Abstract
Accurate assessment of human epidermal growth factor receptor 2 (HER-2) is crucial in selecting patients for targeted therapy. Commonly used methods for HER-2 testing are immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). Here we presented the implementation, optimization and standardization of two FISH protocols using breast cancer samples and assessed the impact of pre-analytical and analytical factors on HER-2 testing. Formalin fixed paraffin embedded (FFPE) tissue samples from 70 breast cancer patients were tested for HER-2 using PathVysion™ HER-2 DNA Probe Kit and two different paraffin pretreatment kits, Vysis/Abbott Paraffin Pretreatment Reagent Kit (40 samples) and DAKO Histology FISH Accessory Kit (30 samples). The concordance between FISH and IHC results was determined. Pre-analytical and analytical factors (i.e., fixation, baking, digestion, and post-hybridization washing) affected the efficiency and quality of hybridization. The overall hybridization success in our study was 98.6% (69/70); the failure rate was 1.4%. The DAKO pretreatment kit was more time-efficient and resulted in more uniform signals that were easier to interpret, compared to the Vysis/Abbott kit. The overall concordance between IHC and FISH was 84.06%, kappa coefficient 0.5976 (p < 0.0001). The greatest discordance (82%) between IHC and FISH was observed in IHC 2+ group. A standardized FISH protocol for HER-2 assessment, with high hybridization efficiency, is necessary due to variability in tissue processing and individual tissue characteristics. Differences in the pre-analytical and analytical steps can affect the hybridization quality and efficiency. The use of DAKO pretreatment kit is time-saving and cost-effective.
Keywords: Breast cancer, HER-2, fluorescent in situ hybridization, FISH, standardization, IHC, immunohistochemistry, concordance
INTRODUCTION
Successful treatment of patients with breast cancer depends on many complex factors, such as: early detection of tumor, tumor biology, pathological prognostic factors, and tumor biomarkers (e.g., hormone receptor status and human epidermal growth factor receptor 2 [HER-2] status) [1]. The amplification of the HER-2 (also known as HER-2) gene and subsequent overexpression of the protein are found in 15-20% of breast cancers; both are negative predictors of survival and are associated with poor prognosis and high metastatic potential [2-4]. The HER-2 gene is a molecular target for specific therapies that are associated with a good clinical outcome [5-7]. Accurate determination of HER-2 status using precise, highly sensitive and specific tests is imperative in selecting patients for targeted therapy [5-7]. The most commonly used methods for HER-2 assessment in breast cancer specimens are immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH), performed on formalin-fixed, paraffin-embedded (FFPE) tissue samples [8-9].
FISH is a molecular cytogenetic technique that enables the detection of genetic abnormalities (e.g., gene amplifications, deletions, and chromosomal rearrangements) in metaphase and interphase cells [2-4,10-11]. This technique is based on the hybridization of labeled probes to the complementary DNA or RNA sequences [11]. In cancer cells, FISH determines the number of copies of the HER-2 gene and centromere of chromosome 17 (CEP17) with fluorescently labeled DNA probes specific for those genomic regions. The stability of DNA makes FISH less sensitive to factors associated with preservation and storage of tissue samples, compared to IHC [5]. FISH is a gold standard for HER-2 assessment and a superior method in selecting patients for the treatment with trastuzumab [4-5,12-14]. However, the use of FFPE tissue specimens encounters a number of technical problems, parallel to those found in IHC, which emphasizes the need for the standardization of protocols for tissue processing [3,11,15,16]. Factors that are important to consider in hybridization methods include: variations in cold ischemia time (CIT), duration of fixation, enzymatic pretreatment, hybridization conditions, and post-hybridization washing [11,15].
Here we presented the implementation, optimization, and standardization of two FISH protocols based on identical HER-2 DNA kit but with two different paraffin pretreatment kits, using FFPE breast cancer samples. In a series of experiments, we assessed the impact of pre-analytical and analytical factors (i.e., fixation, baking, digestion, and post-hybridization washing) on the HER-2 testing.
MATERIALS AND METHODS
Tissue samples
FFPE tissue blocks from 70 patients diagnosed with invasive breast carcinoma of no special type (NST), between 2014 and 2016, were used. We included patients who underwent radical mastectomy and did not receive neoadjuvant therapy. All tissue samples were previously fixed in 10% buffered formalin at room temperature with unknown CIT. Out of 70 samples, 40 samples were received from our hospital and they were fixed overnight (24 hours) at our institute. The remaining 30 samples were obtained from outside hospitals, with fixation of more than 48 hours (48-96 hours).
