Abstract
Hepatitis C virus (HCV) infection is responsible for liver diseases and hepatocellular carcinoma in chronically-infected patients. Owing to high sequence variability in HCV genome, numerous subtypes have emerged. This study determined HCV strains among patients with clinical hepatitis and blood donors in Ibadan. Blood samples were collected from consented 176 subjects who tested positive to HCV IgM antibodies, including 99 patients with clinical hepatitis and 77 apparently healthy blood donors. Viral RNA was extracted from blood samples, while presence of HCV was tested by amplifying the NS5B gene using polymerase chain reaction (PCR). The amplified NS5B gene was sequenced and sequences were aligned on MEGA 7.0. Phylogenetic tree was constructed with Neighbor-Joining method. Data were analyzed using descriptive statistics at P<0 .05. The NS5B gene was amplified in 38 samples, of which 29 were successfully sequenced. Phylogenetic analysis revealed three of seven known genotypes of HCV including genotypes / subtypes 1a (34.5%), 1b (17.2%), 2b (13.8%), 2c (3.6%) and 5a (31.3%). Subtypes 1b and 2b were found among patients with clinical hepatitis, while the single 2c was found among donors. Although subtype 1a was detected among both populations, its rate was higher among blood donors (P = 0 .003). Subtype 5a was found among the two groups (P= 1. 00). HCV subtypes 1a and 5a are the predominant strains in Ibadan. The diversity of HCV observed has implications for treatment of patients and design of a broadly protective vaccine against the virus.
Keywords: Hepatitis C Virus, Patients, Blood donors, Genotypes, Nigeria
INTRODUCTION
Over 185 million people worldwide are infected with hepatitis C virus (HCV) (Mora et al., 2016). An estimated 3–4 million people become infected every year globally (Ashfaq et al., 2011). This accounts for about 3% of the world’s population that are infected with HCV, with most of these cases occurring in Africa (Kapoor et al., 2011). HCV infection is responsible for most deaths emanating from liver failure and liver cancer each year (Simmonds, 2004).
Studies have shown that almost 75% of HCV related deaths occur among adults between the ages of 45 and 64 as a result of long-term infection with HCV that leads to chronic liver disease including liver cirrhosis and hepatocellular carcinoma (Mukherjee, 2012; Dhawan, 2016; Ly et al., 2012; Davis et al., 1989). Chronic infection with HCV has been reported as the main cause of liver disease, and this might be the reason for carrying out most of the Orthotopic Liver Transplantation (OLT) procedures in the USA (Dhawan, 2016; Davis et al., 1989). In sub-Saharan Africa, HCV infection is a major health challenge and has been implicated in liver disease and its complications in chronically-infected-patients (Mora et al., 2016; WHO, 2012; Rao et al., 2015).
Hepatitis C virus is a member of the Flaviviridae family and the only member of the genus Hepacivirus (Simmonds, 2004; WHO, 2012). The virus is a small enveloped, spherical virus with a positive sense, single-stranded RNA genome (Simmonds, 2004). The HCV genome consists of a single, open reading frame (ORF) that is 9600 nucleotide bases long and 2 untranslated, but highly conserved regions namely 5′- UTR and 3′-UTR located at both ends of the genome (Kato, 2000). The genome encodes a single polyprotein starting with the core proteins (structural) and ending with the NS5B protein, a non-structural protein that codes for the RNA polymerase (Lindenbach et al., 2005). The NS5B gene codes for RNA-dependent RNA polymerase (RdRp), an enzyme that is essential for viral maturation and plays an important catalytic role during replication of HCV (Penin et al., 2004). According to Ashfaq et al (2011), this gene represents an ideal target for the development of antiviral drugs.
Genotypes and subtypes of HCV can be differentiated based on the sequences of the NS5B gene, a relatively variable region of HCV genome (Gededzha et al., 2012). This variability results in substitutions as the virus mutates. These nucleotide substitutions during HCV replication has resulted in the emergence of seven major HCV genotypes (1–7), each further divided into subtypes based on their genetic diversity (Simmonds et al., 2005; Ohno, 2007). To date 67 well defined and 20 unconfirmed subtypes have been identified (Messina et al., 2014).
