Graphical abstract
Improved and efficient metabolite extraction method for maximizing the number of metabolites identified from a single biological sample, compatible with DI-FTICR-MS-based metabolomics.
Keywords: FTICR-MS, Vitis vinifera, Metabolomics
Highlights
-
•
An improved metabolite extraction method for DI-FTICR metabolomics was developed.
-
•
The number of identified metabolites from grapevine leaves was maximized.
-
•
The extraction method allowed the extraction of polar and non-polar compounds.
-
•
Identified metabolites covered all major classes found in plants.
Abstract
In metabolomics there is an ever-growing need for faster and more comprehensive analysis methods to cope with the increase of biological studies. Direct infusion Fourier-transform ion cyclotron-resonance mass spectrometry (DI-FTICR-MS) is used in non-targeted metabolomics to obtain high-resolution snapshots of the metabolic state of a system. In any metabolic profiling study, the establishment of an effective metabolite extraction protocol is paramount. We developed an improved metabolite extraction method, compatible with DI-FTICR-MS-based metabolomics, using grapevine leaves. This extraction protocol allowed the extraction of polar and non-polar compounds, covering all major classes found in plants and increasing metabolome coverage.
1. Introduction
Grapevine (Vitis vinifera L.) is the most widely cultivated and economically important fruit crop in the world, mainly due to the wine industry. Many grapevine varieties are also grown for their use as food products, not only for Table grapes, but also for the consumption of their leaves. Due to their astringent and hemostatic properties and phenolic composition, vine leaves are considered a healthy food and are consumed in several countries, including Saudi Arabia, Turkey and Greece [1]. The biochemical composition of both grapes and leaves is determinant for their nutritional value and taste. Furthermore, some authors believe that the most reliable source of biomarkers for resistance or susceptibility against pathogens is the leaf surface and the polar extracts from defatted leaf tissues [2], [3]. Hence, the analysis of the compounds present in leaves is of utmost importance. This is particularly relevant when concerning plants, which are biochemically highly complex and contain a unique metabolome that change with the environment, the development and upon pathogen infections [4].
So far, most of the metabolite studies in grapevine were performed by nuclear magnetic resonance (NMR) spectroscopy and were based on the analysis of a single extract from leaves [5], [6]. In these studies by NMR, larger amounts of initial plant material are required (between 25 and 50 mg), the limit of detection is around 10 μM and even using 1D and 2D NMR techniques, and the number of metabolites identified is usually less than 20. More recently, mass spectrometry coupled to liquid chromatography (LC–MS) has been used in the identification and quantification of grapevine metabolites [7]. Although this methodology is more sensitive, only 135 primary metabolites (sugars, amino acids, organic acids and amines) were identified and quantified in a 30-min hydrophilic interaction LC run coupled to a triple quadrupole mass spectrometer [7].
To achieve higher sensitivity and maximum metabolome coverage, we resort to mass spectrometry using high-resolution and high-mass accuracy instruments, based on Fourier transform technology. The sensitivity of this methodology is much higher (typically ρg level) and different fractions can be analyzed (from aqueous to organic extractions) [8]. One of these instruments, the Fourier-transform ion cyclotron-resonance mass spectrometer (FTICR), provides ultra-high-mass accuracy (below 1 ppm) and the highest mass resolution (more than 1,000,000) [9]. Moreover, using direct infusion coupled to ultra-high-resolution mass spectrometry, metabolites are analyzed in a high-throughput way, providing a rapid analysis of complex metabolite samples, and eliminating the time-consuming separation by liquid chromatography (LC) [10].
In addition to high mass accuracy instruments, efficient sample extraction methodologies are a priority in metabolomics. These are especially critical when working with plant material, where caution must be taken during harvesting, grinding and metabolite extraction, to avoid consequences in the accuracy of results [11].