HER-2 testing by IHC was performed in all cases as part of a routine practice at our institute. Regardless of the IHC results, FISH testing was also performed in all cases, using parallel sections from the same FFPE tissue blocks used for IHC.
Immunohistochemistry
IHC was performed on a BenchMark GX automated staining instrument (Ventana Medical Systems, Inc., USA) using Ventana anti-HER-2/neu rabbit monoclonal primary antibody, clone 4B5 and UltraVIEW universal DAB Detection Kit (Ventana). Briefly, after deparaffinization with EZ Prep (Ventana), the slides were pretreated with Cell Conditioning 1 (Ventana) for 36 minutes at 100°C and then incubated with the anti-HER-2 primary antibody for 20 minutes at 37°C. The antibody was detected with a chromogen, then counterstained with hematoxylin, and after that with bluing reagent, for 4 minutes in both steps. The slides were examined and scored according to the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) criteria [9].
Fluorescent in situ hybridization
We used two FISH protocols for HER-2 detection, with an identical HER-2 DNA probe kit but with two different paraffin pretreatment kits. The PathVysion™ HER-2 DNA Probe Kit (Vysis/Abbott, IL, USA) enables simultaneous detection of the HER-2 and CEP17 with two fluorophore-labeled DNA probes: SpectrumOrange labeled DNA probe for the HER-2 locus and SpectrumGreen labeled DNA probe for the CEP17. The protocol 1 included the Paraffin Pretreatment Reagent Kit (Vysis/Abbott, IL, USA), while for the protocol 2, the Histology FISH Accessory Kit (DAKO, Denmark) was used. To optimize the two FISH protocols, due to different fixation times in the pre-analytical phase, we performed a series of experiments on 5 representative tissue samples and analyzed the effect of the pretreatment, digestion and post-hybridization washing, as presented in Table 1. After the evaluation of the results, optimal intervals for the key steps in both protocols were defined, according to which the remaining tissue samples were processed: 40 samples were processed with protocol 1 and 30 samples with protocol 2 (Table 2 and 3). For each patient, two 4-µm thick tumor sections from the same FFPE tissue block were used: one section for standard hematoxylin and eosin (H&E) staining to identify and mark the lesion of interest, and the other section for FISH analysis.
TABLE 1.
Different experimental steps in two fluorescence in situ hybridization protocols applied to 5 representative formalin fixed paraffin embedded (FFPE) breast cancer samples
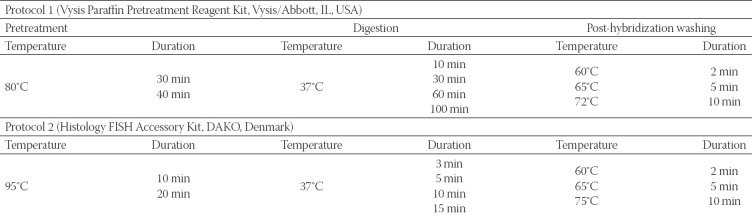
TABLE 2.
Detailed description of analytical steps in two fluorescence in situ hybridization (FISH) protocols
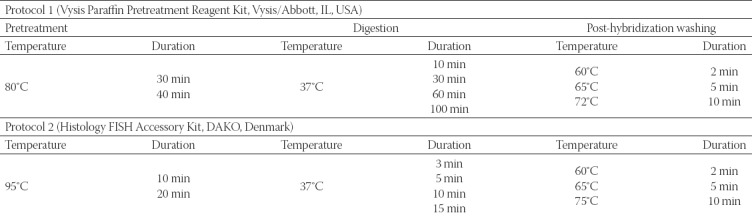
TABLE 3.
Number of formalin fixed paraffin embedded (FFPE) breast cancer samples according to different pre-analytical and analytical steps

From the 40 samples processed with protocol 1, 20 samples that were fixed for 24 hours were mounted on silanized slides (Canelli, Italy) and baked at 56°C overnight. The other 20 samples were fixed for 48-96 hours and mounted on positively charged adhesive slides (Thermo Scientific, Germany). Among these 20 samples, the first 10 slides were baked at 56°C overnight, while the second 10 slides were baked at 70°C for 35 minutes.
Out of the 30 samples processed with protocol 2, 20 samples fixed for 24 hours were mounted on silanized slides, while 10 samples that were fixed for more than 48 hours were mounted on positively charged adhesive slides. All samples in this protocol were baked at 56°C overnight.
