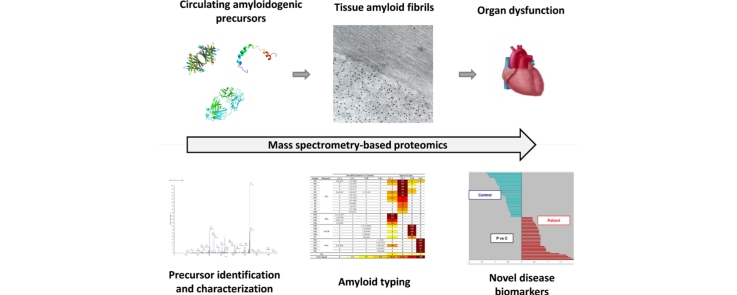
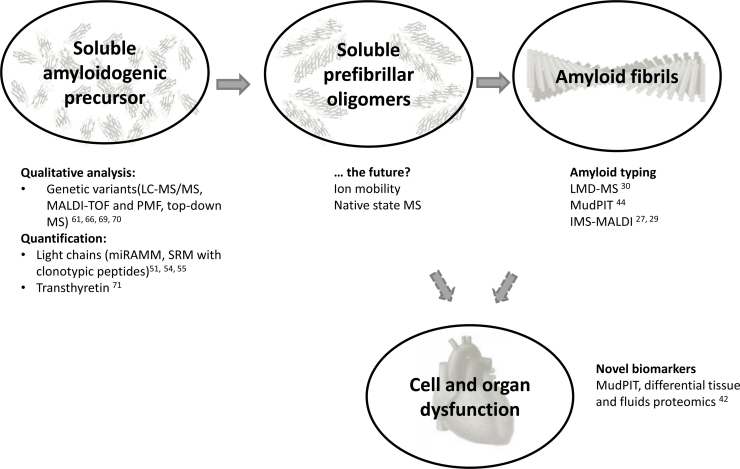
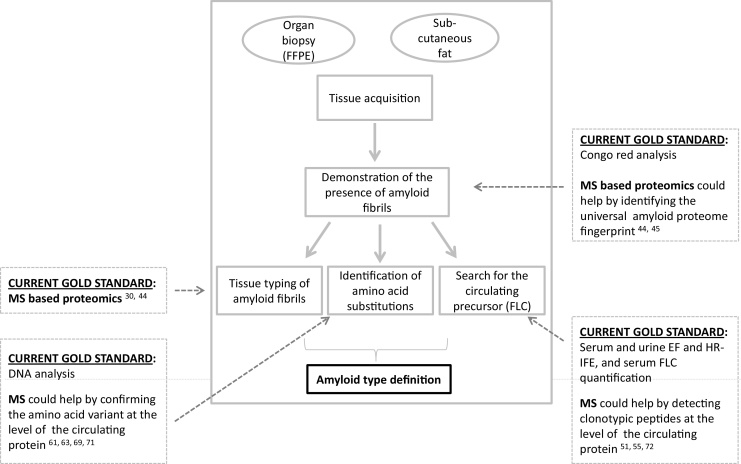
Graphical abstract
Keywords: Amyloidosis, Clinical proteomics, Tissue biomarkers, Amyloidogenic proteins, Proteotoxicity
Highlights
-
•
Proteomics is an established approach for diagnostic amyloid typing.
-
•
Mass spectrometry-based methods to analyze amyloid precursors have been developed.
-
•
Proteomic studies are ongoing to identify novel biomarkers and clarify disease mechanisms.
Abstract
Systemic amyloidoses are caused by misfolding-prone proteins that polymerize in tissues, causing organ dysfunction. Since proteins are etiological agents of these diseases, proteomics was soon recognized as a privileged instrument for their investigation. Mass spectrometry-based proteomics has acquired a fundamental role in management of systemic amyloidoses, being now considered a gold standard approach for amyloid typing. In parallel, approaches for analyzing circulating amyloid precursors have been developed. Moreover, differential and functional proteomics hold promise for identifying novel biomarkers and clarifying disease mechanisms. This review discusses recent proteomics achievements in systemic amyloidoses, providing a perspective on its present and future applications.
Among the examples of proteomics applications in clinical practice, typing of systemic amyloidoses has certainly gained a prominent role. Since the first descriptions of proteomic approaches for fibril identification, approximately a decade ago [1], [2], mass spectrometry (MS)-based amyloid typing has moved to being a unique example of a routinely used, accredited clinical proteomics assay [3]. Systemic amyloidoses have indeed been an especially fertile ground for the application of proteomics, which is not limited to tissue amyloid typing. Mass spectrometry-based analysis, in fact, plays a role in at least three additional aspects: (1) evaluation of amyloid precursors in body fluids; (2) identification of disease biomarkers; (3) study of the mechanisms of disease (Fig. 1). The unique success of clinical and translational proteomics in systemic amyloidoses is largely due to the primary role that proteins play in the pathogenesis of these diseases [4], [5]. Amyloidoses are a group of clinical conditions in which misfolding-prone autologous proteins acquire the ability to aggregate into amyloid fibrils and create insoluble interstitial tissue deposits. In the systemic forms, fibril deposition originates from proteins transported to target organs through blood, is widespread and affects vital organs, such as the hearth, kidney, liver, peripheral and autonomic nervous systems. The classification of amyloidoses is based on which protein originates the fibrils; an increasing number of different species are known to cause amyloid diseases in humans, and a dozen of these can cause systemic forms [5]. The common denominator of amyloidogenic proteins is the ability to acquire, upon loss of the native structure, a fibrillar conformation, and to aggregate into unbranched fibrils, which are remarkably similar, despite the wide differences in folding, function and sequence of the precursors [6]. The most common forms of systemic amyloidoses in industrialized countries are light chain (AL) amyloidosis, caused by the deposition of monoclonal immunoglobulin free light chains (FLC) produced by a bone marrow plasma cell clone [7], [8], [9], and transthyretin (ATTR) amyloidosis, either in its hereditary form (caused by misfolding-prone genetic variants of TTR), or related to deposition of wild type TTR with aging (wild type ATTR amyloidosis, previously known as senile systemic amyloidosis) [10], [11], [12], [13]. Less frequent hereditary and sporadic forms also exist [5]; importantly, the clinical presentation of all forms is largely overlapping, preventing their accurate distinction on a purely clinical basis.
Fig. 1.
Proteomics as a tool of clinical usefulness to investigate the different aspects of systemic amyloid diseases.
