Abstract
Human leukocyte antigen class I (HLA-I) down-regulation has been reported in many human cancers to be associated with poor clinical outcome. However, its connection to tumor-initiating cells (TICs) remains unknown. In this study, we report that HLA-I is down-regulated in a subpopulation of cells that have high tumor initiating capacity in different types of human sarcomas. Detailed characterization revealed their distinct molecular profiles regarding proliferation, apoptosis and stemness programs. Notably, these TICs can be induced to differentiate along distinct mesenchymal lineages, including the osteogenic pathway. The retinoic acid receptor signaling pathway is overexpressed in HLA-1 negative TICs. All-trans retinoic acid treatment successfully induced osteogenic differentiation of this subpopulation, in vitro and in vivo, resulting in significantly decreased tumor formation. Thus, our findings indicate down-regulated HLA-I is a shared feature of TICs in a variety of human sarcomas, and differentiation therapy strategies may specifically target undifferentiated TICs and inhibit tumor formation.
Keywords: Human leukocyte antigen class I, tumor-initiating cells, sarcomas
1. Introduction
Sarcomas represent a family of over 70 malignancies of soft tissue and bone, each with unique genetics and specific clinical behavior. The nature of the tumor initiating cell in sarcomas of soft tissue and bone remains unclear, and may vary from one sarcoma subtype to the next (Matushansky and Maki, 2005). This is made evident clinically with the observation that gastrointestinal stromal tumors only start in the gastrointestinal tract or mesentery, not in other organs; similarly, bona fide conventional chondrosarcomas do not arise outside bony structures. The mesenchymal stem cell (MSC) is an obvious target for such cancers, but data on its role as a tumor initiating cell are limited (Charytonowicz et al., 2009).
Human leukocyte class I (HLA-I) molecules are critical for antigen presentation to CD8+ cytotoxic T-lymphocyte (Garrido et al., 1993), and are understood to be widely expressed on the cell surface of all normal human tissues (Daar et al., 1984). However, HLA-I expression has been reported to be absent in embryonic stem cells, and down-regulated in mesenchymal stem cells (Drukker et al., 2002, Portmann-Lanz et al., 2006).
In a variety of human cancers, down-regulation of HLA-I expression has been observed at high frequency (Chang et al., 2005). Within the same cancer, HLA-I demonstrates heterogeneous expression. HLA-I down-regulated cells have been found to be highly enriched in metastases compared to primary tumors from the same patients, such phenotype being associated with poor clinical outcome (Cordon-Cardo et al., 1991). Collectively, these data suggest that HLA-I negative/low cells have survival and/or proliferative advantages and may function as tumor-initiating cells (TICs). TICs of human sarcomas (cancers of mesenchymal derivation) have been identified by analysis of the side population (Murase et al., 2009) or using stem cell markers, e.g., CD133, CD57, CD117 and Stro-1 (Tirino et al., 2011; Wahl, et al., 2010)
Recently, a subpopulation of prostate cancer cells with tumor initiating capacity have been reported and found to display an HLA-I negative phenotype, whereas the bulk of the differentiated prostate cancer cells in the same lesion expressed HLA-I, and when isolated did not exhibit tumor initiating capacity (Domingo-Domenech et al, 2012). We undertook the present study in order to investigate the question of if down-regulation of HLA-I can be used to identify and functionally characterize TICs from different types of human sarcomas.
2. Materials and methods
2.1 Cell Culture
Sarcoma lines MFH, CW9019, MG63, LPS141 and SKNEP, were from ATCC and cultured as described (Mills et al., 2009). All cell lines have been tested for mycoplasma. Sphere formation assay was performed by culturing cells with serum-free medium supplemented with B27 (1×), N2 (1×), bFGF (20 ng/ml), and EGF (20 ng/ml) in the low-attachment plate. To induce differentiation, HLA-I(−) and HLA-I(+) cells were cultured in hMSC medium (ATCC) to 50% confluence, then switched to osteogenic or adipogenic differentiation medium (ATTC) or ATRA (100 µM, Sigma-Aldrich) for up to 28 days.
2.2 Flow Cytometry
HLA-I(−) and (+) subpopulations were sorted by flow cytometry following standard procedures. Sarcoma cell suspensions were prepared trypsin digestion or by mechanical dissociation. PE conjugated HLA-I antibody (Abcam, ab43545) was used at 1:500 dilution.
2.3 Immunohistochemistry, Immunofluorescence, and Immunoblot
Standard methods were performed on cell lines and FFPE tissue sections. Primary antibodies for HLA class I W6/32 (Abcam, ab23755), CD44 (BD, 550392), Sox-9 (Santa Cruz, sc2009), and actin (Sigma, AC-15) were used.
2.4 Tumor Xenograft Formation and Tumorigenic Capacity Assay
All protocols for mouse experiments were in accordance with institutional guidelines and approved by the Mount Sinai Medical Center Institutional Animal Care and Use Committee. Tumor samples from the patients were immediately dissociated mechanically into single cell suspension. Up to 107 cells were injected for tumor xenograft formation.
