Abstract
Ethyl alcohol is a toxin that, when consumed at high levels, produces organ damage and death. One way to prevent or ameliorate this damage in humans, is to reduce the exposure of organs to alcohol by reducing alcohol ingestion. Both the propensity to consume large volumes of alcohol, and the susceptibility of human organs to alcohol-induced damage, exhibit a strong genetic influence. We have developed an integrative genetic/genomic approach to identify transcriptional networks that predispose complex traits, including propensity for alcohol consumption and propensity for alcohol-induced organ damage. In our approach, the phenotype is assessed in a panel of recombinant inbred (RI) rat strains and quantitative trait locus (QTL) analysis is performed. Transcriptome data from tissues/organs of naïve RI rat strains are used to identify transcriptional networks using Weighted Gene Coexpression Network Analysis (WGCNA). Correlation of the first principal component of transcriptional coexpression modules with the phenotype across the rat strains, and overlap of QTLs for the phenotype and the QTLs for the coexpression modules (module eigengene QTL) provide the criteria for identification of the functionally related groups of genes that contribute to the phenotype (candidate modules). While we previously identified a brain transcriptional module whose QTL overlapped with a QTL for levels of alcohol consumption in HXB/BXH RI rat strains and 12 selected rat lines, this module did not account for all of the genetic variation in alcohol consumption. Our search for QTL overlap and correlation of coexpression modules with phenotype can, however, be applied to any organ in which the transcriptome has been measured, and this represents a holistic approach in the search for genetic contributors to complex traits.
Previous work has implicated liver/brain interactions, particularly involving inflammatory/immune processes, as influencing alcohol consumption levels. We have now analyzed the liver transcriptome of the HXB/BXH RI rat panel in relation to the behavioral trait of alcohol consumption. We used RNA-Seq and microarray data to construct liver transcriptional networks, and identified a liver candidate transcriptional coexpression module that explained 24% of the genetic variance in voluntary alcohol consumption. The transcripts in this module focus attention on liver secretory products that influence inflammatory and immune signaling pathways. We propose that these liver secretory products can interact with brain mechanisms that affect alcohol consumption, and targeting these pathways provides a potential approach to reducing high levels of alcohol intake and also protecting the integrity of the liver and other organs.
Keywords: RNA-Seq, WGCNA transcriptional networks, liver, alcohol drinking, recombinant inbred rats
Introduction
Alcohol (ethanol) is an environmental toxin that can have adverse effects on organs and tissues (as well as cause addiction) in humans. The degree of toxicity caused by alcohol ingestion is dependent on the dose (amount) of alcohol consumed (Rusyn and Bataller 2013; Szabo and Saha 2015). Individuals vary not only in their susceptibility to alcohol-induced toxicity, but also in their level of alcohol exposure, which can be considered a “host factor” that is, at least initially, under partial control by the individual (i.e., is part of the individual’s behavior or lifestyle) (Garte 2008). However, both susceptibility to alcohol-induced organ damage and the predisposition to consume a particular dose of alcohol are also influenced by genetics (Rusyn and Bataller 2013).
Our work focuses not only on the consequences of alcohol ingestion, but also on the genetic basis for the behavior that results in differential levels of alcohol exposure. We have taken a “systems genetic” approach to elucidating the factors that affect alcohol consumption. Systems genetics has evolved into a process by which one can identify functional relationships among gene products (WGCNA, Langfelder and Horvath 2008; Zhang and Horvath 2005), understand the relationships between these gene expression networks and physiologic/pathologic processes (Obeidat et al. 2017; Saba et al. 2015), and understand the relationships between polymorphisms associated with disease in GWAS analysis and causal mechanisms predisposing or progressing the disease state (Civelek et al. 2017). Quantitative trait locus (QTL) analysis of complex traits in plants and non-human animals preceded the advent of GWAS studies with humans (Lander and Kruglyak 1995; Visscher et al. 2012), and was often used to suppose the “candidate genes” that were associated with a particular trait (Abiola et al. 2003). GWAS and QTL studies, if based solely on DNA sequence variants, have a significant drawback if there is no evidence regarding the organ in which particular “candidate genes” are expressed, or the context (network) in which the “candidate genes” function (Civelek and Lusis 2014). In addition, the DNA variants identified in QTL and GWAS studies are often located not within the coding sequences for genes, but in regions of the genome important (but not elucidated) for regulation of gene expression (Schaub et al. 2012). The inclusion of quantitative transcriptome information in systems genetic approaches generates a key intermediate for understanding the relationship between DNA variants and phenotypic traits (Albert and Kruglyak 2015; Saba et al. 2015; Tabakoff et al. 2009). Many of the current systems genetics approaches to understanding the relationship between genomics and biology benefit from the wealth of information that has been collected and catalogued on the genome and transcriptome sequences of species and individuals within those species, and information on the quantitative landscape of the transcriptome in various cells and organs (Hermsen et al. 2015; https://phenogen.ucdenver.edu ; Lappalainen et al. 2013).
We and others have employed an integrative genomic approach that takes advantage of the availability of transcriptional coexpression network and genomic data to identify genetic determinants of complex physiological/behavioral traits (Hasin et al. 2017; Saba et al. 2015; Tabakoff et al. 2009). The concept behind these approaches is that if expression levels of the related transcripts in the network influence a phenotype, then the genomic region that regulates the network transcript expression levels (eQTL) should coincide with the genomic region that regulates the phenotype (QTL). To implement this approach in the present study, the phenotype of voluntary alcohol consumption was assessed in a panel of recombinant inbred rat strains. It should be noted, that the rank order of behavioral phenotypes that have a strong genetic determinant (for alcohol consumption in our rat panel h2=0.46) can be reproduced over many generations in inbred rodents (Wahlsten et al. 2006). Whole transcriptome information was gathered from tissues of naive rats of the same HXB/BXH RI panel using RNA-Seq and exon array technology (the reproducibility of such information is discussed in Marioni et al. (2008)), and transcriptional coexpression networks were generated using Weighted Gene Coexpression Network Analysis (WGCNA) (Langfelder and Horvath 2008; Zhang and Horvath 2005). All raw and processed data are publically available on https://PhenoGen.ucdenver.edu.
While it seems appropriate to assess brain transcriptional coexpression networks when investigating the genetic influence on the predisposition to consume alcohol, the approach that we have developed and implemented does not have to be limited to an organ or tissue that is a priori thought to determine a complex trait. Because a phenotype QTL analysis simply identifies genomic regions associated with a quantitative trait, it is feasible that the identified area of the genome can regulate transcriptional activity in or from any organ. The continued growth of evidence regarding functional consequences of interactions of metabolic processes and products of various organs (liver and brain, GI system and brain and vice-versa, brown adipose tissue and heart and brain, etc.) dictates the use of a holistic approach when considering the possible predisposing or causal genetic factors contributing to a trait of interest. When alcohol is consumed, it enters the gut, is absorbed, passes through the liver, and is then distributed to brain, heart and other organs. Alcohol can alter the gut microbiome and disrupt the intestinal epithelial barrier, resulting in increased intestinal permeability. This effect allows for translocation of microbially-derived products such as LPS into the circulation, to produce inflammatory responses (Szabo 2015). The brain is also exposed to these products, as well as to peripherally-generated hormones, cytokines, activated immune cells and other molecules secreted by the liver, gut, adipose tissue and other organs in response to alcohol consumption.
