Graphical abstract
Keywords: Epithelial mesenchymal transition, Mass spectrometry, Proteomics, Biomarkers
Highlights
-
•
Epithelial mesenchymal transition (EMT) is essential for driving cancer plasticity.
-
•
The EMT-associated program involves the activation of several cellular networks.
-
•
Different mass spectrometry approaches were used to dissect the EMT complexity.
-
•
Potential markers of diagnosis and therapy were identified by MS-technologies.
Abstract
The growing understanding of the molecular mechanisms underlying epithelial-to-mesenchymal transition (EMT) may represent a potential source of clinical markers. Despite EMT drivers have not yet emerged as candidate markers in the clinical setting, their association with established clinical markers may improve their specificity and sensitivity. Mass spectrometry-based platforms allow analyzing multiple samples for the expression of EMT candidate markers, and may help to diagnose diseases or monitor treatment efficiently. This review highlights proteomic approaches applied to elucidate the differences between epithelial and mesenchymal tumors and describes how these can be used for target discovery and validation.
1. The EMT process
A major concern in the management of tumor patients is the presence of metastatic cells in distant organs. Activation of the epithelial-to-mesenchymal transition (EMT) program is commonly observed in human cancers and is closely related to tumor progression and resistance to standard chemotherapeutic drugs and targeted agents. This model proposes morphological changes in the structural organization of epithelial cells, with epithelial polarized cells that lose their apical-basal organization and acquire a fibroblast-like shape, as well as fundamental changes at different genetic and epigenetic levels that can initiate and sustain the process [1].
EMT is a dynamic process that does not occur homogenously across the whole tumor. This is supported by the observation that inside the tumor mass, cells with mesenchymal features reside predominantly at the invasive front at the tumor-stroma interface [2]. Within a tumor, epithelial and mesenchymal cells coexist and cooperate, by direct cell-cell interactions or by diffusible factors, allowing epithelial cells to undergo EMT and increase their ability to locally invade into the surrounding tissues and intravasate into the blood vessels, initiating their spreading to distant organs [3].
After disseminating through the circulation, circulating tumor cells (CTCs) maintain the expression of mesenchymal markers and a molecular profiling of these cells may be carried out to predict disease progression [4], [5]. Although antibodies against epithelial cell adhesion molecule (EpCAM) and Cytokeratins (CK-8, CK-18, CK-19) have been widely used to isolate CTCs, more recent and sensitive methods that include a cocktail of antibodies against mesenchymal markers have been developed [6], [8]. This approach demonstrated clinical validity by its ability to predict more accurately breast cancer patients with a worse prognosis than the evaluation of epithelial antigens [5]. This was also confirmed in other studies where a higher numbers of mesenchymal CTCs were identified in patients in the metastatic stage of the disease in a variety of tumor types [6]. For this reason, multiple devices were developed and functionalized not only with epithelial markers but also with mesenchymal markers combined with cancer-associated antigens, such as HER2 (EGFR2/ERBB2), and epithelial growth factor receptor (EGFR) [4]. Moreover, together with the detection of commonly used EMT molecules, there is also a need to identify new specific CTCs markers. To address this issue, protein-based technologies that rely on specific markers that are not down-regulated during EMT and absent in blood cells were also described and validated [9].
At present, one of the key questions is whether epithelial or mesenchymal detection methods are able to capture the entire spectrum of CTCs. It should be noted that the detection of CTCs based on the expression of a specific set of markers should require an assumption about the nature of these cells, and considering the heterogeneity of CTCs, this cannot be easily predicted. As the EMT features of CTCs can evolve in a spectrum of different phenotypic phases during disease progression or treatment, current efforts to characterise simultaneously CTCs with epithelial and mesenchymal features might be the key not only to improve CTCs capture efficiency but also to increase prognostic and predictive information [7].
At the metastatic site, a reversion to an epithelial morphology, also known as mesenchymal-to-epithelial transition (MET), has been postulated and proposed to be essential for disseminated tumor cells to re-enter a proliferative state and give rise to macrometastatic nodules [10]. The mechanisms underlying this process are still poorly understood. However, an explanation of this process was proposed by Brabletz and involves the differentiated vs undifferentiated status of primary tumors. The dynamic EMT-MET interconversion is an important determinant of metastases characterised by differentiated cells [11]. In this model, cancer cells in primary tumors acquire EMT to increase their metastatic potential, maintain the EMT status in circulation, and undergo a MET at the secondary metastatic sites to promote metastatic colonization. In contrast to the EMT-MET model, undifferentiated metastases derived from primary tumors with intrinsic genetic alterations are in a permanent EMT state and only a weak redifferentiation is necessary to support their growth [11].
When considering the contribution of EMT in cancer progression, it is important to address its role during the first steps of cancer initiation. Although EMT may be principally ascribed to later stages of carcinoma progression, evidences that EMT may occur earlier and be detectable in precancerous lesions are reported. In a mouse model of pancreatic cancer, EMT was identified in premalignant lesions and cells with mesenchymal features were present in the circulation of mice prior to the development of a detectable malignancy [12].
The EMT process has been characterized in different cancer types including breast [13], lung [14], ovarian [15], prostate [16], and liver [17] cancers. The mechanisms by which an epithelial cell is able to acquire a mesenchymal phenotype include the diminished expression of cell–cell adhesion components, elevated expression of proteins involved in cytoskeleton remodeling, and increased motility (Fig. 1). Such mechanisms have important implication for the identification of a set of EMT markers that can be clinically useful to characterize the process. Depending of their specific role, EMT driving factors can be involved in transcriptional regulation, maintenance of epithelial integrity and metabolism.
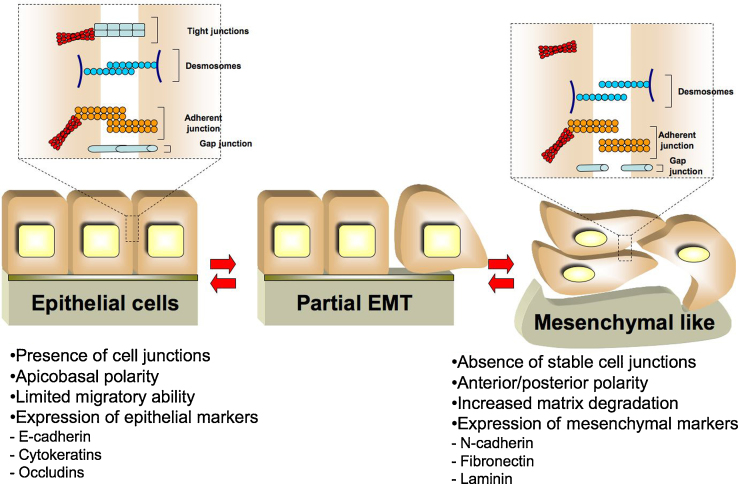
Fig. 1.
Cellular modifications associated with EMT program. After the activation of the EMT program epithelial cells switch off the expression of epithelial markers, such as E-cadherin and Cytokeratins, and turn on mesenchymal markers, including N-cadherin and Fibronectin. After metastatic dissemination mesenchymal cells can redifferentiate into epithelial structures by mesenchymal-epithelial transition. Depending on the tissue and signaling context, epithelial cells may lose only some characteristics or may show some epithelial and mesenchymal properties; this can be considered as a partial EMT.
2. Factors driving EMT activation
EMT results from the coordinated regulation of transcriptional, post-transcriptional, translational and post-translational events. At the transcriptional level, EMT is regulated by several transcription factors including zinc-finger proteins (SNAI1/2 and ZEB1/2) and basic helix-loop-helix proteins (TWIST1/2). Overexpression of these factors has been shown to be sufficient to act as a trigger of the EMT program, increasing cell migration and invasiveness of tumor cells [18].
In addition to the contribution of this transcriptional program, small-non coding RNAs or microRNAs (miRNA or miR) have been found to play a critical role in EMT regulation [19]. MiRNAs can either promote or repressing the EMT program, depending on the different cell contexts. MiR-200 family acts by targeting the EMT factors Zeb1 and Zeb2, thus preventing E-cadherin down-regulation [20], [21]. However, members of this family are also required to promote EMT and metastatization [22]. MiRNAs can cooperate with different EMT-controlling signaling networks. In ovarian cancer, miR-181a can modulate tumor growth factor-β (TGF-β) signaling, increasing cell survival, drug resistance, and tumor dissemination [23]. In breast cancer, ectopic overexpression of miR-374a promoted EMT and metastasis both in vitro and in vivo by targeting several negative regulators of the Wnt/β-catenin signaling cascade [24]. Taken together, miRNAs may regulate EMT working in close collaboration with transcriptional factors or reinforcing EMT signaling network.
Tumor microenvironment including immune cells, tumor stroma, and extracellular matrix can participate in the regulation of EMT through a direct cell contact or by secreting signaling factors such as TGF-β, epidermal growth factor (EGF) or platelet-derived growth factor (PDGF), and hormones [25]. Cancer-associated fibroblasts (CAFs) can promote EMT, by releasing CXCL12 or by activating specific collagen receptors that regulate the stability of EMT transcription factors, respectively [26], [27]. The crucial role of the microenvironment during EMT is also supported by the observation that circulating tumor cells can undergo EMT in response to a physical interaction with platelets. Platelet-derived TGF-β triggers EMT through the synergistic activation of TGF-β and nuclear factor-κB (NF-κB) pathways [28].
