Abstract
This article contains further data and information from our published manuscript [1]. We aim to identify significant transcriptome alterations of vascular smooth muscle cells (VSMCs) in the aortic wall of myocardial infarction (MI) patients. Microarray gene analysis was applied to evaluate VSMCs of MI and non-MI patients. Prediction Analysis of Microarray (PAM) identified genes that significantly discriminated the two groups of samples. Incorporation of gene ontology (GO) identified a VSMCs-associated classifier that discriminated between the two groups of samples. Mass spectrometry-based iTRAQ analysis revealed proteins significantly differentiating these two groups of samples. Ingenuity Pathway Analysis (IPA) revealed top pathways associated with hypoxia signaling in cardiovascular system. Enrichment analysis of these proteins suggested an activated pathway, and an integrated transcriptome-proteome pathway analysis revealed that it is the most implicated pathway. The intersection of the top candidate molecules from the transcriptome and proteome highlighted overexpression.
Specifications Table
| Subject area | Biology |
| More specific subject area | Genomics, Proteomics, Bioinformatics, Cardiovascular |
| Type of data | Tables, figures |
| How data was acquired | Microarray (Gene Titan Instrument, Affymetrix), mass spectrometry (LC-MS/MS system comprised of a Dionex Ultimate 3000 RSLC nano-HPLC system, coupled to an online Q-Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Hudson, NH, USA)), RT-qPCR (QuantStudio™ 12K Flex system (Life Technologies; Thermo Fisher Scientific Inc, USA)) |
| Data format | Raw, analyzed |
| Experimental factors | Laser capture microdissection, total RNA extraction and protein extraction from aortic tissues from surgical patients |
| Experimental features | Data analysis with Principal Component Analysis (PCA), Prediction Analysis of Microarray (PAM), Gene Ontology (GO), Ingenuity Pathway Analysis (IPA) |
| Data source location | Singapore |
| Data accessibility | Data is with this article. |
Value of the data
-
•
Combination of multiple technologies and bioinformatics analysis performed in this study reveals the molecular changes induced by myocardial infarction on aortic smooth cells in humans.
-
•
The alterations of the VSMCs transcriptome are congruent with alterations at the protein levels. Both levels show notably the up-regulation of the superoxide dismutase (SOD) with the activation of superoxide radical degradation pathway.
-
•
Differentially expressed genes and pathways identified in these comparisons may be used in future experiments investigating response in myocardial infarction.
1. Data
1.1. Clinical analysis
The characteristics of the myocardial infarction (MI) and non-MI samples undergoing transcriptomics and proteomics studies are presented in Table 1A, Table 1B respectively. The baseline demographic and clinical characteristics of samples undergoing transcriptomics study were compared with those of the samples from the proteomics study (Table 2). In addition, the characteristics of the transcriptomic MI and non-MI samples with those of the independent cohorts comprising additional MI and non-MI patients undergoing RT-qPCR were compared (Table 3, Table 4).
Table 1A.
Demographic characteristics of MI and non-MI groups undergoing transcriptomics analysis.
| Characteristics | Transcriptomics | Transcriptomics | p-value | |
|---|---|---|---|---|
| MI | Non-MI | |||
| (n=17) | (n=19) | |||
| Ethnic | Chinese | 12 | 10 | 0.557 |
| Malay | 2 | 6 | ||
| Indian | 2 | 2 | ||
| Others | 1 | 1 | ||
| Gender | Male | 14 | 16 | 0.881 |
| Female | 3 | 3 | ||
| Age (Mean ± SD) | 59.53 ± 8.28 | 59.68 ± 8.85 | 0.957 | |
| Ejection Fraction | Good (>45%) | 11 | 13 | 0.292 |
| Fair (30–45%) | 4 | 6 | ||
| Poor (<30%) | 2 | 0 | ||
| Smoking | No | 8 | 9 | 0.985 |
| Yes | 9 | 10 | ||
| Renal Impairment | No | 15 | 19 | 0.124 |
| Yes | 2 | 0 | ||
| Diabetes Mellitus | No | 9 | 7 | 0.332 |
| Yes | 8 | 12 | ||
| Hypertension | No | 1 | 3 | 0.345 |
| Yes | 16 | 16 | ||
| Hyperlipidaemia | No | 0 | 0 | – |
| Yes | 17 | 19 | ||
| Antihyperlipidemic Medication | No | 0 | 0 | – |
| Yes | 17 | 19 | ||
| Troponin I (µg/L) | 12.20 ± 20.86 | 0.01 ± 0.004 | <0.05 | |
| (Mean ± SD) | (n=15) | (n=4) |
Table 1B.
Demographic characteristics of MI and non-MI proteomics groups.
| Characteristics | Proteomics | Proteomics | p-value | |
|---|---|---|---|---|
| MI | Non-MI | |||
| n=25 | n=25 | |||
| Ethnic | Chinese | 11 | 13 | 0.745 |
| Malay | 8 | 7 | ||
| Indian | 5 | 5 | ||
| Others | 1 | 0 | ||
| Gender | Male | 20 | 18 | 0.508 |
| Female | 5 | 7 | ||
| Age (Mean ± SD) | 60.88±12.34 | 61.68±8.26 | 0.789 | |
| Ejection Fraction | Good (>45%) | 14 | 16 | 0.344 |
| Fair (30–45%) | 9 | 9 | ||
| Poor (<30%) | 2 | 0 | ||
| Smoking | No | 12 | 12 | 1 |
| Yes | 13 | 13 | ||
| Renal Impairment | No | 25 | 25 | NA |
| Yes | 0 | 0 | ||
| Diabetes Mellitus | No | 10 | 9 | 0.771 |
| Yes | 15 | 16 | ||
| Hypertension | No | 3 | 1 | 0.297 |
| Yes | 22 | 24 | ||
| Hyperlipidaemia | No | 1 | 0 | 0.312 |
| Yes | 24 | 25 | ||
| Antihyperlipidemic Medication | No | 4 | 1 | 0.157 |
| Yes | 21 | 24 | ||
| Troponin I (µg/L) | 19.54 ± 19.24 | 0.015 ± 0.006 | <0.05 | |
| (Mean ± SD) | (n=22) | (n=9) |
Table 2.
Comparative demographic characteristics of transcriptomics and proteomics groups.
| Characteristics | Transcriptomics | Proteomics | p-value | |
|---|---|---|---|---|
| n=36 | n=50 | |||
| Ethnic | Chinese | 22 | 24 | 0.404 |
| Malay | 8 | 15 | ||
| Indian | 4 | 10 | ||
| Others | 2 | 1 | ||
| Gender | Male | 30 | 38 | 0.41 |
| Female | 6 | 12 | ||
| Age (Mean ± SD) | 59.61 ± 8.47 | 61.28 ± 10.40 | 0.431 | |
| Ejection Fraction | Good (>45%) | 24 | 30 | 0.708 |
| Fair (30–45%) | 10 | 18 | ||
| Poor (<30%) | 2 | 2 | ||
| Smoking | No | 17 | 24 | 0.943 |
| Yes | 19 | 26 | ||
| Renal Impairment | No | 34 | 50 | 0.092 |
| Yes | 2 | 0 | ||
| Diabetes Mellitus | No | 16 | 19 | 0.548 |
| Yes | 20 | 31 | ||
| Hypertension | No | 4 | 4 | 0.624 |
| Yes | 32 | 46 | ||
| Hyperlipidaemia | No | 0 | 1 | 0.393 |
| Yes | 36 | 49 | ||
| Antihyperlipidemic Medication | No | 0 | 5 | 0.051 |
| Yes | 36 | 45 | ||
| Troponin I (µg/L) | 9.63 ±19.09 | 13.87 ± 18.45 | 0.445 | |
| (Mean ± SD) | (n=19) | (n=31) |
Table 3.
Demographic characteristics of MI study group and MI validation group.
| Characteristics | MI Microarray | MI Validation | p-value | |
|---|---|---|---|---|
| (n=17) | (n=20) | |||
| Ethnic | Chinese | 12 | 14 | 0.662 |
| Malay | 2 | 4 | ||
| Indian | 2 | 2 | ||
| Others | 1 | 0 | ||
| Gender | Male | 14 | 17 | 0.828 |
| Female | 3 | 3 | ||
| Age (Mean ± SD) | 59.53 ± 8.28 | 61.40 ± 7.88 | 0.487 | |
| Ejection Fraction | Good (>45%) | 11 | 10 | 0.661 |
| Fair (30–45%) | 4 | 7 | ||
| Poor (<30%) | 2 | 3 | ||
| Smoking | No | 8 | 8 | 0.666 |
| Yes | 9 | 12 | ||
| Renal Impairment | No | 16 | 20 | 0.272 |
| Yes | 1 | 0 | ||
| Diabetes Mellitus | No | 9 | 6 | 0.157 |
| Yes | 8 | 14 | ||
| Hypertension | No | 1 | 4 | 0.211 |
| Yes | 16 | 16 | ||
| Hyperlipidaemia | No | 0 | 1 | 0.35 |
| Yes | 17 | 19 | ||
| Antihyperlipidemic Medication | No | 0 | 1 | 0.35 |
| Yes | 17 | 19 | ||
| Troponin I (µg/L) | 12.20 ± 20.86 | 20.94 ± 27.80 | 0.319 | |
| (Mean ± SD) | (n=15) | (n=17) |
Table 4.
