Abstract
The data presented here are related to the research paper entitled “Study of a Novel Agent for TCA Precipitated Proteins Washing - Comprehensive Insights into the Role of Ethanol/HCl on Molten Globule State by Multi-Spectroscopic Analyses” (Eddhif et al., submitted for publication) [1]. The suitability of ethanol/HCl for the washing of TCA-precipitated proteins was first investigated on standard solution of HSA, cellulase, ribonuclease and lysozyme. Recoveries were assessed by one-dimensional gel electrophoresis, Bradford assays and UPLC-HRMS. The mechanistic that triggers protein conformational changes at each purification stage was then investigated by Raman spectroscopy and spectrofluorometry. Finally, the efficiency of the method was evaluated on three different complex samples (mouse liver, river biofilm, loamy soil surface). Proteins profiling was assessed by gel electrophoresis and by UPLC-HRMS.
Specifications Table
| Subject area | Chemistry |
| More specific subject area | Proteomics, protein purification, protein precipitation, trichloroacetic acid |
| Type of data | Tables, Figures |
| How data was acquired | Raman (LabRAM HR800UV confocal microspectrometer, Horiba Jobin Yvon, Kyoto, Japan) |
| Bradford assay (DC Protein Assay, Biorad) | |
| Electrophoresis (ImageJ software) | |
| UPLC-HRMS (Accela LC pumps, Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer equipped of an ESI source, Thermo Fisher Scientific, Waltham, MA, USA) | |
| MASCOT search engine (Matrix Science, London, UK; version 2.6.0) and Skyline software (MacCoss Lab, Washington, US; version 3.7.0.10940) | |
| ProteomeXchange Consortium with identifier PXD008110 | |
| Data format | Raw, analyzed and processed data |
| Experimental factors | |
| Experimental features | Proteins extraction was performed on 500 mg of soil, 10 mg of biofilm and 15 mg of mouse liver as starting material according to protocols of Chourey et al.[2], Huang et al.[3]and Song et al.[4]respectively. |
| Proteins were precipitated with 25% (w/v) trichloroacetic acid (TCA). | |
| The washing of protein pellet was performed with three different agents (acetone, ethanol, or ethanol/HCl). The mixture was vortexed and kept at −20 °C for 1 h, centrifuged at 16,600 g for 15 min at 4 °C. The resulting pellets were dried in a SpeedVac concentrator, solubilized in a 50 mM of ammonium bicarbonate buffer containing 10 mM of Tris. Proteins were subjected to trypsin digestion for 24 h at 37 °C. Digestion was stopped with formic acid before gel, bradford and mass analysis. | |
| Data source location | Poitiers, France |
| Data accessibility | data are with this article |
Value of the data
-
•
Data show a comprehensive evaluation of protein conformational changes throughout TCA precipitation and one single step purification with various solvents.
-
•
Data highlight the efficiency of ethanol/HCl purification for TCA-precipitated proteins.
-
•
Ethanol/HCl represents a quick and inexpensive purification agent for proteomics studies.
-
•
Presence and variability of proteins are potential values to determine which purification method must be used for proteomics investigation.
1. Data
TCA precipitation is one of the most common and robust technique required for protein analyses [5], [6], [7]. However it leads to molten globule states which hamper the solubilization of proteins in aqueous buffers for mass spectrometry analysis.
1.1. Comparison of washing agents on standard solutions
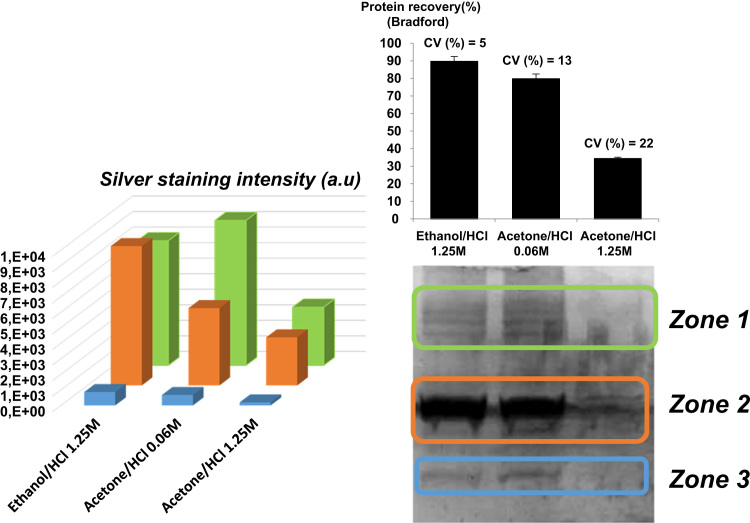
A standard solution of HSA, cellulase (exoglucanases and endoglucanases mixture), lysozyme and ribonuclease A, 35 µg mL−1 each, was prepared in high purified water. Prot eins were precipitated with 25% (w/v) trichloroacetic acid (TCA) (final concentration). The clean-up of protein pellet was performed following three different approaches: ethanol/HCl (1.25 M; 3.8%), acetone/HCl (0.06 M; 0.2%); acetone/HCl (1.25 M; 3.8%) (Fig. 1).
Fig. 1.
Standard proteins quantification by Bradford assay and silver-staining on electrophoresis gel. The thin line bars represent standard deviations at the top of the Bradford histogram. For both methods, histograms were constructed from the mean value of three independent assays.
1.2. Extraction and purification of endogenous proteins from complex sample matrices
See Fig. 2.
Fig. 2.
