Abstract
Chromatin is an intelligent building block that can express either external or internal needs through structural changes. To date, three methods to change chromatin structure and regulate gene expression have been well-documented: histone modification, histone exchange, and ATP-dependent chromatin remodeling. Recently, a growing body of literature has suggested that histone tail cleavage is related to various cellular processes including stem cell differentiation, osteoclast differentiation, granulocyte differentiation, mammary gland differentiation, viral infection, aging, and yeast sporulation. Although the underlying mechanisms suggesting how histone cleavage affects gene expression in view of chromatin structure are only beginning to be understood, it is clear that this process is a novel transcriptional epigenetic mechanism involving chromatin dynamics. In this review, we describe the functional properties of the known histone tail cleavage with its proteolytic enzymes, discuss how histone cleavage impacts gene expression, and present future directions for this area of study.
Keywords: Chromatin, Gene expression, Histone cleavage, Histone modification
INTRODUCTION
In eukaryotic cells, the genomic DNA is packed in the nucleus via its compaction into chromatin. The fundamental unit of chromatin is called the nucleosome, in which approximately 147 base pairs of DNA are wrapped around an octamer of core histone. Histones are highly conserved basic proteins found in eukaryotes and even in some archaea (1–6). They are divided into five major families: H1, H2A, H2B, H3, and H4. Two copies of H2A, H2B, H3, and H4 histones form the core histone octamer, while the linker histone H1 binds to the nucleosome at the site where internucleosomal DNA enters and exits, further contributing to the structural and functional flexibility of chromatin (5, 7, 8).
Chromatin is a dynamic structure, existing either as heterochromatin or euchromatin. Heterochromatin is more compact and transcriptionally inactive since genes are not easily accessible by transcription machinery proteins. In contrast, euchromatin is a decondensed type of chromatin and is transcriptionally active (3, 9). To allow transcription machinery proteins access to compacted DNA, a process known as chromatin remodeling reorganizes the chromatin structure to expose or hide regions of DNA for transcriptional regulation (10). It has been reported that chromatin remodeling occurs in several ways including histone modifications, exchange of histone variants, and ATP-dependent chromatin remodeling (11).
The core histones contain two common domains: histone folding and histone tail. The N-terminal tail of all four core histones and the C-terminal tail of H2A and H2B protrude outside the nucleosome, which undergoes a variety of posttranslational modifications (PTMs) including methylation, acetylation, propionylation, butylation, crontonylation, 2-hydroxyisobutylation, malonylation, succinylation, formylation, ubiquitination, citrullination, phosphorylation, O-GlcNAcylation, and ADP ribosylation (12–22). Functional outcomes of these histone modifications have been investigated. A well-established hypothesis suggests that histone modification may lead to alteration of contacts between different histones in adjacent nucleosomes or the interaction of histones with DNA. Notably, acetylation neutralizes the positive charge of the lysine and weakens the interaction between histone and DNA, resulting in loosening the chromatin structure (13, 23). Another well-characterized hypothesis of histone modification is that distinct patterns of histone modifications, or “histone codes”, provide signals for the recruitment of specific non-histone protein complexes including transcription factors or proteins that modify the chromatin structure (18, 24, 25). Diverse histone modifications within the chromatin act in a coordinated and orderly fashion to regulate gene transcription (26). The precise alteration of chromatin histone modification is regulated by both the chemical and physical changes of core histones. A typical mechanism of chemical modification is the selective enzymatic addition and elimination of specific histone modifications by histone-modifying enzymes (“writers” and “eraser”), which is a reversible chemical change (27, 28). A more extreme mechanism utilizes physical changes of nucleosome so that the ATP-dependent chromatin remodeler catalyzes the exchange of canonical histones with histone variants, resulting in the removal of all pre-existing PTMs (29). An intermediate mechanism between these two mechanisms is the regulated proteolysis of histone tail, which physically and irreversibly removes pre-existing multiple PTMs (30–33).
In contrast to histone modification or histone exchange, the mechanism and the biological role of histone tail cleavage have not been fully understood. Although the presence of a proteolytic enzyme in histone preparations of calf thymus was observed in 1959 (34), and truncated histones were found in calf thymus and Tetrahymena in 1970s (35, 36), few publications were reported until the early 2000s. However, the regulated cleavage of histone has become an active and interesting topic of epigenetic regulation since the late 2000s. Various enzymes responsible for histone clipping and the biological significance associated with histone cleavage have been studied as shown in Table 1. We discuss the identified enzymes, the regulation, and the biological significance of histone cleavage. Moreover, we propose putative mechanisms that suggest how histone cleavage modulates gene expression.
Table 1.