For tumorigenic capacity assay, isolated subpopulations of HLA-I(−) and HLA-I(+) cells were serially diluted, injected subcutaneously into 10-weeks old female NGS mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ). 10 mice were used for every cell dilution. Mice were grouped using a randomized block design.
2.5 Next-generation Sequencing
Total RNA was extracted from using mirVana miRNA Isolation Kit (Life Technologies), treated with DNase I (Invitrogen). cDNA libraries were constructed with TruSeq stranded total RNA (with Robo-Zero) library preparation method. Next-generation sequencing was performed by Illimina HT DNA sequencing HiSeq2500/1500 system for single read clustering and 51 cycles sequencing.
2.6 Statistical Analysis
Experimental data was expressed as means ± SD and analyzed by two-sided Student’s t test. Limiting dilution data was analyzed with a generalized linear model described by Hu and Smyth (2009). Next-generation sequencing data was analyzed by Linear Models for Micro Array Analysis test to identify differentially expressed genes. Differentially expressed genes were subjected to GO or IPA in order to interpret the biological context. The significant GO terms were identified by fisher-exact test with p value of <0.05.
2.7 Data Availability
RNA-Sequencing data have been deposited into the NCBI Sequence Read Archive (SRA) under accession code SRP076437.
3. Results
3.1 A Subpopulation of HLA-I(−) Cells Display Stem-like Characteristics in Human Sarcomas
To investigate the potential presence of an HLA-I negative cell (HLA-I(−)) subpopulation in sarcomas, we examined the expression of HLA-I in formalin fixed paraffin embedded (FFPE) sarcoma samples(n=230 cases) using immunohistochemistry (IHC) analysis. As shown in Figure 1A, HLA-I(−) cells were found in all sarcoma subtypes studied, including clear cell sarcoma, pleomorphic liposarcoma, leiomyosarcoma, malignant peripheral nerve sheath tumor, and dedifferentiated liposarcoma. To further characterize this subpopulation of HLA-I(−) cells, we tested five sarcoma cell lines including MFH (undifferentiated pleomorphic sarcoma), CW9019 (rhabdomyosarcoma), SKNEP (Ewing sarcoma), MG63 (osteosarcoma), and LPS141 (liposarcoma). HLA-I(−) cells were identified in all cell lines, albeit at low frequencies (MFH: 1.6±0.6%; CW9019: 3.1±1.9%; SKNEP: 2.8±1.4%; MG63: 1.2±0.3%; and LPS141: 2.6±1.0%)(Figures 1B and 1C).
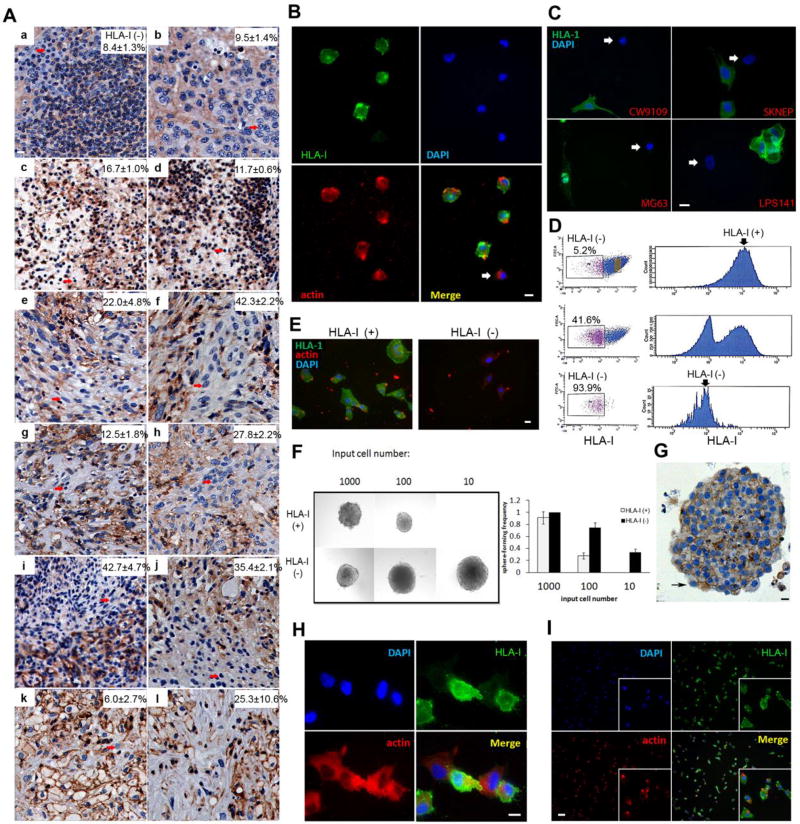
Figure 1. HLA-I(−) Cells Exist in Human Sarcomas and Show Stem Cell-like Properties.