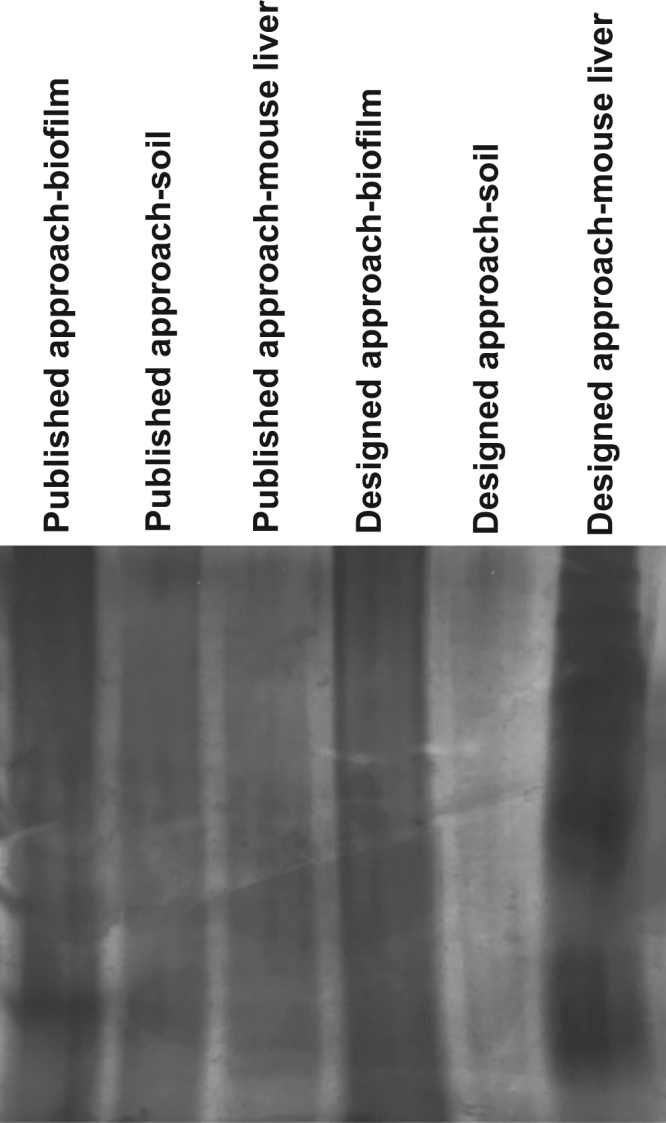
One-dimensional gel electrophoresis of complex matrices (biofilm, soil and mouse liver) after purification following the designed approach versus published protocols on complex matrices. The gel was stained with silver nitrate.
1.3. Effects of successive ethanol/HCl washings on proteins recoveries
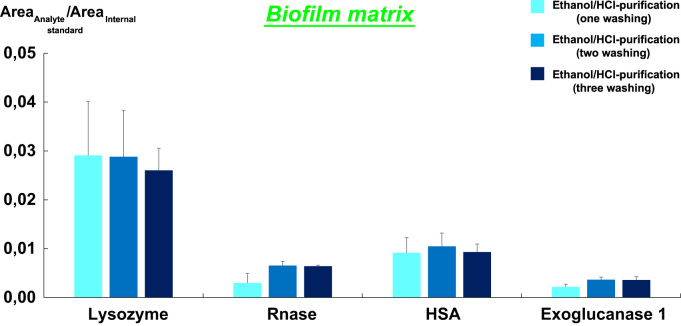
10 mg of biofilm samples were spiked with the standard solution of HSA, exoglucanase 1 from the mix of cellulase, lysozyme, and ribonuclease A (Rnase). Proteins final concentration was 1 µg mg−1 of matrix to enable HRMS detection of the proteins after the whole process. The mixture was vortexed and left during 24 h at room temperature to favor proteins adsorption on the matrix. After extraction following the published protocol of Huang et al. [3], protein pellets were subjected to one, two or three ethanol/HCl washing(s).
They were then dissolved in 50 mM of ammonium bicarbonate containing 10 mM of Tris (pH 8.5), diluted in a ratio of 1:3 using the same buffer and subjected to trypsin digestion.
Experiments were performed in triplicate. Fig. 3 gives the mean protein recoveries following the designed approach (Ethanol/HCl) on biofilm matrix after multiple washing steps.
Fig. 3.
Proteins recoveries following the designed approach on biofilm sample. The thin line bars represent standard deviations at the top of each column. Each bar shows mean±s.e.m. from three independent purification assays. Protein recoveries in Tris buffer were determined by UPLC/HRMS in a full scan mode with a resolution of 70.000 and mass range of 200–3000 m/z.
1.4. Understanding the effect of ethanol/HCl on proteins conformation
1.4.1. Spectrofluorometry
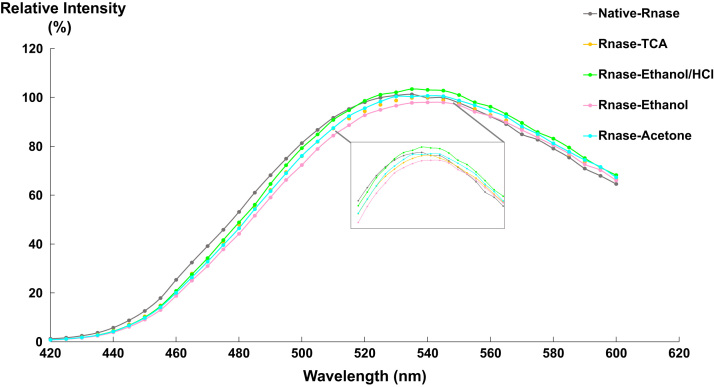
To get insights into the role of ethanol/HCl on proteins solubility, their conformational changes were comprehensively investigated, as an extension of the results reported in Ref. [1]. These measures were performed at each purification stage with two spectroscopic techniques: spectrofluorometry and Raman.
