Graphical abstract
Keywords: Papillary thyroid carcinoma, IQGAP, Proteomics, Mass spectrometry
Highlights
-
•
IQGAP proteins demonstrated differential expression between CV-PTC and FV-PTC.
-
•
Proteome similarities between benign and FV-PTC hampered accurate unsupervised clustering.
-
•
Changes in actin cytoskeleton proteins play predominant roles in thyroid pathology.
Abstract
Introduction
Fine Needle Aspiration Biopsy (FNAB) allows the cytological differentiation of benign and malignant thyroid nodules. However, the method itself is not adequate in determining some cases. For example, the diagnosis of Follicular Variant Papillary Thyroid Carcinoma (FV-PTC) can be challenging. In the current study we investigate the protein profiles of FV-PTC and classical variant PTC (CV-PTC) with no lymph node metastasis and compare it with benign thyroid tissue.
Method
We used CV-PTC (n = 6), FV-PTC (n = 6) and benign thyroid tissues (n = 6) to prepare tissue lysates. Proteins from each group were trypsin and lys-C digested. The samples were analyzed on a Q Exactive Orbitrap mass spectrometer.
Results
We identified 2560 proteins across all 18 specimens. Protein profiles revealed that there was no clear distinction between benign and FV-PTC samples. However, further examination of our data showed that proteins in energy metabolism have altered in FV-PTC. Proteomic pathway analysis showed marked alteration of the actin cytoskeleton proteins, especially several members of Arp2/3 complex were significantly increased in CV-PTC. We made the novel observation that IQGAP1 protein was significantly increased in CV-PTC, whereas IQGAP2 protein was highly expressed in FV-PTC lesions, suggesting differential roles of IQGAP proteins in thyroid pathology.
Conclusion
In the present study, mass spectrometry based label free quantification approach was applied to investigate the protein profiles of FV-PTC, CV-PTC and benign thyroid tissues. This study pointed out that actin cytoskeleton proteins, IQGAP proteins and changes in energy metabolism play predominant roles in thyroid pathology.
1. Introduction
Thyroid carcinoma, most common endocrine malignancy, can arise from follicular and parafollicular cells of the thyroid gland. Papillary (PTC), follicular (FTC) and anaplastic thyroid cancer can originate from the follicular cells in thyroid follicles. During the last decades, the incidence of thyroid cancer sharply increased in many countries [1]. This increase in incidence is commonly attributed to the overdiagnosis of early indolent cancers. The histopathological diagnosis of thyroid nodules is mostly dependent on Fine Needle Aspiration Biopsy (FNAB) as it is a gold standard method due to its reliability and inexpensiveness. This method allows the cytological differentiation of benign and malignant thyroid nodules. Despite the benefits of FNAB based differential diagnosis, it is not helpful in determining some cases of thyroid nodules which then results in unnecessary surgical operation for patients with benign thyroid nodules [2], [3].
PTC has many histological variants arising from different thyroid cells [4]. The classic variant of PTC (CV-PTC)—the most common variant—shows papillary structures with branching and are covered by cells with large nuclei and eosinophilic cytoplasm [5]. In the follicular variant of PTC (FV-PTC) papillary structure is absent and it forms follicles. However, cells that form these follicles have nuclear changes that are typical for papillary carcinoma. In these cases, the absence of papillary structures may obscure the definitive diagnosis of cytological examination of FV, since the core features of PTC are faint/unclear.
FV-PTC is hardly characterized and hence diagnosed with FNAB. Thus, sometimes there is a necessity for a second surgical operation to remove the other thyroid lobe when no certain diagnosis is made for FV-PTC cases. Thereafter, the ongoing discussion in FV-PTC among experts resulted in the revision of the nomenclature for noninvasive FV-PTC [6]. The diagnosis of this type is challenging as it is solely based on the nuclei features due to its non-invasive characteristics [6]. This could result in subjective and controversial diagnosis in thyroid pathology. Recently, noninvasive FV-PTC is termed as “noninvasive follicular thyroid neoplasms with papillary-like nuclear features” (NIFTP) with very low adverse outcome [6].
Additionally, the genetic analysis of FV-PTC lesions showed increased RAS mutations rather than B-type Raf kinase (BRAF) and rearranged in transformation/papillary thyroid carcinoma (RET/PTC) rearrangements, which have been linked with classic PTC [2], [5], [7]. The RAS mutations have also been described in follicular lesions of the thyroid gland such as follicular adenoma and follicular thyroid carcinoma.
Many studies on thyroid pathology used immunohistochemistry and gene-based analysis to shed light on the molecular mechanisms while only limited proteomic studies investigate thyroid pathology. Most of these studies used two dimensional gel electrophoresis and provided valuable but limited information regarding the proteome profiles of thyroid tumors [8], [9], [10]. Recently, Martinez-Aguilar et al. used advanced mass spectrometry technologies and demonstrated that actin cytoskeleton dynamics, differentiation markers and cell contact molecules were highly changed in PTC [11].