Control tissue sections (known cases of positive and negative HER-2 amplification) were used to ensure proper performance of the test.
Before reading the results, the samples were left for 1 hour in a refrigerator at +4°C to stabilize the signals. If not evaluated the same day, the samples were stored in the refrigerator at -20°C and analyzed in the next two days.
Evaluation of the results
For the accurate localization of the invasive component of the cancer, the FISH assays were viewed in conjunction with the H&E sections from the same FFPE tissue block (Figure 1). A 4’,6-diamidino-2-phenylindole (DAPI) counterstain was used to identify the tumor nuclei at ×100 magnification. The signals were analyzed at ×1000 magnification, using the appropriate filter: SpectrumOrange for HER-2 and SpectrumGreen for CEP 17. Only the tumor nuclei with the intact nuclear membrane, that were not overlapped and were approximately of the same diameter, were analyzed (Figure 1). The normal ductal epithelium, stromal cells, and lymphocytes served as internal control (Figure 2). The results were interpreted according to the new recommendations of the ASCO/CAP, where HER-2 status is defined as positive when the HER-2/CEP17 ratio is >2, and negative when the ratio is <2 (Figure 2) [9]. The results were analyzed using the BX43 fluorescence microscope (Olympus Corporation, Japan) equipped with SpectrumOrange, SpectrumGreen and DAPI mono filters and SpectrumOrange/SpectrumGreen double bandpass filter. The slides were photographed and documented with the XM camera (Olympus) and analyzed using the Olympus cellSens Standard software, Version 1.15.
FIGURE 1.
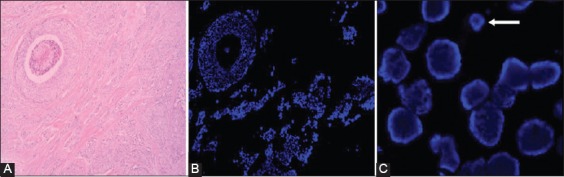
(A) Hematoxylin and eosin (H&E) stained section of a breast carcinoma (H&E, ×100); (B) 4’,6-diamidino-2-phenylindole (DAPI) stained parallel section with invasive (lower right) and in situ breast cancer [upper left] (DAPI counterstain, ×100); (C) Only nuclei with the intact nuclear membrane, of approximately the same size, and that were not overlapped were evaluated. The small nuclei (arrow) were not analyzed due to nuclear truncation (DAPI counterstain, ×1000).
FIGURE 2.
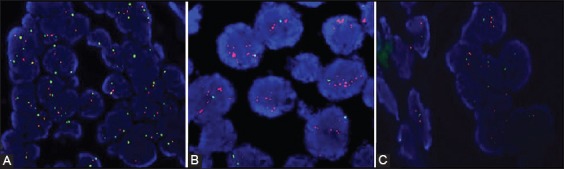
(A) A case with negative HER-2 amplification status, HER-2/CEP17 ratio <2, DAKO Paraffin Pretreatment Kit (4’,6-diamidino-2-phenylindole [DAPI] counterstain,×1000); (B) A case with positive HER-2 amplification status, HER-2/CEP17 > 2, DAKO Paraffin Pretreatment Kit (DAPI counterstain, ×1000); (C) Stromal cells (left half) were used as the internal control to tumor cells (right half), DAKO Paraffin Pretreatment Kit (DAPI counterstain, ×1000).
After evaluating the IHC and FISH results for HER-2, the concordance between the two methods was determined.
Statistical analysis
Statistical analyses were performed using SPSS Statistics for Windows, Version 17.0. (SPSS Inc., Chicago, USA). Data are presented as absolute numbers and percentages. Fisher’s exact two tailed test was used to compare the FISH positive and negative results in relation to the IHC results. A value of p < 0.05 was considered statistically significant. The concordance was defined as the proportion of cases scored IHC 2+/3+ and positive for FISH, or the proportion of cases scored IHC 0/1+ and negative for FISH. Kappa test was used to determine the concordance between the two methods.
RESULTS
Implementation and standardization of FISH protocols
The two FISH protocols were first applied on 5 representative FFPE breast cancer tissues and the effect of the pretreatment, digestion and post-hybridization washing on the efficiency and quality of hybridization was analyzed (Table 4 and 5).
TABLE 4.
The effect of pretreatment and digestion on fluorescence in situ hybridization results
TABLE 5.