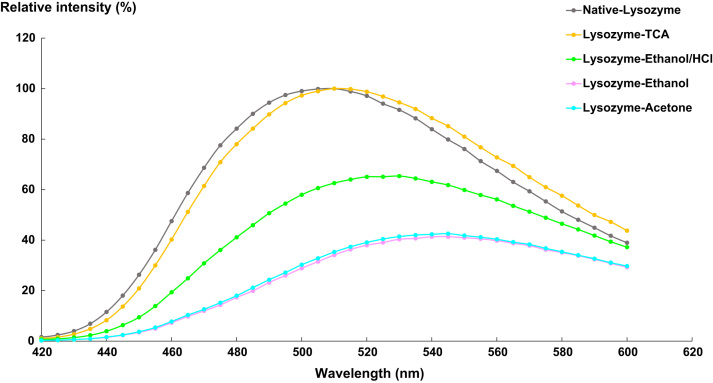
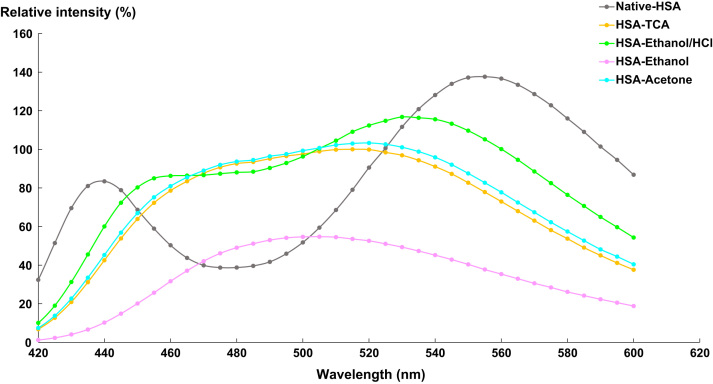
Fig. 4, Fig. 5, Fig. 6 represent the fluorescence emission spectra of lysozyme, HSA and Rnase after TCA precipitation and washing steps (ethanol/HCl, ethanol or acetone).
Fig. 4.
Emission spectra of lysozyme (λexc = 400 nm) at different purification steps. Native lysozyme (grey spectrum); Lysozyme-TCA (orange spectrum); Lysozyme-ethanol/HCl (green spectrum); Lysozyme-ethanol (purple spectrum); Lysozyme-acetone (blue spectrum).
Fig. 5.
Emission spectra of HSA (λexc = 400 nm) at different purification steps. Native HSA (grey spectrum); HSA-TCA (orange spectrum); HSA-ethanol/HCl (green spectrum); HSA-ethanol (purple spectrum); HSA-acetone (blue spectrum).
Fig. 6.
Emission spectra of RNASE (λexc = 400 nm) at different purification steps. Native Rnase (grey spectrum); Rnase-TCA (orange spectrum); Rnase-ethanol/HCl (green spectrum); Rnase-ethanol (purple spectrum); Rnase-acetone (blue spectrum).
1.4.2. Raman microspectroscopy
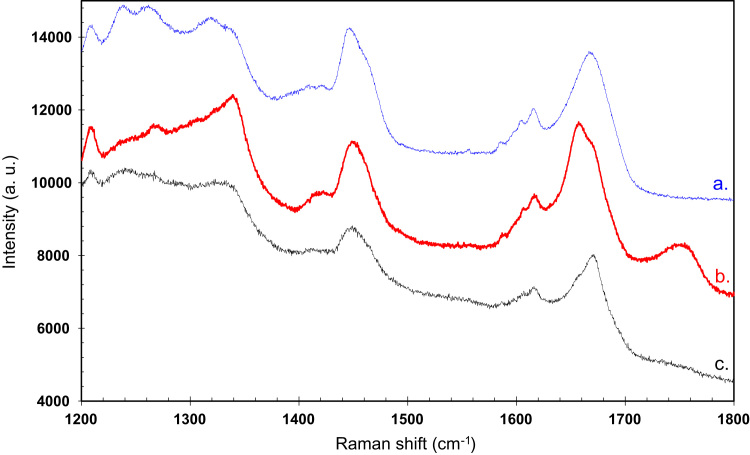
Raman spectrum for Rnase, is presented in Fig. 7. Spectra and curve fitting of the amide I band of proteins corresponding to lysozyme and HSA are presented in Fig. 5, Fig. 6 in Ref. [1], respectively (Fig. 8, Fig. 9, Fig. 10, Fig. 11).
Fig. 7.
Raman spectra of Rnase at different purification steps (range 1200–1800 cm−1). a. Native Rnase (blue spectrum); b. Rnase-TCA (red spectrum) (shifted 1500 arbitrary units (a. u.) downward); c. Rnase-ethanol/HCl (black spectrum) (shifted 600 a. u. upward).
Fig. 8.
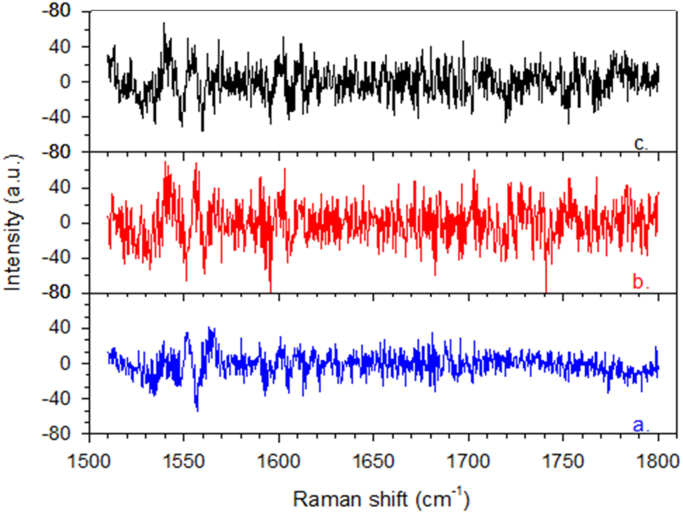
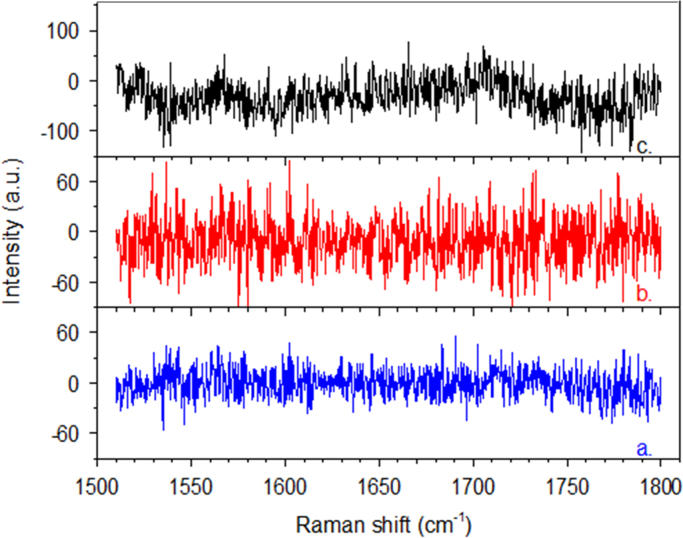
Difference spectra (experimental - fitting curve) after analysis of the amide I Raman bands of lysozyme at different purification steps (Fig. 5, [1]). a. Native lysozyme (blue); b. Lysozyme-TCA (red); c. Lysozyme-ethanol/HCl (black).