The diagnosis of amyloidosis is biopsy-based and requires the demonstration and characterization of amyloid deposits in tissues [7], [14]. Amyloid typing, i.e. definition of the core protein constituent of the fibrils, is crucial, since the various types of amyloidoses differ drastically in terms of pathogenesis, clinical course, prognosis and treatment. In recent years, radical therapeutic progresses have been made for several forms [8], [15], with approval of effective new treatments and regimens that can now change the natural history and improve survival. However, therapeutic orientation and patient management approaches differ completely among the amyloidosis types, and precise diagnostic typing is a crucial prerequisite for correct treatment.
The advent of diagnostic proteomics in the clinical management of systemic amyloidoses has been mostly driven by the need for an unbiased method for identifying the deposited protein, which would allow to overcome the drawbacks (mainly consisting in limited sensitivity and specificity) of traditional, immunohistochemistry (IHC)-based typing techniques. In parallel, a number of proteomics approaches have also been developed for the qualitative and quantitative analysis of the amyloid precursors – in particular TTR and monoclonal FLC – in body fluids. Moreover, the scarce knowledge on the mechanisms of proteotoxicity at the cellular and tissue level has prompted functional, targeted and high-throughput proteomics studies, which have begun to cast new light on the molecular events associated with the disease, and have identified protein candidates to be studied as potential novel tissue biomarkers.
This paper reviews the main achievements in the use of proteomics for the diagnosis and the study of systemic amyloidoses and related aspects, with particular focus on the most recent acquisitions, providing a critical presentation of the available literature and presenting future perspectives.
1. Mass spectrometry-based tissue amyloid typing: a decade of experience
It has been less than a decade since the pioneering studies on the use of MS for identifying amyloid (i.e. for defining the principal amyloid-forming protein) in clinical samples [1], [2]. During this time, proteomics has rapidly gained a critical role in the diagnostic definition of systemic amyloidoses in the routine clinical workup and has been applied for typing thousands of patients’ samples, up to the point that MS-based amyloid identification is now considered a gold standard disease typing approach [3], [16] (Fig. 2).
Fig. 2.
Workflow of amyloid diagnosis and typing on tissues and the role of MS in the various steps.
The advent of MS for the biochemical definition of the nature of amyloid fibrils had initially been advocated as a potential way to overcome the drawbacks of the traditional amyloid typing approaches, i.e. antibody-based methods. Although the use of immuno-electron microscopy [17] and/or performing the immunohistochemistry analysis in referral centers [18], [19] improve the performances of antibody-based methods, it is recognized that attributing the amyloid type on the basis of immunohistochemistry can suffer of limited diagnostic sensitivity and specificity, leading to inaccurate diagnosis and potentially fatal errors [20], [21], [22], [23], [24], [25]. The factors responsible for these drawbacks have been extensively discussed elsewhere [26] and can be largely attributed to the altered conformation and extensive processing of amyloid proteins in the fibrils. MS-based amyloid typing is antibody-independent and is based on a completely distinct concept, providing, as an output, the identity of the proteins present in a sample, including those deposited as fibrils. Specificity of amyloid typing is thus mediated by the specificity of protein identification through bioinformatics, and by the correct interpretation of the protein identification lists, to extract the correct amyloidogenic species.
The information obtained with a single proteomic analysis, however, is not limited to a list of identified proteins; the MS/MS assessment of peptide sequences, in fact, also allows the evaluation of other relevant features, such as amino acid variants and post-translational modifications (PTM), as outlined later. For these characteristics – antibody-independent identification, wealth of information provided, coupled to the rapidity and robustness of the current MS analytical methods – the success of proteomics in this field has been explosive.
Although procedures for amyloid classification based on 2D gels and imaging MS (IMS) have been proposed [2], [27], [28], [29], the methods currently employed in the clinic are based on liquid chromatography coupled to MS (LC–MS), using high-resolution instruments. Two main procedures, in particular, are in use in amyloid centers worldwide. The first was developed by pathologists at Mayo Clinic and can be used for typing amyloid from formalin-fixed, paraffin-embedded (FFPE) specimens [3], [30]. The amyloid deposits are first cut from thick (10 μm) tissue slices mounted on specific slides by laser capture dissection/microdissection (LMD), using an LMD microscope equipped with a fluorescence module that allows visualizing the Congo red positive areas. Laser dissection leads to a strong enrichment of the amyloid deposits over the tissue protein background and provides material suitable for the LC–MS/MS analysis. Assignment of the disease type, from the list of proteins identified in the dissected areas, is based on which amyloid protein is found with greater abundance, on the basis of spectral counting. This method has proven able to define the most frequent, as well as rare forms, on virtually any tissue type [3], [30], [31], [32], [33], [34], [35], [36].
Over time, the accurate proteomic typing of large cohorts of organ biopsies has provided an updated map of the epidemiology of the surgical amyloid types, disclosing sometimes unexpected results. In a large cohort of endomyocardial biopsies characterized by MS, for example, ATTR was the predominant amyloidosis type [37], further underlying the concept that tissue typing should be performed even in presence of monoclonal components. In liver, the recently recognized ALect2 amyloidosis was found to be responsible for up to 25% of cases, besides other common and rare forms [38]. Also in nerve and kidney, other frequent biopsy sites, the origin of amyloidosis could be determined in most cases, even in presence of infrequent or novel forms [31], [32], [33], [39], [40]. Overall, LMD–LC–MS/MS was reported to have 98% sensitivity and specificity for typing [30], comparing favorably with immunohistochemistry: a recent work, analyzing the performances of the two techniques, has found 100% concordance between positive IHC and LMD–LC–MS/MS, whereas the latter increased diagnostic accuracy from 76% to 94% [41]. This LMD–LC–MS/MS proteomic assay, every step of which has been standardized for better quality control, has received regulatory approval for patient diagnosis in the United States, being the first example of a shotgun proteomics test to receive such accreditation [3].
A second approach for amyloid typing by MS, developed by the Pavia amyloid team and currently in clinical use, is based on a different workflow [42], [43], [44]. Upon acquisition of the complete proteome map of unfractionated tissue by shotgun LC–MS/MS analysis, a semiquantitative label-free pairwise comparison of the map of amyloid positive samples against an average map of negative ones is performed. In this case, amyloid identification from the whole tissue proteome is based on calculation of a parameter, called alpha-value [44], representing the normalized relative abundance of each known amyloid protein in patients compared to controls. The method can be applied for typing both fresh fat tissue and FFPE tissue samples, provided that an adequate control map is available. On adipose tissue, this approach, for which concordance with IEM typing was 100% [44], allows amyloid classification in 96% of amyloid-positive samples (Pavia case series, unpublished). The important feature of this shotgun assay is the presence of a control map, which allows to estimate, in particular, the average abundance of carried-over plasma proteins; these, as discussed later, are often confounding factors during interpretation of the identified proteins lists. A shotgun procedure for the analysis of unfixed and unfractionated fat samples was also more recently implemented by Mayo Clinic researchers, enabling classification of amyloid type in 90% of cases when tested as a clinical assay on a wide set of amyloid-positive samples [45].