Effect of post-hybridization washing step on hybridization results
The duration of incubation with the pretreatment solution showed no effect on the efficiency and quality of hybridization, so the shorter periods were chosen as more appropriate, as follows: 30-minute pretreatment incubation at 80°C for the Vysis/Abbott pretreatment kit and 10-minute pretreatment incubation at 90°C for the DAKO pretreatment kit. Regarding the effect of enzymatic digestion, the best results were obtained with 100-minute digestion with Vysis protease buffer at 37°C in protocol 1 and 5-minute digestion with pepsin at 37°C in protocol 2. Digestion for shorter time led to increased DAPI binding, background autofluorescence, incomplete hybridization or unspecific signal at the periphery of the cells, while prolonged digestion caused changes in the nuclear morphology and led to loss of the signal (Figure 3). The post-hybridization washing had a greater effect on the hybridization signals. When the conditions for post-hybridization washing were inappropriate, background autofluorescence or loss of the signals appeared. The optimal results were obtained with 2-minute washing at 72°C using 2×SSC/0.3% NP-40 wash solution (Vysis) in protocol 1, and with 5-minute washing with DAKO stringent wash buffer (Tris/HCl) at 60°C in protocol 2. High temperature of the wash buffers and prolonged washing led to loss of the signals, while the washing for shorter period and at lower temperature gave unspecific signals and background autofluorescence (Figure 3).
FIGURE 3.
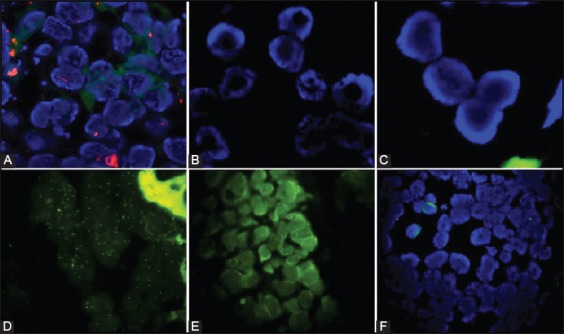
Examples of fluorescence in situ hybridization (FISH) assays that were inadequate for scoring. (A) Increased binding of DAPI, autofluorescence, and absent signals in nuclei due to incomplete hybridization and shorter digestion, Vysis/Abbott pretreatment kit with 10-minute protease digestion (DAPI counterstain, ×1000); (B) Disrupted nuclear morphology, inadequate DAPI binding and loss of signals due to prolonged pepsin digestion, DAKO pretreatment kit with 15-minute pepsin digestion (DAPI counterstain, ×1000); (C) Loss of signals due to prolonged post-hybridization washing with higher temperature, DAKO pretreatment kit, 20×Tris/HCl, 75°C/10 minutes (DAPI counterstain, ×1000); (D) Excessive, non-specific signals due to non-stringent wash, (Spectrum green, ×1000); (E) Background autofluorescence masking the signals, Vysis/Abbott pretreatment kit, 2×SSC/0.3% NP-40, 60°C/5 min (Spectrum Green, ×1000); (F) Unsuccessful hybridization in case of prolonged fixation time, using Vysis/Abbott pretreatment kit (DAPI counterstain, ×1000). SSC: Saline sodium citrate; NP40: Nonionic detergent; DAPI: 4’,6-diamidino-2-phenylindole.
After optimizing the main pre-analytical and analytical steps (pretreatment, enzyme digestion, and post-hybridization washing) for both FISH protocols, 40/70 cases were tested with protocol 1 using the Vysis/Abbott pretreatment kit, while 30/70 cases were tested with DAKO pretreatment kit (protocol 2, Table 2).
Among the 70 cases analyzed by two different FISH protocols and with different fixation times, the overall hybridization efficiency was 98.6% (69/70); the failure rate was 1.4%. The hybridization was not successful only in one tissue sample processed with protocol 1 and with 96-hour fixation. This sample was then processed with protocol 2, but the hybridization was again not achieved.
Greater adherence of tissue to the slides was observed with positively charged silanized slides. Namely, 4 tissue sections mounted on positively charged adhesive slides were wasted versus 2 tissue sections that were lost when using silanized slides. Moreover, the tissue loss was detected only in samples processed with protocol 1 (Vysis/Abbott pretreatment kit). The overnight baking at 56°C led to greater tissue adherence, with only 8.3% (5/60) tissue loss, contrary to 10% (1/10) tissue loss with 35 minute-baking on a hot plate.