To the best of our knowledge, this is the first proteomics study that assesses the proteome profiles of invasive FV-PTC and NIFTP lesions in order to increase the understanding of the biology behind these noninvasive neoplasms. Here we present a label-free quantitative mass spectrometry approach with high resolution and high mass accuracy to investigate invasive FV-PTC and NIFTP proteomes and compare the protein profiles with CV-PTC and benign lesions. Our results led to the identification of over 2500 proteins, providing comprehensive proteome information of FV-PTC, CV-PTC and benign lesions. From our results, we aimed to refine our understanding of biology behind the variants of papillary thyroid carcinoma.
2. Material and methods
2.1. Subjects and clinical materials
Six fresh frozen thyroid tissues from each group (benign, CV-PTC and FV-PTC) were analyzed by mass spectrometry. All samples were obtained from Acibadem Maslak Hospital, Istanbul-Turkey. The tissue samples were pathologically confirmed to be of the CV-PTC and FV-PTC and classified according to the World Health Organization (WHO) Classification of Tumors Pathology and Genetics: Tumors of Endocrine Organs [12]. Each FV-PTC samples were re-reviewed by two independent pathologists to confirm the diagnosis. Based on the consensus diagnostic criteria for NIFTP [6], one FV-PTC sample was re-evaluated as NIFTP. No lymph node metastasis was detected in CV-PTC cases. Benign thyroid tissues were obtained from the subjects with benign diseases that had undergone thyroidectomy. The study was approved by the Ethics Committee of the Acibadem Mehmet Ali Aydinlar University (ATADEK-2014-621). Informed consent was obtained from all subjects.
2.2. Sample preparation
All samples were homogenized with solubilization buffer (150 mM NaCl, 50 mM Tris pH 7.5, 0.3% SDS, 150U/ml Benzonase and EDTA-free protease inhibitor cocktail (Roche)) in a Retsch Mixer Mill MM400 (Retsch GmbH, Haan, Germany) homogenizer. The protein sample was centrifuged for 60 min at 20,000g to pellet any debris. Thereafter, methanol/chloroform precipitation was performed [13] and protein samples were solubilized in denaturation buffer (6 M urea, 2 M thiourea in 10 mM HEPES, pH 8.0). The protein concentration of the samples was determined according to Bradford [14]. The proteins were reduced by adding 350 mM dithiothreitol (DTT) and alkylated by 55 mM iodoacetamide. The protein samples were then treated with 0.5 μg/μl lysyl-endopeptidase (Lys-C) (Wako Chemicals) and 0.5 μg/μl Trypsin (Promega). The protein samples were prepared for LC–MS/MS analysis using Stage tips according to Rappsilber et al. [15].
2.3. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis
Desalted peptides were separated on a 5%–45% acetonitrile gradient (240 min) with 0.1% formic acid at a flow rate of 200 nL/min using the EASY-nLC II system (Thermo Fisher Scientific) on in-house manufactured 25 cm fritless silica micro-columns with an inner diameter of 75 μm. Columns were packed with the ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Entringen, Germany). A Q Exactive plus instrument (Thermo Fisher Scientific) was operated in the data dependent mode with a full scan in the Orbitrap followed by top 10 MS/MS scans using higher-energy collision dissociation (HCD). The full scans were performed with a resolution of 70,000, a target value of 3 × 106 ions and a maximum injection time of 20 ms. The MS/MS scans were performed with a 17,500 resolution, a 1 × 106 target value and a 60 ms maximum injection time.
2.4. Protein identification and label-free quantification
Peak lists were generated from raw data files using MaxQuant software version 1.5.1.2 according to the standard workflow [16], which corrects for systematic inaccuracies of measured peptide masses and corresponding retention times of extracted peptides [17]. Proteins were identified by searching against the Uniprot human reference proteome database.
As described in much detail in [18], in the MaxLFQ label-free quantification method a retention time alignment and identification transfer protocol (“match-between-runs” feature in MaxQuant) is applied and a novel algorithm is used to extract the maximum possible quantification information.
2.5. Differentially expressed proteins and statistical analysis
Data analysis was performed using the freely available software Perseus (version 1.5.8.2) and R [19]. The LFQ intensities of proteins from the MaxQuant analysis were imported and transformed to logarithmic scale with base two. Valid values were considered to have at least 4 out of 6 LFQ intensity values in at least one of the three groups. Missing values were replaced (imputated) with the value of the lowest LFQ intensity in the whole dataset. The protein quantification and calculation of statistical significance was done with a Welch test and correction of the alpha error was performed using the FDR correction described by Benjamini–Hochberg. Protein-protein interactions were visualized by String V10.5 [20].
