorporation confirmed that this treatment significantly decreased the proliferation of both MKN74 (Fig. 5B) and CD44v6 cells (Fig. 5C) on CD44v6-decellularized matrices. In contrast, no effect on cell proliferation was observed on TCPS or Control ECMs, while a slight trend, but no significant difference was noted when GC cells were seeded onto MKN74-decellularized matrices. Similarly, image analysis indicated that MMP inhibition had no significant effect on cell clustering on TCPS, Control and decellularized MKN74-associated matrices, while the same treatment promoted cell clustering on decellularized CD44v6-associated matrices (Fig. 5D). These results imply that MMPs contributed to the observed phenotypic changes of GC cells in response to CD44v6-matrices, whereas possible intrinsic differences in MMP activity between MKN74 and CD44v6 were not a driving factor. Together, these data suggest that ASC-mediated changes in ECM remodeling may contribute to the aggressiveness of GC cells by modulating MMPs.
Fig. 5.
Decellularized CD44v6-associated ECMs increase GC cell proliferation and decrease cell clustering in an MMP-dependent manner. (A) Representative immunofluorescence images of MKN74 and CD44v6 cells after 3 days of culture on decellularized ECMs assembled under different TCM culture conditions and TCPS in the presence and absence of MMPs inhibitor (batimastat). Scale bars = 20 μm. (B and C) BrdU incorporation by MKN74 cells (B) and CD44v6 cells (C) after culture on decellularized ECMs and TCPS in the presence and absence of batimastat as determined by immunofluorescence image analysis (n = 15 images per condition) * p <0.05 from different culture conditions for the same ECM, # and ◆ p <0.05 between ECMs for the same culture condition. (D) Percentage of individual cells and clusters of MKN74 and CD44v6 cells after culture on decellularized ECMs and TCPS in the presence and absence of batimastat as determined by immunofluorescence image analysis (n = 20 images per condition) * p < 0.05.
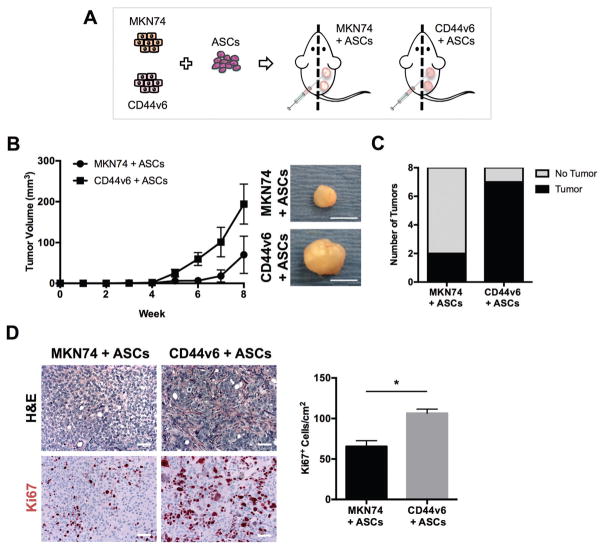
Upon co-injection with ASCs CD44v6 tumors are more proliferative and desmoplastic than MKN74 tumors
To determine the relevance of the above in vitro findings in vivo, MKN74 or CD44v6 cells were injected in combination with ASCs into the subcutaneous space of SCID mice (Fig. 6A). The co-implantation of ASCs and GC cells resulted in a trend for development of larger tumors with CD44v6 cells, when compared to MKN74 cells (Fig. 6B). The lack of statistical significance is likely related to the high heterogeneity in tumor sizes from the study arms. Eight weeks post-injection, tumors were observed in 87% (7 out of 8) of CD44v6 + ASCs co-injections, whereas only 25% (2 out of 8) of MKN74 + ASCs co-injections were able to generate tumors (Fig. 6C). Furthermore, on histological evaluation, tumors derived from CD44v6 + ASCs displayed increased fibrous stroma intersecting packets of malignant epithelial cells (Fig. 6D), in contrast to tumors derived from MKN74 + ASCs. This desmoplastic response was accompanied by increased cell proliferation in CD44v6 + ASCs tumors relative to MKN74 + ASCs tumors, as determined by nuclear immunoreactivity of the cell proliferation marker Ki-67 (Fig. 6D). Notably, this finding corroborates with our in vitro observations on the decellularized ECM model system. Additionally, larger and more pleomorphic nuclei were found in CD44v6 + ASCs tumors than in MKN74 + ASCs tumors suggesting increased malignancy when CD44v6 is expressed (Fig. 6D). Overall, these findings indicate that CD44v6 expression modulates GC malignant behavior in vivo by enhancing ASC-mediated contributions to tumor stroma formation.