TGF-β pathway promotes EMT and also enhances the migratory and invasive properties of cancer cells. The latter events are transiently active only in a small population of cells throughout the tumor where activation of TGF-β signaling drives the expression of genes that promote single cell motility [29]. Proteomic studies provided a systemic view of TGF-β action. The process is regulated via the activation of ECM-receptor interaction, focal adhesion, and actin cytoskeleton proteins and the down-regulation of proteins related to cell cycle inhibition, nucleic acid metabolism, transcription, and regulation of DNA replication and repair. Important effectors of these modifications include the transcriptional regulators SMAD2, SMAD3, SNAIL2, SMAD7, and c-MYC [30].
EMT is regulated by the interplay of different signaling networks, with multiple points of regulation, feedback and cross-talk. Canonical EMT pathways include Ras/MAPK, PI3 K/Akt/GSK, and Wnt/β-catenin. ERK is critical for EMT induction by the hepatocyte growth factor (HGF) [31], TGF-β [32], or EGF [33]. In melanoma cell lines, increased Twist1 mRNA/protein expression was shown to be dependent on the ERK signaling thus promoting invasion, and matrix metalloproteinase-1 expression [34]. Activation of the PI3K and MAPK pathways can also regulate EMT by the suppression of GSK-3β activity and stabilization of Snail. GSK, by phosphorylating Snail at two consensus motifs, regulates Snail degradation and subcellular localization [35].
The Akt family of kinases, comprising Akt-1, Akt-2 and Akt-3, is a major effector of tumor growth and metastasis. Although the signaling through Akt is essential to promote EMT, the functional role and relative contribution of the three different isoforms is still controversial [36]. In breast cancer samples, the ratio of Akt-1 to Akt-2 and the abundance of miR-200 and of the mRNA encoding E-cadherin revealed that the miR-200-E-cadherin axis is under the control of Akt pathway. The comparison of cells reconstituted with Akt-1, or Akt-2 demonstrated that miR-200 family expression appears to depend on the balance between Akt-1 and Akt-2. Akt-1 may regulate Akt2 or it may interfere with the Akt-2-mediated decrease in miR-200 abundance downstream of Akt-2. Akt-1 knockdown, but not down-regulation of Akt-2 or of Akt-1 plus Akt-2, contributes to the activation of EMT induced by TGF-β by decreasing the abundance of the miR-200 family [36]. Activation of Akt drives the acquisition of a mesenchymal phenotype in squamous cell carcinoma lines promoting tissue invasion and metastasis [37]. Hepatocellular carcinoma cells exposure to hypoxic conditions increased HIF-1α expression and induced EMT through a pathway that involves PI3K/Akt [38]. In vivo, high levels of Akt were found to correlate with high levels of Twist1 Ser42 phosphorylation with subsequent decrease of p53 levels and the inhibition of cell cycle arrest and apoptosis [39]. Akt can also regulate EMT through NF-κB. Akt induces NF-κB activity, its nuclear translocation and Snail activation with the consequent repression of the CDH1 gene encoding E-cadherin [40].
Connections between Wnt signaling and EMT is supported by several studies. In the canonical WNT signaling, the binding of WNT ligands to the Frizzled family of protein receptors results in the inhibition of GSK3β activity leading to stabilization of Snail and Slug expression [41], [42]. During EMT, Wnt signaling is implicated in the reprogramming and maintenance of a cancer stem cell (CSC) state. This is obtained through the association of Twist1 with Wnt signaling-related molecules. In particular, EMT activation in tumor cells induces a switch from the β-catenin/E-cadherin/Sox15 complex to the β-catenin/Twist1/TCF4 complex enhancing the transcriptional activity of this group of proteins including the binding to cancer stem cell genes related promoters. Immunohistochemical analysis of lung cancer samples correlated the specific protein signature identified in vitro (nuclear β-cateninHigh/nuclear Twist1High/E-cadherinLow/Sox15Low/CD133High) with clinical features including tumor progression and metastasis. These data indicated the potential role of these proteins as predictors for poor overall patient survival [43].
3. Immune response and EMT
A link between EMT and immunoediting was described and several mechanisms proposed. Tumor cells that evade immunoediting often activate EMT signaling pathways such as BRAF-MAPK, STAT3, and Wnt/β-catenin which trigger multiple immunosuppressive cascades that result in the production of immunosuppressive molecules (e.g., TGF-β, IL-10, IL-6, VEGF, and CCL2) and induction of immunosuppressive immune cells (e.g., regulatory T cells, tolerogenic dendritic cells, and myeloid-derived suppressor cells) [44], [45], [46]. Moreover, during EMT, tumor cells evade the immuno-recognition through the down-regulation of immune receptor ligands that directly stimulate immune cells. In colorectal cancer, the plasma membrane receptor natural killer group 2 member D (NKG2D) expressed in natural killer (NK) and T cells which is involve to recognize malignant cells is under the control of an immunological checkpoint that relies on NKG2D-mediated immune responses in EMT [47]. We demonstrated the down-regulation of Human leukocyte antigen class I antigens (HLA-I) in serous ovarian cancer, which is essential in immune response by presenting antigenic peptides to cytotoxic T lymphocytes. This is done by cleavage of the immunoproteasome 11S [48]. Cleavage of PSME1 (proteasome activator complex subunit 1, 11S regulator complex [syn: PA28 alpha]), an antigen processing machinery, into the Reg-alpha fragment could lead to default self-antigen presentation [48], [49]. Our data describe a high expression level of PA28 in early and late stages ovarian carcinomas, and proposed a role for this protein as a marker for monitoring patient response during therapy [49]. Its alteration by cleavage in ovarian carcinomas may be a mechanism to evade immune recognition. Similar hypothesis has already been proposed in the case of APM chaperones such as TAP, LMP2, LMP10, and tapasin in colon carcinoma, small cell lung carcinoma, and pancreatic carcinoma cell lines. In fact, IFN-© treatment of these carcinoma cell lines corrects the TAP, LMP, and tapasin deficiencies and enhances PA28〈 LMP7, calnexin and calreticulin expression, which is accompanied by increased levels of MHC class 1 antigens. Recently, the PA28 fragment was detected in a MALDI MSI multicentric study and correlated to stroma activation in breast cancer samples [50].
There are data to support the role of soluble molecules expressed or secreted in the tumor microenvironment in the regulation of EMT and immunoediting. Recent data suggest that TGF-β and EGF play a role in regulating HLA-I. The treatment of prostate cancer cells with TGF-β and EGF induced and EMT program and significantly decreased HLA-I expression. This attenuated the cytotoxic T cell mediated lysis of tumor cells through a pathway that involve the up-regulation of Snail [51]. Taken together, it is clear that in EMT hallmarks the immune response take a major place as a therapeutic target.
4. Clinical relevance of genes/proteins associated with EMT
In support to the relevance of EMT, histological analysis of tumor samples suggested that EMT occurs in vivo promoting cancer progression and chemoresistance.
Loss of cell polarity is a main event in several epithelial tumor types, as loss of the junctions mediating tissue architecture could contribute to the formation of tumor masses in vivo [52], or promote the expansion of pre-neoplastic cell [53]. One fundamental event of EMT is the decrease of E-cadherin expression, an adherent junction protein implicated in cell–cell adhesion of epithelial tissues. There are multiple mechanisms by which E-cadherin influences tumor progression, including inhibition of EGF receptor signaling and maintenance of cell–cell adhesion and epithelial cell polarity [54]. Reduced E-cadherin expression is often associated with the release of β-catenin into the cytosol and potentially into the nucleus [54]. Immunohistochemical analysis of breast cancer samples revealed that this association is a characteristic of tumors of triple-negative and basal-like phenotype [55].
Several studies have correlated the down-regulation of E-cadherin with clinicopathological parameters of several human tumor types. In a cohort of squamous cell lung carcinoma patients, high E-cadherin expression can be a positive indicator for overall survival (OS) and disease-free survival (DFS) [56]. Increased cytoplasmic ALCAM/E-cadherin loss was found to represent the most significant adverse prognostic factors for oral squamous cell carcinoma (OSCC) patients [57].
A meta-analysis of 24 studies and 2961 cases suggested that E-cadherin expression is significantly associated with poorer differentiation degree in esophageal cancer [58]. Moreover, abnormal E-cadherin expression emerged as a strong independent prognostic factor for overall survival of gastric cancer patients [59].
Stable loss of E-cadherin can occur through different molecular mechanisms, including promoter hypermethylation and transcriptional control. The transcriptional repressors of E-cadherin expression, Snail, Slug and Twist, are involved in the activation of EMT by binding to E-box elements in the E-cadherin promoter. These factors can regulate EMT in vitro through the repression of E-cadherin, the up-regulation of mesenchymal markers, and the acquisition of stem cell properties [18]. Knockdown of these transcription factors restores the expression of E-cadherin and induces an epithelial phenotype [60]. Although these EMT regulators are thought to function in a redundant manner, several studies reported unique functions suggesting a differential participation in the EMT process [61] with a precise spatial and temporal regulation [61], [64]. In breast cancer patients, Snail levels in the primary tumor predicted for metastasis, while Twist levels and the Twist/Snail ratio in bone marrow micrometastatic tumor cells were found to be highly predictive for distant relapses [62]. In a cohort of patients with invasive ductal carcinoma, Snail expression correlated with β-catenin cytoplasmic and nuclear levels, while Slug correlated with N-cadherin and vimentin protein expression [63]. In melanoma cells, the expression of EMT factors is under the control of oncogenic signaling pathways. During melanomagenesis Snail2 and Zeb2 behave as onco-suppressive proteins in the melanocytic differentiation program, whereas, in response to MEK-ERK pathway activation, Zeb1 and Twist1 are up-regulated promoting dedifferentiation and neoplastic transformation of melanocytes [64].
Snail, Slug and Twist are expressed in the nuclear and cytoplasmic fraction of tumor samples, and correlated with advanced stage, and poor survival in several cancer types [65], [66]. Moreover, there is some evidence of a positive staining of these transcription factors in tumor microenvironment of tumor samples with a worse outcome [67].
