Abstract
Down syndrome (DS) is the most common chromosomal disorder and the leading genetic cause of intellectual disability in humans, which results from the triplication of chromosome 21. DS individuals have an increased risk of developing Alzheimer’s disease (AD)-like pathology and dementia by the age of 40 due to the triplication of several genes involved in the formation of amyloid plaques and tau tangles. Further, DS and AD are characterized by the aberrant accumulation of unfolded/misfolded proteins resulting from over-burdened protein quality control systems. The accumulation of misfolded proteins in the endoplasmic reticulum (ER) triggers a cellular stress response called the unfolded protein response (UPR). Long-term activation of the UPR mediates neuronal dysfunction in AD. We hypothesized that the UPR is impacted in a mouse model of DS. To test this, we performed gene and protein expression analysis of ER stress markers in the Ts65Dn mouse model of DS at 3, 9, and 18 months. We identified activation of the PERK pathway in Ts65Dn DS mice at 3 months of age compared to euploid controls. We also determined that the early and overt UPR activation decreased with age, the UPR signal was significantly reduced by 18 months. Our data suggest that UPR activation in DS mouse models occurs early before consistent brain neurodegeneration and might be an essential contributor to dys-proteostasis.
Keywords: Down syndrome, eif2 alpha, PERK, Ts65Dn, unfolded protein response
INTRODUCTION
Down syndrome (DS) is the most common chromosomal disorder and leading genetic cause of intellectual disability in humans [1, 2]. As individuals with DS age, they have an increased risk for cognitive decline, dementia, and Alzheimer’s disease (AD) [3, 4]. This risk is likely due to the genetic imbalance present in DS: triplication and overexpression of genes located on chromosome 21, such as APP, SOD1, and BACE2 [5]. As is the case for the accelerated onset of cognitive decline and dementia in DS, the etiology and pathogenesis of AD are not yet completely understood. DS and AD neuropathology have many common features including deposition of senile plaques, neurofibrillary tangles, increased oxidative stress, and impairment of protein quality control system (PQC) [5–9].
An increasing number of studies suggest that endoplasmic reticulum (ER) alterations play a pivotal role in cellular dysfunction and death in AD and related dementias [10–13], but ER stress has not been previously investigated in DS. Perturbations of ER homeostasis, triggered by several factors including calcium depletion from the ER, oxidative stress, and secretory pathway alterations are responsible for the accumulation of unfolded/misfolded proteins in its lumen leading to ER stress. To re-establish homeostasis, the unfolded protein response (UPR) is activated [11, 14]. The adaptive effects of the UPR confer protection and survival via clearance of proteins from the ER or attenuation of protein synthesis. However, prolonged UPR activity leads to pro-apoptotic signals, which has been observed in both DS humans and mouse models [15].
Under normal, non-stressed physiological conditions, the ER chaperone glucose-regulated protein 78 (GRP78) binds the ER stress sensor protein kinase RNA (PKR)-like ER kinase (PERK) [16], activating transcription factor 6 (ATF6) and inositol-requiring enzyme-1alpha (IRE1α). Under ER stress, GRP78 dissociates from these sensors and promotes their activation, inducing phosphorylation of PERK and IRE1α and the translocation of ATF6 to the Golgi where it is cleaved into its active form [17]. Once activated, ER stress sensors activate several transcription factors that promote the expression of chaperones as well as other modulators of PQC. Long-term activation of the UPR is associated with neurodegenerative disorders. For example, markers specific for UPR activation, such as GRP78, pPERK, and pIRE1, are increased in AD cortex and hippocampus compared to non-demented control brains [18–21].
We recently showed that GRP78 is oxidized in vivo prior to significant AD pathology in DS brain [22]. Oxidative damage leads to GRP78 dysfunction. Besides these observations, there is little known about ER stress in DS and how DS genetics may alter the UPR. Therefore, we hypothesized that GRP78 oxidation in DS brain unleashed selective activation of the UPR.
The goal of this study is to investigate the role of the UPR in the hippocampus from DS mouse models at different time points. We investigated the potential contribution of the UPR and the role of PERK-mediated disruption of protein synthesis in DS neuropathology. Our data suggest that the UPR is altered in DS.
MATERIALS AND METHODS
Mouse colonies
Ts65Dn (B6EiC3Sn a/A-Ts(1716)65Dn/J), a well-established mouse model of DS, carries a reciprocal translocation that is trisomic for approximately 104 genes (56%) on Mmu16, from Mrpl39 to the distal telomere, with homologues on HSA21 [23]. Mice were generated by repeatedly backcrossing Ts65Dn trisomic females with (B6EiC3SnF1/J) F1 hybrid males; the parental generations were purchased from Jackson Laboratories. These breeding pairs produce litters containing both trisomic (Ts65Dn) and disomic (2N) offspring. The pups were genotyped to establish trisomy using standard PCR, as described by Reinoldt [24]. In addition, the recessive retinal degeneration 1 mutation (Pdebrd1), which segregates in the colony and results in blindness in homozygotes, was analyzed for all trisomic animals used in the present study by using standard PCR. This information is reported in Table 1. Mice were housed in clear Plexiglas cages (20 × 22 × 20 cm) under standard laboratory conditions with a temperature of 22 ± 2°C and 70% humidity, a 12-h light/dark cycle and free access to food and water. Littermates were spliced among age groups to avoid littermates/dam-specific effect. Mice employed for western blot and RT-PCR studies (Ts65Dn and age-matched euploid) were sacrificed by cervical dislocation at 3, 9, and 18mo and the hippocampus was collected and frozen at −80°C.
Table 1.
Mice used
| Age | Genotype | Sample Size |
Sex (M/F) |
Weight (gr ± SD) |
|---|---|---|---|---|
| 3mo | Eu | 6 | 3/3 | 23.6 ± 1.3 |
| Ts65Dn | 6 | 4/2 | 25.0 ± 2.0 | |
| 9mo | Eu | 6 | 4/2 | 33.8 ± 6.1 |
| Ts65Dn | 6 | 3/3 | 37.1 ± 8.1 | |
| 18mo | Eu | 6 | 2/4 | 39.2 ± 6.4 |
| Ts65Dn | 6 | 3/3 | 31.6 ± 7.1 |
All the experiments were performed in strict compliance with the Italian National Laws (DL 116/92) and the European Communities Council Directives (86/609/ EEC). Experimental protocol was approved by Italian Ministry of Health authorization n° 1183/2016-PR. All efforts were made to minimize the number of animals used.
Quantitative real-time RT-PCR
RNA was extracted from the frozen hippocampus of the right hemisphere from Ts65Dn and euploid mice at 3, 9, and 18mo (6 samples for each group) using Tissue Total RNA Kit according to manufacturer’s instructions (Fisher Molecular biology). RNA was quantified using the Biospec Nano spectrophotometer (Shimadzu, Columbia MD) and cDNA was reverse transcribed using the cDNA High Capacity kit (Applied Biosystems, Foster City CA). The standardized samples were added to the High Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA) including reverse transcriptase, random primers and buffer according to manufacturer’s instructions. The cDNA was produced through a series of heating and annealing cycles in the Veriti™ 96-well Thermocycler (Applied Biosystems, Grand Island, NY). Quantitative real-time RT-PCR was achieved using the TaqMan® Array Micro Fluidic card (Life Technologies, Carlsbad, CA); 1 µl of the cDNA (100 ng/µl of standardized RNA samples)was diluted with 49 µl of RNAse-free water, 50 µl of TaqMan Universal Mastermix II and added to each well. Real-time RT-PCR was performed using the ViiA™7 Real Time PCR system (Applied Biosystems, Grand Island, NY). Gene card array was designed to evaluate changes in the expression of genes delineated in Table 2 with their PMID and Taqman ID. Gene expression was evaluated by normalizing to GAPDH expression as an internal control. Fold change was determined using the 2−ΔΔCt method [25].