3. Results
3.1. Proteomic profiles of FV-PTC and CV-PTC showed differential protein expressions
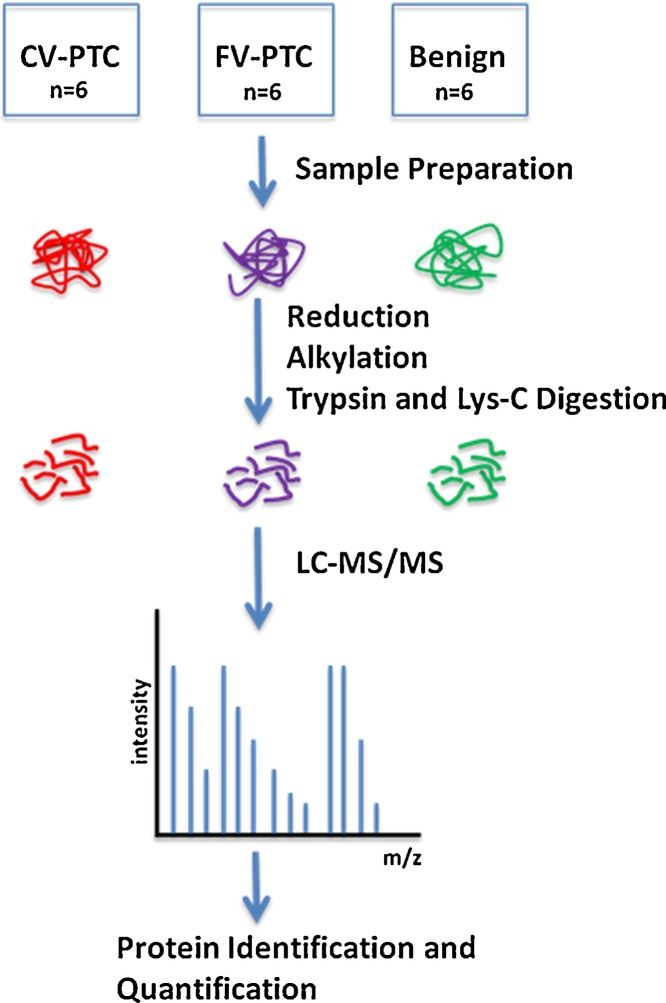
Fig. 1 shows a schematic overview of the sample preparation and LC–MS/MS analysis conducted in this work. We assembled 18 fresh frozen thyroid tissues from CV-PTC (n = 6), FV-PTC (n = 6) and benign thyroid tissues (n = 6) to analyze the proteomic profiles of thyroid tumors. Quantitative analysis revealed 2560 protein identifications across all 18 samples.
Fig. 1.
Schematic representation of experimental workflow. Fresh frozen tissue samples from CV-PTC (n = 6), FV-PTC (n = 6) and benign (n = 6) were lysed and reduced, alkylated and trypsin and lys-C digested. Digested protein samples were then analyzed by LC–MS/MS.
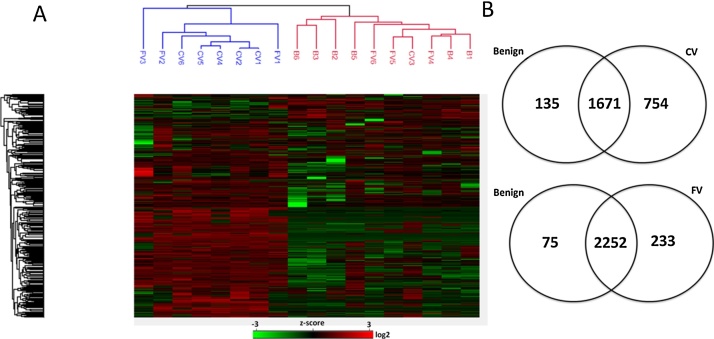
Hierarchical cluster analysis of protein expressions in all sample groups demonstrated a clear separation of CV-PTC from benign tissues whereas FV-PTC lesions showed no obvious distinction between CV-PTC and benign tissues (Fig. 2A). Interestingly, unsupervised clustering analysis demonstrated that the only NIFTP case (denoted as FV1) was clustered with CV-PTC cases. Besides, detailed comparison of FV-PTC and benign proteomes showed remarkable similarities. For example, out of 308 differentially expressed proteins in FV-PTC cases, 233 proteins seemed to be increased (fold-change fc ≥ 2) in FV-PTC. However, after Benjamini Hochberg correction none of these proteins showed differential expression between FV-PTC and benign tissues. The Venn diagram in Fig. 2B shows the intersection of proteins identified in all three groups. The common proteins between benign tissues and FV-PTC (n = 2252) are remarkably higher than common proteins between benign tissues and CV-PTC (n = 1671) (Fig. 2B). Higher number of common proteins identified in FV-PTC and benign cases are consistent with the hierarchical cluster analysis.
Fig. 2.