To investigate a possible peripheral organ-brain interaction that can affect alcohol drinking, we used RNA-Seq and microarray data on transcript expression from liver of non-alcohol-exposed rats of the HXB/BXH RI rat panel to generate liver transcriptional networks associated with the predisposition to alcohol consumption. Our results suggest that liver secretion products, including pro- and anti-inflammatory cytokines, can affect the level of voluntary alcohol consumption by rats.
Materials and Methods
Alcohol Consumption and Alcohol Consumption Quantitative Trait Loci (QTL) (all data are publically available on https://Phenogen.ucdenver.edu)
We have previously reported data on alcohol consumption levels by male rats of the HXB/BXH RI panel (23 RI strains and progenitor strains) (Tabakoff et al. 2009). The rats were given 10% alcohol as their only choice of fluid for one week, and were then given a choice between 10% alcohol and water. Alcohol consumption data (daily average of grams of ethanol consumed per kilogram of body weight) for the second week of the two-bottle choice paradigm were used to identify QTLs for voluntary alcohol consumption in this model (Saba et al. 2015; Tabakoff et al. 2009). QTLs, including genome-wide p-values via permutation, and Bayesian credible intervals, were identified using marker regression with alcohol consumption data represented as strain means (21 RI strains with both alcohol consumption and genotype information, Online Resource 1). A genetic marker set (specifically SNPs) from the STAR consortium (http://www.snp-star.eu/; STAR Consortium 2008) was used for QTL analyses. Probes from the original arrays that determined this marker set were aligned to the RN6 version of the rat genome using BLAT (Kent 2002). Markers were retained for QTL analysis if: 1) their probe sequence aligned both perfectly and uniquely to the genome, 2) genotypes differed between the progenitor strains (BN-Lx/Cub and SHR/Ola/pcv), 3) neither progenitor had a heterozygous genotype, and 4) less than 5% of the RI strains had a missing or heterozygous genotype call. Markers with large genetic distances compared to physical distance (improbable recombination rates, flanked by 10 cM on each side) as well as double recombinant markers were removed. Genetic distances were estimated using the R/qtl package in R (version 1.40–8, Broman et al. 2003).
Many of the adjacent markers (SNPs) display the same genotype pattern among all the RI strains (i.e., no recombination events for any strain between SNPs), which would result in the same level of statistical significance (p-value) for all of these SNPs when performing QTL analysis. Therefore, we reduced the number of association tests, without losing information, by identifying unique strain distribution patterns (SDPs; i.e., the genotypes for all strains at a particular SNP) for the 30 RI strains. Alcohol consumption QTLs were identified with marker regression of the strain means using the R/qtl package in R. Genome-wide p-values were determined using permutation (Churchill and Doerge 1994) and Bayesian credible intervals (Sen and Churchill 2001) were estimated for significant (genome-wide p-value<0.05) and suggestive (genome-wide p-value<0.63, Lander and Kruglyak 1995) QTLs.
Liver RNA Coexpression Networks (data available on https://Phenogen.ucdenver.edu)
RNA Sequencing & Processing
Total RNA isolated from whole liver of 3 alcohol naive biological replicates (males) of each of the two progenitor strains of the HXB/BXH RI panel was previously sequenced (2 × 100 paired-end reads) (Harrall et al. 2016). For the current study, prior to alignment, reads were demultiplexed and trimmed for adaptors and for quality using Cutadapt (version 1.9.1; Martin et al 2011). Reads were eliminated if the trimmed length of either read fragment was less than 20 nucleotides. Reads were then aligned to the RN6 version their respective strain-specific genomes derived from our DNA sequencing (Saba et al. 2015) using Bowtie2/TopHat suite of tools (version 2.1.0, Langmead et al. 2009) with the default settings. A genome- and transcriptome-guided reconstruction was executed for each progenitor strain separately using the Cufflinks algorithm and software (version 2.2.1, Trapnell et al. 2010) and the Ensembl Rat Transcriptome (RGSC 6.0) to guide the reconstruction process. Strain-specific transcriptomes were merged by first limiting to high confidence transcripts (defined as transcripts that were at least 200 nucleotides long and had an average read depth of 10 within a strain (total reads including all biological replicates)). Transcripts were merged across strain-specific transcriptomes if their exon junctions matched precisely or if they were one-exon transcripts that had transcription start sites within 100 base pairs of each other and transcription stop sites within 100 base pairs of each other. Each of the six parental strain samples was re-quantified using the merged transcriptome and RSEM (Li and Dewey 2011). Transcripts were retained in the final merged transcriptome if they had an average read depth of at least 18 reads (total reads including all biological replicates).
Exon Arrays
Gene expression data were generated from whole liver tissue of naïve 10 week-old male rats using Affymetrix Rat Exon Arrays 1.0 ST (Affymetrix, Santa Clara, CA). Transcript expression was measured in rats from 21 HXB/BXH RI strains (Online Resource 1). Two to four rats per strain were used and RNA from each rat was hybridized to a separate array, resulting in a total of 108 individual arrays (Harrall et al 2016).
Prior to normalization, individual probes on the array were removed from consideration if their nucleotide sequence did not uniquely map to a region in the BN rat reference genome (RN6) or if the probe contained a known SNP or indel between the two HXB/BXH RI progenitor strains (Saba et al. 2015). Entire probe sets were removed if less than three probes remained after filtering. The genomic regions where probe sets aligned were compared to the liver reconstructed transcriptome generated from the RNA-Seq data. Probe sets were combined into a gene cluster if the entire probe set was contained within the transcribed region of a single gene identified in the reconstructed transcriptome. Expression values were normalized and summarized into gene level transcripts using robust multichip analysis (RMA) (Irizarry RA, 2003) implemented in Affymetrix Power Tools (version 1.18.0) (http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx) and ComBat was used to adjust for batch effects (sva version 3.20.0, Johnson et al. 2007). The coefficient of determination (R2) from a one-way ANOVA was used to estimate broad-sense heritability for each transcript. Those with low heritability (R2 < 0.20) were not included in further analyses. ComBat batch adjustment, heritability and all further statistical analyses were executed in R (version 3.3.1) unless otherwise specified.
Liver RNA Coexpression Networks (data available on https://Phenogen.ucdenver.edu)
An unsigned weighted gene co-expression network analysis (WGCNA) was performed on the HXB/BXH RI strain mean expression values of the whole liver expression data to identify co-expression modules using the wgcna package in R (version 1.51, Zhang and Horvath 2005). Data from parental strains were excluded to avoid confounding due to population structure. A bi-weight mid-correlation coefficient (robust to outliers) as well as a soft thresholding parameter (β) of 9 (default value suggested based on sample size; https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/faq.html) was used to construct the adjacency and connectivity matrices. The minimum module size (set to 5) and the deepSplit parameter (set to 4) were altered from the default values to allow for the identification of smaller modules. Intramodular connectivity of a gene was calculated as the sum of pairwise connectivity measures between a gene and all other genes within the module. The nomenclature, hub gene, is used for the gene within the module with the highest intramodular connectivity. We used the eigengene, the first principal component of the module, to represent the gene expression profile across strains for each module for correlation and mapping analyses. The quantitative nature of the eigengene allows for the use of QTL analysis, and the module eigengene QTLs (meQTLs) were identified as described earlier for the alcohol consumption QTLs. The meQTL analysis was performed using data from 19 rat strains for which both genotype and liver transcriptome information were available (Online Resource 1).