Table 2.
Target genes for RT-PCR
| Genes of interest | PMID | TagMan ID |
|---|---|---|
| HSPA5 | NM_022310.3 | Mm00517690_g1 |
| EIF2AK3 | NM_010121.2 | Mm00438700_m1 |
| ATF6 | NM_001081304.1 | Mm01295317_m1 |
| ERN1 | NM_023913.2 | Mm00470233_m1 |
| EIF2A | NM_001005509.2 | Mm01289723_m1 |
| PPP1R15A | NM_008654.2 | Mm01205601_g1 |
| ATF4 | NM_009716.3 | Mm00515325_g1 |
| DDIT3 | NM_001290183.1 | Mm01135937_g1 |
| GAPDH | NM_008084.3 | Mm99999915_g1 |
Sample preparation for Western Blot
Approximately 25 mg hippocampus, per sample from 3, 9, and 18mo Ts65Dn and euploid animals (6 samples per group) were homogenized by sonication in RIPA buffer (pH 7.4) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 1 mM PMSF, 1 mM NaF and 1 mM Na3VO4 and centrifuged at 14,000 × g for 30 min to remove cellular debris. The supernatant was extracted to determine the total protein concentration by the BCA method (Pierce, Rockford, IL).
Western Blot
For Western blot, 30 µg of proteins (3, 9, and 18mo Ts65Dn and Euploid) were separated by 7.5% and 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), using Criterion TGX (Tris-Glycine extended) Stain-Free precast gels. Blots were normalized on gel total protein load acquired taking advantage of the Stain free technology on ChemiDoc XP densitometer (Bio-Rad). After image acquisition gels were transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). Membranes were blocked with 7% nonfat dry milk (Lab Scientific) with 0.02% sodium azide and incubated over night at 4 °C with following primary antibodies: pPERK (1:250) and PERK(1:250), CHOP(1:500), GRP78 (1:500), ATF6 (1:500), pIRE1 (1:500), IRE1 (1:500) from Santa Cruz Biotechnology (Santa Cruz, CA, USA), peIF2a (1:500), and eIF2a (1:1000) from Cell Signaling). The membranes were incubated for 1 h at room temperature with secondary antibody horseradish peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat IgG (1:5000); Sigma-Aldrich, St. Louis, MO, USA). Membranes were developed with the Clarity Western ECL Substrate (Bio-Rad), visualized using ChemiDoc XP (Bio-Rad) and analyzed using Image Lab software (Bio-Rad).
Experimental design and statistical analysis
Gene card arrays were performed one time with cDNA from six mice per genotype (Ts65Dn and euploid) for each age group (3, 9, and 18mo). For western blot analysis, we selected six animals per group based on our previous work [26]. Sample size was previously calculated using G-Power 3.1 software with the following parameters: Effect size = 2.5 (expected Mean Difference 0.5 and Standard deviation 0.2); Err- prob = 0.05; Power = 0.95 for a sample size of n = 6 for each group with an actual power of 0.97. Analysis of variance (One way-ANOVA) was used to compare the results obtained from three different ages (3, 9, and 18mo). The t-test was used to evaluate differences between euploid and Ts65dn where p values equal *p=0.05, **p=0.01. Data are expressed as mean ± SD per group. Values above or below two standard deviations of the mean were considered outliers and discarded from the data set. All statistical analysis was performed using Graph Pad Prism 7.0 software.
RESULTS
In order to establish the contribution of ER stress in DS neuropathology, we characterized age-associated modifications of the UPR in a mouse model of DS. We evaluated changes in Ts65Dn hippocampi compared to euploid littermate controls at 3, 9, and 18 months of age by analyzing gene expression and protein levels. We first measured the expression level of genes implicated in the UPR. To this end, we designed a gene card array to measure expression levels of key UPR genes in Ts65Dn brains. Using hippocampal RNA from Ts65Dn mice, we measured changes in expression of Eukaryotic Translation Initiation Factor 2 Alpha Kinase 3 (EIF2AK3, which codes for PERK), Activating Transcription Factor 6 (ATF6), and Endoplasmic Reticulum To Nucleus Signaling (ERN1, which codes for IRE1), EIF2A (which codes for eIF2α), ATF4 (Activating Transcription Factor 4), PPP1R15A (Protein Phosphatase 1 Regulatory Subunit 15A, which codes for Growth Arrest and DNA Damage-Inducible Protein 34, or GADD34), and DDIT3 (which codes for DNA-Damage-Inducible Transcript 3, C/EBP homologous protein, or CHOP, a downstream effector of PERK-mediated apoptosis) (Table 2).
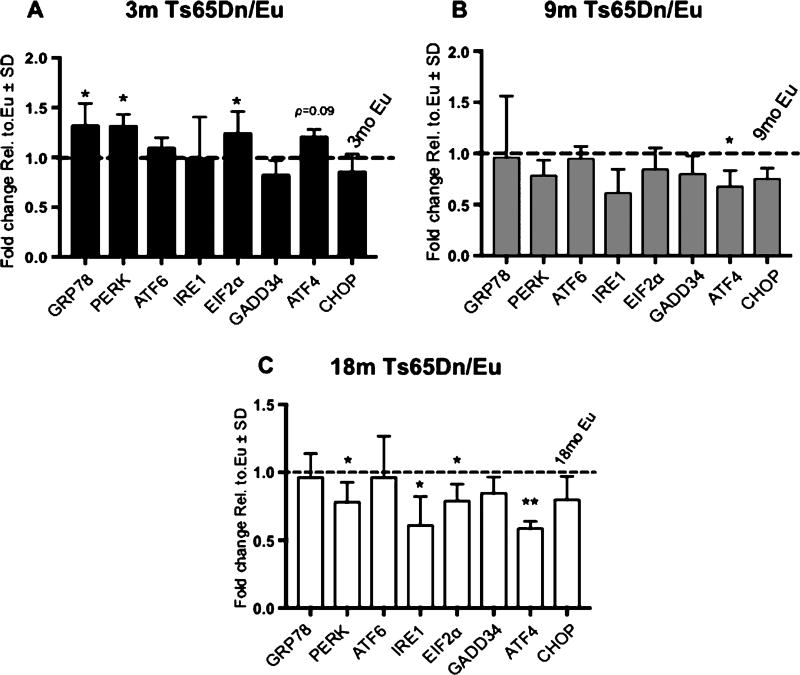
We detected significantly increased expression of GRP78, EIF2AK3, and EIF2A occurring at 3mo; however, ATF6, ERN1, PPP1R15A, ATF6, and DDIT3 were unchanged when compared to the age matched control mice (Fig. 1A, Table 3). Except for a significant decrease in ATF4 expression, these changes did not persist through the 9mo time point (Fig. 1B). Meanwhile, at the 18mo time point, we detected a significant reduction of PERK, ERN1, EIF2A, and ATF4 (Fig. 1C, Table 3). These data allow us to identify progressive phases of UPR activation in DS neuropathology. We identified a consistent and selective activation of the PERK pathway suggesting that ER stress results in the increased expression of the PERK-eIF2α-ATF4-CHOP pathway at 3mo that was decreased at 18mo.