Demographic characteristics of non-MI study group and non-MI validation group.
| Characteristics | Non-MI Microarray | Non-MI Validation | p-value | |
|---|---|---|---|---|
| (n=19) | (n=20) | |||
| Ethnic | Chinese | 10 | 12 | 0.763 |
| Malay | 6 | 6 | ||
| Indian | 2 | 2 | ||
| Others | 1 | 0 | ||
| Gender | Male | 16 | 17 | 0.946 |
| Female | 3 | 3 | ||
| Age (Mean ± SD) | 59.68 ± 8.85 | 59.95 ± 8.34 | 0.924 | |
| Ejection Fraction | Good (>45%) | 13 | 13 | 0.083 |
| Fair (30–45%) | 6 | 3 | ||
| Poor (<30%) | 0 | 4 | ||
| Smoking | No | 9 | 11 9 | 0.634 |
| Yes | 10 | |||
| Renal Impairment | No | 18 | 15 | 0.088 |
| Yes | 1 | 5 | ||
| Diabetes Mellitus | No | 7 | 8 | 0.839 |
| Yes | 12 | 12 | ||
| Hypertension | No | 3 | 4 | 0.732 |
| Yes | 16 | 16 | ||
| Hyperlipidaemia | No | 0 | 0 | – |
| Yes | 19 | 20 | ||
| Antihyperlipidemic Medication | No | 0 | 0 | – |
| Yes | 19 | 20 | ||
| Troponin I (µg/L) | 0.01 ± 0.004 | NA | NA | |
| (Mean ± SD) | (n=4) |
1.2. Gene expression data analysis and class prediction by Prediction Analysis of Microarray (PAM)
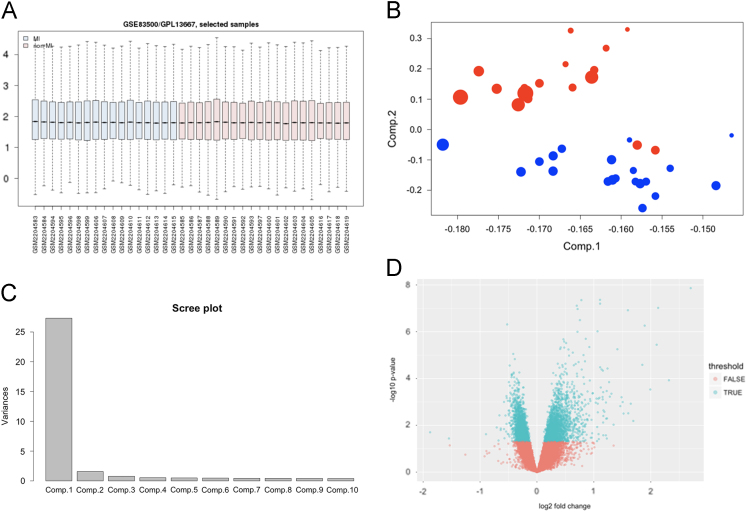
The samples were preprocessed through several steps, including quality assessment and outlier identification, normalization, batch effect correction and evaluation (Fig. 1). To interrogate differentially expressed genes between MI and non-MI we conducted gene-expression profiling using the Affymetrix U219 microarray platform. The R ‘limma’ package (https://www.bioconductor.org/help/workflows/arrays/) identified 4,357 probe sets, selected at a ‘limma’-defined p-value < 0.05. Based on this set of differentially expressed genes (DEGs), we performed principal component analysis (PCA) (Fig. 1).
Fig. 1.
(A) Normalized data. (B) Pseudo three dimensional plot of PCA analysis of the 4,357 DEGs between MI (red) and non-MI (blue). The sizes of the dot represent the loading values of the Comp.3 that perpendicular on the Comp.1 and Comp.2 plane. (C) Scree plot shows the variances explained by the individual principle component. (D) Volcano plot of expression data. Green dot represents differentially expressed genes.
To determine subgroup of genes distinguishing MI from non-MI subjects, we performed supervised PAM [2] and identified a set of differentially expressed genes (DEGs) that discriminated between the two subtypes at Wilcox FDR < 0.1 (Table 5).
Table 5.
List of differentially expressed transcripts.
| Probe ID | Gene symbol | Up/down regulated in MI | wilcox | wilcox FDR |
| 11760204_x_at | CKMT1B | Upregulated inMI | 0.00035204 | 0.00224576 |
| 11760991_a_at | CKMT1B | Upregulated in MI | 0.00101536 | 0.00364739 |
| 11718483_s_at | UBE2N | Upregulated in MI | 0.0001261 | 0.00150508 |
| 11752082_a_at | CDH12 | Upregulated in MI | 0.00010799 | 0.00137774 |
| 11744327_x_at | UBB | Upregulated in MI | 1.4127E-06 | 0.00026136 |
| 11735320_a_at | RBMS3 | Upregulated in MI | 4.8014E-05 | 0.00093501 |
| 11743116_s_at | KPNB1 | Upregulated in MI | 4.2217E-06 | 0.0003841 |
| 11716989_s_at | PNRC2 | Upregulated in MI | 0.00053133 | 0.00252042 |
| 11761378_at | NAALADL2 | Upregulated in MI | 0.0006064 | 0.00276999 |
| 11754075_s_at | KRT222 | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11718123_at | AIMP1 | Upregulated in MI | 0.00010799 | 0.00137774 |
| 11721001_at | HEXIM1 | Upregulated in MI | 0.00456914 | 0.00871434 |
| 11759202_s_at | C9orf41 | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11758784_at | UBE3A | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11764120_at | – | Upregulated in MI | 0.00017084 | 0.0016669 |
| 11729194_s_at | MYNN | Upregulated in MI | 9.2259E-05 | 0.00137774 |
| 11757837_x_at | HNRNPA1 | Upregulated in MI | 0.00078736 | 0.0030992 |
| 11721747_a_at | ANKRD12 | Upregulated in MI | 0.00115015 | 0.00390417 |
| 11753179_s_at | FAM134B | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11757794_s_at | PAPD5 | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11732126_x_at | UBB | Upregulated in MI | 0.00014694 | 0.0016669 |
| 11729661_a_at | GLB1 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11717422_s_at | RBM8A | Upregulated in MI | 0.0001261 | 0.00150508 |
| 11758158_s_at | FOXP1 | Upregulated in MI | 0.00078736 | 0.0030992 |
| 11743098_a_at | TARSL2 | Upregulated in MI | 0.00687991 | 0.01183985 |
| 11715501_s_at | IGFBP7 | Upregulated in MI | 9.2259E-05 | 0.00137774 |
| 11725969_a_at | THUMPD1 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11760913_at | ASAH2 | Upregulated in MI | 2.0029E-05 | 0.00061757 |
| 11718344_a_at | CNOT7 | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11735389_at | CYLC2 | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11747800_a_at | HIF1A | Upregulated in MI | 0.04043046 | 0.0488865 |
| 11721215_a_at | TMEM106B | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11755052_s_at | GYPE | Upregulated in MI | 0.00130072 | 0.00422165 |
| 11717433_a_at | ECHDC1 | Upregulated in MI | 4.8014E-05 | 0.00093501 |
| 11752628_a_at | SYNCRIP | Upregulated in MI | 0.00376444 | 0.00791387 |
| 11747485_a_at | SR140 | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11715657_a_at | GLB1 | Upregulated in MI | 0.00130072 | 0.00422165 |
| 11757402_s_at | MED13 | Upregulated in MI | 0.00026521 | 0.00192411 |
| 11730368_at | ZNF557 | Upregulated in MI | 0.02104398 | 0.028626 |
| 11718577_s_at | NET1 | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11732339_at | BCL11A | Upregulated in MI | 0.02493936 | 0.03283831 |
| 11744873_a_at | KLKP1 | Upregulated in MI | 0.00089487 | 0.00334446 |
| 11763952_at | – | Upregulated in MI | 0.00030585 | 0.00205755 |
| 11726870_at | SETBP1 | Upregulated in MI | 0.0006064 | 0.00276999 |
| 11727433_s_at | NUTF2 | Upregulated in MI | 0.02423264 | 0.03248578 |
| 11726614_at | CDH2 | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11744433_x_at | AMY2B | Upregulated in MI | 3.4121E-05 | 0.00084164 |
| 11720250_a_at | RWDD1 | Upregulated in MI | 0.00010799 | 0.00137774 |
| 11724140_a_at | CRIPAK | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11754192_s_at | SFRS11 | Upregulated in MI | 2.3988E-05 | 0.00068274 |
| 11758666_s_at | LOC284861 | Upregulated in MI | 0.0006064 | 0.00276999 |
| 11764171_s_at | DCUN1D1 | Upregulated in MI | 0.00687991 | 0.01183985 |
| 11751513_a_at | TXNDC6 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11758715_s_at | DEFB126 | Upregulated in MI | 0.00502731 | 0.00926328 |
| 11755681_x_at | HMGB1 | Upregulated in MI | 9.4442E-06 | 0.00038514 |
| 11737234_s_at | LOC162632 | Upregulated in MI | 0.04365745 | 0.05177326 |
| 11751041_x_at | PCMTD2 | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11720954_s_at | RPL30 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11732933_a_at | RUNX1 | Upregulated in MI | 0.00115015 | 0.00390417 |