Liver Modules Associated with Voluntary Alcohol Consumption
For a liver module to be associated with voluntary alcohol consumption it was required to meet several criteria: 1) its module eigengene had to be significantly correlated with voluntary alcohol consumption (g/kg/day) across the RI strains (Pearson Correlation, unadjusted p-value < 0.05, using data from the 19 RI strains with genotype and liver transcriptome information); 2) the module eigengene had to explain at least 50% of module variance; 3) the module had to have a significant (genome-wide p-value < 0.05) meQTL; and 4) the peak position of the module’s maximum meQTL had to fall within the 90% Bayesian credible interval of an alcohol consumption QTL (Saba et al. 2015). These criteria ensured that module eigengene values were genetically driven, were associated with alcohol consumption, and that the module eigengene values were regulated from the same region(s) of the genome as alcohol consumption.
Further Characterization of Candidate Liver Modules
The UCSC Genome Browser (https://genome.ucsc.edu/) was used to annotate genes that were not previously annotated by the Ensembl-guided transcriptome reconstruction. This was accomplished by comparing isoforms from the transcriptome reconstruction to Ensembl and RefSeq-predicted genes in the same genomic location. To protect against associations due solely to co-localization, we examined partial correlations for genes in coexpression modules controlling for the module eigengene QTL using the ppcor package (Version 1.0) in R (Kim 2015). For purposes of examining connectivity between genes both prior to and after partial correlation, an edge represents a pairwise correlation between two genes that is either greater than 0.5 (0.3 for partial correlations) or less than −0.5 (−0.3 for partial correlations). The number of edges in a candidate module after partial correlation had to be at least 50% of the number of edges in the original module.
Liver cell type-specific transcriptome analysis
Liver cells (hepatocytes, hepatic stellate cells, sinusoidal endothelial cells, Kupffer cells) were isolated from livers of alcohol-naïve adult male BN-Lx/CubPrin and SHR/OlaPrin rats (parental strains of the HXB/BXH RI panel) as previously described (Harrall et al. 2016). RNA from 4 rats per strain and cell type was extracted and hybridized to separate Affymetrix Rat Exon Arrays 1.0 ST, and data were analyzed in the same manner as described for whole liver. Expression values were normalized and summarized into genes using RMA, and ComBat was used to adjust for batch effects. The results of this analysis are shown in Online Resource 2.
Results
Identification of Candidate Liver Modules
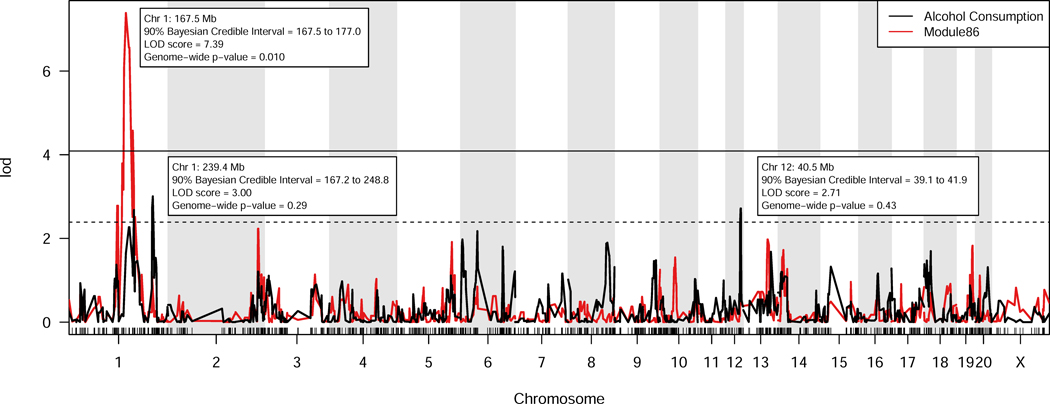
A total of 735 transcriptional coexpression modules were identified from the liver data. The eigengene values of 29 of these modules were significantly correlated (p< 0.05) with the phenotype of alcohol consumption. One of these modules (“module 86”, correlation coefficient 0.49, p-value = 0.032) had a significant meQTL genome-wide LOD score, and a maximum meQTL that overlapped the Bayesian credible interval of a previously-identified alcohol consumption QTL on chromosome 1 (Module 86, Figure 1). Sixty percent of the variance in this module was explained by the module eigengene. The chromosome 1 alcohol consumption QTL included two peaks within the 90% Bayesian credible interval (Sen and Churchill 2001) (Figure 1). To determine if this region represented two independent QTLs, the QTLs were recalculated after adjustment for their respective maximum peaks. This analysis indicated that the two peaks in the chromosome 1 QTL are likely due to linkage disequilibrium between the markers (Online Resource 3). Independently, the liver module eigengene explained 24% of the genetic variance in alcohol consumption and the previously-identified brain module eigengene (Saba et al. 2015) explained 35% of the genetic variance, calculated using linear regression of strain means.
Figure 1. Alcohol consumption QTLs and liver module 86 eigengene QTL (meQTL).
Strain means and R/qtl were used to calculate both the QTLs for voluntary alcohol consumption (Saba et al. 2015), and meQTL for module 86. The horizontal lines represent genome-wide significant (p<0.05, solid line) and genome-wide suggestive (p<0.63) LOD scores (Lander and Kruglyak 1995). The location and significance of the module 86 meQTL (red) and the alcohol consumption QTLs (black) are shown.
Characterization of the Liver Candidate Module (Module 86)
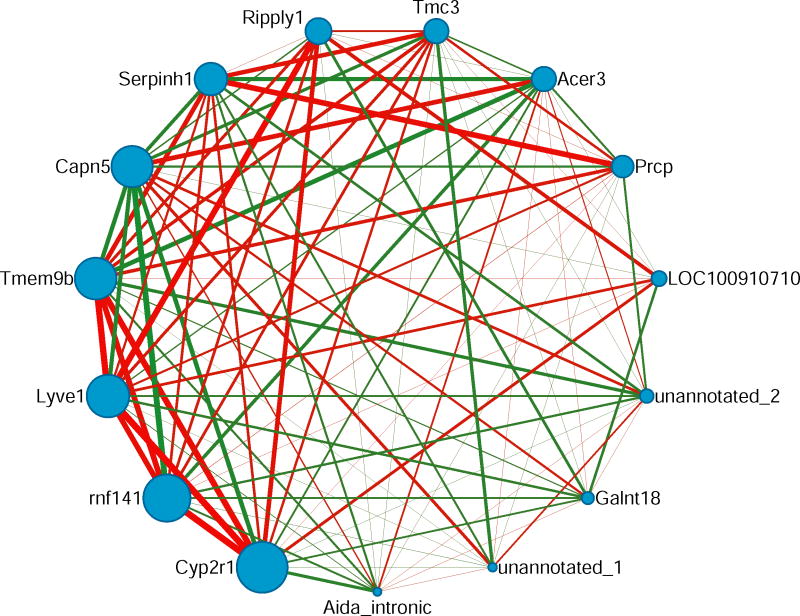
The module contained 15 transcripts that are listed in Table 1. A network diagram of the module was generated using gene connectivity and pairwise correlations between genes within the module (https://www.ncbi.nlm.nih.gov/pubmed/14597658) (Figure 2). Ten transcripts in the module were co-localized to chromosome 1 (Table 1), so a partial correlation analysis was performed (see Methods) to determine if the correlation among transcripts was only a result of independent causal loci in linkage disequilibrium. After the partial correlation analysis, there were 41 edges in the module, compared with 74 edges in the original module. Thirty-three of the edges after the partial correlation were also in the original network, and 8 new edges were detected after the partial correlation (Online Resource 4). These results strongly suggest that, although linkage disequilibrium may significantly contribute to the coexpression of transcripts in the module, the coexpression relationships that are retained within the module after accounting for this linkage disequilibrium (i.e., partial correlation) likely represent biological relationships (Dobrin et al. 2009; Doss et al. 2005), that go beyond simple linkage disequilibrium.