Fig. 1.
Ts65Dn mice display differential age-dependent activation of the UPR. A–C) Gene expression changes of UPR-related genes in 3 (A), 9 (B), and 18 (C) month-old Ts65Dn mice compared to euploid (Eu) control mice. All gene expression data were normalized to GAPDH expression. Fold change was determined using 2(−ΔΔCt) method. Fold change for (A-C) was calculated from the corresponding age matched controls, which are indicated by the dashed line on each graph set to 1. Sample size was six mice per group. Statistical significance was determined using Student t-test analysis (*p < 0.05, **p < 0.01).
Table 3.
Fold increase values for RT-PCR
| Gene | Single age comparison | Age dependent comparison | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Ratio Ts65Dn:Eu | 9mo:3mo | 18mo:3mo | |||||
|
|
|
|
|||||
| 3mo | 9mo | 18mo | Eu | Ts65Dn | Eu | Ts65Dn | |
| HSPA5 | 0.30* | 0.04 | –0.04 | 1.22** | 0.68 | 0.57* | 0.42* |
| EIF2AK3 | 0.31* | −0.02 | –0.22* | 0.39 | –0.25* | 0.69** | –0.08 |
| ATF6 | 0.09 | −0.05 | –0.04 | 0.07 | 0.03 | 0.13 | 0.21 |
| ERN1 | –0.02 | −0.39 | –0.39* | 0.58 | –0.18 | 0.70* | –0.27 |
| EIF2A | 0.31* | −0.16 | –0.21* | 0.61* | 0.01 | 0.54** | –0.18 |
| PPP1R15A | –0.18 | −0.20 | –0.15 | 0.10 | –0.03 | 0.10 | –0.04 |
| ATF4 | 0.20 | −0.33* | –0.41* | 0.31 | –0.29* | 0.55* | –0.18* |
| DDIT3 | –0.15 | −0.25 | –0.2 | 0.56* | –0.07 | 0.37* | –0.27* |
Statistical significance was determined using Student t-test analysis (*p < 0.05, **p < 0.01).
We then investigated whether changes in expression translated into differences in protein levels in 3mo mice. As in the gene expression assays (Fig. 1), we evaluated the three major arms of the UPR and the canonical downstream target of the PERK pathway using western blots. A major objective of this study was to evaluate the extent to which the UPR contributes to abnormalities resulting from hippocampal lesions. Since the Ts65Dn mice have a robust genetic change, we were concerned that any unintended effects of the genetic alteration could impact levels of individual proteins such as traditional gel loading markers. To accurately establish changes in protein levels, we normalized our protein loading to a total protein load using stain free technology. In doing so, we can carefully detect discreet changes in protein levels independent of changes in actin, GAPDH, tubulin, or other more traditional loading markers.
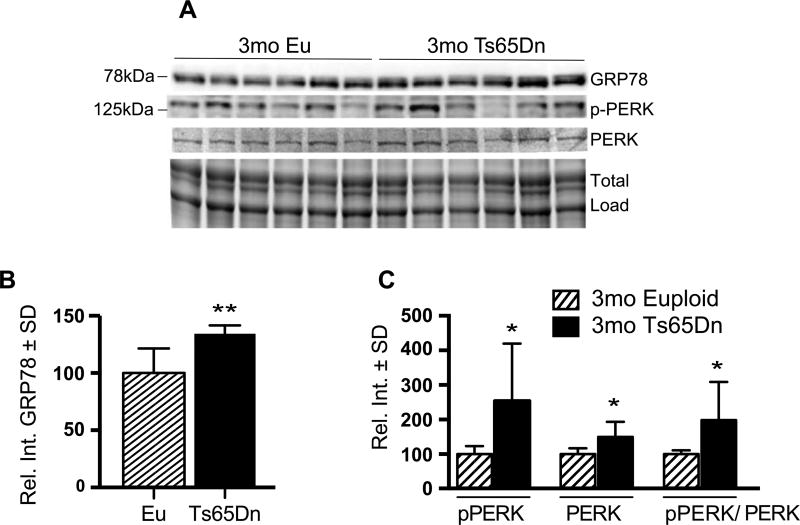
We measured protein levels using hippocampal tissue from Ts65Dn mice and euploid controls and determined that Grp78 levels were significantly increased by 35% in the 3mo Ts65Dn brains compared to euploid controls (35% p = 0.004; Fig. 2A, B). These results suggest that activation of the UPR is an early phenomenon resulting from DS progression. As expected from the gene expression data (Fig. 1A), total PERK levels were also increased in the Ts65Dn brains (Fig. 2A). Importantly, we identified that the active form of PERK, phosphorylated PERK or pPERK (pT981), was also significantly increased in Ts65Dn brains. The ratio of phospho- to total PERK showed a 77% (p = 0.0367) increase in 3mo Ts65Dn mice compared to controls (Fig. 2C).
Fig. 2.
Active PERK levels are present in 3mo Ts65Dn mice brains. A) Representative western blot showing increased GRP78 and pPERK (pT981). B) Quantification of panel A showing significantly increased GRP78 levels in Ts65Dn brains. C) Quantification of panel A showing levels of pPERK, PERK, and the pPERK to total PERK ratio. Densitometric values shown in the bar graph are the mean of 6 samples per each group normalized per total load and are given as percentage of Euploid mice, set as 100%. Statistical significance was determined using Student t-test analysis (*p < 0.05, **p < 0.01).