List of histone proteases identified and their roles
| Histone | Protease | Cleavage site(s) | Model | Biological significance of activity | Reference |
|---|---|---|---|---|---|
| H2A | Histone H2A specific protease (H2Asp) | Asn90-Asp91 | Chicken liver extract | ? | 47 |
| Neutrophil Elastase | Val114-Leu115 | Neutrophil | NET (neutrophil extracellular trap) formation | 44 | |
| H2A protease | Val114-Leu115 | Calf thymus | ? | 38 | |
| H2B | Tryptase | ? | Mouse mast cells | Mast cell differentiation | 48, 49 |
| H3 | MMP-9 | Lys18-Gln19 | Bone marrow macrophages | Osteoclastogenesis | 53 |
| Cathepsin L | Ala21-Thr22, Thr22-Lys23, Lys23-Ala24 Ala24-Ala25, Arg26-Lys27, Lys27-Ser28 | Mouse ESCs | ESC differentiation | 51 | |
| Human ESC protease | Ala21-Thr22, Arg26-Lys27, Ala31-Thr32 | Human ESCs | ESC differentiation | 50 | |
| Glutamate dehydrogenase | Lys23-Ala24, Lys27-Ser28 | Chicken liver extracts | ? | 58 | |
| Yeast endopeptidase | Ala21-Thr22 | S. cerevisiae | Induced under nutrient deprivation and sporulation | 55 | |
| Vacuolar protease B (Prb1) | Lys23-Ala24 | S. cerevisiae | ? | 69 | |
| Foot-and-mouth disease virus 3C protease (FMDV 3C protease) | Leu20-Ala21 | Infected BHK-21 cells | Host cell transcription shut off | 61 | |
| JMJD5 | Lys9-Ser10 | Lung cancer cells | Induced under DNA damage | 56 | |
| ? | ? | Tetrahymena micronuclei | ? | 60 | |
| Cathepsin D | Lys23-Ala24 | Mouse mammary gland | Involution mammary gland | 71 | |
| Tryptase | Mouse mast cells | Mast cell differentiation | 48, 49 | ||
| H4 | Granzyme A | ? | B-lymphoid | Staurosporine-induced cell death | 76 |
| Histone H1 | ? | H1 processed into three different components (α, β, γ). | Tetrahymena micronuclei | Cell division | 75 |
HISTONE H2A AND H2B CLEAVAGE
In 1976, a protease associated with purified calf thymus chromatin was found to act specifically upon histone H2A, yielding a C-terminal truncated H2A. This fragment was derived from the removal of fifteen amino acids from the C-terminal H2A, of which Val114 was the new C-terminal residue (36). The responsible enzyme was further investigated and named the H2A specific protease (H2Asp) (37, 38). The truncation of H2A was also observed in both myeloid and lymphatic leukemia cells (39–41) and retinoic acid-induced differentiation of THP-1 promonocytes into macrophages (42, 43). Later, it was argued that H2Asp might be neutrophil elastase (44). Neutrophil elastase is a neutrophil-specific protease required for neutrophil extracellular trap (NET) formation. It has been reported that, upon reactive oxygen species production, the neutrophil elastase translocates from azurophilic granules into the nucleus and degrades specific histones, leading to chromatin decondensation (45). However, the biological significance of the C-terminal clipped H2A in NET formation remains unclear.
Volger et al. showed that cells expressing C-terminally truncated H2A (amino acids 1–114) were vulnerable to cellular stress, implying a pivotal role of this tail in cellular homeostasis (46). They then demonstrated that the H2A C-terminal tail is important in nucleosome stability and mobility in vivo and in vitro. Furthermore, they showed that this C-terminal H2A tail is implicated in regulating chromatin remodeling process by ISWI-type remodelers and functions as a recognition module for histone H1. These results suggest that H2A C-terminal tail (amino acids 115–129) stabilizes the nucleosomal core particle, while mediating the protein interactions regulating chromatin dynamics.
In 1993, a second H2A specific protease was found in a chicken liver nuclear extract (47). It was argued that by using a protease inhibition assay, this H2Asp is an aspartic acid-like protease. The H2Asp generated a single clipped H2A product (H2AGlu91) in the in vitro cleavage assay. Interestingly, it was found that the expression and the activity of H2Asp were only detected in liver nuclear extract (tissue-specific). However, the function of the C-terminal deleted H2A and the characterization of the responsible protease remains elusive.
