SUMMARY
Mammalian oocytes have the ability to reset the transcriptional program of differentiated somatic cells into that of totipotent embryos through somatic cell nuclear transfer (SCNT). However, the mechanisms underlying SCNT-mediated reprogramming are largely unknown. To understand the mechanisms governing chromatin reprogramming during SCNT, we profiled DNase I hypersensitive sites (DHSs) in donor cumulus cells and one-cell stage SCNT embryos. To our surprise, the chromatin accessibility landscape of the donor cells is drastically changed to recapitulate that of the in vitro fertilization (IVF)-derived zygotes within 12 hr. Interestingly, this DHS reprogramming takes place even in the presence of a DNA replication inhibitor, suggesting that SCNT-mediated DHS reprogramming is independent of DNA replication. Thus, this study not only reveals the rapid and drastic nature of the changes in chromatin accessibility through SCNT but also establishes a DNA replication-independent model for studying cellular reprogramming.
In Brief
Djekidel et al. used low-input DNase-seq to map the chromatin accessibility dynamics of donor cells and SCNT one-cell embryos. They revealed a drastic and fast global DHS reprogramming of donor cells in a DNA replication-independent manner.
INTRODUCTION
Among the currently available systems for cell fate reprogramming, somatic cell nuclear transfer (SCNT) is the only one capable of reprogramming terminally differentiated cells to a toti-potent state (Jullien et al., 2011; Mitalipov and Wolf, 2009). SCNT therefore provides an excellent model for understanding how cell memory can be fully reprogrammed to generate totipotent cells, and thus can provide important clues on how to improve other reprogramming systems. However, despite more than 50 years after the first successful cloning by SCNT (Gurdon, 1962), the molecular mechanisms underlying SCNT-mediated reprogramming are almost completely unknown.
Reprogramming requires change to the chromatin, epigenetic, and transcriptional landscapes of somatic cells. Many studies have been performed to characterize these changes during the induced pluripotent stem cell (iPSC) reprogramming process. These studies used different assays including RNA sequencing (RNA-seq), chromatin immunoprecipitation sequencing (ChIP-seq), assays for transposase-accessible chromatin using sequencing (ATAC-seq), Hi-C, and proteomics analyses (Hussein et al., 2014; Knaupp et al., 2017; Koche et al., 2011; Krijger et al., 2016; Li et al., 2017; Sridharan et al., 2013; Stadhouders et al., 2018). The studies revealed the dynamic nature of the chromatin, epigenetics, and transcriptome during the iPSC generation process and identified important factors and molecular events that facilitate or impede the reprogramming process. While such multi-dimensional analyses have been applied to the iPSC reprogramming system, only transcriptome analyses have been performed for SCNT reprogramming (Chung et al., 2015; Hormanseder et al., 2017; Inoue et al., 2015; Matoba et al., 2014). Although these studies revealed that a donor cell transcriptional program is largely reprogrammed to an embryonic program by the time of zygotic genome activation (ZGA), with the exception of reprogramming-resistant regions (Chung et al., 2015; Matoba et al., 2014), its molecular basis is still unknown and further study of the chromatin landscape changes during the reprogramming process is necessary.
Chromatin accessibility is a good indicator of transcriptional regulatory elements and can serve as a predictor of gene transcription activity. It can be identified genome-wide by DNase I sequencing or ATAC-seq (Boyle et al., 2008; Buenrostro et al., 2013). Recent refinements to these techniques have allowed the profiling of the open chromatin landscape using limited number of cells by low-input DNase I sequencing (liDNase-seq) or at the single-cell level by ATAC-seq (Buenrostro et al., 2015; Jin et al., 2015; Lu et al., 2016), thereby facilitating the study of chromatin accessibility in mouse preimplantation embryos (Inoue et al., 2017; Lu et al., 2016; Wu et al., 2016). In this work, we used liDNase-seq to study chromatin accessibility changes during SCNT reprogramming, which revealed the quick and DNA replication-independent nature of the reprogramming process.