Here we present an efficient metabolite extraction protocol for grapevine leaves, suitable for the characterization of the V. vinifera metabolome by direct infusion Fourier-transform ion cyclotron-resonance mass spectrometry (DI-FTICR-MS).
2. Materials and methods
2.1. Plant material
V. vinifera cv Pinot noir young leaves were harvested from five different plants (three biological replicates were considered), at the Portuguese Grapevine Germplasm Bank at INIA—Estação Vitivinícola Nacional (Dois Portos), immediately frozen in liquid nitrogen and stored at −80 °C. Leaves were ground in liquid nitrogen and used for metabolite extraction.
2.2. Metabolite extraction
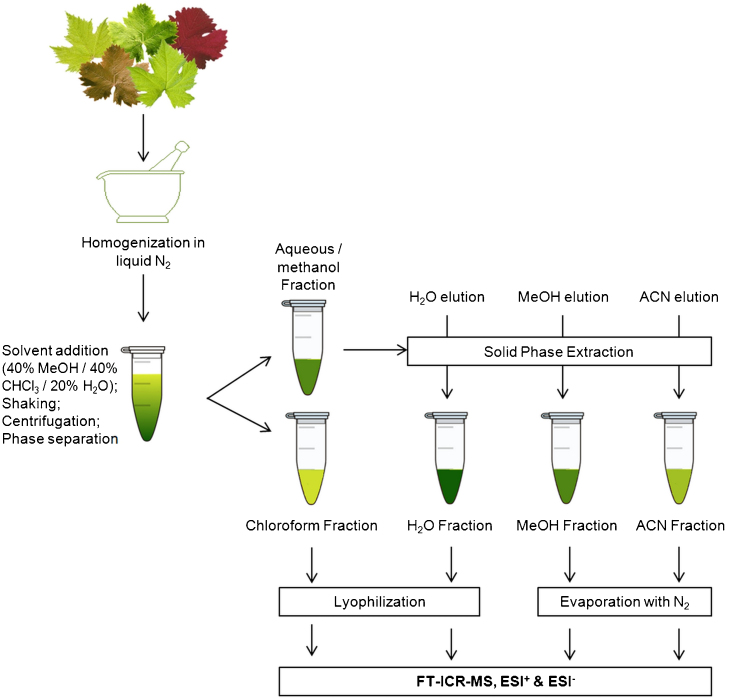
Metabolite extraction from grapevine leaves was performed using different solvent systems coupled to solid phase extraction (SPE) fractionation. We used the mixture 40% methanol (LC–MS grade, Merck)/40% chloroform (Sigma Aldrich)/20% water (v/v/v) as previously described for grapes [12], but the ratio was 0.1 g of grinded leaves to 1 mL of solvent. Samples were vortexed for 1 min and maintained in an orbital shaker for 15 min at room temperature. Samples were centrifuged at 1000g for 10 min for phase separation: the lower chloroform fraction and the upper aqueous/methanol fraction. The chloroform fraction (C) was further centrifuged for 5 min at 10,000g to remove debris and lyophilized at −40 °C. The aqueous/methanol layer was further processed by SPE using Merck LiChrolut RP-18 columns, pre-equilibrated with methanol. Metabolite fractions were collected by vacuum through sequential elution with 1 mL of water (W), methanol (M) and acetonitrile (A, LC–MS grade, Merck). The water fraction was lyophilized at −40 °C, while both methanol and acetonitrile fractions were evaporated under a nitrogen stream. A workflow of the experimental procedure is shown in Fig. 1.
Fig. 1.
Experimental procedure for metabolite extraction from grapevine leaves compatible with FTICR-based metabolomics.
