Abstract
Epidermal specific deletion of the homeobox transcription regulator DLX3 disrupts keratinocyte differentiation and results in an IL-17-linked psoriasis-like skin inflammation. To identify the epidermal initiating signals produced by DLX3-null keratinocytes, we performed acute deletion of DLX3 in adult epidermis using a tamoxifen-inducible Krt14-cre/ERT system. K14CreERT;DLX3fl/fl (icKO) skin exhibited dysregulated expression of differentiation-associated genes, upregulation of proinflammatory cytokines, and accumulation of Langerhans cells and macrophages within 3 days of tamoxifen-induced DLX3 ablation. We also observed increased accumulation of IL-17A-secreting Vγ4 γδ T-cells and heightened levels of IL-17 and IL-36 family of cytokines starting one week after DLX3 deletion. Interestingly, transcriptome profiling of icKO epidermis at 3 days identified activated STAT3 as a transcriptional regulator and revealed differential expression of STAT3 signaling-related genes. Furthermore, activation of STAT3 was strongly increased in icKO skin, and topical treatment with an inhibitor of STAT3 activation attenuated the immune phenotype. RNA-seq analysis of vehicle and STAT3 inhibitor treated icKO skin identified differentially expressed genes associated with inhibition of leukocyte infiltration. Collectively, our results show that DLX3 is a critical regulator of STAT3 signaling network that maintains skin homeostasis.
INTRODUCTION
The epidermal barrier acts as a first line of defense by providing protection against environmental stimuli (Candi et al., 2005, Simpson et al., 2011). Disruption of epidermal barrier integrity is associated with, and can initiate, inflammatory responses. Inflammation is in part orchestrated through secretion of IL-1 family proinflammatory cytokines, and antimicrobial peptides (S100A8 and S100A9) (Akitsu et al., 2015, Eckert et al., 2004, Nestle et al., 2009, Shornick et al., 1996). Hyperproliferation, aberrant differentiation of keratinocytes and cytokine- and chemokine-mediated infiltration of immune effector cells are characteristics of lesional skin in psoriasis and atopic dermatitis (Albanesi et al., 2007, Monteleone et al., 2011). The regulation of skin inflammatory circuits involved in these diseases have been studied utilizing genome wide association and transgenic mouse models (Ellinghaus et al., 2013, Gudjonsson et al., 2007, Swindell et al., 2011, Tsoi et al., 2012). Mouse models of psoriasis have addressed the involvement of key signaling pathways including AP1, PPARβ/δ, STAT3, TGFβ, NFKβ, WNT/β-catenin, VEGF and cytokines including IL-1, IL-12, IL-23, IL-17 and TNFα in pathomechanisms of chronic inflammation (Augustin et al., 2013, Gudjonsson et al., 2007, Swindell et al., 2011). However, the cell-intrinsic contributions of keratinocytes in human inflammatory skin diseases are less well characterized.
Transcription factors, including AP1, ETS, Klf4, p63 and DLX3 regulate the highly-orchestrated terminal differentiation program that results in the formation of the epidermal barrier (Botchkarev, 2015, Eckert and Welter, 1996, Hwang et al., 2011, Sinha et al., 2000). The homeodomain transcription factor DLX3 is crucial for keratinocyte differentiation and maintenance of the epidermal barrier (Hwang et al., 2011; Palazzo et al., 2016, 2017). DLX3 is expressed in the differentiated layers of epidermis and in hair follicles (Hwang et al., 2008). Autosomal dominant mutations in DLX3 cause an ectodermal dysplasia termed Tricho-Dento-Osseous syndrome which is characterized by teeth, bone and hair defects (Duverger et al., 2008, Price et al., 1998, Wright et al., 2008). Misexpression of DLX3 in the basal layer results in decreased keratinocyte proliferation and premature terminal differentiation (Morasso et al., 1996). More recently, we have shown that DLX3 regulates keratinocytes proliferation and differentiation by controlling p53 downstream targets and cell cycle exit (Palazzo et al., 2016).
Epidermal-specific ablation of DLX3 in murine skin causes complete alopecia, hyperproliferation of keratinocytes, impaired terminal differentiation and development of IL-17-dependent skin inflammation (Hwang et al., 2011). Downregulation of DLX3 was also observed in human psoriatic lesions, indicating that a loss or decrease of DLX3 function in keratinocytes correlates with a psoriatic phenotype (Hwang et al., 2011).
The present study aimed to define keratinocyte-intrinsic DLX3-dependent signals that lead to the IL-17-associated psoriatic-like skin inflammation. To identify inflammation triggering signals from DLX3-null keratinocytes and the reciprocal signaling crosstalk between epidermis and dermis that induced infiltration of antigen-presenting cells and IL-17 producing T cells into the dermis, we acutely deleted DLX3 in adult epidermal keratinocytes using a tamoxifen-inducible K14Cre-ERT system (K14CreERT; Dlx3f/f). Our results demonstrate a direct link between DLX3 ablation and phosphorylation of STAT3 that results in rapid immune cell infiltration and development of skin inflammation.
