Graphical abstract
Keywords: Peptidomics, Heparin, Hemorrhagic shock, Proteolysis, Mass spectrometry
Highlights
-
•
A protocol for peptidome plasma analysis has been set up.
-
•
The presence of heparin did not affect peptides detection in plasma specimens.
-
•
Plasma displays an increase in peptides after hemorrhagic shock.
Abstract
A preliminary mass spectrometry based shotgun protocol was set up to compare the peptidome of plasma samples from healthy and hemorrhagic shock rats with the aim of verifying the possible role of uncontrolled proteolytic activity in circulatory shock. Since the hemorrhagic shock model requires heparin as anticoagulant, a preliminary experiment using plasma sample obtained in the presence/absence of heparin from healthy rats was performed to determine whether its presence is fully compatible with the peptidomic protocol proposed. The entire protocol was tested in a pilot experiment to compare the peptidome of healthy or heparin-anticoagulated rats subjected to hemorrhagic shock.
Circulatory shock is a frequent cause of death and a most important unresolved medical problem [1], [2]. Several studies have found that intestine failure triggers a diffuse inflammatory state [3], which can contribute to the development of multiple organ dysfunction (MODS) and post-HS death.
In hemorrhage and sepsis, the intestinal mucosal barrier may become severely damaged due to hypoperfusion. As a consequence many enteral digestive enzymes, such as proteases and lipases may escape from the intestinal lumen and reach the systemic circulation, resulting in global cellular and organ dysfunction [4].
To test the hypothesis that in hemorrhagic shock there is an increase of proteases activity, we use for the first time, a high-throughput quantitative mass spectrometry-based approach to analyze the peptidome of rat plasma collected after hemorrhagic shock (HS) and plasma of healthy control rats (CTRL) in order to quantify the relative changes. The experimental hemorrhagic shock model (HS) requires the use of heparin as anticoagulant. Due to the peculiar chemical properties of this highly acidic polymeric compound, preliminary experiments need to be performed to determine whether its presence is fully compatible with the analytical approach proposed since, to the best of our knowledge, shot-gun nanoLC-ESI–MS/MS has never been used to investigate heparin-containing plasma peptidome.
1 mL venous blood sample was collected from healthy animals in a vial after the addition of a protease inhibitor solution (cOmplete™ Protease Inhibitor Cocktail, Roche) with or without 1 unit heparin/mL (EDTA + heparin) or (EDTA), respectively. Due to the high complexity and wide dynamic range of plasma peptides and proteins, we applied a two-step enrichment method to selectively extract peptides and low molecular weight components, which are low abundant in plasma. 500 μL of each plasma sample were diluted with an equal volume of 32% (v/v) acetic acid to favor disruption of peptide/protein interactions and ultra-filtrated with a 10 kDa cut-off filter (Centricon 10, Millipore) to deplete the high molecular weight proteins. The filtrate was then precipitated with two volumes of cold acetonitrile containing 0.1% of trifluoroacetic acid to complete the removal of any residual high molecular weight component. The supernatant from each sample was then dried (Speed Vacuum), dissolved in 1% (v/v) formic acid, desalted (Zip-Tip C18, Millipore) and directly analyzed by nanoLC-ESI MS/MS.
Mass spectrometry analysis was performed using a LTQ Orbitrap Velos (Thermo Fisher Scientific) equipped with a Dionex UltiMate 3000HPLC System as reported in [5]. Data acquisition was controlled by Xcalibur 2.0 and Tune 2.4 software. Mass spectra were analyzed using MaxQuant software (version 1.3.0.5). The initial maximum allowed mass deviation was set to 15 ppm for monoisotopic precursor ions and 0.5 Da for MS/MS peaks. Enzyme specificity was set to unspecific, a maximum of two missed cleavages were allowed, N-terminal acetylation, methionine oxidation, and asparagine/glutamine deamidation were set as variable modifications. The spectra were searched by the Andromeda search engine in MaxQuant [6] against the rat Uniprot sequence database (release 03.12.2014) setting required false positive rate both to 5% or 1% at the peptide and protein level (in order to detect potential effects of the content of the starting sample on the final identification confidence) and the minimum required peptide length to 6 amino acids. The reversed sequences of the target database were used as decoy database. Comparative analyses were performed using the Perseus software (version 1.4.0.6, http://coxdocs.org/doku.php?id=perseus:start).