Recently, an enzyme for an H2B tail cleavage was reported by Pejler et al. They demonstrated that tryptase clipped off the N-terminal tails of histone H2B as well as histone H3 (48, 49). Mast cells contain a high content of cytoplasmic secretory granules densely packed with bioactive substances, including a mast cell specific protease, tryptase, which performs pro-inflammatory functions. Unexpectedly, they revealed that the tryptase relocalizes from granules into the nucleus during the cell death process and cleaves core histones. The tryptase-dependent truncation of Histone H2B and H3 also occurred in mast cell differentiation. They also found that the tryptase is mostly localized to the heterochromatin, and chromatin fraction from tryptase deficient cells is more resistant to micrococcal nuclease than that from wild type. This implies that tryptase may control the chromatin structure by promoting the formation of euchromatin over heterochromatin. In their subsequent study (49), however, they demonstrated that the absence of tryptase leads to the age-dependent accumulation of H2BK5ac, which was associated with an extensive upregulation of markers of non–mast cell lineages. These results suggest that tryptase can positively or negatively modulate gene expression depending on the chromatin state of the mast cells.
HISTONE H3 CLEAVAGE
The clipping of Histone H3 has been investigated more intensively than that of other histones. Histone H3 has numerous sites that are susceptible to several proteases (33) and the N-terminal cleavage of H3 has been reported in many cellular processes, including mouse embryonic stem cell (ESC) differentiation, viral infection, aging, yeast sporulation, senescence, DNA damaging stress, and osteoclastogenesis (50–56). Additionally, H3 protease activity has been found in Tetrahymena micronuclei, avian liver tissues, human ESCs, and mouse mast cells (48, 54, 57–59).
In 1980, Allis et al. reported that micronuclei of Tetrahymena contained two electrophoretically distinct forms of histone H3; the fast migrating form (H3F) and the slow migrating form (H3S). The fast migrating form, H3F was derived from H3S by a physiologically regulated, proteolytic processing event in the condition of cell growth and/or division, but not in non-growth and starvation, indicating that H3 proteolytic cleavage may occur regularly each generation at a specific point in the cell cycle (60).
Several studies have shown the effect of viral infection on the integrity of histones. It was reported that infection of BHK cells with foot-and-mouth disease virus (FMDV) caused the loss of histone H3 and the simultaneous formation of a new chromatin-associate protein (Pi) which migrates between histones H2A and H4 on SDS-polyacrylamide gels (52). Subsequently, Falk et al. showed that Pi is the truncated H3 form lacking the 20 N-terminal residues by proteolytic cleavage during infection (61). Also, they found that the transition of histone H3 into Pi was catalyzed by the FMDV 3C protease, which had been known to be related in the proteolytic processing of the viral polyprotein into the mature gene products. Another group reported that FMDV protease 3C inhibits gene transcription and its inhibitory mechanism might involve the truncation of histone H3 (62). These results suggested that the FMDV virus controls the host cell’s transcription by modulating histone H3 N-terminal cleavage.
Cathepsin L, a lysosomal protease, is a member of the papain-like super family of cysteine protease. Cathepsin L plays an important role in extracellular matrix degradation related to cancer, bone remodeling, and cardiovascular disease, and functions as an immune modulator via the generation of peptide antigens for MHC class II presentation (63, 64). Although Cathepsin L is mainly localized within lysosome, it is also found in the nucleus, and the functions of Cathepsin L in the nucleus have been reported (65–68). In 2008, Duncan et al. demonstrated that Cathepsin L proteolytically processes histone H3 during mouse ESC differentiation (51), in the first report presenting the implication of histone cleavage in the alteration of epigenetic signatures upon differentiation. Histone H3 was cleaved at the N-terminal tail during ESC differentiation, with alanine 21 being the primary site of cleavage, while Cathepsin L was identified as a protease responsible for H3 N-terminal tail clipping. They also suggested that this cleavage activity might be modulated by covalent modifications such as H3K18ac or H3K27me2 via in vitro cleavage assays. Moreover, this removal of histone H3 N-terminal tail inhibited the ability of CBX27 to bind to H3K27 methylation, indicating the histone H3 cleavage by Cathepsin L could lead to significant downstream effects. The biological significances of the Cathepsin L-mediated H3 cleavage during ESC differentiation remain elusive.
The proteolytic processing of histone H3 tail by Cathepsin L was also observed in models of oncogene-induced and replicative senescence (50). It was found that unlike ESC differentiation, H3.3 is preferentially cleaved over H3.1 during cellular senescence. RNA-sequencing studies revealed that the overexpression of the cleaved H3.3 in fibroblasts results in transcriptional downregulation of cell cycle genes, with significant overlap of genes that lose H3K4me3 during cellular senescence. It was suggested that H3.3 tail cleavage plays a key role in silencing the transcription of cell cycle-promoting genes by removing H3K4me3.