HCV genotypes and subtypes are distributed differently throughout the world (Ramia and Eid-Fares 2006). Divergent strains of genotypes 1 and 2 have been shown to be endemic in West African countries including Burkina Faso, Ghana, Guinea Bissau, Benin Republic and Nigeria (Forb et al., 2012; Markov et al., 2009), genotypes 3 is found in South Asia, genotype 4, 5 and 6 are more predominant in central Africa and Middle East, South-east Asia, Northern region of South Africa and Belgium respectively (Markov et al., 2009). Genotype 7 has been reported only in central African immigrants in Canada (Messina et al., 2014; Murphy et al., 2015).
Information regarding HCV diversity in Nigeria is limited and this has resulted in limited number of HCV sequences from Nigeria that are available in nucleotide databases. In the past, most studies on HCV were mainly serology-based (Nwankiti et al., 2009; Adewole et al., 2009; Balogun et al., 2010) and did not provide information about the molecular epidemiology of HCV in Nigeria. Due to endemic nature of HCV in Africa (Mustapha et al., 2007; Laraba et al., 2007; Forbi et al., 2012), good knowledge of genetic diversity of HCV in the region is very critical for understanding the epidemiology of the virus, its evolutionary dynamics and design of effective universal vaccine against HCV infection and management of the infection with currently available antiviral drugs.
Response to HCV treatment is known to be largely dependent on the infecting genotype (Le Guillou-Guillemette et al., 2007), and due to geographical variations in HCV distribution, genotyping of infecting HCV becomes very important. The objective of this study therefore, was to determine the genetic diversity of hepatitis C virus isolates circulating in Ibadan, in order to form care and management of HCV infection in Nigeria.
MATERIALS AND METHODS
Study Population and Sample Collection
This was a cross-sectional study in which blood samples were collected from 176 subjects with positive HCV IgM antibodies (99 patients with clinical hepatitis and 77 blood donors). Ethical approval for the study was obtained from University of Ibadan/ University College hospital Ethical Review Committee and Oyo State Ministry of Health before commencement of study. Samples were collected from patients referred from different clinics to the Department of Virology, College of Medicine, University of Ibadan and blood donors at Blood Bank, University College Hospital, Ibadan (UCH). The mean age of the participants was 42.2 years (age range 3 months to 83 years). There were 139 males and 37 females included in the study.
HCV RNA Extraction and Amplification of NS5B gene
RNA was extracted from plasma samples using a commercially available kit, according to manufacturer’s (Jena Bioscience total RNA Purification kit) instructions. Reverse-transcription (first strand cDNA synthesis) of the extracted RNA was performed using random hexamer and specific primers using Script cDNA synthesis kit (Jena Bioscience), in a final volume of 20-μl. The synthesis conditions were 42o C for 10min followed by incubation at 50oC for 45min. The NS5B gene fragment located at positions 8275–8618 of the virus was then amplified using a nested PCR protocol. The PCR reaction mix constituted of 2.0 μl of the cDNA, 2.5-μl of Taq polymerase, 0.5-μl each of forward and reverse primers and 7.0-μl of nuclease-free water in a final volume of 12.5-μl reaction. Details of the primers and cycling conditions used are shown in the table 1 below. The amplified gene fragments were visualised using gel electrophoresis in 1.5% agarose gel (Forbi et al., 2012).
Table 1.
PCR primers and cycling conditions
| Nested PCR | Primer Sequences | Cycling Conditions | Size of Amplicons |
|---|---|---|---|
| 1st Round | Forward: 5′ TGGGGATCCCGTATGATACCCGCTGCTTTGA Reverse: 5′GGCGGAATTCCTGGTCATAGCCTCCGTGAA |
95°C for 5min, 94°C for 30sec, 50°C for 30sec, 72°C for 45sec,72° C for 10min, for 30cycles | 400bp |
| 2nd Round | F2- CTCAACCGTCACTGAGAGAGACAT R2- GCTCTCAGGCTCGCCGCGTCCTC |
95°C for 5min, 94°C for 30sec, 50°C for 30sec, 72°C for 45sec,72°C for 10min, for 45cycles | 300bp |
Sequencing of and Phylogenetic Analysis
PCR amplicons were purified with Exo SAP-it Amplicon Purification kit and sequenced with ABI V3.1 Big dye terminator according to manufacturer’s instructions. Sequencing was done in both directions using the inner PCR primers F2 and R2. The chromatograph of each sequence was inspected and edited using Bio Edit software version 7.0.5. Each consensus sequence was blasted in NCBI to determine HCV reference sequences with closest matching identity or relatedness to the study sequences. Study sequences were identified using country of origin and population studied.