Fig. 6.
Upon co-injection with ASCs CD44v6 tumors are more proliferative and desmoplastic than MKN74 tumors. (A) Schematic showing the study design for subcutaneous xenograft co-injections of MKN74 and CD44v6 cells with ASCs in SCID mice. (B) Measurement of tumor volume from MKN74 with ASCs (n = 3) and CD44v6 with ASCs (n = 8) subcutaneous xenografts over eight weeks of tumor growth with representative gross images of tumors explanted at the end of the experiment. Scale bars = 5 mm. (C) Comparison of the number of tumors formed per total number of subcutaneous implantations between experimental conditions. Analysis was performed 8 weeks after subcutaneous injection. (D) Representative photomicrographs of H&E stained sections of tumors collected 8 weeks after subcutaneous implantation of MKN74 cells + ASCs and CD44v6 cells + ASCs. Comparison of GC cell proliferation between conditions as quantified by immunohistological analysis of Ki67 positive cells (n = 7). Scale bars = 50 μm. * p < 0.05.
Discussion
This study elucidates that CD44v6 overexpression may enhance GC malignant behavior by modulating ASC-derived ECM remodeling. Specifically, our results indicate that soluble factors secreted by CD44v6 expressing GC cells increase fibrotic/desmoplastic ECM deposition and remodeling by promoting pro-fibrotic differentiation of ASCs. These ECM changes, in turn, alter the behavior of GC cells towards a more aggressive phenotype including increased proliferative and proteolytic activity as well as reduced cell–cell interactions, which are features reminiscent of aggressive tumor cells in patients.
We have previously demonstrated that CD44v6 is progressively expressed during malignant transformation of gastric pre-neoplastic lesions to advanced carcinoma.13 However, the question remained whether CD44v6 plays a role in the clinicopathological features of GC. In diffuse-type GC, the neoplastic cells are poorly cohesive and rapidly invade the gastric wall, which becomes stiffer due to an extensive desmoplastic (fibrotic) response.54 In contrast, intestinal-type GC from which the MKN74 cell line lacking endogenous CD44v6 expression was derived as a model, is not generally associated with desmoplasia. Accordingly, we demonstrated that MKN74 xenografts are characterized by a very mild desmoplastic response. However, when forced to overexpress CD44v6, MKN74 cells are able to promote fibrotic remodeling and tumor growth and our results imply that the resulting changes in ECM remodeling may play a role in this process. In fact, in a variety of human malignancies such as stomach, breast, and colorectal cancers, the expression of CD44v6 is also associated with aggressive behavior of the disease.11,45 Furthermore, CD44v6 has been shown to be involved in cell–cell and cell–ECM interactions,5,7 and appears to be an important factor driving crosstalk between tumor cells and surrounding stromal cells, thus supporting the formation of a tumor-promoting microenvironment by increasing desmoplasia. Nevertheless, future studies will be needed to test the functional relevance of our findings in an orthotopic mouse model of GC and determine consequences on GC invasion and metastasis.
In the context of GC, adipose tissue constitutes a major component of tumor stroma that GC cells encounter as they progressively invade and therefore, ASCs may play a role in the formation of a tumor-promoting microenvironment.35 Previous reports have shown that direct cell–cell interaction between GC cells and stromal cells contributes to the aggressive phenotype of GC.32–34 Using conditioned media-experiments, we now show that soluble factors from CD44v6 expressing cells may promote protumorigenic GC-stromal cell interactions by stimulating ASCs to proliferate and undergo differentiation into myofibroblasts. This was confirmed by the higher proliferative capacity of ASCs determined through BrdU staining, as well as increased levels of α-SMA, fibronectin, and type I collagen, whose up-regulation is known to accompany the neoplastic transformation of the stomach.57 Our findings are consistent with previous reports27–29,34 and suggest that CD44v6 expression up-regulates TGF-β secretion,50,51 which may function in a paracrine manner to stimulate the production of ECM components58,59 and modulate the levels of α-SMA independent of pre-existing or newly deposited ECM.60 Nevertheless, further investigation is warranted to understand the functional link between TGF-β and CD44v6 expression and to identify additional growth factors and cytokines that may contribute to CD44v6-mediated changes in ASC-associated ECM deposition and remodeling.