A. Unsupervised hierarchical cluster analysis of proteins in CV-PTC, FV-PTC and benign samples. FV1 is the only NIFTP case in FV-PTC group, which was clustered with CV-PTC samples. B. Venn Diagrams of proteins identified across all three groups. The proteome profiling of all three groups contributed to 2.560 protein identifications (p < 0.05). 65% of proteins were common in benign and CV-PTC, whereas 88% of proteins found to be identical in benign and FV-PTC. In both hierarchical clustering and Venn diagram; FV denotes FV-PTC, CV denotes CV-PTC.
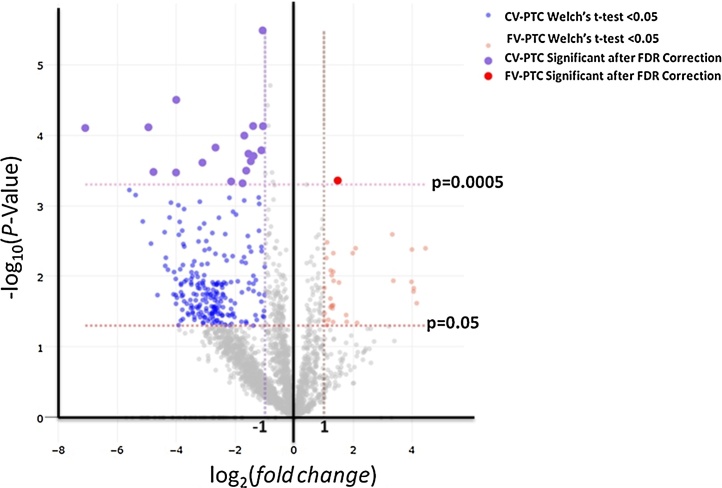
Comparison of CV-PTC and benign proteomes resulted in 296 differentially expressed proteins (fc ≥ 2, FDR p-value <0.0005). Additionally, comparison of FV-PTC with CV-PTC samples resulted in 27 differentially expressed proteins (FDR p-value <0.0005) (Table 1). To display this quantitative data, volcano plot −log10 (p value) versus log2 (fold change) was constructed (Fig. 3). Points above FDR-p < 0.0005 shows significantly and differentially altered proteins in FV-PTC and CV-PTC. Interestingly, out of a total of 27 differentially expressed proteins in FV-PTC and CV-PTC, only 2 proteins were increased in FV-PTC while 25 proteins showed increased expression in CV-PTC.
Table 1.
List of proteins with largest increased expression levels in CV-PTC and FV-PTC.
| Largest Increase in Expression Levels in CV-PTC | |||
|---|---|---|---|
| UniProt ID | Protein Name | Fold Changea | FDR corrected P-valueφ |
| ARPC3 | Actin-related protein 2/3 complex subunit 3 | 1,07 | 7,47E-05 |
| ANXA1 | Annexin A1 | 2,15 | 0,0005 |
| WARS | Tryptophan–tRNA ligase, cytoplasmic;T1-TrpRS;T2-TrpRS | 1,64 | 0,0003 |
| STAT1 | Signal transducer and activator of transcription 1-alpha/beta | 4,00 | 3,16E-05 |
| ACTR3 | Actin-related protein 3 | 1,12 | 0,0002 |
| ACTR2 | Actin-related protein 2 | 1,08 | 3,29E-06 |
| CAP1 | Adenylyl cyclase-associated protein 1 | 0,82 | 1,98E-05 |
| MVP | Major vault protein | 2,68 | 0,0002 |
| PLEC | Plectin | 0,76 | 0,0004 |
| ARPC1B | Actin-related protein 2/3 complex subunit 1B | 1,39 | 0,0002 |
| ARPC2 | Actin-related protein 2/3 complex subunit 2 | 0,89 | 7,37E-05 |
| TYMP | Thymidine phosphorylase | 3,12 | 0,0002 |
| FLNA | Filamin-A | 0,68 | 0,0005 |
| PSM8 | Proteasome subunit beta type | 1,56 | 0,0002 |
| SERPINB1 | Leukocyte elastase inhibitor | 0,73 | 0,0005 |
| CORO1A | Coronin-1A;Coronin | 4,02 | 0,0003 |
| RECQL | ATP-dependent DNA helicase Q1 | 0,98 | 0,0004 |
| ANXA11 | Annexin A11 | 0,91 | 3,79E-05 |
| ARHGDIB | Rho GDP-dissociation inhibitor 2 | 1,48 | 0,0002 |
| S100A10 | Protein S100-A10 | 1,76 | 0,0005 |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein | 1,40 | 7,44E-05 |
| FERMT3 | Fermitin family homolog 3 | 4,95 | 7,74E-05 |
| MZB1 | Marginal zone B- and B1-cell-specific protein | 7,09 | 7,93E-05 |
| PARP4 | Poly [ADP-ribose] polymerase 4 | 4,78 | 0,0003 |
| SAMHD1 | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 1,70 | 0,0001 |
| Largest Increase in Expression Levels in FV-PTC | |||
|---|---|---|---|
| UniProt ID | Protein Name | Fold Changea | FDR corrected P-valueφ |
| IQGAP2 | Ras GTPase-activating-like protein IQGAP2 | 1,47 | 0,0004 |
| PGK1 | Phosphoglycerate kinase 1 | 0,41 | 0,0005 |
aFold change of protein expression level (log2).