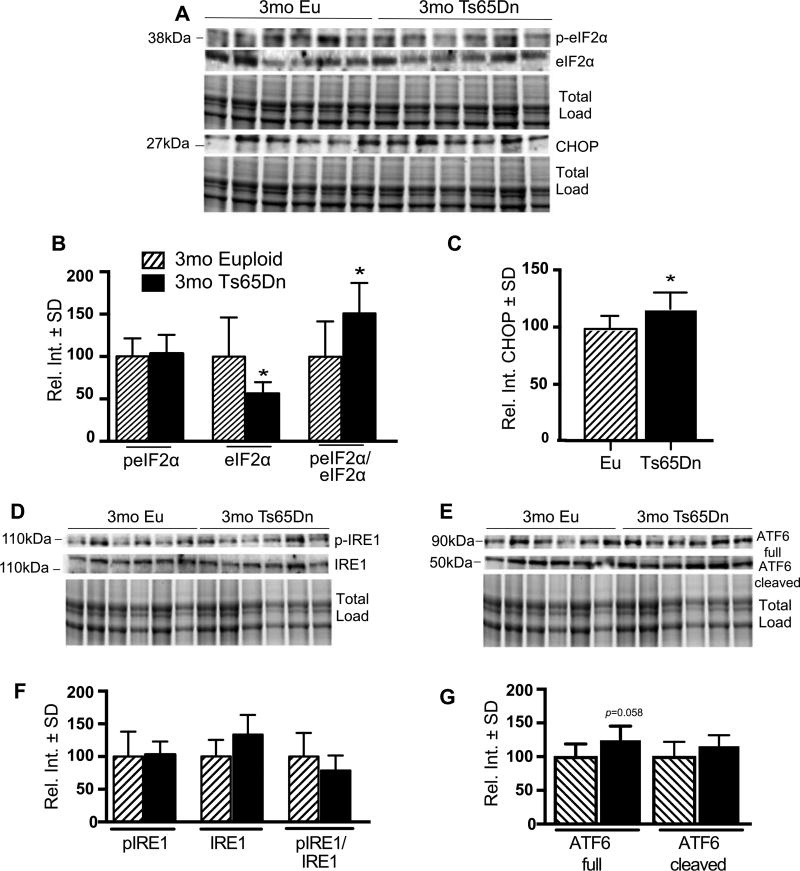
PERK phosphorylates the eukaryotic initiation factor 2α (eIF2α), which attenuates RNA translation [16]. This effect reduces input of nascent proteins to the ER. However, chronic attenuation of translation is detrimental to the cell, and neurons are particularly susceptible to reduced protein synthesis via the PERK pathway. We identified significant down regulation of total eIF2α level, and, a large proportion of the total eIF2α was phosphorylated (Ser51) making the ratio of phospho- to total eIF2α significantly increased (51%, p = 0.0363) in 3mo Ts65Dn (Fig. 3A, B).
Fig. 3.
3 mo Ts65Dn mice show PERK-dependent CHOP activation. A) Representative western blot showing increased peIF2α (pS51) and CHOP. B) Quantification of panel A showing levels of peIF2α, eIF2α, and the peIF2α (pS51) to total eIF2α ratio. C) Quantification of panel A showing significantly increased CHOP levels in Ts65Dn brains. D,E) Representative western blot showing no change in pIRE1 (S724) and ATF6. F) Quantification of panel A showing levels of pIRE1, IRE1, and the pIRE1 to total IRE1 ratio. G) Quantification of panel E showing levels of ATF6 (Full) and ATF6 (Cleaved) form. Densitometric values shown in the bar graph are the mean of 6 samples per each group normalized per total load and are given as percentage of Euploid mice, set as 100%. Statistical significance was determined using Student t-test analysis (*p < 0.05).
We also investigated activation of CHOP, a downstream effector of PERK-mediated apoptosis. We observed a 16% increase in CHOP levels in 3mo Ts65Dn mice (16%, p = 0.0371) (Fig. 3A–C). This suggests that ER stress results in the increased expression of CHOP via the PERK-eIF2α pathway. These studies also revealed no significant differences in the activation or abundance of IRE1 (and pIRE1) and ATF6 (full length and cleaved) (Fig. 3D–G).
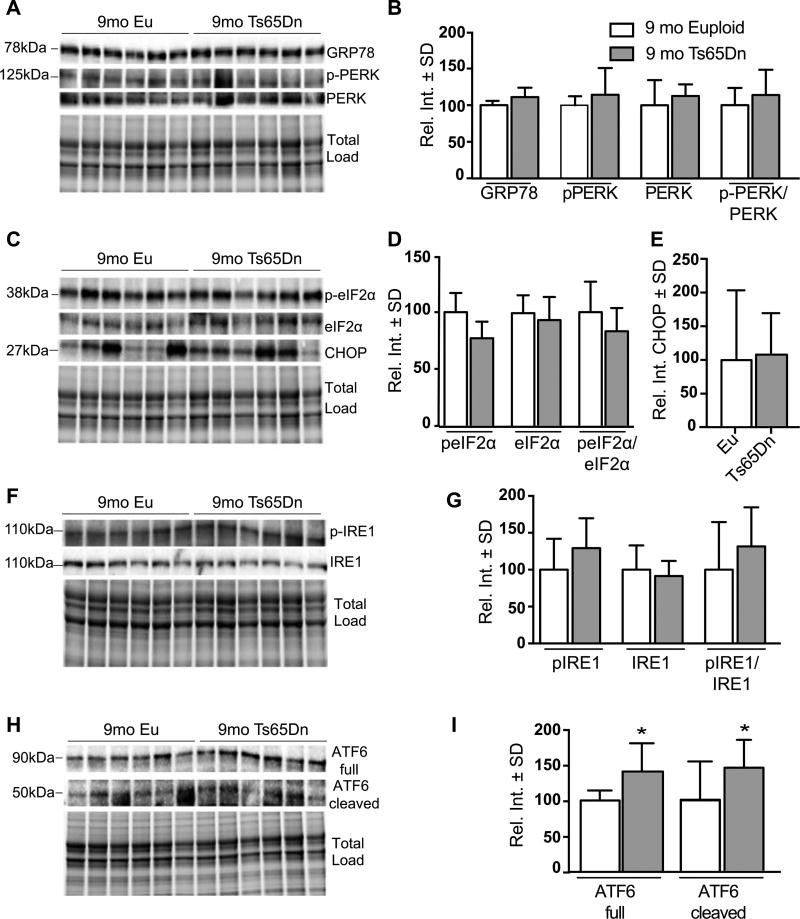
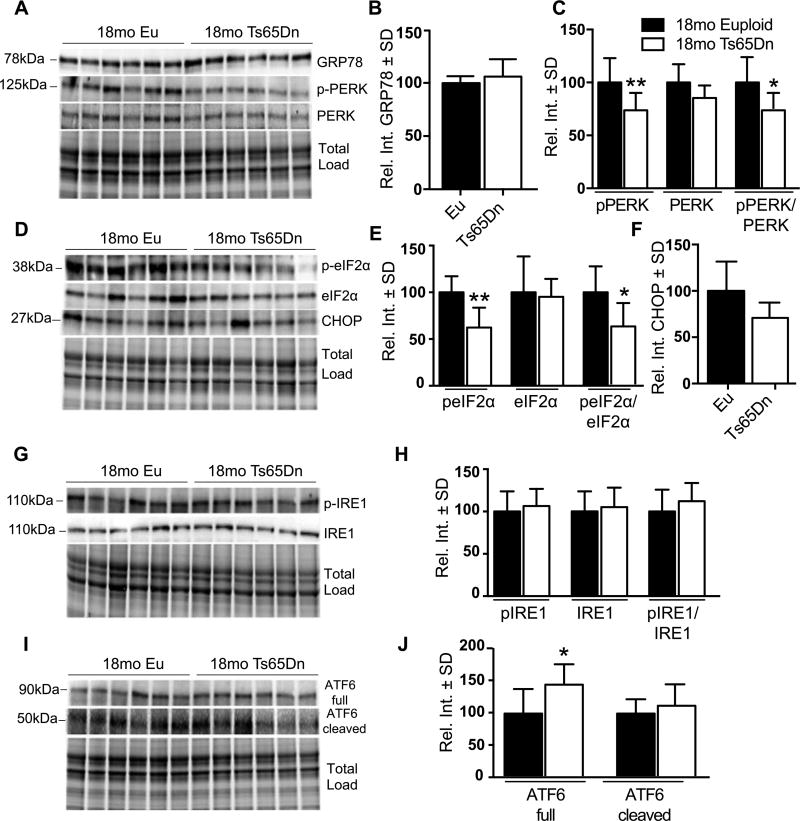
We observed a general restoration of the UPR at 9mo: GRP78, pPERK/PERK (Fig. 4A, B), peIF2α/eIF2α (Fig. 4C, D), CHOP (Fig. 4C, E), and pIRE1/IRE1 (Fig. 4F, G) were unchanged. However, we found that compared to euploid, the levels of ATF6 and its cleaved form were significantly increased in Ts65Dn hippocampi 52% (p = 0.0045) and 46% (p = 0.0473), respectively (Fig. 4H, I). During the UPR, ATF6 is cleaved and the N-terminal domain translocates into the nucleus [17]. There, it activates transcription of ER chaperones including Grp78, Grp94, and calnexin, to promote protein folding in the ER lumen. By 18mo, we observed a clear reduction in the pPERK/PERK ratio namely due to a 37% reduction of pPERK (p = 0.0458) (Fig. 5A, C). The peIF2α/eIF2α ratio was also decreased; this was mostly due to a stark reduction of peIF2α (38% p = 0.0321) since total eIF2α levels were similar to controls (Fig. 5D, E). Interestingly, Grp78 (Fig. 5A, B), CHOP (Fig. 5D, F) pIRE1/IRE1 (Fig. 5G, H), and cleaved ATF6 (Fig. 5I, J) were unchanged compared to controls. Full lengthATF6was the only protein that increased at 18mo in Ts65Dn mice (46%, p = 0.0413; Fig. 5I, J) suggesting that ATF6 promotes an adaptive response despite an overall attenuation of UPR activity.