High expression of nuclear Snail predicts poor survival in nasopharyngeal carcinoma and basal-like breast cancer [65], [66]. Twist over-expression was observed in patients with head and neck squamous cell carcinoma, clear cell carcinoma of the ovary, advanced oral squamous cell carcinoma, colorectal carcinoma, breast carcinoma and associated with aggressive tumor properties and poor survival [68], [69], [70], [71]. The prognosis of patients with Twist and Snail co-expression is worse in tumor samples compared with samples where only one marker is expressed, supporting the hypothesis that these two transcription factors may cooperate to promote EMT [72].
N-cadherin up-regulation frequently follows E-cadherin down-regulation with a concomitant increase of cell motility and migration [73]. N-cadherin over-expression stimulates mammary tumor metastasis in the MMTV-NeuNT mouse model, and induces up-regulation of FGFR expression and phosphorylation, and of Snail and Slug expression in a FGFR-dependent manner [74]. N-cadherin expression is positively correlated with clinical parameters in several cancer types. In lung cancer tissues, overexpression of N-cadherin is associated with advanced TNM stage, poor differentiation and reduced overall survival. Moreover, a significant correlation was found between Twist and N-cadherin expression by Spearman correlation analysis [75]. Moreover, N-cadherin expression was found to correlate with superficial urothelial tumor progression and with poor histological differentiation of oral squamous cell carcinomas [76], [77].
Vimentin is the major intermediate filament protein of mesenchymal/stromal cells, including fibroblasts, endothelial cells and peripheral blood mononuclear cells [78]. High-throughput approaches reported altered expression levels for vimentin in numerous cancer types, correlating its overexpression with the aggressive behavior and metastatic potential of these tumors [79], [80], [81], [82], [83], [84], [85], [86]. In epithelial tumors, expression of vimentin alone or in combination with other mesenchymal markers is a useful predictor of cancer progression, chemoresistance, high histological grade, and in general associated with poor prognosis. Vimentin immunoreactivity has been observed in both stromal and tumor cells where it is expressed at the invasive front [85].
High expression of vimentin was observed in triple-negative breast cancer compared with other breast tumour subtypes, and correlated with younger age, high nuclear grade, high Ki67 expression, and poor prognosis in terms of both recurrence-free survival and overall survival [87], [88]. Vimentin is a predictive biomarker in patients with non-small cell lung cancer treated with Erlotinib as second- or third-line therapy [89], and a negative indicator for overall survival in squamous cell lung carcinoma patients [56].
4.1. Clinical significance of combined detection of FDA serum tumor markers and EMT markers
The potential uses of cancer markers are for screening, diagnosis, prognosis, prediction of response to therapy, monitoring of patients with diagnosed disease. Since EMT markers have demonstrated clinical value in detection and monitoring of a disease, their expression may improve the sensibility and specificity of most traditionally used clinical markers. Moreover, despite their prevalent tissue-based location, EMT markers can also be found in other biological fluids, including serum, increasing their potential clinical utility. So far, only few EMT markers have been proposed in correlation with US Food and Drug Administration (FDA)-approved biomarkers.
Alpha-fetoprotein (AFP) is an indicator of several malignant and chronic conditions, including hepatocellular carcinoma (HCC) [90]. The expression of AFP in HCC models is regulated by the miR122 [91], which is associated with EMT and spontaneous HCC formation [92]. Reduced expression of miR122 in HCC cells contributes to elevated AFP expression through a pathway involving CUX1/miR214/ZBTB20; this evidence suggests that serum AFP levels in HCC patients may be a surrogate marker for deregulated intracellular miR122-dependent signaling pathways in HCC tissues.
Vimentin is an EMT markers consistently over-expressed in HCCs compared to cirrhotic and normal liver tissues [93]. This over-abundance was more marked in smaller carcinomas (≤2 cm). In the serum of HCC patients, vimentin expression as detected by ELISA described a significant difference between non-neoplastic controls and HCCs of all sizes and a specificity of 87.5% (95% CI: 76.85–94.45%) in separating non-neoplastic subjects from those with HCCs of small size. Importantly, vimentin was proposed as a better marker compared with the conventional serum AFP for detecting small tumors and that when combined with AFP, this detection sensitivity and specificity can be considerably enhanced.
Mucins are a large family of transmembrane glycoproteins, and are frequently over-expressed and aberrantly glycosylated in cancer. Mucins are secreted from and/or localized to the apical borders of normal epithelial cell sheets. Over-expression of MUC1, MUC4, and MUC16 has diagnostic and prognostic significance in different cancer types [94].
MUC16 is a high molecular weight membrane associated-mucin, which is over-expressed in advanced serous epithelial ovarian cancers [95]. CA125 is a marker that allows the detection of circulating MUC16 antigen, which is increased in approximately 80% of all epithelial ovarian cancers (EOC) and in only 50% of stage I EOC and associated with a significantly longer progression-free survival and overall survival [96]. CA125 serum levels directly correlate with the levels of protein production in tumor cells suggesting that the up-regulation of this protein at the tissue level may have a role in promoting cancer progression and recurrence. Knockdown of MUC16 decreased the ability of tumor cells to form colony in soft agar and prevented the formation of subcutaneous tumors in nude mice. In contrast, tumor cells with an ectopic expression of MUC16 showed a decrease of E-cadherin and a gain of N-cadherin and vimentin expression together with an enhanced tumorigenic phenotype [97]. Although this data suggests a functional role of MUC16 in regulating EMT, results from another study report an inverse correlation between CA125 serum levels and the expression of mesenchymal markers at tissue level. The study of Tothill et al. described that preoperative serum CA125 levels are lower in ovarian cancer tissues with a mesenchymal gene signature compared with tissues characterized by a different genomic profile [98]. These data suggest that MUC16 expression may be needed to promote the onset of EMT but not to maintain a mesenchymal phenotype.
MUC1 is approved as a biomarker for breast cancer in combination with diagnostic imaging, patient history, and physical examination during active cancer therapy to monitor metastatis [99]. Two serum assays, CA15.3 and CA27.29, were approved by the FDA for the detection of secreted MUC1 in breast cancer patients. Moreover, MUC1 is aberrantly over-expressed in greater than 95% of metastatic pancreatic cancer and associated with poor prognosis [100]. Over-expression of MUC1 can induce EMT by β-catenin-MUC1 interaction and translocation to the nucleus, leading to activation of Snail and Slug [101]. Moreover, MUC1 stimulates the over-expression of Zeb1 and the down-regulation of miR-200c, with consequent induction of EMT [102].
In 1965, the oncofetal antigen carcinoembryonic antigen (CEA) was found for the first time in extracts of colonic adenocarcinomas [103]. CEA belongs to a large family of carcinoembryonic antigen cell adhesion molecules (CEACAMs) proteins that mediate homophilic and heterophilic cellular interactions [104]. The utilization of CEA in clinical applications is mainly aimed at the management of gastrointestinal malignancies, especially colorectal carcinomas. It is also detected in various epithelial tumors such as lung, small cell lung cancer, pancreas, gallbladder, urinary bladder, mucinous ovarian, and endometrium [105]. In a cohort of 99 pancreatic tumor samples, CEACAM6 (CD66c) expression was correlated with clinicopathological characteristics including tumor differentiation and lymph node status, and inversely correlated with E-cadherin expression. Silencing of CEACAM6 in pancreatic CFPAC-1 cells led to a transition from mesenchymal to epithelial morphology with a concomitant up-regulation of E-cadherin and down-regulation of vimentin [106]. In breast cancer samples, the combined analysis of CEA and E-cadherin expression showed a 3.6 times higher risk of relapse for patients with elevated expression of CEA, regardless of E-cadherin expression, compared with patients with below-median CEA and above-median E-cadherin tumour expression [107].
5. EMT markers in biological fluids
Despite extensive investigations, there are no currently approved applications for canonical EMT markers in the clinical setting. This is in contrast with the importance of EMT in the context of cancer biology and with their correlation with a series of clinical variables as supported by recent experimental studies.
There are multiple reasons why EMT markers have not gone through the clinic and they are to some extent related to the dynamic nature of the process. As described above, primary tumors are characterized by the expression of EMT markers whose detection is complicated by a specific spatio-temporal regulation. Hence, only a particular marker may be expressed at any time point with the unavoidable consequence that tissue slices should be probed with several different antibodies, which may lead to an increase of healthcare cost.
The detection of tumor markers in the blood is a highly active area of research that will extend the use of EMT markers in the clinic providing a shift from the classical histological diagnosis based on the anatomical site. A proteomic signature of breast cancer biomarkers released in proximal fluids was assessed by liquid chromatography mass spectrometry (LC–MS/MS). Authors analysed the proximal fluids of seven breast cancer cell lines with different EMT features, and identified a group of proteins whose expression varied significantly among cell models [108]. A set of three markers, cathepsin D, fructose 1,6-bisphosphate aldolase, and keratin 19, was required to categorize breast models as HER2 positive, HER2 negative and hormone receptor positive, and triple negative. Other authors investigated by 2DE-DIGE and LC–MS/MS the secretome of MDCK and Ras transformed MDCK cells [109]. Ras transformed cells exhibit typical EMT features including an increased migratory capacity and an up-regulation of extracellular proteases and factors promoting migration. Among the proteins identified, kallikrein 6 (KLK6) was six folds up-regulated in the secretome of Ras transformed MDCK cells. KLK6 may represent a promising biomarker, because this protein belongs to a family of serine proteases, which includes also the clinical biomarker KLK3 (known as prostate-specific antigen, PSA). However, its role in the EMT scenario is not totally clear, because a recent study unrevealed an inhibitory function of KLK6 on tumor cell proliferation and mobility and an inverse correlation to EMT [110].