In 2009, Santos-Rosa et al. used a yeast model and detected the endopeptidase activity of nuclei preparations toward histone H3, which was low in cells growing exponentially but increased in cells grown into stationary phase or shifted to sporulation (55). They found that the H3 endopeptidase is a serine protease clipping histone H3 after alanine 21, but were unable to further define a specific protease among the 24 serine proteases of S. cerevisiae. They also showed that loss of the H3 N-terminal tail correlates with the induction of gene expression that precedes H3 displacement. To test if the H3 tail-cleavage is prerequisite for gene induction, Santos-Rosa et al. used a yeast strain having the mutation in the histone H3 cleavage site which is unable to cleave the H3 tail. This mutant showed impaired expression of genes activated on the stationary phase and sporulation, indicating that the cleavage of the H3 tail at gene promoters contributes to the proper induction of gene expression. After 5 years, it was reported the vacuolar proteinase B (Prb1) has a cleavage activity toward histone H3 N-terminus in both whole cell extracts and nuclear extracts of S. cerevisiae. They demonstrated that the purified Prb1 can cleave the H3 N terminus between Lys23 and Ala24 in vitro (69).
Mandal et al. reported that a cysteine protease activity in chicken liver is specific for histone H3 (57). Later, they revealed via mass spectrometric analysis that this protease is glutamate dehydrogenase (58). Glutamate dehydrogenase was found to bind to four core histones but cleaving only the histone H3 tail in its free and chromatin context. Additionally, the truncation of H3 was observed in quail liver during the aging process, although the responsible enzyme was not identified (54, 70).
In the involuting mammary gland, the nuclear translocation of Cathepsin D caused the proteolytic cleavage of H3 at its amino terminal tail (71). They showed that H3 was preferentially cleaved by Cathepsin D between lysine 23 and alanine 24. Interestingly, they found that the nuclear translocation of Cathepsin D was mediated by its posttranslational modification, tyrosine nitration.
Recently, our group found that matrix metalloproteinase 9 (MMP-9) is the principal protease of the histone H3 N-terminal tail during osteoclast differentiation (53). It has been shown that MMPs are a family of zinc-dependent endopeptidases that remodel the pericellular space primarily through the cleavage of extracellular matrix proteins. Although MMP-9 was known as a secretory protein, we detected the nuclear accumulation of MMP-9 during osteoclastogenesis through biochemical studies (74). We found that MMP-9 specifically cleaved H3K18-Q19 in vitro and in vivo. Furthermore, our results also showed that MMP-9 enzymatic activity toward H3 N-terminal tail is significantly facilitated by H3K18 acetylation and that p300/CBP is the histone acetyl transferases responsible for H3K18ac observed in osteoclast cells. Importantly, our findings indicated that MMP-9 was necessary for H3 tail cleavage near transcription start sites or gene bodies of a set of osteoclastogenic genes and concurrent gene activation during osteoclast differentiation, suggesting that H3 N-terminal proteolysis induces gene activation.
Shen et al. demonstrated that JMJD5 mediates histone H3 N-tail proteolytic cleavage at the carboxyl side of H3K9me1 under stress conditions such as a DNA damage response (56). JMJD5, a Jumonji C (JmjC) domain-containing protein, was originally known as histone H3 lysine 36 dimethylation (H3K36me2), responsible for gene expression. In A549 cells, under cell stress conditions, JMJD5 bound to gene promoters and cleaved H3K9me1, suggesting JMJD5-mediated histone cleavage may be responsible for gene transcription regulation.
As previously mentioned, mast cell tryptase functioned as a clippase of histone H3 as well as H2B and, in aging mast cells, the partial loss of tryptase expression was accompanied by a decrease of H3 and H2B clipping (48, 49). While tryptase-deficiency did not affect the posttranslational modifications of histone H3, it caused an increase of H2BK5ac. Therefore, the biological significance of histone H3 clipping in mast cells still needs to be determined.
OTHER HISTONES CLEAVAGE
A limited number of studies have reported the conversion of histone H1 or H4 into a smaller form via a regulated proteolysis. Nevertheless, the degradation of histone H1 or H4 has been shown in several biological processes including the macronuclear degradation of Tetrahymena and staurosporine-induced cell death (75–77). Interestingly, it was shown that histone H1 of Tetrahymena micronuclei was processed into smaller forms. Tetrahymena contains two different nuclei: a macronucleus and a micronucleus. Micronucleus-specific histone H1 differs from macronuclear H1. Micronucleus-specific histone H1 was larger and is proteolytically processed into different components (α, β, γ and δ). This proteolytic processing of H1 histone was regulated in the pathway of micronuclear differentiation (75). Lee et al. demonstrated that histone H4 is cleaved by granzyme A under apoptotic process in B-lymphocytes (76). Granzyme A is a serine protease in the cytotoxic granules found in natural killer cells and cytotoxic T cells. They suggested that Granzyme A-mediated H4 tail cleavage contributes to the disintegration of the chromatin architecture during the cell death process (76).
