Abstract
The remarkable progress in the field of ionic liquids (ILs) in the last two decades has involved investigations on different aspects of ILs in various conditions. The nontoxic and biocompatible nature of ILs makes them a suitable substance for the storage and application of biomolecules. In this regard, the aqueous IL solutions have attracted a large number of studies to comprehend the role of water in modulating various properties of biomolecules. Here, we review some of the recent studies on aqueous ILs that concern the role of water in altering the behavior of ILs in general and in case of biomolecules solvated in ILs. The different structural and dynamic effects caused by water have been highlighted. We discuss the different modes of IL interaction that are responsible for stabilization and destabilization of proteins and enzymes followed by examples of water effect on this. The role of water in the case of nucleic acid storage in ILs, an area which has mostly been underrated, also has been emphasized. Our discussions highlight the fact that the effects of water on IL behavior are not general and are highly dependent on the nature of the IL under consideration. Overall, we aim to draw attention to the significance of water dynamics in the aqueous IL solutions, a better understanding of which can help in developing superior storage materials for application purposes.
Keywords: IL structure, IL dynamics, IL protein interactions, IL DNA interaction
Introduction
The impact of ionic liquids (ILs) in the field of materials science can be realized by the growing number of new research articles published in this area in the recent past. Since the first report of room temperature ILs by Wilkes and Zaworotko (1992), several studies have focused on developing new ILs while others have concentrated on understanding properties of ILs for its applications. Today, ILs find their applications in the field of biotechnology (Lee et al. 2007; Attri et al. 2011; Dominguez de Marıá 2012; Jeong et al. 2012), chemical synthesis and catalysis (Welton 1999; Wasserscheid and Keim 2000; Sheldon 2001; Pârvulescu and Hardacre 2007; van Rantwijk and Sheldon 2007; Weingärtner 2008), electrochemistry, (Zein El Abedin and Endres 2006, Fedorov and Kornyshev 2008; Armand et al. 2009; Torriero 2015a, 2015b), and several other fields (Dupont and Scholten 2010; Tang et al. 2012; Chatel and MacFarlane 2014; Lei et al. 2014). All of these fields have been explored in so much detail that each of these areas can be a separate topic of review articles. Some of the recent reviews in the field of ILs discuss its prospect in pharmaceutics and medicine (Egorova et al. 2017), in extraction and separation of biomolecules (Ventura et al. 2017), and in improving enzymatic organic synthesis (Itoh 2017). In this review, we mainly focus on the aspects of ILs in combination with biomolecules.
Several key aspects of ILs make them highly suitable for their use in biotechnology applications. Some of these features include their non-toxicity, low vapor pressure, low cost, and high stability (Huddleston et al. 2001; He et al. 2006) which make ILs a better replacement for the volatile organic solvents. The properties of ILs can be manipulated fairly easily by altering their constituent ions (Earle Martyn and Seddon Kenneth 2000; Wasserscheid and Keim 2000). These properties make the ILs suitable as reaction media for biocatalysis studies (Erbeldinger et al. 2000; Kim et al. 2001) and for the separation and purification of biomolecules (Zhao et al. 2005; Domínguez-Pérez et al. 2010; Freire et al. 2010; Tomé et al. 2010; Ventura et al. 2011). Several studies have focused on the structure and dynamic features of proteins and enzymes solvated in the IL solutions. While the structural aspects of proteins in IL solutions have been compiled in a recent review by Smiatek (Jens 2017), studies on the structure and dynamics of several kinds of biomolecules have been presented in the review by Benedetto and Ballone (2016). Considering the advantages of ILs in the field of biotechnological applications, the role of water in the IL solutions is crucial to understand for the growth in this field. Therefore, we focus here on the aqueous IL solution for the review.
The studies of biomolecules and water remain an active area of research even today. The unique interaction of water with both ILs and biomolecules makes them an interesting entity in the aqueous IL solutions. The wide range of applications of ILs resulted in several experimental and theoretical studies of the effect of water in these solutions. Here, we aim to discuss some of these findings in the context of ILs’ application in biotechnology. We first discuss different studies pertinent to the structure and dynamics of IL cations and anions in the presence of water in bulk aqueous IL solution. Then, we move to the studies of proteins and enzymes and discuss some of the current findings on the behavior of these molecules in aqueous IL solutions. This is followed by the studies on IL and nucleic acid studies. Our aim in this review is to provide an overview of the importance of water in this area.
IL structure and dynamics in the presence of water
Before going into the discussions on biomolecules solvated in aqueous IL solution, we briefly discuss the structural and dynamical changes in bulk IL-water solution. The hygroscopic nature of ILs makes the water molecules an integral component of these systems. Hence, knowing the exact effect of these water molecules is crucial for elucidating the IL behavior in general.