Regardless of the approach used, it is important to emphasize that these proteomics methods provide information on the composition, not on the conformation of proteins in the samples; detection of fibrils still relies on imaging and on use of specific staining. Nevertheless, proteomic analysis can provide indirect information on the presence of amyloid deposits, through detection of amyloid associated proteins. These species (which include serum amyloid P, apolipoprotein E, apolipoprotein A-IV, clusterin, vitronectin, HSPG etc), are associated with amyloid fibrils regardless of the disease type, creating a sort of “amyloid proteome signature” both in organ biopsies and in subcutaneous abdominal fat aspirates [44], [45]. Indeed, the contemporaneous identification of multiple amyloid-associated proteins (in particular apolipoprotein E, serum amyloid P-component and apoliprotein A-IV) has recently been proposed as an indirect diagnostic index of the presence of amyloid deposits [45], which could be of support for diagnostic characterization.
As mentioned earlier, tissue proteomic analysis can also provide a wealth of additional information useful for diagnostic purposes. MS and MS/MS analysis, in particular, can provide sequence information useful for detecting and locating amino acid variants on amyloidogenic proteins. As discussed above, amyloidogenic proteins have the peculiarity of possessing primary sequences that often differ from wild-type ones, as in the case of proteins responsible for hereditary forms. The mutation usually shifts the mass of the affected peptides, and the variant proteins can be identified, provided that the peptide is visible by MS and the mutated sequence is present in the database. In fact, in order to maximize the information obtainable with traditional searching algorithms and increase the rate of assignment of peptides containing the amino acid substitution, standard protein sequence databases must be supplemented with specific sequences of known or predicted mutated amyloidogenic proteins [46]. Recently, a bioinformatics platform has been implemented by Mayo Clinic researchers in order to test the ability of clinical proteomics to detect known as well as novel amyloidogenic mutations in proteins responsible for hereditary forms [46]. The known mutations are identified through classical searching algorithms on an augmented database, whereas the novel variants are identified by matching the MS/MS spectra against wild type protein sequences, using a sequence tagging search strategy configured to look for unanticipated mutations. The sensitivity and specificity of these workflows are high (respectively, 92% and 100% for the known mutation detection and 82% and 99% for novel mutation detection), indicating that tissue proteomics is a useful method for rapidly and efficiently confirming the deposition of a pathogenic variant.
The data sets from patients with AL amyloidosis can instead be interrogated by bioinformatics to recovery information on the deposited FLC. Immunoglobulin light chains present specific issues during proteomic analysis, related to the sequence diversity of the variable region. A complex combination of genetic VJC genes rearrangement and somatic hypermutation, in fact, translates in the fact that each light chains’s variable region possesses a unique primary sequence: this leads to the frequent unassignment of variable domain peptide ions, given the lack of a corresponding sequence in databases. Recently, a novel informatics workflow has been proposed [47], employing an augmented database search with known light chains variable region sequence templates, to detect the clonotypic peptides from patient MS/MS data. Even though the method cannot sequence the whole pathogenic light chain, mapping the detected peptides to the corresponding variable region gene loci has proven useful to determine the germ line gene, and represents an interesting and promising alternative to genetic sequencing.
Overall, although available only in specialized centers (due to the need for specific equipment and especially trained personnel), proteomics has become a reference method for amyloid identification worldwide. Indeed, it would be misleading to believe it to be completely free from specific issues, which need instead to be considered in the course of result interpretation [26], [48]. A first issue relates to the already mentioned variability of amyloid proteins compared to their normal counterpart. Whereas supplemented databases and advanced bioinformatics can cope with this issue in most cases of hereditary forms, sequence heterogeneity still remains a major problem in the case of light chains, in which peptides from the V region are often unassigned, affecting protein identification and quantification. A second significant issue stands in the possible carryover of blood proteins. Given that almost all species responsible for systemic amyloidoses are, in their non-pathogenic form, normal constituents of human plasma, it is clear that identifying potential amyloidogenic precursors from incompletely washed samples can lead to incorrect or equivocal typing. Some solutions to this issue have been proposed, employing mathematical corrections and/or control samples to compensate for background plasma contaminants, and to identify amyloid proteins present with the greatest abundance [44], [49].
More recently, novel methods, based on imaging MS, have been applied to systemic amyloid typing and analysis [27], [28], [50] on solid organ biopsies. Although these approaches have not yet been developed as clinical grade assays, their ability to couple MS with histology examination makes them promising new tools.
2. MS analysis of circulating amyloidogenic proteins
2.1. Immunoglobulin light chains and transthyretin
Proteomic analysis of circulating precursors can provide important information of potential clinical relevance from either a qualitative or a quantitative point of view. In practice, MS assessment of amyloidogenic proteins in patients’ plasma or serum has been mainly used to investigate TTR and immunoglobulin light chains [51], [52], [53], [54], [55] and, to a lesser extent, other proteins such as serum amyloid A [56]. In the case of AL amyloidosis, the investigation must be focused specifically on the free form of light chains (FLC) [57], [58], i.e. not bound to the heavy chain as in an intact immunoglobulin.
Although precursor analysis has not yet entered the clinical routine as it has happened for tissue proteomics, several of the developed approaches show potential as new means to identify destabilizing mutations at the protein level, as a complement to genetic analysis, or to quantify the pathogenic proteins, in alternative to immunometric tests. As described later, compared to the current clinical-grade assays, MS offers unique perspectives in terms of specificity towards particular isoforms or pathogenic subpopulations.
In most proteomic approaches aiming at the circulating precursors, MS analysis is preceded by strategies to enrich the protein of interest and reduce the interfering effects of blood constituents. A widely used strategy is based on capturing the protein using antibodies [51], [59], [60], either off-line or on-line with MS [52], [59]. For assessing FLCs, it is necessary to employ antibodies directed specifically against light chains not bound to the heavy chain, in order not to precipitate the abundant pool of polyclonal immunoglobulins normally present in blood. Our group employed a strategy based on the use of anti-FLC antibodies covalently linked to agarose beads in microcentrifuge tubes [51]. In alternative to immunoprecipitation, in the case of TTR, chip-based enrichment in the context of SELDI-TOF MS-based methods have been described [61], [62]. Recently, a method that couples capillary zone electrophoresis with MS has been reported for screening of TTR variants in serum samples [63]. MS-based strategies are also being developed to analyze monoclonal light chains from intact serum antibodies, which can be useful when the patient’s clone produces an intact monoclonal immunoglobulin besides the FLC excess [54], [64], [65]. In this instance, light chains are studied once detached from the heavy chain through reduction of the disulfide bonds.