EPIGENETIC REGULATORY MECHANISM BY HISTONE CLEAVAGE
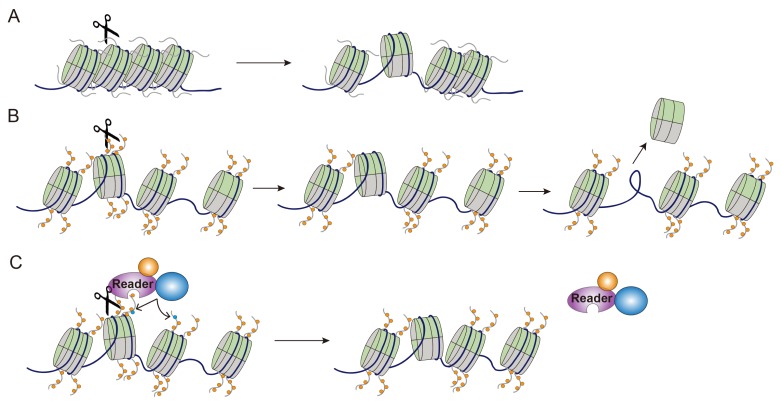
The proteolytic cleavage of histone is an interesting and active topic of epigenetic modifications. Despite the increasing number of studies that demonstrate novel enzymes and cellular processes related to histone cleavages, the mechanism by which cleaved histones regulate gene expressions still needs to be explained. Here, we discuss recent findings, focusing on how histone cleavage impacts gene expression (Fig. 1).
Fig. 1.
Schematic model showing the functional roles of histone cleavage in gene expression. (A) Influence of genomic DNA or chromatin compaction on accessibility. Histone tail cleavages by protease might generate open chromatin structures and increase DNA accessibility for transcription factors, thereby contributing to gene activation. (B) Induction of histone eviction. The truncated histone by protease is unstable in the nucleosome. Histone tail cleavage may induce histone eviction, allowing easy access of transcription factors to DNA elements during gene activation. (C) Removal of PTMs and interacting proteins. Histone tail cleavage may lead to the radical removal of multiple modifications, resulting in blockage of the recruitment of effector proteins (“Reader” protein) or other PTM cascades on histone tails.
Influence on chromatin compaction
Eukaryotic DNA is packed into chromatin, which enables the genomic DNA to fit in the limited space of the nucleus. However, chromatin limits the accessibility of transcription factors and RNA polymerase to gene promoters. Thus, chromatin conformation plays an important role in gene expression (55).
Tailless nucleosomes have been used to investigate the role of histone tails (78, 79). Using tailless nucleosomes treated with trypsin to cleave off the core histone tail domains, it was reported that the histone tails participate in inter- or intra-nuclesomal interactions and play a significant role in regulating the accessibility of genomic DNA (79). Also, nucleosomes containing truncated H3 and/or truncated H4 were utilized to clarify the role of flexible tails of H3 and H4 histones. The N-terminal regions of histone H3 and H4 were implicated in the intranucleosomal interactions by restricting the DNA breathing motion and compacting the nucleosome (78). Therefore, histone tail truncations by specific protease might increase DNA accessibility and generate open conformation of nucleosomes, leading to gene activation.
Linking to histone eviction
Histones are evicted at many promoters during gene activation to allow access to the transcription machinery (80, 81). It was also suggested that the entire nucleosome is at least sometimes disassembled and reassembled during the process of RNA polymerase passage (80).
As mentioned previously, Santos-Rosa et al. identified a histone H3 endopeptidase in S.cerevisiae, which cleaves the H3 N-terminal tail at the promoter sites of genes following the induction of transcription (55). Notably, H3 clipping precedes the process of histone eviction, resulting in the localized clearing of repressive signals during the induction of gene expression. They suggested the possible role of a clipped H3 to mark the nucleosomes that are to be displaced prior to gene induction. Taken together with previous studies, histone tail cleavage may induce histone eviction, allowing easy access of transcription machinery to DNA during gene activation.
Removal of PTMs and interacting proteins
Diverse PTMs are found primarily on the tails of histones that extend distinctively from the nucleosome core, which is sensitive to proteases. To date, two characterized models have been used to consider the outcome of histone tail modifications. The first model suggests that histone tail modifications induce changes in the physical properties of the nucleosome that reduces inter- or intra-nucleosomal contacts, producing a more relaxed chromatin fiber. Of all known modifications, histone acetylation via the neutralization of the positive charge of lysine, and histone phosphorylation via the addition of negative charge, may cause decondensation of the chromatin structure (13). The second model suggests that modifications function as recognition marks to mediate the recruitment of effector proteins. Numerous chromatin-associated factors have been shown to specifically interact with modified histones through many distinct domains such as bromodomain, PHD finger, chromodomain, Tudor domain, PWWP domain, and MBT domain (62–64). These histone tail modifications are modulated by specific enzymes that add or remove modifications, respectively. Although specific modifications can be removed by specific enzymes, histone clipping may be an efficient way to erase multiple modifications of histone tails concurrently. That is, histone tail cleavage could cause the radical removal of multiple modifications, thereby blocking the recruitment of effector proteins or other PTM cascades on histone tails.