Table 1.
Transcripts in the Liver Coexpression Module
| Gene Symbol | Gene Name | Location |
Connectivity within module 86 |
|---|---|---|---|
| Cyp2r1 | Cytochrome P450, family 2, subfamily r, polypeptide 1 | Chr1:184.1 Mb | 1.006 |
| Rnf141 | Ring finger protein 141 | Chr1:175.6 Mb | 0.925 |
| Lyve1 | Lymphatic vessel endothelial hyaluronan receptor 1 | Chr1:175.7 Mb | 0.823 |
| Tmem9b | TMEM9 domain family, member B | Chr1:174.4 Mb | 0.803 |
| Capn5 | Calpain 5 | Chr1:163.1 Mb | 0.793 |
| Serpinh1 | Serpin family H member 1 (Hsp47) | Chr1:164.3 Mb | 0.600 |
| Ripply1 | Ripply transcriptional repressor 1 | ChrX:111.1 Mb | 0.458 |
| Tmc3 | Transmembrane channel-like 3 | Chr1:145.7 Mb | 0.422 |
| Acer3 | Alkaline ceramidase 3 | Chr1:163.1 Mb | 0.421 |
| Prcp | Prolylcarboxypeptidase | Chr1:157.7 Mb | 0.365 |
| LOC100910710 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex 12-like | Chr6:192.9 Mb | 0.215 |
| Galnt18 | Polypeptide N-acetylgalactosaminyl-transferase 18 | Chr1:176.3 Mb | 0.151 |
| Unannotated2 | 0.183 | ||
| Unannotated1 | 0.070 | ||
| Aida (intronic) | Axin interactor, dorsalization associated | Chr13:101.7 Mb | 0.057 |
Figure 2. Liver transcriptional coexpression module associated with alcohol consumption.
The figure shows pairwise connections between gene products in Module 86. A green edge represents a negative correlation between two nodes, and a red edge represents a positive correlation between two nodes. The thickness of each edge represents the strength of each correlation (i.e., a correlation with a larger magnitude has a thicker line), and edges are only visible if their associated correlation coefficient is greater than |0.50|. Cyp2r1 is the hub (most connected) gene product and its expression levels are positively correlated with alcohol consumption.
Sample Size and Statistical Considerations and Caveats
When calculating the voluntary alcohol consumption QTL, we used all strains with both genotype and phenotype information (21 strains), and we mapped strain means rather than phenotypic measures from individual animals. The use of strain means significantly reduces the influence of non-genetic variance on our phenotype estimates. With an RI panel, fewer loci are expected to contribute a higher proportion of genetic variance than with an outbred population. Both factors increase power to detect QTL. Transcriptome data were also available on 21 strains and this information was used to identify modules by WGCNA. The significant caveat is that the same 21 strains were not available for both phenotypic QTL calculations and WGCNA. When examining the association between the module and the phenotype, and performing meQTL analysis, we used data from the 19 strains that had genotype, phenotype and RNA expression information. If we limited our phenotypic QTL analysis to these 19 strains, we retained both phenotypic QTLs and did not identify any additional phenotypic QTLs.
The standard recommendation for a minimum sample size for identifying robust modules in WGCNA is 20 (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/faq.html), with a similar recommendation for co-expression networks in general (Ballouz et al. 2015). In our WGCNA analysis, we used within-strain means from the 21 strains of the HXB/BXH RI panel that had transcriptome data. Again, by using strain means, we reduced the non-genetic variance that often contributes to spurious correlations and reduces robustness of co-expression networks. In our experience, 20 strains, with multiple biological replicates within each strain, is the minimum number of strains needed for our approach (e.g., Saba et al 2015). An increase in sample size used for WGCNA has not produced a systematic difference in the number of co-expression modules identified.
With regard to statistical significance of correlations and associations in our analysis, a formal strategy for multiple testing correction is not as straightforward as in other types of analyses. This is a function of the fact that we require the convergence of evidence across several different analyses (correlation between module and phenotype; significant module eigengene QTL; overlap of phenotypic QTL and module eigengene QTL; retention of connections after partial correlation), where each of these analyses is not necessarily independent. The application of this series of filters to our data provides confidence that the resulting genetically-controlled gene expression module is functionally relevant to the genetically-controlled phenotype.
Discussion
Using the criteria that we have established to identify modules of co-expressed groups of genes that are associated with a phenotype, we have identified a liver transcriptional module associated with a predisposition to consume varying levels of alcohol. It is important to reiterate that the transcriptional coexpression module and the transcripts included in the module were identified by an analysis with no input about behavioral phenotype, i.e., this module is an inherent characteristic of the rat liver. It is the overlap of the module eigengene QTL with the phenotypic QTL, and the correlation of the module eigengene with the phenotype across the strains of the RI rat panel, that allows us to propose a functional relationship between transcript expression levels and phenotype. The analysis performed for this study represents a “data mining” experiment, in which we used our database of rat liver transcriptional modules to discover the association of a liver coexpression module with the phenotype of voluntary alcohol consumption. These results well illustrate how, using catalogued genome and transcriptome data, one can uncover novel possibilities for functional relationships between genome, transcriptome and a complex trait phenotype.
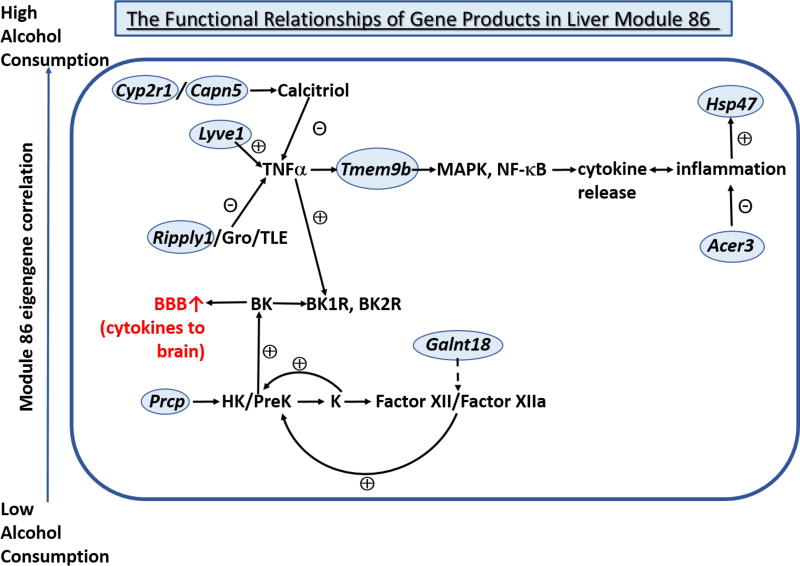
We used a modified version of the Formal Concept Analysis (Saba et al. 2015) to investigate the functional relationships among the coexpressed gene products, and how these relationships may influence the phenotype of voluntary alcohol consumption (Figure 3). The function of the transcripts in this module can be primarily related to inflammatory processes. The most connected transcript (hub gene) in the module is Cyp2r1, with expression levels positively correlated with alcohol consumption. This gene codes for a cytochrome P450 enzyme that converts cholecalciferol to 25-OH vitamin D3, the major circulating form of the vitamin. This intermediate is then converted to the biologically active calcitriol in various organs and cells, including brain and immune cells (Schuster 2011). Calcitriol can be formed in macrophages, and interacts with the vitamin D receptor to produce complex effects on the inflammatory responses of monocytes and macrophages, with an anti-inflammatory effect in mature macrophages (Di Rosa et al. 2012). In these macrophages, calcitriol delays activation of IL-6, TNFα and toll-like receptors (TLRs) during the inflammatory process (Di Rosa et al. 2012). The activity of calcitriol depends on the dimerization of the vitamin D receptor with the retinoid receptor (RXR) (Haussler et al. 1997), and the product of another transcript in the module, Capn5 (Calpain 5), cleaves the amino terminus of RXRα, reducing its nuclear localization (Casas et al. 2003), thus modifying the biologic impact of calcitriol. Calcitriol can also act in the hypothalamus to lower food intake (Sisley et al. 2016). Thus calcitriol represents a secretory product of liver that can affect both appetite and inflammation. Very recently, it has been reported that vitamin D levels are low in alcohol-dependent subjects, and this difference was suggested to contribute to alcohol “craving” (Schuster et al. 2017).