(A) HLA-I(−) cells (arrows) were found in a human sarcomas by IHC. (a,b) clear cell sarcoma; (c,d) pleomorphic liposarcoma; (e,f) leiomyosarcoma; (g,h) malignant peripheral nerve sheath tumor; (i,j) liposarcoma, not otherwise specified; (k,l) dedifferentiated liposarcoma. (B) HLA-I(−) cells were detected by immunofluorescence in cell line MFH. (C) HLA-I(−) cells were detected in sarcoma lines: CW9019, SKNEP, MG63, and LPS141 by immunofluorescence. (D) The subpopulation of HLA-I(−) cells was isolated from cell line MFH by flow cytometry with a double-sort method. From top to bottom: first sort, second sort, and purity check. (E) Isolated HLA-I(+) and (−) subpopulations from MFH were cultured overnight. The enrichment of HLA-I(−) cells were verified. (F) Sphere formation assay showed that as few as 10 HLA-I(−) cells were able to form sarcoma spheres. Left: representative pictures of sarcoma spheres; Right: Sphere-forming frequency, the mean ± SD. (G) HLA-I expression in sphere cultured HLA-I(−) cells. (H) After 48 hours, HLA-I(+) cells were observed in HLA-I(−) cell cultures. Scale bar (B,C,E,G):10 µm (I) After 5 days, a cell culture starting from isolated HLA-I(−) cells now had a majority of cells that were HLA-I(+); a lower magnification (5×) was used to show a representative picture of HLA-I expression in an HLA-I(−) cell culture at 5 days. Scale bar (I): 100 µm.
To further characterize this HLA-I(−) subpopulation, a double sorting method by flow cytometry was used to isolate HLA-I(−) cells from MFH (Figure 1D). The phenotype of the isolated HLA-I(−) cells was further verified by immunofluorescence, western blot and quantitative RT-PCR (Figure 1E, S1A and S1B). Down-regulated HLA-1 at both protein and mRNA level in HLA-I(−) cells were observed. Such observation suggests that lacking HLA-I on the cell surface is mainly regulated at the expression level of HLA-1 genes.
Sphere formation assays were performed as indicators of self-renewal. Limiting dilution analysis was performed to determining sphere-forming potential. HLA-I(−) cells were able to form spheres with the initial input of as fewer as 10 cells (Figure 1F). The frequencies of sphere-forming cells, calculated based on a Poisson probability distribution, were 1 in 567.2 HLA-I positive cells (HLA-I(+)) (95% confidence interval (CI): 1/841.7-1/382.3) and 1 in 41.2 HLA-I(−) cells (95% CI: 1/61.8-1/27.5). Thus, sphere-forming cells were 13.8-fold enriched in cells displaying an HLA-I(−) phenotype. HLA-I(−) cells isolated from three additional cell lines, CW9019, SKNEP, and MG63, showed significantly higher sphere-forming efficiency than their HLA-I(+) counterparts (Figure S1D). Expression of stem cell markers, such as Oct4, Nanog, and Myc, was found at higher levels in HLA-I(−) cells than HLA-I(+) cells (Figure S1E). Interestingly, we observed that daughter cells of HLA-I(−) cells were mostly HLA-I(+), suggesting an asymmetrical division process. After 48 hours of culturing isolated HLA-I(−) cells, we found that most of the cells were HLA-I(+) cells (Figure 1H) (HLA-I(−) cells 36.8±6.2%). After 5 days in culture the majority of the cells were HLA-I(+) (Figure 1I); and by day 10 the frequency of HLA-I(−) cells was only 1.03±0.79% (Figure S1C). Interestingly, HLA-I(+) daughter cells were also observed in spheroid culture of HLA-I(−) cells (Figure 1G). Taken together, data from above studies support that HLA-I(−) sarcoma cells have high self-renewal capacity, and that they are capable of generating HLA-I(+) cells, suggesting the capacity of asymmetrical division.
3.2 Sarcoma cells with an HLA-I Negative Phenotype Display High Tumorigenic Efficiency
To evaluate the tumorigenic capacity of sarcoma cells with a HLA-I(−) versus HLA-I(+) phenotypes, we serially diluted HLA-I(−) and HLA-I(+) cells (from 105 to 103 cells), and then injected these final cellular products subcutaneously into NGS mice (n=10). Two sarcoma cell lines, MFH and CW9019, were used for these studies. The calculated tumor initiating cell (TIC) frequencies of HLA-I(−) cells were 1/621 for MFH, and 1/9204 for CW9019; substantially higher (95-and 14-fold, respectively) than their HLA-I(+) counterparts (Figure 2A). Thus, TICs are significantly associated within the subpopulation of sarcoma cells displaying the HLA-I negative phenotype.