In parallel to the evolution of MS instrumentation and methods, the analytical approaches and goals of the proteomic analysis of precursors have also evolved. The main focus of former studies was directed towards qualitative characterization, especially for detecting amyloidogenic mutations. The developments of diagnostic methods has been particularly fertile in ATTR amyloidosis, in which MS has proved informative to evidence mass shifts in the digested [60], [66], [67] or in the intact protein [61], [62], [68], [69], [70]. While several past studies used MALDI-TOF and peptide mass fingerprinting for identifying mutations-bearing peptides [66], [67], more recent approaches confirm and locate the variant by MS/MS sequencing or top-down analysis, using high-resolution instruments [69].
In parallel, quantitative MS-based methods have begun to emerge. In a recent work, a methodology for quantification of serum TTR, as well as for quantification of its PTMs, has been described, involving MS analysis of the intact protein and targeted LC-MS analysis of peptides carrying the PTMs of interest [71]. In AL amyloidosis, quantitation of the pathogenic monoclonal free light chain using MS has been advocated as a way to increase specificity over the polyclonal background [54], [64]. Indeed, given the presence of the variable region, FLC have very unique sequences; this translates in the fact that each full length protein has a peculiar mass, and that the peptides from each light chain are not only protein-specific, but also specific for the patient’s clone [51], [72]. These properties have been exploited to develop MS-based methods that monitor monoclonal components using the mass of the full length light chain (in the miRAMM approach: monoclonal immunoglobulin Rapid Accurate Molecular Mass) [54], [64] or its “clonotypic” peptides [72]. Whereas these methods, so far applied to multiple myeloma patients, hold great promise for increasing the specificity of pathogenic light chain quantification, significant uncertainty still concerns their sensitivity. A combination of polyclonal immunoglobulin depletion and SRM-based quantification using labeled light chain peptide standards has recently been proposed for quantifying total serum FLC, showing concordance with results obtained by nephelometry, excellent linearity and good sensitivity [55]. This area of research is expanding very rapidely, and ad hoc studies will need to be designed to precisely define the clinical performances of these promising methods, compared to current diagnostic approaches.
3. Translational proteomics: novel information to improve disease knowledge
A most exciting application of proteomics to systemic amyloidoses consists in its use for studying the molecular mechanisms of disease at the cell and tissue level. Although protein misfolding and aggregation are necessary components of the pathogenetic cascade, the proteotoxic events in target organs are still largely unknown. The availability of validated model systems and of human disease-specific, proteomics-grade biobanks has been the basis for a number of recent studies investigating, from a high-throughput point of view, the biological bases of misfolded proteins toxicity. Mass spectrometry-based proteomics, either employed in the context of systems biology analyses [2], [42], [73], or aimed to study specific aspects as protein PTMs, has indeed already provided important clues on the events occurring in vivo and in vitro.
Our team has employed a MudPIT-based differential proteomics approach for identifying proteins changing in abundance in patients’ subcutaneous abdominal fat tissue [42]. This label-free differential analysis, coupled to computational studies, has led to the identification of tissue-resident species, biological pathways and functions quantitatively affected in relation to amyloid deposition. These proteome changes concern both the intra- and the extracellular compartments and are involved in a variety of processes; prominent features include subversion of the matrix composition, with increase of collagen and heparan sulphate proteoglycans, changes in the cytoskeleton, in species involved in chaperone activity and protein processing, tissue-specific metabolism and mitochondrial energetics. The specific role of these alterations in the disease pathogenesis can now be assessed in a targeted manner with functional studies; moreover, the differentially represented proteins, as single analytes or in combination, could be assessed as novel potential biomarkers of amyloid-associated tissue damage [74].
Proteomic analysis of human tissues has also been used for studying the biochemical features of amyloid fibrils, under the hypothesis that peculiar modifications of the amyloidogenic precursor, such as truncation or other PTMs, might be pro-amyloidogenic factors [75], [76]. Using 2D-PAGE coupled to MALDI-TOF MS, the amyloid deposits of AL, ATTR and β-2 microglobulin were studied in detail. In all cases, fibrils were shown to contain a mixture of pI isoforms of full-length precursors and fragments thereof [2]. The pI heterogeneity can be referred to the presence of post-translational modifications, such as oxidation and deamidation, whereas fragmentation is likely due to the activity of still undefined proteases, acting in the peculiar physical context of target tissues [77], and appears to be connected with amyloid deposition [2], [76], [78]. Fragmentation is especially prominent in AL amyloidosis, in which C-terminally truncated species constitute a large fraction of the total deposited proteins. However, the presence of the full length light chain, besides fragments, was clearly demonstrated by proteomics [2], [30], supporting biochemical and biophysical studies that have demonstrated the critical role of the constant region in amyloidogenesis [79].
Mass spectrometry has also been used as an instrument to characterize the features of the circulating amyloidogenic precursors, on which it allowed demonstrating and locating the presence of known and unexpected PTMs. Extensive tryptophan oxidation, N-terminal pyroglutamate and unexpected S-cysteinylation of internal cysteines have been detected by MS/MS on serum light chains [51], whereas cysteinylation of the C-terminal cysteine has been shown in urinary kappa light chains [80]. Variant amyloidogenic TTR and wtTTR from patients affected by hereditary and wild type ATTR amyloidosis have been shown to contain heterogeneous PTMs at the Cys-10 residue, consisting of mixed disulfides (S-sulfonation, S-glycinylcysteinylation, S-cysteinylation and S-glutathionylation) [51], [71], [81], [82]. A method for their targeted quantitation in patients has recently been proposed [71], under the rationale that these PTMs may play an important biological role in protein destabilization and in the onset of the disease.
The most innovative application of MS in the evaluation of amyloidogenic proteins consists in the study of folding and quaternary structure. Using native and ion mobility MS, the formation of oligomers and variant conformational states has been explored, especially in the case of β-2 microglobulin [83], [84], [85] and serum transthyretin [86].