| 11750545_a_at | CNOT7 | Upregulated in MI | 0.01229819 | 0.01880302 |
| 11716615_s_at | REEP5 | Upregulated in MI | 0.01477913 | 0.02163607 |
| 200037_PM_s_at | CBX3 | Upregulated in MI | 0.00561109 | 0.01012733 |
| 11749445_a_at | ARHGAP15 | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11719713_a_at | PPM1B | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11725073_s_at | PHF17 | Upregulated in MI | 0.02104398 | 0.028626 |
| 11715490_s_at | AMY1A | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11757108_a_at | GSTTP1 | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11758637_x_at | AMY1A | Upregulated in MI | 0.0147944 | 0.02163607 |
| 11743386_s_at | PRPF40A | Upregulated in MI | 0.00010799 | 0.00137774 |
| 11719932_x_at | KIAA0319L | Upregulated in MI | 0.01229819 | 0.01880302 |
| 11750815_s_at | DDX5 | Upregulated in MI | 0.03190866 | 0.04002103 |
| 11761866_at | NCOA7 | Upregulated in MI | 0.01229819 | 0.01880302 |
| 11762842_s_at | PLEKHB2 | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11758021_s_at | DDX3× | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11755779_a_at | ADAMTS20 | Upregulated in MI | 0.00294638 | 0.00689976 |
| 11734919_s_at | TCEA1 | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11741476_x_at | MAP3K7 | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11745795_s_at | DDX5 | Upregulated in MI | 0.01018421 | 0.01603471 |
| 11746794_a_at | C5orf33 | Upregulated in MI | 0.01617252 | 0.0232834 |
| 11758181_s_at | HMGB1 | Upregulated in MI | 0.01119828 | 0.01740908 |
| 11758811_x_at | HNRNPA1 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11761671_a_at | ETV7 | Upregulated in MI | 0.00186355 | 0.00526347 |
| 11758000_s_at | CXADR | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11733216_s_at | USP53 | Upregulated in MI | 0.01348981 | 0.02012593 |
| 200012_PM_x_at | RPL21 | Upregulated in MI | 5.1905E-06 | 0.0003841 |
| 11722616_at | UBLCP1 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11743186_a_at | KIAA1430 | Upregulated in MI | 0.01018421 | 0.01603471 |
| 11758697_x_at | RPL10A | Upregulated in MI | 0.00294638 | 0.00689976 |
| 11758663_s_at | MATR3 | Upregulated in MI | 0.00062978 | 0.00284168 |
| 11739605_a_at | CCDC88A | Upregulated in MI | 0.00839186 | 0.01392372 |
| 11718654_s_at | PKD2 | Upregulated in MI | 0.01119828 | 0.01740908 |
| 11740007_at | POLR3G | Upregulated in MI | 0.00839186 | 0.01392372 |
| 11723448_x_at | MALL | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11725386_a_at | HOMER1 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11721237_a_at | LHFPL2 | Upregulated in MI | 0.00925052 | 0.01488126 |
| 11740255_x_at | UBE2NL | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11763843_a_at | UACA | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11755332_a_at | TJAP1 | Upregulated in MI | 0.00019528 | 0.0016669 |
| 11731400_s_at | TMCO1 | Upregulated in MI | 0.00089487 | 0.00334446 |
| 11755203_x_at | RPL21 | Upregulated in MI | 7.8646E-05 | 0.00137774 |
| 11748647_a_at | PTPRR | Upregulated in MI | 0.02710422 | 0.03543661 |
| 11751975_a_at | SGIP1 | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11739563_a_at | ITPR1 | Upregulated in MI | 0.00010799 | 0.00137774 |
| 11719614_a_at | LARP4 | Upregulated in MI | 0.00030585 | 0.00205755 |
| 11722359_x_at | EPB41L2 | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11754410_s_at | APLNR | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11739606_x_at | CCDC88A | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11737468_a_at | PDC | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11732569_at | SLCO1B3 | Upregulated in MI | 0.04365745 | 0.05177326 |
| 11733180_a_at | ETV1 | Upregulated in MI | 0.02493936 | 0.03283831 |
| 11742053_a_at | COG5 | Upregulated in MI | 0.05074665 | 0.05885975 |
| 11732720_a_at | EREG | Upregulated in MI | 0.00376444 | 0.00791387 |
| 11722645_s_at | ZNHIT6 | Upregulated in MI | 0.02104398 | 0.028626 |
| 11722842_s_at | ENAH | Upregulated in MI | 1.145E-05 | 0.00038514 |
| 11746051_a_at | HP1BP3 | Upregulated in MI | 0.00666163 | 0.01173715 |
| 11758520_s_at | GUCY1A3 | Upregulated in MI | 0.00209437 | 0.00574011 |
| 11751517_a_at | PRKAA1 | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11757817_s_at | BASP1 | Upregulated in MI | 0.00165565 | 0.00486183 |
| 11719053_s_at | CEP350 | Upregulated in MI | 0.0062173 | 0.0110067 |
| 11763534_x_at | CNTNAP3 | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11715329_at | SLC6A15 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11761881_at | ZNF33A | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11723314_x_at | PXMP2 | Upregulated in MI | 2.476E-07 | 9.1611E-05 |
| 11758802_a_at | ENY2 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11758108_s_at | EFEMP1 | Upregulated in MI | 0.0027968 | 0.00667624 |
| 11739011_s_at | PAFAH1B1 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11749062_a_at | ERG | Upregulated in MI | 0.00103627 | 0.00368673 |
| 11761958_s_at | TRA@ | Upregulated in MI | 0.01348981 | 0.02012593 |
| 11753646_x_at | CFL1 | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11723447_at | MALL | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11725729_s_at | C1orf56 | Upregulated in MI | 0.00040442 | 0.00241348 |
| 11725658_a_at | MTFR1 | Upregulated in MI | 0.00925052 | 0.01488126 |
| 11725496_a_at | AGPAT9 | Upregulated in MI | 0.00687991 | 0.01183985 |
| 11735535_at | ZNF660 | Upregulated in MI | 0.00186355 | 0.00526347 |
| 11754898_a_at | ZNF573 | Upregulated in MI | 0.00839186 | 0.01392372 |
| 11719509_a_at | CSRP2BP | Upregulated in MI | 0.00039848 | 0.00241348 |
| 11729721_s_at | LILRB3 | Upregulated in MI | 5.1905E-06 | 0.0003841 |
| 11728429_a_at | LCOR | Upregulated in MI | 4.053E-05 | 0.00088212 |
| 11750795_a_at | KLHL1 | Upregulated in MI | 1.145E-05 | 0.00038514 |
| 11754487_x_at | C5orf33 | Upregulated in MI | 0.01018421 | 0.01603471 |
| 11727856_s_at | NUP50 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11732303_a_at | CREB1 | Upregulated in MI | 0.00035204 | 0.00224576 |
| 11738720_s_at | OR2T3 | Upregulated in MI | 0.00209437 | 0.00574011 |
| 11718993_at | CRKL | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11727390_a_at | STEAP2 | Upregulated in MI | 0.00121855 | 0.00406184 |
| 11728769_at | ST6GALNAC3 | Upregulated in MI | 0.00760327 | 0.0128457 |
| 11742385_s_at | OR8B2 | Upregulated in MI | 0.00240361 | 0.00617593 |
| 11756285_s_at | IGF2BP3 | Upregulated in MI | 0.02292193 | 0.03084041 |
| 11722814_s_at | RANBP2 | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11751269_a_at | SUPT3H | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11763119_x_at | – | Upregulated in MI | 0.01617252 | 0.0232834 |