Alignment of the study sequences with prototype strain H77 and other sequences from different continents spanning 300–310nt of the NS5B gene {HCV H77 position, 8275–8618 (GenBank, NC.004102.1)} obtained from HCV sequence Database [Markov et al., 2009], was done on MEGA 7.0 version software [Kumar et al., 2015]. Phylogenetic trees were constructed using the Neighbor-Joining method [Saitou and Nei, 1987]. Test of Phylogeny that is, the percentage replicate trees in which the associated taxa clustered together was performed with Bootstrap replication of 1000 and branch support values of >60%.
Statistical Analysis
Statistical differences were evaluated by the Chi-Squares test using SPSS version 20 program. Results were expressed as percentages at P = 0.05.
RESULTS
The NS5B gene was amplified in 38 of the 176 specimens giving an amplification success rate of 22%. Twenty-nine of the PCR positive samples were successfully sequenced and analyzed. Phylogenetic analysis of the sequences showed three of the seven known HCV genotypes (genotypes 1, 2, 5) are co-circulating in Ibadan, Nigeria (Figure 1).
Fig. 1.

Distribution of HCV subtypes among the study participants
Table 2 shows the distribution of HCV genotypes among study participants as revealed by the phylogenetic analyses in figures 3 and 4. The rate of occurrence of genotypes 1a, 1b, 2c, 3a and 5a among study participants was 34.5%, 17.2%, 13.8%, 3.6 and 31.1% respectively. Subtypes 1a and 5a were the predominant strains in the study. Subtypes 1b and 2b were found among patients with clinical hepatitis; while the single subtype 2c detected was among blood donors as shown in Figure 2.
Table 2.
Distribution of Study HCV Genotypes
| Genotype/Subtypes | Frequency (%) |
|---|---|
| 1a | 10 (34.5) |
| 1b | 5 (17.2) |
| 2b | 4 (13.8) |
| 2c | 1 (3.6) |
| 5a | 9 (31.4) |
| Total | 29 |
Fig. 3.
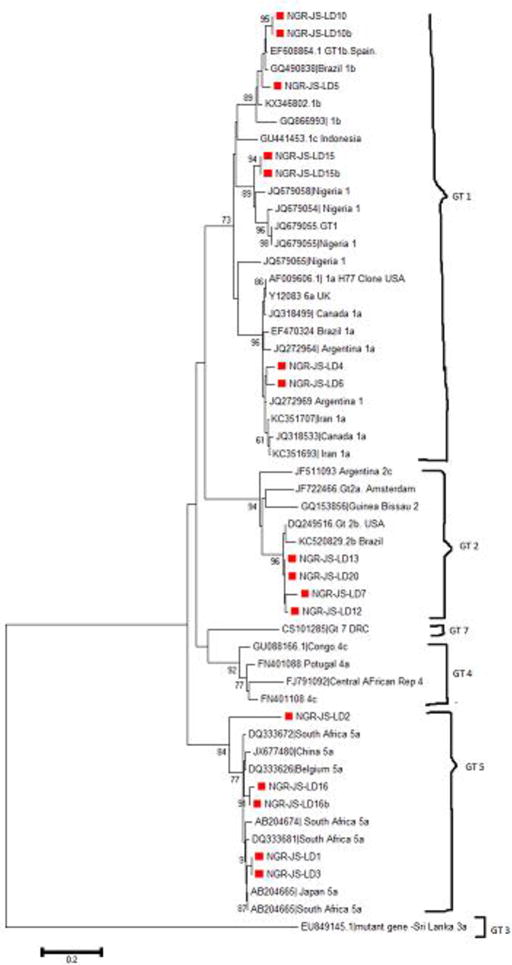
Phylogenetic tree of HCV NS5B genes in Patients with clinical hepatitis showing sequences from this study (marked with red blocks). Tree was constructed using Neighbor-Joining Method with bootstrap value of 1000 replicates. Arrows indicate where study genotypes/subtypes 1a, 1b, 2b & 5a are located on the tree.
Figure 4.