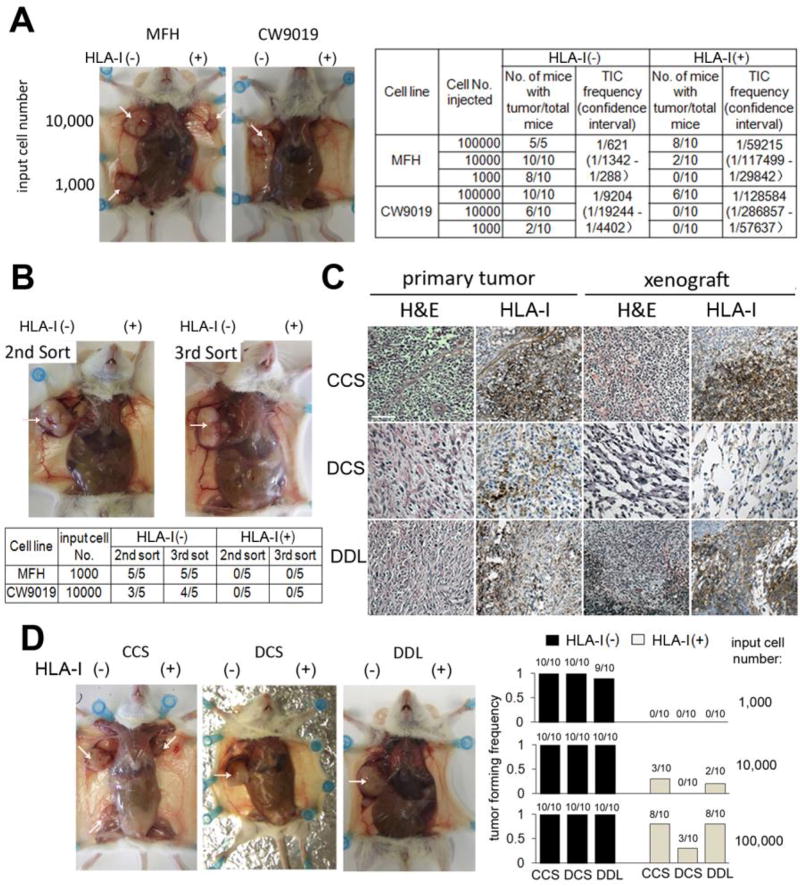
Figure 2. TICs Are Highly Enriched in the HLA-I(−) Subpopulation.
(A) Left: Representative pictures showed tumors formed from 104 and 103 cells of MFH (left) and CW9019 (right). Bulk tumors are indicated by arrows. Right: Tumor incidence and TIC frequency. (B) HLA-I(−) cells serially passed by tumor formation. Only HLA-I(−) cells were able to form tumor xenografts (arrows). (C) Tumor xenografts were histologically similar with the parental tumor (H&E stain) and showed cellular heterogeneity in HLA-I expression by IHC. Scale bar: 100 µm. (D) HLA-I(−) cells isolated from the three primary sarcoma xenografts were highly tumorigenic. Left: representative pictures of the tumor formed by HLA-I(−) and (+) cells from CCS, DCS and DDL. CCS: two doses, 104 and 103 CCS cells, were injected in each mouse; DCS and DDL: a single dose of 103 cells was injected in each mouse. Right: tumor forming frequency. Each bar represents 10 transplantations.
The in vivo self-renewal capacity of HLA-I(−) cells was tested by serial passages of HLA-I(−) cells from tumor xenografts. Subpopulations of HLA-I(−) and HLA-I(+) cells were isolated from tumor xenografts formed by HLA-I(−) cells, and re-injected into secondary and tertiary recipients (103 input cells). For every passage, only sarcoma cells displaying the HLA-I(−) phenotype were able to form tumors (Figure 2B). Additionally, these tumors contained both HLA-I(−) and (+) cell subpopulations in similar percentages, recapitulating the cellular phenotype of the parental tumors (Figure S2A & B).
We next extended these in vitro and in vivo analyses into human primary sarcoma tissue samples. Out of 7 human sarcomas tested, 3 formed xenografts for further studies, including a clear cell sarcoma (CCS), a dedifferentiated chondrosarcoma (DCS), and a dedifferentiated liposarcoma (DDL). These tumor xenografts were histologically similar to their parental primary tumors, all displaying both HLA-I(−) and (+) cell subpopulations (Figure 2C). Sarcoma cells with HLA-I(−) and (+) phenotypes were further isolated using flow cytometry assays, and their tumorigenic capacity also analyzed. Once more, HLA-I(−) cells from all three patient-derived tumor xenografts generated significantly higher tumor formation capacity than their HLA-I(+) counterparts. Using 103 injected sarcoma cells, those displaying the HLA-I(−) phenotype from CCS, DCS, and DDL formed tumors at high frequencies (10/10, 10/10, and 8/10, respectively); while HLA-I(+) cells failed to form tumors (Figure 2D). Additionally, tumors formed by HLA-I(−) cells contained both HLA-I(−) and (+) subpopulations in similar percentages (Figure S2). These features were retained for at least 3 passages when tumors formed by HLA-I(−) cells were dissociated for cell sorting to isolated HLA-I(−) and (+) subpopulations for subsequent tumor formation. For each passage, percentage of HLA-I(−) cells increased which may resulting from continuous selection of HLA-I(−) cells. Thus, TICs are characterized by an HLA-I(−) phenotype when tested in different human sarcoma histological subtypes.