RESULTS AND DISCUSSION
Fast DNase I Hypersensitive Site Reprogramming upon SCNT
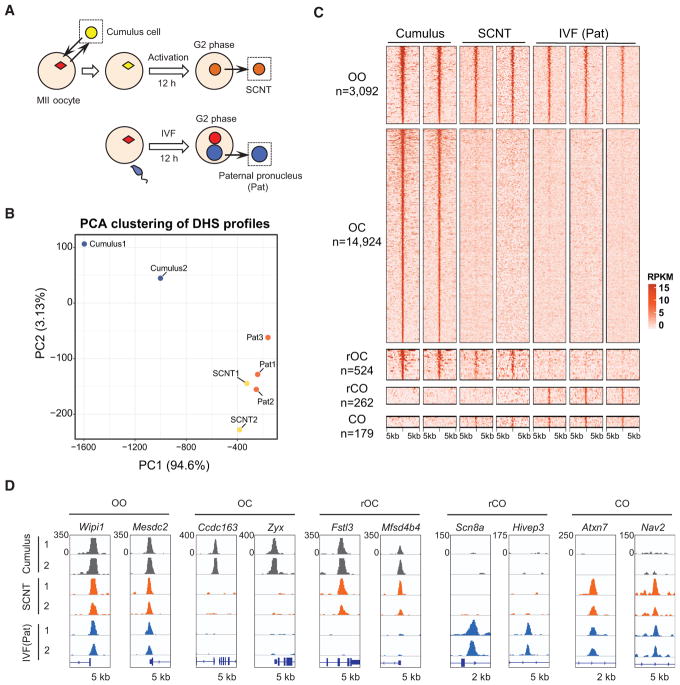
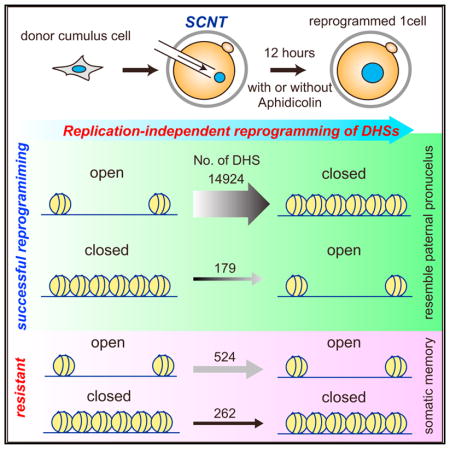
To understand how the chromatin accessibility of somatic donor cells is reprogrammed to that of the totipotent one-cell embryo, we attempted to generate the DNase I hypersensitive site (DHS) map of SCNT one-cell embryos. To this end, we collected mouse cumulus cells to serve as somatic donor cells and performed SCNT. Twelve hours post-activation (hpa), pseudopronuclei were isolated from SCNT one-cell embryos for liDNase-seq (Figure 1A) with biological duplicates for both the donor cells and one-cell SCNT embryos (Figures S1A and S1B). Since sperm chromatin is reprogrammed under physiological conditions upon fertilization (Inoue et al., 2017), we used the DHS map of paternal pronuclei (Pat) of 12 hr post-fertilization (hpf) zygotes as a control (Figure 1A). Using stringent criteria for peak calling and reproducibility (irreproducibility discovery rate [IDR] < 0.05, mean reads per kilobase million [RPKM] > 2, RPKM in all replicate > 1, sex chromosomes were excluded), we identified 23,353, 3,005, and 3,610 DHSs in donor cumulus cells, SCNT one-cell embryo, and Pat, respectively (Table S1). Principal-component analysis (PCA) indicates that the overall DHS landscape of SCNT embryos is similar to that of Pat (Figure 1B), suggesting that SCNT-mediated reprogramming of chromatin accessibility is largely complete by 12 hr after activation.
Figure 1. Rapid Reprogramming of Donor Cell Chromatin Accessibility in SCNT.
(A) Schematic illustration of the experimental design for studying the chromatin accessibility dynamics in SCNT and that of the paternal pronucleus from in vitro-fertilized (IVF) zygotes. The collected samples for liDNase-seq analysis are shown inside dotted boxes.
(B) PCA of the genome-wide DHS profile of the cumulus, IVF(Pat), and one-cell SCNT samples. Each dot represents one sample.