2.3. Metabolite analysis by FTICR-MS
W and C fractions were reconstituted in methanol/water (1:1), while M and A fractions were suspended in the respective pure solvent. For the analysis of metabolites, all fractions were diluted 1000-fold in the appropriate solvent: M and A fractions were diluted in the same solvent for positive- (ESI+) and negative-ion (ESI−) mode analysis; W and C fractions were diluted in methanol for ESI+ or in methanol/water (1:1) for ESI−. The standard leucine enkephalin (YGGFL, Sigma Aldrich) was added to all samples at a concentration of 0.5 μg/mL, and was used as a standard for control and quality assessment of analytical precision ([M + H]+ = 556.276575 Da or [M-H]− = 554.260925 Da), through the determination of the relative standard peak deviation and internal calibration. For the analysis in ESI+, formic acid (final concentration 0.1% (v/v), Sigma Aldrich, MS grade) was added to all samples. Extracted metabolites were analysed by direct infusion in the Apex Qe 7-Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (FTICR-MS, Brüker Daltonics), with a flow rate of 240 μL h−1. Between each sample run, the ESI source was cleaned with methanol or acetonitrile for 10–15 min and the spectrum was collected. Mass spectra were acquired with an acquisition size of 512k, in the mass range between 100 and 1000 Da (with a resolution of 130,000 at 400 m/z), and 50 scans were accumulated for each sample. In ESI+, the nebulizer gas flow rate was set to 2.0 L/min and the dry gas flow rate to 4.0 L/min, at a temperature of 180 °C. The capillary voltage was set to 4500 V and the spray shield voltage was 4000 V. In ESI−, the nebulizer gas flow rate was 2.5 L/min and the dry gas flow rate was set to 4.0 L/min, at a temperature of 220 °C. The capillary voltage was 4300 V and the spray shield voltage was set to 3800 V. In both ionization modes, ions were accumulated in the collision cell for 1.0 s, and a time of flight of 1.0 ms was used prior to their transfer to the ICR cell.
2.4. Data analysis and metabolite identification
Using the Data Analysis 4.1 software package (Brüker Daltonics, Bremen, Germany), the resulting mass spectra were internally calibrated using leucine enkephalin at both ESI modes; external calibration was performed with cyclopamine (m/z 412.32100, [M + H]+) in all 4 fractions at ESI+, whereas at ESI−, hexadecanoic acid (m/z 255.23295, [M+H]−) was used to calibrate M, A and C fractions, while W fraction calibration was calibrated with glutathione (m/z 306.07653, [M+H]−). Peak height lists were then exported as ASCII files, setting at a signal-to-noise ratio at 4. The total number of identified ions (peaks) ranged between 1049 and 1346 for ESI− and 10202 to 11444 for ESI+ in 3 different biological replicates. The peak lists were combined to a peak matrix with an error of 1.0 ppm, as described by [13], implemented in a Python script based on the Pandas library for data analysis. Peaks with just 1 non-zero intensity (single mass events) were removed from the matrix as well as peaks that were detected in less than 50% of all biological replicates. Overall, 1018 peaks for ESI− and 6266 peaks for ESI+ remained after all filtration processes.
For metabolite identification, the mass list was submitted to the MassTrix 3 server (http://masstrix3.helmholtz-muenchen.de/masstrix3/, [14]) server selecting V. vinifera as organism, considering possible adducts M + H and M + Na for ESI+, and M-H and M + Cl for ESI− data, with a maximum error acceptance of 3 ppm. A total of 221 masses were annotated for ESI− and 1366 for ESI+. A manual curation for compounds with biological role was done by searching the annotated metabolites in the public databases PubChem (http://pubchem.ncbi.nlm.nih.gov/, [15]), KNApSAcK (http://kanaya.naist.jp/KNApSAcK/, [16]), KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/kegg2.html, [17]), Lipid Maps (http://www.lipidmaps.org/, [18]) and Metabolomics workbench (http://www.metabolomicsworkbench.org/, [19]).
3. Results and discussion
The analysis of unknown metabolites and the biological interpretation of their relationships represent a very important basis for the profiling of unique metabolic systems and the comparison of such profiles in different phenotypes. To ensure meaningful results and high data quality it is important to have a thorough experimental design and an efficient extraction protocol, specifically designed to be used with an accurate analytical technique.