3.3 Gene Expression Profiling of TICs Displays Molecular Characteristics of Stem Cells
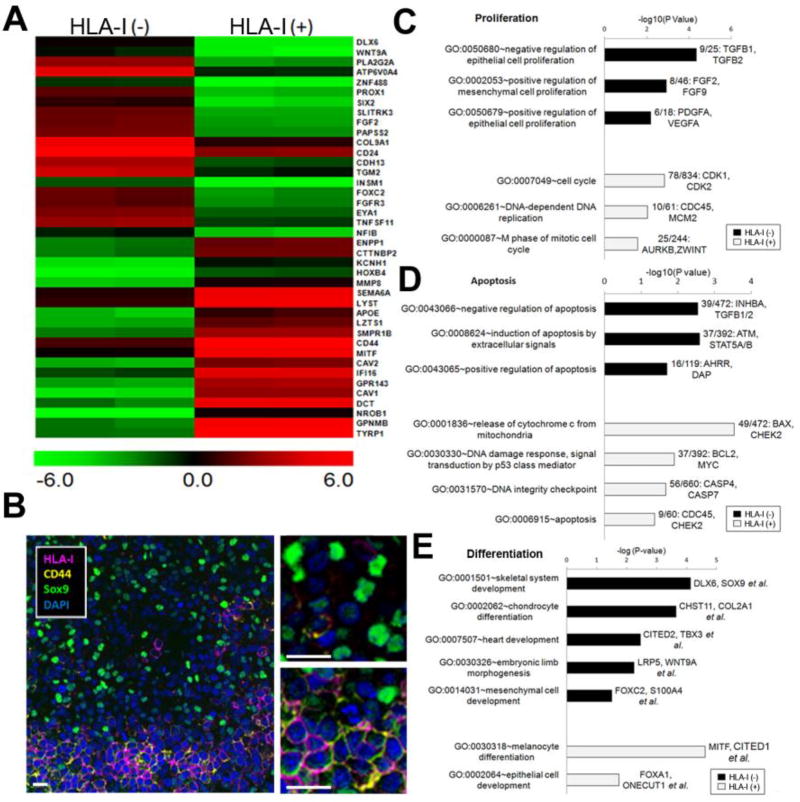
To further delineate the molecular profile of HLA-I(−) TICs, and to design effective therapeutic strategies to target these cell subpopulation, we designed a series of gene expression profile analysis using RNA sequencing, comparing the obtained signatures to those from HLA-I(+) non-TICs. The clear cell sarcoma xenograft model was chosen for this experiment due to a relatively higher percentage of HLA-I(−) cells (26.7±8.3%). Differential gene expression analysis revealed distinct signatures corresponding to 1,214 over-expressed and 1,293 under-expressed genes in HLA-I(−) TICs when compared to HLA-I(+) non-TICs from the same tumor (>1.5 fold, P<0.05). The top 40 differentially expressed genes related to cell differentiation are shown in Figure 3A. Some of these genes were verified by multicolor immunofluorescence. CD44, a widely studied biomarker (Du et al., 2008), was found specifically to co-localize with HLA-I antigen, being expressed in the non-TIC subpopulation. Sox-9, a developmental gene reported to play important roles in other cancer stem cells such as in breast carcinoma (Guo et al., 2012), was specifically expressed in the HLA-I(−), TIC subpopulation (Figure 3B). Clear cell sarcoma HLA-I(−) TICs display a Sox-9 positive/CD44 negative phenotype. Gene ontogeny (GO) enrichment analysis of proliferation-related and apoptosis-related genes revealed that cell cycle regulatory genes were expressed at lower levels in the HLA-I(−) TICs (Figure 3C & 3D). Ingenuity pathway analysis (IPA) also revealed significantly down-regulated apoptosis genes in the HLA-I(−) TIC subpopulation.
Figure 3. TICs Are Characterized by Gene Expression and Functional Assays.
(A) A heat map showed top 40 differentially expressed genes between TICs and non TICs. (B) CCS tumors were tested by immunofluorescence. (C,D,E) GO analysis indicated the proliferation regulatory genes were expressed by TICs and cell cycle genes were highly expressed in the non-TICs, apoptosis genes were expressed at higher level in non-TICs. And TICs cells expressed skeleton system development and mesenchymal cell related genes. Non-TICs expressed melanocyte differentiation and epithelial cell genes.