Structural effects of water in ILs
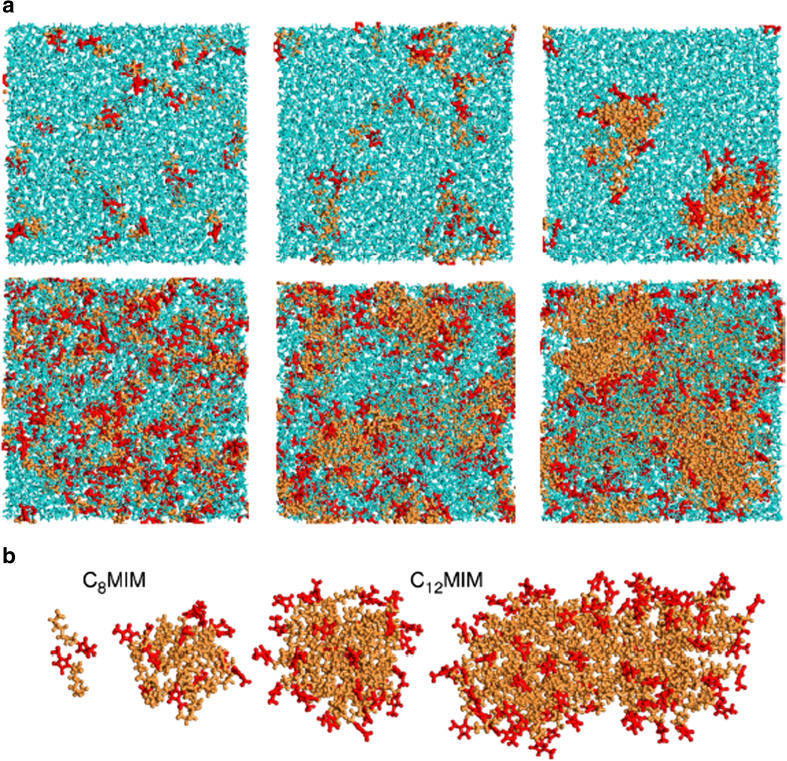
ILs in their pure form is not always found to form homogeneous mixtures. A primary source for the heterogeneity in these mixtures is the hydrophobic alkyl chains of the cations. It has been observed that in the pure form the cations with short alkyl chains form nearly homogeneous mixtures while those with longer alkyl chains often show phase separation in the mixture (Triolo et al. 2007, Hayes et al. 2015; Bruce et al. 2017). The water in the ILs is known to alter the nanostructure (Jiang et al. 2007) of these ions. A number of reports have pointed out the existence of micelle formation in the IL-water mixtures. Different studies have found the cation size to be a major player in the formation of these micelles. Blesic et al. (2007) employed interfacial measurement, fluorescence, and NMR measurements to show that the ILs [Cnmim]Cl showed self-aggregation in the bulk phase when n > 8 whereas ILs with shorter chain lengths such as n = 4 or 6 did not show any tendency to form such aggregates. The transitional ILs with n = 6 form a monolayer at the interface between the aqueous solution and air with no self-aggregation in the bulk phase. Molecular dynamics (MD) simulations have been very effective in finding out the structure that these IL forms in water. While Liu et al. (2015) showed rod-like micelle formation for imidazolium-based ILs, Vicent-Luna et al. (2017) showed that the critical concentration required for the micelle formation lowered with an increase in the anisotropy for the imidazolium-based IL systems. The general pattern of these micelles is shown in Fig. 1.
Fig. 1.
a Representative snapshots from MD simulation of IL/water mixtures at low (30 IL ion pair) and high (200 IL ion pair) concentration in the top and bottom panel, respectively. The systems shown here are (left) [C4mim]+ which does not show aggregation at both low and high concentration, (middle) [C8mim]+ which remains as monomers at low concentrations and aggregate at higher concentrations, and (right) [C12mim]+ which can be seen to aggregate at both low and high concentrations. b The aggregated forms of [C8mim]+and [C12mim]+cations. The orange color represents the tail of the cations, red color represents the head of the cations and water is represented in blue color. Reprinted with permission from Vicent-Luna et al. 2017. Copyright 2017 American Chemical Society
The modulation of IL structure in water has been explained through investigation of the IL-water interactions where the general trend is that the interactions are predominantly stronger with the anions (Cammarata et al. 2001). The study by Cammarata et al. (2001) have shown that water molecules remain H-bonded to the anions in 1:2 ratios by making anion…HOH…anion structures. With anions of stronger basicity, the absorbed water content has been found to increase in the IL mixture. The transmission IR spectroscopy study revealed the strength of the H-bonds between IL anions and water to be in the following order: [PF6]− < [SbF6]− < [BF4]− < [(CF3SO2)2N]− < [ClO4]− < [CF3SO3]− < [NO3]− < [CF3CO2]−(Cammarata et al. 2001). Different simulation studies also reveal similar conclusions that the water cluster size inside the ILs are independent of the cation chain length, although the hydrophobicity of the anions are crucial for determining the miscibility of the ILs (Méndez-Morales et al. 2011). Similar conclusions have been drawn in the article by Kohno and Ohno (2012) where it was asserted that the hydrated hydrophobic ions undergo a dynamic phase transition from being a homogeneous mixture to phase-separated states with increasing temperature. The water on the surface of the ILs also alters the orientations of the ions at the interface. The origin of these effects stems from the difference in activity of water in hydrophilic ILs compared to that in hydrophobic ones (Anthony et al. 2001). Since in the case of hydrophilic ILs the water molecules are stabilized through favorable H-bonding and dipole-dipole forces (Anthony et al. 2001, Cammarata et al. 2001), the water molecules are likely to remain in the bulk phase of ILs to gain proper solvation. However, for the water immiscible ILs, a dramatic reorientation at the gas-liquid interface can be observed(Rivera-Rubero and Baldelli 2004). These interactions of water with ILs result in the difference in the properties of the solutions. For example, the surface tension of the hydrophobic IL [BMIM] [BF4] was found to be 35 mJ/m2 in water (Bowers et al. 2004) whereas in the absence of water, the values were reported to be around 41 (Law and Watson 2001) and 46.6 (Huddleston et al. 2001) mJ/m2. On the contrary, changes are significantly smaller (36.8 mJ/m2 with water and 37.5 mJ/m2 in the dry state (Huddleston et al. 2001)) for the hydrophilic [BMIM][imide].