6. Using proteomics to dissect EMT complexity
A differential proteomic analysis was performed to investigate the proteome modifications after the induction or knockdown of EMT markers, or to compare cell and tissue models with different EMT features. Other approaches have been used to expand the information about EMT markers including immunoprecipitation of protein complexes followed by MS/MS.
Advances in MS-based proteomics provided a systems view of functional networks modified after EMT induction [111]. To address this issue and to enable the study of global proteome modifications, different fractionation methods were used. This approach includes the analysis of membrane, cytosol, and nuclear proteins, and the isolation of phosphopeptides that were enriched by direct affinity chromatography. This strategy was evaluated in an inducible model of EMT that was obtained using tumor cell lines in which the expression of Snail and Zeb1 was under the control of a doxycycline inducible promoter. Cells were treated with TGF-β for one week, and lysed to fractionate cellular specific components that were analysed by LC–MS/MS. High-throughput proteomics revealed modifications in a wide range of cellular networks and alterations in the expression of cell–cell junctional proteins, an increased production of proteases, an up-regulation of proinflammatory cytokines, and a reduced need of metabolic pathways required for macromolecular biosynthesis. These modifications correlate with the modulation of specific transcription nodes including NFκB2/RelA, ATF3/ATF2, Myc, FOXA1 and 2, ETS1, NFE2L2/L3, MEF2C, Snail/Slug, and Zeb1 and 2.
As discussed above, several signaling pathways can induce EMT and many EMT inducers have been identified including HGF. Shotgun proteomics combined with SILAC, for the quantitative comparison of proteomic differences, was applied to identify HGF-induced expression changes in MDCK cells. Six protein clusters modified their expression in a time-dependent manner after different time points of stimulation. In particular, the expression of proteins that regulate cell cycle, cell migration, integrin signaling, ubiquitination, and transcription increased over 24 h of HGF exposure, in contrast, metabolic enzymes, pro-apoptotic, and transcriptional suppressors decreased over the same period. Moreover, members of HIPPO/MST2 and ISG15 pathways were modified after HGF-treatment at the protein level by ubiquitin ligases. Modulation of these pathways seems to have a major role in regulating cell scattering and morphology [112].
Cells undergoing EMT acquire resistance to anticancer agents and contribute in vivo to the generation of chemoresistant metastasis [113], [114]. In pancreatic adenocarcinoma, intrinsic gemcitabine-resistant and -sensitive human cell lines were compared by a label free quantification strategy to identify an EMT-proteomic signature associated with the resistant phenotype [115]. The list of identified proteins includes the EMT markers ANPEP, ALCAM, DSG3, DSG2, KRT14, KRT19, KRT8, CLDN1, VIM, CDH1, SDC1, CD44, ITGB1, NT5E, and DSP, suggesting that the acquisition of a mesenchymal phenotype is involved in gemcitabine resistance. As many of these proteins are closely associated with cytoskeletal reorganization, the acquisition of a more aggressive and invasive phenotype may be important in conferring drug resistance against gemcitabine.
Breast cancer models have been widely used to study EMT. Based on the established molecular classification, breast tumors can be divided into several subtypes with specific epithelial and mesenchymal features. In this classification, triple-negative breast cancers (TNBC), represent a specific group enriched for the expression of mesenchymal markers. Compared to HER2 cancer samples, TNBC display a proteomic profile characterised by the over-expression of breast cancer stem cells markers, such as ALDH1A1, and drug-resistance proteins including Hsp70, Periostin precursor, RhoA, Actinin α4, and Annexin1 [116]. Annexins belong to a family of calcium/phospholipids-binding and actin regulatory proteins, and play an important role in the regulation of EMT. In mesenchymal breast cancer cells, Annexin1 was found differentially expressed at protein and mRNA level [117], and identified as a marker for breast cancer outcome prediction and treatment response [118]. Recently, the deeper proteomic classification performed by Lawrence and colleagues highlighted the remarkable differences between TNBC and luminal tumors. This result supported the metastatic potential of mesenchymal tumors, which express high level of signaling proteins associated with metastasis, including ECM receptor interaction, cell adhesion, and angiogenesis [119].
A hallmark of EMT is the down-regulation of cell–cell junctions, including adherens junctions. In our group we used a proteomic approach to identify proteins modified after the knockdown of E-cadherin in breast cancer cells. Experimental data were obtained from two different models, MCF-7 and MDA-415 cells stably infected with a lentivirus for the expression of specific E-cadherin shRNAs. Bioinformatics analysis of this protein dataset revealed that different protein networks were modified after E-cadherin knockdown, including cell invasion, viability, survival, and metabolism [120]. This expands the role of E-cadherin to a possible regulation of other pathways not strictly related to cell adhesion, and highlights the complexity of the E-cadherin interactome. Immunoprecipation experiments using MS provided a sensitive way of characterising specific molecules that directly are influenced by E-cadherin. One of these methodologies relies on the application of biotin-streptavidin affinity chromatography and LC–MS/MS to identify proteins associated with an engineered form of E-cadherin, which expresses a biotin ligase to biotinylate proteins based on its proximity [121]. Biotinylated proteins were then isolated by streptavidin affinity chromatography, digested into peptides, and analyzed by high-resolution MS. In the immunoprecitated of E-cadherin, 561 proteins were identified and classified in different functional categories based on their gene ontology. The largest group was that of adaptor proteins, the remaining categories included transmembrane proteins and proteins involved in transcription, translation, trafficking, proteolysis and metabolism.
7. Conclusions
Data discussed in the above sections describe the biological complexity of EMT and report the role of high-throughput technologies to provide insight into this complexity (Table 1). Although the molecular details that underlie the process still need to be completely clarified, these results highlight how EMT takes advantage by the activation of different cellular pathways that work in coordination to sustain cells adaptation to a different molecular program. Besides the role of classical EMT markers such as cell–cell adhesions proteins and transcription factors, other potential markers were identified by proteomics. Considering the patient-to-patient biological variation, large multi-center studies are required to prove the validity of these molecules as clinical biomarkers. Moreover, extensive experimental evidence indicates the need to shift the focus from a single or few biomarkers towards large panels of EMT markers that can better describe the molecular dynamism of a tumor. In this direction, the usefulness of EMT markers alone or in combinations still remains to be determined, although initial studies using with multi-marker combinations are promising [122]. To achieve this goal, proteomics technologies can now support the screening of a large set of samples with great sensitivity also in combination with targeted data acquisition strategy. An example of this approach was recently carried out for the discovery and validation of breast cancer biomarkers. In this work, a list of N-glycosylated proteins was obtained from breast cancer samples with different clinical features. Then, a list of putative biomarkers was selected and quantified by LC-SRM (single reaction monitoring) leading to the validation of 10 proteins differentially expressed in tumor samples [123]. More recently, analyses performed on quadrupole-orbitrap mass spectrometers operated in hyper reaction monitoring (HRM) enables comprehensive recording of a large number of peptide ions with high precision and reproducibility bypassing the drawbacks associated with SRM [124]. The approach was used to investigate the effect of acetaminophen (APAP) on the proteome of liver microtissues generating a spectral library of more than 22.454 peptides [125]. This further expands the role of proteomics in the clinical investigation and validation of biomarkers, with new opportunities also in the field of EMT.
Table 1.
Proteomic studies applied to study the EMT complexity.