Fig. 9.
Difference spectra (experimental – fitting curve) after analysis of the amide I Raman bands of HSA at different purification steps (Fig. 6, [1]). a. Native HSA (blue); b. HSA-TCA (red); c. HSA-ethanol/HCl (black).
Fig. 10.
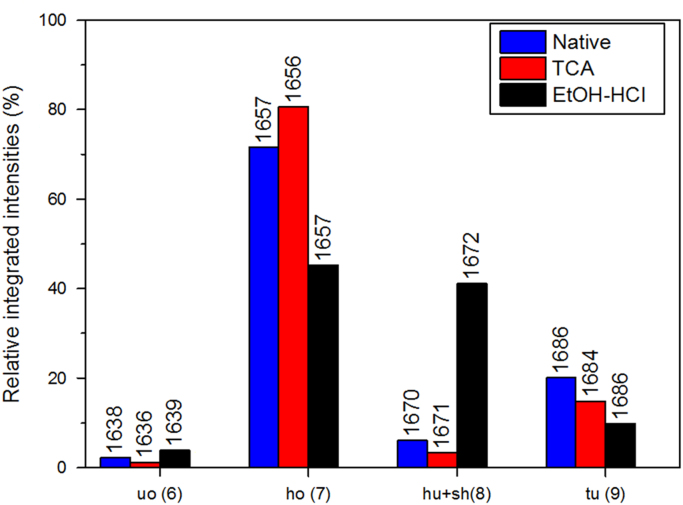
Relative integrated intensities of lysozyme amide I contribution from peak #6 assigned to unordered structures (uo), peak#7 (ordered α helices, ho), peak#8 (unordered α helices and β sheets, hu+sh), and peak #9 (turns, tu) as obtained after profile fitting of amide I region of the Raman spectra (Fig. 5, Ref. [1]). Values on top of each bar correspond to the Raman shift on which the contribution peak was centred at the end of the fitting.
Fig. 11.
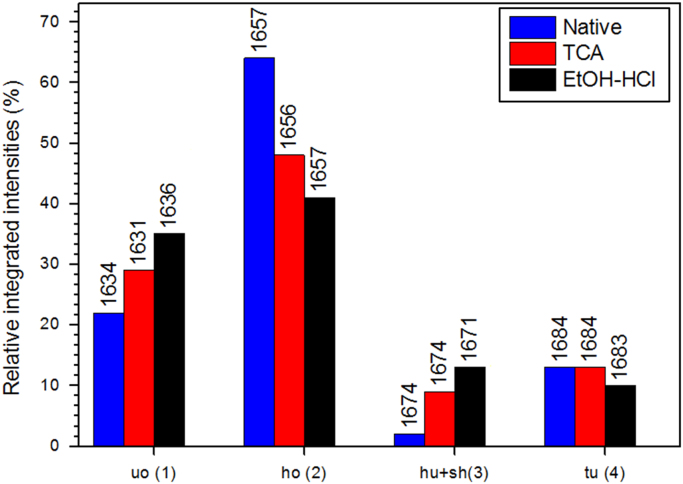
Relative integrated intensities of HSA amide I contribution from peak #1 assigned to unordered structures (uo), peak#2 (ordered α helices, ho), peak#3 (unordered α helices and β sheets, hu+sh), and peak #4 (turns, tu) as obtained after profile fitting of amide I region of the Raman spectra shown in Fig. 6[1]. Values on top of each bar correspond to the Raman shift on which the contribution peak was centred at the end of the fitting.
The unfolding or aggregation of proteins usually involves some dynamic changes in their secondary structures. These changes are mainly monitored by the analysis of the amide I region (1600–1690 cm−1) which is assumed to be sensitive to α-helical secondary structures [8].
1.5. Extraction and purification of proteins from complex samples: LC-HRMS analysis
We present processed data of UPLC- HRMS analysis of proteins from different samples (mouse liver, river biofilm, soil) after TCA precipitation and solvent purification. The datasets in XML format can be used to evaluate ethanol/HCl purification for proteins profiling. Table 1 gives the HRMS features of peptides targeted for the standard proteins after in silico tryptic digestion. Table 2 presents endogenous proteins identified in soil, biofilm and mouse liver samples after purification following either the designed approach or published protocols (Mascot identification). Table 3 presents endogenous proteins detected in the mouse liver sample and quantified through Skyline with corresponding peptides and transitions for PRM. Table 4 presents endogenous proteins detected in the biofilm sample and quantified through Skyline with corresponding peptides and transitions for PRM (Table 5).
Table 1.