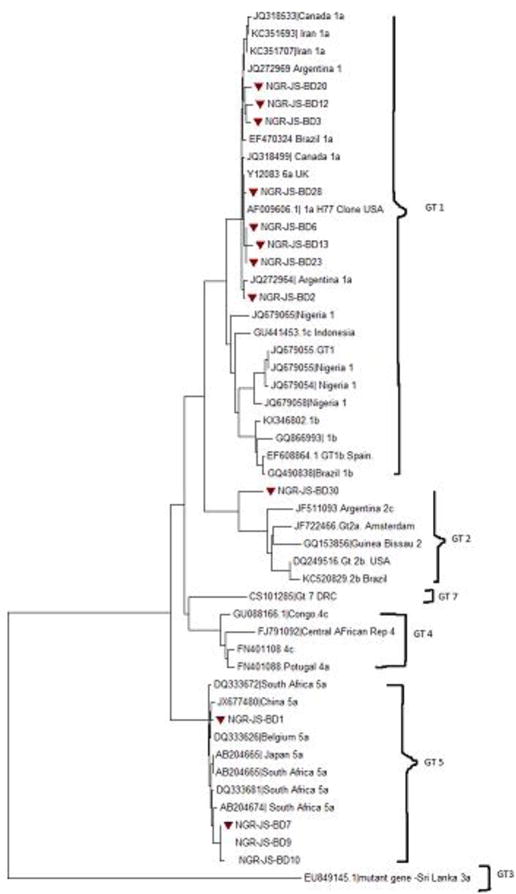
Molecular Phylogeny of HCV NS5B genes in Blood Donors showing study sequences marked with red blocks. Tree was constructed using Neighbor-Joining Method with bootstrap value of 1000 replicates. Arrows indicate where study genotypes/subtypes (1a, 2c and 5a) are located on the phylogenetic tree.
Fig. 2.

Distribution of HCV Subtypes compared between Patients with Clinical Hepatitis and Blood donors
Although subtype 1a was detected among both population, the rate was higher among blood donors (P = 0 .003), as indicated in Table 3. Subtype 5a was found among all study participants (P= 1.000). Subtype 5a is being reported for the first time in Nigeria and from West Africa in this study. The phylogenetic trees of HCV sequences, showing each study genotype/subtypes among patients with clinical hepatitis is shown in figure 3 while the tree showing genotypes/subtypes among blood donors is shown in figure 4. Figure 5 shows the amino acid alignment of study HCV sequences with reference to HCV H77 prototype strain. The conserved amino acid positions on NS5B gene are marked with red dots while variable sites are undotted.
Table 3.
Distribution of HCV Genotypes by clinical status
| Genotype/Subtypes | Patients with clinical hepatitis (LD) N= 16 (%) Occurrence |
Blood Donors (BD) N= 13(%) Occurrence |
Total N=29 |
p |
|---|---|---|---|---|
| 1a | 2 (20%) | 8 (80%) | 10 | 0.03 |
| 1b | 5 (100%) | 0 (0%) | 5 | 0.03 |
| 2b | 4 (100%) | 0 (0%) | 4 | 0.03 |
| 2c | 0 (0%) | 1 (100%) | 1 | 0.03 |
| 5a | 5 (55.6%) | 4 (44.4%) | 9 | 1. 00 |
Figure 5.

Amino acid alignment of all study HCV sequences in comparison with the HCV prototype strain H77 indicating conserved amino acid positions (with red dots) and variable sites (undotted)
DISCUSSION
Our study has shown that multiple HCV genotypes (1, 2 and 5) are co-circulating in Ibadan, Nigeria. Here, we report for the first time, the presence of genotype 5 (subtype a) in Nigeria and from West Africa circulating in both patients with clinical hepatitis and blood donors. The findings from this study are in support of previous report by Okwuraiwe et al., (2014), in which multiple HCV genotypes were found except genotypes 5 and 7 in Lagos, Nigeria. Prior to their work, genotypes 1 and 4 were reported in 1996 by Oni and Harrison (1996). Several years after, genotypes 1 and 2 were identified in two remote villages in North-Central Nigeria in the work done by Forbi et al., (2012). These show that even within regions in Nigeria, HCV genotypes are differentially distributed and this has serious implication for treatment of HCV infection in the country. The presence of diverse forms of HCV in Africa with emphasis on Nigeria lacking adequate data was reported by Markov et al., (2009). This diverse HCV types could lead to high treatment failure as well as high prevalence of liver disease including hepatocellular carcinoma (liver cancer) in chronically infected patients (Mustapha et al., 2007; Laraba et al., 2010; Forbi et al., 2012). It may also constitute a serious obstacle to designing a universal vaccine against HCV infection.