| 11721835_s_at | TMEM14B | Upregulated in MI | 0.00186355 | 0.00526347 |
| 11736501_x_at | SS18 | Upregulated in MI | 0.02292193 | 0.03084041 |
| 11717574_s_at | PFN1 | Upregulated in MI | 0.00165565 | 0.00486183 |
| 11757274_s_at | ARGLU1 | Upregulated in MI | 0.00030585 | 0.00205755 |
| 11729916_s_at | ARL5B | Upregulated in MI | 0.01929797 | 0.0270464 |
| 11721216_s_at | TMEM106B | Upregulated in MI | 0.00017084 | 0.0016669 |
| 11759049_at | ACSS3 | Upregulated in MI | 0.08324897 | 0.09305777 |
| 11736190_a_at | OGN | Upregulated in MI | 0.01929797 | 0.0270464 |
| 11734314_at | SPTA1 | Upregulated in MI | 0.00089487 | 0.00334446 |
| AFFX-HUMGAPDH/M33197_ | GAPDH | Upregulated in MI | 0.02942452 | 0.03767153 |
| 11753680_x_at | RPL21 | Upregulated in MI | 4.053E-05 | 0.00088212 |
| 11738693_at | OR13C3 | Upregulated in MI | 0.01348981 | 0.02012593 |
| 11746970_at | NPY6R | Upregulated in MI | 0.04365745 | 0.05177326 |
| 11722149_a_at | YTHDC2 | Upregulated in MI | 0.05462796 | 0.06316358 |
| 11748766_a_at | FBXW7 | Upregulated in MI | 0.00022952 | 0.00176918 |
| 11744244_a_at | MASP1 | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11759047_x_at | ABCB1 | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11733995_x_at | C5orf33 | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11719660_at | ATP1A2 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11732982_at | OR2J2 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11720945_x_at | SNRPA1 | Upregulated in MI | 0.05534295 | 0.06379094 |
| 11759697_at | SLITRK3 | Upregulated in MI | 0.00209437 | 0.00574011 |
| 11757257_at | PISRT1 | Upregulated in MI | 0.04365745 | 0.05177326 |
| 11728052_s_at | FAM126B | Upregulated in MI | 0.01929797 | 0.0270464 |
| 11728076_at | HDAC9 | Upregulated in MI | 0.01980258 | 0.02764888 |
| 11742048_at | ITGB1 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11758636_s_at | ASPN | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11756080_s_at | NUS1 | Upregulated in MI | 0.05074665 | 0.05885975 |
| 11730843_a_at | MXI1 | Upregulated in MI | 0.00265809 | 0.00638632 |
| 11739731_s_at | CSNK1G1 | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11719476_at | C20orf108 | Upregulated in MI | 0.02837207 | 0.03696361 |
| 11722845_s_at | UBE2R2 | Upregulated in MI | 0.03066104 | 0.03911925 |
| 11752838_s_at | CIDEB | Upregulated in MI | 0.05074665 | 0.05885975 |
| 11753282_a_at | CMTM4 | Upregulated in MI | 0.02493936 | 0.03283831 |
| 11759361_at | SHOX | Upregulated in MI | 0.00030585 | 0.00205755 |
| 11742378_a_at | AKR1B10 | Upregulated in MI | 0.01348981 | 0.02012593 |
| 11757489_x_at | RPL22 | Upregulated in MI | 0.00078736 | 0.0030992 |
| 11720443_s_at | BAZ1A | Upregulated in MI | 0.03740276 | 0.04597682 |
| 11756351_x_at | SOD1 | Upregulated in MI | 0.00069158 | 0.00284317 |
| 11716368_x_at | PRR13 | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11741875_x_at | AKTIP | Upregulated in MI | 0.02104398 | 0.028626 |
| 11749630_a_at | KRR1 | Upregulated in MI | 0.02493936 | 0.03283831 |
| 11758560_s_at | HERC4 | Upregulated in MI | 0.00101536 | 0.00364739 |
| 11738204_a_at | SPAM1 | Upregulated in MI | 0.07779419 | 0.08775564 |
| 11757384_x_at | UROD | Upregulated in MI | 0.00022952 | 0.00176918 |
| 11721520_at | ZDHHC17 | Upregulated in MI | 0.04709299 | 0.05549174 |
| 11753061_a_at | SLFN5 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11728110_at | GRIP1 | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11722667_a_at | MAPT | Upregulated in MI | 0.00687991 | 0.01183985 |
| 11718781_s_at | SSBP2 | Upregulated in MI | 2.865E-05 | 0.00075718 |
| 11750759_at | CES4 | Upregulated in MI | 0.00760327 | 0.0128457 |
| 11737583_s_at | SGCD | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11727015_s_at | PAPPA | Upregulated in MI | 0.00186355 | 0.00526347 |
| 11728682_at | KRR1 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11737293_at | TACR3 | Upregulated in MI | 0.02104398 | 0.028626 |
| 11746047_x_at | KGFLP2 | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11734530_x_at | HLA-F | Upregulated in MI | 9.4442E-06 | 0.00038514 |
| 11723507_s_at | ZNF609 | Upregulated in MI | 0.05074665 | 0.05885975 |
| 11742902_s_at | AP3S1 | Upregulated in MI | 0.08324897 | 0.09305777 |
| 11741095_at | CORO2A | Upregulated in MI | 0.00839186 | 0.01392372 |
| 11724290_x_at | ZNF641 | Upregulated in MI | 0.01119828 | 0.01740908 |
| 11738606_a_at | KCTD16 | Upregulated in MI | 0.00115015 | 0.00390417 |
| 11735483_at | LANCL3 | Upregulated in MI | 0.00925052 | 0.01488126 |
| 11739334_a_at | PTPRC | Upregulated in MI | 0.0062173 | 0.0110067 |
| 11735206_at | MMAA | Upregulated in MI | 0.00053077 | 0.00252042 |
| 11741856_s_at | LOC653501 | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11735379_a_at | KIAA1009 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11719268_at | TNNC1 | Upregulated in MI | 0.03456513 | 0.04306093 |
| 11763439_s_at | HNRNPU | Upregulated in MI | 0.00040442 | 0.00241348 |
| 11732224_a_at | ZNF664 | Upregulated in MI | 0.02942452 | 0.03767153 |
| 11730746_s_at | PAIP2 | Upregulated in MI | 0.00035204 | 0.00224576 |
| 11744413_x_at | HSPA1A | Upregulated in MI | 0.01348981 | 0.02012593 |
| 11744535_s_at | TCEB1 | Upregulated in MI | 0.00925052 | 0.01488126 |
| 11728719_a_at | LTBP1 | Upregulated in MI | 0.0357177 | 0.04434747 |
| 11741101_a_at | ZNF655 | Upregulated in MI | 0.00505725 | 0.00926328 |
| 11723042_at | UBE2D1 | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11716750_a_at | CD99L2 | Upregulated in MI | 0.01348981 | 0.02012593 |
| 11754732_a_at | CNDP1 | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11740403_at | C12orf69 | Upregulated in MI | 0.0176765 | 0.02515502 |
| 11761250_x_at | ARL5A | Upregulated in MI | 0.00455191 | 0.00871434 |
| 11763585_s_at | TMPO | Upregulated in MI | 0.08900096 | 0.09859388 |
| 11738200_a_at | CCDC102B | Upregulated in MI | 0.02942452 | 0.03767153 |
| 11718702_a_at | ARFIP1 | Upregulated in MI | 0.0631128 | 0.0718515 |
| 11747428_a_at | CDK20 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11745483_s_at | BECN1 | Upregulated in MI | 0.00026521 | 0.00192411 |
| 11730250_a_at | LNX1 | Upregulated in MI | 0.03190866 | 0.04002103 |
| 11716587_at | AXL | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11758860_at | HNRNPU | Upregulated in MI | 0.01229819 | 0.01880302 |
| 11734908_a_at | CADPS2 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11734244_a_at | ATG10 | Upregulated in MI | 0.01617252 | 0.0232834 |
| 11728312_at | ZNF148 | Upregulated in MI | 0.03190866 | 0.04002103 |
| 11739159_at | FAM8A1 | Upregulated in MI | 0.00146865 | 0.0044541 |
| 11740664_a_at | GPRC6A | Upregulated in MI | 0.00209437 | 0.00574011 |
| 11757874_x_at | PFDN1 | Upregulated in MI | 0.00760327 | 0.0128457 |