HRMS features of peptides targeted for the four standard proteins after in silico tryptic digestion.
| Protein name | Peptide sequence | [M+H]1+ | [M+2H]2+ | [M+3H]3+ | [M+4H]4+ |
|---|---|---|---|---|---|
| LYSO-1 | FESNFNTQATNR | 714.8288 | 476.8883 | ||
| LYSO-2 | HGLDNYR | 874.4166 | 437.7119 | 292.1437 | |
| RNASE-1 | CKPVNTFVHESLADVQAVCS QK | 839.7457 | 630.0611 | ||
| RNASE-2 | HIIVACEGNPYVPVHFDASV | 1112.5464 | 742.0334 | ||
| RNASE-3 | YPNCAYK | 915.4029 | 458.2051 | ||
| HSA-1 | AVMDDFAAFVEK | 671.8210 | 448.2164 | ||
| HSA-2 | LVAASQAALGL | 1013.5990 | 507.3031 | ||
| HSA-3 | YLYEIAR | 927.4934 | 464.2504 | 309.8360 | |
| EXO-1 | GSCSTSSGVPAQVESQSPNA K | 1039.4764 | 693.3200 | ||
| EXO-2 | YGTGYCDSQCPR | 732.2876 | 488.5275 | ||
| EXO-3 | VTFSNIK | 808.4563 | 404.7282 |
Table 2.
Endogenous proteins identified in soil, biofilm and mouse liver after purification following either the designed approach or the published protocols.
| Sample | Location | Protein name | Phylogenetic origin | Protein coverage (%) |
Scorea |
GRAVY | MW (Da)b | ||
|---|---|---|---|---|---|---|---|---|---|
| The designed approach | Published protocol | The designed approach | Published protocol | ||||||
| Soil | Extracellular region | Endoglucanase EG-II | Hypocrea jecorina | 18 | 19 | 161 | 251 | −0.19 | 44883 |
| Extracellular region | Xyloglucanase | Hypocrea jecorina | 1 | 1 | 76 | 114 | −0.21 | 87307 | |
| Biofilm | Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | C-phycoerythrin alpha chain | Microchaete diplosiphon | 29 | 29 | 269 | 239 | −0.15 | 17786 |
| chloroplast thylakoid membrane ; Peripheral membrane protein By similarity; Stromal side | R-phycoerythrin alpha chain | Porphyra purpurea | 20 | 17 | 168 | 119 | −0.19 | 17972 | |
| Cellular thylakoid membrane; Peripheral membrane protein ; Cytoplasmic side | C-phycocyanin-1 alpha chain | Synechococcus sp, | 17 | 17 | 181 | 177 | −0.11 | 17335 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | C-phycoerythrin alpha chain | Synechocystis sp, | 20 | 20 | 209 | 176 | −0.12 | 17756 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | Allophycocyanin alpha chain 1 | Microchaete diplosiphon | 11 | 11 | 76 | 84 | −0.14 | 17411 | |
| chloroplast thylakoid membrane ; Peripheral membrane protein ; Stromal side | B-phycoerythrin beta chain | Porphyridium purpureum | 21 | 20 | 117 | 183 | 0.25 | 18884 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | C-phycoerythrin beta chain | Microchaete diplosiphon | 21 | 16 | 138 | 85 | 0.21 | 19568 | |
| chloroplast thylakoid membrane ; Peripheral membrane protein ; Stromal side | R-phycoerythrin beta chain | Pyropia haitanensis | 23 | 28 | 129 | 144 | 0.26 | 18810 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | C-phycocyanin-1 beta chain | Microchaete diplosiphon | 16 | 12 | 64 | 122 | 0.17 | 18080 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | Allophycocyanin subunit alpha 1 | Nostoc sp, | 17 | 19 | 99 | 112 | −0.09 | 17392 | |
| chloroplast thylakoid membrane ; Peripheral membrane protein ; Stromal side | C-phycocyanin beta chain | Aglaothamnion neglectum | 11 | 12 | 112 | 111 | 0.09 | 18290 | |
| NI | Ribulose bisphosphate carboxylase large chain | Trichodesmium erythraeum | 5 | 8 | 90 | 122 | −0.32 | 53615 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | Allophycocyanin alpha chain | Anabaena cylindrica | 6 | 11 | 84 | 83 | 0.01 | 17128 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | C-phycoerythrin alpha chain | Pseudanabaena tenuis | 18 | 18 | 144 | 126 | −0.24 | 17780 | |
| chloroplast thylakoid membrane ; Multi-pass membrane protein | Photosystem II CP47 reaction center protein | Odontella sinensis | 8 | 8 | 117 | 114 | 0.08 | 56436 | |
| NI | Ribulose bisphosphate carboxylase large chain | Cyanothece sp, | 9 | 6 | 94 | 89 | −0.27 | 53531 | |
| chloroplast | Ribulose bisphosphate carboxylase large chain (Fragment) | Calyptrosphaera sphaeroidea | 5 | 9 | 90 | 107 | −0.10 | 50919 | |
| chloroplast | Ribulose bisphosphate carboxylase large chain | Gracilaria tenuistipitata var, liui | 8 | 10 | 111 | 132 | −0.10 | 54442 | |
| chloroplast | Ribulose bisphosphate carboxylase large chain | Cylindrotheca sp, | 6 | 6 | 109 | 108 | −0.12 | 54400 | |