In this study, subtypes 1b and 2b were found only in patients with clinical hepatitis. This finding supports a study in which HCV subtype 1b was found as the prevalent HCV subtype among patients with hepatocellular carcinoma (Levrero, 2006). Presence of these subtypes in the patients with clinical disease may be indicative of the virulence of these viral strains, in addition to hosts and environmental factors that contribute to disease outcome in the patients.
Contrary to previous studies in West African regions, that have reported genotypes 1, 2 and 4 as the circulating strains in Guinea Bissau, Central African Republic, and North-Central Nigeria (Markov et al., 2009; Forbi et al., 2012), this study has shown that genotype 5 also circulate in Ibadan, Nigeria. Of importance, is its circulation among patients with clinical hepatitis and blood donors, although with no statistical difference in its prevalence among the two groups (P= 1.00). However, there are more conserved regions in genotype 5 in blood donors while many variable sites exist among patients with clinical disease as shown in Figure 5. But whether this variation is responsible for clinical symptoms in patients is not known. In comparing the amino acid translation of the study genotypes to HCV prototype strain H77; it was observed that HCV genotypes in our study have their amino acid conserved at some positions. Other positions have lots of substitutions with other amino acids that have probably affected the viral protein confirming the diversity observed in this study (Figure 5).
Genotype 1 (subtype 1a); is widely distributed and it accounts for most HCV infections all over the world (WHO, 2012). Its response to combination therapy with Peg-Interferon and Ribavirin is said to be low, than those of genotypes 2 and 3 (Le Guillou-Guillemette et al., 2007). HCV subtype 1a according to this study circulates in blood donors more than in patients with clinical hepatitis (P = 0.03). Infection in blood donors is asymptomatic and so this finding may imply that HCV subtype 1a is mostly found in asymptomatic infection than the other HCV strains. This may explain the sharp contrast in the subtype distribution in patients that are already having clinical disease (hepatitis) from those of blood donors. The variation observed between study subgroups may have resulted from differences in the route of acquisition of infection in addition host and viral factors.
Phylogenetic analysis of the NS5B gene sequences of study isolates showed that the different genotypes circulating among patients clustered with similar strains from other countries in Africa and sequences obtained previously from Nigeria as shown on the phylogenetic trees in Figure 3. The same was obtained for genotypes circulating in blood donors in Figure 4. In both, distantly-related isolates were seen to be farther away on the phylogenetic tree. Based on the bootstrap replication of 1000, it is clear to note that these sequences that clustered together are closely related and also might have emanated from the same ancestor. For instance, genotype1 (subtype a) isolates obtained in this study clustered with HCV prototype reference H77 clone and reference sequences from Canada and Argentina (Figure 3). With the references from Africa (Figure 3), these same isolates clustered together especially with sequences from Nigeria showing their origin from a common ancestor. For those that are far-branched, the divergence might have resulted evolution in those lineages. This study has several implications for treatment and prevention of HCV infection in Nigeria. Presence of diverse forms of the virus is a serious consideration with regard to the period and type of therapy to administer to patients with chronic HCV infections, especially in areas where the use of direct acting antivirals (DAA), which are reported to have high cure rate is not yet licensed. DAAs such as Sofosbuvir, a NS5B polymerase inhibitor that suppresses NS5B replication is not widely used for treatment in Nigeria. Interferon and Ribavirin are the standard therapy for HCV infection in Nigeria for now, involving about 48 weeks or more for difficult-to-treat strains of HCV. Furthermore, additional studies are required to understand the immunological interactions between these diverse strains of HCV and their hosts with regards to disease progression, as well as to determine pre-treatment drug-resistant markers in the diverse HCV strains in Nigeria.
In conclusion, this study has shown that diverse forms of HCV circulate in Ibadan, with predominant genotypes as 1a and 5a. This has implications for therapy and management of HCV infection in Nigeria. Based on our findings, there is need to genotype infecting HCV in patients and blood donors before commencement of therapy.