(C) Heatmap showing the DHSs clustered according to their combinatorial enrichment in cumulus, one-cell SCNT, and IVF(Pat) samples. Each row represents a locus (DHS center ± 5 kb), and the red gradient color indicates the liDNase-seq signal intensity. OO, open in cumulus cells, SCNT, and IVF(Pan); OC, open in cumulus cells and closed in SCNT and IVF(Pat); rOC, open in cumulus cells and closed in IVF(Pat), but failed to be closed in SCNT; rCO, closed in cumulus cells and open in IVF(Pat), but failed to be opened in SCNT; CO, closed in cumulus cells and open in IVF(Pat) and SCNT.
(D) Genome browser views showing the normalized coverage (per million mapped reads) at representative loci of the different DHS categories.
See also Figure S1.
To closely examine the DHS dynamics, we classified the detected DHSs into five groups using the following criteria (3-fold difference between DHS-negative and -positive categories, mean RPKM > 2 and RPKM in all replicates > 1 in DHS-positive categories). Groups 1 to 5, respectively, represent DHSs detected in all samples (open to open [OO], n = 3,092), those only detected in cumulus cells (open to closed [OC], n = 14,924), those detected in both cumulus and SCNT samples (resistant to open to closed [rOC], n = 524), those detected in both SCNT and in vitro fertilization (IVF) (Pat) (closed to open [CO], n = 179), and those specifically detected in IVF(Pat) (resistant to closed to open [rCO], n = 262) (Figures 1C, 1D, and S1C). The great majority of DHSs were in the OC category (78.6%; Figure 1C), suggesting that SCNT-mediated DHS reprogramming is accompanied with a global loss of DHSs.
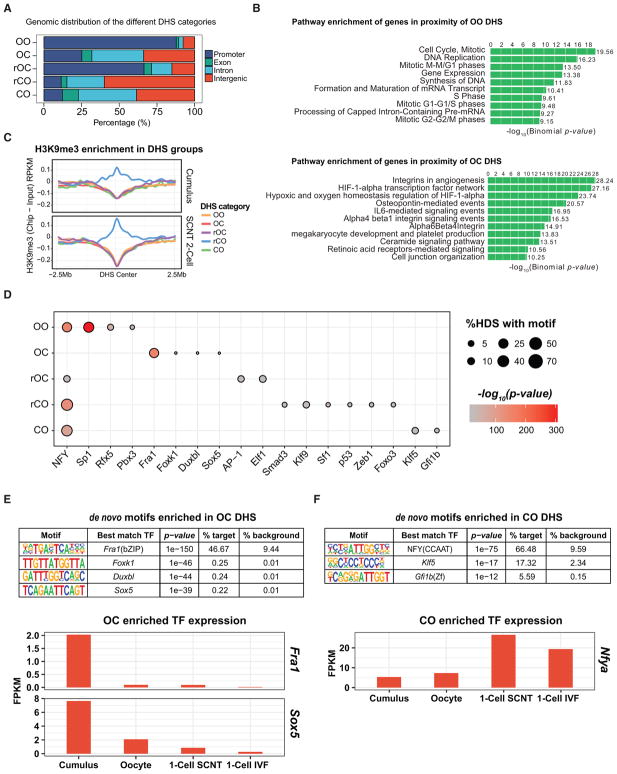
The DHS categories show an interesting genomic distribution, among the non-reprogrammed DHSs, the majority of OO (87.8%) and rOC (66.4%) categories are located in promoters (transcription start site [TSS] ± 1 kb) (Figure 2A), while only 11.5% of the rCO category are located near promoter regions. In contrast, the majority of reprogrammed DHSs, the OC (74.2%) and CO (87.7%) categories, are located outside the promoter region (Figure 2A). These results suggest that distal regulatory elements are prone to be reprogrammed upon SCNT, while failure to close accessible somatic promoters or to open distal regulatory regions required for differentiation program may be the major reprogramming barriers. This observation is similar to recent ATAC-seq analysis of chromatin reprogramming, which demonstrated that accessible chromatin at promoter regions is relatively stable during transcription factor (TF)-mediated iPSC generation (Knaupp et al., 2017; Li et al., 2017). Biological pathway and gene ontology (GO) enrichment analysis of the genes associated with OO and OC DHSs revealed that the OO-associated genes are enriched in ubiquitous cellular functions, such as cell cycle and DNA replication, while OC-associated genes are enriched in specific somatic cell programs such as angiogenesis and HIF1-alpha network (Figures 2B, S2A, and S2B). No specific GO or pathway enrichment in the other DHS categories was found (data not shown). These data suggest that SCNT reprogramming involves a specific loss of somatic cell memory while maintaining ubiquitous cellular functions.