φp value determined from Welch’s t-test (FDR corrected P-value <0.0005).
Fig. 3.
Volcano Plot shows differentially abundant proteins in CV-PTC and FV-PTC. The plot constructed using −log10 (p value) against the log2 (fold change).The non-axial vertical lines represent ±1-fold change. The non-axial horizontal lines show p = 0.05 and p = 0.0005 (Benjamini-Hochberg corrected P value).
3.2. Expressional changes in IQGAP proteins in CV-PTC and FV-PTC
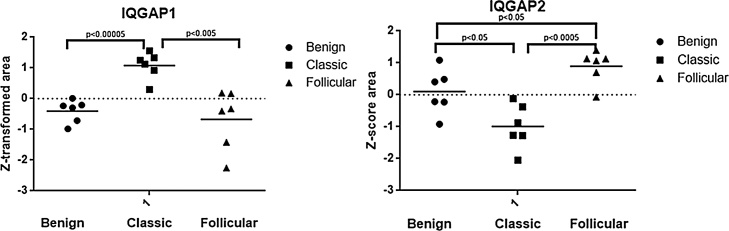
CV-PTC and FV-PTC samples shared dysregulation of evolutionary conserved Ras GTPase-activating-like proteins (IQGAPs) (Fig. 4). IQGAP1 was found at increased levels in CV-PTC (fc ≥ 2), while the expression of this protein was not changed in benign and FV-PTC lesions. On the other hand, IQGAP2 protein was one of the two proteins that showed increased expression in FV-PTC lesions (fc ≥ 2) and statistically significant decreased expression in CV-PTC, suggesting different mechanism of actions of these scaffold proteins in thyroid tumor biology.
Fig. 4.
Expressional levels of IQGAP1 and IQGAP2 proteins were compared between CV-PTC, FV-PTC and benign samples. IQGAP1 expression level was higher in CV-PTC and relatively lower in FV-PTC and benign samples. Conversely, CV-PTC and benign samples showed significantly lower levels of IQGAP2 protein.
3.3. Molecular pathway analysis revealed cytoskeleton related protein changes in CV-PTC
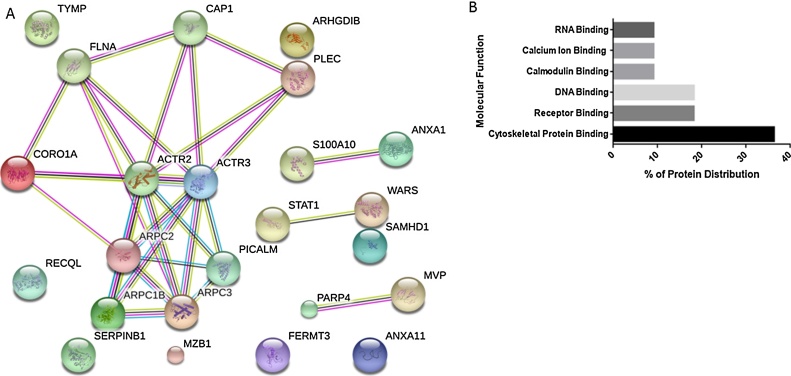
Differentially expressed proteins in CV-PTC (n = 25, FDR p-value <0.0005) were further investigated using STRING database to establish known and predicted protein interactions. These interactions indicated enrichment of actin cytokeleton proteins (Fig. 5A) and additionally supported by the increase in Rho related proteins (fc ≥ 2), which are known to regulate actin cytoskeleton. For example, several members of actin-related protein 2/3 (Arp 2/3), filamin-a, coronin-1a and plectin were overexpressed in CV-PTC. Moreover, gene ontology analysis based on the molecular functions of proteins that are up-regulated in CV-PTC showed that 45% of these proteins have binding properties. Apart from cytoskeletal binding proteins, DNA and receptor binding proteins showed increased expressions in CV-PTC (Fig. 5B).
Fig. 5.
A. Protein interaction network generated with String that contains 25 significantly and differentially expressed proteins in CV-PTC. Major cluster of interacting proteins belong to Arp 2/3 complex. B. Binding protein distribution in CV-PTC (n = 11, p < 0.0005). Gene ontology analysis revealed that 45% of proteins that are significanty and differentially expressed in CV-PTC have binding properties.