RESULTS
Acute epidermal deletion of DLX3 leads to abnormal epidermal proliferation, differentiation and barrier dysfunction
To delete DLX3 in epithelial keratinocytes, dorsal skin of K14-CreERT2;Dlx3fl/fl (icKO) mice and wild-type (WT) littermates was treated topically with tamoxifen for five consecutive days, followed by analysis at early (3 day) and late stages (1 and 2 weeks) after treatment (Figure 1a). Treatments were initiated at postnatal day 42–46 (P42-46) while the hair follicles were in synchronized telogen. Deletion of epidermal DLX3 was validated by RNA-Seq (Figure 1b). Control skin keratinocytes prominently expressed DLX3, while expression of the second and third exons was not detected in icKO epidermis, confirming successful deletion (Figure 1b). Hematoxylin & eosin (H&E)-stained sections of icKO skin did not reveal overt changes in skin and hair follicles in comparison to WT counterpart at any time points (Supplementary Figure S1a), and no significant changes in epidermal thickness were observed (Supplementary Figure S1b). There were no significant differences in the expression pattern of the epidermal differentiation marker Keratin 10 (K10) between WT and icKO mice (Supplementary Figure S1c). However, significant increases in the numbers of Ki67+ proliferating keratinocytes were detected in icKO epidermis at 1 and 2 weeks after DLX3 deletion (Supplementary Figure S1d).
Figure 1. Acute epidermal deletion of DLX3 in adult skin induces altered keratinocyte differentiation and barrier defects.
(a) Schematic representation of topical tamoxifen treatment and sample collection post-tamoxifen (TAM) treatment. (b) RNA Sequencing tracks for the DLX3 locus shows distinct peaks for three exons in WT (DLX3fl/fl) and absence of peaks at exon 2 and 3 in icKO (K14-CreERT2;Dlx3fl/fl) confirming conditional inducible deletion of DLX3. (c) Log2 fold changes between DLX3 WT and icKO groups were used for Gene Ontology (GO) terms. The graph summarizes all the biological processes detected by GO enrichment analysis at 3 days, 1 week and 2 weeks in the epidermal transcriptome. Antilog of p-values was used for graphical representation. (p <0.05). (d) Heatmap showing differential expression of epidermal differentiation complex genes (EDC) at 3 days and 1 week. Values represent normalized log2 RPKM (n = 3 per group). (e) Immunohistochemistry of skin sections with antibodies for Keratin 6 (K6) and differentiation marker Filaggrin at 3 days and 1 week after TAM treatment. All sections were counterstained with Hoechst to establish landmarks and visualize tissue morphology and nuclei. The white dotted line demarcates epidermis from dermis. Scale bar = 100um
To characterize the molecular changes due to DLX3 deletion in an unbiased fashion, we performed transcriptomic analysis of icKO epidermal and dermal compartments using RNA-seq. To corroborate the purity of epidermal and dermal fractions, we verified the specific expression of Keratin 14 and vimentin genes in epidermal and dermal samples respectively (Supplementary Figure S1e). GO enrichment analyses of differentially expressed transcripts showed cell proliferation, differentiation, migration and inflammation as significantly enriched biofunctions in icKO epidermis (Figure 1c), while the dermal transcriptome highlighted regulation of immune system by MHC class I antigen presentation and T-cell mediated cytotoxicity (Supplementary Figure S2). Analysis of the epidermal transcriptome also showed pronounced changes in the transcription of Epidermal Differentiation Complex (EDC) genes (Figure 1d). LCE and SPRR family members were differentially upregulated as early as 3 days after DLX3 deletion (Figure 1d). The expression of these EDC genes is known to be upregulated in skin with altered barrier formation (de Cid et al., 2009, Koch et al., 2000). Several proinflammatory members of the S100 family (Broome et al., 2003, Eckert et al., 2004) were also upregulated in icKO epidermis (Figure 1d). Interestingly, significant upregulation in expression of keratins K6b and K16 was also detected. K6b was increased in icKO epidermis as early as 3 days post-treatment (Figure 1e). K6 is a marker of hyperproliferation that is strongly induced during wound healing and in lesional psoriatic epidermis (Rothnagel et al., 1999). By 1 week, many LCE (10 out of 16 genes) and SPRR (10 out of 15 genes) family members were differentially regulated, indicating altered cornification and barrier function in icKO skin (Figure 1d). We also observed down-regulation of the epidermal differentiation marker filaggrin (Figure 1e). Alterations in filaggrin expression have been observed in several skin inflammatory diseases (Sandilands et al., 2009). Collectively, our results show that acute deletion of DLX3 in adult epidermis alters keratinocyte differentiation and induces dysregulated expression of genes associated with epidermal barrier function.