Figure 2. Transcription Factor Network Switch Likely Accompanies DHS Reprogramming.
(A) Bar graph showing the genomic distribution of the different DHS categories. DHS peaks within TSS ± 1 kb are considered promoter DHS, and those not located in promoters, exons, or introns are labeled as intergenic.
(B) Bar plot showing the top ten enriched pathways of OO- or OC-associated genes using GREAT enrichment (McLean et al., 2010).
(C) Averaged H3K9me3 ChIP-seq signal profile normalized to that of the input in cumulus and two-cell stage SCNT embryos in a DHS ± 2.5-Mb window.
(D) Dot plot showing the enriched de novo TFs motifs (x axis) in the different DHS categories (y axis). Circle size represents the proportion of DHS having the de novo TF motif, and the gradient red color indicates the p value enrichment. The names of TFs with motif enriched in the different DHS categories are indicated.
(E and F) TFs with binding motif enriched in the OC (E) and CO (F) DHSs and their relative expression levels in donor cells, oocytes, one-cell SCNT, and IVF embryos.
See also Figure S2.
It has been shown that H3K9me3 in donor cells functions as an epigenetic barrier preventing SCNT-mediated reprogramming (Chung et al., 2015; Matoba et al., 2014). Thus, we hypothesized that reprogramming-resistant regions, particularly the rCO category, should be enriched for H3K9me3 in donor cells and SCNT embryos. Analysis of a public H3K9me3 ChIP-seq dataset in cumulus cells and SCNT two-cell embryos (Liu et al., 2016) indeed revealed that H3K9me3 is specifically enriched in rCO sites in cumulus cells and SCNT two-cell embryos (Figure 2C). This result provides another piece of evidence supporting the role of H3K9me3 in preventing SCNT-mediated reprogramming.
TF Network Switch Likely Accompanies DHS Reprogramming
Data presented in Figure 2C indicate that OC loci are devoid of H3K9me3 despite lack of chromatin accessibility in SCNT embryos. This observation suggests that global loss of DHSs at the OC loci is not due to gain of H3K9me3 but by the displacement of TFs from the transplanted somatic cell chromatin. To assess this possibility, we performed TF motif enrichment analysis for each DHS category (p value < 0.001, fragments per kilo-base of transcript per million mapped reads [FPKM] > 1 in cumulus, SCNT, or IVF, and at least 2-fold enrichment compared to background) (Figure 2D). Interestingly, we found that 46.7% of the OC DHSs contain the Fra1 binding motif (Figure 2E), which does not show enrichment in any other DHS category (Figures 2D and 2E). Fra1 is known to regulate the specific transcriptional program in cumulus cells (Sharma and Richards, 2000) and is very lowly expressed in oocytes (Xue et al., 2013) or one-cell SCNT and IVF embryos (Matoba et al., 2014) (Figure 2E). Similar expression pattern was also observed for Sox5, a TF also with binding motif enriched in the OC loci (Figure 2E). Thus, lack of Fra1 and Sox5 binding to chromatin in SCNT embryos at least partly explains the dramatic loss of DHS in the OC category. These observations suggest that loss of donor cell-specific TF occupancy may contribute to the global loss of DHSs in SCNT embryos.
In addition to the large number of OC category, which is likely responsible for the loss of donor cell chromatin identity, the other reprogrammed CO category could be important for acquiring totipotency. Analysis of the CO loci identified NFY as the most enriched motif (Figures 2D and 2F). Since Nfya, a subunit of the NFY complex, has been shown to be involved in ZGA (Lu et al., 2016), it is possible that the NFY complex also contributes to SCNT-mediated DHS reprogramming. Indeed, Nfya is expressed at a relatively lower level in cumulus cells while higher in one-cell SCNT and IVF embryos (Figure 2F). Another interesting finding from the motif enrichment analysis comes from the rOC category. Although NFY motif was enriched in rOC category as in the CO category, we found that the AP-1(Jun) and Elf1 motifs were exclusively enriched in this category (Figures 2D and S2C). Interestingly, it has been recently shown that chromatin binding of AP-1 in differentiated cells functions as a reprogramming barrier preventing iPSC generation (Li et al., 2017). This suggests that failure of specific somatic cell TFs to dissociate from chromatin can also be a barrier in SCNT reprogramming. Taken together, our data support the notion that the donor cell-specific TF network is quickly lost and a new zygotic TF network is established upon SCNT within 12 hr.