PERSPECTIVES
Although a growing body of studies has revealed histone tail cleavages in diverse cellular processes and identified specific proteases responsible for histone cleavages, these findings have raised many questions.
In particular, it would be interesting to determine the genes that are regulated by histone cleavage. Our integrated genome-wide analysis of ChIP-seq and RNA-seq data revealed that MMP-9 cleaved histone tails in approximately 8% of all protein-coding genes during osteoclastogenesis and many osteoclast-related genes were regulated (53). Additionally, JMJD5-mediated H3 tail clipping affected cell cycle-related gene transcription (56). However, the genes that are directly affected by protease-specific histone cleavage still need to be investigated.
It is also important to determine how proteases localize at specific genes under specific conditions. It is suggested that specific histone modifications could recruit histone proteases. We observed that H3K27me1 is necessary for the localization and function of MMP-9 at osteoclastogenic genes as a protease catalyzing H3 tail proteolysis (unpublished data). Alternatively, a crosstalk or interaction between proteases and other histone modifying enzymes might exist that enables histone proteases to localize at specific sites (30).
Finally, a way of controlling protease activity needs to be investigated. Protease can be post-translationally regulated. For example, the nuclear translocation of Cathepsin D was regulated by its post-translational modification, tyrosine nitration (71). Also, histone tail modifications affect the cleavage activity. It has been suggested that Cathepsin L cleavage activity might be modulated by covalent modifications such as H3K18ac or H3K27me2 (51). Moreover, CBP/p300-mediated acetylation of H3K18 facilitated MMP-9-dependent H3 proteolysis (53). Nonetheless, it will be interesting to determine the upstream effectors that stimulate or inhibit protease activity in signaling pathways.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Research Foundation (NRF) of Korea (2015R1A4A1041869 and 2017R1C1B2008017 to K.K.; 2016R1A6A3A11935271 to S.J.Y.).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Bentley GA, Lewit-Bentley A, Finch JT, Podjarny AD, Roth M. Crystal structure of the nucleosome core particle at 16 A resolution. J Mol Biol. 1984;176:55–75. doi: 10.1016/0022-2836(84)90382-6. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Reeve JN, Sandman K, Daniels CJ. Archaeal histones, nucleosomes, and transcription initiation. Cell. 1997;89:999–1002. doi: 10.1016/S0092-8674(00)80286-X. [DOI] [PubMed] [Google Scholar]
- 7.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 8.Thomas JO. Histone H1: location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 9.Wolffe AP, Kurumizaka H. The nucleosome: a powerful regulator of transcription. Prog Nucleic Acid Res Mol Biol. 1998;61:379–422. doi: 10.1016/S0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 10.Demeret C, Vassetzky Y, Mechali M. Chromatin remodelling and DNA replication: from nucleosomes to loop domains. Oncogene. 2001;20:3086–3093. doi: 10.1038/sj.onc.1204333. [DOI] [PubMed] [Google Scholar]
- 11.Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 12.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin. 2013;6:24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck HC, Nielsen EC, Matthiesen R, et al. Quantitative proteomic analysis of post-translational modifications of human histones. Mol Cell Proteomics. 2006;5:1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Sprung R, Tang Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai L, Peng C, Montellier E, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 17.Fierz B, Muir TW. Chromatin as an expansive canvas for chemical biology. Nat Chem Biol. 2012;8:417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Dai J, Dai L, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young NL, Dimaggio PA, Garcia BA. The significance, development and progress of high-throughput combinatorial histone code analysis. Cell Mol Life Sci. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 26.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azad GK, Tomar RS. Proteolytic clipping of histone tails: the emerging role of histone proteases in regulation of various biological processes. Mol Biol Rep. 2014;41:2717–2730. doi: 10.1007/s11033-014-3181-y. [DOI] [PubMed] [Google Scholar]
- 31.Dhaenens M, Glibert P, Meert P, Vossaert L, Deforce D. Histone proteolysis: a proposal for categorization into ‘clipping’ and ‘degradation’. Bioessays. 2015;37:70–79. doi: 10.1002/bies.201400118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osley MA. Epigenetics: how to lose a tail. Nature. 2008;456:885–886. doi: 10.1038/456885a. [DOI] [PubMed] [Google Scholar]