Acute loss of epidermal DLX3 triggers skin inflammation
To characterize the effect of acute epidermal DLX3 ablation in adult skin, we analyzed transcriptomes of epidermis and dermis of WT and icKO mice. Inflammatory mediators including IL-1α and Defb3 were upregulated in icKO epidermis as early as 3 days post DLX3 deletion (Figure 2a). The related IL-1 family members, IL-36α, IL-36β and IL- 36γ and other proinflammatory cytokines IL-11, IL-17 and IL-24 that are linked to cutaneous disorders (Blumberg et al., 2007, Sabat et al., 2007) were up-regulated in icKO epidermis at later time points (Figure 2b). Examination of the cytokine and chemokine profile in icKO dermis showed increased expression of only a few genes at 3 days, and at 1 week (Figure 2a and b). Cytokines that are produced by keratinocytes and non-epithelial immune cells residing in the epidermis may constitute initiating signals that lead to the infiltration of antigen presenting innate immune cells in the dermis. Langerhans cells are primary antigen presenting cells in the epidermis, and first to respond to barrier perturbations (Kubo et al., 2009). We observed significantly increased numbers of Langerin+ Langerhans cells in icKO epidermis at 3 days and 1 week post DLX3 deletion (Figure 2c). We also found significantly heightened number of macrophages and CD3+ cells in icKO dermis at 3 days and 1 weeks (Figure 2d and Supplementary Figure S3a). Immunohistological analysis revealed significant increase of CD4+ T cell in the icKO skin at the 1 week time point (Figure 2e).
Figure 2. Inducible DLX3 deletion in keratinocytes triggers proinflammatory cytokine expression and infiltration of T cells, primarily IL-17A-secreting T-cells.
(a) Heatmaps displaying cytokines upregulated and downregulated in epidermis and dermis 3 days after TAM treatment. (b) Upregulation of subsets of cytokines and chemokines in epidermis and dermis 1 week after DLX3 deletion by TAM treatment. (c) Immunofluorescent labeling of Langerhans cells using langerin antibody. Bottom panel, Bar graph depicting significant changes in total numbers of Langerhans cells in icKO epidermis in comparison to WT. (d) Representative images of immunofluorescence staining shows increases in CD3+ T cells in icKO versus WT skin at 3 days and 1 week after DLX3-ablation. Bar graph indicates numbers of CD3+ T cells in WT and icKO skin. (e) Immunolabeling of CD4+ T cells subpopulation in WT and icKO skin at 3 days and 1 week. Hoechst was used as a counterstain. Graph represents numbers of CD4 + cells in the dermal compartment. The white dotted line demarcates epidermis from dermis. Scale bar = 100um (f) Flow cytometry analysis characterized the total and specific subpopulations of IL-17A secreting T cells at 3 days. (g) Increased total number of IL- 17A producing cells and their sub-populations 1 week after TAM treatment. p-value significance *>0.05 and **>0.01.
Flow cytometric analysis of single cell suspensions from icKO skin showed that numbers of CD45+, CD3+ and Class II+ cells were elevated by 2-fold at 3 days, with additionally increased accumulation at 1 week and 2 weeks (Supplementary Figure S3a). The accumulation of leukocytes and macrophages was also assessed by immunohistochemistry (Supplementary Figure S3b). Total numbers of IL-17A+ T cells were increased by 2-fold at 3 days, with further increases at 1 week (3-fold) and 2 weeks (4-fold) (Figure 2f and 2g and Supplementary Figure S3c). Major sources of IL-17A were TCRγδ+ and Vγ4+ T-cells at 1 week and 2 weeks (Figure 2g and Supplementary Figure S3c). Collectively, our results show that DLX3 deletion in epidermis triggers dermal accumulation of immune and inflammatory cells, in particular IL-17A producing Vγ4-bearing γδT cells.
Stat3 signaling contributes to skin inflammation in icKO keratinocytes
Upstream regulator analysis via IPA identified cytokines including IL-13, IL-17a, IL-15, IL-2, IL-1b and TNF, as well as the transcription regulator STAT3 as the top activated pathways at 3 days (Figure 3a). The causal analysis compares the known targets of each upstream regulator present in the Ingenuity Knowledge Base to the observed changes in the user’s dataset and identify the relevant upstream regulatory molecules associated with the transcriptomic changes (Kramer et al., 2014). STAT3 activation has a significant overlap p-value of 9.07 and activation z-Score of 3.5 which indicate statistically significant overlap between the dataset genes and the genes that are regulated by activated form of STAT3. (Figure 3a). We also assessed changes in gene expression downstream of STAT3 activation (Figure 3a). To determine if STAT3 activation was induced in icKO skin, we assessed levels of phosphorylated STAT3 and detected significant increases in nuclear staining of activated STAT3 (pSTAT3) in icKO when compared to control tissues from day 3 onward (Figure 3b). Correspondingly, western blot analyses showed increased levels of phosphorylated STAT3 and no changes in total STAT3 (Figure 3c). Analysis of RNA-Seq expression data from WT and icKO epidermis revealed several STAT3 signaling pathway genes were differentially expressed in icKO skin (Supplementary Figure S4).