Loss of Donor Cell DHSs Is Associated with Downregulation of Somatic Cell Transcription Program
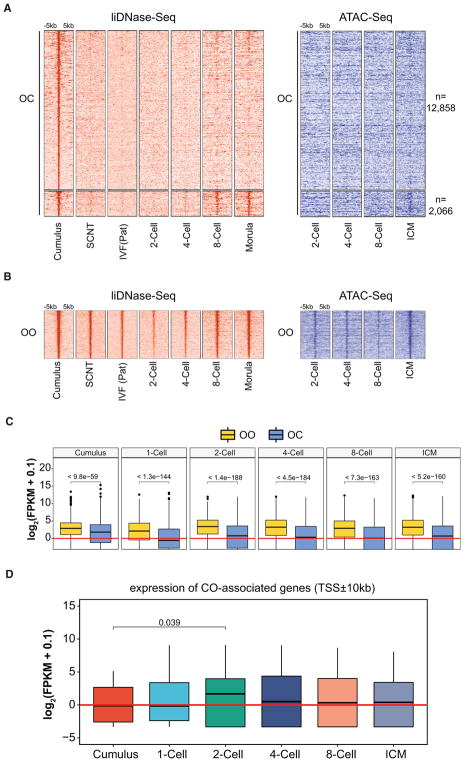
To understand the consequences of DHS reprogramming, we asked whether the OC loci maintain inaccessible states or become accessible later during preimplantation development. To this end, we analyzed publicly available liDNase-seq and ATAC-seq datasets at each preimplantation embryo development stages (Lu et al., 2016; Wu et al., 2016). Interestingly, both datasets consistently revealed that most of the OC loci maintain their inaccessibility throughout preimplantation development (Figure 3A). However, a small number of OC loci regain accessibility at the eight cell-to-morula stages in the liDNase-seq dataset (morula/cumulus > 3, RPKM > 1, and mean RPKM > 2 in morula) and at the inner cell mass (ICM) stage in the ATAC-seq dataset (Figure 3A). This chromatin accessibility dynamics is strikingly different from the OO loci, which remain accessible throughout preimplantation development (Figure 3B). These data suggest that, once lost upon SCNT, the majority of DHSs do not reappear during preimplantation development.
Figure 3. Loss of Donor Cell DHSs Is Associated with Downregulation of Somatic Cell Transcription Program.
(A and B) Heatmap showing the dynamics of OC (A) and OO (B) DHSs during preimplantation development analyzed by liDNase-seq (left) and ATAC-seq (right). Each row represents a locus (DHS center ± 5 kb), and the red and blue gradient colors represent the liDNase-seq and ATAC-seq signal intensity, respectively. OC DHSs were separated into two groups based on whether they reappear at the morula stage (FC[morula/cumulus] > 3, RPKM > 1, and mean RPKM > 2 in morula embryos). Peaks were ordered based on the signal intensity in cumulus cells.
(C) Averaged gene expression levels of the OO- or OC-associated genes (TSS ± 10 kb) in cumulus cells and preimplantation embryos. The p values were calculated using the Wilcoxon rank sum test.
(D) Averaged gene expression levels of the CO-associated genes (TSS ± 10 kb) in cumulus cells and preimplantation embryos. The p values were calculated using the paired t test.
See also Figure S3.