| 11756150_at | B2M | Upregulated in MI | 0.02493936 | 0.03283831 |
| 11757936_s_at | GCSH | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11715593_s_at | YWHAH | Upregulated in MI | 0.04043046 | 0.0488865 |
| 11729688_s_at | LYRM7 | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11742446_s_at | FOXD4L2 | Upregulated in MI | 0.00186355 | 0.00526347 |
| 11759938_a_at | ITFG2 | Upregulated in MI | 0.0062173 | 0.0110067 |
| 11754680_a_at | MAP3K2 | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11717578_a_at | VPS26A | Upregulated in MI | 0.03190866 | 0.04002103 |
| 11744671_x_at | CTBP2 | Upregulated in MI | 0.0631128 | 0.0718515 |
| 11751946_a_at | ARHGAP21 | Upregulated in MI | 0.00130072 | 0.00422165 |
| 11729687_at | LYRM7 | Upregulated in MI | 0.02104398 | 0.028626 |
| 11750160_a_at | LSM14A | Upregulated in MI | 0.02942452 | 0.03767153 |
| 11727125_a_at | PVRL3 | Upregulated in MI | 0.03456513 | 0.04306093 |
| 11722445_a_at | TRAPPC5 | Upregulated in MI | 1.145E-05 | 0.00038514 |
| 11757916_s_at | TBX3 | Upregulated in MI | 0.0036725 | 0.00780935 |
| 11739487_s_at | SUZ12 | Upregulated in MI | 0.00395253 | 0.00813896 |
| 11756151_x_at | B2M | Upregulated in MI | 0.01477913 | 0.02163607 |
| 11743427_at | AHCYL1 | Upregulated in MI | 0.03190866 | 0.04002103 |
| 11733078_a_at | PPP1R12A | Upregulated in MI | 0.00925052 | 0.01488126 |
| 11760812_at | C10orf46 | Upregulated in MI | 0.04709299 | 0.05549174 |
| 11758911_at | DYRK2 | Upregulated in MI | 0.08900096 | 0.09859388 |
| 11724018_a_at | LDHC | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11753232_a_at | NDRG1 | Upregulated in MI | 0.00115015 | 0.00390417 |
| 11729110_s_at | ADAMDEC1 | Upregulated in MI | 0.05074665 | 0.05885975 |
| 11757659_x_at | RPL12 | Upregulated in MI | 0.00101536 | 0.00364739 |
| 11754221_s_at | FAM104B | Upregulated in MI | 0.00235021 | 0.00608096 |
| 11753454_a_at | PRKG1 | Upregulated in MI | 0.03740276 | 0.04597682 |
| 11715662_x_at | TMEM189 | Upregulated in MI | 0.00409148 | 0.00813896 |
| 11760194_at | LRRIQ3 | Upregulated in MI | 0.00329182 | 0.00729325 |
| 11761070_at | MRPL20 | Upregulated in MI | 0.01018421 | 0.01603471 |
| 11750330_a_at | MYOCD | Upregulated in MI | 0.01477913 | 0.02163607 |
| 11728272_x_at | ZNF330 | Upregulated in MI | 0.00561109 | 0.01012733 |
| 11743661_a_at | MBNL1 | Upregulated in MI | 0.00414689 | 0.00820508 |
| 11731899_s_at | PPAT | Upregulated in MI | 0.08883741 | 0.09859388 |
| 11725485_at | DIRC2 | Upregulated in MI | 0.0176765 | 0.02515502 |
| 11758273_s_at | ARF6 | Upregulated in MI | 0.00046374 | 0.00245118 |
| 11751364_a_at | SLC47A1 | Upregulated in MI | 0.0026334 | 0.00636836 |
| 11740734_a_at | A2BP1 | Upregulated in MI | 0.02942452 | 0.03767153 |
| 11743792_a_at | TTC39C | Upregulated in MI | 0.02710422 | 0.03543661 |
| AFFX-r2-Ec-bioB-5_at | – | Upregulated in MI | 0.00019823 | 0.0016669 |
| 11752951_x_at | RPL15 | Upregulated in MI | 0.04365745 | 0.05177326 |
| 11720802_s_at | BIN3 | Downregulatedin MI | 9.2259E-05 | 0.00137774 |
| 11760353_at | SAFB2 | Downregulatedin MI | 0.00687991 | 0.01183985 |
| 11754373_a_at | MARK3 | Downregulatedin MI | 0.00046374 | 0.00245118 |
| 11730830_x_at | AMPD2 | Downregulatedin MI | 0.00505725 | 0.00926328 |
| 11737523_x_at | VSIG8 | Downregulatedin MI | 1.145E-05 | 0.00038514 |
| 11759492_at | RPS27L | Downregulatedin MI | 0.00235021 | 0.00608096 |
| 11728009_at | VPREB3 | Downregulatedin MI | 0.00146865 | 0.0044541 |
| 11730408_a_at | C19orf33 | Downregulatedin MI | 0.0385022 | 0.04717157 |
| 11728044_a_at | C14orf126 | Downregulatedin MI | 0.00875884 | 0.01446773 |
| 11763618_a_at | SNAPC4 | Downregulatedin MI | 0.00026521 | 0.00192411 |
| 11716415_s_at | NDUFB8 | Downregulatedin MI | 0.00165565 | 0.00486183 |
| 11737238_s_at | EOMES | Downregulatedin MI | 0.0062173 | 0.0110067 |
| 11760953_x_at | UBXN11 | Downregulatedin MI | 0.00022952 | 0.00176918 |
| 11746163_a_at | LARP4B | Downregulatedin MI | 0.04043046 | 0.0488865 |
| 11724561_x_at | TSR2 | Downregulatedin MI | 0.02292193 | 0.03084041 |
| 11735855_at | TNP2 | Downregulatedin MI | 0.00409148 | 0.00813896 |
| 11757649_a_at | TIMM16 | Downregulatedin MI | 7.8646E-05 | 0.00137774 |
| 11732524_a_at | CHKB-CPT1B | Downregulatedin MI | 0.00358451 | 0.00780935 |
| 11736945_a_at | HIPK4 | Downregulatedin MI | 0.00040442 | 0.00241348 |
| 11716547_s_at | TLR9 | Downregulatedin MI | 0.01477913 | 0.02163607 |
| 11746635_a_at | LEF1 | Downregulatedin MI | 0.00089487 | 0.00334446 |
| 11744612_a_at | NUDCD1 | Downregulatedin MI | 0.00121855 | 0.00406184 |
| 11731356_a_at | IP6K3 | Downregulatedin MI | 0.00078736 | 0.0030992 |
| 11728896_a_at | LRP8 | Downregulatedin MI | 0.01018421 | 0.01603471 |
| 11759458_at | LOC285501 | Downregulatedin MI | 0.01617252 | 0.0232834 |
| 11762010_a_at | GRAMD1B | Downregulatedin MI | 0.00019823 | 0.0016669 |
| 11757381_x_at | GTF2A2 | Downregulatedin MI | 0.0728868 | 0.0824713 |
| 11743193_a_at | PARD6G | Downregulatedin MI | 0.00019823 | 0.0016669 |
| 11726367_a_at | ERICH1 | Downregulatedin MI | 0.03740276 | 0.04597682 |
| 11750342_a_at | FRMPD1 | Downregulatedin MI | 0.08324897 | 0.09305777 |
| 11744231_a_at | MAPK7 | Downregulatedin MI | 0.00294638 | 0.00689976 |
| 11724255_a_at | OAS1 | Downregulatedin MI | 0.0176765 | 0.02515502 |
| 11737305_a_at | FAM166A | Downregulatedin MI | 0.00561109 | 0.01012733 |
| 11733958_a_at | GTPBP3 | Downregulatedin MI | 0.00089487 | 0.00334446 |
| 11739429_a_at | ZDHHC24 | Downregulatedin MI | 0.01900683 | 0.02694455 |
| 11751172_a_at | TRIB3 | Downregulatedin MI | 0.05732237 | 0.06586732 |
| 11737677_at | BTBD18 | Downregulatedin MI | 0.02104398 | 0.028626 |
| 11725393_s_at | MAK16 | Downregulatedin MI | 0.0631128 | 0.0718515 |
| 11744029_a_at | BBS4 | Downregulatedin MI | 0.04043046 | 0.0488865 |
| 11743134_x_at | FKBP8 | Downregulatedin MI | 0.00022952 | 0.00176918 |
| 11745187_a_at | BET1L | Downregulatedin MI | 0.00925052 | 0.01488126 |
| 11737856_a_at | OPCML | Downregulatedin MI | 0.00101536 | 0.00364739 |
| 11759126_a_at | THRA | Downregulatedin MI | 0.04365745 | 0.05177326 |
| 11722129_at | FAM102B | Downregulatedin MI | 0.00165565 | 0.00486183 |
| 11762149_at | C18orf45 | Downregulatedin MI | 0.00115015 | 0.00390417 |
| 11734407_a_at | MATN4 | Downregulatedin MI | 0.00760327 | 0.0128457 |
| 11730872_x_at | RASSF5 | Downregulatedin MI | 0.00409148 | 0.00813896 |
| 11753413_x_at | DLK1 | Downregulatedin MI | 0.07262653 | 0.08242889 |
Gene Ontology (GO) analysis of the DEGs was performed using DAVID Bioinformatics tools [3] (http://david.abcc.ncifcrf.gov/). The GO results for the down-regulated transcripts were not enriched for any GO terms. The GO analysis revealed biological processes (Table 6).
Table 6.
Gene ontology of upregulated genes.