Both protocols resulted in good hybridization quality, i.e., preserved nuclear morphology, scorable signals and little or no autofluorescence. The green signals were slightly stronger and more prominent over the red signals when using the Vysis/Abbott pretreatment kit, while both signals were more uniform and in some cases easier to interpret with the DAKO pretreatment kit (Figure 4).
FIGURE 4.
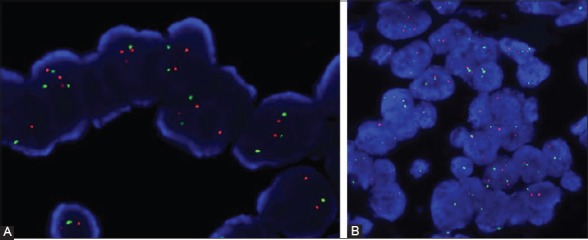
(A) Strong, uniform signals easy to interpret obtained with DAKO pretreatment kit (4’,6-diamidino-2-phenylindole [DAPI] counterstain, ×1000); (B) Strong scorable signals and predominant green signals obtained with Vysis/Abbott pretreatment kit (DAPI counterstain, ×1000).
Correlation of IHC and FISH results for HER-2
Overall, the successful hybridization was achieved in 69/70 analyzed cases (98.6%). The one sample in which the hybridization was not successful was excluded from the correlation analysis.
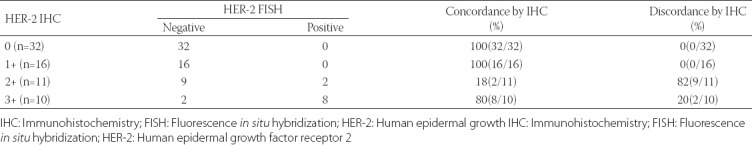
Out of the 69 cases, 10 (14.50%) were FISH positive and 59 (85.50%) were FISH negative. The IHC results for HER-2 showed that 32 (46.38%) cases were scored as IHC 0, followed by 16 (23.19%) scored as IHC 1+, 11 (15.94%) scored as IHC 2+, and 10 (14.50%) scored as IHC 3+. None of the cases scored as IHC 0 and 1+ were FISH positive. Two of the 11 IHC 2+ cases and 8/10 IHC 3+ cases were FISH positive. As indicated in Table 6, the concordance rate for IHC 0 and 1+ cases was 100%, for IHC 2+ 18%, and for IHC 3+ it was 80%. When IHC 2+ and IHC 3+ cases were grouped together, the overall concordance between the two methods was 84.06% (58/69), and the kappa coefficient was 0.5976 (p < 0.0001).
TABLE 6.
Comparison of IHC and FISH results for HER-2
DISCUSSION
The accurate and reproducible assessment of HER-2 status using standardized tests requires strict adherence to the quality control standards [2,16-17]. Factors that may affect FISH testing include: CIT, fixation time, tissue processing, quality of the probe, and DNA accessibility [1,10,14,18-19]. The use of FISH technique with archived tissue blocks can be especially challenging, primarily because the formalin fixation affects the accessibility of the target DNA and an additional optimization of the pretreatment and hybridization steps is required [1,10,14,18-19]. To optimize two FISH protocols we performed a series of experiments on 5 representative samples with different fixation times, evaluating the effect of pretreatment, digestion and post-hybridization washing on the efficiency and quality of hybridization. Pretreatment reagents increase the efficiency of probe hybridization by increasing the accessibility of the target DNA and reducing background autofluorescence [1,11]. In our study, the hybridization was not affected by the incubation time of pretreatment solutions in the two protocols. On the contrary, the enzymatic digestion altered the hybridization results, so optimizing this step in FISH analysis appears to be important. Namely, the longer incubation with pepsin/protease caused changes in the nuclear morphology, inadequate DAPI binding and loss of the signal, while shorter digestion resulted in increased DAPI absorption, autofluorescence or absent signals. Unspecific signals may be seen at the periphery of the cells in some cases, as a result of capturing the probe between the cells due to insufficient permeabilization.
We achieved the best results with 100-minute incubation with Vysis protease and 5-minute incubation with DAKO pepsin. Changes in the nuclear morphology observed after enzymatic digestion maybe related to the tissue characteristics or previous steps in tissue processing. In addition, we found that inappropriate post-hybridization washing significantly affected the intensity and stability of signals, especially the green signals. Thus, the temperature should be adjusted in relation to the fixation time and age of archived tissue samples [1,11]. The best results for Vysis wash buffer were obtained with washing at 72°C for 2 minutes, and for DAKO stringent wash buffer with 5-minute washing at 60°C.