Acknowledgments
This study was supported by Medical Education Partnership Initiative Nigeria (MEPIN) through National Institute of Health (NIH) USA grant funded by Fogarty International Centre, the office of AIDS Research and National Human Genome Research Institute of NIH, the Health Resources and Services Administration (HRSA) and the Office of the U.S. Global AIDS Coordinator under award number R24TW008878. Our appreciation goes to the patients and blood donors that participated in this study and also to all the laboratory technologists in the department that assisted during this work. Data analysis and writing of this paper was supported by the University of Ibadan Medical Education Partnership Initiative Junior Faculty Training Programme (UI-MEPI-J) project funded by Fogarty International Center, National Institute of Health under Award Number D43TW010140.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations. Our appreciation also goes to the patients and blood donors that participated in this study and the entire laboratory Technologists in the Department of Virology who assisted assisted during this work
References
- Adewole OO, Anteyi E, Ajuwon Z, Wada I, Elegba F, Ahmed P, Betiku Y, Okpe A, Eze S, et al. Hepatitis B and C co-infection in Nigerian patients with HIV infection. J Infect Dev Ctries. 2009;3:369–375. doi: 10.3855/jidc.245. [DOI] [PubMed] [Google Scholar]
- Ashfaq UA, Javed T, Rehman S, Nawaz Z, Riazuddin S. An overview of HCV molecular biology, replication and immune responses. Virology Journal. 2011;8:161. doi: 10.1186/1743-422X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogun TM, Emmanuel S, Wright KO. Hepatitis C virus co-infection in HIV positive patients. Nig QJ Hosp Med. 2010;20:117–120. [PubMed] [Google Scholar]
- Davis GL, Balart LA, Schiff ER, et al. Treatment of chronic hepatitis c with recombinant interferon alfa. A multicenter randomized, controlled trial. Hepatitis Intervention Therapy Group. N Engl J Med. 1989;321(22):151–6. doi: 10.1056/NEJM198911303212203. 1989 Nov. [DOI] [PubMed] [Google Scholar]
- Dhawan VK. Hepatitis C. Medscape. 2016;2016 Update, p1-48 www.emedicine.medscape.com. [Google Scholar]
- Forbi JC, Purdi MA, Campo DS, Vaughan G, Dimitrova GE, Ganova-Raeva LM, Xia GL, Khudyakov YE. Epidemic history of hepatitis C virus infection in two remote communities in Nigeria, West Africa. J Gen Virol. 2012;93(Pt 7):1410–21. doi: 10.1099/vir.0.042184-0. 2012 Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gededzha MP, Selabe SG, Kyaw T, Rakgole JN, Blackard JT, Mphahlele MJ. Introduction of new subtypes and variants of hepatitis C virus genotype 4 in South Africa. J Med Virol. 2012;84(4):601–7. doi: 10.1002/jmv.23215. 2012 Apr. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. Characterization of a canine homolog of hepatitis C. 2011 doi: 10.1073/pnas.1101794108. Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N. Genome of Human hepatitis virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5(3):129–51. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- Kazmierczak J, Pawelczyk A, Cortes KC, Radkowski M. Seronegative Hepatitis C Virus Infection. Arch Immunol Ther Exp. 2013 doi: 10.1007/s00005-013-0257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura MEGA 7: Molecular Evolutionary Genetic Analysis (MEGA 7) 2015 Version 7.0. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054. 2016 July. Epub 2016 Mar 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laraba A, Wadzali G, Sunday B, Abdulfatai O, Fatai S. Hepatitis C virus infections in Nigerians with chronic liver disease. The Internet Journal of Gastroenterology. 2010;9 [Google Scholar]
- Le Guillou-Guillemette H, Vallet S, Gaudy-Graffin C, Payan C, Pivert A, Goudeau A, Lunel-Fabiani F. Genetic diversity of the hepatitis C vrus: Impact and issues in the antiviral therapy. World J Gastroenterol 2007. 2007;13(17):2416–2426. doi: 10.3748/wjg.v13.i17.2416. http://www.jgnet.com/1007-9327/13/2416.asp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene (2006) 2006;25:3834–3847. doi: 10.1038/sj.onc.1209562. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C Virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. http://dx.doi.org. [DOI] [PubMed] [Google Scholar]
- Ly KN, Xing J, Klevens RM, Jiles RB, Ward JW, Holmberg SD. The increasing burden of mortality from viral hepatitis in the United States between 1999–2007. Ann Intern Med. 2012;156(4):271–8. doi: 10.7326/0003-4819-156-4-201202210-00004. 2012 Feb. [DOI] [PubMed] [Google Scholar]