Figure 3. Functional relationships among transcripts in the liver transcriptional coexpression module associated with alcohol consumption (Module 86).
Each of the transcripts in the coexpression module is circled in blue. The ⨁ and ⊝ signs indicate the effects of the transcript products on the activity of the indicated pathway. The module eigengene is positively correlated with the phenotype of alcohol consumption across the HXB/BXH RI strains, as indicated by the arrow on the left. The correlations among the transcripts are indicated in Figure 2. ↑BBB = increase in permeability of the blood-brain barrier; PreK = prekallikrein; K = kallikrein; BK = bradykinin; BK1R and BK2R = bradykinin 1 and 2 receptors, respectively.
Prcp codes for an enzyme in endothelial cells (prolylcarboxypeptidase) that regulates the levels of various hormones by cleaving a C-terminal Pro-X bond (Wang et al. 2014). Plasma prekallikrein is a glycoprotein that is mainly synthesized in liver (Bjorkqvist et al. 2013). Prekallikrein circulates bound to high molecular weight kininogen (HK), and prekallkrein is brought to the surface of endothelial cells when they bind HK (Joseph and Kaplan 2005). Prolylcarboxypeptidase (the protein product of Prcp) activates prekallikrein when it is bound to HK on endothelial cells (Shariat-Madar et al. 2004). The kallikrein formed from this reaction activates Factor XII to Factor XIIa, which further increases activation of prekallikrein. Kallikrein also cleaves HK to release bradykinin, an inflammatory mediator that activates constitutive bradykinin BK2 receptors and the inducible BK1 receptors to release nitric oxide and prostaglandins, as well as activate the MAPK and NFκB pathways (Bjorkqvist et al. 2013; Dutra 2017). Bradykinin, when administered in combination with the angiotensin converting inhibitor, captopril (which increases bradykinin levels by inhibition of kininase II), has been reported to reduce alcohol intake and increase water intake by rats (Robertson et al. 1993). It is also of particular interest that bradykinin can disrupt the function of the blood brain barrier (Easton and Abbott 2002). Therefore, bradykinin derived from prekallikrein via an initial action of the prolylcarboxypeptidase can potentially affect alcohol consumption by its own central actions, and/or by increasing inflammation and enhancing the ability of cytokines and hormones released from the liver to enter the brain. The BK1 receptor has also been implicated in regulation of energy balance and food intake (Mori et al. 2008). These findings suggest that bradykinin, like calcitriol, can influence both inflammatory processes and appetite/food intake.
Some of the other correlated transcripts in the module can also be related to prekallikrein metabolism. The product of Galnt18 (polypeptide N-acetylgalactosaminyltransferase 18) is one of a large family of enzymes that control mucin-type O-glycosylation, the step where the first glycan is attached to a serine or threonine residue in a protein, forming an N-acetylgalactosamine (GalNAc) α1-O-serine/threonine linkage in O-glycoproteins (Bennett et al. 2012; Kong et al. 2015). These reactions take place in the Golgi apparatus (Bennett et al. 2012). There are up to 20 GalNAc-Ts that all catalyze the initiation step where GalNAc is attached to serine and threonine residues. These enzymes have been divided into 9 families based on phylogenetic and genomic analysis (Bennett et al. 2012), and they have different, but overlapping substrate specificities (Schjoldager and Clausen 2012). It appears that the in vivo GalNAc O-glycosylation of proteins may be more dependent on the enzymes that are expressed in a particular cell than on substrate specificity of the enzymes as assessed in vitro (Kong et al. 2015). Our data show that Galnt18 is expressed at the highest level in liver sinusoidal endothelial cells, similar to Prcp (Online Resource 2). There is little or no information available regarding substrate specificity of GalNAc-T18 (Kong et al. 2015), however this enzyme does have an identified activity (Raman et al. 2012), and it has been characterized as a chaperone-like protein that can modulate the activity of GalNAc-T2, which is highly expressed in liver (Bennett et al. 2012). Factor XII, which is involved in the formation of bradykinin from prekallikrein, is subject to mucin-type Threonine 309 glycosylation, and could be a substrate of GalNAc-T2 and/or –T18.
Another member of the module, Serpinh1, also known as Hsp47, is an endoplasmic reticulum (ER)-resident collagen-specific chaperone that binds to procollagen in the ER, and is involved in the transport of procollagen to the Golgi apparatus (Miyata et al. 2013). Our data show a high level of Hsp47 expression in hepatic stellate cells (Online Resource 2), and increased expression of Hsp47 in hepatic stellate cells has been implicated in liver fibrosis (Ishida and Nagata 2011). Inflammation and cell death are key processes in the development of liver fibrosis, involving the inflammasome response and activation of hepatic stellate cells (Alegre et al. 2017). Expression levels of Hsp47 may therefore reflect the baseline degree of liver inflammation (and predisposition to the development of fibrosis) in the rats of the RI panel. Hsp47 expression levels are also affected by the level of O-glycosylation in the Golgi apparatus, where O-glycosylation is associated with procollagen maturation and is necessary for glycoprotein trafficking (Miyata et al. 2013). This relationship suggests a functional interaction between Hsp47 and Galnt18, which is also expressed in hepatic stellate cells (Online Resource 2).
Several of the other transcripts in the module can be linked to inflammatory processes through the activity of TNFα. Tmem9b is a highly connected transcript that codes for a lysosomal transmembrane protein that is required for activation of the NFκB and MAPK pathways by TNFα (Dodeller et al. 2008). This activation leads to the production of the cytokine IL-6, which has context-dependent pro- and anti-inflammatory effects, and the inflammatory chemokine, IL-8 (Dodeller et al. 2008; Dong and Zheng 2015; Rothaug et al. 2016). Lyve1 (lymphatic vessel endothelial hyaluronan receptor 1) is another highly connected transcript associated with TNFα activity. This transcript codes for a hyaluronan-binding protein that is expressed in liver sinusoidal endothelial cells (Online Resource 2) and in activated macrophages (Arimoto et al. 2010; Pinto et al. 2012). The high molecular weight forms of the hyaluronan polymer are anti-inflammatory, while the lower molecular weight forms are pro-inflammatory and lead to the release of TNFα (Jackson 2009). The conversion of the longer forms of hyaluronan to the shorter forms depends on internalization of hyaluronan into lymphatic endothelial cells, and Lyve1 is the likely protein that mediates this internalization (Jackson 2009). Expression of Lyve1 is positively correlated with expression of Tmem9b, suggesting a cooperative interaction in regulating the activity of TNFα.