There is no doubt that mass spectrometry (MS) in metabolomics has facilitated the simultaneous detection and quantification of a large number of metabolites within a large dynamic range. Additionally, it provides structural information through fragmentation experiments in the absence of commercially available standards [20]. Within the several MS instruments available, FTICR is the most powerful tool to fingerprint complex samples, due to its extremely high resolution and high mass accuracy, often better than 1 ppm [21], [22], having therefore a huge potential in the screening of different samples (as discussed in [22], [23]). However, there are very few studies that use FT-MS in plant metabolomics [20], [24], [25], [26], [27]. Most of these studies are based on the analysis of a single extract from the plant material, either using a single solvent, normally methanol or acetonitrile, or a mixture of solvents, either methanol/water or methanol/water/chloroform. The later has several advantages in metabolite extraction yield and reproducibility, by allowing the simultaneous extraction of polar and non-polar metabolites [28]. In addition, chloroform contributes to protein denaturation, therefore preventing the occurrence of biochemical reactions during the extraction step [29].
With the goal of maximizing the number of metabolites identified from a single biological sample, particularly from plant secondary metabolism, and to take advantage of the high-resolution and high-mass accuracy of the FTICR, we developed an improved and efficient metabolite extraction method. We used V. vinifera cv Pinot noir leaves and started with a mixture of methanol/chloroform/water, as previously described for grapes [12]. Then, a sequential extraction using solid phase fractionation was followed, and metabolites were eluted with water, methanol and acetonitrile. In total, we obtained four fractions (chloroform, water, methanol and acetonitrile), all analysed by direct infusion-FTICR (DI-FTICR), using electrospray ionization (ESI) in both positive and negative modes. Direct infusion coupled to ultra-high resolution MS provides a rapid analysis of complex metabolite mixtures, eliminating the chromatographic separation, which can be very time-consuming [10]. Indeed, the LC step in Theodoridis et al. method increases in 60 min each analysis [12]. On the other hand, in this study, using the same solvent mixture of 40% chloroform/40% methanol/20% water (v/v/v), about 4500 peaks were obtained [12], whereas using our workflow the number of identified ions (peaks) reached over 11000. We identified 719 unique masses in leaves of V. vinifera cv Pinot noir using DI-FTICR, corresponding to 1383 putative metabolites (since the same mass value can be attributed to more than one compound, particularly isomers), excluding drugs and pesticides. In a previous work using grapevine leaves, only 96 were identified in acetone and butanol extracts by GC–MS [2]. More recently, methanol extracts from V. vinifera leaves were analysed by FTICR and 40 compounds were detected [20]. In our work, we identified 158 unique masses only in the methanol fraction in both ionization modes, highlighting the importance of the extraction method for FTICR in non-targeted metabolomics. In potato tubers (Solanum tuberosum), the use of FTICR allowed the identification of at least 152 different compounds only in mitochondria [27], whereas in full potato extracts around 150 were detected by GC–MS [30].
