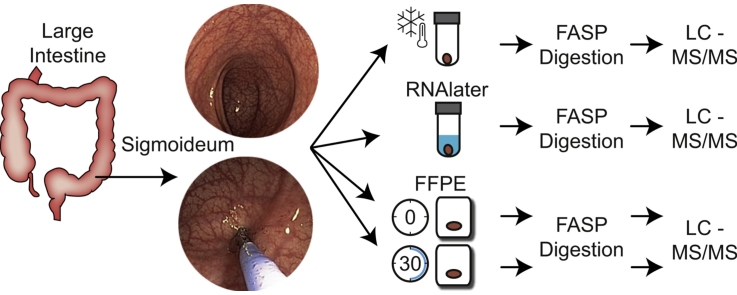
Graphical abstract
Abbreviations: CAN, acetonitrile; FA, formic acid; FDR, false discovery rate; DF, directly-frozen; FASP, filter-aided sample preparation; FFPE, formalin-fixed; HLA-A class I, histocompatibility antigen A-23 alpha chain; HLA-DRB1 class II, histocompatibility antigen DRB1-4 beta chain; LFQ, label-free quantification; iFFPE, immediately formalin-fixed; PCA, principle component analysis; PSM, peptide spectral match; PTM, post-translational modification; s, standard deviation; sFFPE, stored for 30 min prior to formalin-fixed; SDC, sodium deoxycholate; SDS, sodium dodecyl sulfate; TEAB, triethylammonium bicarbonate
Keywords: Proteomics, RNAlater, Formalin-fixed, Paraffin-embedded, Human colon mucosa, Preservation, Mass spectrometry
Highlights
-
We evaluated proteome analyses of snap frozen, RNAlater, and FFPE preserved samples.
-
Reliable proteome studies can be conducted on RNAlater, and FFPE preserved samples.
-
RNAlater sample preservation is highly usable for proteomics studies.
Abstract
Large biobanks exist worldwide containing formalin-fixed, paraffin-embedded samples and samples stored in RNAlater. However, the impact of tissue preservation on the result of a quantative proteome analysis remains poorly described.
Human colon mucosal biopsies were extracted from the sigmoideum and either immediately frozen, stabilized in RNAlater, or stabilized by formalin-fixation. In one set of biopsies, formalin stabilization was delayed for 30 min. The protein content of the samples was characterized by high throughput quantitative proteomics.
We were able to identify a similar high number of proteins in the samples regardless of preservation method, with only minor differences in protein quantitation.
1. Introduction
Clinical proteomic research is dependent on the availability of clinical samples. Proteomics can provide information concerning disease etiology, biomarkers for diagnosis, response to therapy and novel drug discovery [1]. A rapid stabilization of the sample with elimination of enzymatic and cell activity is critical to preserve the biological state of the material. A commonly used method for sample stabilization is by directly freezing (DF) the samples with liquid nitrogen at ⿿196 °C or dry ice at ⿿79 °C, which preserves the sample with minimal introduction of chemical modifications. Biobanks containing DF samples, formalin-fixed, paraffin-embedded (FFPE) tissue samples, and samples stored in RNAlater constitute vast sources of samples, whereof especially the latter remains largely unexploited for proteomics analysis [2], [3]. Due to standard hospital and clinic protocols a delay between sample extraction and sample stabilization can be expected for most samples in biobanks [4]. However, the impact of RNAlater preservation on human tissue samples on the result of a quantative proteome study, as well as the impact of delaying tissue stabilization, remains poorly described.
RNAlater is primarily used to stabilize the RNA content of samples for clinical genomic and transcriptomic analysis, and samples stored in RNAlater have been used extensively in DNA and RNA studies. The RNAlater solution contains high concentration of quaternary ammonium sulfates and cesium sulfate which denature proteins, including DNases, RNases, and proteases, thereby stabilizing the DNA, RNA, and protein content [5], [6]. A few studies have investigated the feasibility in retrieving proteins from samples stored in RNAlater [5], [7], [8], [9], [10]. Saito et al.have demonstrated the feasibility of retrieving proteins from bacteria stored in RNAlater with a high number of identified proteins, and a similar relative protein abundance compared with frozen bacteria [9]. Han et al. [10] have performed a protein-based biomarker discovery study on human tissue stored in RNAlater identifying hundreds of different proteins. However, no studies have investigated the impact on the extracted protein abundances using human tissue samples, nor the impact on the post-translational modifications (PTMs) of preserving samples in RNAlater, which is critical for the subsequent data analysis. The feasibility in conducting a reliable proteome analysis of tissue stored in RNAlater would make coupled transcriptomics and proteomics analysis accessible for rigorous and comprehensive molecular assessment [9].
The stability of FFPE tissue mainly arises due to molecular crosslinking of proteins which are established during formalin-fixation [11], [12]. The crosslinks arise as the product of a three step modification: (1) formalin reacts with the amino or thiol groups on the amino acids leading to methylol additions. (2) The methylol adduct on the primary amino groups is partially dehydrated leading to the formation of labile Schiff bases, (3) which can form crosslinks between several amino acids, i.e., arginine, asparagine, cysteine, glutamine, histidine, tryptophan, and tyrosine [11], [12]. FFPE samples are highly stable and the histological and morphological architecture of the tissue is preserved [2], [13]. As a result, samples are routinely acquired as clinical diagnostic biopsies and large repositories have been generated worldwide [2], [13]. Several proteomic studies have demonstrated the feasibility in extracting and identifying proteins from FFPE samples using proteomics techniques [2], [3], [14], [15], [16], [17], [18], [19], [20], [21].
The aims of the study were to investigate the impact on the result of a quantative proteome study of (1) preserving human tissue samples in RNAlater compared to DF and FFPE, and (2) delaying tissue stabilization for 30 min, in terms of protein abundances, protein modifications, and protein denaturation. We, therefore, examined methods for protein extraction from tissue preserved in RNAlater solution or by FFPE, with DF tissue as a control, comparing the result of a LC⿿MS/MS-based proteome analysis. We focused our analysis on a set of soft tissue colon biopsies extracted for the purpose of this study to demonstrate the potentials in biobank analysis with the suggested protocols. The filter-aided sample preparation (FASP) method is commonly used for preparing samples prior to bottom-up proteomics analysis. Molecular cutoff spin filters are central in the FASP protocol, which facilitates efficient and easily conducted buffer changes, beneficial in relation to protein extraction from tissue in RNAlater solution to remove salts [22], [23], [24]. For FFPE samples we adapted the de-crosslinking method from Wakabayashi et al. [16] for use in a FASP protocol. Additionally, we implemented a buffer optimization recommended by Kawashima et al. [25] to enhance the yield of protein extraction.
2. Materials and methods
2.1. Collection of sample material
Colon mucosal biopsies were sampled from the sigmoideum of two gastroenterological healthy persons, by endoscopy at Hospital of Southern Jutland, Aabenraa, Denmark. Twelve biopsies were extracted from each person approximately 40 cm from the anus, kept constant for each person. All biopsies had an approximate size of 1⿿2 mm3, and the biopsies were preserved by four different methods. Directly frozen biopsies (DF) were immediately transferred to individual cryotubes and within 10⿿20 s snap frozen with liquid nitrogen followed by storage at ⿿80 °C. RNAlater biopsies were immediately transferred to individual cryotubes prefilled with 0.5 mL RNAlater (Life Technologies, Carlsbad, CA, USA), stored at room temperature for 24 h followed by storage at ⿿80 °C, according to manufacturer⿿s instructions. FFPE biopsies were following extraction within 10⿿20 s placed in preparation cartridges. Biopsies were either immediately (iFFPE) stabilized in 4% formaldehyde, or stored for 30 min at ambient temperature before stabilization with 4% formaldehyde (sFFPE) to simulate a clinical situation. Paraffin embedding was performed after a week at Department of Pathology, Aalborg University Hospital, Denmark, according to current standards. All samples were stored for a total of one month prior to proteomics sample preparation and analysis.