| chloroplast thylakoid membrane ; Peripheral membrane protein ; Stromal side | Allophycocyanin beta chain | Cyanidium caldarium | 13 | 16 | 94 | 83 | −0.04 | 17574 | |
| chloroplast | Ribulose bisphosphate carboxylase small chain | Antithamnion sp, | 5 | 5 | 72 | 72 | −0.58 | 16247 | |
| NI | Carbon dioxide-concentrating mechanism protein CcmK homolog 1 | Synechocystis sp, | 18 | 29 | 71 | 72 | −0.19 | 11128 | |
| chloroplast thylakoid membrane ; Peripheral membrane protein ; Stromal side | R-phycoerythrin beta chain | Aglaothamnion neglectum | 7 | 7 | 100 | 69 | 0.27 | 18710 | |
| chloroplast | Ribulose bisphosphate carboxylase large chain (Fragment) | Haptolina hirta | 9 | 10 | 141 | 139 | −0.11 | 51098 | |
| chloroplast | Ribulose bisphosphate carboxylase large chain | Antithamnion sp, | 7 | 7 | 117 | 113 | −0.12 | 54372 | |
| Cellular thylakoid membrane ; Peripheral membrane protein ; Cytoplasmic side | Allophycocyanin beta chain | Thermosynechococcus elongatus | 18 | 18 | 103 | 121 | 0.10 | 17462 | |
| Cell inner membrane ; Multi-pass membrane protein | Photosystem I P700 chlorophyll a apoprotein A2 | Gloeobacter violaceus | 2 | 2 | 78 | 75 | 0.15 | 96126 | |
| chloroplast thylakoid membrane; Peripheral membrane protein; Stromal side | Phycobiliprotein ApcE | Aglaothamnion neglectum | 1 | 1 | 73 | 72 | −0.23 | 101319 | |
| NI | Ribulose bisphosphate carboxylase large chain | Synechocystis sp, | 6 | 6 | 120 | 117 | −0.29 | 53084 | |
| chloroplast thylakoid membrane; Peripheral membrane protein; Stromal side | Allophycocyanin beta chain | Galdieria sulphuraria | 16 | 16 | 96 | 73 | 0.02 | 17536 | |
| Mouse liver | Nucleus, Mitochondrion | Carbamoyl-phosphate synthase | Mus musculus | 39 | 33 | 1637 | 1268 | −0.12 | 165711 |
| Cytoplasm | Arginase-1 | Mus musculus | 29 | 35 | 300 | 310 | −0.19 | 34957 | |
| Cytosol, Nucleus,Membrane | Selenium-binding protein | Mus musculus | 31 | 28 | 526 | 405 | −0.31 | 53147 | |
| Cytoplasm | Argininosuccinate synthase | Mus musculus | 32 | 15 | 429 | 191 | −0.11 | 46840 | |
| Mitochondrion | Glyceraldehyde-3-phosphate dehydrogenase | Mus musculus | 31 | 32 | 321 | 298 | −0.04 | 36072 | |
| cytosol | Cytosolic 10-formyltetrahydrofolate dehydrogenase | Mus musculus | 9 | 17 | 139 | 361 | −0.36 | 99502 | |
| Extracellular region | 3-ketoacyl-CoA thiolase, mitochondrial | Mus musculus | 10 | 20 | 137 | 216 | −0.38 | 42260 | |
| Nucleus, Cytoskeleton,Cytosol | Serum albumin | Mus musculus | 15 | 18 | 327 | 349 | −0.09 | 70700 | |
| Cytoplasm | Alcohol dehydrogenase 1 | Mus musculus | 19 | 29 | 161 | 212 | 0.20 | 40601 | |
| membrane | Aspartate aminotransferase, mitochondrial | Mus musculus | 15 | 16 | 231 | 215 | −0.23 | 47780 | |
| Endoplasmic reticulum | Carboxylesterase 3B | Mus musculus | 12 | 14 | 201 | 183 | −0.12 | 63712 | |
| Cytoplasm | Glycine N-methyltransferase | Mus musculus | 29 | 19 | 131 | 127 | −0.25 | 33110 | |
| membrane | Cytochrome P450 2D10 | Mus musculus | 9 | 2 | 100 | 123 | −0.06 | 57539 | |
| Cytoplasm | Aspartate aminotransferase, cytoplasmic | Mus musculus | 7 | 13 | 112 | 115 | −0.25 | 46504 | |
| Cytoplasm | Adenosylhomocysteinase | Mus musculus | 27 | 14 | 335 | 120 | −0.07 | 47780 | |
| Cytosol | Fructose-1,6-bisphosphatase 1 | Mus musculus | 12 | 16 | 117 | 120 | −0.12 | 37288 | |
| Endoplasmic reticulum | Carboxylesterase 3A | Mus musculus | 13 | 9 | 220 | 139 | −0.12 | 63677 | |
| Mitochondrion | Sarcosine dehydrogenase, mitochondrial | Mus musculus | 8 | 6 | 182 | 209 | −0.25 | 102644 | |
| membrane | UDP-glucuronosyltransferase 1-1 | Mus musculus | 4 | 8 | 94 | 141 | 0.09 | 60749 | |
| Cytosol | Hemoglobin subunit beta-1 | Mus musculus | 16 | 24 | 111 | 105 | 0.08 | 15944 | |
| Peroxisome | Peroxisomal bifunctional enzyme | Mus musculus | 3 | 2 | 98 | 78 | −0.12 | 78822 | |
| membrane | Microsomal glutathione S-transferase | Mus musculus | 17 | 21 | 80 | 87 | 0.15 | 17597 | |
| membrane | Cytochrome P450 2F2 | Mus musculus | 6 | 7 | 128 | 130 | −0.13 | 56141 | |
| NI | Pyrethroid hydrolase Ces2a | Mus musculus | 9 | 5 | 100 | 76 | _ | 57539 | |
| Extracellular region | Homogentisate 1,2-dioxygenase | Mus musculus | 6 | 6 | 81 | 114 | −0.34 | 50726 | |