Regarding the analysis of the proteotoxic mechanisms at the cellular level, functional proteomics has been a powerful approach for exploring novel experimental possibilities. In a recent work, we followed the hypothesis that the interaction of amyloidogenic FLC with specific protein partners in target cardiac cells might mediate cardiac damage, through perturbation of the interactors’ function and biological activity [87]. Using a functional proteomics-based approach, we identified a subset of proteins, with mitochondrial (OPA1, VDAC and ACAD9) and peroxisomal (ACOX1) localization, interacting specifically with cardiotoxic light chains. The occurrence of the interactions has been verified in cardiac cells, along with alterations of the morphology and protein expression of mitochondria. This proteomic analysis has opened new perspectives on the pathogenesis of AL cardiac toxicity, and serves as a basis to specifically study the functional role of single molecules in cell damage.
4. Conclusions and perspectives
The introduction of proteomics has had a profound impact on clinical management and research in the field of systemic amyloidosis. Amyloid typing by MS has already achieved regulatory approval in the United States and is now routinely used as gold standard diagnostic tool in clinical practice. In the perspective of offering proteomic amyloid typing as an health care service, by those centers where this technique is available, it is also of critical importance to achieve CE accreditation for in-vitro diagnostics. In this way, mass spectrometry-based typing will possess all the formal requisites to be used in the clinical setting throughout European institutions. The availability of curated, proteomics-grade biobanks in major specialized centers has granted a collection of optimal material for human fluid and tissue proteomic analysis, as well as for experimental proteomics studies. Applied to the field of basic research on the mechanisms of disease, proteomics has disclosed involved molecules and affected pathways, with promising translational applications. Overall, a great wealth of proteomic data sets from tissues and experimental models is being collected, holding the promise to serve as a gold mine from which other important information could be extracted in the future, in parallel to the development of novel search tools and experimental queries.
Acknowledgements
This work was supported by the Italian Ministry of Health (GR-2010-2317596), Associazione Italiana per la Ricerca sul Cancro special program “5 per mille” (N° 9965), Fondazione Cariplo (2013-0964), Amyloidosis Foundation and Fondazione Mintas, Ghislieri College, Pavia.
References
- 1.Murphy C.L., Wang S., Williams T., Weiss D.T., Solomon A. Characterization of systemic amyloid deposits by mass spectrometry. Methods Enzymol. 2006;412:48–62. doi: 10.1016/S0076-6879(06)12004-2. [DOI] [PubMed] [Google Scholar]
- 2.Lavatelli F., Perlman D.H., Spencer B. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol. Cell. Proteom. 2008;7(8):1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theis J.D., Dasari S., Vrana J.A., Kurtin P.J., Dogan A. Shotgun proteomics-based clinical testing for diagnosis and classification of amyloidosis. J. Mass Spectrom. 2013;48(10):1067–1077. doi: 10.1002/jms.3264. [DOI] [PubMed] [Google Scholar]
- 4.Merlini G., Seldin D.C., Gertz M.A. Amyloidosis: pathogenesis and new therapeutic options. J. Clin. Oncol. 2011;29(14):1924–1933. doi: 10.1200/JCO.2010.32.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sipe J.D., Benson M.D., Buxbaum J.N. Nomenclature 2014: amyloid fibril proteins and clinical classification of the amyloidosis. Amyloid. 2014;21(4):221–224. doi: 10.3109/13506129.2014.964858. [DOI] [PubMed] [Google Scholar]
- 6.Merlini G., Bellotti V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003;349(6):583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 7.Obici L., Perfetti V., Palladini G., Moratti R., Merlini G. Clinical aspects of systemic amyloid diseases. Biochim. Biophys. Acta. 2005;1753(1):11–22. doi: 10.1016/j.bbapap.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Merlini G., Wechalekar A.D., Palladini G. Systemic light chain amyloidosis: an update for treating physicians. Blood. 2013;121(26):5124–5130. doi: 10.1182/blood-2013-01-453001. [DOI] [PubMed] [Google Scholar]
- 9.Merlini G., Palladini G. Light chain amyloidosis: the heart of the problem. Haematologica. 2013;98(10):1492–1495. doi: 10.3324/haematol.2013.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saraiva M.J. Hereditary transthyretin amyloidosis: molecular basis and therapeutical strategies. Expert Rev. Mol. Med. 2002;4(12):1–11. doi: 10.1017/S1462399402004647. [DOI] [PubMed] [Google Scholar]
- 11.Sekijima Y. Transthyretin (ATTR) amyloidosis: clinical spectrum, molecular pathogenesis and disease-modifying treatments. J. Neurol. Neurosurg. Psychiatry. 2015;86(9):1036–1043. doi: 10.1136/jnnp-2014-308724. [DOI] [PubMed] [Google Scholar]
- 12.Planté-Bordeneuve V., Kerschen P. Transthyretin familial amyloid polyneuropathy. Handb. Clin. Neurol. 2013;115:643–658. doi: 10.1016/B978-0-444-52902-2.00038-2. [DOI] [PubMed] [Google Scholar]
- 13.Tanskanen M., Peuralinna T., Polvikoski T. Senile systemic amyloidosis affects 25% of the very aged and associates with genetic variation in alpha2-macroglobulin and tau: a population-based autopsy study. Ann. Med. 2008;40(3):232–239. doi: 10.1080/07853890701842988. [DOI] [PubMed] [Google Scholar]
- 14.Picken M.M. Modern approaches to the treatment of amyloidosis: the critical importance of early detection in surgical pathology. Adv. Anat. Pathol. 2013;20(6):424–439. doi: 10.1097/PAP.0b013e3182a92dc3. [DOI] [PubMed] [Google Scholar]
- 15.Ueda M., Ando Y. Recent advances in transthyretin amyloidosis therapy. Transl. Neurodegener. 2014;3:19. doi: 10.1186/2047-9158-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seldin D.C., Sanchorawala V. Amyloidomics comes of age. Blood. 2012;119(8):1795–1796. doi: 10.1182/blood-2011-10-381178. [DOI] [PubMed] [Google Scholar]