The project was approved by The Regional Scientific Ethical Committee (S-20120204) and the Danish Data Protection Agency (2008-58-035), and all participants had given informed consent to participate in the study.
2.2. Proteomics sample preparation
We utilized a modified FASP tryptic protein digestion protocol for the sample preparation, with ethyl acetate phase inversion to facilitate surfactant removal [22], [23], [24], [26], [27]. Wakabayashi et al. [16] utilized a lysis buffer with 100 mM Tris⿿HCl for the protein extraction. However, Kawashima et al. [25] found that increasing the concentration of Tris⿿HCl in the lysis buffer to 300 mM significantly improved the efficiency of the protein extraction, which we implemented in the FASP protocol.
RNAlater and DF preserved samples were homogenized in 0.5 mL lysis buffer (12 mM sodium deoxycholate (SDC), 12 mM sodium dodecyl sulfate (SDS) in 300 mM Tris/HCl, pH 9.0) with steel beads, using a Bullet Blender Gold power-setting 10 for 5 min (Next Advance Inc., Averill Park, NY, USA). The homogenized samples were incubated at 95 °C for 10 min and sonicated for 10 min.
FFPE tissues were extracted using a scalpel, deparaffinized and rehydrated by washing in xylene (3ÿ), and in 100% ethanol (2ÿ), 96% ethanol (2ÿ), 70% ethanol (2ÿ), water. The samples were homogenized in 0.5 mL lysis buffer with steel beads, using a Bullet Blender Gold power-setting 10 for 5 min (Next Advance Inc., Averill Park, NY, USA). The homogenized samples were incubated at 95 °C for 60 min for de-crosslinking formalin fixation, and sonicated for 10 min [16].
The total lysate protein concentration was determined using a bicinchoninic acid assay (BCA) for normalization of sample material with BSA as standard, as well as absorbance at 280 nm (A280) using a NanoDrop 1000 UV⿿vis Spectrophotometer (Thermo Scientific, Waltham, MA, USA). For each sample, a volume corresponding to 100 μg protein was transferred to individual YM-30 kDa spin filters for digestion (Millipore, Billerica, MA, USA) and centrifuged. All centrifugation steps were performed at 14,000 g for 15 min at 4 °C. Protein disulfide bonds were reduced with 12 mM tris(2-carboxyethyl) phosphine (Thermo Scientific, Waltham, MA, USA) for 30 min at 37 °C, and alkylated with 50 mM chloroacetamide (Sigma⿿Aldrich, St. Louis, MO, USA) for 20 min at 37 °C, and centrifuged after each step. The cysteine alkylation was done using chloroacetamide instead of iodoacetamide specified in the original protocol [24]. The protocol modification was introduced as specificity issues have been reported with iodoacetamide alkylation, and chloroacetamide has been suggested as an alternative [28], [29]. The reducing and alkylating agents were dissolved in 120 mM SDC in 50 mM triethylammonium bicarbonate (TEAB), pH 8.5. In preparation for digestion, 400 μL digestion buffer (12 mM SDC in 50 mM TEAB) was added to the spin filter and centrifuged. A 1:50 (w/w) trypsin:protein ratio dissolved in 50 μL digestion buffer was added to the spin filter, and the samples were digested overnight at 37 °C. The flow-through containing the peptides was retrieved by addition of 50 μL digestion buffer and centrifugation. To facilitate SDC removal, a phase separation was performed with 3:1 (v/v) ethyl acetate:sample and acidified by addition of formic acid (FA) to a final concentration of 0.5%. Total phase separation was achieved by 2 min agitation followed by centrifugation. The aqueous phase was collected and vacuum centrifuged overnight and the dry peptide product was stored at ⿿80 °C until time of analysis.
2.3. Mass spectrometry analysis
The samples were resuspended in 2% acetonitrile (ACN), 0.1% FA, briefly sonicated, and 5 μg total peptide material was analyzed per LC⿿MS analysis, in a random sample order [30]. The samples were analyzed using a UPLC-nanoESI MS/MS setup with an NanoRSLC system (Dionex, Sunnyvale, CA, USA). The system was coupled online with an emitter for nanospray ionization (New objective picotip 360-20-10) to a Q Exactive Plus mass spectrometer (Thermo Scientific, Waltham, USA). The peptide material was loaded onto a 2 cm trapping reversed phase Acclaim PepMap RSLC C18 column (Dionex), and separated using an analytical 50 cm reversed phase Acclaim PepMap RSLC C18 column (Dionex). Both columns were kept at 40 °C. The sample was eluted with a gradient of 96% solvent A (0.1% FA) and 4% solvent B (0.1% FA in ACN), which was increased to 10% solvent B on a 1 min ramp gradient at a constant flow rate of 300 nL/min. Subsequently, the gradient was raised to 30% solvent B, on a 180 min ramp gradient. The mass spectrometer was operated in positive mode, selecting up to 12 precursor ions with a mass window of m/z 1.6 based on highest intensity for HCD fragmenting, at a normalized collision energy of 27. Selected precursors were dynamically excluded for fragmentation for 30 s.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002029 [31], [32].
2.4. Peptide modification analysis
We conducted a PTM analysis with the purpose of identifying the most commonly single observed peptide modifications, based on the data from the colon biopsies. Mascot generic format files were generated from the raw data-files in Proteome Discoverer 1.4 (Thermo Scientific, Waltham, USA). The Mascot generic format files were searched individually using ProteinPilot 4.5 (Rev. 1656, Paragon algorithm 4.5.0.0 [33]) (SCIEX, Framingham, USA) against the Uniprot Homo sapiensreference proteome (UP000005640, last modified 2015-01-16, protein count 68,015). The files were searched in ⿿thorough⿿ mode with a focus on biological modifications to include 303 different PTMs. To give a representation of the global PTM distribution the search result was analyzed using ProteinPilot Descriptive Statistics Template version 3.001 (SCIEX) according to manufacturers⿿ instructions. The statistics template included the first 20,000 peptide spectral matches (PSMs) resulting in the identification of peptides with <1% local peptide false discovery rate (FDR). A stricter filtering setting than the standard <5% local peptide FDR was applied for included peptides, to ensure that only high confidence data was included in the analysis.
2.5. Protein identification and quantitation data analysis
A label-free relative quantitation analysis was performed in MaxQuant 1.5.1.2. The rawfiles were searched against the previously mentioned H. sapiens Uniprot database [34], [35]. All standard settings were employed with carbamidomethyl (C) as a static peptide modification, and deamidation (NQ), oxidation (M), formylation (N-terminal and K), and protein acetylation (N-terminal) as variable modifications. The output containing the list of proteins identified below 1% FDR and their abundances was further filtered and processed in Perseus v1.5.0.31. Initially, all reverse hits and proteins tagged as contaminants were removed from further analysis, and the data was log 2-transformed. Two unique peptides or more was required for a protein quantitation. Additionally, a non-zero quantitation value in at least two of the six biopsies from minimum one preservation method was required for the quantifiable proteins. To characterize proteins unique to a given preservation method, Gene Ontology-annotations were imported from Uniprot knowledgebase when available for all proteins using STRAP v1.5 [36]. A principle component analysis (PCA) was performed in Perseus, with all measured protein abundances as input. As PCA does not allow missing values (i.e., proteins where a quantitation value was not obtained for a given replicate analysis), missing values were replaced with values from a normal distribution (width 0.3 and down shift 1.8) to simulate signals from low abundant proteins [37]. The grouping of the replicates on the PCA scores plots was investigated.