3.4. FV-PTC showed protein expression changes in energy metabolism
The comparison of FV-PTC with benign tissues showed increased expression (fc ≥10) of the proteins regulating the energy metabolism. For example; the expressional magnitudes of pyruvate carboxylase, members of branched-chain alpha-keto dehydrogenase complex, NADPH:adrenodoxin oxidoreductase and ADP/ATP translocase 2 were significantly higher in FV-PTC lesions (p < 0.05, fc ≥10), suggesting a change in energy metabolism. However, the fact that the expression of most proteins remained unchanged between FV-PTC and benign tissues underlies the proteome similarities and clearly demonstrates the controversial diagnosis of FV-PTC.
4. Discussion
The mass spectrometric assessment of protein changes in benign, CV-PTC and FV-PTC samples correlated with histological and pathological features of thyroid tumors. More than 2500 protein identifications in 18 samples form detailed proteomic analysis of thyroid tumors. Ongoing discussions about FV-PTC and reclassification of noninvasive FV-PTC as “NIFTP” attracted much attention in thyroid pathology. Our mass spectrometric analysis on this subject provides the most comprehensive understanding of FV-PTC cases to date. Additionally, all the CV-PTC samples included in our study were free of lymph node metastasis. This is important as our results can help to explain the less aggressive characteristics of CV-PTC when compared with the FV-PTC and benign tissues.
A remarkable result of our proteomic analysis revealed that FV-PTC cannot be accurately clustered due to the proteome similarity between benign and FV-PTC. Our finding is highly consistent with the key publication of Nikiforov et al., where an international panel of pathologists and clinicians reclassified noninvasive FV-PTC as “non-cancer” to reflect its noninvasiveness and low recurrence risk [6]. Interestingly, some proteins regulating the energy metabolism showed increased expression (fc ≥10) in FV-PTC lesions when compared to the benign tissues. Cellular proliferation relies on the energy and biomass production from tricarboxylic acid (TCA) cycle. In order to fulfill the demands of TCA cycle, pyruvate is converted to oxaloacetate. The enzyme pyruvate carboxylase catalyzes this reaction and replenishes mitochondrial oxaloacetate levels [21]. Additionally, branched chain amino acid catabolism provides major nitrogen source for glutamine, an important amino acid for the production of proteins, hexosamines and macromolecules. In fact, the role of glutamine and glucose in cancer and cellular proliferation has already been under spotlights (reviewed in Pavlova and Thompson et al. [21]). Although these lesions are considered less aggressive, the increase in pyruvate carboxylase and members of branched-chain amino acid dehydrogenase in FV-PTC highlight the change in energy metabolism. Moreover, up-regulated expression of mitochondrial proteins also indicated the higher demand for energy production in FV-PTC lesions.
The proteome comparison between CV-PTC and FV-PTC showed that the expression of 2 proteins was remarkably high in all FV-PTC samples (p < 0.005). Phosphoglycerate kinase-1 (PGK1), an important glycolytic enzyme that has dual roles in tumorigenesis—regulation of cancer cell metabolism and gene transcription—was upregulated in FV-PTC samples. PGK1 has previously been reported in gastric, colon and pancrease cancer [22], [23], [24], [25]. Recently, Ralhan et al. applied immunohistochemistry to examine the potential of seven proteins including PGK1 to discriminate thyroid cancer from benign [26]. Their results demonstrated that loss of PGK1 in combination with an increase in Galectin-3 could distinguish benign from malignant thyroid nodules. Although their study set contained 33 FV-PTC cases, there was no specific information on how PGK1 expression was changed in FV-PTC. However, increased expression of PGK1 in FV-PTC and decreased expression of this protein in CV-PTC in our work, are consistent with the findings of Ralhan et al. [26].
Another protein that showed increased expression in FV-PTC lesions was IQGAP2. To the best of our knowledge, structural and functional information regarding IQGAP2 is very limited [27]. Taken together with the features of FV-PTC and new classification of noninvasive FV-PTC as NIFTP lesions, higher expression of this protein in these samples can indicate a potential protective role. Notably, this potential protective role of IQGAP2 has also been shown in gastric cancer cell lines and primary gastric cancer tissues. Jin et al. studied IQGAP2 silencing through methylation and showed inverse correlation between IQGAP2 expression and gastric cancer progression [28]. Same hypothesis is supported by Tamura et al., in which hormone-refractory prostate cancer demonstrated decreased expression of IQGAP2 [29]. Altogether, these studies highlight the potential of IQGAP2 as a tumor suppressor. Although evolutionary conserved IQGAP proteins share similar structure and sequence homology, IQGAP1 has been proposed to be an oncogene [27]. It has been well known that IQGAP1, which has important roles in regulation and modulation of cytoskeleton and participate in cell adhesion by binding to specific molecules, is over-expressed in certain cancer types [30], [31]. Several reports suggest the involvement of IQGAP1 in thyroid tumorigenesis [31], [32], [33]. For example, Liu et al. assessed IQGAP1 copy number gain in different types of thyroid cancer, suggesting that IQGAP1 copy gain has a crucial role in thyroid tumorigenesis [34]. Our results indicated that IQGAP1 was found at increased levels in CV-PTC (fc ≥ 2), while the expression of this protein was not changed in benign and FV-PTC samples. Generally speaking, increased level of IQGAP1 protein in CV-PTC can support the oncogenicity; while the higher levels of IQGAP2 protein in FV-PTC favor the tumor suppressor features. However, there seems to be a gap in the literature between the mechanism that either triggers the up-regulation of IQGAP1 and down-regulation of IQGAP2 in different neoplasms [27].