Ripply1 (ripply1 homolog (zebrafish)) codes for a protein that associates with a family of proteins that act as transcriptional repressors, the Groucho/Transducin-Like Enhancer of split (Gro/TLE) co-repressor proteins (Kaul et al. 2015; Kawamura et al. 2008). These proteins do not directly bind DNA, but are recruited by DNA-bound repressor proteins. Ripply family proteins act as specific adaptors that recruit the global Gro/TLE proteins and can convert transcription factors from activators to repressors (Kawamura et al. 2008). TLE1 is highly expressed in macrophages, and inhibits LPS induction of TNFα, i.e., has anti-inflammatory effects (De Paoli et al. 2016). In addition to the full-length Gro/TLE proteins, mouse and human AES (amino terminal enhancer of split) and ESG (enhancer of split groucho) proteins have been identified, and protein-protein interactions between AES and the p65 subunit of NFκB have been identified (Tetsuka et al. 2000). This interaction inhibited TNFα–dependent activation of the NFκB and MAPK pathways. The positive correlation of Ripply and Tmem9b expression levels suggests that the products of these transcripts can generate a feedback loop to control the activity of TNFα.
The module also contains Acer3 (alkaline ceramidase 3), which is one of a heterogeneous family of ceramidases, with the major function of hydrolyzing ceramides to generate sphingosine which is phosphorylated to sphingosine-1 phosphate. Ceramides are bioactive lipids that mediate many cellular processes, and are increased in cells exposed to cytokines (TNFα, IL-1β (Hanna et al. 2001; Rolz et al. 2003) and vitamin D (Mao and Obeid 2008)), among other stresses. Acer3 is active under alkaline conditions and its activity is stimulated by Ca2+. The enzyme efficiently hydrolyzes long-chain unsaturated ceramides, phytoceramides or dihydroceramide (Mao and Obeid 2008). It has been shown that Acer3 affects the immune response by regulating the levels of C18:1 ceramide in macrophages and other cells of the innate immune system. In response to LPS, expression of Acer3 is downregulated, leading to increased levels of C18:1 ceramide and expression of pro-inflammatory cytokines (Wang et al. 2016). These findings demonstrate that the enzyme encoded by Acer3 per se can have an anti-inflammatory effect in macrophages, and interestingly, Acer3 expression is strongly negatively correlated with Tmem9b, Lyve1, Serpinh1 and Prcp, the products of which are associated with pro-inflammatory effects.
The function of the remaining annotated transcripts in the module (Tmc3, rnf141, LOC100910710) is, at this time, less clear, but their presence in the liver coexpression module (Module 86) may suggest “guilt by association” for their involvement in inflammatory processes in the liver and/or liver/brain communication.
The functions of the products of the transcripts in Module 86 indicate that this module is a component of inflammatory processes active in liver. There is a plethora of information available regarding ethanol potentiating or suppressing liver inflammation (Lowe, et al. 2017). The role of inflammation and innate immune signaling in alcohol consumption has also recently been the subject of investigation. Several studies with mice have demonstrated that various chemokines, chemokine receptors, cytokines and inflammatory pathways have effects on alcohol intake (Karlsson et al. 2016; Robinson et al. 2014; Truitt et al. 2016). It is particularly noteworthy that many of the transcripts and signaling pathways represented in our candidate coexpression module have opposing effects, i.e., are pro- or anti-inflammatory. This result suggests that it is the balance among these opposing forces (e.g., differences in level of expression of the “opposing” transcripts) in each rat strain that influences the variation in predisposition to consume alcohol and may, as well, influence liver inflammation in response to alcohol ingestion. The products of two of the transcripts, Cyp2r1 and Prcp, also implicate Module 86 in pathways related to food intake and energy metabolism. We and others have noted that the caloric content of alcohol is an important contributor to alcohol intake by rats, and that peripherally-produced peptides that regulate appetite and consummatory behavior also influence the intake of alcohol and other drugs (Engel and Jerlhag 2014; Tabakoff et al. 2009).
The central nervous system is usually considered as the source of motivation to consume alcohol. However, there is evidence that cytokines can mediate communication between the periphery and brain (D'Mello and Swain 2014). Such communication has been invoked to explain how peripheral activation of TL4 by LPS can influence alcohol consumption (Mayfield et al. 2013). The production of cytokines in the brain (microglia) can be induced by peripheral immune or inflammatory signals, including bradykinin, which can generate neuroinflammation through interaction with the BK1 receptor (Asraf et al. 2017; D'Mello and Swain 2011). Our systems genetic approach provides evidence for how peripheral inflammation, as well as liver secretory products that may affect appetite and energy metabolism, can influence brain function and alcohol consumption. These results emphasize the importance of considering the whole body to understand the genetic factors that affect the predisposition to consume alcohol, and potentially the effects of alcohol exposure that contribute to organ (liver) damage.
It has to be noted that, for the current analysis, both transcriptome and behavioral phenotype data are from adult male rats. The age range and gender were chosen to reflect these parameters used in a majority of phenotypic studies with rats. This may, however, limit the generality of the results, for instance, it is known that female rodents have QTLs for alcohol consumption that differ from males (Vanderlinden et al. 2015; Vendruscolo et al. 2006), and rats of different ages also exhibit differences in drinking behavior (Wang et al. 2003). Nevertheless, because of the nature of the RI panel (genetically identical animals over generations), and the fact that we identify genetically-controlled transcriptional networks that are associated with the predisposition to consume alcohol in sexually mature males, the current data could be used as the foundation to compare and contrast the genetically-mediated networks that predispose voluntary alcohol consumption across characteristics such as sex and age, as well as the influence that environmental factors may have on the modulation of the alcohol drinking phenotype.
In summary, while many studies have been performed that implicate alcohol-induced inflammation as leading to organ damage, particularly alcoholic liver disease (Lowe et al. 2017), less emphasis has been placed on the role of individual differences in immunity or inflammation that may contribute to an individual’s predisposition to consume alcohol. Our findings suggest that targeting inflammatory processes in the periphery may be a novel approach to reducing alcohol consumption and alcohol-induced organ damage.
Supplementary Material
Acknowledgments
This work was supported in part by National Institute on Alcohol Abuse and Alcoholism/ National Institutes of Health (R24 AA013162) and the Banbury Fund. We thank Yinni Yu and Adam Chapman for expert technical assistance with microarrays and RNA-Seq assays, and Laura Breen and Joseph Gatewood for technical assistance in maintaining the BN-Lx, SHR and HXB/BXH RI rat colonies at the University of California, San Diego, under the supervision of Dr. Morton Printz. Dr. Michal Pravenec, Institute of Physiology, Czech Academy of Sciences, Prague, Czechoslovakia, provided tissue from BN-Lx and SHR rats for liver RNA-Seq analysis. The University of Nevada, Las Vegas National Supercomputing Institute provided access to high-performance computing and networking resources for this research.