Fig. 4.
UPR proteins are unchanged in 9mo Ts65Dn mice. A) Representative western blot showing no change in GRP78 pPERK (pT981) levels in 9 months old Ts65Dn mice. B) Quantification of panel A showing no significant difference in GRP78 levels and levels of pPERK, PERK, and the pPERK to total PERK ratio. C) Representative western blot showing no change in peIF2α (pS51) and in CHOP levels. D) Quantification of panel D showing levels of peIF2α, eIF2α, and the peIF2α (pS51) to total eIF2α ratio. E) Quantification of panel C showing no change in CHOP levels. F) Representative western blot showing significantly increased in ATF6 Full protein and no change in the cleaved form. G) Quantification of panel F showing levels of ATF6 (Full) and ATF6 (Cleaved) forms. H) Representative western blot showing no change in pIRE1 (S724). I) Quantification of panel G showing levels of pIRE1, IRE1, and the pIRE1 to total IRE1 ratio. Densitometric values shown in the bar graph are the mean of 6 samples per each group normalized per total load and are given as percentage of Euploid mice, set as 100%. Statistical significance was determined using Student t-test analysis (*p < 0.05).
Fig. 5.
PERK-eIF2α pathway is decreased in 18mo Ts65Dn mice. A) Representative western blot showing no change in GRP78 levels at 18 months and decreased in pPERK (pT981) levels. B) Quantification of panel A showing no significant difference in GRP78 levels. C) Quantification of panel A showing levels of pPERK, PERK, and the pPERK to total PERK ratio. D) Representative western blot showing decreased in peIF2α (pS51) and no change in CHOP levels. E) Quantification of panel D showing levels of peIF2α, eIF2α, and the peIF2α (pS51) to total eIF2α ratio. F) Quantification of panel D showing no change in CHOP levels. G) Representative western blot showing significantly increased in ATF6 Full protein and no change in the cleaved form. H) Quantification of panel G showing levels of ATF6 (Full) and ATF6 (Cleaved) forms. I) Representative western blot showing no change in pIRE1 (S724). J) Quantification of panel I showing levels of pIRE1, IRE1, and the pIRE1 to total IRE1 ratio. Densitometric values shown in the bar graph are the mean of 6 samples per each group normalized per total load and are given as percentage of Euploid mice, set as 100%. Statistical significance was determined using Student t-test analysis (*p < 0.05, **p < 0.01).
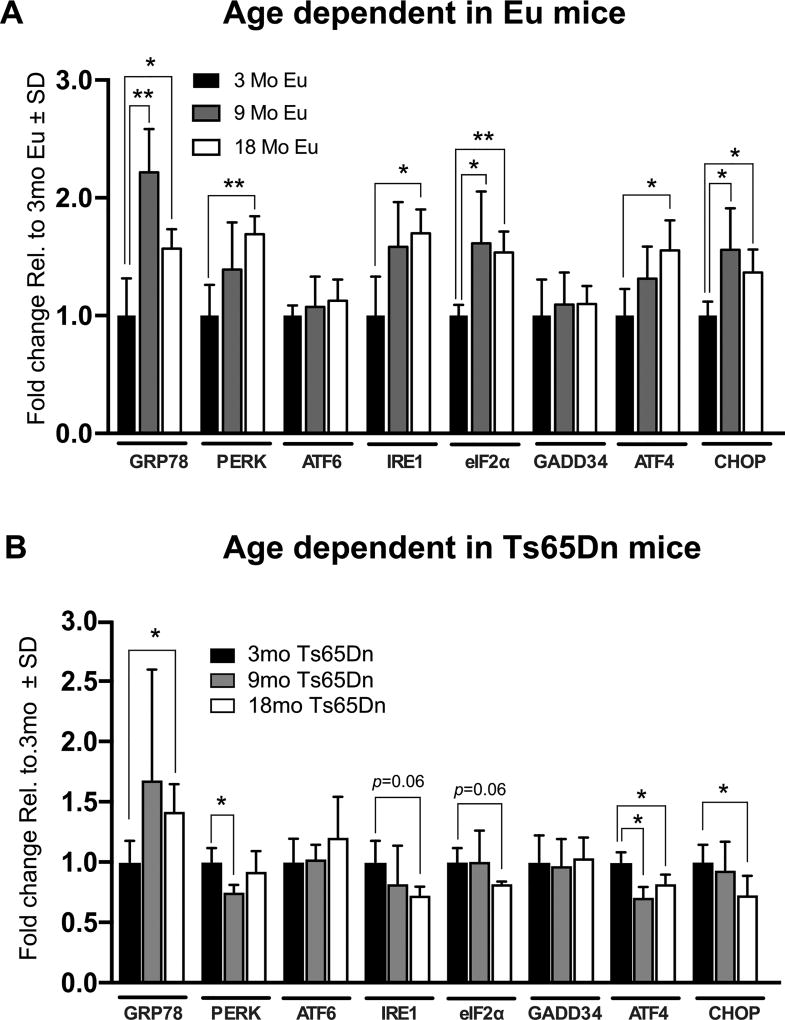
We then investigated persistence of UPR activation over time in euploid and Ts65Dn mice. In euploid controls only, gene card array analyses showed overall increased expression of UPR genes when referenced to the 3mo time point (Fig. 6A). Specifically, GRP78, PERK, IRE1, eIF2α, ATF4, and CHOP were significantly increased at 9mo and/or 18mo compared to 3mo. In contrast, this trend was reversed in most genes of Ts65Dn mice (Fig. 6B). The most salient decrease was evident in ATF4 and CHOP expression, which was significantly reduced over time. Interestingly, GRP78 expression continued increasing over time suggesting that the UPR was still promoting protein folding in the ER. Interestingly, total PERK expression decreased at 9mo but increased to an intermediate level between 3mo and 9mo. These results are summarized in Table 3.