After optimizing the pretreatment, digestion and post-hybridization washing conditions for the two FISH protocols, we processed all 70 FFPE breast cancer samples (40 samples were treated with the Vysis/Abbott pretreatment kit and 30 samples with DAKO pretreatment kit). The efficiency of the hybridization and the quality of the signals were analyzed and the results of the two FISH protocols were compared. Our study group was relatively homogenous considering the type and age of tissue samples, but relatively heterogeneous with regard to the fixation time (range: 24-96 hours).
The failure rate for FISH test was reported as 5% [20]. Using different incubation times for tissue fixation and two commercial kits for paraffin pretreatment, the overall hybridization efficiency in our study was 98.6%. It also appears that the different fixation conditions did not affect the overall efficiency and quality of hybridization. The failure rate of only 1.4% in our study may be the result of prolonged fixation and CIT. Our results are in agreement with previous studies, which indicate that FISH is less sensitive to tissue fixation methods owing to relative stability of DNA [5]. Considering the quality of fluorescence signals, we obtained the optimal results with 24-hour fixation. Inadequate formalin fixation before tissue processing leads to the fixation of tissue in alcohol in the processor, contributing to background autofluorescence. On the other hand, overfixation results in extensive cross-linking of proteins that will require more aggressive pretreatment, and may lead to the damage of nuclear morphology and loss of signal [1,10,14,18-19]. In our study, the tissue samples mounted on silanized slides showed good adherence, regardless of the FISH protocol used for sample processing. The overnight baking at 56°C resulted in greater tissue adherence, compared to 35-minute baking on a hot plate. However, we should emphasize here that all samples with shorter baking time were mounted on positively charged adhesive slides and processed with more aggressive Vysis/Abbott pretreatment kit, so we cannot exclude the impact of the pretreatment or type of slides that were used. Also the number of samples with different characteristics in each group was small in our study. We did not find other studies analyzing the effect of different types of microscopic slides or the backing process on FISH testing, so additional research is needed to confirm our findings. The duration of pretreatment is approximately 4.5 hours with Vysis/Abbott pretreatment kit, and approximately 2.5 hours with DAKO pretreatment kit. The signals obtained after DAKO pretreatment were more uniform, easier to interpret, and with higher reproducibility. In cases where we had weak green signals after DAKO pretreatment, the Vysis pretreatment kit gave much better results.
As recommended in the literature [1,3,21], the FISH results should not be analyzed if there is excessive background fluorescence that may mask the signal in more than 10% of cells, if the signals are weak and not uniform in more than 25% of the cells, if autofluorescence is high or if the nuclear morphology is not clearly visible. FISH signals fade over time, so it is recommended to make representative images from each case whenever possible. The slides can be stored at -20°C for at least 12 months [19].
When using a conventional camera for recording the results of FISH test the signals must be clear and with strong intensity, especially when using more than one fluorochrome, because less light passes through the combined filters, and parts of the signals are lost [11]. This problem can be overcome by using the appropriate software for data analysis [11]. We used the Olympus cellSens Standard, Version 1.15 software, which helps to reduce nonspecific background staining and improve the intensity of signals.
Finally, we correlated the FISH and IHC results for HER-2 and determined the concordance between the two methods. The concordance rate for IHC 0 and 1+ cases was 100%, while for IHC 2+ and 3+ was 18.18% and 80%, respectively. The overall concordance in our study was 84.06%. Using 50 breast cancer specimens Sui et al. [22] reported overall concordance of 82%, with FISH positivity in 5/26 of IHC 0/1+ cases, 7/10 of IHC 2+ cases, and 13/14 of IHC 3+ cases [22]. The same concordance rate was reported by Dybdal et al. [23]. The authors showed the agreement between FISH and IHC method to be 97%, 93%, 24% and 89% for IHC 0, 1+, 2+ and 3+, respectively. Similar to our results, the discordance rate was the highest in IHC 2+ group [23]. The discordance rate observed in our study is probably due to a lack of standardization in tissue processing (i.e., CIT and fixation conditions). Other authors reported higher concordance rates that were related to improved specimen handling, test standardization, and experience in the interpretation of HER-2 IHC [24].