Figure 3. Activation of STAT3 signaling in DLX3 deleted icKO skin.
(a) Upstream regulators of 3 days gene dataset by IPA (left panel) and the differentially observed genes related to the potential upstream regulator of STAT3 activation (right panel). (b) Immunofluorescence for phosphorylated STAT3 (p-STAT3) showed increased detection in 3 days and 1 week icKO epidermis post-TAM treatment. Percentage of positive p-STAT3 cells in the epidermis are presented in the bar graph. The white dotted line indicates epidermal-dermal boundary. Scale bar = 100um (c) Western blot analysis of p-STAT3 and total STAT3. RPL11 was used as an internal control for normalization. Quantitative analysis of western blot results showed increased expression of p-STAT3 with respect to total STAT3 in icKO skin at 3 days and 1 week.
Reversal of the inflammatory response in icKO skin after treatment with STAT3 Inhibitor
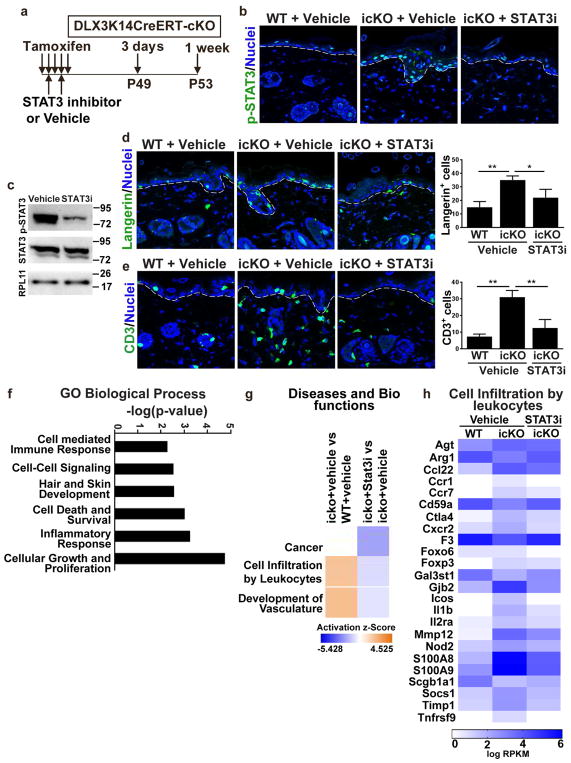
To assess the linkage between DLX3 function, STAT3 phosphorylation and skin immune homeostasis, we blocked STAT3 activation in icKO using the small molecule inhibitor cryptotanshinone. Cryptotanshinone inhibits STAT3 phosphorylation by binding to the SH2 domain, inhibiting nuclear translocation and binding to its target genes for transactivation (Lu et al., 2013). Treating the icKO mice with two systemic doses (IP injections) of cryptotanshinone reduced phosphorylated STAT3 levels, but not total STAT3, in icKO epidermis (Figure 4a–c). Inhibition of STAT3 phosphorylation led to a significant decrease in the number of Langerhans cells and CD3+ T-cells in icKO epidermis (Figure 4d and 4e). Gene ontology enrichment analysis of transcriptome profiles of icKO skin treated with vehicle and cryptotanshinone showed genes related to inflammatory response, specifically inhibition of cell infiltration by leukocytes and development of vasculature, were significantly enriched in vehicle treated skin (Figure 4f and g). Heatmaps associated with cell infiltration by leukocytes showed the upregulation of genes in icKO skin treated with vehicle whereas, there was downregulation of a subset of those genes in icKO skin treated with cryptotanshinone (Figure 4h).
Figure 4. Treatment with STAT3 specific inhibitor cryptotanshinone reduced inflammation in icKO skin.
(a) Schematic representation of TAM and cryptotanshinone (STAT3i) or Vehicle treatment and subsequent sample collection after treatment. (b) Immunolabeling of p-STAT3 in vehicle and STAT3i treated skin. (c) Western blot analysis showed reduction of p-STAT3 expression after STAT3i treatment in icKO skin. (d) Langerin staining and quantitation of Langerin-positive Langerhans cells in icKO skin after vehicle and STAT3i treatment. WT with vehicle treatment was used as control. (e) Immunofluoresecence with CD3 antibody of vehicle and STAT3i treated WT and icKO skin. Bar graph demonstrates decrease of CD3+ T cells after STAT3i treatment. p-value significance *>0.05 and **>0.01. (f) GO functional classification of differentially expressed genes. (g) Z-score identifies the activation or inhibition of biological functions. (h) Gene expression in icKO vehicle and cryptotanshinone treated samples. Color key: red represents upregulated and green downregulated expression.