Fig. 6.
Differential UPR activation in Eu and Ts65Dn mice. A) Gene expression of UPR-related genes plotted over time against Eu mice. B) Gene expression changes of UPR-related gene expression in 3, 9, and 18mo. All gene expression data were normalized to GAPDH expression. Fold change was determined using 2(−ΔΔCt) method. Fold change for (A–B) was calculated from the 3mo Ts65Dn mice (A) and from the 3mo Eu (B) which are set to 1. Sample size was six mice per group. Statistical significance was determined using Student t-test analysis (*p < 0.05, **p < 0.01).
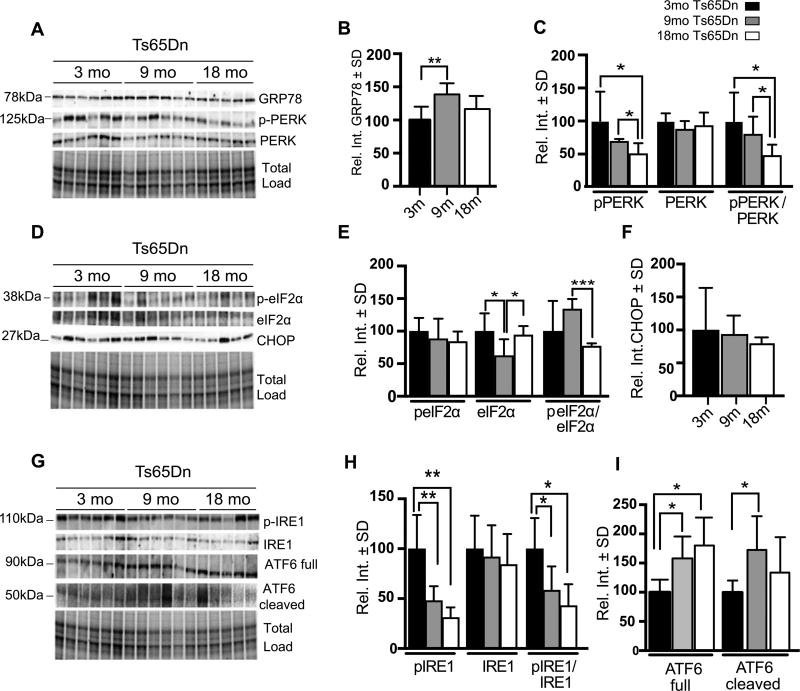
We evaluated changes in protein levels corresponding to Grp78, PERK (phospho- and total), eIF2α (phospho- and total), CHOP, IRE1, and ATF6 (total and cleaved) in Ts65Dn brains (Fig. 7A–I). As expected from the gene expression assays (Fig. 6B), GRP78 protein levels were increased at 9mo compared to 3mo Ts65Dn mice (39%; p = 0.0048), but its levels decreased to an intermediate point at 18mo (Fig. 7A, B). In contrast, pPERK levels decreased significantly over time in Ts65Dn mice (Fig. 7A, C). At 18mo, identified a 49% (p = 0.0369) reduction in pPERK levels when compared to the 3mo time point; we also detected a 19% reduction (p = 0.0409) when we compared 9mo to 18mo Ts65Dn mice. Moreover, the pPERK/PERK ratio steadily declined over time (Fig. 7A, C).
Fig. 7.
UPR activity promotes adaptive pro-survival response. A) Representative western blot showing increased GRP78 and pPERK (pT981) levels. B) Quantification of panel A showing significantly increased GRP78 levels in 9m Ts65Dn brains with respect to 3m Ts65Dn. C) Quantification of panel A showing levels of pPERK, PERK, and the pPERK to total PERK ratio. D) Representative western blot showing increased levels of peIF2α (pS51) at 9m compared to 18m old Ts65Dn mice and no change in CHOP levels. E) Quantification of panel D showing levels of p eIF2α, eIF2α, and the peIF2α (pS51) to total eIF2α ratio. F) Quantification of panel D showing CHOP levels in Ts65Dn brains. G) Representative western blot showing significantly increased pIRE1 (S724) in 3mo TS65Dn mice and significantly increased ATF6 at 9 and 18 months and significantly increased of cleaved ATF6 at 9 months. H) Quantification of panel G showing levels of pIRE1, IRE1, and the pIRE1 to total IRE1 ratio. I) Quantification of panel G showing levels of ATF6 (Full) and ATF6 (Cleaved) forms. Densitometric values shown in the bar graph are the mean of 6 samples per group normalized to total load and are given as percentage of 3mo Ts65Dn mice, set as 100%. Statistical significance was determined using Student t-test analysis. (*p < 0.05, **p < 0.01, ***p < 0.001).
As we continued to dissect the PERK cascade, we focused on eIF2α. Total eIF2α gene expression at 9mo was similar to the 3mo time point (Fig. 7E), yet the protein levels were significantly decreased at 9mo compared to both 3mo and 18mo time points. As in the 3mo time point (Fig. 3A, B), the marked decrease in total eIF2α influenced the ratio of phospho- to total eIF2α (Fig. 7E) such that there was an increased ratio at 9mo that declined significantly at 18mo (57%; p = 0.0002).
We also measured changes in CHOP, a mediator of UPR-related apoptosis. We found that total CHOP levels did not change over time in the Ts65Dn brains (Fig. 7D–F). This suggests that ER-mediated apoptosis is not increased in the surviving cells at these time points, which could indicate persistence of the UPR’s adaptive response.
We then evaluated changes in the levels of the other two major pathways of the UPR: IRE1 and ATF6. Although IRE1 (phospho- and total) levels did not change when comparing Ts65Dn to euploid controls (Figs. 3 and 5), there was a stark and progressive reduction of pIRE1 similar to what we observed for pPERK (Fig. 7G, H). In addition, total IRE1 did not change between the three time points thereby driving the pIRE1:IRE1 ratio to an overall reduction. A major contrast to these results was ATF6, which showed an increase in total protein as well as the cleaved and active species (Fig. 7G–I). Therefore, ATF6 expression and cleavage suggest that the ER is undergoing an adaptive phase for survival. These data suggest that there is an overall age-dependent reduction of the PERK and IRE1 pathways, and that this is especially marked in the context of an apoptotic response per unchanged CHOP levels.
DISCUSSION
After the fifth decade of life, the majority of DS individuals develop neuropathology characteristic of AD such as amyloid plaques and neurofibrillary tau tangles. Besides these major disease hallmarks, AD is also characterized by chronic UPR activation [13, 19]. However, activation of the UPR in DS has not been previously reported. Our study provides evidence that UPR is altered in DS mice at different time points. To decipher the impact of the UPR in DS, we characterized age-associated modifications of the UPR in Ts65Dn mice.
Previous studies show that GRP78, pPERK, pIRE1, and peIF2α levels are increased in AD brain [18–21, 27, 28]. Moreover, GRP78 and pPERK levels directly correlate with Braak stage [18]. These observations support a role for UPR alteration in the development of AD. Consistent with these reports, our data show the aberrant activation of UPR pathway in DS mice, since early stages of pathology (Figs. 1 and 2).