Footnotes
Conflict of interest: On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, Blankenhorn EP, Blizard DA, Bolivar V, Brockmann GA, Buck KJ, Bureau JF, Casley WL, Chesler EJ, Cheverud JM, Churchill GA, Cook M, Crabbe JC, Crusio WE, Darvasi A, de Haan G, Dermant P, Doerge RW, Elliot RW, Farber CR, Flaherty L, Flint J, Gershenfeld H, Gibson JP, Gu J, Gu W, Himmelbauer H, Hitzemann R, Hsu HC, Hunter K, Iraqi FF, Jansen RC, Johnson TE, Jones BC, Kempermann G, Lammert F, Lu L, Manly KF, Matthews DB, Medrano JF, Mehrabian M, Mittlemann G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Mountz JD, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Paigen B, Palmer AA, Peirce JL, Pomp D, Rosemann M, Rosen GD, Schalkwyk LC, Seltzer Z, Settle S, Shimomura K, Shou S, Sikela JM, Siracusa LD, Spearow JL, Teuscher C, Threadgill DW, Toth LA, Toye AA, Vadasz C, Van Zant G, Wakeland E, Williams RW, Zhang HG, Zou F Complex Trait C. The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- Alegre F, Pelegrin P, Feldstein AE. Inflammasomes in Liver Fibrosis. Semin Liver Dis. 2017;37:119–127. doi: 10.1055/s-0037-1601350. [DOI] [PubMed] [Google Scholar]
- Arimoto J, Ikura Y, Suekane T, Nakagawa M, Kitabayashi C, Iwasa Y, Sugioka K, Naruko T, Arakawa T, Ueda M. Expression of LYVE-1 in sinusoidal endothelium is reduced in chronically inflamed human livers. J Gastroenterol. 2010;45:317–325. doi: 10.1007/s00535-009-0152-5. [DOI] [PubMed] [Google Scholar]
- Asraf K, Torika N, Danon A, Fleisher-Berkovich S. Involvement of the Bradykinin B1 Receptor in Microglial Activation: In Vitro and In Vivo Studies. Front Endocrinol (Lausanne) 2017;8:82. doi: 10.3389/fendo.2017.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballouz S, Verleyen W, Gillis J. Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. 2015;31:2123–2130. doi: 10.1093/bioinformatics/btv118. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkqvist J, Jamsa A, Renne T. Plasma kallikrein: the bradykinin-producing enzyme. Thromb Haemost. 2013;110:399–407. doi: 10.1160/TH13-03-0258. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Wu Y, Pan C, Raulerson CK, Ko A, He A, Tilford C, Saleem NK, Stancakova A, Scott LJ, Fuchsberger C, Stringham HM, Jackson AU, Narisu N, Chines PS, Small KS, Kuusisto J, Parks BW, Pajukanta P, Kirchgessner T, Collins FS, Gargalovic PS, Boehnke M, Laakso M, Mohlke KL, Lusis AJ. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am J Hum Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello C, Swain MG. Liver-brain inflammation axis. Am J Physiol Gastrointest Liver Physiol. 2011;301:G749–761. doi: 10.1152/ajpgi.00184.2011. [DOI] [PubMed] [Google Scholar]
- D'Mello C, Swain MG. Liver-brain interactions in inflammatory liver diseases: implications for fatigue and mood disorders. Brain Behav Immun. 2014;35:9–20. doi: 10.1016/j.bbi.2013.10.009. [DOI] [PubMed] [Google Scholar]
- De Paoli F, Copin C, Vanhoutte J, Derudas B, Vinod M, Zawadzki C, Susen S, Pattou F, Haulon S, Staels B, Eeckhoute J, Chinetti-Gbaguidi G. Transducin-like enhancer of split-1 is expressed and functional in human macrophages. FEBS Lett. 2016;590:43–52. doi: 10.1002/1873-3468.12029. [DOI] [PubMed] [Google Scholar]
- Di Rosa M, Malaguarnera G, De Gregorio C, Palumbo M, Nunnari G, Malaguarnera L. Immuno-modulatory effects of vitamin D3 in human monocyte and macrophages. Cell Immunol. 2012;280:36–43. doi: 10.1016/j.cellimm.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, Schadt EE. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10:R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodeller F, Gottar M, Huesken D, Iourgenko V, Cenni B. The lysosomal transmembrane protein 9B regulates the activity of inflammatory signaling pathways. J Biol Chem. 2008;283:21487–21494. doi: 10.1074/jbc.M801908200. [DOI] [PubMed] [Google Scholar]
- Dong R, Zheng S. Interleukin-8: A critical chemokine in biliary atresia. J Gastroenterol Hepatol. 2015;30:970–976. doi: 10.1111/jgh.12900. [DOI] [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra RC. Kinin receptors: Key regulators of autoimmunity. Autoimmun Rev. 2017;16:192–207. doi: 10.1016/j.autrev.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Easton AS, Abbott NJ. Bradykinin increases permeability by calcium and 5-lipoxygenase in the ECV304/C6 cell culture model of the blood-brain barrier. Brain Res. 2002;953:157–169. doi: 10.1016/s0006-8993(02)03281-x. [DOI] [PubMed] [Google Scholar]
- Engel JA, Jerlhag E. Role of appetite-regulating peptides in the pathophysiology of addiction: implications for pharmacotherapy. CNS Drugs. 2014;28:875–886. doi: 10.1007/s40263-014-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garte S. Individual susceptibility and gene-environment interaction. In: Wild CP, Vincis P, Garte S, editors. Molecular Epidemiology of Chronic Diseases. John Wiley & Sons, Ltd; 2008. pp. 55–69. [Google Scholar]
- Hanna AN, Berthiaume LG, Kikuchi Y, Begg D, Bourgoin S, Brindley DN. Tumor necrosis factor-alpha induces stress fiber formation through ceramide production: role of sphingosine kinase. Mol Biol Cell. 2001;12:3618–3630. doi: 10.1091/mbc.12.11.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrall KK, Kechris KJ, Tabakoff B, Hoffman PL, Hines LM, Tsukamoto H, Pravenec M, Printz M, Saba LM. Uncovering the liver's role in immunity through RNA co-expression networks. Mamm Genome. 2016;27:469–484. doi: 10.1007/s00335-016-9656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R, de Ligt J, Spee W, Blokzijl F, Schafer S, Adami E, Boymans S, Flink S, van Boxtel R, van der Weide RH, Aitman T, Hubner N, Simonis M, Tabakoff B, Guryev V, Cuppen E. Genomic landscape of rat strain and substrain variation. BMC Genomics. 2015;16:357. doi: 10.1186/s12864-015-1594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://phenogen.ucdenver.edu
- Ishida Y, Nagata K. Hsp47 as a collagen-specific molecular chaperone. Methods Enzymol. 2011;499:167–182. doi: 10.1016/B978-0-12-386471-0.00009-2. [DOI] [PubMed] [Google Scholar]
- Jackson DG. Immunological functions of hyaluronan and its receptors in the lymphatics. Immunol Rev. 2009;230:216–231. doi: 10.1111/j.1600-065X.2009.00803.x. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Joseph K, Kaplan AP. Formation of bradykinin: a major contributor to the innate inflammatory response. Adv Immunol. 2005;86:159–208. doi: 10.1016/S0065-2776(04)86005-X. [DOI] [PubMed] [Google Scholar]
- Karlsson C, Schank JR, Rehman F, Stojakovic A, Bjork K, Barbier E, Solomon M, Tapocik J, Engblom D, Thorsell A, Heilig M. Proinflammatory signaling regulates voluntary alcohol intake and stress-induced consumption after exposure to social defeat stress in mice. Addict Biol. 2016 doi: 10.1111/adb.12416. [DOI] [PubMed] [Google Scholar]