| Category | Term | Count | % | p-value | Genes | List Total | Pop Hits | Pop Total | Fold Enrichmen | tBonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0070647~protein modification by small protein conjugation or removal | 12 | 4.109589 | 4.96E-05 | ENY2, UBE2N, SUZ12, SUPT3H, ATG10, FBXW7, UBE3A, UBB, UBE2D1, TMEM189, FBXW11, LNX1 | 216 | 160 | 13528 | 4.697222222 | 0.08427547 | 0.084275 | 0.084054 |
| GOTERM_BP_FAT | GO:0032446~protein modification by small protein conjugation | 10 | 3.4246575 | 2.56E-04 | UBE2N, SUZ12, ATG10, FBXW7, UBE3A, UBB, UBE2D1, TMEM189, FBXW11, LNX1 | 216 | 132 | 13528 | 4.744668911 | 0.365690828 | 0.203565 | 0.433849 |
| GOTERM_BP_FAT | GO:0016567~protein ubiquitination | 9 | 3.0821918 | 6.19E-04 | UBE2N, SUZ12, FBXW7, UBE3A, UBB, UBE2D1, TMEM189, FBXW11, LNX1 | 216 | 119 | 13528 | 4.736694678 | 0.666812995 | 0.30674 | 1.044247 |
| GOTERM_BP_FAT | GO:0006940~regulation of smooth muscle contraction | 5 | 1.7123288 | 0.00299912 | TACR3, MYOCD, GUCY1A3, ATP1A2, SOD1 | 216 | 38 | 13528 | 8.240740741 | 0.995162875 | 0.736278 | 4.964734 |
| GOTERM_BP_FAT | GO:0051147~regulation of muscle cell differentiation | 5 | 1.7123288 | 0.00330086 | TBX3, MYOCD, EREG, UBB, HDAC9 | 216 | 39 | 13528 | 8.029439696 | 0.99717351 | 0.690793 | 5.451184 |
| GOTERM_BP_FAT | GO:0006414~translational elongation | 7 | 2.3972603 | 0.00544585 | RPL30, RPL22, RPL21, RPL15, UBB, RPL10A, RPL12 | 216 | 101 | 13528 | 4.3406674 | 0.999938274 | 0.801202 | 8.842269 |
| GOTERM_BP_FAT | GO:0006937~regulation of muscle contraction | 6 | 2.0547945 | 0.00571597 | TACR3, MYOCD, TNNC1, GUCY1A3, ATP1A2, SOD1 | 216 | 72 | 13528 | 5.219135802 | 0.999961887 | 0.766265 | 9.261108 |
| GOTERM_BP_FAT | GO:0048742~regulation of skeletal muscle fiber development | 4 | 1.369863 | 0.00703496 | TBX3, MYOCD, UBB, HDAC9 | 216 | 25 | 13528 | 10.02074074 | 0.999996388 | 0.791204 | 11.28038 |
| GOTERM_BP_FAT | GO:0007507~heart development | 10 | 3.4246575 | 0.00750053 | CRKL, TBX3, MYOCD, HEXIM1, TNNC1, PKD2, HDAC9, CXADR, ITGB1, FOXP1 | 216 | 215 | 13528 | 2.913006029 | 0.999998429 | 0.773463 | 11.98298 |
| GOTERM_BP_FAT | GO:0016202~regulation of striated muscle tissue development | 5 | 1.7123288 | 0.00807194 | TBX3, MYOCD, UBB, HDAC9, CXADR | 216 | 50 | 13528 | 6.262962963 | 0.999999435 | 0.762736 | 12.83814 |
| GOTERM_BP_FAT | GO:0048634~regulation of muscle development | 5 | 1.7123288 | 0.00865211 | TBX3, MYOCD, UBB, HDAC9, CXADR | 216 | 51 | 13528 | 6.140159768 | 0.9999998 | 0.753948 | 13.69842 |
| GOTERM_BP_FAT | GO:0048534~hemopoietic or lymphoid organ development | 11 | 3.7671233 | 0.00874561 | PTPRC, CRKL, RPL22, BCL11A, TCEA1, SPTA1, HDAC9, SOD1, RUNX1, ITGB1, FOXP1 | 216 | 260 | 13528 | 2.6497151 | 0.999999831 | 0.727281 | 13.83632 |
| GOTERM_BP_FAT | GO:0030036~actin cytoskeleton organization | 10 | 3.4246575 | 0.01017938 | EPB41L2, PFN1, CALD1, CFL1, PAFAH1B1, ARF6, SPTA1, PRKG1, ITGB1, KLHL1 | 216 | 226 | 13528 | 2.77122255 | 0.999999987 | 0.752662 | 15.92501 |
| GOTERM_BP_FAT | GO:0048641~regulation of skeletal muscle tissue development | 4 | 1.369863 | 0.01066911 | TBX3, MYOCD, UBB, HDAC9 | 216 | 29 | 13528 | 8.638569604 | 0.999999995 | 0.743329 | 16.62747 |
| GOTERM_BP_FAT | GO:0045935~positive regulation of nucleobase, nucleoside, nucleotide and nuc | 19 | 6.5068493 | 0.01072364 | ENY2, BMP3, PTPRC, TBX3, GRIP1, CREB1, MED13, DDX5, CNOT7, UBE2N, YWHAH, HIF1A, MYOCD, EREG, ZNF148, GU | 216 | 624 | 13528 | 1.906991928 | 0.999999995 | 0.720797 | 16.70534 |
| GOTERM_CC_FAT | GO:0030529~ribonucleoprotein complex | 19 | 6.7857143 | 9.63E-04 | KRR1, SNRPA1, ERG, RPL15, SYNCRIP, HSPA1A, DDX5, HNRNPA1, HNRNPU, MRPL20, RPL30, RBM8A, RPL22, PNRC2, R | 197 | 515 | 12782 | 2.393750924 | 0.246766301 | 0.246766 | 1.275853 |
| GOTERM_CC_FAT | GO:0005829~cytosol | 36 | 12.857143 | 9.94E-04 | NAMPT, ENAH, UBE3A, GRIP1, RPL15, MAP3K7, RPL30, MAP3K2, MAPT, GSTZ1, GUCY1A3, PAFAH1B1, PPP3CA, RPL12 | 197 | 1330 | 12782 | 1.756238312 | 0.253459145 | 0.135974 | 1.315771 |
| GOTERM_CC_FAT | GO:0031981~nuclear lumen | 37 | 13.214286 | 0.00232476 | ENY2, SUPT3H, HMGB1, SYNCRIP, ZNF655, CNOT7, ZNF330, CORO2A, DDX3×, RBM8A, ZNF148, NUP50, TCEA1, UBE2D | 197 | 1450 | 12782 | 1.655641519 | 0.495544858 | 0.203949 | 3.053045 |
| GOTERM_CC_FAT | GO:0005794~Golgi apparatus | 25 | 8.9285714 | 0.00378136 | CCDC88A, AIMP1, BECN1, PTPRR, AP3S1, ARF6, ARFIP1, CXADR, PRKG1, GCC2, TJAP1, B2M, ARHGAP21, PNPLA8, ZDHH | 197 | 872 | 12782 | 1.860184883 | 0.671699641 | 0.243049 | 4.921769 |
| GOTERM_CC_FAT | GO:0005654~nucleoplasm | 25 | 8.9285714 | 0.00436162 | ENY2, HMGB1, SUPT3H, SYNCRIP, CNOT7, CORO2A, DDX3×, RBM8A, NUP50, TCEA1, UBE2D1, KPNB1, PRPF40A, POLR | 197 | 882 | 12782 | 1.839094352 | 0.72338246 | 0.22665 | 5.656882 |
| GOTERM_CC_FAT | GO:0030864~cortical actin cytoskeleton | 4 | 1.4285714 | 0.0087726 | EPB41L2, CALD1, CFL1, SPTA1 | 197 | 28 | 12782 | 9.269035533 | 0.925019331 | 0.350631 | 11.07554 |
| GOTERM_CC_FAT | GO:0005635~nuclear envelope | 9 | 3.2142857 | 0.01377595 | UACA, NUP50, CBX3, PAFAH1B1, TMPO, RANBP2, KPNB1, MATR3, ITPR1 | 197 | 205 | 12782 | 2.848532871 | 0.983063526 | 0.441562 | 16.87269 |
| GOTERM_CC_FAT | GO:0070013~intracellular organelle lumen | 39 | 13.928571 | 0.02000434 | ENY2, SUPT3H, HMGB1, SYNCRIP, ZNF655, CNOT7, ZNF330, MRPL20, CORO2A, DDX3×, RBM8A, ZNF148, NUP50, TCEA | 197 | 1779 | 12782 | 1.42239837 | 0.997370329 | 0.524131 | 23.60067 |
| GOTERM_CC_FAT | GO:0044451~nucleoplasm part | 16 | 5.7142857 | 0.02385422 | ENY2, POLR3G, SUPT3H, CTBP2, CREB1, YY1, MED13, CNOT7, SUZ12, CORO2A, PHF17, HIF1A, DDX3×, RBM8A, HDAC9 | 197 | 555 | 12782 | 1.870508072 | 0.99917336 | 0.545557 | 27.50364 |
| GOTERM_CC_FAT | GO:0031965~nuclear membrane | 5 | 1.7857143 | 0.02568978 | CBX3, PAFAH1B1, TMPO, MATR3, ITPR1 | 197 | 73 | 12782 | 4.444058132 | 0.999524672 | 0.534736 | 29.29883 |