- 33.Zhou P, Wu E, Alam HB, Li Y. Histone cleavage as a mechanism for epigenetic regulation: current insights and perspectives. Curr Mol Med. 2014;14:1164–1172. doi: 10.2174/1566524014666141015155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips DM, Johns EW. A study of the proteinase content and the chromatography of thymus histones. Biochem J. 1959;72:538–544. doi: 10.1042/bj0720538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eickbush TH, Watson DK, Moudrianakis EN. A chromatin-bound proteolytic activity with unique specificity for histone H2A. Cell. 1976;9:785–792. doi: 10.1016/0092-8674(76)90141-0. [DOI] [PubMed] [Google Scholar]
- 36.Gorovsky MA, Keevert JB. Absence of histone F1 in a mitotically dividing, genetically inactive nucleus. Proc Natl Acad Sci U S A. 1975;72:2672–2676. doi: 10.1073/pnas.72.7.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eickbush TH, Godfrey JE, Elia MC, Moudrianakis EN. H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J Biol Chem. 1988;263:18972–18978. [PubMed] [Google Scholar]
- 38.Watson DK, Moudrianakis EN. Histone-dependent reconstitution and nucleosomal localization of a nonhistone chromosomal protein: the H2A-specific protease. Biochemistry. 1982;21:248–256. doi: 10.1021/bi00531a008. [DOI] [PubMed] [Google Scholar]
- 39.Okawa Y, Takada K, Minami J, Aoki K, Shibayama H, Ohkawa K. Purification of N-terminally truncated histone H2A-monoubiquitin conjugates from leukemic cell nuclei: probable proteolytic products of ubiquitinated H2A. Int J Biochem Cell Biol. 2003;35:1588–1600. doi: 10.1016/S1357-2725(03)00140-7. [DOI] [PubMed] [Google Scholar]
- 40.Pantazis P, Sarin PS, Gallo RC. Detection of the histone-2A related polypeptide in differentiated human myeloid cells (HL-60) and its distribution in human acute leukemia. Int J Cancer. 1981;27:585–592. doi: 10.1002/ijc.2910270504. [DOI] [PubMed] [Google Scholar]
- 41.Simpkins H, Mahon K. The histone content of chromatin preparations from leukaemic cells. Br J Haematol. 1977;37:467–473. doi: 10.1111/j.1365-2141.1977.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 42.Glibert P, Vossaert L, Van Steendam K, et al. Quantitative proteomics to characterize specific histone H2A proteolysis in chronic lymphocytic leukemia and the myeloid THP-1 cell line. Int J Mol Sci. 2014;15:9407–9421. doi: 10.3390/ijms15069407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minami J, Takada K, Aoki K, et al. Purification and characterization of C-terminal truncated forms of histone H2A in monocytic THP-1 cells. Int J Biochem Cell Biol. 2007;39:171–180. doi: 10.1016/j.biocel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Dhaenens M, Glibert P, Lambrecht S, et al. Neutrophil Elastase in the capacity of the “H2A-specific protease”. Int J Biochem Cell Biol. 2014;51:39–44. doi: 10.1016/j.biocel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogler C, Huber C, Waldmann T, et al. Histone H2A C-terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet. 2010;6:e1001234. doi: 10.1371/journal.pgen.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panda P, Chaturvedi MM, Panda AK, Suar M, Purohit JS. Purification and characterization of a novel histone H2A specific protease (H2Asp) from chicken liver nuclear extract. Gene. 2013;512:47–54. doi: 10.1016/j.gene.2012.09.098. [DOI] [PubMed] [Google Scholar]
- 48.Melo FR, Vita F, Berent-Maoz B, Levi-Schaffer F, Zabucchi G, Pejler G. Proteolytic histone modification by mast cell tryptase, a serglycin proteoglycan-dependent secretory granule protease. J Biol Chem. 2014;289:7682–7690. doi: 10.1074/jbc.M113.546895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melo FR, Wallerman O, Paivandy A, et al. Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells. J Allergy Clin Immunol. 2017;140:474–485. doi: 10.1016/j.jaci.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 50.Duarte LF, Young AR, Wang Z, et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun. 2014;5:5210. doi: 10.1038/ncomms6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duncan EM, Muratore-Schroeder TL, Cook RG, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grigera PR, Tisminetzky SG. Histone H3 modification in BHK cells infected with foot-and-mouth disease virus. Virology. 1984;136:10–19. doi: 10.1016/0042-6822(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 53.Kim K, Punj V, Kim JM, et al. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016;30:208–219. doi: 10.1101/gad.268714.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahendra G, Kanungo MS. Age-related and steroid induced changes in the histones of the quail liver. Arch Gerontol Geriatr. 2000;30:109–114. doi: 10.1016/S0167-4943(00)00042-X. [DOI] [PubMed] [Google Scholar]