ECM provides critical biochemical and biophysical cues that direct cell growth, motility, survival, and differentiation, both in physiological and pathological conditions such as cancer.16 Nevertheless, the mechanisms and cues driving tumorigenesis are typically studied with oversimplified and artificial systems that do not resemble the native architecture and biochemical composition of human tumor-associated ECM. In the past years, more sophisticated in vitro systems such multicellular spheroids,61 organotypic constructs,62 and cell-derived 3D matrices42 have been developed to better recapitulate various aspects of the in vivo microenvironment and to bridge the gap between in vitro and in vivo systems. Still, the vast majority of previous reports regarding CD44 and its isoforms have been performed on conventional 2D substrates. Yet engineered biomaterials provide an effective alternative to dissect biophysical cues that influence CD44 alternative splicing in GC cells.39 Decellularized matrices, in particular, can accurately mimic the composition and architecture of native ECMs, making them attractive biomimetic platforms to unravel the effect of ECM on tumor cell behavior.41,43 Hence, we used this approach to study the role of CD44v6-induced changes in ECM physicochemical characteristics on GC cell behavior. Notably, our findings illustrate that decellularized CD44v6-associated ECMs increased the malignant behavior of human GC cells by increasing their proliferation and decreasing cell clustering – a change in cellular phenotype that is associated with a more aggressive phenotype of GC.56 These findings are highly relevant as perturbations in epithelial cell junctions and individualization of epithelial cells are important steps in the initiation and progression of cancer.56 The decreased ability of both GC cell lines to form multicellular clusters, regardless of CD44v6 expression, on decellularized matrices with denser and thicker fiber networks, suggests crosstalk between matrix structural characteristics and GC cell behavior. Nevertheless, the specific mechanisms underlying these matrix-induced differences in GC cell behavior will need to be uncovered in future studies.
Various mechanisms may have collectively contributed to our finding that CD44v6 expression promotes malignancy by altering ASC-derived ECM deposition. As CD44v6-associated ECMs exhibit structural and compositional alterations (denser matrix, thicker fibers) known to alter cell–matrix interactions, these alterations may differentially activate mechanosignaling. In fact, thicker fibers are stiffer and may alter ligand density, which in turn, would change focal adhesion formation and downstream cellular behavior.22 Furthermore, CD44v6 can activate the mitogen-activated protein kinase (MAPK) pathway, which plays an important role in translating extracellular stimuli to altered cellular behavior including increased tumor cell proliferation.63 Additionally, unregulated proliferation of cancer cells can be achieved by means in which MMPs may also play a role: (i) by cleaving and activating precursor proteins or releasing cleavage fragments with new bioactivities; (ii) by shedding transmembrane precursors of some growth factors; and (iii) by regulating the ECM composition, which indirectly promotes proliferation through interactions between ECM molecules and integrins.20,22,64 In our studies, ECM-induced changes in tumor cell behavior are decreased when GC cells are treated with a broad spectrum MMP inhibitor (batimastat). This suggests that CD44v6-associated changes in ECM remodeling modulate tumor cell behavior due in part to altered MMP activity. It is possible that the proteolytic CD44v6-associated ECM remodeling resulted in the release of breakdown products with biologic activity that may have contributed to our observations. For example, the MMP-dependent cleavage of native fibrillar collagen exposes cryptic RGD sites that can then be ligated by αvβ3 integrin, and this interaction promotes cell proliferation.65 Although previous reports may support our hypothesis, future mechanistic studies will be necessary to further confirm our findings. Another aspect worthy to be addressed is whether CD44v6-mediated changes in ECM remodeling have an effect on ASCs themselves and whether potential changes in ASC paracrine signaling may further modulate GC cell behavior. Another aspect to consider in future work is which ECM components may underlie the detected changes in GC cells. Although fibronectin and type I collagen have been characterized in this study, as they are described as the most abundant fibrillar ECM proteins found in cancer-associated ECM,24 other ECM components including hyaluronic acid should also be considered.
Using ASC-derived decellularized ECMs, this study highlights that CD44v6 expression by GC cells is functionally linked to desmoplastic stroma remodeling and consequential changes in malignancy. Importantly, our in vitro results are supported by in vivo studies, highlighting the relevance of decellularized matrices to be used as reliable biomimetic platforms to unravel the role of ECM changes in tumor cell malignant behavior. Likewise, the decellularized matrices explored in this study may also be useful in future studies to identify novel strategies that reduce GC malignancy by targeting CD44v6.