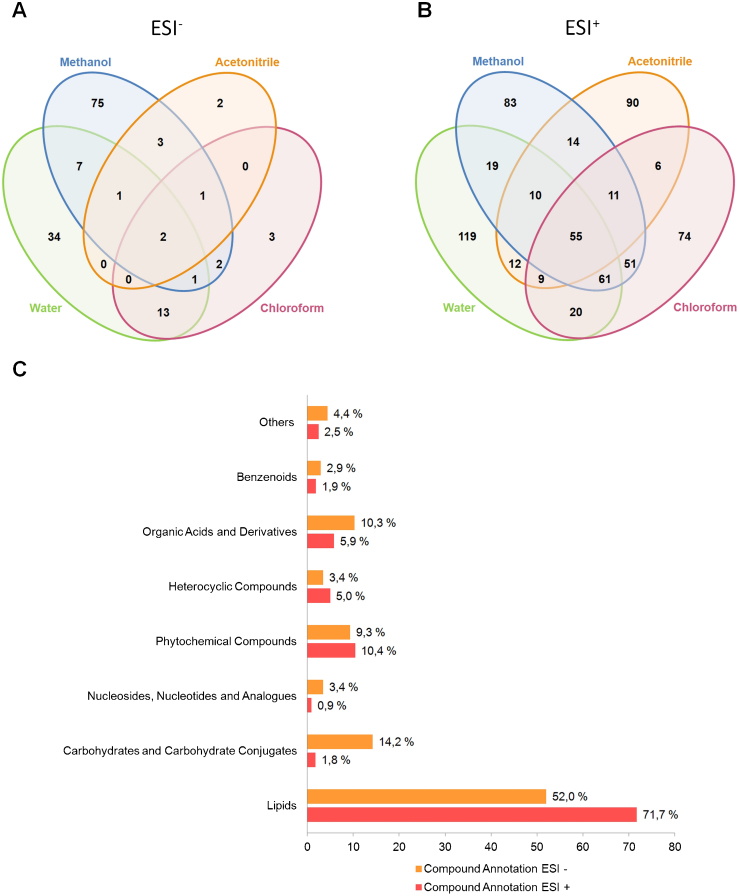
In order to increase metabolome coverage, each V. vinifera fraction was analyzed in ESI+ and ESI−. We identified 144 unique masses by ESI− and 634 by ESI+ (Fig. 2, A and B, respectively), with only 59 masses common to both ionization modes. In ESI−, most of the compounds identified were extracted with water and methanol. In the ESI− mode, compounds as carboxylic acids (e.g citric acid cycle) are preferentially identified [27]. In V. vinifera we found both malate (m/z 133.01403, [M-H]−) and citrate and/or isocitrate (m/z 191.01990, [M-H]−). Other carboxylic acids, particularly from the glucuronic acid pathway, were also found in our extracts exclusively in the ESI− analysis, including glucaric acid (m/z 209.03011, [M−H]−), glucuronic acid (m/z 193.03549, [M−H]−), gluconic acid (m/z 195.05114, [M−H]−), and ascorbic acid (m/z 175.02494, [M−H]−). Several flavonoids were also identified in negative ionization mode, but most of them were also present in the ESI+ analysis. When combining the information from both ionization modes, we increase the certainty of the presence of this subclass of metabolites, even if they are in minority in the total extract [31]. Examples include the flavonoids quercetin (m/z 447.09278, [M−H]− and m/z 449.10824, [M+H]+), quercetin 3-O-glucoside (m/z 463.08800, [M−H]− and m/z 465.10312, [M+H]+), and kaempferol 3-O-beta-D-glucosylgalactoside (m/z 609.14538, [M-H]− and m/z 633.14392, [M+Na]+), among others. A highly relevant phenolic compound, caffeic acid (m/z 179.03516, [M−H]−), and its derivative caffeic acid 3-glucoside (m/z 341.08732, [M−H]−), which confer resistance to pathogenic fungi in V. vinifera [6], were exclusively detected in ESI−.
Fig. 2.
Grapevine metabolite count and annotation. Four-way Venn diagram summarizing the number of shared metabolites in each fraction in positive (A) and negative (B) ionization modes; compound annotation by major classes, in both ionization modes (C).
It is clear that most of the compounds were identified in the positive ionization mode. In this mode, 55 compounds were common to all fractions, being the water and methanol fractions the ones with most compounds, 305 and 304, respectively, followed by the chloroform fraction where 287 compounds were identified. In fact, water, methanol and chloroform are the most commonly used solvents in metabolomics [29]. However, 90 unique masses were detected exclusively in the acetonitrile fraction using ESI+ (Fig. 2B). Among the detected compounds, we highlight the alkaloid valeroidine (m/z 242.17446, [M+H]+), previously identified in barley as a resistance-related constitutive metabolite in the defence against Fusarium graminearum [32]. Our results demonstrate that the extraction method that we developed increases the number of extracted metabolites, allowing higher metabolome coverage.