- 55.Santos-Rosa H, Kirmizis A, Nelson C, et al. Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen J, Xiang X, Chen L, et al. JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep. 2017;18:2131–2143. doi: 10.15252/embr.201743892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandal P, Azad GK, Tomar RS. Identification of a novel histone H3 specific protease activity in nuclei of chicken liver. Biochem Biophys Res Commun. 2012;421:261–267. doi: 10.1016/j.bbrc.2012.03.149. [DOI] [PubMed] [Google Scholar]
- 58.Mandal P, Verma N, Chauhan S, Tomar RS. Unexpected histone H3 tail-clipping activity of glutamate dehydrogenase. J Biol Chem. 2013;288:18743–18757. doi: 10.1074/jbc.M113.462531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vossaert L, Meert P, Scheerlinck E, et al. Identification of histone H3 clipping activity in human embryonic stem cells. Stem Cell Res. 2014;13:123–134. doi: 10.1016/j.scr.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 61.Falk MM, Grigera PR, Bergmann IE, Zibert A, Multhaup G, Beck E. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol. 1990;64:748–756. doi: 10.1128/jvi.64.2.748-756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tesar M, Marquardt O. Foot-and-mouth disease virus protease 3C inhibits cellular transcription and mediates cleavage of histone H3. Virology. 1990;174:364–374. doi: 10.1016/0042-6822(90)90090-E. [DOI] [PubMed] [Google Scholar]
- 63.Sudhan DR, Siemann DW. Cathepsin L targeting in cancer treatment. Pharmacol Ther. 2015;155:105–116. doi: 10.1016/j.pharmthera.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turk V, Stoka V, Vasiljeva O, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 20121824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goulet B, Baruch A, Moon NS, et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. doi: 10.1016/S1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 66.Goulet B, Sansregret L, Leduy L, et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. 2007;5:899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 67.Goulet B, Truscott M, Nepveu A. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol Chem. 2006;387:1285–1293. doi: 10.1515/BC.2006.159. [DOI] [PubMed] [Google Scholar]
- 68.Grotsky DA, Gonzalez-Suarez I, Novell A, et al. BRCA1 loss activates cathepsin L-mediated degradation of 53BP1 in breast cancer cells. J Cell Biol. 2013;200:187–202. doi: 10.1083/jcb.201204053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xue Y, Vashisht AA, Tan Y, Su T, Wohlschlegel JA. PRB1 is required for clipping of the histone H3 N terminal tail in Saccharomyces cerevisiae. PLoS One. 2014;9:e90496. doi: 10.1371/journal.pone.0090496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mishra RN, Kanungo MS. Alterations in histones of the liver and oviduct of Japanese quail during aging. Mol Biol Rep. 1994;20:15–18. doi: 10.1007/BF00999850. [DOI] [PubMed] [Google Scholar]
- 71.Khalkhali-Ellis Z, Goossens W, Margaryan NV, Hendrix MJ. Cleavage of Histone 3 by Cathepsin D in the involuting mammary gland. PLoS One. 2014;9:e103230. doi: 10.1371/journal.pone.0103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fanjul-Fernandez M, Folgueras AR, Cabrera S, Lopez-Otin C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim Biophys Acta. 20101803:3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 74.Xie Y, Mustafa A, Yerzhan A, et al. Nuclear matrix metalloproteinases: functions resemble the evolution from the intracellular to the extracellular compartment. Cell Death Discov. 2017;3:17036. doi: 10.1038/cddiscovery.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allis CD, Allen RL, Wiggins JC, Chicoine LG, Richman R. Proteolytic processing of h1-like histones in chromatin: a physiologically and developmentally regulated event in Tetrahymena micronuclei. J Cell Biol. 1984;99:1669–1677. doi: 10.1083/jcb.99.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee PY, Park BC, Chi SW, et al. Histone H4 is cleaved by granzyme A during staurosporine-induced cell death in B-lymphoid Raji cells. BMB Rep. 2016;49:560–565. doi: 10.5483/BMBRep.2016.49.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin R, Cook RG, Allis CD. Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymena macronuclear development. Genes Dev. 1991;5:1601–1610. doi: 10.1101/gad.5.9.1601. [DOI] [PubMed] [Google Scholar]
- 78.Nurse NP, Jimenez-Useche I, Smith IT, Yuan C. Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys J. 2013;104:1081–1088. doi: 10.1016/j.bpj.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polach KJ, Lowary PT, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- 80.Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 20131819:332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]