One indication of the involvement of ER stress in DS comes from our previous work using a redox proteomics approach in DS brain [22]. We identified unique proteins differentially expressed in DS brains, among which was GRP78. Our data also demonstrated that GRP78 was oxidized in vivo thereby suggesting a relationship between oxidative stress and protein misfolding in DS brain. Oxidation of GRP78 suggests that it would lose its chaperone activity causing ER stress. A maladaptive response, such as chronic UPR activation, provides a link between deficits in molecular chaperones, accumulation of misfolded proteins, and neurotoxicity.
The first evidence of maladaptive chronic UPR activity in our study was the unexpected increase in gene and protein expression of GRP78, PERK, and eIF2α in 3mo Ts65Dn mice (Figs. 1A and 3A, B) compared to euploid controls (Fig. 2). Overexpression of these proteins suggests that the PERK pathway is selectively active in early stages of the disease phenotype. As mentioned previously, increased GRP78 activates the UPR promoting release and consequent PERK phosphorylation. Active PERK phosphorylates eIF2α thereby inhibiting delivery of methionyl-tRNA to the ribosome; the result is attenuation of translation [16, 29]. Only transcripts containing internal ribosomal site (IRES) elude this mechanism and are effectively translated despite eIF2α phosphorylation. These transcripts lead to synthesis of several UPR proteins including ATF4 and another ER chaperone, GRP94 [16, 30, 31].
We found non-significant increased gene expression of downstream PERK pathway intermediates such as ATF4, GADD34, and CHOP in 3mo Ts65Dn mice compared to the euploid (Fig. 1A). These data suggest that despite PERK activation at 3mo, downstream consequences of the PERK pathway are not yet widespread in the hippocampus. Therefore, these data establish an effective therapeutic window to target PERK as a potential target.
CHOP, a pro-apoptotic transcription factor, is expressed by the PERK and IRE1 pathways of the UPR [32, 33]. CHOP protein levels were increased in 3mo Ts65Dn mice compared to the age-matched euploid (Fig. 3G). We hypothesize that CHOP upregulation is due to peIF2α phosphorylation with direct CHOP transcriptional induction by ATF4, and this is supported by the lack of IRE1 activation at any time point compared to controls (Figs. 3–5). Surprisingly, pPERK levels and its pathway intermediates do not change until the mice reach 18mo (Figs. 5 and 7). Moreover, 18 mo Ts65Dn mice show reduced PERK and eIF2α activation (Fig. 5C, E), strengthening the idea that an increase in the PERK pathway could be specific in the pre-symptomatic stages of DS pathology of Ts65Dn mice. Furthermore, UPR activation in the age dependent studies allowed us to identify three different phases along DS progression in Ts65Dn mice: a first phase at 3 mo characterized by a selective UPR activation, a second phase at 9mo where we observed its restoration to basal 9mo controls, and a third phase (18mo) with a consistent decrease of the UPR. Previous studies show that apoptosis is implicated in cell death of AD-like mechanisms in DS aging brains [15]. The impact of apoptosis in Ts65Dn mice at different ages demonstrated a proapoptotic phenotype [15], which in agreement with the steady increase in CHOP levels, despite an overall reduction of UPR intermediates over time, suggests that the UPR might mediates apoptosis in this model.
Disturbances in protein degradation pathways may contribute to ER stress and UPR activation. Decreased proteasomal activity occurs in AD brains [34], and neurons affected by neurofibrillary pathology accumulate immature forms of autophagosomes with autophagy impairment [35]. Consistent with these studies, we previously showed decreased proteosomal activity in DS brain [22, 36]. This is associated with a significant decrease in a marker of autophagosome formation (LC3 II/I) both in DS and in the Ts65Dn mouse at 6mo [9, 37]. In addition, our group and others demonstrated that the overexpression of some of the genes onHSA21(chromosome 16 in mice), such as SOD1, APP, or bach1, might lead to increased “subchronic” levels of oxidative stress in DS individuals as well as in mouse models of DS/AD pathology triggering UPR activation [31, 38, 39].
Our data show a significant increase of ATF6 expression over time (Fig. 7I). ATF6 is an important transcription factor involved in secretion [40], ER-associated degradation [41], and induction of stress response. This increase could be linked to a cellular mechanism to promote survival under the molecular consequences of DS neuropathology in Ts65Dn mice.
Under basal conditions, we observed increased expression of GRP78, PERK, eIF2α, ATF4, IRE1, and CHOP in aged euploid mice from 3 mo to 18 mo, whereas the expression of ATF6 and GADD34 is unchanged (Fig. 6A). The UPR operates as a central player to maintain ER homeostasis or the induction of cell death of chronically damaged cells. Our results also place ER stress as a driver of brain aging. However, it is important to note that Ts65Dn mice and its derivative strains carry an additional copy of part of mouse chromosome 17 that is not comparable to any part of human chromosome 21 [42]. The triplication of these additional genes may contribute to UPR alterations thereby diverging DS model mice data from DS in humans.
In conclusion, this is a characterization study in which our data suggest that UPR undergoes early and selective over activation in Ts65Dn mice contributing to the failure of PQC and to the development of neurodegenerative pathways. Within this context, PERK could represent a valuable target for a future therapeutic intervention in DS. Interventions targeting the UPR, and PERK in particular in different models of the disease, are currently underway.
Acknowledgments
We acknowledge the University of Kentucky Alzheimer’s Disease Center (UK-ADC), which is supported by NIH/NIA P30 AG028383, and the University of Kentucky Microscopy Core, which is supported by NIH/NINDS P30NS051220. JFA, SNF, AI, RAC. SEM, EM, DL, and GKN, were supported by NIH/NINDS 1R01 NS091329-01, Alzheimer’s Association NIRG-14-322441, NIH/NCATS 5UL1 TR000117-04, NIH/NIGMS 5P30GM110787, Department of Defense AZ140097, the University of Kentucky Epilepsy Center (EpiC), NIH/NIA P30 AG028383, NIH/NIMHD L32 MD009205-01 and NIH/NICHHD R01HD064993. This work was also supported by Fondi di Ateneo and SIR program of the Italian Ministry of Education, University and Research (RBSI144MTL) to F.D.D.
Footnotes
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/17-0617r2).