Concerning compound annotation, the detected metabolites were divided in eight different major classes: lipids; carbohydrates and carbohydrates conjugates; nucleosides, nucleotides and analogues; phytochemical compounds; heterocyclic compounds; organic acids and derivatives; benzenoids; and others (compounds with unknown annotation and organonitrogen, organooxygen and organophosphorus compounds), (Fig. 2C). The most represented class in V. vinifera cv Pinot noir is the Lipids class, defining more than half of the identified compounds, both in positive (71.7%) and negative (52.0%) ionization modes. This is not surprising, since we used organic solvents for metabolite extraction, and in fact lipidomics is a major sub-area inside metabolomics [33]. Phytochemical compounds and organic acids (and derivatives) correspond to 10.4% and 5.9% of the total identified metabolites in positive ionization mode, and 9.3% and 10.3% in negative ion mode, respectively.
Concerning metabolite intracellular concentration, we detected not only compounds present in higher levels, but also those found in lower amounts in plants. Among the metabolites present in high levels in plants [34], we identified in V. vinifera malate (m/z 133.01403, [M-H]−), citrate (m/z 191.01990, [M-H]−), sucrose (m/z 341.10899, [M-H]−) and the hexoses glucose and/or fructose, and/or galactose (m/z 179.05621, [M-H]−), and derivatives. Regarding compounds present in very low concentrations, which is the case for phytohormones, we were able to identify the jasmonic acid derivatives methyljasmonate (m/z 247.12980, [M+Na]+) and dihydrojasmonic acid (m/z 235.13018, [M+Na]+), acetylsalicylic acid (m/z 179.03516, [M-H]−), abscisic acid (m/z 265.14381, [M+H]+), the gibberellins A20 (m/z 355.15096, [M+Na]+) and allogibberic acid (m/z 307.12989, [M+Na]+), and brassinolide (m/z 503.33533, [M+Na]+). These results demonstrate that the proposed extraction method was able to extract a wide range of compounds, even those present in low amounts.
In our study, we were also able to detect several pesticides in grapevine leaves. These compounds were manually curated and excluded from the metabolome annotation. However, for vine leaves production and commercialization, we believe that quality assessment is important and the identification of pesticides is highly relevant for producers to authenticate the quality of the leaves. We were able to detect pesticides such as the insecticide fenthion (m/z 312.98909, [M+Cl]−) and the herbicides bromacil (m/z 283.00551, [M+Na]+) and terbacil (m/z 217.07368, [M+H]+), which are widely used in vineyards to control parasites, insects and fungi. These compounds have serious negative effects in our health, not only by environmental contamination and accidental or intentional poisonings [35], but also by their presence in processed products like grapes and wine [36]. Hence, the development of new techniques able to detect pesticides in food products are much needed and the method described here may be a good starting point.
4. Concluding remarks
In conclusion, we developed a metabolite extraction methodology suitable to use with DI-FTICR analysis for untargeted metabolomics. With our extraction protocol, we increased the extraction of polar and non-polar compounds, covering all major classes found in plants. We were able to identify 719 unique masses and also some pesticides in V. vinifera cv Pinot noir leaves.
Conflict of interest
None.
Acknowledgments
This work was supported by projects EXPL/BBB-BIO/0439/2013, REDE/1501/REM/2005, UID/MULTI/00612/2013, PEst-OE/BIA/UI4046/2014 and grant SFRH/BPD/99712/2014 from Fundação para a Ciência e Tecnologia (Portugal), and by the European FP7 project PERSSILAA (grant agreement 610359).
Contributor Information
Andreia Figueiredo, Email: aafigueiredo@fc.ul.pt.