To investigate the measured protein abundances across the different preservation methods, the protein abundances were combined method-wise by the mean. The data was investigated by scatterplots and Pearson⿿s correlation coefficients were calculated in Perseus. Protein physicochemical properties were calculated using ProtParam on the ExPASy Server for physicochemical bias analysis [38].
3. Results
3.1. Peptide modification analysis
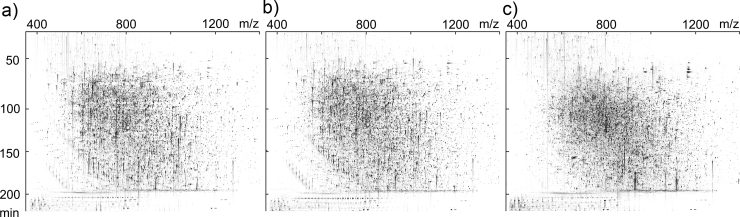
All samples were analyzed by LC⿿MS without contaminants interrupting the stable spray, such as incompletely removed salts. We generated and inspected 2D LC⿿MS heat maps (Fig. 1) and no repetitive intense signals could be observed, which would have indicated remaining detergents or polymers in the samples. To facilitate the identification of modified peptides and elucidate tentative preservation method specific modifications, we performed a peptide PTM analysis of the data from the 24 colon biopsies (six from each preservation method) in ProteinPilot, which included 303 different modifications. The analysis elucidated the presence of PTMs on the peptide level from the DF, RNAlater, iFFPE, and sFFPE preserved biopsies (Table 1). For all four preservation methods, at least half of all peptides were identified in a modified state. As expected, the lowest number of modified peptides were identified in the DF preserved samples. A similar ratio of modified peptides was found in the iFFPE and sFFPE preserved tissues, and the highest ratio of modified peptides was found in the RNAlater preserved tissue. Formylated N-terminals and formylated lysines (K) were expected artifacts in the FFPE preservation methods as they are introduced during the formalin fixation. Accordingly, a higher ratio of peptides carrying the modifications was found in the iFFPE and sFFPE preserved samples, compared to RNAlater and DF (Table 2). Finally, an increased number of deamidated peptides (N and Q) were identified in the FFPE preserved biopsies compared to DF or RNAlater. Based on the PTM analysis, carbamidomethyl (C), oxidation (M), deamidated (N or Q), formyl (N-term or K) and acetyl (protein N-term) modification were included in the subsequent data analysis.
Fig. 1.
Representative two-dimensional LC⿿MS heat maps from the analysis of the colon biopsies preserved by (a) direct freezing (DF), (b) RNAlater, or (c) immediate formalin-fixed, paraffin-embedded (iFFPE).
Table 1.
Peptide properties. Analysis of the first 20,000 peptide spectral matches (PSMs) resulting in the identification of peptides with <1% local peptide false discovery rate (resulting peptide confidence listed). DF: directly frozen biopsies, RNAlater: biopsies preserved directly in RNAlater, iFFPE: immediate formalin-fixed, paraffin-embedded biopsies, sFFPE: biopsies stored for 30 min prior to formalin-fixation, paraffin-embedding. The termini of the identified peptides is given, with expecting termini being tryptic. Carbamidomethylated cysteines are not counted as a PTM, as the modification is deliberately introduced prior to digestion with trypsin. Standard deviations are given (±s).
| DF (%) | RNAlater (%) | iFFPE (%) | sFFPE (%) | |
|---|---|---|---|---|
| Unmodified peptides | 55.5 ± 2.5 | 56.0 ± 0.6 | 50.7 ± 3.8 | 49.9 ± 2.2 |
| Modified peptides | 44.5 ± 2.5 | 44.0 ± 0.6 | 49.3 ± 3.8 | 50.1 ± 2.2 |
| Peptide confidence | 97.7 ± 0.39 | 98.0 ± 0.25 | 98.1 ± 0.3 | 98.0 ± 0.3 |
| Tryptic termini | 95.0 ± 0.4 | 94.9 ± 0.4 | 95.5 ± 0.5 | 95.8 ± 0.3 |
| Semi-specific (only one tryptic terminus) | 5.0 ± 0.4 | 5.0 ± 0.4 | 4.4 ± 0.5 | 4.1 ± 0.3 |
| Non-specific (neither terminus tryptic) | 0.0 ± 0.1 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 |
Table 2.
Residue and terminal specific modifications. Method-vise pooled top ten peptide modifications in biopsies preserved by direct freezing (DF), RNAlater, immediate formalin-fixed, paraffin-embedded (iFFPE), or 30 min stored formalin-fixed, paraffin-embedded (sFFPE). Modified amino acid is given by one-letter code. The first 20,000 peptide spectral matches (PSMs) resulting in the identification of peptides with <1% local peptide false discovery rate were included in the analysis. PTM peptides of possible is the percentage of peptides found in a given modified state, in relation to all peptides which could have the specific modification. Standard deviations are given (±s).
| Feature | ο mass (Da) | PTM peptides of possible |
|||
|---|---|---|---|---|---|
| DF (%) | RNAlater (%) | iFFPE (%) | sFFPE (%) | ||
| Carbamidomethyl (C) | 57.0215 | 100 ± 0.0 | 100 ± 0.0 | 100 ± 0.0 | 100 ± 0.0 |
| Oxidation (M) | 15.9949 | 29.3 ± 3.9 | 33.7 ± 1.8 | 45.4 ± 12.3 | 49.7 ± 16.9 |
| Protein terminal acetyl@N-term | 42.0106 | 34.4 ± 4.7 | 34.1 ± 2.1 | 39.0 ± 3.0 | 39.2 ± 2.9% |
| Gln- > pyro-Glu@N-term | ⿿17.0265 | 27.0 ± 6.4 | 24.0 ± 2.2 | 29.0 ± 1.8 | 29.0 ± 4.6% |
| Deamidated (N) | 0.984 | 11.7 ± 0.5 | 11.8 ± 0.5 | 17.0 ± 1.0 | 16.0 ± 0.8% |
| Deamidated (Q) | 0.984 | 6.2 ± 0.1 | 6.2 ± 0.1 | 7.6 ± 0.4 | 7.4 ± 0.5% |
| Formyl@N-term | 27.9949 | 5.1 ± 3.6 | 2.8 ± 0.6 | 6.4 ± 2.4 | 7.3 ± 3.2% |
| Oxidation (P) | 15.9949 | 1.3 ± 0.3 | 1.1 ± 0.6 | 1.8 ± 0.3 | 1.7 ± 0.4% |
| Dioxidation (M) | 31.9898 | 0.7 ± 0.2 | 0.8 ± 0.1 | 1.5 ± 0.6 | 1.6 ± 0.8 |
| Met- > Hcy (M) | ⿿14.0157 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.1 |
| Formyl (K) | 27.9949 | 0.4 ± 0.5 | 0.2 ± 0.1 | 3.7 ± 0.5 | 3.3 ± 0.4% |
| Methyl (K) | 14.0157 | 0.0 ± 0.0 | 0.0 ± 0.0 | 2.5 ± 0.3 | 2.3 ± 0.3% |
3.2. Preparation method specific proteins
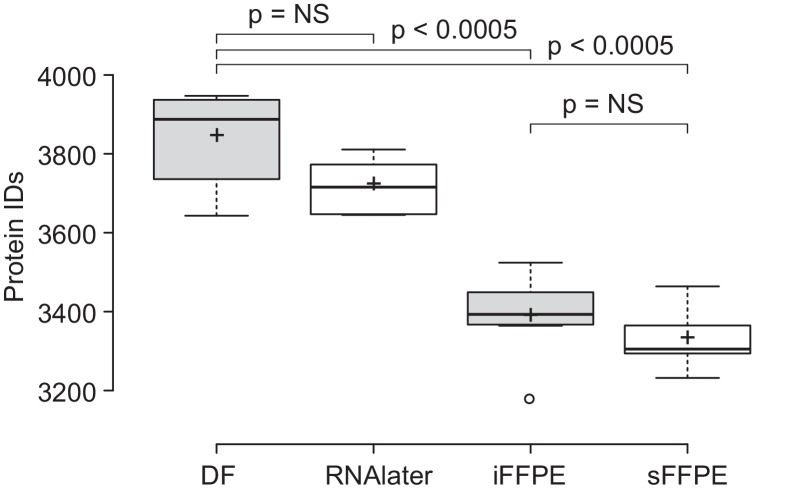
The LC⿿MS raw-data from the colon biopsies were processed in MaxQuant to identify and quantify proteins. A similar mean number of proteins were identified from the biopsies preserved by DF and RNAlater, namely 3840 and 3718 proteins (Fig. 2). Biopsies preserved by iFFPE and sFFPE yielded a statistically significant lower number of identified proteins (p < 0.05) as determined by a two-sample t-tests, compared to DF and RNAlater, namely 3384 and 3328 proteins. The difference in mean number of identified proteins between iFFPE and sFFPE was minor and not found to be statistically significant (p > 0.05). This indicated that the 30 min stored prior to stabilization of the sFFPE preserved colon biopsies did not result in protein degradation to an extend that had an impact on the number of identifiable proteins.