| GOTERM_CC_FAT | GO:0043233~organelle lumen | 39 | 13.928571 | 0.0276102 | ENY2, SUPT3H, HMGB1, SYNCRIP, ZNF655, CNOT7, ZNF330, MRPL20, CORO2A, DDX3×, RBM8A, ZNF148, NUP50, TCEA | 197 | 1820 | 12782 | 1.39035533 | 0.999733881 | 0.526841 | 31.13293 |
| GOTERM_CC_FAT | GO:0030054~cell junction | 15 | 5.3571429 | 0.02854188 | ARHGAP21, PTPRC, ENAH, CTBP2, CADPS2, GRIP1, PVRL3, CD99L2, ABCB1, CDH2, HOMER1, CXADR, ITGB1, RIMS1, TJA | 197 | 518 | 12782 | 1.878858554 | 0.999799239 | 0.508085 | 32.00679 |
| GOTERM_CC_FAT | GO:0044445~cytosolic part | 7 | 2.5 | 0.02964735 | RPL30, PFDN1, UACA, RPL22, RPL21, GUCY1A3, UBB | 197 | 152 | 12782 | 2.988044349 | 0.999856352 | 0.4937 | 33.03034 |
| GOTERM_CC_FAT | GO:0005912~adherens junction | 7 | 2.5 | 0.03218777 | PTPRC, ENAH, PVRL3, ABCB1, CDH2, CXADR, ITGB1 | 197 | 155 | 12782 | 2.930211233 | 0.999933536 | 0.496948 | 35.32875 |
| GOTERM_CC_FAT | GO:0031974~membrane-enclosed lumen | 39 | 13.928571 | 0.03603007 | ENY2, SUPT3H, HMGB1, SYNCRIP, ZNF655, CNOT7, ZNF330, MRPL20, CORO2A, DDX3×, RBM8A, ZNF148, NUP50, TCEA | 197 | 1856 | 12782 | 1.363387231 | 0.999979362 | 0.512871 | 38.66672 |
| GOTERM_MF_FAT | GO:0003723~RNA binding | 23 | 8.2142857 | 0.0016071 | KRR1, SNRPA1, AIMP1, CPEB2, RPL15, SYNCRIP, MBNL1, IGF2BP3, DDX5, HNRNPA1, HNRNPU, MRPL20, RPL30, LARP4 | 201 | 718 | 12983 | 2.069104339 | 0.477004104 | 0.477004 | 2.220802 |
| GOTERM_MF_FAT | GO:0016881~acid-amino acid ligase activity | 10 | 3.5714286 | 0.00396374 | UBE2N, AKTIP, UBE3A, HERC4, UBE2NL, UBE2D1, TMEM189, FBXW11, LNX1, UBE2R2 | 201 | 201 | 12983 | 3.213534318 | 0.798217111 | 0.550797 | 5.394694 |
| GOTERM_MF_FAT | GO:0019787~small conjugating protein ligase activity | 9 | 3.2142857 | 0.00418513 | UBE2N, AKTIP, UBE3A, UBE2NL, UBE2D1, TMEM189, FBXW11, LNX1, UBE2R2 | 201 | 166 | 12983 | 3.501978061 | 0.815507665 | 0.43072 | 5.687888 |
| GOTERM_MF_FAT | GO:0003702~RNA polymerase II transcription factor activity | 11 | 3.9285714 | 0.00457311 | SUPT3H, ETV7, HIF1A, TBX3, ZNF148, CREB1, TCEA1, MED13, TCEB1, LCOR, FOXP1 | 201 | 244 | 12983 | 2.911936221 | 0.842320571 | 0.36985 | 6.199674 |
| GOTERM_MF_FAT | GO:0016879~ligase activity, forming carbon-nitrogen bonds | 10 | 3.5714286 | 0.00955683 | UBE2N, AKTIP, UBE3A, HERC4, UBE2NL, UBE2D1, TMEM189, FBXW11, LNX1, UBE2R2 | 201 | 231 | 12983 | 2.796192199 | 0.979140063 | 0.538828 | 12.54851 |
| GOTERM_MF_FAT | GO:0003735~structural constituent of ribosome | 8 | 2.8571429 | 0.01527537 | RPL30, RPL22, RPL21, RPL15, UBB, RPL10A, RPL12, MRPL20 | 201 | 168 | 12983 | 3.075811419 | 0.997977614 | 0.644387 | 19.34096 |
| GOTERM_MF_FAT | GO:0016566~specific transcriptional repressor activity | 4 | 1.4285714 | 0.01770409 | HEXIM1, YY1, HDAC9, FOXP1 | 201 | 36 | 12983 | 7.176893311 | 0.999252415 | 0.642414 | 22.07479 |
| GOTERM_MF_FAT | GO:0030528~transcription regulator activity | 34 | 12.142857 | 0.02428221 | ENY2, SUPT3H, ETV7, GRIP1, CBX3, CNOT7, MXI1, MYOCD, HEXIM1, ZNF148, BCL11A, ETV1, TCEA1, ERG, ZNF33A, SSB | 201 | 1512 | 12983 | 1.452466503 | 0.99995015 | 0.710127 | 29.05338 |
| GOTERM_MF_FAT | GO:0003712~transcription cofactor activity | 12 | 4.2857143 | 0.02515659 | ENY2, SUPT3H, CTBP2, MYOCD, GRIP1, YY1, CREB1, BCL11A, MED13, HDAC9, DDX5, MXI1 | 201 | 363 | 12983 | 2.135274043 | 0.999965267 | 0.680458 | 29.936 |
| GOTERM_MF_FAT | GO:0004842~ubiquitin-protein ligase activity | 7 | 2.5 | 0.0262357 | UBE2N, UBE3A, UBE2NL, UBE2D1, FBXW11, LNX1, UBE2R2 | 201 | 147 | 12983 | 3.075811419 | 0.999977772 | 0.657477 | 31.01122 |
| GOTERM_MF_FAT | GO:0008134~transcription factor binding | 15 | 5.3571429 | 0.02754274 | ENY2, SUPT3H, HMGB1, CTBP2, GRIP1, YY1, CREB1, MED13, MXI1, DDX5, HIF1A, MYOCD, BCL11A, LCOR, HDAC9 | 201 | 513 | 12983 | 1.888656135 | 0.999987063 | 0.640565 | 32.29303 |
| GOTERM_MF_FAT | GO:0003779~actin binding | 11 | 3.9285714 | 0.02969715 | EPB41L2, PFN1, CORO2A, ENAH, YWHAH, CCDC88A, TNNC1, CALD1, CFL1, SPTA1, KLHL1 | 201 | 326 | 12983 | 2.179486006 | 0.999994708 | 0.636668 | 34.3577 |
| GOTERM_MF_FAT | GO:0019899~enzyme binding | 15 | 5.3571429 | 0.03171181 | PTPRC, CCDC88A, PTPRR, CBX3, CDH2, SOD1, RIMS1, PFN1, YWHAH, HIF1A, MAPT, PKD2, PAFAH1B1, RANBP2, HDAC9 | 201 | 523 | 12983 | 1.852544163 | 0.99999771 | 0.631749 | 36.23541 |
| GOTERM_MF_FAT | GO:0008092~cytoskeletal protein binding | 14 | 5 | 0.04821415 | ENAH, CCDC88A, TNNC1, CALD1, KLHL1, EPB41L2, PFN1, CORO2A, YWHAH, MAPT, CFL1, PKD2, PAFAH1B1, SPTA1 | 201 | 504 | 12983 | 1.794223328 | 0.999999998 | 0.758878 | 49.84218 |
| GOTERM_MF_FAT | GO:0016564~transcription repressor activity | 10 | 3.5714286 | 0.05535048 | CTBP2, TBX3, HEXIM1, ZNF148, YY1, BCL11A, CBX3, HDAC9, MXI1, FOXP1 | 201 | 316 | 12983 | 2.044051892 | 1 | 0.783426 | 54.84568 |
Clustering of genes were done by two methods, hierarchical and k-mean clustering. Hierarchical clustering with multiscale bootstrap resampling was done by Pvclust, an R statistical software package [4]. The Pvclust is an R package for assessing the uncertainty in hierarchical cluster analysis. For each cluster in hierarchical clustering, quantities called p-values are calculated via multiscale bootstrap resampling. The parameters (https://cran.r-project.org/web/packages/pvclust/pvclust.pdf) used here were 10000 bootstrap replications, cluster method: Ward algorithm and distance method: Euclidean. For the heat maps plot, we used log2 scale.
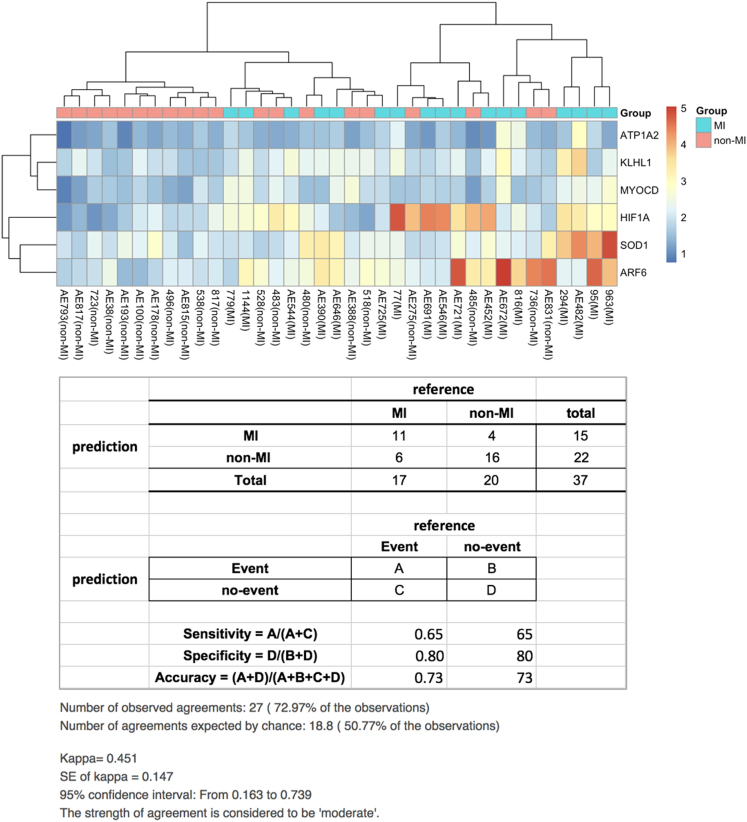
The k-mean clustering was performed by R (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/kmeans.html), showing that the selection of features gave a higher accuracy than PAM alone. The genes were discriminated between the MI and non-MI vascular smooth muscle cells (VSMCs) samples (Table 7). A clustered result is shown in Fig. 2 of Ref. [1].
Table 7.