DISCUSSION
A tightly regulated immune microenvironment in skin relies on extensive cross talk between epithelial keratinocytes, dermal fibroblast and immune cells (Pasparakis et al., 2014). Dysregulation of this finely tuned interaction likely contributes to the pathogenesis of chronic inflammatory disorders including atopic dermatitis and psoriasis, yet the keratinocyte-derived molecular signals for immune activation in hyperproliferative skin are incompletely characterized. (Lowes et al., 2008, Monteleone et al., 2011, Weidinger and Novak, 2016). In the present study, we acutely deleted DLX3 in adult epidermis to identify keratinocyte-specific DLX3-dependent signal(s) that regulate the physiological skin immune environment.
Here we report that in vivo abrogation of DLX3 function in adult murine keratinocytes results in impaired barrier function and triggers infiltration of immune cells including Langerhans cells, macrophages and T-cells. Langerhans cells are primary antigen presenting dendritic cells in epidermis and their increased presence has been shown in inflammatory skin disorders (Ginhoux and Merad, 2010). Previously published data also shows that barrier disruption through tape stripping or administration of proinflammatory cytokines such as IL-1 promotes Langerhans cell activation (Kubo et al., 2009, Wang et al., 1999). Consistent with these studies, the increased expression of IL-1α by DLX3-deficient epidermis triggered an increased number of Langerhans cells in icKO skin, which subsequently may induce dermal infiltration of antigen presenting cells such as macrophages. In psoriasis, prominent infiltration by macrophages is a characteristic feature of the disorder (Schon et al., 2000).
Current concepts of inflammatory skin disorders emphasize important roles for T cells in disease pathogenesis (Lowes et al., 2008). Indeed, we observed a significant recruitment of CD3/CD4 positive T cells in DLX3-icKO skin in the early time point of analysis (3 days). FACS profiling of mutant adult DLX3 epithelium also revealed infiltration of specific γδ TCR+ T-cell populations. Unlike the vast majority of mature αβ T lymphocytes, which reside is secondary lymphoid organs and are recruited upon immune response, subsets of γδ T cells preferentially reside in epithelia including the epidermis, gut, lung, or the genitourinary tract (Weaver et al., 2013). Moreover, correlating with our experimental model, depletion of Langerhans cells in a psoriatic mouse model suppresses infiltration of IL-17A producing cells followed by decreased levels of IL-17 cytokines (Yoshiki et al., 2014). In aggregate, our results demonstrate that an epidermal-specific DLX3-dependent signal is necessary and sufficient for the development of a skin inflammatory response.
As early as 3 days after epidermal DLX3 deletion, we identified activated STAT3 as a candidate upstream regulator that influences the downstream gene expression changes we observed. STAT3 signaling affects the survival of keratinocyte stem cells, which are important for adult epidermis renewal and maintenance (Sano et al., 2005, Sano et al., 2008). Overexpression of active-Stat3 also augmented the development of spontaneous psoriasis-like skin disease in mice (Sano et al., 2005, Sano et al., 2008). We documented activation of STAT3 in icKO skin via detection of active phosphorylated form of STAT3. Absence of DLX3 function led to increased STAT3 activation through increased phosphorylation of STAT3 and inflammation in DLX3-icKO skin that was inhibited via treatment with a specific STAT3 inhibitor. Our findings document a crucial DLX3- STAT3 network that regulates normal skin immune homeostasis.
Material and Methods
Mice and genotyping
Mouse strains DLX3Kin/+ (LacZ knock-in heterozygote), DLX3f/f (floxed homozygote), and K14-CreERT2 (Jax5107) were used to generate K14-CreERT2;Dlx3fl/fl (icKO) mice and genotyped as reported previously (Hwang et al., 2008). Upon tamoxifen treatment Cre is expressed in epithelial basal cells as well as appendages of the skin. K14-CreERT2;Dlx3fl/fl mice were born at Mendelian frequency and after birth were indistinguishable from their wild-type littermates (Dlx3 fl/fl). The dorsal skin was shaved 2 days prior to application of 4-hydroxy-tamoxifen (4-OH tamoxifen, Sigma-Aldrich Co., St. Louis, MO). 5 mg/ml of 4-OH tamoxifen dissolved in ethanol 200 ul was topically applied to 6-weeks-old mice dorsal skin for 5 consecutive days. Treatment of Cryptotanshinone was done twice of 40ug/mg before sample collection. We treated the animals according to protocol approved by the Animal Use and Care Committee at the National Institute of Arthritis and Musculoskeletal and Skin.