- 17.Fernández de Larrea C., Verga L., Morbini P. A practical approach to the diagnosis of systemic amyloidoses. Blood. 2015;125(14):2239–2244. doi: 10.1182/blood-2014-11-609883. [DOI] [PubMed] [Google Scholar]
- 18.Schönland S.O., Hegenbart U., Bochtler T. Immunohistochemistry in the classification of systemic forms of amyloidosis: a systematic investigation of 117 patients. Blood. 2012;119(2):488–493. doi: 10.1182/blood-2011-06-358507. [DOI] [PubMed] [Google Scholar]
- 19.Linke R.P. On typing amyloidosis using immunohistochemistry. Detailled illustrations, review and a note on mass spectrometry. Prog. Histochem. Cytochem. 2012;47(2):61–132. doi: 10.1016/j.proghi.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Picken M.M. Amyloidosis—where are we now and where are we heading? Arch. Pathol. Lab. Med. 2010;134(4):545–551. doi: 10.5858/134.4.545. [DOI] [PubMed] [Google Scholar]
- 21.Lachmann H.J., Booth D.R., Booth S.E. Misdiagnosis of hereditary amyloidosis as AL (primary) amyloidosis. N. Engl. J. Med. 2002;346(23):1786–1791. doi: 10.1056/NEJMoa013354. [DOI] [PubMed] [Google Scholar]
- 22.Solomon A., Murphy C.L., Westermark P. Unreliability of immunohistochemistry for typing amyloid deposits. Arch. Pathol. Lab. Med. 2008;132(1):14. doi: 10.5858/2008-132-14b-IR. author reply 14–15. [DOI] [PubMed] [Google Scholar]
- 23.Solomon A., Murphy C.L., Westermark P. Misclassification of amyloidosis is unwarranted. Blood. 2006;108(2):776. doi: 10.1182/blood-2006-02-005462. author reply 776–777. [DOI] [PubMed] [Google Scholar]
- 24.Satoskar A.A., Efebera Y., Hasan A. Strong transthyretin immunostaining: potential pitfall in cardiac amyloid typing. Am. J. Surg. Pathol. 2011;35(11):1685–1690. doi: 10.1097/PAS.0b013e3182263d74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoskar A.A., Burdge K., Cowden D.J., Nadasdy G.M., Hebert L.A., Nadasdy T. Typing of amyloidosis in renal biopsies: diagnostic pitfalls. Arch. Pathol. Lab. Med. 2007;131(6):917–922. doi: 10.5858/2007-131-917-TOAIRB. [DOI] [PubMed] [Google Scholar]
- 26.Lavatelli F., Vrana J.A. Proteomic typing of amyloid deposits in systemic amyloidoses. Amyloid. 2011;18(4):177–182. doi: 10.3109/13506129.2011.630762. [DOI] [PubMed] [Google Scholar]
- 27.Casadonte R., Kriegsmann M., Deininger S.O. Imaging mass spectrometry analysis of renal amyloidosis biopsies reveals protein co-localization with amyloid deposits. Anal. Bioanal. Chem. 2015;407(18):5323–5331. doi: 10.1007/s00216-015-8689-z. [DOI] [PubMed] [Google Scholar]
- 28.Winter M., Tholey A., Krüger S., Schmidt H., Röcken C. MALDI—mass spectrometry imaging identifies vitronectin as a common constituent of amyloid deposits. J. Histochem. Cytochem. 2015;63(10):772–779. doi: 10.1369/0022155415595264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanishi T., Ito M., Nirasawa T., Tsuji M., Takubo T. Topologies of amyloidogenic proteins in Congo red-positive sliced sections of formalin-fixed paraffin embedded tissues by MALDI-MS imaging coupled with on-tissue tryptic digestion. Clin. Biochem. 2013;46(15):1595–1600. doi: 10.1016/j.clinbiochem.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 30.Vrana J.A., Gamez J.D., Madden B.J., Theis J.D., Bergen H.R., Dogan A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114(24):4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 31.Sethi S., Vrana J.A., Theis J.D., Dogan A. Mass spectrometry based proteomics in the diagnosis of kidney disease. Curr. Opin. Nephrol. Hypertens. 2013;22(3):273–280. doi: 10.1097/MNH.0b013e32835fe37c. [DOI] [PubMed] [Google Scholar]
- 32.Said S.M., Sethi S., Valeri A.M. Renal amyloidosis: origin and clinicopathologic correlations of 474 recent cases. Clin. J. Am. Soc. Nephrol. 2013;8(9):1515–1523. doi: 10.2215/CJN.10491012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein C.J., Vrana J.A., Theis J.D. Mass spectrometric-based proteomic analysis of amyloid neuropathy type in nerve tissue. Arch. Neurol. 2011;68(2):195–199. doi: 10.1001/archneurol.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Said S.M., Reynolds C., Jimenez R.E. Amyloidosis of the breast: predominantly AL type and over half have concurrent breast hematologic disorders. Mod. Pathol. 2013;26(2):232–238. doi: 10.1038/modpathol.2012.167. [DOI] [PubMed] [Google Scholar]
- 35.Erickson L.A., Vrana J.A., Theis J. Analysis of amyloid in medullary thyroid carcinoma by mass spectrometry-based proteomic analysis. Endocr. Pathol. 2015 doi: 10.1007/s12022-015-9390-7. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 36.D'Souza A., Theis J., Quint P. Exploring the amyloid proteome in immunoglobulin-derived lymph node amyloidosis using laser microdissection/tandem mass spectrometry. Am. J. Hematol. 2013;88(7):577–580. doi: 10.1002/ajh.23456. [DOI] [PubMed] [Google Scholar]
- 37.Maleszewski J.J., Murray D.L., Dispenzieri A. Relationship between monoclonal gammopathy and cardiac amyloid type. Cardiovasc. Pathol. 2013;22(3):189–194. doi: 10.1016/j.carpath.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Mereuta O.M., Theis J.D., Vrana J.A. Leukocyte cell-derived chemotaxin 2 (LECT2)-associated amyloidosis is a frequent cause of hepatic amyloidosis in the United States. Blood. 2014;123(10):1479–1482. doi: 10.1182/blood-2013-07-517938. [DOI] [PubMed] [Google Scholar]
- 39.Sethi S., Theis J.D., Shiller S.M. Medullary amyloidosis associated with apolipoprotein A-IV deposition. Kidney Int. 2012;81(2):201–206. doi: 10.1038/ki.2011.316. [DOI] [PubMed] [Google Scholar]
- 40.Sethi S., Vrana J.A., Theis J.D. Laser microdissection and mass spectrometry-based proteomics aids the diagnosis and typing of renal amyloidosis. Kidney Int. 2012;82(2):226–234. doi: 10.1038/ki.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbertson J.A., Theis J.D., Vrana J.A. A comparison of immunohistochemistry and mass spectrometry for determining the amyloid fibril protein from formalin-fixed biopsy tissue. J. Clin. Pathol. 2015;68(4):314–317. doi: 10.1136/jclinpath-2014-202722. [DOI] [PubMed] [Google Scholar]