| Technical platform | Quantification | Comments | Main results | Ref. |
|---|---|---|---|---|
| LC–MS/MS | Label-free quantification by MaxQuant, SILAC analysis of phosphopeptides | Analysis of the proteomic and phosphoproteomic changes of cultured human keratinocytes undergoing EMT in response to stimulation with TGF-β. Authors quantified significant changes in 2079 proteins and 2892 phosphorylation sites regulated by TGF-β | Authors performed a networks and pathways analysis of TGF-β regulated proteins which revealed significant differences in the abundance of proteins associated with EMT and cell proliferation. A set of upstream transcription regulators induced by TGF-β treatment was also identified | [30] |
| LC–MS/MS | Label-free quantification by Scaffold software | Authors identified and characterized the most abundant secreted proteins from a panel of cancer human cell lines: HER2 positive, HER2 negative and hormone receptor positive and triple negative (HER2−, ER−, PR−) | Bioinformatics analysis classified HER2 positive, HER2 negative and triple negative models based on the expression of only two proteins, muscle fructose 1,6-bisphosphate aldolase and keratin 19 | [108] |
| 2-D DIGE and LC–MS/MS | 2-DE image analysis software and mRNA and western blot validation | Authors compared the secretome of MDCK cells that undergo EMT following transformation with oncogenic Ras | DIGE analysis identified 47 proteins differentially regulated in MDCK cells after Ras-induced EMT. Proteins involved in cell migration and matrix degradation were enriched in this network | [109] |
| LC–MS/MS | iTRAQ | EMT was induced in a tumor cell model stably transfected with doxycycline-inducible Zeb1 or Snail cDNAs or after the exposed to exogenous TGF-β⋅ Proteomic changes were investigated after cellular fractionation of membrane, nuclear, and cytosolic proteins. Phosphopeptides were also isolated by directed affinity chromatography | Four functional groups of proteins were modified after EMT activation: cell adhesion and migration, metabolism, transcription nodes and proliferation/survival networks | [111] |
| LC–MS/MS | SILAC | Authors performed a quantitative proteomic analysis of MDCK cells treated with HGF at different time points | After HGF exposure, MDCK cells expressed higher levels of proteins associated with the ubiquitination machinery, whereas expression of proteins regulating apoptotic pathways was suppressed. Hippo/MST2 and ISG15 pathways are key determinants of HGF-induced EMT alterations | [112] |
| LC–MS/MS | Label-free quantification by Scaffold software | Authors performed a comparative proteomic analysis of pancreatic cancer cell lines with a different sensitivity to gemcitabine | Bioinformatics analysis identified 13 EMT-related proteins that were closely associated with drug resistance including CAV1, IQGAP1, ITGB4, ITGA6, CTNNB1, ACTN4, FLNA, FLNB, KRT18, MYH14, MYH9, MYL6, and PXN | [115] |
| LC–MS/MS | Label-free quantification by Scaffold software | Authors performed a comparative proteomic profiling of HER2 positive and triple negative breast cancer tissues | Galectin-3-binding protein and ALDH1A1 were found preferentially elevated in TNBC, whereas CK19, transferrin, transketolase, and thymosin β4 and β10 were elevated in HER2-positive cancers | [116] |
| 2-DE and TOF/TOF | 2-DE image analysis software and mRNA and western blot validation | In this paper authors performed a comparative proteomic analysis of two breast cancer cell lines with epithelial and mesenchymal features | 28 proteins were identified as significantly up- and down-regulated. Proteins that were differentially expressed by these cell lines were enriched for metabolic, mobility, and signaling functions | [117] |
| LC–MS/MS | iBAQ approach | This paper is a large-scale proteomic characterization of triple negative breast cancer cell lines and tissues using MS. Results of this study are freely available at the website (https://zucchini.gs.washington.edu/BreastCancerProteome/) | PCA analysis classified tumor samples into different groups. Luminal-like cells expressed higher levels of pathways associated with proliferation, such as cell cycle, growth factor signaling, metabolism, and DNA damage repair mechanisms. TNBC cell types, expressed higher levels of pathways associated with metastasis, such as ECM-receptor interaction, cell adhesion, and angiogenesis | [119] |
| 2-DE and TOF/TOF | 2-DE image analysis software and western blot validation | Proteomic analysis of breast cancer cell lines after shRNA knockdown of E-cadherin | 81 spots differentially expressed between scramble and shEcad cells, 54 proteins identified by MS/MS. Proteins involved in the regulation of actin cytoskeleton and cellular metabolism were enriched in this dataset | [120] |
| Immunoprecipitation and LC–MS/MS | iBAQ approach | Authors used immunoprecipitation and LC–MS/MS to identify 561 E-cadherin interactome components | More than 50% of the 561 identified proteins belong to six main groups including adaptor proteins, transmembrane proteins, guanosine triphosphatase (GTPase) regulators, kinases and phosphatases, actin dynamics regulators, and cytoskeleton structural and motor proteins | [121] |
Conflict of interest
All authors have contributed to, and read, the paper and have given permission for their name to be included as a co-author. The manuscript, including figures and tables, has not been previously published and is not under consideration elsewhere.
Authors declare that there is no conflict of interest.
Acknowledgements
We are grateful to the managers of ASL_LE, Dr. Giovanni Gorgoni, Dr. Antonio Sanguedolce, and Dr. Vito Gigante for their support to the Lab of Clinical Proteomics. This work was supported by the PON project 254/Ric. “Implementation of human and environment health research center” cod. PONa3_00334, PONa3_00334 “Research Center for Environment and Human health”, PRIN 2010FPTBSH “NANO Molecular tEchnologies for Drug delivery—NANOMED”.
We gratefully acknowledge funding from the Apulia Regional Cluster project SISTEMA.
Contributor Information
Daniele Vergara, Email: danielevergara@libero.it.
Michele Maffia, Email: michele.maffia@unisalento.it.
References
- 1.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(November (5)):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Nassar A., Radhakrishnan A., Cabrero I.A., Cotsonis G.A., Cohen C. Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Appl. Immunohistochem. Mol. Morphol. 2010;18(October (5)):433–441. doi: 10.1097/PAI.0b013e3181dddb20. [DOI] [PubMed] [Google Scholar]
- 3.Celià-Terrassa T., Meca-Cortés O., Mateo F., de Paz A.M., Rubio N., Arnal-Estapé A., Ell B.J., Bermudo R., Díaz A., Guerra-Rebollo M., Lozano J.J., Estarás C., Ulloa C., Álvarez-Simón D., Milà J., Vilella R., Paciucci R., Martínez-Balbás M., de Herreros A.G., Gomis R.R., Kang Y., Blanco J., Fernández P.L., Thomson T.M. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J. Clin. Invest. 2012;122(May (5)):1849–1868. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T., Isakoff S.J., Ciciliano J.C., Wells M.N., Shah A.M., Concannon K.F., Donaldson M.C., Sequist L.V., Brachtel E., Sgroi D., Baselga J., Ramaswamy S., Toner M., Haber D.A., Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(February (6119)):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradilone A., Raimondi C., Nicolazzo C., Petracca A., Gandini O., Vincenzi B., Naso G., Aglianò A.M., Cortesi E., Gazzaniga P. Circulating tumour cells lacking cytokeratin in breast cancer: the importance of being mesenchymal. J. Cell. Mol. Med. 2011;15(May (5)):1066–1070. doi: 10.1111/j.1582-4934.2011.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S., Liu S., Liu Z., Huang J., Pu X., Li J., Yang D., Deng H., Yang N., Xu J. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One. 2015;10(April (4)):e0123976. doi: 10.1371/journal.pone.0123976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit S., van Dalum G., Lenferink A.T., Tibbe A.G., Hiltermann T.J., Groen H.J., van Rijn C.J., Terstappen L.W. The detection of EpCAM(+) and EpCAM(?) circulating tumor cells. Sci. Rep. 2015;5(July):12270. doi: 10.1038/srep12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Punnoose E.A., Atwal S.K., Spoerke J.M., Savage H., Pandita A., Yeh R.F., Pirzkall A., Fine B.M., Amler L.C., Chen D.S., Lackner M.R. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5(September (9)):e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokobori T., Iinuma H., Shimamura T., Imoto S., Sugimachi K., Ishii H., Iwatsuki M., Ota D., Ohkuma M., Iwaya T., Nishida N., Kogo R., Sudo T., Tanaka F., Shibata K., Toh H., Sato T., Barnard G.F., Fukagawa T., Yamamoto S., Nakanishi H., Sasaki S., Miyano S., Watanabe T., Kuwano H., Mimori K., Pantel K., Mori M. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013;73(April (7)):2059–2069. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 10.Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(December (6)):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabletz T. To differentiate or not–routes towards metastasis. Nat. Rev. Cancer. 2012;12(May (6)):425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 12.Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H., Leach S.D., Stanger B.Z. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(January (1–2)):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foroni C., Broggini M., Generali D., Damia G. Epithelial-mesenchymal transition and breast cancer: role, molecular mechanisms and clinical impact. Cancer Treat Rev. 2012;38(October (6)):689–697. doi: 10.1016/j.ctrv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Sato M., Shames D.S., Hasegawa Y. Emerging evidence of epithelial-to-mesenchymal transition in lung carcinogenesis. Respirology. 2012;17(October (7)):1048–1059. doi: 10.1111/j.1440-1843.2012.02173.x. [DOI] [PubMed] [Google Scholar]