Cell Culture and adenoviral transduction
Primary keratinocytes were isolated from Dlx3f/f mice, and were plated in low-calcium medium (10% chelated FCS, 0.05 mM Ca2+). Following day after plating, cells were transduced with adenovirus expressing Cre-recombinase or GFP (Vector Biolabs) at 5 MOI in infection media (S-MEM with 4 μg/mL of polybrene and 0.05mM Ca2+) for 30 min at 37C. The cells were subsequently cultured in low-calcium medium for 24 hr and lysed in RIPA Buffer containing 20 mM Tris-HCl (pH 7.5) 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 1 mM Na3VO4,1 mM PMSF and 1X of Protein Inhibitor cocktail (Roche).
Tissue preparation, histology and confocal microscopy
Tissues were collected at 3 days, 1 week and 2 weeks after treatment. Samples were fixed overnight at 4°C paraformaldehyde in 1x PBS, dehydrated and embedded in paraffin blocks. 5 μm thick sections were stained with hematoxylin and eosin or used for immunohistochemistry after incubation with primary antibodies as follows: anti-DLX3 (1:250, abcam), anti-F4/80 (1:100, Serotec), anti-CD45 (1:100, BD pharmingen), CD3 (1:500, Serotec), CD4 (1:2000, abcam), p-STAT3 (1:1000, Cell Signaling), STAT3 (1:1000, Cell Signaling) , RPL11 (1:1000, Bethyl) and secondary antibodies Alexa Fluor 488 donkey IgG (1:500; Molecular Probes) or Alexa Fluor 547 goat IgG (1:500; Molecular Probes). Image analysis was performed using Zeiss LSM 510 META laser-scanning confocal microscope.
Western Blot Analysis
Dorsal mouse skins were collected and treated with trypsin overnight for epidermis and dermis separation. Epidermis was sonicated for lysis in denaturing buffer composed of 300 mM NaCl, 2 mM EDTA, 20 mM Hepes, 1% SDS,0.1 mM hemin chloride, 5 mM NEM, 20 mM NaF, 100mM PMSF. Lysates were centrifuged to remove DNA and cell debris. Protein concentrations were measured by the BCA protein assay (Thermo Scientific Inc., Rockford, IL). 30 ug of lysate protein were loaded for electrophoresis on 4–12% SDS-PAGE gels (Invitrogen, Carlsbad, CA, USA). After electrophoresis, transfer to PVDF membrane (Invitrogen, Carlsbad, CA, USA) and incubating with blocking solution (5% Nonfat Milk) and subsequently with primary antibody was followed. Blots were then rinsed in TBST and incubated in peroxidase-conjugated secondary antibodies, respectively. Proteins were visualized using Bio-Rad Chemi Doc XRS with ECL (Pierce Biotech, Waltham, MA, USA). Antibodies used: anti-DLX3 (Morasso lab; (Hwang et al., 2008), anti-CD3 (BD Bioscience), anti-CD4 (Abcam), anti-Filaggrin, (Covance), anti-K6 (Covance), anti-K10 (Covance), anti-Ki67 (Biolegend), anti-langerin (Novus Biologicals), anti-pSTAT3 (abcam), anti-STAT3 (Cell Signaling), anti-RPL11 (Bethyl) and anti-Vinculin (abcam).
Single cell suspension and flow cytometry analysis
Hair was removed from euthanized mice with clippers and a 3.5 x 2 cm piece of dorsal skin was excised, washed in PBS and incubated with 0.4mg/ml Liberase RPMI at 37°C for 1 hour. 0.05% DNaseI was added for 15 min at 37°C. Single cell suspensions were prepared by 60 cc syringe, followed by sequential filtration through 100, 70, 40 μm nylon mesh. Cells were stained with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen) to exclude dead cells and incubated with Fc Block (eBioscience) for 15 min at 4°C. Cells were incubated with fluorochrome-conjugated antibody for 30 min at 4°C. For intracellular cytokine detection in T cells, cells were stimulated with 50ng/ml of PMA (Sigma) and 500 ng/ml of ionomycin (Calbiochem) in the presence of Brefeldin A (BD pharmingen) for 4 hours. After surface staining, fixation and permeabilization was performed with BD cytofix/cytoperm kit and anti- IL-17A antibody was added for 30 min at 4°C.
RNA preparation and RNA-seq data analysis
Total RNA was prepared from the epidermis and dermis using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, and RNA was evaluated using a Bioanalyzer (Agilent Technologies). RNA sequencing was performed using the Illumina Hiseq 200 at the NIAMS Genome Analysis Core Facility in NIH. The Illumina TruSeq RNA sample preparation kit (Illumina) was used according to the manufacturer’s protocol. In brief, poly-A containing mRNA molecules were purified from 2 ug total RNA using poly-T oligo attached magnetic beads using two rounds of purification. First strand cDNA was synthesized using random hexamers to eliminate the general bias towards the 3’ end of the transcripts. The cDNA was further subjected to end repair, A-tailing, and adapter ligation with barcoded adapters in accordance with the Illumina protocol. Purified cDNA templates were enriched by 15 cycles of PCR for 10 s at 98°C, 30 s at 60°C, and 30 s at 72°C. The samples were cleaned using AMPure XP Beads (Beckman Coulter, Indianapolis IN) and eluted in 30 ul Resuspension Buffer as instructed. Purified cDNA libraries were quantified using Bioanalyzer.