Conclusions
By using a combination of media conditioning experiments and decellularized matrices in conjunction with in vivo studies, our findings revealed that CD44v6 overexpression contributes to the desmoplastic response of GC. Further, the data presented here indicate that these changes in ECM remodeling directly enhance the malignant behavior of GC cells. Collectively, our findings improve understanding of the role of ASC-mediated ECM remodeling in GC and highlight a potential new mechanism by which the protumorigenic CD44v6 isoform regulates gastric tumorigenesis. Overall, CD44v6 isoform emerges as an attractive and specific molecule that can be explored as a potential therapeutic target in the surveillance and treatment of GC patients.
Supplementary Material
Insight, innovation, integration.
Fibrotic extracellular matrix (ECM) remodeling is a hallmark of aggressive cancer, but its role in gastric cancer (GC) remains elusive. Similarly, the hyaluronic acid receptor CD44 variant 6 (CD44v6) has been associated with invasion and metastasis in several cancers and is expressed de novo in more than 60% of GCs. However, it is unclear whether GC expression of CD44v6, fibrotic ECM remodeling, and GC cell aggressiveness are functionally linked. By integrating media-conditioning experiments, decellularized matrices, and in vivo experiments, our studies reveal that induced expression of CD44v6 in GC cells stimulates fibrotic stroma and ECM remodeling. These differences, in turn, enhance the malignant potential of GC cells due to varied ECM remodeling opening new opportunities for CD44v6-targeted therapies of GC.
Acknowledgments
This work was supported by: (1) the Center on the Physics of Cancer Metabolism through Award Number 1U54CA210184-01 and (2) R01CA185293, both from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. (3) Cornell BRC – Biotechnology Resource Center Imaging Facility (grant number NIH-S10RR025502); (4) European Regional Development Fund (ERDF) through the COMPETE 2020 – Operational Programme for Competiveness and Internationalization (POCI), Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, and (5) Portuguese funds through the Portuguese Foundation for Science and Technology (FCT) in the framework of the projects: “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274), NORTE-01-0145-FEDER-000029, “PEst-C/SAU/LA0003/2013”, and NORTE-01-0145-FEDER-000012 – Structured program on bioengineered therapies for infectious diseases and tissue regeneration, and FCT Fellowships to Bianca N. Lourenço (PhD grant ref. SFRH/BD/87400/2012) and to Daniel Ferreira (PhD grant ref. PD/BD/105976/2014). IPATIMUP and INEB integrate the i3S Research Unit, which is partially supported by FCT. Illustrations were produced using Servier Medical Art (www.servier.com), for which the authors would like to acknowledge Servier.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ib00179g
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
BNL, PLG, and CF conceptualized the project. BNL, NLS, and DF performed experiments and formal analysis. BNL wrote the original draft. NLS, DF, CO, PLG, and CF reviewed and edited the manuscript. PLG, CO, and CF acquired funding and supervised the project.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Ca-Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ, Smith DB, Langley RE, Verma M, Weeden S, Chua YJ, Participants MT. N Engl J Med. 2006;355:11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 3.Orian-Rousseau V. Eur J Cancer. 2010;46:1271–1277. doi: 10.1016/j.ejca.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 4.Louderbough JM, Schroeder JA. Mol Cancer Res. 2011;9:1573–1586. doi: 10.1158/1541-7786.MCR-11-0156. [DOI] [PubMed] [Google Scholar]
- 5.Ponta H, Sherman L, Herrlich PA. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 6.Xu YY, Guo M, Yang LQ, Zhou F, Yu C, Wang A, Pang TH, Wu HY, Zou XP, Zhang WJ, Wang L, Xu GF, Huang Q. Oncotarget. 2017;8:45848–45861. doi: 10.18632/oncotarget.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer B, Jauch KW, Gunthert U, Figdor CG, Schildberg FW, Funke I, Johnson JP. Lancet. 1993;342:1019–1022. doi: 10.1016/0140-6736(93)92879-x. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi T, Kondo K, Hino N, Uyama T, Monden Y. Clin Cancer Res. 1997;3:1289–1297. [PubMed] [Google Scholar]