References
- 1.Lott IT. The neurology of Alzheimer disease in Down syndrome. Prog Clin Biol Res. 1992;379:1–14. [PubMed] [Google Scholar]
- 2.Lejeune J, Turpin R, Gautier M. Mongolism; a chromosomal disease (trisomy) Bull Acad Natl Med. 1959;143:256–265. [PubMed] [Google Scholar]
- 3.Johannsen P, Christensen JE, Mai J. The prevalence of dementia in Down syndrome. Dementia. 1996;7:221–225. doi: 10.1159/000106883. [DOI] [PubMed] [Google Scholar]
- 4.Schupf N, Sergievsky GH. Genetic and host factors for dementia in Down’s syndrome. Br J Psychiatry. 2002;180:405–410. doi: 10.1192/bjp.180.5.405. [DOI] [PubMed] [Google Scholar]
- 5.Head E, Lott IT, Wilcock DM, Lemere CA. Aging in Down syndrome and the development of Alzheimer’s disease neuropathology. Curr Alzheimer Res. 2016;13:18–29. doi: 10.2174/1567205012666151020114607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterfield DA, Di Domenico F, Swomley AM, Head E, Perluigi M. Redox proteomics analysis to decipher the neurobiology of Alzheimer-like neurodegeneration: Overlaps in Down’s syndrome and Alzheimer’s disease brain. Biochem J. 2014;463:177–189. doi: 10.1042/BJ20140772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lott IT, Head E, Doran E, Busciglio J. Beta-amyloid, oxidative stress and down syndrome. Curr Alzheimer Res. 2006;3:521–528. doi: 10.2174/156720506779025305. [DOI] [PubMed] [Google Scholar]
- 8.Nixon RA, Yang DS. Autophagy failure in Alzheimer’s disease–locating the primary defect. Neurobiol Dis. 2011;43:38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perluigi M, Di Domenico F, Butterfield DA. mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol Dis. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Abisambra JF, Jinwal UK, Blair LJ, O’Leary JC, 3rd, Li Q, Brady S, Wang L, Guidi CE, Zhang B, Nordhues BA, Cockman M, Suntharalingham A, Li P, Jin Y, Atkins CA, Dickey CA. Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. J Neurosci. 2013;33:9498–9507. doi: 10.1523/JNEUROSCI.5397-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell MC, Meier SE, Ingram AL, Abisambra JF. PERK-opathies: An endoplasmic reticulum stress mechanism underlying neurodegeneration. Curr Alzheimer Res. 2016;13:150–163. doi: 10.2174/1567205013666151218145431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Scheper W. Activation of the unfolded protein response is an early event in Alzheimer’s and Parkinson’s disease. Neurodegener Dis. 2012;10:212–215. doi: 10.1159/000334536. [DOI] [PubMed] [Google Scholar]
- 13.Scheper W, Hoozemans JJ. Endoplasmic reticulum protein quality control in neurodegenerative disease: The good, the bad and the therapy. Curr Med Chem. 2009;16:615–626. doi: 10.2174/092986709787458506. [DOI] [PubMed] [Google Scholar]
- 14.Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 15.Tramutola A, Pupo G, Di Domenico F, Barone E, Arena A, Lanzillotta C, Broekaart D, Blarzino C, Head E, Butterfield DA, Perluigi M. Activation of p53 in Down syndrome and in the Ts65Dn mouse brain is associated with a proapoptotic phenotype. J Alzheimers Dis. 2016;52:359–371. doi: 10.3233/JAD-151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 17.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W. The unfolded protein response is activated in pretangle neurons in Alzheimer’s disease hippocampus. Am J Pathol. 2009;174:1241–1251. doi: 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- 20.Stutzbach LD, Xie SX, Naj AC, Albin R, Gilman S, Group PSPGS. Lee VM, Trojanowski JQ, Devlin B, Schellenberg GD. The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathol Commun. 2013;1:31. doi: 10.1186/2051-5960-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unterberger U, Hoftberger R, Gelpi E, Flicker H, Budka H, Voigtlander T. Endoplasmic reticulum stress features are prominent in Alzheimer disease but not in prion diseases in vivo. J Neuropathol Exp Neurol. 2006;65:348–357. doi: 10.1097/01.jnen.0000218445.30535.6f. [DOI] [PubMed] [Google Scholar]
- 22.Di Domenico F, Coccia R, Cocciolo A, Murphy MP, Cenini G, Head E, Butterfield DA, Giorgi A, Schinina ME, Mancuso C, Cini C, Perluigi M. Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer’s disease neuropathology: Redox proteomics analysis of human brain. Biochim Biophys Acta. 2013;1832:1249–1259. doi: 10.1016/j.bbadis.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 24.Reinholdt LG, Ding Y, Gilbert GJ, Czechanski A, Solzak JP, Roper RJ, Johnson MT, Donahue LR, Lutz C, Davisson MT. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm Genome. 2011;22:685–691. doi: 10.1007/s00335-011-9357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Di Domenico F, Pupo G, Mancuso C, Barone E, Paolini F, Arena A, Blarzino C, Schmitt FA, Head E, Butterfield DA, Perluigi M. Bach1 overexpression in Down syndrome correlates with the alteration of the HO-1/BVR-a system: Insights for transition to Alzheimer’s disease. J Alzheimers Dis. 2015;44:1107–1120. doi: 10.3233/JAD-141254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang RC, Suen KC, Ma CH, Elyaman W, Ng HK, Hugon J. Involvement of double-stranded RNA-dependent protein kinase and phosphorylation of eukaryotic initiation factor-2alpha in neuronal degeneration. J Neurochem. 2002;83:1215–1225. doi: 10.1046/j.1471-4159.2002.01237.x. [DOI] [PubMed] [Google Scholar]
- 28.Hamos JE, Oblas B, Pulaski-Salo D, Welch WJ, Bole DG, Drachman DA. Expression of heat shock proteins in Alzheimer’s disease. Neurology. 1991;41:345–350. doi: 10.1212/wnl.41.3.345. [DOI] [PubMed] [Google Scholar]
- 29.Vannuvel K, Renard P, Raes M, Arnould T. Functional and morphological impact of ER stress on mitochondria. J Cell Physiol. 2013;228:1802–1818. doi: 10.1002/jcp.24360. [DOI] [PubMed] [Google Scholar]
- 30.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Suntharalingam A, Abisambra JF, O’Leary JC, 3rd, Koren J, 3rd, Zhang B, Joe MK, Blair LJ, Hill SE, Jinwal UK, Cockman M, Duerfeldt AS, Tomarev S, Blagg BS, Lieberman RL, Dickey CA. Glucose-regulated protein 94 triage of mutant myocilin through endoplasmic reticulum-associated degradation subverts a more efficient autophagic clearance mechanism. J Biol Chem. 2012;287:40661–40669. doi: 10.1074/jbc.M112.384800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 35.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 36.Tramutola A, Di Domenico F, Barone E, Arena A, Giorgi A, di Francesco L, Schinina ME, Coccia R, Head E, Butterfield DA, Perluigi M. Polyubiquitinylation profile in Down syndrome brain before and after the development of Alzheimer neuropathology. Antioxid Redox Signal. 2017;26:280–298. doi: 10.1089/ars.2016.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tramutola A, Lanzillotta C, Arena A, Barone E, Perluigi M, Di Domenico F. Increased mammalian target of rapamycin signaling contributes to the accumulation of protein oxidative damage in a mouse model of Down’s syndrome. Neurodegener Dis. 2016;16:62–68. doi: 10.1159/000441419. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed MM, Sturgeon X, Ellison M, Davisson MT, Gardiner KJ. Loss of correlations among proteins in brains of the Ts65Dn mouse model of down syndrome. J Proteome Res. 2012;11:1251–1263. doi: 10.1021/pr2011582. [DOI] [PubMed] [Google Scholar]
- 39.Choi JH, Berger JD, Mazzella MJ, Morales-Corraliza J, Cataldo AM, Nixon RA, Ginsberg SD, Levy E, Mathews PM. Age-dependent dysregulation of brain amyloid precursor protein in the Ts65Dn Down syndrome mouse model. J Neurochem. 2009;110:1818–1827. doi: 10.1111/j.1471-4159.2009.06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 41.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckley F. Modelling Down syndrome. Downs Syndr Res Pract. 2008;12:98–102. doi: 10.3104/updates.2054. [DOI] [PubMed] [Google Scholar]