- 42.Brambilla F., Lavatelli F., Di Silvestre D. Shotgun protein profile of human adipose tissue and its changes in relation to systemic amyloidoses. J. Proteome Res. 2013;12(12):5642–5655. doi: 10.1021/pr400583h. [DOI] [PubMed] [Google Scholar]
- 43.Brambilla F., Lavatelli F., Merlini G., Mauri P. Clinical proteomics for diagnosis and typing of systemic amyloidoses. Proteom. Clin. Appl. 2013;7(1–2):136–143. doi: 10.1002/prca.201200097. [DOI] [PubMed] [Google Scholar]
- 44.Brambilla F., Lavatelli F., Di Silvestre D. Reliable typing of systemic amyloidoses through proteomic analysis of subcutaneous adipose tissue. Blood. 2012;119(8):1844–1847. doi: 10.1182/blood-2011-07-365510. [DOI] [PubMed] [Google Scholar]
- 45.Vrana J.A., Theis J.D., Dasari S. Clinical diagnosis and typing of systemic amyloidosis in subcutaneous fat aspirates by mass spectrometry-based proteomics. Haematologica. 2014;99(7):1239–1247. doi: 10.3324/haematol.2013.102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasari S., Theis J.D., Vrana J.A. Clinical proteome informatics workbench detects pathogenic mutations in hereditary amyloidoses. J. Proteome Res. 2014;13(5):2352–2358. doi: 10.1021/pr4011475. [DOI] [PubMed] [Google Scholar]
- 47.Dasari S., Theis J.D., Vrana J.A. Proteomic detection of immunoglobulin light chain variable region peptides from amyloidosis patient biopsies. J. Proteome Res. 2015;14(4):1957–1967. doi: 10.1021/acs.jproteome.5b00015. [DOI] [PubMed] [Google Scholar]
- 48.Kaul E., Pilichowska M., Vullaganti M., Madan N., Comenzo R.L. Twists and turns of determining amyloid type and amyloid-related organ damage: discordance and clinical skepticism in the era of proteomic typing. Amyloid. 2014;21(1):62–65. doi: 10.3109/13506129.2013.856779. [DOI] [PubMed] [Google Scholar]
- 49.Sun W., Sun J., Zou L. The successful diagnosis and typing of systemic amyloidosis using a microwave-assisted filter-aided fast sample preparation method and LC/MS/MS analysis. PLoS One. 2015;10(5):e0127180. doi: 10.1371/journal.pone.0127180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakanishi T., Yoshioka M., Moriuchi K., Yamamoto D., Tsuji M., Takubo T. S-Sulfonation of transthyretin is an important trigger step in the formation of transthyretin-related amyloid fibril. Biochim. Biophys. Acta. 2010;1804(7):1449–1456. doi: 10.1016/j.bbapap.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Lavatelli F., Brambilla F., Valentini V. A novel approach for the purification and proteomic analysis of pathogenic immunoglobulin free light chains from serum. Biochim. Biophys. Acta. 2011;1814(3):409–419. doi: 10.1016/j.bbapap.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Bergen H.R., Abraham R.S., Johnson K.L., Bradwell A.R., Naylor S. Characterization of amyloidogenic immunoglobulin light chains directly from serum by on-line immunoaffinity isolation. Biomed. Chromatogr. 2004;18(3):191–201. doi: 10.1002/bmc.323. [DOI] [PubMed] [Google Scholar]
- 53.Botz C.M., Barnidge D.R., Murray D.L., Katzmann J.A. Detecting monoclonal light chains in urine: microLC-ESI-Q-TOF mass spectrometry compared to immunofixation electrophoresis. Br. J. Haematol. 2014;167(3):437–438. doi: 10.1111/bjh.13003. [DOI] [PubMed] [Google Scholar]
- 54.Barnidge D.R., Dasari S., Botz C.M. Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J. Proteome Res. 2014;13(3):1419–1427. doi: 10.1021/pr400985k. [DOI] [PubMed] [Google Scholar]
- 55.VanDuijn M.M., Jacobs J.F., Wevers R.A., Engelke U.F., Joosten I., Luider T.M. Quantitative measurement of immunoglobulins and free light chains using mass spectrometry. Anal. Chem. 2015;87(16):8268–8274. doi: 10.1021/acs.analchem.5b01263. [DOI] [PubMed] [Google Scholar]
- 56.Corlin D.B., Sen J.W., Ladefoged S., Lund G.B., Nissen M.H., Heegaard N.H. Quantification of cleaved beta2-microglobulin in serum from patients undergoing chronic hemodialysis. Clin. Chem. 2005;51(7):1177–1184. doi: 10.1373/clinchem.2005.049544. [DOI] [PubMed] [Google Scholar]
- 57.Bradwell A.R., Carr-Smith H.D., Mead G.P. Highly sensitive, automated immunoassay for immunoglobulin free light chains in serum and urine. Clin. Chem. 2001;47(4):673–680. [PubMed] [Google Scholar]
- 58.Mead G.P., Carr-Smith H.D., Bradwell A.R. Free light chains. Ann. Clin. Biochem. 2008;45(Pt. 4):444. doi: 10.1258/acb.2008.072541. [DOI] [PubMed] [Google Scholar]
- 59.Bergen H.R., Zeldenrust S.R., Naylor S. An on-line assay for clinical detection of amyloidogenic transthyretin variants directly from serum. Amyloid. 2003;10(3):190–197. doi: 10.3109/13506120308999000. [DOI] [PubMed] [Google Scholar]
- 60.Théberge R., Connors L.H., Skinner M., Costello C.E. Detection of transthyretin variants using immunoprecipitation and matrix-assisted laser desorption/ionization bioreactive probes: a clinical application of mass spectrometry. J. Am. Soc. Mass Spectrom. 2000;11(2):172–175. doi: 10.1016/S1044-0305(99)00136-1. [DOI] [PubMed] [Google Scholar]
- 61.Tasaki M., Ueda M., Obayashi K. Rapid detection of wild-type and mutated transthyretins. Ann. Clin. Biochem. 2015 doi: 10.1177/0004563215605541. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 62.Ueda M., Misumi Y., Mizuguchi M. SELDI-TOF mass spectrometry evaluation of variant transthyretins for diagnosis and pathogenesis of familial amyloidotic polyneuropathy. Clin. Chem. 2009;55(6):1223–1227. doi: 10.1373/clinchem.2008.118505. [DOI] [PubMed] [Google Scholar]
- 63.Pont L., Benavente F., Barbosa J., Sanz-Nebot V. Analysis of transthyretin in human serum by capillary zone electrophoresis electrospray ionization time-of-flight mass spectrometry. Application to familial amyloidotic polyneuropathy type I. Electrophoresis. 2015;36(11–12):1265–1273. doi: 10.1002/elps.201400590. [DOI] [PubMed] [Google Scholar]
- 64.Mills J.R., Barnidge D.R., Murray D.L. Detecting monoclonal immunoglobulins in human serum using mass spectrometry. Methods. 2015;81:56–65. doi: 10.1016/j.ymeth.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 65.Barnidge D.R., Krick T.P., Griffin T.J., Murray D.L. Using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to detect monoclonal immunoglobulin light chains in serum and urine. Rapid Commun. Mass Spectrom. 2015;29(21):2057–2060. doi: 10.1002/rcm.7314. [DOI] [PubMed] [Google Scholar]