Interestingly, genes related with melanocyte differentiation, a characteristic of clear cell sarcoma, were highly expressed in HLA-I(+) non-TICs (GO:0030318, P<0.001), including key transcription factors, such as MITF (30.4-fold) and its downstream genes TYRP1, GPNMB, DCT, CAV1, and CAV2 (Figure 3A & 3E). Moreover, HLA-I(+) non-TICs were also found to express high levels of other differentiation genes, including FOXA1, ONECUT1. In contrast, HLA-I(−) TICs display high levels of genes related to mesenchymal development (GO:0001501, P<0.0001) and embryonic morphogenesis (GO:0030326, P<0.01); such as FOXC2 and S100A4 genes (Figure 4A), consistent with the reported mesenchymal properties of TICs (Polyak and Weinberg, 2009).
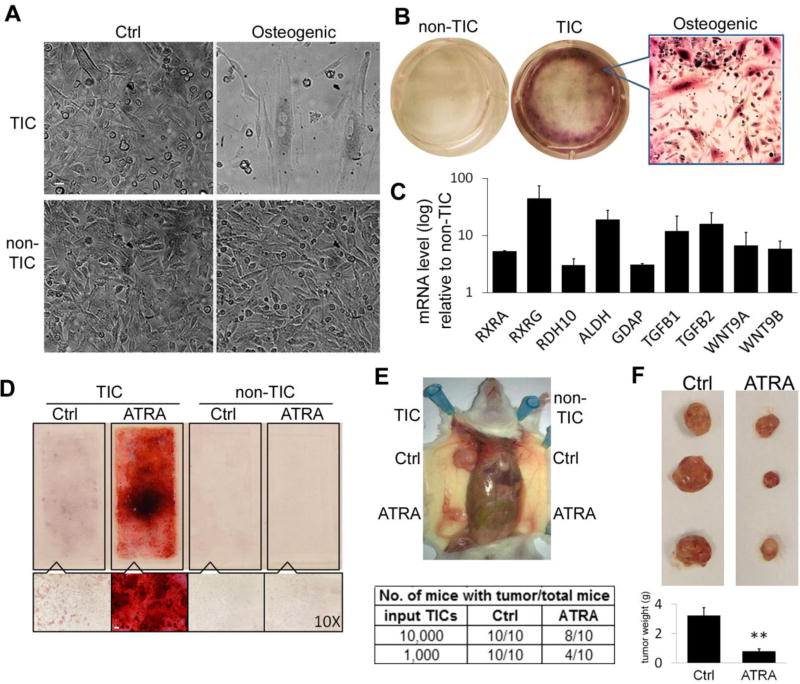
Figure 4. TICs Undergo Osteogenic Diffe rentiation by ATRA Treatment with Decreased Malignancy.
(A) Under osteogenic differentiation conditions, morphological alterations were observed in TICs in 10 days. (B) Osteogenic differentiated HLA-I(−) TICs showed strong positive Alizarin-Red-S staining. (C) Quantitative RT-PCR results showed highly expressed RAR pathways genes in HLA-I(−) TICs compared to non-TICs. Data represent the mean ± SD. (D) ATRA treatment induced osteogenic differentiation of TICs in vitro. Top: 4-well chamber slide stained with Alizarin-red-S; Bottom: bright field microscope (10×). Scale bar: (A,B):10 µm; (D):100 µm (E) TICs and non-TICs were treated in vitro with ATRA before transplanted into NGS mice. Tumor formation by ATRA treated TICs were significantly decreased. (F) Mice transplanted with TICs cells were treated orally with ATRA or DMSO every other day for 6 weeks. ATRA treatment inhibited the tumor formation by TICs in vivo. Top: Representative tumors formed by HLA-I(−) cells; Bottom: average tumor mass (error bars represent the standard errors, **: P<0.01).
We then assessed if HLA-I(−) cells could differentiate along mesenchymal phenotypes. HLA-I(−) and HLA-I(+) cells were cultured under well-described osteogenic and adipogenic differentiation conditions, as reported by our group (Matushansky et al., 2008). After 10 days, we observed phenotypic changes in the morphology of the HLA-I(−) subpopulation, which became large and osteoclast-like cells with relatively slower cell population growth (Figure 4A). Matrix mineralization (detected by a strong Alizarin-S-red staining) (Figure 4B) and increased expression of osteogenic differentiation markers (Figure S3B), suggested differentiation along osteogenic pathway. Following adipogenic differentiation conditions, HLA-I(−) cells showed weak Oil-Red-O staining which suggested only limited differentiation potential along adipogenic pathway (Figure S3A). In contrast, HLA-I(+) cells showed no response under neither of these mesenchymal differentiation conditions, as predicted by their already executed differentiation phenotype and further revealed by expression profiling and GO analysis.