| Cytoplasm | Regucalcin | Mus musculus | 4 | 13 | 72 | 112 | −0.28 | 33899 | |
| Peroxisome | 3-ketoacyl-CoA thiolase B, peroxisomal | Mus musculus | 13 | 8 | 116 | 84 | 0.05 | 44481 | |
| membrane | Sorbitol dehydrogenase | Mus musculus | 6 | 6 | 90 | 89 | 0.06 | 38795 | |
| membrane | ATP synthase subunit f, mitochondrial | Mus musculus | 26 | 26 | 70 | 71 | −0.30 | 10394 | |
| membrane | ATP synthase subunit alpha, mitochondrial | Mus musculus | 14 | 10 | 193 | 160 | −0.10 | 59830 | |
| Cytosol | Urocanate hydratase | Mus musculus | 2 | 1 | 100 | 76 | −0.14 | 75227 | |
| Extracellular region | Fumarylacetoacetase | Mus musculus | 3 | 6 | 75 | 74 | −0.21 | 46488 | |
| Mitochondrion; Peroxisome | Uricase | Mus musculus | 17 | 11 | 157 | 97 | −0.46 | 35245 | |
| Cytoskeleton | Fructose-bisphosphate aldolase B | Mus musculus | 15 | 13 | 180 | 119 | −0.26 | 39938 | |
| membrane | UDP-glucuronosyltransferase 2B17 | Mus musculus | 11 | 6 | 104 | 96 | −0.03 | 61386 | |
| NI | Pyrethroid hydrolase | Mus musculus | 9 | 7 | 108 | 89 | −0.08 | 62356 | |
| Cytoplasm | 3-hydroxyanthranilate 3,4-dioxygenase | Mus musculus | 9 | 6 | 90 | 87 | −0.55 | 32955 | |
| Mitochondrion | Hydroxymethylglutaryl-CoA synthase, mitochondrial | Mus musculus | 7 | 6 | 86 | 70 | −0.34 | 57300 | |
| Mitochondrion | Trifunctional enzyme subunit alpha, mitochondrial | Mus musculus | 9 | 7 | 90 | 81 | −0.10 | 83302 | |
| Endoplasmic reticulum | Microsomal triglyceride transfer protein large subunit | Mus musculus | 1 | 1 | 74 | 80 | −0.16 | 99664 | |
| membrane | Cytochrome b-c1 complex subunit 2, mitochondrial | Mus musculus | 4 | 4 | 73 | 76 | −0.06 | 48262 | |
MASCOT score greater than 67.
MW: Molecular weight.
Table 3.
Endogenous peptides and transitions for PRM methods.
| PRM |
||||
|---|---|---|---|---|
| Protein name | Abreviattion | Peptide | Precursor(m/z) | Product (m/z) |
| Carbamoyl-phosphate synthase | CPSM | TAVDSGIALLTNFQVTK | 898.4844 | 950.5306 |
| 837.4465 | ||||
| 736.3988 | ||||
| VLGTSVESIMATEDR | 804.4009 | 1051.4725 | ||
| 722.3138 | ||||
| 591.2733 | ||||
| AFAMTNQILVER | 696.8688 | 972.5473 | ||
| 516.3140 | ||||
| 403.2300 | ||||
| GQNQPVLNITNR | 677.3653 | 926.5418 | ||
| 617.3365 | ||||
| 390.2096 | ||||
| AADTIGYPVMIR | 653.8448 | 835.4495 | ||
| 615.3647 | ||||
| 472.2402 | ||||
| EPLFGISTGNIITGLAAGAK | 644.0263 | 801.4829 | ||
| 688.3988 | ||||
| 587.3511 | ||||
| IALGIPLPEIK | 582.3735 | 696.4291 | ||
| 355.2340 | ||||
| 468.3180 | ||||
| VMIGESIDEK | 560.7814 | 890.4466 | ||
| 777.3625 | ||||
| 231.1162 | ||||
| SVGEVMAIGR | 509.7711 | 832.4345 | ||
| 646.3705 | ||||
| 547.3021 | ||||
| Argininosuccinate synthase | ASSY | EQGYDVIAYLANIGQK | 891.4571 | 977.5415 |
| 743.4410 | ||||
| 630.3570 | ||||
| FELTCYSLAPQIK | 785.4027 | 1085.4972 | ||
| 556.3453 | ||||
| 485.3082 | ||||
| QHGIPIPVTPK | 593.8508 | 921.5768 | ||
| 751.4713 | ||||
| 541.3344 | ||||
| NQAPPGLYTK | 544.7904 | 846.472 | ||
| 775.4349 | ||||
| 314.1459 | ||||
| YLLGTSLARPCIAR | 530.9643 | 657.8692 | ||
| 601.3271 | ||||
| 277.1547 | ||||
| Selenium-binding protein 2 | SBP2 | GSFVLLDGETFEVK | 770.8983 | 1037.515 |
| 924.4309 | ||||
| 809.404 | ||||
| EEIVYLPCIYR | 727.871 | 984.4971 | ||
| 821.4338 | ||||
| 708.3498 | ||||
| LTGQIFLGGSIVR | 680.901 | 848.4989 | ||
| 701.4304 | ||||
| 588.3464 | ||||
| IYVVDVGSEPR | 617.3273 | 957.5 | ||
| 858.4316 | ||||
| 545.2678 | ||||
| IFVWDWQR | 575.2956 | 889.4315 | ||
| 790.3631 | ||||
| 261.1598 | ||||
| VIEASEIQAK | 544.3033 | 875.4469 | ||
| 746.4043 | ||||
| 675.3672 | ||||
| Glyceraldehyde-3-phosphate dehydrogenase | G3P | VPTPNVSVVDLTCR | 778.9087 | 1259.6412 |
| 949.4771 | ||||
| 630.3243 | ||||
| WGEAGAEYVVESTGVFTTMEK | 764.3561 | 912.4495 | ||
| 892.4123 | ||||
| 756.3597 | ||||
| GAAQNIIPASTGAAK | 685.3753 | 815.4621 | ||
| 702.3781 | ||||
| 668.3726 | ||||
| LISWYDNEYGYSNR | 593.9373 | 1021.4625 | ||
| 539.2572 | ||||
| 376.1939 | ||||
| Arginase-1 | ARGI1 | VMEETFSYLLGR | 722.8607 | 1214.6052 |
| 855.4723 | ||||
| 708.4039 | ||||
| EGLYITEEIYK | 679.3479 | 1058.5405 | ||
| 895.4771 | ||||
| 782.3931 | ||||
| VSVVLGGDHSLAVGSISGHAR | 673.3641 | 866.9581 | ||
| 817.4239 | ||||
| 760.8819 | ||||
| SLEIIGAPFSK | 581.3293 | 606.3246 | ||
| 556.3341 | ||||
| 478.266 | ||||
Table 4.