Contingency table of prediction results from 21 genes.
| Reference | ||||
| MI | Non-MI | Total | ||
| Prediction | MI | 16 | 2 | 18 |
| Non-MI | 1 | 18 | 19 | |
| Total | 17 | 20 | 37 | |
| Reference | ||||
| Event | No-Event | Total | ||
| Prediction | Event | A | B | A+B |
| No-Event | C | D | C+D | |
| Total | A+C | B+D | A+B+C+D | |
| Sensitivity = A/(A+C) | 0.941176 | 94 | ||
| Specificity = D/(B+D) | 0.9 | 90 | ||
| Accuracy = (A+D)/(A+B+C+D) | 0.918919 | 92 | ||
Fig. 2.
Hierarchical clustering on six RT-qPCR-based validation genes.
1.3. Protein processing, electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and LC-MS/MS analysis using Q-Exactive mass spectrometer
Differential expressed proteins identified are shown in Table 8. Only peptides identified with strict spectral false discovery rate of less than 1% (q-value ≤ 0.01) were considered.
Table 8.
Differentially expressed proteins.
 |
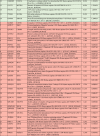 |
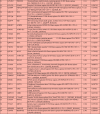 |
1.4. Hierarchical cluster analysis of RT-qPCR-based detected genes
Using six RT-qPCR-supported genes as a representative gene classifier characterizing the differences between MI and non-MI aortic samples, hierarchical clustering was performed with multiscale bootstrap resampling by Pvclust. The result is shown in Fig. 2.
1.5. Transcriptomic and proteomic pathways analysis
Systemic evaluation was performed using IPA (www.ingenuity.com) to identify transcriptomic and proteomic pathways, and significantly enriched canonical pathways are shown in Table 9. An integrated transcriptome-proteome correlation is performed to identify common enriched pathway and molecule (Table 10).
Table 9.
Pathway mapping of 370 transcripts (highlighted in light blue) and 94 proteins (highlighted in yellow).
 |
Table 10.
Pathway mapping of combined 21 gene signature and 94 proteins.
| Ingenuity Canonical Pathways | -log(p-value) | Ratio | Molecules |
|---|---|---|---|
| Superoxide Radicals Degradation | 3.1 | 0.4 | CAT,SOD1 |
| RhoGDI Signaling | 2.11 | 0.046 | ITGB1,RACK1,GDI2,ACTG1 |
| Acetyl-CoA Biosynthesis III (from Citrate) | 2.04 | 1 | ACLY |
| Clathrin-mediated Endocytosis Signaling | 1.9 | 0.04 | ITGB1,UBB,ARF6,ACTG1 |
| Amyotrophic Lateral Sclerosis Signaling | 1.82 | 0.052 | CAT,BID,SOD1 |
| Paxillin Signaling | 1.82 | 0.052 | ITGB1,ARF6,ACTG1 |
| Regulation of eIF4 and p70S6K Signaling | 1.69 | 0.035 | ITGB1,EIF3G,EIF3F,RPS4X |
| Actin Cytoskeleton Signaling | 1.64 | 0.033 | ITGB1,PFN1,SSH3,ACTG1 |
| Mitochondrial Dysfunction | 1.63 | 0.033 | NDUFA9,NDUFV2,CAT,COX5A |
| Crosstalk between Dendritic Cells and Natural Killer Cells | 1.61 | 0.074 | CAMK2D,ACTG1 |
| NRF2-mediated Oxidative Stress Response | 1.57 | 0.032 | UBB,CAT,SOD1,ACTG1 |
2. Experimental design, materials and methods
2.1. Sample collection
Aortic tissue samples were obtained from patients who presented with coronary artery disease undergoing coronary artery bypass graft (CABG) surgery at the National University Hospital of Singapore from 2009 to 2013. Patients underwent CABG either after a recent myocardial infarction (MI group) or as stable angina patients (non-MI group). An aortic punch tissue was collected at the time of proximal anastomosis between the aorta and saphenous vein grafts. The tissues from the aortic punch were immediately preserved on dry ice, and stored in liquid nitrogen tank. The study was approved by the National Healthcare Group Domain Specific Review Board (Tissue Bank registration: NUH/2009-0073), and written informed consent was obtained from all patients. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
2.2. Sample grouping
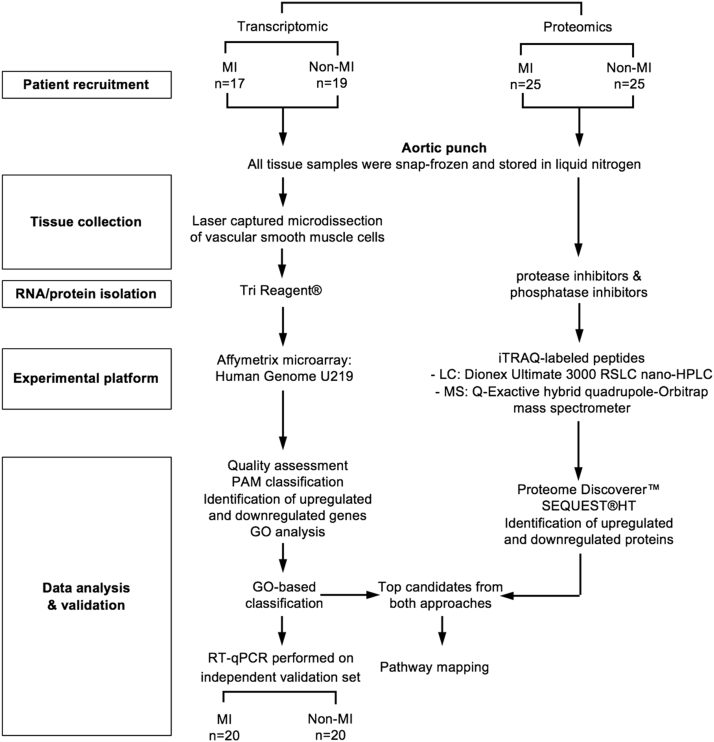
17 MI and 19 non-MI samples were recruited for laser capture microdissection (LCM) and microarray profiling. The proteomic study included 25 MI and 25 non-MI samples. Four MI and six non-MI samples overlapped between the microarray and proteomic studies. RT-qPCR was done on an independent cohort of samples, including an additional 20 MI and 20 non-MI samples. A schematic of the design and workflow is presented in Fig. 3.
Fig. 3.
Work flow and study design.
2.3. Sample processing
The protocols for (1) cryosectioning and staining of aortic tissue, (2) LCM of VSMCs, total RNA isolation and complementary DNA (cDNA) synthesis, and (3) protein processing, ERLIC and LC-MS/MS analysis using Q-Exactive mass spectrometer are described in our manuscript [1].
2.4. RT-qPCR on an independent cohort of MI and non-MI samples
The RT-qPCR protocol is described in our manuscript [1]. The primers for ARF6, ATP1A2, GUCY1A3, HIF-1A, KLHL1, MYOCD, SOD1, and UBB were obtained from the Primer Bank: ARF6 forward primer 5′-GGGAAGGTGCTATCCAAAATCTT-3′ and reverse primer 5′-CACATCCCATACGTTGAACTTGA-3′; ATP1A2 forward primer 5′-TCTATCCACGAGCGAGAAGAC-3′ and reverse primer 5′-CCATGTAGGCATTTTGAAAGGC-3′; GUCY1A3 forward primer 5′-TCAGCCCTACTTGTTGTACTCC-3′ and reverse primer 5′-CAGAATAGCGATGTGGGAATCAC-3′; HIF-1A forward primer 5′-GAACGTCGAAAAGAAAAGTCTCG-3′ and reverse primer 5′-CCTTATCAAGATGCGAACTCACA-3′; KLHL1 forward primer 5′-TCAGGCTCTGGGCGAAAAG-3′ and reverse primer 5′-AAAGTGCTCACACCGCTTCTC-3′; MYOCD forward primer 5′-ACGGATGCTTTTGCCTTTGAA-3′ and reverse primer 5′- AACCTGTCGAAGGGGTATCTG-3′; SOD1 forward primer 5′-AAAGATGGTGTGGCCGATGT-3′ and reverse primer 5′-CAAGCCAAACGACTTCCAGC-3′; UBB forward primer 5′-GGTCCTGCGTCTGAGAGGT-3′ and reverse primer 5′-GGCCTTCACATTTTCGATGGT-3′.
Acknowledgements
This work was supported by the National University Health System Clinician Scientist Program, Singapore and Biomedical Institutes of A*STAR, Singapore. We thank Ms Chan Yang Sun (Genomic Institute of Singapore, A*STAR, Singapore) for laboratory support and tissue processing. We thank Dr. Zhiqun Tang (Bioinformatics Institute, A*STAR, Singapore) for bioinformatics suggestions. We also thank Dr. Yenamandra S.P. for useful discussion of the experimental design and optimization of the experimental study protocols.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2018.01.108.
Transparency document. Supplementary material
Supplementary material
References
- 1.T. Wongsurawat, C.C. Woo, A. Giannakakis, X.Y. Lin, E.S.H. Cheow, C.N. Lee, M. Richards, S.K. Sze, I. Nookaew, V.A. Kuznetsov, V. Sorokin, Distinctive molecular signature and activated signaling pathways in aortic smooth muscle cells of patients with myocardial infarction, Atherosclerosis (2018), 10.1016/j.atherosclerosis.2018.01.024. [DOI] [PubMed]
- 2.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki R., Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material