3.4 Differentiation The rapy Successfully Targeted the HLA-I Negative Sarcoma Cell Subpopulation
Since sarcoma cells displaying HLA-I(−) phenotype and tumor initiation characteristics were also capable of osteogenic maturation, we next explored if this subpopulation of cells could efficiently be the target of a differentiation therapy strategy. All-trans retinoic acid (ATRA) is one of the best studied and successful molecules used in differentiation-induced treatment, achieving long-term remissions and even cures in treating certain human cancers, including acute promyelocytic leukemia (Altucci and Gronemeyer, 2001). More importantly, the osteogenic differentiation effect of ATRA has been reported in both mouse and human (Wan et al., 2006; Zhang et al., 2010). Interestingly, our expression profiling and GO analyses revealed that HLA-I(−) cells over-expressed RA responsive genes (GO:0032526, P<0.02), a subset of which were further confirmed by qRT-PCR studies (Figure 4C). IPA network analysis also showed that retinoid acid receptor activation related genes were elevated in HLA-I(−) cells (Figure S4, P<0.001).
We then designed an experimental protocol that allowed us to study the effects of ATRA-induced osteogenic differentiation both in vitro and in vivo. Subpopulations of HLA-I(−) TICs and HLA-I(+) non-TICs isolated from tumor xenografts were cultured in vitro and treated with 100 µM ATRA. Mineralization was detected by both morphological changes as well as clearly positive Alizarin-red-S staining observed in the HLA-I(−) TICs, but not in the HLA-I(+) non-TICs (Figure 4D). Expression of osteogenic differentiation genes, including SPP1 (osteopontin), BMP2 (bone morphogenetic protein 2) and ALPL (alkaline phosphatase), were significantly increased in ATRA-treated HLA-I(−) TICs. In contrast, these genes were expressed at significantly lower levels in HLA-I(+) non-TICs, which did not displayed phenotype changes by ATRA treatment (Figure S3B). Most importantly, differentiation therapy with ATRA increased HLA class I mRNA expression in the HLA-I(−) TICs treated cultures (Figure S3B). We followed these studies by injecting differentiated HLA-I(−) TICs induced by ATRA into NGS mice in order to assess their tumor formation capacity. Compared with DMSO vehicle treated control cells, ATRA treatment significantly inhibited the tumor formation of HLA-I(−) TICs (Figure 4E). Furthermore, we tested the tumor formation inhibition effect of ATRA in vivo. NGS mice injected with HLA-I(−) TICs were treated with oral ATRA or DMSO control 5 days a week. After 4 weeks of treatment, tumors formed in oral ATRA treatment group were significantly smaller (>75% decrease in tumor mass) than the control group (Figure 4F). Thus, tumors formed by HLA-I(−) TICs were induced to differentiate along the osteogenic pathway by ATRA, resulting in significantly decreased tumorigenicity.
4. Discussion
Herein we report the identification and characterization of a cell subpopulation displaying a HLA-I(−) phenotype and tumorigenic capacity in all distinct sarcoma subtypes analyzed. This subpopulation of sarcoma cells share many commonalities with the previously reported HLA-I(−) TIC subpopulation in human prostate cancer. Furthermore, these HLA-I(−) sarcoma TICs display certain stem-like properties, including a molecular signature rich in developmental and embryonic genes, in addition to self-renewal capacity and asymmetrical division. This later characteristic is supported by data revealing that undifferentiated, HLA-I(−) cells can generate HLA-I(+), differentiated cells as illustrated in Figures 1 and 2. More interestingly, our results also suggest a means to restore HLA-I expression and differentiation programs in this subpopulation of HLA-I(−) TICs. This is supported by both in vitro and in vivo studies, in which we observed induced osteogenic differentiation of HLA-I(−) TICs using either osteogenic medium or ATRA treatment. Exposure of HLA-I(−) TICs to such conditions resulted in significant increase of HLA-I expression, differentiation and decreased tumorigenic capacity. Taken together, these findings support that differentiation therapies, or potentially certain epigenetic approaches, can restore HLA-I expression and strikingly decrease tumor formation and tumor progression. These data allow us to further postulate that suppression of HLA-I expression is a critical mechanism used by different types of cancer to escape host immune surveillance, and that differentiation therapy may allow cancer stem cells to be recognized by the host immune system.
Our results underscore the importance of a common therapeutic goal in medical oncology, namely the identification of therapies that target not only differentiated tumor cells, but also undifferentiated subpopulations that may represent the cancer stem cell compartment, possibly responsible for treatment resistance and metastatic spread. In addition to differentiation therapeutic strategy, HLA-I-independent tumor cell killing by cytokine-induced killer cell may be another promising method to target these HLA-I(−) cells (Giraudo et al., 2015). Our future efforts are focused on the characterization of the similarities and differences of the HLA-I enriched population in other sarcomas, given the breadth of biologies they represent, as well as the mechanisms at hand in the parallel differentiation and loss of neoplastic potential of HLA-I(−) cells in different cancer subtypes.
Supplementary Material
Highlights.
Identification of human sarcomas stem cells by a HLA-I (−) phenotype
Molecular and functional characterization of HLA-I (−) cells
Target HLA-I (−) sarcoma stem cells by differentiation therapy
Acknowledgments
This research was supported by NCI-P01-CA087497 (C.C.-C., D.H.); NIH-U54-0OD020353 (C.C.-C., D.H., J.D.-D.), Agilent Though Leader Award (C.C.-C.), and Martel Foundation (C.C.-C., J.D.-D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no competing financial interests.