- 66.Lim A., Prokaeva T., McComb M.E. Characterization of transthyretin variants in familial transthyretin amyloidosis by mass spectrometric peptide mapping and DNA sequence analysis. Anal. Chem. 2002;74(4):741–751. doi: 10.1021/ac010780+. [DOI] [PubMed] [Google Scholar]
- 67.Tachibana N., Tokuda T., Yoshida K. Usefulness of MALDI/TOF mass spectrometry of immunoprecipitated serum variant transthyretin in the diagnosis of familial amyloid polyneuropathy. Amyloid. 1999;6(4):282–288. doi: 10.3109/13506129909007341. [DOI] [PubMed] [Google Scholar]
- 68.Nepomuceno A.I., Mason C.J., Muddiman D.C., Bergen H.R., Zeldenrust S.R. Detection of genetic variants of transthyretin by liquid chromatography-dual electrospray ionization fourier-transform ion-cyclotron-resonance mass spectrometry. Clin. Chem. 2004;50(9):1535–1543. doi: 10.1373/clinchem.2004.033274. [DOI] [PubMed] [Google Scholar]
- 69.Théberge R., Infusini G., Tong W., McComb M.E., Costello C.E. Top-down analysis of small plasma proteins using an LTQ-Orbitrap. Potential for mass spectrometry-based clinical assays for transthyretin and hemoglobin. Int. J. Mass Spectrom. 2011;300(2–3):130–142. doi: 10.1016/j.ijms.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Brien J.F., Bergen H.R. Transthyretin mass determination for detection of transthyretin familial amyloid. Methods Mol. Biol. 2009;492:353–365. doi: 10.1007/978-1-59745-493-3_21. [DOI] [PubMed] [Google Scholar]
- 71.Vilà-Rico M., Colomé-Calls N., Martín-Castel L. Quantitative analysis of post-translational modifications in human serum transthyretin associated with familial amyloidotic polyneuropathy by targeted LC–MS and intact protein MS. J. Proteom. 2015;127(Pt. B):234–246. doi: 10.1016/j.jprot.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 72.Robert Bergen H., Dasari S., Dispenzieri A. Clonotypic light chain peptides identified for monitoring minimal residual disease in multiple myeloma without bone marrow aspiration. Clin. Chem. 2015 doi: 10.1373/clinchem.2015.242651. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.da Costa G., Ribeiro-Silva C., Ribeiro R. Transthyretin amyloidosis: chaperone concentration changes and increased proteolysis in the pathway to disease. PLoS One. 2015;10(7):e0125392. doi: 10.1371/journal.pone.0125392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lavatelli F., Albertini R., Di Fonzo A., Palladini G., Merlini G. Biochemical markers in early diagnosis and management of systemic amyloidoses. Clin. Chem. Lab. Med. 2014;52(11):1517–1531. doi: 10.1515/cclm-2014-0235. [DOI] [PubMed] [Google Scholar]
- 75.Giorgetti S., Stoppini M., Tennent G.A. Lysine 58-cleaved beta2-microglobulin is not detectable by 2D electrophoresis in ex vivo amyloid fibrils of two patients affected by dialysis-related amyloidosis. Protein Sci. 2007;16(2):343–349. doi: 10.1110/ps.062563507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stoppini M., Mangione P., Monti M. Proteomics of beta2-microglobulin amyloid fibrils. Biochim. Biophys. Acta. 2005;1753(1):23–33. doi: 10.1016/j.bbapap.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 77.Marcoux J., Mangione P.P., Porcari R. A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 2015;7(10):1337–1349. doi: 10.15252/emmm.201505357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mangione P.P., Porcari R., Gillmore J.D. Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. U. S. A. 2014;111(4):1539–1544. doi: 10.1073/pnas.1317488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klimtchuk E.S., Gursky O., Patel R.S. The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry. 2010;49(45):9848–9857. doi: 10.1021/bi101351c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim A., Wally J., Walsh M.T., Skinner M., Costello C.E. Identification and location of a cysteinyl posttranslational modification in an amyloidogenic kappa1 light chain protein by electrospray ionization and matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 2001;295(1):45–56. doi: 10.1006/abio.2001.5187. [DOI] [PubMed] [Google Scholar]
- 81.Kingsbury J.S., Théberge R., Karbassi J.A., Lim A., Costello C.E., Connors L.H. Detailed structural analysis of amyloidogenic wild-type transthyretin using a novel purification strategy and mass spectrometry. Anal. Chem. 2007;79(5):1990–1998. doi: 10.1021/ac061546s. [DOI] [PubMed] [Google Scholar]
- 82.Kingsbury J.S., Klimtchuk E.S., Théberge R., Costello C.E., Connors L.H. Expression, purification, and in vitro cysteine-10 modification of native sequence recombinant human transthyretin. Protein Expr. Purif. 2007;53(2):370–377. doi: 10.1016/j.pep.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith D.P., Radford S.E., Ashcroft A.E. Elongated oligomers in beta2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2010;107(15):6794–6798. doi: 10.1073/pnas.0913046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leney A.C., Pashley C.L., Scarff C.A., Radford S.E., Ashcroft A.E. Insights into the role of the beta-2 microglobulin D-strand in amyloid propensity revealed by mass spectrometry. Mol. Biosyst. 2014;10(3):412–420. doi: 10.1039/c3mb70420c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith D.P., Giles K., Bateman R.H., Radford S.E., Ashcroft A.E. Monitoring copopulated conformational states during protein folding events using electrospray ionization-ion mobility spectrometry-mass spectrometry. J. Am. Soc. Mass Spectrom. 2007;18(12):2180–2190. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pont L., Benavente F., Vilaseca M., Giménez E., Sanz-Nebot V. Characterisation of serum transthyretin by electrospray ionisation-ion mobility mass spectrometry: application to familial amyloidotic polyneuropathy type I (FAP-I) Talanta. 2015;144:1216–1224. doi: 10.1016/j.talanta.2015.07.079. [DOI] [PubMed] [Google Scholar]
- 87.Lavatelli F., Imperlini E., Orrù S. Novel mitochondrial protein interactors of immunoglobulin light chains causing heart amyloidosis. FASEB J. 2015;29(11):4614–4628. doi: 10.1096/fj.15-272179. [DOI] [PubMed] [Google Scholar]