Endogenous peptides and transitions for PRM methods.
| PRM |
||||
|---|---|---|---|---|
| Protein name | Abreviattion | Peptide | Precursor(m/z) | Product (m/z) |
| R-phycoerythrin alpha chain, Porphyra purpurea | PHEA_PORPU | SVITTTISAADAAGR | 717.3834 | 1134.5749 |
| 1033.5273 | ||||
| 374.2146 | ||||
| FPSSSDLESVQGNIQR | 588.6235 | 715.3846 | ||
| 621.2515 | ||||
| 587.3260 | ||||
| NPGEAGDSQEK | 566.2493 | 920.3956 | ||
| 663.2944 | ||||
| 491.2460 | ||||
| C-phycocyanin-1 alpha chain, Synechococcus sp. | PHCA1_SYNP6 | TPLTEAVAAADSQGR | 743.8784 | 1175.5651 |
| 945.4748 | ||||
| 775.3693 | ||||
| FLSSTELQVAFGR | 727.8855 | 1194.6113 | ||
| 1107.5793 | ||||
| 790.457 | ||||
| C-phycoerythrin alpha chain, Synechocystis sp. | PHEA_SYNY1 | TLGLPTAPYVEALSFAR | 602.6647 | 1152.6048 |
| 793.4203 | ||||
| 664.3777 | ||||
| FPSTSDLESVQGSIQR | 584.2917 | 688.3737 | ||
| 635.2671 | ||||
| 560.3151 | ||||
| C-phycoerythrin alpha chain, Microchaete diplosiphon | PHEA_MICDP | SVVTTVIAAADAAGR | 701.3834 | 1116.6008 |
| 815.437 | ||||
| 374.2146 | ||||
| ALGLPTAPYVEALSFAR | 592.6612 | 1152.6048 | ||
| 793.4203 | ||||
| 664.3777 | ||||
| FPSTSDLESVQGSIQR | 584.2917 | 688.3737 | ||
| 635.2671 | ||||
| 560.3151 | ||||
Table 5.
Total spectrum, peptide and protein counts after purification by our approach versus published protocols on complex matrices.
| Total spectrum count | Peptide count | Protein count | |
|---|---|---|---|
| Biofilm-published approacha | 932 | 585 | 195 |
| Biofilm-our approacha | 937 | 424 | 163 |
| Mouse liver-published approacha | 1122 | 1408 | 416 |
| Mouse liver-our approacha | 959 | 1205 | 355 |
| Soil-published approachb | 946 | 293 | 72 |
| Soil-our approachb | 932 | 488 | 128 |
Data from the ProteomeXchange Consortium via the PRIDE [10] repository with the dataset identifier PXD0081110 and 10.6019/PXD008110.
Average of three replicates.
Counts of a single replicate.
2. Experimental design, materials and methods
Experimental design and materials and methods have been reported previously [1].
Acknowledgments
This research was carried out with the financial support of the French Ministère de l'Enseignement Supérieur et de la Recherche (5HU66) and the Ligue contre le Cancer (Maj 06-12-2016)
References
- 1.Eddhif B., Lange J., Guignard N., Batonneau Y., Papot S., Geffroy-Rodier C., Poinot P. Study of a novel agent for TCA precipitated proteins washing - comprehensive insights into the role of ethanol/HCl on molten globule state by multi-spectroscopic analyses. J. Proteomics. 2018;173:77–88. doi: 10.1016/j.jprot.2017.11.016. (submitted for publication) [DOI] [PubMed] [Google Scholar]
- 2.Chourey K., Jansson J., VerBerkmoes N., Shah M., Chavarria K.L., Tom L.M., Brodie E.L., Hettich R.L. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J. Proteome. Res. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- 3.Huang H.-J., Chen W.-Y., Wu J.-H. Total protein extraction for metaproteomics analysis of methane producing biofilm: the effects of detergents. Int. J. Mol. Sci. 2014;15:10169–10184. doi: 10.3390/ijms150610169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song S., Hooiveld G.J., Zhang W., Li M., Zhao F., Zhu J., Xu X., Muller M., Li C., Zhou G. Comparative proteomics provides insights into metabolic responses in rat liver to isolated soy and meat proteins. J. Proteome Res. 2016;15:1135–1142. doi: 10.1021/acs.jproteome.5b00922. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L., He L., Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J. Chromatogr. A. 2004;1023:317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Fic E., Kedracka-Krok S., Jankowska U., Pirog A., Dziedzicka-Wasylewska M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 2010;31:3573–3579. doi: 10.1002/elps.201000197. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs D.I., van Rijssen M.S., van der Heijden R., Verpoorte R. Sequential solubilization of proteins precipitated with trichloroacetic acid in acetone from cultured Catharanthus roseus cells yields 52% more spots after two-dimensional electrophoresis. Proteomics. 2001;1:1345–1350. doi: 10.1002/1615-9861(200111)1:11<1345::AID-PROT1345>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Rygula A., Majzner K., Marzec K.M., Kaczor A., Pilarczyk M., Baranska M. Raman spectroscopy of proteins: a review: raman spectroscopy of proteins. J. Raman Spectrosc. 2013;44:1061–1076. [Google Scholar]
- 9.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]