CONCLUSION
A standardized FISH protocol for HER-2 assessment, with high hybridization efficiency, is necessary due to variability between individual tissue samples. Differences in the pre-analytical and analytical steps can affect the hybridization quality and efficiency. Optimization of digestion and post-hybridization washing procedures appears to be important for achieving optimal hybridization conditions. The duration of tissue fixation has no major effect on the hybridization efficiency, but the optimal signal quality was achieved with overnight (24 hours) fixation. Good tissue adherence was obtained with overnight baking at 56°C using positively charged silanized slides. Although both protocols showed high hybridization efficiency, the pretreatment with DAKO Histology kit is more preferable because of the shorter procedure, uniform signals and lower cost. The overall concordance between the IHC and FISH in our study was 84.06%, with the highest concordance rate for negative IHC and the lowest for equivocal IHC results.
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
REFERENCES
- 1.Hicks DG, Tubbs RR. Assessment of the HER2 status in breast cancer by fluorescencein situhybridization: A technical review with interpretive guidelines. Hum Pathol. 2005;36(3):250–61. doi: 10.1016/j.humpath.2004.11.010. https://doi.org/10.1016/j.humpath.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: Ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14(4):320–68. doi: 10.1634/theoncologist.2008-0230. https://doi.org/10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 3.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Richard J, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18–43. doi: 10.5858/2007-131-18-ASOCCO. DOI: 10.1043/1543-2165(2007)131[18:ASOCCO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 4.Stocker A, Hilbers ML, Gauthier C, Grogg J, Kullak-Ublick GA, Seifert B, et al. HER2/CEP17 ratios and clinical outcome in HER2-positive early breast cancer undergoing trastuzumab-containing therapy. PLoS One. 2016;11(7):e0159176. doi: 10.1371/journal.pone.0159176. https://doi.org/10.1371/journal.pone.0159176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for human epidermal growth factor receptor 2 testing: Biologic and methodologic considerations. J Clin Oncol. 2009;27(8):1323–33. doi: 10.1200/JCO.2007.14.8197. https://doi.org/10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 6.Sapino A, Goia M, Recupero D, Marchiò C. Current challenges for HER2 testing in diagnostic pathology: State of the art and controversial issues. Front Oncol. 2013;3:129. doi: 10.3389/fonc.2013.00129. https://doi.org/10.3389/fonc.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lottner C, Schwarz S, Diermeier S, Hartmann A, Knuechel R, Hofstaedter F, et al. Simultaneous detection of HER2/neu gene amplification and protein overexpression in paraffin-embedded breast cancer. J Pathol. 2005;205(5):577–84. doi: 10.1002/path.1742. https://doi.org/10.1002/path.1742. [DOI] [PubMed] [Google Scholar]
- 8.Dowsett M, Hanna WM, Kockx M, Penault-Llorca F, Ruschoff J, Gutjahr T, et al. Standardization of HER2 testing: Results of an international proficiency-testing ring study. Mod Pathol. 2007;20(5):584–91. doi: 10.1038/modpathol.3800774. https://doi.org/10.1038/modpathol.3800774. [DOI] [PubMed] [Google Scholar]
- 9.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/College of American pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. https://doi.org/10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 10.Andersen CL, Hostetter G, Grigoryan A, Sauter G, Kallioniemi A. Improved procedure for fluorescencein situhybridization on tissue microarrays. Cytometry. 2001;45(2):83–6. doi: 10.1002/1097-0320(20011001)45:2<83::aid-cyto1149>3.0.co;2-p. https://doi.org/10.1002/1097-0320(20011001)45:2<83:AID-CYTO1149>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 11.Petersen BL, Sorensen MC, Pedersen S, Rasmussen M. Fluorescencein situhybridization on formalin-fixed and paraffin- embedded tissue: Optimizing the method. Appl Immunohistochem Mol Morphol. 2004;12(3):259–65. doi: 10.1097/00129039-200409000-00013. https://doi.org/10.1097/00129039-200409000-00013. [DOI] [PubMed] [Google Scholar]