Fig. 2.
Number of identified proteins in the individual colon biopsies, using the direct freezing (DF), RNAlater, immediate formalin-fixed, paraffin-embedded (iFFPE), and 30 min stored formalin-fixed, paraffin-embedded (sFFPE) preparation protocols. Significant changes detected by two-sample t-tests and represented by p-values. NS: not significant, p < 0.05 were considered significant.
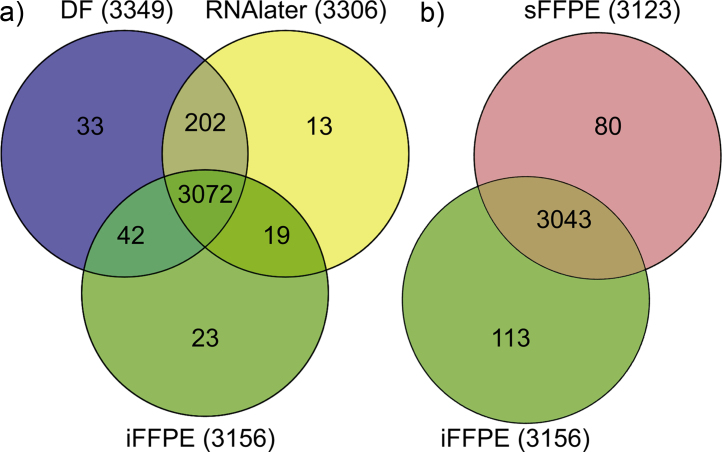
The mean protein abundances were calculated for each preservation method, and the overlap between quantifiable proteins were investigated (Fig. 3). A higher number of proteins were quantifiable in biopsies preserved by DF and RNAlater compared to iFFPE, and 5.9% (202) of the proteins were not found with iFFPE. However, 90.2% (3072) of all quantifiable proteins were found using either method.
Fig. 3.
Number of proteins uniquely quantified in the colon biopsies preserved by (a) direct freezing (DF), RNAlater, or immediate formalin-fixed, paraffin-embedded (iFFPE), and (b) iFFPE or 30 min stored formalin-fixed, paraffin-embedded (sFFPE) preservation protocols.
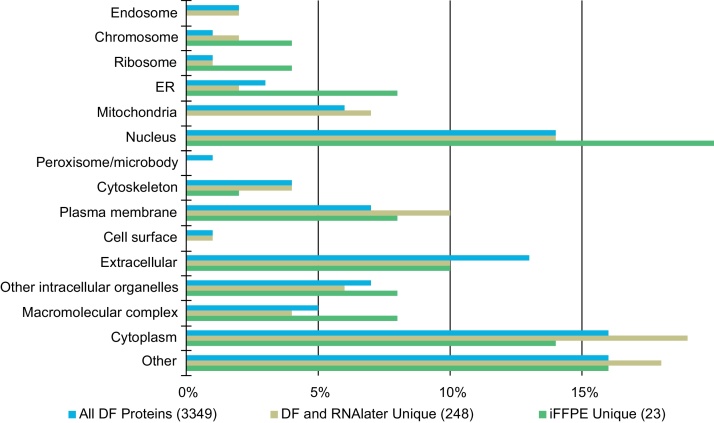
We next investigated if any of the methods systematically enriched or categorically lost specific protein groups, based on the proteins unique to any methods. All quantifiable proteins were classified by subcellular location using available data from Uniprot Knowledgebase Gene Ontology (Fig. 4). We choose to compare proteins, which were uniquely quantifiable in the iFFPE preserved biopsies, to the combined list of proteins unique in the RNAlater or the DF preserved biopsies. RNAlater and DF unique proteins were combined into one group as the lists of quantified proteins using the two methods were nearly identical sharing a 96.8% (3274) overlap. The analysis revealed only minor differences in subcellular location of the proteins unique to DF and RNAlater compared to all proteins quantified in the DF preserved biopsies. Biopsies preserved by iFFPE appeared to be enriched for ER and nucleus proteins, compared to the DF preserved biopsies. However, only 23 proteins were unique to the iFFPE preserved biopsies, and 98.7% (3114) of the proteins quantified in iFFPE were also quantified in the biopsies preserved by the DF preserved biopsies as well, so the differences are minor.
Fig. 4.
Comparative cellular compartment Gene Ontology annotation of quantified proteins in the colon biopsies to investigate potential method biases toward specific protein types. Top-bar: all quantifiable proteins in the directly frozen (DF) biopsies; middle-bar: proteins uniquely quantified in the DF and RNAlater preserved biopsies; lower-bar: proteins uniquely quantifiable in the immediate formalin-fixed, paraffin-embedded (iFFPE) biopsies. The annotations have been normalized to 100%, and number of included proteins are given for each preservation method.
We next investigated the molecular weight and isoelectric point of the method specific proteins. The mean molecular weight and standard deviation (s) were 68,183 Da (s = 85,398 Da) for all DF proteins, 64,950 Da (s = 59957 Da) for proteins unique to DF and RNAlater, and 92,626 Da (s = 78,930 Da) for iFFPE unique proteins. The 23 iFFPE unique proteins have a higher mean molecular weight than the DF proteins, but, the variance is likewise higher. Likewise, the mean isoelectric points of the proteins was 6.72 (s = 1.63) for all DF proteins, 6.71 (s = 1.59) for DF and RNAlater unique proteins, and 6.89 (s = 1.81) for iFFPE unique proteins. As such, only minor differences could be detected in the molecular weight and the isoelectric point between the proteins unique to any method compared to all DF proteins.
3.3. Principle component analysis
We performed a PCA to investigate how the measured protein abundances vary between the differently preserved biopsies. A PCA is a statistical analysis technique that allows for reducing a large number of variables to a smaller number of groups (principle components). The data can be visualized on scores plots based on the principle components, and e.g., be used to interpret how samples in a dataset are separated/grouped based on the variance of all measured protein abundances. In effect, a PCA can be used to interpret the variance in a highly complex dataset, such as a high throughput proteomics dataset [27], [39].