- 9.Kaufmann M, Heider KH, Sinn HP, von Minckwitz G, Ponta H, Herrlich P. Lancet. 1995;345:615–619. doi: 10.1016/s0140-6736(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 10.Zeimet AG, Widschwendter M, Uhl-Steidl M, Muller-Holzner E, Daxenbichler G, Marth C, Dapunt O. Br J Cancer. 1997;76:1646–1651. doi: 10.1038/bjc.1997.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wielenga VJM, Heider KH, Offerhaus GJA, Adolf GR, Vandenberg FM, Ponta H, Herrlich P, Pals ST. Cancer Res. 1993;53:4754–4756. [PubMed] [Google Scholar]
- 12.Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H. Genes Dev. 2002;16:3074–3086. doi: 10.1101/gad.242602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Cunha CB, Oliveira C, Wen X, Gomes B, Sousa S, Suriano G, Grellier M, Huntsman DG, Carneiro F, Granja PL, Seruca R. Lab Invest. 2010;90:1604–1614. doi: 10.1038/labinvest.2010.155. [DOI] [PubMed] [Google Scholar]
- 14.Bissell MJ, Radisky D. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiteside TL. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes RO. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu P, Weaver VM, Werb Z. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egeblad M, Werb Z. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 19.Jodele S, Blavier L, Yoon JM, DeClerck YA. Cancer Metastasis Rev. 2006;25:35–43. doi: 10.1007/s10555-006-7887-8. [DOI] [PubMed] [Google Scholar]
- 20.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin JW, Verbeke CS, Allison KH, Mooney DJ. Nat Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 24.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 25.Cox TR, Erler JT. Dis Models & Mech. 2011;4:165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, Tycko B, Wang TC. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandler EM, Seo BR, Califano JP, Andresen Eguiluz RC, Lee JS, Yoon CJ, Tims DT, Wang JX, Cheng L, Mohanan S, Buckley MR, Cohen I, Nikitin AY, Williams RM, Gourdon D, Reinhart-King CA, Fischbach C. Proc Natl Acad Sci U S A. 2012;109:9786–9791. doi: 10.1073/pnas.1121160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo BR, Bhardwaj P, Choi S, Gonzalez J, Andresen Eguiluz RC, Wang K, Mohanan S, Morris PG, Du B, Zhou XK, Vahdat LT, Verma A, Elemento O, Hudis CA, Williams RM, Gourdon D, Dannenberg AJ, Fischbach C. Sci Transl Med. 2015;7:301ra130. doi: 10.1126/scitranslmed.3010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song YH, Warncke C, Choi SJ, Choi S, Chiou AE, Ling L, Liu HY, Daniel S, Antonyak MA, Cerione RA, Fischbach C. Matrix Biol. 2017;60–61:190–205. doi: 10.1016/j.matbio.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittmann K, Fischbach C. ACS Biomater Sci Eng. 2016;3:1483–1493. doi: 10.1021/acsbiomaterials.6b00328. [DOI] [PubMed] [Google Scholar]
- 31.Guo X, Oshima H, Kitmura T, Taketo MM, Oshima M. J Biol Chem. 2008;283:19864–19871. doi: 10.1074/jbc.M800798200. [DOI] [PubMed] [Google Scholar]
- 32.Semba S, Kodama Y, Ohnuma K, Mizuuchi E, Masuda R, Yashiro M, Hirakawa K, Yokozaki H. Br J Cancer. 2009;101:1365–1373. doi: 10.1038/sj.bjc.6605309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yashiro M, Hirakawa K. Cancer Microenviron. 2010;3:127–135. doi: 10.1007/s12307-010-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomoto-Kojima N, Aoki S, Uchihashi K, Matsunobu A, Koike E, Ootani A, Yonemitsu N, Fujimoto K, Toda S. Cell Tissue Res. 2011;344:287–298. doi: 10.1007/s00441-011-1144-3. [DOI] [PubMed] [Google Scholar]
- 35.Nieman KM, Romero IL, Van Houten B, Lengyel E. Biochim Biophys Acta. 2013;1831:1533–1541. doi: 10.1016/j.bbalip.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 37.Hutmacher DW. Nat Mater. 2010;9:90–93. doi: 10.1038/nmat2619. [DOI] [PubMed] [Google Scholar]