- Kaul AK, Schuster EF, Jennings BH. Recent insights into Groucho co-repressor recruitment and function. Transcription. 2015;6:7–11. doi: 10.1080/21541264.2014.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura A, Koshida S, Takada S. Activator-to-repressor conversion of T-box transcription factors by the Ripply family of Groucho/TLE-associated mediators. Mol Cell Biol. 2008;28:3236–3244. doi: 10.1128/MCB.01754-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Commun Stat Appl Methods. 2015;22:665–674. doi: 10.5351/CSAM.2015.22.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Joshi HJ, Schjoldager KT, Madsen TD, Gerken TA, Vester-Christensen MB, Wandall HH, Bennett EP, Levery SB, Vakhrushev SY, Clausen H. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology. 2015;25:55–65. doi: 10.1093/glycob/cwu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MRt, Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Geuvadis C, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe PP, Gyongyosi B, Satishchandran A, Iracheta-Vellve A, Ambade A, Kodys K, Catalano D, Ward DV, Szabo G. Alcohol-related changes in the intestinal microbiome influence neutrophil infiltration, inflammation and steatosis in early alcoholic hepatitis in mice. PLoS One. 2017;12:e0174544. doi: 10.1371/journal.pone.0174544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J, Ferguson L, Harris RA. Neuroimmune signaling: a key component of alcohol abuse. Curr Opin Neurobiol. 2013;23:513–520. doi: 10.1016/j.conb.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Mizuno T, Koyama Y, Katayama T, Tohyama M. The endoplasmic reticulum-resident chaperone heat shock protein 47 protects the Golgi apparatus from the effects of O-glycosylation inhibition. PLoS One. 2013;8:e69732. doi: 10.1371/journal.pone.0069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Araujo RC, Reis FC, Sgai DG, Fonseca RG, Barros CC, Merino VF, Passadore M, Barbosa AM, Ferrari B, Carayon P, Castro CH, Shimuta SI, Luz J, Bascands JL, Schanstra JP, Even PC, Oliveira SM, Bader M, Pesquero JB. Kinin B1 receptor deficiency leads to leptin hypersensitivity and resistance to obesity. Diabetes. 2008;57:1491–1500. doi: 10.2337/db07-1508. [DOI] [PubMed] [Google Scholar]
- Obeidat M, Nie Y, Chen V, Shannon CP, Andiappan AK, Lee B, Rotzschke O, Castaldi PJ, Hersh CP, Fishbane N, Ng RT, McManus B, Miller BE, Rennard S, Pare PD, Sin DD. Network-based analysis reveals novel gene signatures in peripheral blood of patients with chronic obstructive pulmonary disease. Respir Res. 2017;18:72. doi: 10.1186/s12931-017-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, Paolicelli R, Salimova E, Gospocic J, Slonimsky E, Bilbao-Cortes D, Godwin JW, Rosenthal NA. An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One. 2012;7:e36814. doi: 10.1371/journal.pone.0036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases: completion of the family tree. Glycobiology. 2012;22:768–777. doi: 10.1093/glycob/cwr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, Harding S, Grupp LA. Bradykinin suppresses alcohol intake and plays a role in the suppression produced by an ACE inhibitor. Pharmacol Biochem Behav. 1993;46:751–758. doi: 10.1016/0091-3057(93)90197-2. [DOI] [PubMed] [Google Scholar]
- Robinson G, Most D, Ferguson LB, Mayfield J, Harris RA, Blednov YA. Neuroimmune pathways in alcohol consumption: evidence from behavioral and genetic studies in rodents and humans. Int Rev Neurobiol. 2014;118:13–39. doi: 10.1016/B978-0-12-801284-0.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolz W, Xin C, Ren S, Pfeilschifter J, Huwiler A. Interleukin-1beta inhibits ATP-induced protein kinase B activation in renal mesangial cells by two different mechanisms: the involvement of nitric oxide and ceramide. Br J Pharmacol. 2003;138:461–468. doi: 10.1038/sj.bjp.0705064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothaug M, Becker-Pauly C, Rose-John S. The role of interleukin-6 signaling in nervous tissue. Biochim Biophys Acta. 2016;1863:1218–1227. doi: 10.1016/j.bbamcr.2016.03.018. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Bataller R. Alcohol and toxicity. J Hepatol. 2013;59:387–388. doi: 10.1016/j.jhep.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba LM, Flink SC, Vanderlinden LA, Israel Y, Tampier L, Colombo G, Kiianmaa K, Bell RL, Printz MP, Flodman P, Koob G, Richardson HN, Lombardo J, Hoffman PL, Tabakoff B. The sequenced rat brain transcriptome--its use in identifying networks predisposing alcohol consumption. FEBS J. 2015;282:3556–3578. doi: 10.1111/febs.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Clausen H. Site-specific protein O-glycosylation modulates proprotein processing - deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta. 2012;1820:2079–2094. doi: 10.1016/j.bbagen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Schuster I. Cytochromes P450 are essential players in the vitamin D signaling system. Biochim Biophys Acta. 2011;1814:186–199. doi: 10.1016/j.bbapap.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Schuster R, Koopmann A, Grosshans M, Reinhard I, Spanagel R, Kiefer F. Association of plasma calcium concentrations with alcohol craving: New data on potential pathways. Eur Neuropsychopharmacol. 2017;27:42–47. doi: 10.1016/j.euroneuro.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat-Madar Z, Mahdi F, Schmaier AH. Recombinant prolylcarboxypeptidase activates plasma prekallikrein. Blood. 2004;103:4554–4561. doi: 10.1182/blood-2003-07-2510. [DOI] [PubMed] [Google Scholar]
- Sisley SR, Arble DM, Chambers AP, Gutierrez-Aguilar R, He Y, Xu Y, Gardner D, Moore DD, Seeley RJ, Sandoval DA. Hypothalamic Vitamin D Improves Glucose Homeostasis and Reduces Weight. Diabetes. 2016;65:2732–2741. doi: 10.2337/db16-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148:30–36. doi: 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Saha B. Alcohol's Effect on Host Defense. Alcohol Res. 2015;37:159–170. [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL State WISo, Trait Markers of A. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsuka T, Uranishi H, Imai H, Ono T, Sonta S, Takahashi N, Asamitsu K, Okamoto T. Inhibition of nuclear factor-kappaB-mediated transcription by association with the amino-terminal enhancer of split, a Groucho-related protein lacking WD40 repeats. J Biol Chem. 2000;275:4383–4390. doi: 10.1074/jbc.275.6.4383. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt JM, Blednov YA, Benavidez JM, Black M, Ponomareva O, Law J, Merriman M, Horani S, Jameson K, Lasek AW, Harris RA, Mayfield RD. Inhibition of IKKbeta Reduces Ethanol Consumption in C57BL/6J Mice. eNeuro. 2016;3 doi: 10.1523/ENEURO.0256-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Bennett B, Hoffman PL, Tabakoff B. Influence of sex on genetic regulation of "drinking in the dark" alcohol consumption. Mamm Genome. 2015;26:43–56. doi: 10.1007/s00335-014-9553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo LF, Terenina-Rigaldie E, Raba F, Ramos A, Takahashi RN, Mormede P. Evidence for a female-specific effect of a chromosome 4 locus on anxiety-related behaviors and ethanol drinking in rats. Genes Brain Behav. 2006;5:441–450. doi: 10.1111/j.1601-183X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc Natl Acad Sci U S A. 2006;103:16364–16369. doi: 10.1073/pnas.0605342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Matafonov A, Madkhali H, Mahdi F, Watson D, Schmaier AH, Gailani D, Shariat-Madar Z. Prolylcarboxypeptidase independently activates plasma prekallikrein (fletcher factor) Curr Mol Med. 2014;14:1173–1185. doi: 10.2174/1566524014666141015153519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Xu R, Snider AJ, Schrandt J, Li Y, Bialkowska AB, Li M, Zhou J, Hannun YA, Obeid LM, Yang VW, Mao C. Alkaline ceramidase 3 deficiency aggravates colitis and colitis-associated tumorigenesis in mice by hyperactivating the innate immune system. Cell Death Dis. 2016;7:e2124. doi: 10.1038/cddis.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is involved in ethanol preference and its age-dependent decline in mice. Proc Natl Acad Sci U S A. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.