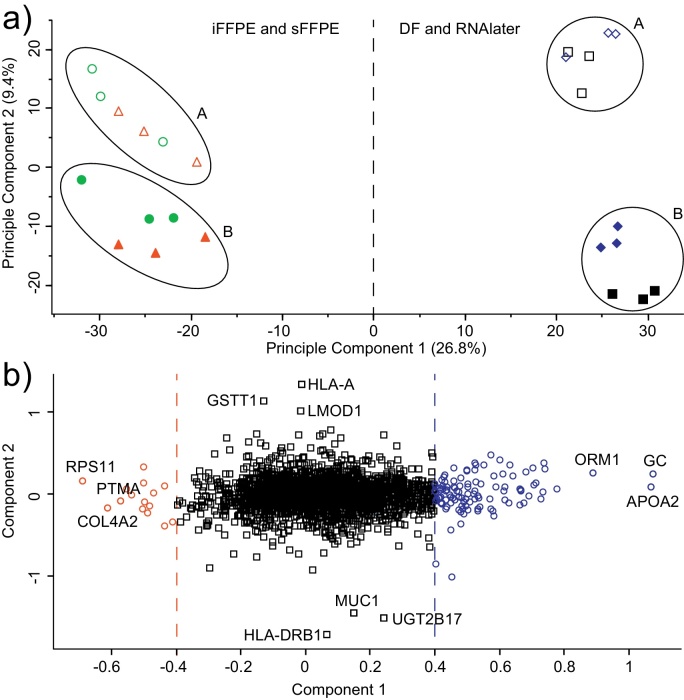
All measured protein abundances in all colon biopsy samples were used as input for the PCA, and a two-dimensional scores plot was constructed (Fig. 5a). A scores plot describe how the biopsies group relative to one another, based on the differences in measured protein abundances of all proteins. Biopsies in which similar protein abundances have been measured will be close in space on the scores plot, relative to the other biopsies. Principal component 1 and principle component 2 represent the largest and second largest variance in the protein abundance dataset, respectively, and explained 34.4% of the variance in the protein abundance in this dataset. In all cases, the biopsies from the individual participants grouped together. Principal component 1 mainly separated the DF and RNAlater preserved biopsies from the FFPE preserved biopsies, whereas principle component 2 mainly separated the two participants from which the biopsies originated.
Fig. 5.
Principle component analysis (PCA) scores plot and loadings plot based on principle component 1 and 2, with all protein abundances in the differently preserved colon biopsies from two participants as input. (a) Scores plot of colon biopsies from participant A (filled symbols) and participant B (hollow symbols) were stabilized by direct frozen (DF) ( and ), RNAlater ( and
and  ), immediate formalin-fixed, paraffin-embedded (iFFPE) (
), immediate formalin-fixed, paraffin-embedded (iFFPE) ( and
and  ), or 30 min stored formalin-fixed, paraffin-embedded (sFFPE) (
), or 30 min stored formalin-fixed, paraffin-embedded (sFFPE) ( and
and  ) preservation protocol. (b) Loadings plot with all quantifiable proteins. Proteins with ±0.4 were analyzed (
) preservation protocol. (b) Loadings plot with all quantifiable proteins. Proteins with ±0.4 were analyzed ( and
and  ) were chosen for further analysis. The gene names of the top three extreme proteins on each axis are given. (For interpretation of the references to color in the text, the reader is referred to the web version of this article.)
) were chosen for further analysis. The gene names of the top three extreme proteins on each axis are given. (For interpretation of the references to color in the text, the reader is referred to the web version of this article.)
The impact of each protein on the separation of the biopsies on the scores plot, can be visualized on a loadings plot. To further identify the main differences in the dataset leading to the separation of DF and RNAlater stabilized samples and FFPE samples, the loading plot of principle component 1 was investigated. The isoelectric point and molecular weight of proteins having a loadings score greater than ±0.4 were investigated (Fig. 5b, blue and red circles). The mean molecular weight for the 107 proteins with greater than 0.4 loading score was 34,414 Da, and 55,226 Da for the 15 protein with less than ⿿0.4 loading score. The mean molecular weight of all DF proteins was 68,183 Da. Likewise, the mean isoelectric point of proteins with greater than 0.4 loading score was 6.52 (s = 1.69), and for protein with less than ⿿0.4 was 8.14 (s = 2.80), compared to 6.72 (s = 1.63) for all DF proteins.
We next investigated the proteins mainly contributing to the separation of the two participant. In our dataset this was based on PCA principle component 2. The two proteins with the highest impact were HLA class I histocompatibility antigen A-23 alpha chain (HLA-A) and HLA class II histocompatibility antigen DRB1-4 beta chain (HLA-DRB1). Both are well described in the literature and known to exist in high genetic diversity. To ensure that the two proteins were represented similarly using all tissue preservation methods, we calculated the mean fold abundance change between participant A and B, of HLA-DRB1 for each method. HLA-DRB1 was found in all biopsies and the fold change of the protein between participant B and A was 5.00 in DF biopsies, 4.32 in RNAlater biopsies, 5.66 in iFFPE biopsies, and 5.55 in sFFPE biopsies. HLA-A was found in all biopsies from participant A and none from participant B, regardless of method of sample stabilization. The sensitivity of the assay thereby seems retained regardless of method of sample stabilization for these proteins.
3.4. Protein abundance scatterplots
To investigate preservation method induced variations across all protein abundances, we investigated scatterplots of the protein abundances. Scatterplots compare the quantitative value of every protein between two preservation methods, plotting the protein abundance of different methods on the x and y-axis, respectively. Ideally, the different tissue preservation methods should yield identical protein abundances, represented by a Pearson⿿s correlation coefficient of one, indicating a perfect correlation. The measured protein abundance in the three biological replicates were combined preservation method wise by the mean, and scatterplots were generated comparing the RNAlater, iFFPE, and sFFPE preservation methods to DF, as well as iFFPE to sFFPE (Fig. 6).
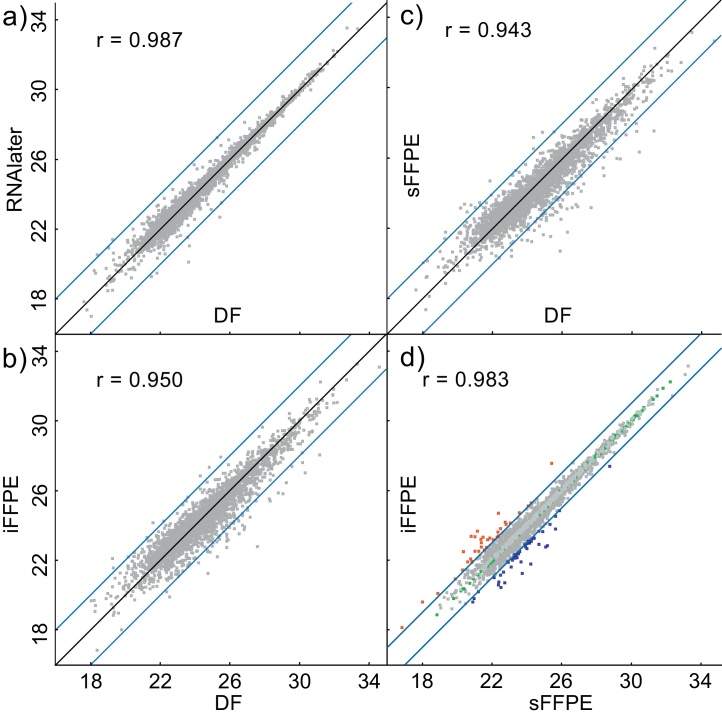
Fig. 6.