- 38.Bidarra SJ, Oliveira P, Rocha S, Saraiva DP, Oliveira C, Barrias CC. Sci Rep. 2016;6:27072. doi: 10.1038/srep27072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunha CB, Klumpers DD, Koshy ST, Weaver JC, Chaudhuri O, Seruca R, Carneiro F, Granja PL, Mooney DJ. Biomaterials. 2016;98:152–162. doi: 10.1016/j.biomaterials.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Barney LE, Dandley EC, Jansen LE, Reich NG, Mercurio AM, Peyton SR. Integr Biol. 2015;7:198–212. doi: 10.1039/c4ib00218k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 42.Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Am J Pathol. 2005;167:475–488. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reichert JC, Quent VMC, Burke LJ, Stansfield SH, Clements JA, Hutmacher DW. Biomaterials. 2010;31:7928–7936. doi: 10.1016/j.biomaterials.2010.06.055. [DOI] [PubMed] [Google Scholar]
- 44.Castello-Cros R, Cukierman E. Methods Mol Biol. 2009;522:275–305. doi: 10.1007/978-1-59745-413-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu YJ, Yan PS, Li J, Jia JF. World J Gastroenterol. 2005;11:6601–6606. doi: 10.3748/wjg.v11.i42.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalluri R, Zeisberg M. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 47.Wang K, Andresen Eguiluz RC, Wu F, Seo BR, Fischbach C, Gourdon D. Biomaterials. 2015;54:63–71. doi: 10.1016/j.biomaterials.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jotzu C, Alt E, Welte G, Li J, Hennessy BT, Devarajan E, Krishnappa S, Pinilla S, Droll L, Song YH. Cell Oncol. 2011;34:55–67. doi: 10.1007/s13402-011-0012-1. [DOI] [PubMed] [Google Scholar]
- 49.Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. J Cell Biol. 1993;122:103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Q, Stamenkovic I. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Xiao L, Luo CH, Zhou H, Zeng L, Zhong J, Tang Y, Zhao XH, Zhao M, Zhang Y. Mol Med Rep. 2015;11:3505–3510. doi: 10.3892/mmr.2015.3145. [DOI] [PubMed] [Google Scholar]
- 52.Chandler EM, Saunders MP, Yoon CJ, Gourdon D, Fischbach C. Phys Biol. 2011;8:015008. doi: 10.1088/1478-3975/8/1/015008. [DOI] [PubMed] [Google Scholar]
- 53.Grigioni WF, Derrico A, Fortunato C, Fiorentino M, Mancini AM, Stetlerstevenson WG, Sobel ME, Liotta LA, Onisto M, Garbisa S. Mod Pathol. 1994;7:220–225. [PubMed] [Google Scholar]
- 54.Carneiro F, Lauwers GY. Morson and Dawson’s Gastrointestinal Pathology. Vol. 13. Wiley-Blackwell; 2013. pp. 180–222. [DOI] [Google Scholar]
- 55.Song YH, Shon SH, Shan MR, Stroock AD, Fischbach C. Integr Biol. 2016;8:205–215. doi: 10.1039/c5ib00277j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiery JP. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 57.David L, Nesland JM, Holm R, Sobrinho-Simoes M. Cancer. 1994;73:518–527. doi: 10.1002/1097-0142(19940201)73:3<518::aid-cncr2820730305>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 58.Mahara K, Kato J, Terui T, Takimoto R, Horimoto M, Murakami T, Mogi Y, Watanabe N, Kohgo Y, Niitsu Y. Br J Cancer. 1994;69:777–783. doi: 10.1038/bjc.1994.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida K, Yokozaki H, Niimoto M, Ito H, Ito M, Tahara E. Int J Cancer. 1989;44:394–398. doi: 10.1002/ijc.2910440303. [DOI] [PubMed] [Google Scholar]
- 60.Huang XW, Yang NH, Fiore VF, Barker TH, Sun Y, Morris SW, Ding Q, Thannickal VJ, Zhou Y. Am J Respir Cell Mol Biol. 2012;47:340–348. doi: 10.1165/rcmb.2012-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charoen KM, Fallica B, Colson YL, Zaman MH, Grinstaff MW. Biomaterials. 2014;35:2264–2271. doi: 10.1016/j.biomaterials.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fatehullah A, Tan SH, Barker N. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 63.Marhaba R, Bourouba M, Zoller M. Cell Signalling. 2005;17:961–973. doi: 10.1016/j.cellsig.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 64.Grant MB, Caballero S, Bush DM, Spoerri PE. Diabetes. 1998;47:1335–1340. doi: 10.2337/diab.47.8.1335. [DOI] [PubMed] [Google Scholar]
- 65.Petitclerc E, Strömblad S, von Schalscha TL, Mitjans F, Piulats J, Montgomery AM, Cheresh DA, Brooks PC. Cancer Res. 1999;59:2724–2730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.