Marta Sousa Silva, Email: mfsilva@fc.ul.pt.
References
- 1.Kosar M., Kupeli E., Malyer H., Uylaser V., Turkben C., Baser K.H. Effect of brining on biological activity of leaves of Vitis vinifera L: (Cv. Sultani Cekirdeksiz) from Turkey. J Agric Food Chem. 2007;55:4596–4603. doi: 10.1021/jf070130s. [DOI] [PubMed] [Google Scholar]
- 2.Batovska D.I., Todorova I.T., Parushev S.P., Nedelcheva D.V., Bankova V.S., Popov S.S., Ivanova I.I., Batovski S.A. Biomarkers for the prediction of the resistance and susceptibility of grapevine leaves to downy mildew. J. Plant Physiol. 2009;166:781–785. doi: 10.1016/j.jplph.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Batovska D.I., Todorova I.T., Nedelcheva D.V., Parushev S.P., Atanassov A.I., Hvarleva T.D., Djakova G.J., Bankova V.S., Popov S.S. Preliminary study on biomarkers for the fungal resistance in Vitis vinifera leaves. J. Plant Physiol. 2008;165:791–795. doi: 10.1016/j.jplph.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Fernie A.R., Trethewey R.N., Krotzky A.J., Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 5.Ali K., Maltese F., Figueiredo A., Rex M., Fortes A.M., Zyprian E., Pais M.S., Verpoorte R., Choi, YH Alterations in grapevine leaf metabolism upon inoculation with Plasmopara viticola in different time-points. Plant Sci. 2012;191–192:100–107. doi: 10.1016/j.plantsci.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Figueiredo A., Fortes A.M., Ferreira S., Sebastiana M., Choi Y.H., Sousa L., Acioli-Santos B., Pessoa F., Verpoorte R., Pais M.S. Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J. Exp. Bot. 2008;59:3371–3381. doi: 10.1093/jxb/ern187. [DOI] [PubMed] [Google Scholar]
- 7.Gika H.G., Theodoridis G.A., Vrhovsek U., Mattivi F. Quantitative profiling of polar primary metabolites using hydrophilic interaction ultrahigh performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2012;1259:121–127. doi: 10.1016/j.chroma.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Pan Z., Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 9.Han J., Danell R.M., Patel J.R., Gumerov D.R., Scarlett C.O., Speir J.P., Parker C.E., Rusyn I., Zeisel S., Borchers C.H. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 2008;4:128–140. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirwan J.A., Weber R.J., Broadhurst D.I., Viant M.R. Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci. Data. 2014;1:140012. doi: 10.1038/sdata.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H.K., Verpoorte R. Sample preparation for plant metabolomics. Phytochem. Anal. 2009;21:4–13. doi: 10.1002/pca.1188. [DOI] [PubMed] [Google Scholar]
- 12.Theodoridis G.A., Gika H.G., Franceschi P., Caputi L., Arapitsas P., Scholz M., Masuero D., Wehrens R., Vrhovsek U., Mattivi F. LC-MS based global metabolite profiling of grapes: solvent extraction protocol optimization. Metabolomics. 2012;8:175–185. [Google Scholar]
- 13.Lucio M., Fekete A., Weigert C., Wagele B., Zhao X., Chen J., Fritsche A., Haring H.U., Schleicher E.D., Xu G., Schmitt-Kopplin P., Lehmann R. Insulin sensitivity is reflected by characteristic metabolic fingerprints—a Fourier transform mass spectrometric non-targeted metabolomics approach. PLoS One. 2010;5:e13317. doi: 10.1371/journal.pone.0013317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suhre K., Schmitt-Kopplin P. MassTRIX: mass translator into pathways. Nucleic Acids Res. 2008;36:481–484. doi: 10.1093/nar/gkn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afendi F.M., Okada T., Yamazaki M., Hirai-Morita A., Nakamura Y., Nakamura K., Ikeda S., Takahashi H., Altaf-Ul-Amin M., Darusman L.K., Saito K., Kanaya S. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahy E., Sud M., Cotter D., Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., Edison A., Fiehn O., Higashi R., Nair K.S., Sumner S., Subramaniam S. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1042. (First published online October 13, 2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker L., Poutaraud A., Hamm G., Muller J.F., Merdinoglu D., Carre V., Chaimbault P. Metabolic study of grapevine leaves infected by downy mildew using negative ion electrospray—Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chim. Acta. 2013;795:44–51. doi: 10.1016/j.aca.2013.07.068. [DOI] [PubMed] [Google Scholar]
- 21.Brown S.C., Kruppa G., Dasseux J.L. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom. Rev. 2005;24:223–231. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 22.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 23.Villas-Boas S.G., Mas S., Akesson M., Smedsgaard J., Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- 24.Aharoni A., Ric de Vos C.H., Verhoeven H.A., Maliepaard C.A., Kruppa G., Bino R., Goodenowe D.B. Nontargeted metabolome analysis by use of fourier transform ion cyclotron mass spectrometry. OMICS. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- 25.Motohashi R., Satou M., Myouga F., Oikawa A., Ohta D. Arabidopsis metabolome analysis using infusion ESI FT-ICR/MS. Bio-protocol. 2015;5:e1463. [Google Scholar]
- 26.Oikawa A., Nakamura Y., Ogura T., Kimura A., Suzuki H., Sakurai N., Shinbo Y., Shibata D., Kanaya S., Ohta D. Clarification of pathway-specific inhibition by Fourier transform ion cyclotron resonance/mass spectrometry-based metabolic phenotyping studies. Plant Physiol. 2006;142:398–413. doi: 10.1104/pp.106.080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques A.P., Serralheiro M.L., Ferreira A.E., Freire A.P., Cordeiro C., Sousa Silva M. Metabolomics for undergraduates: identification and pathway assignment of mitochondrial metabolites. Biochem. Mol. Biol. Educ. 2016;44:38–54. doi: 10.1002/bmb.20919. [DOI] [PubMed] [Google Scholar]
- 28.Wu H., Southam A.D., Hines A., Viant M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008;372:204–212. doi: 10.1016/j.ab.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Villas-Boas S.G. Sampling and sample preparation. In: Villas-Boas S.G., Nielsen J., Smedsgaard J., Hansen M.A.E., Roessner-Tunali U., editors. Metabolome Analysis: An Introduction. John Wiley & Sons; New Jersey: 2007. pp. 39–82. [Google Scholar]
- 30.Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 31.Cuyckens F., Claeys M. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectrom. 2004;39:1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- 32.Bollina V., Kushalappa A.C., Choo T.M., Dion Y., Rioux S. Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol. Biol. 2011;77:355–370. doi: 10.1007/s11103-011-9815-8. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths W.J., Ogundare M., Williams C.M., Wang Y. On the future of omics: lipidomics. J. Inherit. Metab. Dis. 2011;34:583–592. doi: 10.1007/s10545-010-9274-4. [DOI] [PubMed] [Google Scholar]
- 34.Roessner U. Plant metabolomics. In: Villas-Boas S.G., Nielsen J., Smedsgaard J., Hansen M.A.E., Roessner-Tunali U., editors. Metabolome Analysis: An Introduction. John Wiley & Sons; New Jersey: 2007. pp. 215–238. [Google Scholar]
- 35.WHO . Preventing disease through healthy environments. In: WHO, editor. Public Health and Environment. World Health Organization; Geneva: 2010. p. 6. [Google Scholar]
- 36.Cabras P., Angioni A. Pesticide residues in grapes, wine, and their processing products. J. Agric. Food Chem. 2000;48:967–973. doi: 10.1021/jf990727a. [DOI] [PubMed] [Google Scholar]