To gain insight into the functional significance of the loss of DHSs, we examined whether OC-associated genes are transcriptionally turned off in preimplantation embryos. To this end, we analyzed RNA-seq datasets of preimplantation embryos (Wu et al., 2016) and cumulus cells (Matoba et al., 2014). Comparative analyses between OC- and OO-associated genes (DHS within TSS ± 10 kb) (Tables S2 and S3) revealed that the gene expression level of OC-associated genes (median, ~1 FPKM) is significantly lower than that of OO-associated genes (median, ~4 FPKM) in all stages (Figure 3C). These data indicate that the OC-associated genes are expressed at a higher level in cumulus cells than in preimplantation embryos. A similar result was obtained when analyzing the genes harboring promoter DHSs (TSS ± 1 kb) (Figure S3A), excluding the possibility that these results could account for the difference in genomic distribution between OC and OO DHSs (Figure 2A). This observation was further confirmed by comparing the expression levels of the OC-associated genes between cumulus and preimplantation embryos (Figure S3B). Taken together, these results suggest that SCNT-mediated loss of DHS may be required for turning off the somatic cell transcriptional program.
We also analyzed the expression of CO-associated genes. Interestingly, this group of genes exhibits an expression pattern that peaks at the two-cell stage consistent with ZGA (Figure 3D). Indeed, 12 out of the 52 CO-proximal genes (23.08%) are activated during the one-cell to two-cell transition (FC > 3, mean FPKM in two cell > 2) (Figure S3C).
DHS Reprogramming in SCNT Is Independent of DNA Replication
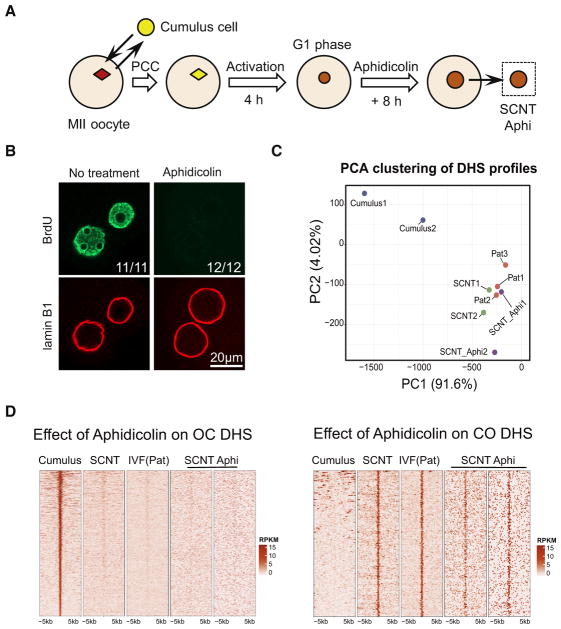
SCNT embryos complete the first round of DNA replication within 12 hpa. Previous studies indicated that DNA replication is required for somatic cell reprogramming in a heterokaryon-mediated reprogramming setting (Tsubouchi et al., 2013). Given that TF-mediated iPSC generation generally takes 1 week, DNA replication must be required. To determine whether DNA replication is also required for SCNT-mediated DHS reprogramming, we treated SCNT embryos with a potent DNA replication inhibitor, aphidicolin, from 4 to 12 hpa (Figure 4A). We first confirmed that aphidicolin treatment effectively blocked DNA replication in SCNT embryos using a bromodeoxyuridine (BrdU) incorporation assay (Figure 4B). We then collected pseudopronuclei from aphidicolin-treated SCNT embryos at 12 hpa and performed liDNase-seq with biological duplicates (Figure S4A). Hierarchical clustering and PCA revealed that the DHS landscape of aphidicolin-treated SCNT embryos is much more similar to those of the non-treated SCNT and IVF(Pat) than to that of the cumulus cells (Figures 4C and S4B). A closer examination of the reprogrammed DHSs (CO and OC, Figure 1C) showed a limited effect of aphidicolin treatment on DHS reprogramming compared to that of the change from cumulus to SCNT (Figures 4D, S4C, and S4D). These results suggest that SCNT-mediated DHS reprogramming is largely DNA replication independent.
Figure 4. DHS Reprogramming in SCNT Is Independent of DNA Replication.
(A) Schematic illustration of the experiment to examine the effect of aphidicolin on SCNT-mediated DHS reprogramming.
(B) Representative images of SCNT one-cell embryos immunostained with BrdU and lamin B1 antibodies. No BrdU signal was detected in aphidicolin-treated embryos. The number of SCNT embryos exhibiting the staining pattern and the total number of embryos analyzed are shown, respectively. Scale bar: 20 μm.