As indicated by the pathway analysis, differentially expressed proteins in CV-PTC (n = 25, FDR p-value <0.0005) showed an enrichment of actin cytoskeleton and cytoskeleton regulatory proteins. Specifically, there was strong expression of several members of Arp 2/3 complex in CV-PTC. Arp 2/3 complex is a major regulator of actin polymerization and increased expression of Arp 2/3 members were detected in several cancer types [35]. Our finding is supported by a very recent publication by Martinez-Aguilar et al., in which the authors used data-independent acquisition mass spectrometry to show that proteins regulating the cell to cell contact and actin cytoskeleton dynamics—especially members of Arp 2/3 complex—were altered in papillary thyroid carcinoma [11].
In summary, we examined protein profiles of CV-PTC, FV-PTC and benign thyroid tissues by mass spectrometry. Remarkably, we have found that actin cytoskeleton proteins are highly altered and reorganization of the cytoskeleton plays predominant role in the progression of papillary thyroid carcinoma. Additionally, although there was no clear distinction between and benign and FV-PTC samples, these less aggressive samples demonstrated changes in the energy metabolism. Finally, expressional change in IQGAP proteins in CV-PTC and FV-PTC samples needs to be further investigated to address the explicit roles of IQGAP proteins in thyroid pathology.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by TUBITAK (Scientific and Technological Research Council of Turkey) 1001 grant, 214S012 and International Cost Project BM 1403 Native Mass Spectrometry and Related Methods for Structural Biology, 110664. The authors gratefully acknowledge the assistance of Cavit Kerem Kayhan, Irep Uras, and Muazzez Ceren Yilmaz for their support in sample preparations.
References
- 1.Kitahara C.M., Sosa J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016;12:646–653. doi: 10.1038/nrendo.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr. Rev. 2007;28:742–762. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 3.Cheng S., Serra S., Mercado M., Ezzat S., Asa S.L. A high-throughput proteomic approach provides distinct signatures for thyroid cancer behavior. Clin. Cancer Res. 2011;17:2385–2394. doi: 10.1158/1078-0432.CCR-10-2837. [DOI] [PubMed] [Google Scholar]
- 4.Fischer S., Asa S.L. Application of immunohistochemistry to thyroid neoplasms. Arch. Pathol. Lab. Med. 2008;132:359–372. doi: 10.5858/2008-132-359-AOITTN. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd R.V., Buehler D., Khanafshar E. Papillary thyroid carcinoma variants. Head Neck Pathol. 2011;5:51–56. doi: 10.1007/s12105-010-0236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikiforov Y.E., Seethala R.R., Tallini G. Nomenclature revision for encapsulated follicular variant of papillary thyroid carcinoma: a paradigm shift to reduce overtreatment of indolent tumors. JAMA Oncol. 2016;2:1023–1029. doi: 10.1001/jamaoncol.2016.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grogan R.H., Mitmaker E.J., Clark O.H. The evolution of biomarkers in thyroid cancer-from mass screening to a personalized biosignature. Cancers (Basel) 2010;2:885–912. doi: 10.3390/cancers2020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown L.M., Helmke S.M., Hunsucker S.W., Netea-Maier R.T., Chiang S.A., Heinz D.E., Shroyer K.R., Duncan M.W., Haugen B.R. Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Mol. Carcinog. 2006;45:613–626. doi: 10.1002/mc.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Netea-Maier R.T., Hunsucker S.W., Hoevenaars B.M., Helmke S.M., Slootweg P.J., Hermus A.R., Haugen B.R., Duncan M.W. Discovery and validation of protein abundance differences between follicular thyroid neoplasms. Cancer Res. 2008;68:1572–1580. doi: 10.1158/0008-5472.CAN-07-5020. [DOI] [PubMed] [Google Scholar]
- 10.Sofiadis A., Becker S., Hellman U., Hultin-Rosenberg L., Dinets A., Hulchiy M., Zedenius J., Wallin G., Foukakis T., Höög A., Auer G., Lehtiö J., Larsson C. Proteomic profiling of follicular and papillary thyroid tumors. Eur. J. Endocrinol. 2012;166:657–667. doi: 10.1530/EJE-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Aguilar J., Clifton-Bligh R., Molloy M.P. Proteomics of thyroid tumours provides new insights into their molecular composition and changes associated with malignancy. Sci. Rep. 2016;6:23660. doi: 10.1038/srep23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eng C., DeLellis R.A., Lloyd R.V., Heitz P.U. 2004. WHO Classification of Tumors Pathology and Genetics of Tumors of Endocrine Organs. [Google Scholar]