- Markov PV, Pepin J, Frost E, Deslandes S, Labbe A, Pybus OG. Phylogeography and Molecular Epidemiology of Hepatitis C Virus genotype 2 in Africa. Journal of General Virology (2009) 2009;90:2086–2096. doi: 10.1099/vir.0.011569-0. [DOI] [PubMed] [Google Scholar]
- Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61(1):77–87. doi: 10.1002/hep.27259. 2015 Jan. Epub 2014 July 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd HK, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis c virus infections: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 2013. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- Mora N, Adams WH, Kleithermes S, Dugas L, Balagubramanian N, Sandhu J, Nde H, Small C, Jose J, Scanglione S, Layden JE. A Synthesis of Hepatitis C prevalence estimate in Sub-Saharan Africa: 2000–2013. BMC Infect Dis. 2016;13–160 doi: 10.1186/s12879-016-1584-1. 2016 June. 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukerjee S. Hepatitis C. Medscape emedicine.com. 2012 August 2012. [Google Scholar]
- Murphy DG, Sablon E, Chamberland J, Fournier E, Dandavino R, Tremblay CL. Hepatitis C virus genotype 7, a new genotype originating from Central Africa. J Clin Microbiol (2015) 2015;53(3):967–972. doi: 10.1128/JCM.02831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha SK, Bolori MT, Ajayi NA, Nggada HA, Pindiga UH, Gashau W, Khalil MAI. Hepatitis C virus antibodies in Nigerians with hepatocellular carcinoma. The Internet Journal of Oncology. 2006;4(2) [Google Scholar]
- Naamani KAI, Sinani SAI, Deschenes M. Epidemiology and Treatment of Hepatitis C genotypes 5–6. Can J Gastroenterol. 2013;27(1) doi: 10.1155/2013/624986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwankiti OO, Ndako JA, Echeonu GO, Olabode AO, Nwosuh CI, Onovoh EM, Okeke LA, Akinola JO, Duru BN, et al. Hepatitis C virus infection in apparently healthy individuals with family history of diabetes in Vom, Plateau state Nigeria. Virol J. 2009;6:110. doi: 10.1186/1743-422X-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes Ia, 1b, 2a, 2b,3a, 3b 4,5 and 6a. J Clin Microbiol. 2007;35(1):201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okwuraiwe AP, Salu OB, Anomneze E, Audu RA, Ujah IAO. Hepatitis C virus genotypes and viral ribonucleic acid titers in Nigeria. J Gastroenterology-African Journals. 2012 online. [Google Scholar]
- Onakewhor JU, Okonofua FE. Seroprevalence of Hepatitis c viral antibodies in pregnancy in a tertiary health facility in Nigeria. Niger J Clin Pract. 2009;12:65–73. [PubMed] [Google Scholar]
- Oni AO, Harrison TJ. Genotype of hepatitis C Virus in Nigeria. J Med Virol. 1996;49:178–186. doi: 10.1002/(SICI)1096-9071(199607)49:3<178::AID-JMV4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Otegbayo JA, Taiwo BO, Akingbola TS, Odaibo GN, Adedapo KS, Penugonda S, Adewole FI, Olaleye DO, Murphy R, Kanki P. Prevalence of hepatitis B and C Seropositivity in Nigerian Cohort of HIV-infected Patients. Annals of Hepatology 2008. 2008 Apr-Jun;7(2):152–156. [PubMed] [Google Scholar]
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- Ramia S, Eid-Fares J. Distribution of hepatitis C virus genotypes in the Middle East. Int J Infect Dis 2006. 2006;10(4):272–277. doi: 10.1016/j.ijid.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Rao IB, Johari N, du cros P, Messina J, Ford N, Cooke GS. Hepatitis C Seroprevalence &HIV Co-infection in Sub-Saharan Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2015;15(7):819–824. doi: 10.1016/S1473-3099(15)00006-7. 2015 Jul. 8. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining Method-a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sarrazin C, Hezode C, Zeuzem S, Pawlotsky JM. Antiviral strategies in hepatitis C virus infection. J Hepatol. 2012;56(Suppl 1):S88–S100. doi: 10.1016/S0168-8278(12)60010-5. [DOI] [PubMed] [Google Scholar]
- Simmonds P. The origin and evolution of hepatitis viruses in humans. 2000 Fleming Lecture. Journal of General Virology (2001) 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Genetic diversity and evolution of hepatitis C virus-15 years on. Journal of General Virology, (2004) 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. http://vir.sgmjournals. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin I, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpazees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences (USA) 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Viral Hepatitis Factsheet. 2012;2012 [Google Scholar]