(C) PCA of the genome-wide DHS profile of cumulus, IVF(Pat), SCNT, and aphidicolin-treated SCNT samples. Each dot represents an independent sample.
(D) Heatmap showing liDNase-seq enrichment signal in the OC and CO DHSs after aphidicolin treatment. Each row represents a locus (DHS center ± 5 kb), and the red gradient color indicates the signal intensity.
See also Figure S4.
By profiling and comparing the chromatin accessibility of donor cells, SCNT, and IVF one-cell embryos, we revealed that DHS reprogramming is largely completed within 12 hr and that this reprogramming takes place in a DNA replication-independent manner. This appears to be different from the cell fusion-mediated reprogramming and the TF-mediated iPSC reprogramming systems that require DNA replication or cell division, suggesting that SCNT reprogramming might be mechanistically different. The fast DHS reprogramming without a single-cell division allows the separation of reprogramming events from developmental processes such as major ZGA, which does not take place until the two-cell stage (Aoki et al., 1997). This finding is important as it identified the time window to which future mechanistic studies should be focused on. The global loss of DHSs is likely due to the displacement of TFs from the transplanted somatic cell chromatin (Figure 2). Given that most TFs except the pioneer factors are believed to be displaced from chromatin at the mitotic metaphase (Iwafuchi-Doi and Zaret, 2014), the SCNT-mediated loss of DHSs might be triggered by premature chromosome condensation (PCC) of donor chromatin that is induced by M-phase-promoting factors in oocyte cytoplasm soon after microinjection. The large amount of TFs stored in the oocyte cytoplasm, which is about 1,000 times larger than most somatic cells in volume (diameter, 70–80 μm in oocyte versus 5–10 μm in somatic cells), may explain the fast and drastic replacement of TFs in donor somatic nuclei. Once donor cell DHSs are lost upon SCNT, the majority of these loci remain inaccessible and their associated genes are transcriptionally silenced during pre-implantation development (Figure 3). TF motif analyses revealed that some maternal factors might be important for SCNT-mediated DHS reprogramming (Figure 2). Future studies should test whether they are required for this process using oocyte-specific knockout approaches.
EXPERIMENTAL PROCEDURES
Collection of Mouse Oocytes
All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee of Harvard Medical School. The procedures of oocyte collection and IVF were described previously (Inoue et al., 2017). The mouse strain used in this study was B6D2F1/J (BDF1) (The Jackson Laboratory; 100006).
SCNT
The procedures of SCNT using cumulus cells as donors were described previously (Matoba et al., 2011). Briefly, MII oocytes were collected from superovulated 8- to 11-week-old BDF1 females. Cumulus cells were removed from oocytes by treatment with 300 U/mL bovine testicular hyaluronidase (Calbiochem). MII oocytes were enucleated in HEPES-buffered KSOM containing 7.5 μg/mL cytochalasin B (CB) (Chalbiochem; 250233). Nuclei of cumulus cells were injected into the enucleated oocytes using a Piezo-driven micromanipulator (Primetech). After 1-hr incubation in KSOM, SCNT oocytes were activated by Ca-free KSOM containing 3 mM strontium chloride (SrCl2) and 5 μg/ml CB for 1 hr and further cultured in KSOM with CB for 4 hr. At 5 hpa, embryos were washed in KSOM. To block DNA replication, embryos were transferred to KSOM containing 3 μg/mL aphidicolin (Sigma-Aldrich) at 4 hpa until sample collection at 12 hpa.
Whole-Mount Immunostaining
The procedure for BrdU labeling was described previously (Shen et al., 2014). Briefly, SCNT embryos were cultured in 100 μM BrdU from 5 hpa and fixed with 3.7% paraformaldehyde for 20 min at 9 hpa. After permeabilization with 0.5% Triton X-100 for 15 min, embryos were treated with 4 N HCl for 30 min followed by neutralization with 100 mM Tris-HCl (pH 8.0) for 15 min. The primary antibodies against BrdU (1/200; Roche Diagnostic) and lamin B1 (1/2,000; Santa Cruz; sc-6217) were incubated for 1 hr in 1% BSA/PBS at room temperature. The secondary antibodies used were Alexa Flour 488 donkey anti-mouse IgG (Life Technologies) and Alexa Flour 568 donkey anti-goat IgG. The embryos were mounted on a glass slide in Vecta-shield anti-bleaching solution with DAPI (Vector Laboratories). Fluorescence was detected under Zeiss LSM800.
liDNase-Seq
The procedures of liDNase-seq were described previously (Inoue et al., 2017).