Computational analysis
RNA-Seq expression fold changes were calculated using Partek Genomics Suite (http://www.partek.com). GO Biological Pathway enrichment of significantly modulated genes (q<0.05 and Max RPKM>0) was generated through DAVID Functional Annotation Tools (https://david.ncifcrf.gov/) and Panther GO Enrichment Analysis (http://geneontology.org/page/go-enrichment-analysis). Ingenuity Pathway analysis was used for identification of upstream regulator and canonical pathways.
Accession numbers
Raw and analyzed RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) site, accession number GSE104022.
Statistical analysis
All experiments were performed on at least three control and three knockout mice (Mean±S.D) with Prism 5 statistical software (GraphPad Software Inc., San Diego, CA). Statistical significant level was determined using ANOVA. A difference of *P<0.05 and **P<0.01 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (M.I.M ZIA AR041124) and the Center for Cancer Research, National Cancer Institute of the National Institutes of Health. We thank members of the Laboratory of Skin Biology (LSB) for helpful suggestions and discussions. We also thank Gustavo Gutierrez-Cruz, Stefania Dell’Orso and members of the NIAMS Genome Analysis Core Facility and the NIAMS Light Imaging Core Facility.
Footnotes
Conflict of Interest
Dr. Mark Udey has a perceived conflict of interest because he is the Editor of the Journal of Investigative Dermatology. He addressed this by recusing himself from all aspects of the review and editorial processes related to this submission. All other authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akitsu A, Ishigame H, Kakuta S, Chung SH, Ikeda S, Shimizu K, et al. IL-1 receptor antagonist-deficient mice develop autoimmune arthritis due to intrinsic activation of IL-17-producing CCR2(+)Vgamma6(+)gammadelta T cells. Nature communications. 2015;6:7464. doi: 10.1038/ncomms8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi C, De Pita O, Girolomoni G. Resident skin cells in psoriasis: a special look at the pathogenetic functions of keratinocytes. Clinics in dermatology. 2007;25(6):581–8. doi: 10.1016/j.clindermatol.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Augustin I, Gross J, Baumann D, Korn C, Kerr G, Grigoryan T, et al. Loss of epidermal Evi/Wls results in a phenotype resembling psoriasiform dermatitis. The Journal of experimental medicine. 2013;210(9):1761–77. doi: 10.1084/jem.20121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. The Journal of experimental medicine. 2007;204(11):2603–14. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA. Integration of the Transcription Factor-Regulated and Epigenetic Mechanisms in the Control of Keratinocyte Differentiation. The journal of investigative dermatology Symposium proceedings /the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research. 2015;17(2):30–2. doi: 10.1038/jidsymp.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome AM, Ryan D, Eckert RL. S100 protein subcellular localization during epidermal differentiation and psoriasis. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2003;51(5):675–85. doi: 10.1177/002215540305100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6(4):328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41(2):211–5. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, et al. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento- Osseous syndrome. The Journal of biological chemistry. 2008;283(29):20198–208. doi: 10.1074/jbc.M709562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. The Journal of investigative dermatology. 2004;123(1):23–33. doi: 10.1111/j.0022-202X.2004.22719.x. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Welter JF. Transcription factor regulation of epidermal keratinocyte gene expression. Mol Biol Rep. 1996;23(1):59–70. doi: 10.1007/BF00357073. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodriguez E, Matanovic A, Marenholz I, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet. 2013;45(7):808–12. doi: 10.1038/ng.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Merad M. Ontogeny and homeostasis of Langerhans cells. Immunology and cell biology. 2010;88(4):387–92. doi: 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Johnston A, Dyson M, Valdimarsson H, Elder JT. Mouse models of psoriasis. The Journal of investigative dermatology. 2007;127(6):1292–308. doi: 10.1038/sj.jid.5700807. [DOI] [PubMed] [Google Scholar]