- 15.Vergara D., Merlot B., Lucot J.P., Collinet P., Vinatier D., Fournier I., Salzet M. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291(May (1)):59–66. doi: 10.1016/j.canlet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Nauseef J.T., Henry M.D. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat. Rev. Urol. 2011;8(June (8)):428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogunwobi O.O., Liu C. Therapeutic and prognostic importance of epithelial-mesenchymal transition in liver cancers: insights from experimental models. Crit. Rev. Oncol. Hematol. 2012;83(September (3)):319–328. doi: 10.1016/j.critrevonc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Tam W.L., Lu H., Buikhuisen J., Soh B.S., Lim E., Reinhardt F., Wu Z.J., Krall J.A., Bierie B., Guo W., Chen X., Liu X.S., Brown M., Lim B., Weinberg R.A. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24(September (3)):347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullock M.D., Sayan A.E., Packham G.K., Mirnezami A.H. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progressiono. Biol. Cell. 2012;104(January (1)):3–12. doi: 10.1111/boc.201100115. [DOI] [PubMed] [Google Scholar]
- 20.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(April (7)):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson E.L., Kazenwadel J., Bert A.G., Khew-Goodall Y., Ruszkiewicz A., Goodall G.J. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15(February (2)):180–191. doi: 10.1593/neo.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Y., Liu Y.M., Li L.C., Wang L.L., Wu X.L. microRNA-503 inhibits gastric cancer cell growth and epithelial-to-mesenchymal transition. Oncol. Lett. 2014;7(April (4)):1233–1238. doi: 10.3892/ol.2014.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parikh A., Lee C., Joseph P., Marchini S., Baccarini A., Kolev V., Romualdi C., Fruscio R., Shah H., Wang F., Mullokandov G., Fishman D., D’Incalci M., Rahaman J., Kalir T., Redline R.W., Brown B.D., Narla G., Difeo A. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat. Commun. 2014;7(January (5)):2977. doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai J., Guan H., Fang L., Yang Y., Zhu X., Yuan J., Wu J., Li M. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 2013;123(February (2)):566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing Y., Han Z., Zhang S., Liu Y., Wei L. Epithelial-mesenchymal transition in tumor microenvironment. Cell Biosci. 2011;1(August (1)):29. doi: 10.1186/2045-3701-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung Y., Kim J.K., Shiozawa Y., Wang J., Mishra A., Joseph J., Berry J.E., McGee S., Lee E., Sun H., Wang J., Jin T., Zhang H., Dai J., Krebsbach P.H., Keller E.T., Pienta K.J., Taichman R.S. Recruitment of mesenchymal stem cells into prostate tumours promotes metastasis. Nat. Commun. 2013;4:1795. doi: 10.1038/ncomms2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Corsa C.A., Ponik S.M., Prior J.L., Piwnica-Worms D., Eliceiri K.W., Keely P.J., Longmore G.D. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 2013;15(June (6)):677–687. doi: 10.1038/ncb2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labelle M., Begum S., Hynes R.O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(November (5)):576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giampieri S., Manning C., Hooper S., Jones L., Hill C.S., Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 2009;11(November (11)):1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Souza R.C., Knittle A.M., Nagaraj N., van Dinther M., Choudhary C., ten Dijke P., Mann M., Sharma K. Time-resolved dissection of early phosphoproteome and ensuing proteome changes in response to TGF-β. Sci. Signal. 2014;7(July (335)):rs5. doi: 10.1126/scisignal.2004856. [DOI] [PubMed] [Google Scholar]
- 31.Tanahashi T., Osada S., Yamada A., Kato J., Yawata K., Mori R., Imai H., Sasaki Y., Saito S., Tanaka Y., Nonaka K., Yoshida K. Extracellular signal-regulated kinase and Akt activation play a critical role in the process of hepatocyte growth factor-induced epithelial-mesenchymal transition. Int. J. Oncol. 2013;42(February (2)):556–564. doi: 10.3892/ijo.2012.1726. [DOI] [PubMed] [Google Scholar]
- 32.Xie L., Law B.K., Chytil A.M., Brown K.A., Aakre M.E., Moses H.L. ation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6(September–October (5)):603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vergara D., Valente C.M., Tinelli A., Siciliano C., Lorusso V., Acierno R., Giovinazzo G., Santino A., Storelli C., Maffia M. Resveratrol inhibits the epidermal growth factor-induced epithelial mesenchymal transition in MCF-7 cells. Cancer Lett. 2011;310(November (1)):1–8. doi: 10.1016/j.canlet.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Weiss M.B., Abel E.V., Mayberry M.M., Basile K.J., Berger A.C., Aplin A.E. TWIST1 is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res. 2012;72(December (24)):6382–6392. doi: 10.1158/0008-5472.CAN-12-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., Hung M.C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004;6(October (10)):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 36.Iliopoulos D., Polytarchou C., Hatziapostolou M., Kottakis F., Maroulakou I.G., Struhl K., Tsichlis P.N. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci. Signal. 2009;2(October (92)):ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grille S.J., Bellacosa A., Upson J., Klein-Szanto A.J., van Roy F., Lee-Kwon W., Donowitz M., Tsichlis P.N., Larue L. The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 2003;63(May (9)):2172–2178. [PubMed] [Google Scholar]
- 38.Jiao M., Nan K.J. Activation of PI3 kinase/Akt/HIF-1α pathway contributes to hypoxia-induced epithelial-mesenchymal transition and chemoresistance in hepatocellular carcinoma. Int. J. Oncol. 2012;40(February (2)):461–468. doi: 10.3892/ijo.2011.1197. [DOI] [PubMed] [Google Scholar]
- 39.Vichalkovski A., Gresko E., Hess D., Restuccia D.F., Hemmings B.A. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29(June (24)):3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- 40.Julien S., Puig I., Caretti E., Bonaventure J., Nelles L., van Roy F., Dargemont C., de Herreros A.G., Bellacosa A., Larue L. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26(November (53)):7445–7456. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 41.Yook J.I., Li X.Y., Ota I., Fearon E.R., Weiss S.J. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 2005;280(March (12)):11740–11748. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z.Q., Li X.Y., Hu C.Y., Ford M., Kleer C.G., Weiss S.J. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast cancer 1, early onset (BRCA1) repression. Proc. Natl. Acad. Sci. U. S. A. 2012;109(October (41)):16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang Y.W., Su Y.J., Hsiao M., Wei K.C., Lin W.H., Liang C.L., Chen S.C., Lee J.L. Diverse targets of β-catenin during the epithelial-mesenchymal transition define cancer stem cells and predict disease relapse. Cancer Res. 2015;75(August (16)):3398–3410. doi: 10.1158/0008-5472.CAN-14-3265. [DOI] [PubMed] [Google Scholar]
- 44.Iwata-Kajihara T., Sumimoto H., Kawamura N., Ueda R., Takahashi T., Mizuguchi H., Miyagishi M., Takeda K., Kawakami Y. Enhanced cancer immunotherapy using STAT3-depleted dendritic cells with high Th1-inducing ability and resistance to cancer cell-derived inhibitory factors. J. Immunol. 2011;187(July (1)):27–36. doi: 10.4049/jimmunol.1002067. [DOI] [PubMed] [Google Scholar]
- 45.Yaguchi T., Goto Y., Kido K., Mochimaru H., Sakurai T., Tsukamoto N., Kudo-Saito C., Fujita T., Sumimoto H., Kawakami Y. Immune suppression and resistance mediated by constitutive activation of Wnt/β-catenin signaling in human melanoma cells. J. Immunol. 2012;189(September (5)):2110–2117. doi: 10.4049/jimmunol.1102282. [DOI] [PubMed] [Google Scholar]
- 46.Oosterhoff D., Lougheed S., van de Ven R., Lindenberg J., van Cruijsen H., Hiddingh L., Kroon J., van den Eertwegh A.J., Hangalapura B., Scheper R.J., de Gruijl T.D. Tumor-mediated inhibition of human dendritic cell differentiation and function is consistently counteracted by combined p38 MAPK and STAT3 inhibition. Oncoimmunology. 2012;1(August (5)):649–658. doi: 10.4161/onci.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.López-Soto A., Zapico L.H., Acebes-Huerta A., Rodrigo L., Gonzalez S. Regulation of NKG2D signaling during the epithelial-to-mesenchymal transition. Oncoimmunology. 2013;2(September (9)):e25820. doi: 10.4161/onci.25820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemaire R., Desmons A., Tabet J.C., Day R., Salzet M., Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 2007;6(April (4)):1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 49.Longuespée R., Boyon C., Castellier C., Jacquet A., Desmons A., Kerdraon O., Vinatier D., Fournier I., Day R., Salzet M. The C-terminal fragment of the immunoproteasome PA28S (Reg alpha) as an early diagnosis and tumor-relapse biomarker: evidence from mass spectrometry profiling. Histochem. Cell Biol. 2012;138(July (1)):141–154. doi: 10.1007/s00418-012-0953-0. [DOI] [PubMed] [Google Scholar]
- 50.Dekker T.J., Balluff B.D., Jones E.A., Schöne C.D., Schmitt M., Aubele M., Kroep J.R., Smit V.T., Tollenaar R.A., Mesker W.E., Walch A., McDonnell L.A. Multicenter matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) identifies proteomic differences in breast-cancer-associated stroma. J. Proteome Res. 2014;13(November (11)):4730–4738. doi: 10.1021/pr500253j. [DOI] [PubMed] [Google Scholar]
- 51.Chen X.H., Liu Z.C., Zhang G., Wei W., Wang X.X., Wang H., Ke H.P., Zhang F., Wang H.S., Cai S.H., Du J. TGF-β and EGF induced HLA-I downregulation is associated with epithelial-mesenchymal transition (EMT) through upregulation of snail in prostate cancer cells. Mol. Immunol. 2015;65(May (1)):34–42. doi: 10.1016/j.molimm.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Royer C., Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18(September (9)):1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung C.T., Brugge J.S. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature. 2012;482(February (7385)):410–413. doi: 10.1038/nature10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeanes A., Gottardi C.J., Yap A.S. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27(November (55)):6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geyer F.C., Lacroix-Triki M., Savage K., Arnedos M., Lambros M.B., MacKay A., Natrajan R., Reis-Filho J.S. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol. 2011;24(February (2)):209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H., Liu J., Yue D., Gao L., Wang D., Zhang H., Wang C. Clinical significance of E-cadherin, β-catenin, vimentin and S100A4 expression in completely resected squamous cell lung carcinoma. J. Clin. Pathol. 2013;66(November (11)):937–945. doi: 10.1136/jclinpath-2013-201467. [DOI] [PubMed] [Google Scholar]