Scatterplots of the log 2 transformed protein abundances of all quantifiable proteins in the colon biopsies, combined by the mean preservation method wise. The protein abundances are plotted against one another on the x- and y-axes, respectively. Correlations between measured protein abundances using the different methods are represented by Pearson⿿s correlation coefficients (r), where r = 1 signifies a perfect correlation. Protein abundances in directly frozen (DF) colon biopsies was compared to (a) RNAlater, (b) immediate formalin-fixed, paraffin-embedded (iFFPE), or (c) 30 min stored formalin-fixed, paraffin-embedded (sFFPE), and (d) iFFPE and sFFPE are compared. In all cases r-values > 0.94 were calculated indicating a good correlation. Lines indicates a four fold protein abundance difference in (a⿿c), and a two fold protein abundance difference in (d).
The lowest correlation coefficient (0.943) was calculated comparing DF preserved tissue to sFFPE preserved tissue. The highest correlation (0.987) was found between the DF and RNAlater preserved tissues, which was similar to the coefficient between the iFFPE and sFFPE (0.983) protein quantitations. In all cases, the Pearson⿿s correlation coefficients were close to one, indicating a good correlation.
The high Pearson⿿s correlation coefficient between iFFPE and sFFPE indicates that delaying tissue biopsy stabilization with 30 min only had a minor impact on the measured global protein abundances. To further investigate the differences between iFFPE and sFFPE and the impact of the delayed sample stabilization, we added protein instability index scores from ExPASy for changing proteins [38]. The instability index score provides an estimation of a proteins⿿ in vivo stability [40]. An instability index score below 40 estimates the protein to be stable [41]. We assumed that proteins prone to in vivo degradation, would be partly degraded in the 30 min delay in stabilization in the sFFPE preserved biopsies. Therefore, we choose to investigate proteins with a greater than two fold abundance decrease in sFFPE biopsies compared to iFFPE. This came to 48 proteins and 113 iFFPE unique proteins, and the mean protein instability index score for these proteins was calculated to be 42.0 (s = 13.3). The score was compared to the mean protein instability index score for proteins with similar abundances between sFFPE and iFFPE, which we assumed to be more in vivostable. We choose to include all proteins with less than 0.05 mean fold change difference between iFFPE and sFFPE (fold change 1.05⿿0.95). This came to 677 proteins with a mean protein instability index score 43.8 (s = 10.7). Considering the standard deviation, the two stability indexes are highly similar. We additionally investigated the molecular weight and isoelectric point of these protein groups, and similar properties were found (data not shown). This indicates that the protein instability index score isoelectric point and molecular weight cannot account for the abundance change of the 161 proteins (Supplementary list 2) found with lower abundance in the sFFPE biopsies.
4. Discussion
We examined the impact of sample preservation on a discovery-based proteome analysis, in order to enable reliable proteome analysis of differently preserved samples. Human colon mucosa biopsies were extracted and immediately preserved in RNAlater or by FFPE. To simulate a clinical scenario, one set of biopsies were stored for 30 min (sFFPE) at ambient temperature before sample stabilization with formalin. The samples were compared to snap frozen biopsies (DF), where the introduction of chemical artifacts, which might interfere with a proteome analysis, is minimal. The main finding of the study was that biological samples can be stored in RNAlater and preserved by FFPE with a minimal impact on the result of a quantitative proteome analysis compared to DF preserved samples. Additionally, delaying the formalin sample stabilization for 30 min only had a minor impact in our dataset. Similar results and conclusions regarding pathway regulations can be reached for the samples as compared to DF preserved samples. Especially RNAlater preservation was found to be a promising alternative to snap freezing samples for proteomic studies.
We performed a peptide PTM analysis to investigate the efficiency of the de-crosslinking protocol and to identify modifications introduced by the preservation methods. The PTM analysis revealed that FFPE preservation of colon tissue caused an increase in the overall number of peptides identified with a modification compared to DF and RNAlater. The analysis of specific peptide modifications revealed an increased ratio of formylated peptides (K- and N-term) in the FFPE preserved biopsies compared to RNAlater and DF. This was an expected artifact from the FFPE crosslinking. Additionally, an increased number of methylated lysine-containing peptides was identified in the FFPE preserved samples, in accordance with findings in a recent study which gives validation to the applied methodology [42]. However, less than 10% of the PSMs resulted in identification of peptides with a formyl modification, indicating that the applied de-crosslinking protocol is effective. An increased number of deamidated peptides were identified in biopsies preserved with the FFPE protocols compared to the DF and RNAlater protocols. The modification was likely introduced during the exposure to non-physiological pH and elevated temperatures during the sample preparation (alkaline pH) which was prolonged in the FFPE protocol [43], [44]. Only minor differences could be observed in the PTM frequencies between DF and RNAlater stabilized tissue, indicating that the introduction of identifiable chemical artifacts with the two methods is similar.
The alkylation of cysteine-containing peptides with chloroacetamide was demonstrated, and 100% of cysteine-residue containing peptides were found in an alkylated state. Additionally, no peptide alkylation artifacts were observed in this study with frequencies of more than 0.6% of possibly modifiable peptides, such as carbamidomethylated N-terminal and non-cysteine residues. In a recent study of porcine synovial fluid using the FASP protocol with iodoacetamide alkylation, 2.7% of all peptides were identified with a carbamidomethylated N-terminal, and 1.9% of the cysteine- and lysine-containing peptides were identified with an carbamidomethylated lysine [45]. The analyzed samples in the mentioned study are very different from the samples analyzed in this study. Nonetheless, the comparison points to an improved alkylation specificity when using the protocol with chloroacetamide compared to iodoacetamide, in agreement with a previous study [29].
The differences in frequencies of the various modifications demonstrate that the majority of the PTMs are introduced during the sample preservation. As such, the PTM-table can be used to choose relevant modifications for database searching.
We were able to identify a similar number of proteins in tissue preserved by DF and RNAlater. Other proteomic studies have reported a similar or slightly increased number of proteins identified from bacteria, yeast, and fish preserved in RNAlater compared to DF, in agreement with our findings on human tissue [5], [7], [9]. We identified a statistically significantly lower number of proteins in the FFPE preserved tissue compared to DF and RNAlater preserved biopsies. These findings are in agreement with what other proteomic studies on human samples have reported [2], [3], [14], [15], [16], [17], [18], [19], [20], [21]. Even though the number of quantified proteins was lower in the FFPE biopsies, 92.3% of the proteins quantified in the DF preserved biopsies were also quantified in the FFPE preserved biopsies. The high overlap demonstrates the feasibility of conducting proteomics on FFPE samples.
We conducted a Gene Ontology-based analysis of proteins only found in the iFFPE preserved biopsies, compared to the RNAlater and DF preserved. This was done to investigate if the unique proteins originated from a specific cellular location. The analysis revealed that proteins unique to a given method did not belong to a specific subcellular location. The analysis of the isoelectric point and molecular weight of the unique protein groups also did not reveal any determining differences. This indicates that no systematic loss or enrichment of proteins from a specific cellular location, nor proteins with specific physiological properties, is taking place using either of the preservation methods, and thus an unbiased analysis regardless of sample preservation method.