Data Analysis of liDNase-Seq
Reads were trimmed then mapped to the mm9 genome. PCR duplicates and multi-mapped reads were removed. The IDR method was used to select replicable DHS (Li et al., 2011). DHS detected in cumulus, one-cell SCNT, paternal pronuclei, and the SCNT+aphidicolin were merged and used in the downstream analysis.
Additional Public Data
Publicly available ATAC-seq data (Wu et al., 2016), RNA-seq data (Matoba et al., 2014; Wu et al., 2016; Xue et al., 2013), and H3K9me3 ChIP-seq data (Liu et al., 2016) were mapped to the mm9 genome (Supplemental Experimental Procedures).
Statistical Methods
DHS IDR was estimated using the IDR method. Motif p values were calculated using HOMER (Heinz et al., 2010) with background regions having similar GC-bias. GO and pathway p values were estimated using the binomial test in GREAT. Wilcoxon rank sum test (Figures 3C and S3A), paired t test (Figures 3D and S3B), and Pearson’s correlation were done using R (http://www.r-project.org/).
Supplementary Material
Highlights.
Genome-wide DHS maps for donor cumulus cells, one-cell SCNT, and IVF embryos
Rapid and DNA replication-independent DHS reprogramming in SCNT embryos
Transcription factor network switch accompanies DHS reprogramming in SCNT embryos
Loss of donor cell DHSs correlates with suppression of somatic transcription program
Acknowledgments
We thank Dr. Luis Tuesta for critical reading of the manuscript. This project was supported by NIH (R01HD092465) and the Howard Hughes Medical Institute (HHMI). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the datasets reported in this study (summarized in Table S4) is GEO: GSE110851.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.036.
AUTHOR CONTRIBUTIONS
Y.Z. conceived the project. A.I., S.M., T.S., C.Z., and F.L. performed the experiments. M.N.D. and L.J. analyzed sequencing datasets. M.N.D., A.I., and Y.Z. interpreted the data and wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, Zhang Y. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell. 2015;17:758–766. doi: 10.1016/j.stem.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormanseder E, Simeone A, Allen GE, Bradshaw CR, Figlmuller M, Gurdon J, Jullien J. H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell. 2017;21:135–143. e6. doi: 10.1016/j.stem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee DS, Cloonan N, et al. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- Inoue K, Oikawa M, Kamimura S, Ogonuki N, Nakamura T, Nakano T, Abe K, Ogura A. Trichostatin A specifically improves the aberrant expression of transcription factor genes in embryos produced by somatic cell nuclear transfer. Sci Rep. 2015;5:10127. doi: 10.1038/srep10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol. 2011;12:453–459. doi: 10.1038/nrm3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp AS, Buckberry S, Pflueger J, Lim SM, Ford E, Larcombe MR, Rossello FJ, de Mendoza A, Alaei S, Firas J, et al. Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell. 2017;21:834–845. e6. doi: 10.1016/j.stem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PHL, Di Stefano B, de Wit E, Limone F, van Oevelen C, de Laat W, Graf T. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–1779. [Google Scholar]
- Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S, et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–833. e6. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W, et al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell. 2016;165:1375–1388. doi: 10.1016/j.cell.2016.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci USA. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Wolf D. Totipotency, pluripotency and nuclear reprogramming. Adv Biochem Eng Biotechnol. 2009;114:185–199. doi: 10.1007/10_2008_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SC, Richards JS. Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells. Relation of JunD and Fra2 to terminal differentiation. J Biol Chem. 2000;275:33718–33728. doi: 10.1074/jbc.M003555200. [DOI] [PubMed] [Google Scholar]
- Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and hetero-chromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, Gomez A, Collombet S, Berenguer C, Cuartero Y, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell. 2013;152:873–883. doi: 10.1016/j.cell.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.