- 57.Kaur J., Sawhney M., DattaGupta S., Shukla N.K., Srivastava A., Walfish P.G., Ralhan R. Clinical significance of altered expression of β-catenin and E-cadherin in oral dysplasia and cancer: potential link with ALCAM expression. PLoS One. 2013;8(June (6)):e67361. doi: 10.1371/journal.pone.0067361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X.L., Ling Z.Q., Chen S.Z., Li B., Ji W.H., Mao W.M. The impact of E-cadherin expression on the prognosis of esophageal cancer: a meta-analysis. Dis. Esophagus. 2014;27(January (1)):79–86. doi: 10.1111/dote.12024. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Chen C.Q., He Y.L., Cai S.R., Yang D.J., He W.L., Xu J.B., Zan W.H. Abnormal expression of E-cadherin in tumor cells is associated with poor prognosis of gastric carcinoma. J. Surg. Oncol. 2012;106(September (3)):304–310. doi: 10.1002/jso.23008. [DOI] [PubMed] [Google Scholar]
- 60.Fu J.1, Qin L., He T., Qin J., Hong J., Wong J., Liao L., Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21(February (2)):275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Bueno G., Cubillo E., Sarrió D., Peinado H., Rodríguez-Pinilla S.M., Villa S., Bolós V., Jordá M., Fabra A., Portillo F., Palacios J., Cano A. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 2006;66(October (19)):9543–9556. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- 62.Tran D.D., Corsa C.A., Biswas H., Aft R.L., Longmore G.D. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Mol. Cancer Res. 2011;9(December (12)):1644–1657. doi: 10.1158/1541-7786.MCR-11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubois-Marshall S., Thomas J.S., Faratian D., Harrison D.J., Katz E. Two possible mechanisms of epithelial to mesenchymal transition in invasive ductal breast cancer. Clin. Exp. Metastasis. 2011;28(December (8)):811–818. doi: 10.1007/s10585-011-9412-x. [DOI] [PubMed] [Google Scholar]
- 64.Caramel J., Papadogeorgakis E., Hill L., Browne G.J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J.H., Ansieau S., Tulchinsky E. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24(October (4)):466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 65.Luo W.R., Li S.Y., Cai L.M., Yao K.T. High expression of nuclear Snail, but not cytoplasmic staining, predicts poor survival in nasopharyngeal carcinoma. Ann. Surg. Oncol. 2012;19(September (9)):2971–2979. doi: 10.1245/s10434-012-2347-x. [DOI] [PubMed] [Google Scholar]
- 66.Muenst S., Däster S., Obermann E.C., Droeser R.A., Weber W.P., von Holzen U., Gao F., Viehl C., Oertli D., Soysal S.D. Nuclear expression of snail is an independent negative prognostic factor in human breast cancer. Dis. Markers. 2013;35(5):337–344. doi: 10.1155/2013/902042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jouppila-Mättö A., Närkiö-Mäkelä M., Soini Y., Pukkila M., Sironen R., Tuhkanen H., Mannermaa A., Kosma V.M. Twist and snai1 expression in pharyngeal squamous cell carcinoma stroma is related to cancer progression. BMC Cancer. 2011;11(August):350. doi: 10.1186/1471-2407-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gasparotto D., Polesel J., Marzotto A., Colladel R., Piccinin S., Modena P., Grizzo A., Sulfaro S., Serraino D., Barzan L., Doglioni C., Maestro R. Overexpression of TWIST2 correlates with poor prognosis in head and neck squamous cell carcinomas. Oncotarget. 2011;2(December (12)):1165–1175. doi: 10.18632/oncotarget.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Silva S.D., Alaoui-Jamali M.A., Soares F.A., Carraro D.M., Brentani H.P., Hier M., Rogatto S.R., Kowalski L.P. TWIST1 is a molecular marker for a poor prognosis in oral cancer and represents a potential therapeutic target. Cancer. 2013;(October) doi: 10.1002/cncr.28404. [DOI] [PubMed] [Google Scholar]
- 70.Gomez I., Peña C., Herrera M., Muñoz C., Larriba M.J., Garcia V., Dominguez G., Silva J., Rodriguez R., Garcia de Herreros A., Bonilla F., Garcia J.M. TWIST1 is expressed in colorectal carcinomas and predicts patient survival. PLoS One. 2011;6(March (3)):e18023. doi: 10.1371/journal.pone.0018023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wushou A., Hou J., Zhao Y.J., Shao Z.M. Twist-1 up-regulation in carcinoma correlates to poor survival. Int. J. Mol. Sci. 2014;15(November (12)):21621–21630. doi: 10.3390/ijms151221621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jouppila-Mättö A., Närkiö-Mäkelä M., Soini Y., Pukkila M., Sironen R., Tuhkanen H., Mannermaa A., Kosma V.M. Twist and snai1 expression in pharyngeal squamous cell carcinoma stroma is related to cancer progression. BMC Cancer. 2011;11(August):350. doi: 10.1186/1471-2407-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cavallaro U., Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer. 2004;4(February (2)):118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 74.Qian X., Anzovino A., Kim S., Suyama K., Yao J., Hulit J., Agiostratidou G., Chandiramani N., McDaid H.M., Nagi C., Cohen H.W., Phillips G.R., Norton L., Hazan R.B. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene. 2014;33(June (26)):3411–3421. doi: 10.1038/onc.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hui L., Zhang S., Dong X., Tian D., Cui Z., Qiu X. Prognostic significance of twist and N-cadherin expression in NSCLC. PLoS One. 2013;8(April (4)):e62171. doi: 10.1371/journal.pone.0062171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lascombe I., Clairotte A., Fauconnet S., Bernardini S., Wallerand H., Kantelip B., Bittard H. N-cadherin as a novel prognostic marker of progression in superficial urothelial tumors. Clin. Cancer Res. 2006;12(May (9)):2780–2787. doi: 10.1158/1078-0432.CCR-05-2387. [DOI] [PubMed] [Google Scholar]
- 77.Domenico M D.I., Pierantoni G.M., Feola A., Esposito F., Laino L., Rosa D.E., Rullo A., Mazzotta M., Martano M., Sanguedolce F., Perillo L., D'Angelo L., Papagerakis S., Tortorella S., Bufo P., Lo Muzio L., Pannone G., Santoro A. Prognostic significance of N-cadherin expression in oral squamous cell carcinoma. Anticancer Res. 2011;31(December (12)):4211–4218. [PubMed] [Google Scholar]
- 78.Vergara D., Chiriacò F., Acierno R., Maffia M. Proteomic map of peripheral blood mononuclear cells. Proteomics. 2008;8(May (10)):2045–2051. doi: 10.1002/pmic.200700726. [DOI] [PubMed] [Google Scholar]
- 79.Singh S., Sadacharan S., Su S., Belldegrun A., Persad S., Singh G. Overexpression of vimentin: role in the invasive phenotype in an androgen-independent model of prostate cancer. Cancer Res. 2003;63(May (9)):2306–2311. [PubMed] [Google Scholar]
- 80.Vasko V., Espinosa A.V., Scouten W., He H., Auer H., Liyanarachchi S., Larin A., Savchenko V., Francis G.L., de la Chapelle A., Saji M., Ringel M.D. Gene expression and functional evidence of epithelial-to-mesenchymal transition in papillary thyroid carcinoma invasion. Proc. Natl. Acad. Sci. U. S. A. 2007;104(February (8)):2803–2808. doi: 10.1073/pnas.0610733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Craven R.A., Stanley A.J., Hanrahan S., Dods J., Unwin R., Totty N., Harnden P., Eardley I., Selby P.J., Banks R.E. Proteomic analysis of primary cell lines identifies protein changes present in renal cell carcinoma. Proteomics. 2006;6(May (9)):2853–2864. doi: 10.1002/pmic.200500549. [DOI] [PubMed] [Google Scholar]
- 82.Wu M., Bai X., Xu G., Wei J., Zhu T., Zhang Y., Li Q., Liu P., Song A., Zhao L., Gang C., Han Z., Wang S., Zhou J., Lu Y., Ma D. Proteome analysis of human androgen-independent prostate cancer cell lines: variable metastatic potentials correlated with vimentin expression. Proteomics. 2007;7(June (12)):1973–1983. doi: 10.1002/pmic.200600643. [DOI] [PubMed] [Google Scholar]
- 83.El Ayed M., Bonnel D., Longuespée R., Castelier C., Franck J., Vergara D., Desmons A., Tasiemski A., Kenani A., Vinatier D., Day R., Fournier I., Salzet M. MALDI imaging mass spectrometry in ovarian cancer for tracking, identifying, and validating biomarkers. Med. Sci. Monit. 2010;16(August (8)):BR233–45. [PubMed] [Google Scholar]
- 84.Otsuki S., Inokuchi M., Enjoji M., Ishikawa T., Takagi Y., Kato K., Yamada H., Kojima K., Sugihara K. Vimentin expression is associated with decreased survival in gastric cancer. Oncol. Rep. 2011;25(May (5)):1235–1242. doi: 10.3892/or.2011.1185. [DOI] [PubMed] [Google Scholar]
- 85.Satelli A., Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011;68(September (18)):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu L.K., Jiang X.Y., Zhou X.X., Wang D.M., Song X.L., Jiang H.B. Upregulation of vimentin and aberrant expression of E-cadherin/beta-catenin complex in oral squamous cell carcinomas: correlation with the clinicopathological features and patient outcome. Mod. Pathol. 2010;23(February (2)):213–224. doi: 10.1038/modpathol.2009.160. [DOI] [PubMed] [Google Scholar]
- 87.Yamashita N., Tokunaga E., Kitao H., Hisamatsu Y., Taketani K., Akiyoshi S., Okada S., Aishima S., Morita M., Maehara Y. Vimentin as a poor prognostic factor for triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 2013;139(May (5)):739–746. doi: 10.1007/s00432-013-1376-6. [DOI] [PubMed] [Google Scholar]
- 88.Karihtala P., Auvinen P., Kauppila S., Haapasaari K.M., Jukkola-Vuorinen A., Soini Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res. Treat. 2013;138(February (1)):81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 89.Richardson F., Young G.D., Sennello R., Wolf J., Argast G.M., Mercado P., Davies A., Epstein D.M., Wacker B. The evaluation of E-Cadherin and vimentin as biomarkers of clinical outcomes among patients with non-small cell lung cancer treated with erlotinib as second- or third-line therapy. Anticancer Res. 2012;32(February (2)):537–552. [PubMed] [Google Scholar]