The label-free relative quantitation allowed us to assess the impact of the biopsy preservation on the protein abundances. The PCA scores plot of principle component 1 and 2 described the largest variance in protein abundances. The PCA scores plot did not separate DF and RNAlater preserved colon tissue, nor the colon tissue preserved by iFFPE and sFFPE. This demonstrates that the biological variance in the measured proteome of the two participants is greater than the variance introduced by the preservation in DF and RNAlater, as well as iFFPE and sFFPE tissue, respectively. Samples preserved by RNAlater can thus be compared to sample preserved by DF, as the introduction of proteome changes is minimal. The same can be said for iFFPE and sFFPE preserved samples. It should be emphasized that the colon biopsies originated from two participants without gastroenterological findings. The difference in protein abundances between the two sets of biopsies can, therefore, be expected to be less than what is measured in disease studies, e.g., of inflammed and non-inflammed colon biopsies [27]. Even so, we were still able to separate biopsies from the two participants. The findings from the PCA are supported by the scatterplots, where protein abundances across the different methods were investigated. The calculated Pearson⿿s correlation coefficients of RNAlater, iFFPE, sFFPE, and DF preserved colon biopsies were all greater than 0.94. This demonstrates the good correlation in measured protein abundances between the different methods, i.e., a low impact of the method of preservation on the overall measured protein abundances [27], [46]. The result indicates that the largest difference is between the DF and FFPE methods, in agreement with the PCA.
Additionally, the slightly better correlation of DF and RNAlater preserved samples compared to iFFPE and sFFPE, indicates that the impact on the proteome of the RNAlater treatment is less than the impact of a 30 min storage at room temperature. As RNAlater samples are stored at 24 h at room temperature prior to storage at ⿿80 °C, the result indicates that RNAlater efficiently inhibits in vivobiological activity in the biopsies. The similar relative amounts measured using either method, demonstrates that all four preservation methods can be used to stabilize tissues prior to proteomic analysis. This is supported by the PCA showing the ability to separate the participants by genomic diverse proteins such as the two HLA protein classes.
The collection of samples for research at hospitals and clinics should not interfere with the standard clinical protocols, which means that a delay before a sample can be stabilized is in many cases unavoidable. It has been reported that the average time from specimen extraction to processing in a surgical department is 19.3 min [4]. The time can be expected to vary between different hospitals/departments. Therefore, we chose to store samples for 30 min prior to formalin stabilization (sFFPE). We did not find a significant decrease in the number of proteins we were able to recover from the sFFPE samples compared to iFFPE. Nor did we find vast changes in the overall protein abundances by scatterplots, nor changes in the protein stability index scores caused by the 30 min delayed sample stabilization. Our findings thereby strongly support that clinically obtained tissue biopsies can be used for quantative proteomics research, even when stabilization has been delayed. We did not focus our analysis on the impact of unstable protein modifications, such as protein phosphorylation or proteins with short in vivo lifetime/stability. Several studies have reported changes in the phosphorylation-patterns of several stress-related proteins following tissue extraction, when sample stabilization was delayed even a few minutes [4], [47], [48]. Clinical samples obtained with a delay might, therefore, constitute suitable material for global quantative proteome research focusing on protein abundances, but be poorly suitable for e.g., protein phosphoprotein studies. We have included the list of 48 proteins with a greater than two-fold abundances difference between iFFPE and sFFPE preserved biopsies and the 113 iFFPE unique proteins (Supplementary Table 2). It is possible that these proteins display a lower in vivo stability than represented by the protein instability index scores.
Other studies have demonstrated that RNAlater can be used to stabilize bacteria, fungus, and human cervical swabs prior to proteome analysis [5], [8], [9]. Combined with the findings in other studies performed on bacteria and yeast, our findings demonstrate that RNAlater can be used for uniform preservation of a wide range of biological material prior to proteomic, transcriptomic, and genomic studies. The need for snap freezing samples can be reduced, which can be impractical especially in clinical situations. Additionally, RNAlater and FFPE sample preservation removes the need for quick sample handling. This is in contrast to DF preserved tissue where the protective effect of freezing only persists until the sample is thawed, making a rapid sample handling prior to protease inhibition critical [5]. The possibility to utilize established biobanks for proteome analysis constitutes a vast source of well-characterized biological material for clinical proteomics research.
5. Conclusion
This is the first study to investigate the impact of RNAlater and FFPE stabilization of human tissues on PTMs and protein quantitation. Using 24 human colon biopsies from two participants, we have demonstrated that human tissue samples can be stabilized and preserved by RNAlater or FFPE as alternatives to snap freezing with a minimal impact on the quality of protein quantifications. Especially RNAlater preservation was found to be a promising alternative to snap freezing samples for proteomic studies. Comparable proteomics pathway information can be extracted from tissue preserved with either method. Additionally, delaying tissue sample stabilization with formalin for 30 min to simulate a clinical situation, only resulted in a minor impact on the quality of the protein quantitations. Our findings thereby demonstrate that biobanks containing RNAlater preserved samples, FFPE preserved samples, and samples obtained with a delay in stabilization, can be used for proteome analysis. Similar result and conclusions can be obtained on the global proteome level as when studying snap frozen samples. The suggested protocols can thus be used, e.g. to provide retrospective information concerning diagnosis, response to therapy and novel drug discovery.
Conflict of interest
Vibeke Andersen receives compensation as a consultant and advisory board member for MSD (Merck) and Janssen.
Supporting information
Supplementary Table 1 contains the full list of PTMs for each biopsy. Supplementary Table 2 contains the list of proteins displaying a greater than 2 fold change between iFFPE and sFFPE. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002029 [31], [32]. Bennike et al. [49] contains an expanded description of all deposited proteomics data files and Supplementary Table 1 in [49] contains the full list of identified proteins (<1% FDR) in the human colon biopsies.
Acknowledgements
We would like to thank the two participants for taking part in the study. Department of Pathology at Aalborg University Hospital, Denmark is acknowledged for performing the FFPE sample preservation. Knud and Edith Eriksens Memorial Foundation and Ferring are acknowledged for grants, enabling the collection of the biological sample material (VA grant). The Obelske family foundation, the Svend Andersen Foundation and the SparNord foundation are acknowledged for grants to the analytical platform, enabling this study (AS grants). The Lundbeck Foundation and Carlsberg Foundation are acknowledged for grants enabling the analysis (TBB grants). The staff at the Hospital of Southern Jutland are thanked for excellent technical assistance. Finally, the authors would like to thank the PRIDE team for making the proteomics data publically available.