As shown in Table 1, 138 and 104 peptides were identified in plasma with (EDTA + heparin) and without heparin (EDTA), respectively, at 1% FDR; among them 37 peptides were present in both conditions. Thus, the analysis allows detecting a similar number of peptides, suggesting that heparin is indeed compatible with the proposed protocol for plasma peptide enrichment and identification. In accordance, the fraction of peptides identified with 1% or 5% FDR is similar for both samples, implying that the presence of heparin does not alter the peptide identification confidence obtained through the nanoLC-ESI–MS/MS analysis. Finally, analysis of the C-terminal residue indicates that 58% or 67% of the peptides identified in the absence or presence of heparin, respectively, were potentially generated by chymotrypsin-like enzymes, while 37% or 32% of the peptides identified in the absence or presence of heparin, respectively, were potentially generated by trypsin-like enzymes. In conclusion, the presence of heparin does not hamper the detection of peptides originated from any particular protease.
Table 1.
Number of the peptides and proteins from which the peptides originate identified in EDTA + Heparin and EDTA plasma samples with FDR 5% and FDR 1%, respectively.
| EDTA + Heparin |
EDTA |
|||
|---|---|---|---|---|
| FDR 5% | FDR 1% | FDR 5% | FDR 1% | |
| No. of peptides | 184 | 138 | 153 | 104 |
| No. of proteins | 177 | 128 | 150 | 99 |
It was then verified whether the protocol is suited to analyze real samples, i.e. plasma from heparin-anti-coagulated rats subjected to hemorrhagic shock (HS), an essential step preliminary to a more comprehensive analysis that will be carried out on a larger number of control and shock animals.
Two male Wistar rats (300–450 g, Harlan Laboratories, Inc., Indianapolis, IN) were assigned to either a control (CTRL) or a hemorrhagic shock (HS) group. Both HS and CTRL animals were heparinized (porcine heparin) following an initial stabilization after the induction of anesthesia (1 unit heparin/mL total blood volume, estimated at 6% of the body weight). Hemorrhage was induced by blood withdrawal from the femoral vein (0.5 mL/min) to a target mean arterial pressure (MAP) of 35 mmHg. Hypovolemia was maintained for two hours, at which point the shed blood was reinfused (0.5 mL/min). Samples were then centrifuged at 1300 rpm for 10 min and the supernatant analyzed following the protocol described above. The animal protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego and conforms to the Guide for the Care and Use of Laboratory Animals by the United States National Institutes of Health (NIH Publication No. 85–23, 1996).
The peptidome of plasma samples from healthy and hemorrhagic shock rats was compared to identify and quantify the peptides present in plasma. Four technical replicate MS analyses were performed for each sample, CTRL and HS. Supplementary Material Fig. S1 compares the distribution of relevant parameters (precursor charge and molecular mass and mass error) relative to peptides identified in the analysis of CTRL and HS. The average molecular mass of peptides identified in CTRL and HS was 1908 and 1913 Da, respectively.
Only peptides present in at least 2 out of 4 technical replicates were considered as positively identified and quantified. Peptides were considered increased or decreased if they were present only in either the healthy or hemorrhagic animal or if they showed 50% fold change differences (>1.5-fold increase or <0.67-fold decrease) in HS compared to CTRL, as calculated from the ratio of mean XIC-based label free quantification [7].
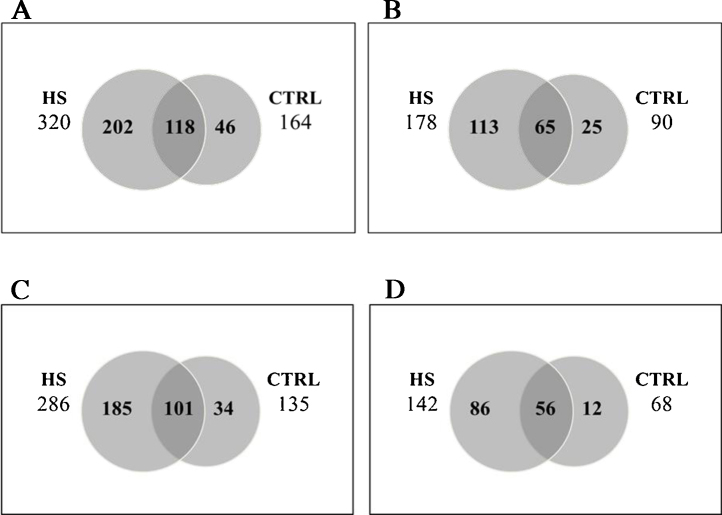
Fig. 1 A-D shows the number of peptides and proteins from which peptides originate identified in CTRL and HS at 5% (A and B) or 1% (C and D) FDR. Again, the fraction of peptides identified at 1% compared to 5% FDR is similar for the two samples.