- 90.Johnson P.J., Melia W.M., Palmer M.K., Portmann B., Williams R. Relationship between serum alpha-foetoprotein, cirrhosis and survival in hepatocellular carcinoma. Br. J. Cancer. 1981;44(October (4)):502–505. doi: 10.1038/bjc.1981.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kojima K., Takata A., Vadnais C., Otsuka M., Yoshikawa T., Akanuma M., Kondo Y., Kang Y.J., Kishikawa T., Kato N., Xie Z., Zhang W.J., Yoshida H., Omata M., Nepveu A., Koike K. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat. Commun. 2011;2(June):338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- 92.Tsai W.C., Hsu S.D., Hsu C.S., Lai T.C., Chen S.J., Shen R., Huang Y., Chen H.C., Lee C.H., Tsai T.F., Hsu M.T., Wu J.C., Huang H.D., Shiao M.S., Hsiao M., Tsou A.P. MicroRNA-plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 2012;122(August (8)):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun S., Poon R.T., Lee N.P., Yeung C., Chan K.L., Ng I.O., Day P.J., Luk J.M. Proteomics of hepatocellular carcinoma: serum vimentin as a surrogate marker for small tumors (< or = 2 cm) J. Proteome Res. 2010;9(April (4)):1923–1930. doi: 10.1021/pr901085z. [DOI] [PubMed] [Google Scholar]
- 94.Kufe D.W. Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer. 2009;9(December (12)):874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bast R.C., Jr., Klug T.L., St John E., Jenison E., Niloff J.M., Lazarus H., Berkowitz R.S., Leavitt T., Griffiths C.T., Parker L., Zurawski V.R., Jr., Knapp R.C. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N. Engl. J. Med. 1983;309(October (15)):883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- 96.Zurawski V.R., Jr., Knapp R.C., Einhorn N., Kenemans P., Mortel R., Ohmi K., Bast R.C., Jr., Ritts R.E., Jr., Malkasian G. An initial analysis of preoperative serum CA 125 levels in patients with early stage ovarian carcinoma. Gynecol. Oncol. 1988;30(May (1)):7–14. doi: 10.1016/0090-8258(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 97.Thériault C., Pinard M., Comamala M., Migneault M., Beaudin J., Matte I., Boivin M., Piché A., Rancourt C. MUC16 (CA125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis6 (CA125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis. Gynecol. Oncol. 2011;121(June (3)):434–443. doi: 10.1016/j.ygyno.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 98.Tothill R.W., Tinker A.V., George J., Brown R., Fox S.B., Lade S., Johnson D.S., Trivett M.K., Etemadmoghadam D., Locandro B., Traficante N., Fereday S., Hung J.A., Chiew Y.E., Haviv I., Australian Ovarian Cancer Study Group, Gertig D., DeFazio A., Bowtell D.D. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008;14(August (16)):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 99.Mall A.S. Analysis of mucins: role in laboratory diagnosis. J. Clin. Pathol. 2008;1(September (9)):1018–1024. doi: 10.1136/jcp.2008.058057. [DOI] [PubMed] [Google Scholar]
- 100.Wang S., Chen X., Tang M. Quantitative assessment of the diagnostic role of MUC1 in pancreatic ductal adenocarcinoma. Tumour Biol. 2014;35(September (9)):9101–9109. doi: 10.1007/s13277-014-2186-4. [DOI] [PubMed] [Google Scholar]
- 101.Roy L.D., Sahraei M., Subramani D.B., Besmer D., Nath S., Tinder T.L., Bajaj E., Shanmugam K., Lee Y.Y., Hwang S.I., Gendler S.J., Mukherjee P. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30(March (12)):1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rajabi H., Alam M., Takahashi H., Kharbanda A., Guha M., Ahmad R., Kufe D. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33(March (27)):1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gold P., Freedman S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and adsorption techniques. J. Exp. Med. 1965;121(March (1)):439–462. doi: 10.1084/jem.121.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beauchemin N., Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(December (3–4)):643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 105.Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999;9(April (2)):67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 106.Chen J., Li Q., An Y., Lv N., Xue X., Wei J., Jiang K., Wu J., Gao W., Qian Z., Dai C., Xu Z., Miao Y. CEACAM6 induces epithelial-mesenchymal transition and mediates invasion and metastasis in pancreatic cancer. Int. J. Oncol. 2013;43(September (3)):877–885. doi: 10.3892/ijo.2013.2015. [DOI] [PubMed] [Google Scholar]
- 107.Saadatmand S., de Kruijf E.M., Sajet A., Dekker-Ensink N.G., van Nes J.G., Putter H., Smit V.T., van de Velde C.J., Liefers G.J., Kuppen P.J. Expression of cell adhesion molecules and prognosis in breast cancer. Br. J. Surg. 2013;100(January (2)):252–260. doi: 10.1002/bjs.8980. [DOI] [PubMed] [Google Scholar]
- 108.Whelan S.A., He J., Lu M., Souda P., Saxton R.E., Faull K.F., Whitelegge J.P., Chang H.R. Mass spectrometry (LC–MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid. J. Proteome Res. 2012;11(October (10)):5034–5045. doi: 10.1021/pr300606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mathias R.A., Wang B., Ji H., Kapp E.A., Moritz R.L., Zhu H.J., Simpson R.J. Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J. Proteome Res. 2009;8(June (6)):2827–2837. doi: 10.1021/pr8010974. [DOI] [PubMed] [Google Scholar]
- 110.Schrader C.H., Kolb M., Zaoui K., Flechtenmacher C., Grabe N., Weber K.J., Hielscher T., Plinkert P.K., Hess J. Kallikrein-related peptidase 6 regulates epithelial-to-mesenchymal transition and serves as prognostic biomarker for head and neck squamous cell carcinoma patients. Mol. Cancer. 2015;14(May (1)):107. doi: 10.1186/s12943-015-0381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomson S., Petti F., Sujka-Kwok I., Mercado P., Bean J., Monaghan M., Seymour S.L., Argast G.M., Epstein D.M., Haley J.D. A systems view of epithelial-mesenchymal transition signaling states. Clin. Exp. Metastasis. 2011;28(February (2)):137–155. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Farrell J., Kelly C., Rauch J., Kida K., García-Muñoz A., Monsefi N., Turriziani B., Doherty C., Mehta J.P., Matallanas D., Simpson J.C., Kolch W., von Kriegsheim A. HGF induces epithelial-to-mesenchymal transition by modulating the mammalian hippo/MST2 and ISG15 pathways. J. Proteome Res. 2014;13(June (6)):2874–2886. doi: 10.1021/pr5000285. [DOI] [PubMed] [Google Scholar]
- 113.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R.F., Vahdat L.T., Altorki N.K., Mittal V., Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;(November) doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;(November) doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim Y., Han D., Min H., Jin J., Yi E.C., Kim Y. Comparative proteomic profiling of pancreatic ductal adenocarcinoma cell lines. Mol. Cells. 2014;37(December (12)):888–898. doi: 10.14348/molcells.2014.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He J., Whelan S.A., Lu M., Shen D., Chung D.U., Saxton R.E., Faull K.F., Whitelegge J.P., Chang H.R. Proteomic-based biosignatures in breast cancer classification and prediction of therapeutic response. Int. J. Proteomics. 2011;2011:896476. doi: 10.1155/2011/896476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vergara D., Simeone P., del Boccio P., Toto C., Pieragostino D., Tinelli A., Acierno R., Alberti S., Salzet M., Giannelli G., Sacchetta P., Maffia M. Comparative proteome profiling of breast tumor cell lines by gel electrophoresis and mass spectrometry reveals an epithelial mesenchymal transition associated protein signature. Mol. Biosyst. 2013;9(June (6)):1127–1138. doi: 10.1039/c2mb25401h. [DOI] [PubMed] [Google Scholar]
- 118.Sobral-Leite M., Wesseling J., Smit V.T., Nevanlinna H., van Miltenburg M.H., Sanders J., Hofland I., Blows F.M., Coulson P., Patrycja G., Schellens J.H., Fagerholm R., Heikkilä P., Aittomäki K., Blomqvist C., Provenzano E., Ali H.R., Figueroa J., Sherman M., Lissowska J., Mannermaa A., Kataja V., Kosma V.M., Hartikainen J.M., Phillips K.A., kConFab/AOCS Investigators, Couch F.J., Olson J.E., Vachon C., Visscher D., Brenner H., Butterbach K., Arndt V., Holleczek B., Hooning M.J., Hollestelle A., Martens J.W., van Deurzen C.H., van de Water B., Broeks A., Chang-Claude J., Chenevix-Trench G., Easton D.F., Pharoah P.D., García-Closas M., de Graauw M., Schmidt M.K. Annexin A1 expression in a pooled breast cancer series: association with tumor subtypes and prognosis. BMC Med. 2015;13(July):156. doi: 10.1186/s12916-015-0392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lawrence R.T., Perez E.M., Hernández D., Miller C.P., Haas K.M., Irie H.Y., Lee S.I., Blau C.A., Villén J. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11(April (4)):630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vergara D., Simeone P., Latorre D., Cascione F., Leporatti S., Trerotola M., Giudetti A.M., Capobianco L., Lunetti P., Rizzello A., Rinaldi R., Alberti S., Maffia M. Proteomics analysis of E-cadherin knockdown in epithelial breast cancer cells. J. Biotechnol. 2015;202(May):3–11. doi: 10.1016/j.jbiotec.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 121.Guo Z., Neilson L.J., Zhong H., Murray P.S., Zanivan S., Zaidel-Bar R. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal. 2014;7(December (354)):rs7. doi: 10.1126/scisignal.2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan T.Z., Miow Q.H., Miki Y., Noda T., Mori S., Huang R.Y., Thiery J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014;6(September (10)):1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sjöström M., Ossola R., Breslin T., Rinner O., Malmström L., Schmidt A., Aebersold R., Malmström J., Niméus E. A Combined shotgun and targeted mass spectrometry strategy for breast cancer biomarker discovery. J. Proteome Res. 2015;14(July (7)):2807–2818. doi: 10.1021/acs.jproteome.5b00315. [DOI] [PubMed] [Google Scholar]
- 124.Law K.P., Lim Y.P. Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev. Proteomics. 2013;10(December (6)):551–566. doi: 10.1586/14789450.2013.858022. [DOI] [PubMed] [Google Scholar]
- 125.Bruderer R., Bernhardt O.M., Gandhi T., Miladinović S.M., Cheng L.Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., Vitek O., Rinner O., Reiter L. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14(May (5)):1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]