Footnotes
Significance: We have demonstrated the feasibility in conducting proteome analysis of samples stabilized in RNAlater and formalin fixed, paraffin-embedded samples, and propose analysis strategies for both. Especially RNAlater preservation was found to be a promising alternative to snap freezing samples for proteomics studies, making a simple and uniform sample preservation possible for proteomic, transcriptomic, and genomic studies. Delaying tissue stabilization with formalin-fixation for 30 min only had a minor impact on the result of the analysis. Our study demonstrated the feasibility in conducting analysis of samples stored in biobanks to extrapolate retrospective information for studies in diagnosis, response to therapy, and novel drug discovery.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.euprot.2015.10.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bennike T., Birkelund S., Stensballe A., Andersen V. Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J. Gastroenterol. WJG. 2014;20:3231–3244. doi: 10.3748/wjg.v20.i12.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanca A., Abbondio M., Pisanu S., Pagnozzi D., Uzzau S., Addis M.F. Critical comparison of sample preparation strategies for shotgun proteomic analysis of formalin-fixed, paraffin-embedded samples: insights from liver tissue. Clin. Proteomics. 2014;11:28. doi: 10.1186/1559-0275-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steiner C., Ducret A., Tille J.-C., Thomas M., McKee T.A., Rubbia-Brandt L. Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues. Proteomics. 2014;14:441–451. doi: 10.1002/pmic.201300311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espina V., Edmiston K.H., Heiby M., Pierobon M., Sciro M., Merritt B. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol. Cell. Proteomics. 2008;7:1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Eijsden R.G.E., Stassen C., Daenen L., Mulders S.E.V., Bapat P.M., Siewers V. A universal fixation method based on quaternary ammonium salts (RNAlater) for omics-technologies: Saccharomyces cerevisiae as a case study. Biotechnol. Lett. 2013;35:891–900. doi: 10.1007/s10529-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 6.Drakulovski P., Locatelli S., Butel C., Pion S., Krasteva D., Mougdi-Pole E. Use of RNAlater® as a preservation method for parasitic coprology studies in wild-living chimpanzees. Exp. Parasitol. 2013;135:257–261. doi: 10.1016/j.exppara.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbaraju N.V., Cai Y., Rees B.B. Protein recovery and identification from the gulf killifish, Fundulus grandis: comparing snap-frozen and RNAlater® preserved tissues. Proteomics. 2011;11:4257–4261. doi: 10.1002/pmic.201100328. [DOI] [PubMed] [Google Scholar]
- 8.Rader J.S., Malone J.P., Gross J., Gilmore P., Brooks R.A., Nguyen L. A unified sample preparation protocol for proteomic and genomic profiling of cervical swabs to identify biomarkers for cervical cancer screening. Proteomics Clin. Appl. 2008;2:1658–1669. doi: 10.1002/prca.200780146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito M.A., Bulygin V.V., Moran D.M., Taylor C., Scholin C. Examination of microbial proteome preservation techniques applicable to autonomous environmental sample collection. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han N.-Y., Choi W., Park J.-M., Kim E.H., Lee H., Hahm K.-B. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J. Dig. Dis. 2013;14:166–174. doi: 10.1111/1751-2980.12035. [DOI] [PubMed] [Google Scholar]
- 11.Metz B., Kersten G.F.A., Baart G.J.E., de Jong A., Meiring H., Hove ten J. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjugate Chem. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 12.Metz B. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 2003;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 13.Fox C.H., Johnson F.B., Whiting J., Roller P.P. Formaldehyde fixation. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 14.Avaritt N.L. Decoding the proteome in formalin-fixed paraffin-embedded (FFPE) tissues. J. Proteomics Bioinf. 2014:07. [Google Scholar]
- 15.Magdeldin S., Yamamoto T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics. 2012;12:1045–1058. doi: 10.1002/pmic.201100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakabayashi M., Yoshihara H., Masuda T., Tsukahara M., Sugiyama N., Ishihama Y. Phosphoproteome analysis of formalin-fixed and paraffin-embedded tissue sections mounted on microscope slides. J. Proteome Res. 2014;13:915–924. doi: 10.1021/pr400960r. [DOI] [PubMed] [Google Scholar]
- 17.Paulo J.A., Lee L.S., Banks P.A., Steen H., Conwell D.L. Proteomic analysis of formalin-fixed paraffin-embedded pancreatic tissue using liquid chromatography tandem mass spectrometry. Pancreas. 2012;41:175–185. doi: 10.1097/MPA.0b013e318227a6b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostasiewicz P., Zielinska D.F., Mann M., Wiſniewski J.R. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J. Proteome Res. 2010;9:3688–3700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 19.Scicchitano M.S., Dalmas D.A., Boyce R.W., Thomas H.C., Frazier K.S. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J. Histochem. Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Addis M.F., Tanca A., Pagnozzi D., Rocca S., Uzzau S. 2-D PAGE and MS analysis of proteins from formalin-fixed, paraffin-embedded tissues. Proteomics. 2009;9:4329–4339. doi: 10.1002/pmic.200900010. [DOI] [PubMed] [Google Scholar]
- 21.Crockett D.K., Lin Z., Vaughn C.P., Lim M.S., Elenitoba-Johnson K.S. Identification of proteins from formalin-fixed paraffin-embedded cells by LC⿿MS/MS. Lab. Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 22.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 23.Manza L.L., Stamer S.L., Ham A.-J.L., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 24.Leon I.R., Schwammle V., Jensen O.N., Sprenger R.R. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol. Cell. Proteomics. 2013;12:2992–3005. doi: 10.1074/mcp.M112.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawashima Y., Kodera Y., Singh A., Matsumoto M., Matsumoto H. Efficient extraction of proteins from formalin-fixed paraffin-embedded tissues requires higher concentration of tris(hydroxymethyl) aminomethane. Clin. Proteomics. 2014;11:4. doi: 10.1186/1559-0275-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda T., Tomita M., Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 27.Bennike T.B., Carlsen T.G., Ellingsen T., Bonderup O.K., Glerup H., Bøgsted M. Neutrophil extracellular traps in ulcerative colitis: a proteome analysis of intestinal biopsies. Inflammatory Bowel Dis. 2015 doi: 10.1097/MIB.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen M.L., Vermeulen M., Bonaldi T., Cox J., Moroder L., Mann M. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 29.Dahl K.H., McKinley-McKee J.S. The reactivity of affinity labels: a kinetic study of the reaction of alkyl halides with thiolate anions⿿a model reaction for protein alkylation. Bioorg. Chem. 1981;10:329–341. [Google Scholar]
- 30.G.C. Urbaniak, S. Plous (2013). Research Randomizer (Version 4.0) [Computer software]. Available: http://www.randomizer.org/ (accessed 22.06.13).
- 31.Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcaíno J.A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics MCP. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.A., ndromeda A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 35.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 36.Bhatia V.N., Perlman D.H., Costello C.E., McComb M.E. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal. Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deeb S.J., Souza R.C.J.D⿿, Cox J., Schmidt-Supprian M., Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics. 2012;11:77–89. doi: 10.1074/mcp.M111.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de C., astro E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao P.K., Li Q. Principal component analysis of proteome dynamics in iron-starved Mycobacterium tuberculosis. J. Proteomics Bioinf. 2009;2:19–31. doi: 10.4172/jpb.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guruprasad K., Reddy B.V., Pandit M.W. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. Springer; 2005. Protein Identification and Analysis Tools on the ExPASy Server. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Muller M., Xu B., Yoshida Y., Horlacher O., Nikitin F. Unrestricted modification search reveals lysine methylation as major modification induced by tissue formalin fixation and paraffin embedding. Proteomics. 2015;15:2568–2579. doi: 10.1002/pmic.201400454. [DOI] [PubMed] [Google Scholar]
- 43.Krokhin O.V., Antonovici M., Ens W., Wilkins J.A., Standing K.G. Deamidation of -Asn-Gly- sequences during sample preparation for proteomics: consequences for MALDI and HPLC-MALDI analysis. Anal. Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- 44.Dick L.W., Kim C., Qiu D., Cheng K.-C. Determination of the origin of the N-terminal pyro-glutamate variation in monoclonal antibodies using model peptides. Biotechnol. Bioeng. 2007;97:544–553. doi: 10.1002/bit.21260. [DOI] [PubMed] [Google Scholar]
- 45.Bennike T., Ayturk U., Haslauer C.M., Froehlich J.W., Proffen B.L., Barnaby O. A normative study of the synovial fluid proteome from healthy porcine knee joints. J. Proteome Res. 2014;13:4377–4387. doi: 10.1021/pr500587x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics MCP. 2012;11 doi: 10.1074/mcp.M111.014050. doi:10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., Friedman A.B., Roh M.-S., Jope R.S. Anesthesia and post-mortem interval profoundly influence the regulatory serine phosphorylation of glycogen synthase kinase-3 in mouse brain. J. Neurochem. 2005;92:701–704. doi: 10.1111/j.1471-4159.2004.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Gould T.D., Yuan P., Manji H.K., Chen G. Post-mortem interval effects on the phosphorylation of signaling proteins. Neuropsychopharmacology. 2002 doi: 10.1038/sj.npp.1300112. [DOI] [PubMed] [Google Scholar]
- 49.T.B. Bennike, K. Kastaniegaard, S. Padurariu, M. Gaihede, S. Birkelund, V. Andersen, et al. (2015). Proteome stability analysis of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human colon mucosal biopsies (Data in Brief; submitted). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.