- 12.Pauletti G, Dandekar S, Rong H, Ramos L, Peng H, Seshadri R, et al. Assessment of methods for tissue-based detection of the HER-2/neu alteration in human breast cancer: A direct comparison of fluorescencein situhybridization and immunohistochemistry. J Clin Oncol. 2000;18(21):3651–64. doi: 10.1200/JCO.2000.18.21.3651. https://doi.org/10.1200/JCO.2000.18.21.3651. [DOI] [PubMed] [Google Scholar]
- 13.Gokhale S, Gatalica Z, Mohammad A, Rampy AI, Velagaleti Gopalrao VN. FISH for HER-2/neu in breast cancer: Standardization makes the difference! Indian J Cancer. 2004;41(4):152–8. [PubMed] [Google Scholar]
- 14.Mass RD, Press MF, Anderson S, Cobleigh MA, Vogel CL, Dybdal N, et al. Evaluation of clinical outcomes according to HER2 detection by fluorescencein situhybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer. 2005;6(3):240–6. doi: 10.3816/CBC.2005.n.026. https://doi.org/10.3816/CBC.2005.n.026. [DOI] [PubMed] [Google Scholar]
- 15.Mascarello JT, Hirsch B, Kearney HM, Ketterling RP, Olson SB, Quigley DI, et al. Section E9 of the American College of Medical Genetics technical standards and guidelines: Fluorescencein situhybridization. Genet Med. 2011;13(7):667–75. doi: 10.1097/GIM.0b013e3182227295. https://doi.org/10.1097/GIM.0b013e3182227295. [DOI] [PubMed] [Google Scholar]
- 16.Walker RA, Bartlett JM, Dowsett M, Ellis IO, Hanby AM, Jasani B, et al. HER2 testing in the UK: Further update to recommendations. J Clin Pathol. 2008;61(7):818–24. doi: 10.1136/jcp.2007.054866. https://doi.org/10.1136/jcp.2007.054866. [DOI] [PubMed] [Google Scholar]
- 17.Perez EA, Cortes J, Gonzalez-Angulo AM, Bartlett JM. HER2 testing: Current status and future directions. Cancer Treat Rev. 2014;40(2):276–84. doi: 10.1016/j.ctrv.2013.09.001. https://doi.org/10.1016/j.ctrv.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Hyytinen E, Visakorpi T, Kallioniemi A, Kallioniemi OP, Isola JJ. Improved technique for analysis of formalin-fixed, paraffin-embedded tumors by fluorescencein situhybridization. Cytometry. 1994;16(2):93–9. doi: 10.1002/cyto.990160202. https://doi.org/10.1002/cyto.990160202. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett JM, Starczynski J, Atkey N, Kay E, O’Grady A, Gandy M, et al. HER2 testing in the UK: Recommendations methods for breast and gastricin-situhybridization. J Clin Pathol. 2011;64(8):649–53. doi: 10.1136/jcp.2011.089847. https://doi.org/10.1136/jcp.2011.089847. [DOI] [PubMed] [Google Scholar]
- 20.Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291(16):1972–7. doi: 10.1001/jama.291.16.1972. https://doi.org/10.1001/jama.291.16.1972. [DOI] [PubMed] [Google Scholar]
- 21.Provenzano E, Johnson N. Overview of recommendations of HER2 testing in breast cancer. Diagn Histopathol. 2009;15(10):478–84. https://doi.org/10.1016/j.mpdhp.2009.07.006. [Google Scholar]
- 22.Sui W, Ou M, Chen J, Wan Y, Peng H, Qi M, et al. Comparison of immunohistochemistry (IHC) and fluorescencein situhybridization (FISH) assessment for her-2 status in breast cancer. World J Surg Oncol. 2009;7:83. doi: 10.1186/1477-7819-7-83. https://doi.org/10.1186/1477-7819-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dybdal N, Leiberman G, Anderson S, McCune B, Bajamonde A, Cohen RL, et al. Determination of HER2 gene amplification by fluorescencein situhybridization and concordance with the clinical trials immunohistochemical assay in women with metastatic breast cancer evaluated for treatment with trastuzumab. Breast Cancer Res Treat. 2005;93(1):3–11. doi: 10.1007/s10549-004-6275-8. https://doi.org/10.1007/s10549-004-6275-8. [DOI] [PubMed] [Google Scholar]
- 24.Sarode VR, Xiang QD, Christie A, Collins R, Rao R, Leitch AM, et al. Evaluation of HER2/neu status by immunohistochemistry using computer-based image analysis and correlation with gene amplification by fluorescencein situhybridization assay: A 10-year experience and impact of test standardization on concordance rate. Arch Pathol Lab Med. 2015;139(7):922–8. doi: 10.5858/arpa.2014-0127-OA. https://doi.org/10.5858/arpa.2014-0127-OA. [DOI] [PubMed] [Google Scholar]