Fig. 1.
A) Venn diagram of the peptides identified in hemorrhagic (HS) and healthy (CTRL) rats with FDR 5%. B) Venn diagram of the proteins from which the identified peptides originate in hemorrhagic (HS) and healthy (CTRL) rats with FDR 5%. C) Venn diagram of the peptides identified in hemorrhagic (HS) and healthy (CTRL) rats with FDR 1%. D) Venn diagram of the proteins from which the identified peptides originate in hemorrhagic (HS) and healthy (CTRL) rats with FDR 1%.
164 and 320 peptides were identified in the CTRL and HS, respectively, at 5% FDR (Fig. 1A); among the 118 peptides present in both CTRL and HS, 50 increased (>1.5 fold-change) and 52 decreased (<0.67 fold change) in concentration in HS. Overall, the analysis identified 252 peptides, which were increased or present only in HS while 98 peptides were decreased or present only in the control clearly showing a significantly increased number of peptides possibly derived from an intrinsic proteolytic activity in HS samples compared to CTRL. Comparison of the number of peptides identified in two similar samples, i.e. healthy animals (164, Fig. 1A) and plasma from healthy animals with added heparin (184, EDTA + Heparin, Table 1), suggests that the overall protocol is reproducible.
The analysis at the protein level identified 139 proteins which underwent higher proteolysis (>1.5 fold-change, 36 proteins) or proteolyzed only in hemorrhagic shock (HS, 113 proteins) while 46 proteins were less proteolysed in HS (<0.67 fold change, 21 proteins) or proteolyzed only in the control (CTRL, 25 proteins) (Fig. 1B).
The main finding of this work is that heparin is an anticoagulant agent compatible with the proposed peptidomic protocol. Our preliminary data support and quantitately suggest an increase in peptides possibly generated by digestive enzymes after hemorrhagic shock confirming that proteolytic activity takes place after HS in accordance with the “Autodigestion Hypothesis” [4]. Complete confirmation of the above hypothesis, including a statistically supported list of the specific peptides generated during shock, however requires validation using a higher number of animals.
This high-throughput mass spectrometry-based approach appears ideal to expand our understanding of postshock pathophysiology and to aid in the identification of causative agents.
Thanks to its simplicity, the experimental conditions of sample treatment and analysis of plasma peptides set up in the present communication will be helpful to extend our studies to different animal models and different types of shock (i.e., cardiogenic shock, septic shock etc.).
Conflict of interest
The authors confirm that there are no conflict of interests.
Acknowledgments
This research was funded by the “ShockOmics” grant #602706 of the European Union. We thank Francesca Grassi Scalvini and Nicoletta Leveratto for their skillful technical assistance.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.euprot.2016.03.004.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Vincent J.L., De Backer D. Circulatory shock. N. Engl. J. Med. 2013;369(18):1726–1734. doi: 10.1056/NEJMra1208943. [DOI] [PubMed] [Google Scholar]
- 2.Martin G.S. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert Rev. Anti Infect. Ther. 2012;10(6):701–706. doi: 10.1586/eri.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lillehei R.C. The intestinal factor in irreversible hemorrhagic shock. Surgery. 1957;42:1043–1054. [PubMed] [Google Scholar]
- 4.Schmid-Schönbein G.W., Chang M. The autodigestion hypothesis for shock and multi-organ failure. Ann. Biomed. Eng. 2014;42(2):405–414. doi: 10.1007/s10439-013-0891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamplenizza M., Lenardi C., Maffioli E., Nonnis S., Negri A., Forti S., Sogne E., De Astis S., Matteoli M., Schulte C., Milani P., Tedeschi G. Nitric oxide synthase mediates PC12 differentiation induced by the surface topography of nanostructured TiO2. J. Nanobiotechnol. 2013;11:35. doi: 10.1186/1477-3155-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 7.Luerman G.C., Nguyen C., Samaroo H., Loos P., Xi H., Hurtado-Lorenzo A., Needle E., Stephen Noell G., Galatsis P., Dunlop J., Geoghegan K.F., Hirst W.D. Phosphoproteomic evaluation of pharmacological inhibition of leucine-rich repeat kinase 2 reveals significant off-target effects of LRRK-2-IN-1. J. Neurochem. 2014;128:161–176. doi: 10.1111/jnc.12483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.