- Hwang J, Kita R, Kwon HS, Choi EH, Lee SH, Udey MC, et al. Epidermal ablation of Dlx3 is linked to IL-17-associated skin inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11566–71. doi: 10.1073/pnas.1019658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development. 2008;135(18):3149–59. doi: 10.1242/dev.022202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch PJ, de Viragh PA, Scharer E, Bundman D, Longley MA, Bickenbach J, et al. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. The Journal of cell biology. 2000;151(2):389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Nagao K, Yokouchi M, Sasaki H, Amagai M. External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. The Journal of experimental medicine. 2009;206(13):2937–46. doi: 10.1084/jem.20091527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. The Journal of investigative dermatology. 2008;128(5):1207–11. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- Lu L, Li C, Li D, Wang Y, Zhou C, Shao W, et al. Cryptotanshinone inhibits human glioma cell proliferation by suppressing STAT3 signaling. Mol Cell Biochem. 2013;381(1–2):273–82. doi: 10.1007/s11010-013-1711-x. [DOI] [PubMed] [Google Scholar]
- Monteleone G, Pallone F, MacDonald TT, Chimenti S, Costanzo A. Psoriasis: from pathogenesis to novel therapeutic approaches. Clinical science. 2011;120(1):1–11. doi: 10.1042/CS20100163. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Markova NG, Sargent TD. Regulation of epidermal differentiation by a Distal-less homeodomain gene. The Journal of cell biology. 1996;135(6 Pt 2):1879–87. doi: 10.1083/jcb.135.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nature reviews Immunology. 2009;9(10):679–91. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo E, Kellett M, Cataisson C, Gormley A, Bible PW, Pietroni V, et al. The homeoprotein DLX3 and tumor suppressor p53 co-regulate cell cycle progression and squamous tumor growth. Oncogene. 2016;35(24):3114–24. doi: 10.1038/onc.2015.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Haase I, Nestle FO. Mechanisms regulating skin immunity and inflammation. Nature reviews Immunology. 2014;14(5):289–301. doi: 10.1038/nri3646. [DOI] [PubMed] [Google Scholar]
- Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Human molecular genetics. 1998;7(3):563–9. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- Rothnagel JA, Seki T, Ogo M, Longley MA, Wojcik SM, Bundman DS, et al. The mouse keratin 6 isoforms are differentially expressed in the hair follicle, footpad, tongue and activated epidermis. Differentiation; research in biological diversity. 1999;65(2):119–30. doi: 10.1046/j.1432-0436.1999.6520119.x. [DOI] [PubMed] [Google Scholar]
- Sabat R, Philipp S, Hoflich C, Kreutzer S, Wallace E, Asadullah K, et al. Immunopathogenesis of psoriasis. Experimental dermatology. 2007;16(10):779–98. doi: 10.1111/j.1600-0625.2007.00629.x. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. Journal of cell science. 2009;122(Pt 9):1285–94. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nature medicine. 2005;11(1):43–9. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- Sano S, Chan KS, DiGiovanni J. Impact of Stat3 activation upon skin biology: a dichotomy of its role between homeostasis and diseases. J Dermatol Sci. 2008;50(1):1–14. doi: 10.1016/j.jdermsci.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Schon M, Denzer D, Kubitza RC, Ruzicka T, Schon MP. Critical role of neutrophils for the generation of psoriasiform skin lesions in flaky skin mice. The Journal of investigative dermatology. 2000;114(5):976–83. doi: 10.1046/j.1523-1747.2000.00953.x. [DOI] [PubMed] [Google Scholar]
- Shornick LP, De Togni P, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, et al. Mice deficient in IL-1beta manifest impaired contact hypersensitivity to trinitrochlorobenzone. The Journal of experimental medicine. 1996;183(4):1427–36. doi: 10.1084/jem.183.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12(9):565–80. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Degenstein L, Copenhaver C, Fuchs E. Defining the regulatory factors required for epidermal gene expression. Molecular and cellular biology. 2000;20(7):2543–55. doi: 10.1128/mcb.20.7.2543-2555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Carbajal S, Han G, Wohn C, Lu J, et al. Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. PloS one. 2011;6(4):e18266. doi: 10.1371/journal.pone.0018266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44(12):1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Amerio P, Sauder DN. Role of cytokines in epidermal Langerhans cell migration. Journal of leukocyte biology. 1999;66(1):33–9. doi: 10.1002/jlb.66.1.33. [DOI] [PubMed] [Google Scholar]
- Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annual review of pathology. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S, Novak N. Atopic dermatitis. The Lancet. 2016;387(10023):1109–22. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- Wright JT, Hong SP, Simmons D, Daly B, Uebelhart D, Luder HU. DLX3 c. 561_562delCT mutation causes attenuated phenotype of tricho-dento-osseous syndrome. American journal of medical genetics Part A. 2008;146A(3):343–9. doi: 10.1002/ajmg.a.32132. [DOI] [PubMed] [Google Scholar]
- Yoshiki R, Kabashima K, Honda T, Nakamizo S, Sawada Y, Sugita K, et al. IL-23 from Langerhans cells is required for the development of imiquimod-induced psoriasis-like dermatitis by induction of IL-17A-producing gammadelta T cells. The Journal of investigative dermatology. 2014;134(7):1912–21. doi: 10.1038/jid.2014.98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.