References
- Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- Cabrera T, Collado A, Fernandez MA, Ferron A, Sancho J, Ruiz-Cabello F, Garrido F. High frequency of altered HLA class I phenotypes in invasive colorectal carcinomas. Tissue Antigens. 1998;52:114–123. doi: 10.1111/j.1399-0039.1998.tb02274.x. [DOI] [PubMed] [Google Scholar]
- Chang CC, Campoli M, Ferrone S. Classical and nonclassical HLA class I antigen and NK Cell-activating ligand changes in malignant cells: current challenges and future directions. Adv Cancer Res. 2005;93:189–234. doi: 10.1016/S0065-230X(05)93006-6. [DOI] [PubMed] [Google Scholar]
- Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer Lett. 2009;279:126–36. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Fuks Z, Drobnjak M, Moreno C, Eisenbach L, Feldman M. Expression of HLA-A,B,C antigens on primary and metastatic tumor cell populations of human carcinomas. Cancer Res. 1991;51:6372–6380. [PubMed] [Google Scholar]
- Daar AS, Fuggle SV, Fabre JW, Ting A, Morris PJ. The detailed distribution of HLA-A, B, C antigens in normal human organs. Transplantation. 1984;38:287–292. doi: 10.1097/00007890-198409000-00018. [DOI] [PubMed] [Google Scholar]
- Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, Bonal DM, Charytonowicz E, Gladoun N, de la Iglesia-Vicente J, Petrylak DP, Benson MC, Silva JM, Cordon-Cardo C. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22:373–288. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker M, Katz G, Urbach A, Schuldiner M, Markel G, Itskovitz-Eldor J, Reubinoff B, Mandelboim O, Benvenisty N. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99:9864–9869. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y, Chen Q. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunol Today. 1993;14:491–499. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- Garrido F, Ruiz-Cabello F, Cabrera T, Pérez-Villar JJ, López-Botet M, Duggan-Keen M, Stern PL. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- Giraudo L, Gammaitoni L, Cangemi M, Rotolo R, Aglietta M, Sangiolo D. Immunotherapy. 2015;7:999–1010. doi: 10.2217/imt.15.61. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Mills J, Matos T, Charytonowicz E, Hricik T, Castillo-Martin M, Remotti F, Lee FY, Matushansky I. Characterization and comparison of the properties of sarcoma cell lines in vitro and in vivo. Hum Cell. 2009;22:85–93. doi: 10.1111/j.1749-0774.2009.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matushansky I, Maki RG. Mechanisms of sarcomagenesis. Hematol Oncol Clin North Am. 2005;19:427–49. doi: 10.1016/j.hoc.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Matushansky I, Hernando E, Socci ND, Matos T, Mills J, Edgar MA, Schwartz GK, Singer S, Cordon-Cardo C, Maki RG. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol. 2008;172:1069–80. doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase M, Kano M, Tsukahara T, Takahashi A, Torigoe T, Kawaguchi S, Kimura S, Wada T, Uchihashi Y, Kondo T, et al. Side population cells have the characteristics of cancer stem-like cells/cancer-initiating cells in bone sarcomas. Br. J. Cancer. 2009;101:1425–1432. doi: 10.1038/sj.bjc.6605330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Portmann-Lanz CB, Schoeberlein A, Huber A, Sager R, Malek A, Holzgreve W, Surbek DV. Placental mesenchymal stem cells as potential autologous graft for pre- and perinatal neuroregeneration. Am J Obstet Gynecol. 2006;194:664–673. doi: 10.1016/j.ajog.2006.01.101. [DOI] [PubMed] [Google Scholar]
- Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, Fazioli F, Pirozzi G, Papaccio G. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011;25:2022–2030. doi: 10.1096/fj.10-179036. [DOI] [PubMed] [Google Scholar]
- Wahl J, Bogatyreva L, Boukamp P, Rojewski M, van Valen F, Fiedler J, Hipp N, Debatin KM, Beltinger C. Ewing’s sarcoma cells with CD57-associated increase of tumorigenicity and with neural crest-like differentiation capacity. Int. J. Cancer. 2010;127:1295–1307. doi: 10.1002/ijc.25163. [DOI] [PubMed] [Google Scholar]
- Wan DC, Shi YY, Nacamuli RP, Quarto N, Lyons KM, Longaker MT. Osteogenic differentiation of mouse adipose-derived adult stromal cells requires retinoic acid and bone morphogenetic protein receptor type IB signaling. Proc Natl Acad Sci U S A. 2006;103:12335–12340. doi: 10.1073/pnas.0604849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Deng ZL, Chen L, Zuo GW, Luo Q, Shi Q, Zhang BQ, Wagner ER, Rastegar F, Kim SH, et al. Retinoic acids potentiate BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2010;5:e11917. doi: 10.1371/journal.pone.0011917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.