- 13.Wessel D., Flügge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 14.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 16.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 17.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 18.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team . R Foundation for Statistical Computing; 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 20.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2017;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zieker D., Konigsrainer I., Weinreich J., Beckert S., Glatzle J., Nieselt K., Buhler S., Loffler M., Gaedcke J., Northoff H., Mannheim J.G., Wiehr S., Pichler B.J., von Weyhern C., Brucher B.L.D.M., Konigsrainer A. Phosphoglycerate kinase 1 promoting tumor progression and metastasis in gastric cancer – detected in a tumor mouse model using positron emission tomography/magnetic resonance imaging. Cell. Physiol. Biochem. 2010;26:147–154. doi: 10.1159/000320545. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad S.S., Glatzle J., Bajaeifer K., Buhler S., Lehmann T., Konigsrainer I., Vollmer J.-P., Sipos B., Ahmad S.S., Northoff H., Konigsrainer A., Zieker D. Phosphoglycerate kinase 1 as a promoter of metastasis in colon cancer. Int. J. Oncol. 2013;43:586–590. doi: 10.3892/ijo.2013.1971. [DOI] [PubMed] [Google Scholar]
- 24.Zieker D., Buhler S., Ustundag Z., Konigsrainer I., Manncke S., Bajaeifer K., Vollmer J., Fend F., Northoff H., Konigsrainer A., Glatzle J. Induction of tumor stem cell differentiation–novel strategy to overcome therapy resistance in gastric cancer. Langenbeck’s Arch. Surg. 2013;398:603–608. doi: 10.1007/s00423-013-1058-5. [DOI] [PubMed] [Google Scholar]
- 25.Jung Y., Shiozawa Y., Wang J., Wang J., Wang Z., Pedersen E.A., Lee C.H., Hall C.L., Hogg P.J., Krebsbach P.H., Keller E.T., Taichman R.S. Expression of PGK1 by prostate cancer cells induces bone formation. Mol. Cancer Res. 2009;7:1595–1604. doi: 10.1158/1541-7786.MCR-09-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralhan R., Veyhl J., Chaker S., Assi J., Alyass A., Jeganathan A., Somasundaram R.T., MacMillan C., Freeman J., Vescan A.D., Witterick I.J., Walfish P.G. Immunohistochemical subcellular localization of protein biomarkers distinguishes benign from malignant thyroid nodules: potential for fine-Needle aspiration biopsy clinical application. Thyroid. 2015;25:1224–1234. doi: 10.1089/thy.2015.0114. [DOI] [PubMed] [Google Scholar]
- 27.White C.D., Brown M.D., Sacks D.B. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S.-H., Akiyama Y., Fukamachi H., Yanagihara K., Akashi T., Yuasa Y. IQGAP2 inactivation through aberrant promoter methylation and promotion of invasion in gastric cancer cells. Int. J. Cancer. 2008;122:1040–1046. doi: 10.1002/ijc.23181. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Furihata M., Tsunoda T., Ashida S., Takata R., Obara W., Yoshioka H., Daigo Y., Nasu Y., Kumon H., Konaka H., Namiki M., Tozawa K., Kohri K., Tanji N., Yokoyama M., Shimazui T., Akaza H., Mizutani Y., Miki T., Fujioka T., Shuin T., Nakamura Y., Nakagawa H. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 30.Briggs M.W., Sacks D.B. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson M., Sharma M., Henderson B.R. IQGAP1 regulation and roles in cancer. Cell Signal. 2009;21:1471–1478. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Su D., Liu Y., Song T. Knockdown of IQGAP1 inhibits proliferation and epithelial–mesenchymal transition by Wntβ-catenin pathway in thyroid cancer. Onco Targets Ther. 2017;10:1549–1559. doi: 10.2147/OTT.S128564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J.-K., Ma L., Song W.-H., Lu B.-Y., Huang Y.-B., Dong H.-M., Ma X.-K., Zhu Z.-Z., Zhou R. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed. Pharmacother. 2016;83:1–7. doi: 10.1016/j.biopha.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z., Liu D., Bojdani E., El-Naggar A.K., Vasko V., Xing M. IQGAP1 plays an important role in the invasiveness of thyroid cancer. Clin. Cancer Res. 2010;16:6009–6018. doi: 10.1158/1078-0432.CCR-10-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goley E.D., Welch M.D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]