Effect of water on the dynamics of ILs
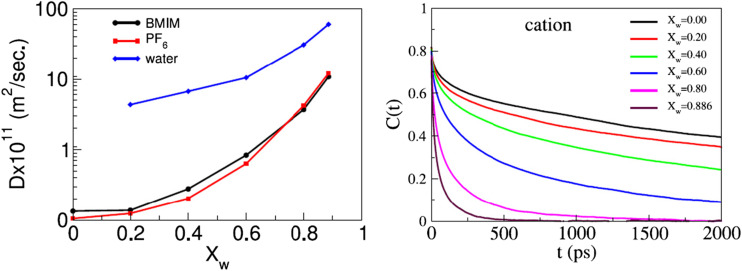
In general, ILs are known to be viscous, and diffusion coefficients of the constituent ions are a few orders of magnitude lower compared to water even at elevated temperatures (Kowsari et al. 2008). Addition of water in ILs is, therefore, expected to alter the dynamical properties of the ions due to the structural changes taking place in ILs. The effect of water on the dynamics of ILs has been measured mainly through the studies of translational diffusion and reorientation dynamics of the IL ions. The recent MD study by Sharma and Ghorai (2016) indicated that the number of dissociated ion pairs has a minor effect on the water concentration. The changes in local structure result in an increase of the self-diffusion coefficient while the reorientation timescale decreases with the increase in water content in [BMIM][PF6] (Sharma and Ghorai 2016). This is summarized in Fig. 2 where the diffusion coefficients of cation, anion, and water are shown at different water contents. The rotational motions are also shown by indicating the cation’s reorientational time correlation function of second order at different water content in Fig. 2 (Sharma and Ghorai 2016).
Fig. 2.
(Left) The self-diffusion coefficient of [bmim]+ cation, [PF6]− anion and water molecules at different mole fraction of water(XW). The figure on the right shows the reorientational time correlation function of second order (l = 2) for [bmim]+ cation at different compositions. Reprinted with permission from Sharma and Ghorai 2016. Copyright 2016 American Institute of Physics
Similar conclusions were drawn in another study by Price et al. (2017) by combining quasi-elastic neutron scattering (QENS) and MD simulation where the cation diffusion was found to increase by a factor of seven while the rotational motions slowed down by a factor of three. Other studies probed the dynamics of solutes inserted in ILs (Kaintz et al. 2013; Araque et al. 2015, 2016). It was found that the dynamics of a small neutral solute and a charged solute have very different behavior in ILs. While the former showed a large positive deviation from the Stokes-Einstein behavior, the latter underwent a long caging dynamics through jump events triggered by the loss and availability of counter ions (Araque et al. 2015).
Proteins and ILs
With the major interest of biotechnological applications involving proteins and ILs, a plethora of studies have been devoted to various aspects of structure and activity of proteins and enzymes in IL solutions. Some recent reviews have comprehensively highlighted the developments in experimental and simulation studies involving ILs and proteins (Jha and Venkatesu 2015; Jens 2017; Schröder 2017). With the significant progress in this area, it will not be possible for us to include all the examples available in the literature that studies proteins and IL mixtures. Since the present review concerns the general behavior of aqueous ILs and its impact on biomolecules, we restrict our discussions to a few examples on the behavior of proteins and enzymes in aqueous IL solutions to get some idea on the role of water in these mixtures. Therefore, we first discuss the general behavior of proteins in neat ILs before going to the discussion of aqueous IL solutions.
Interactions of ILs with proteins
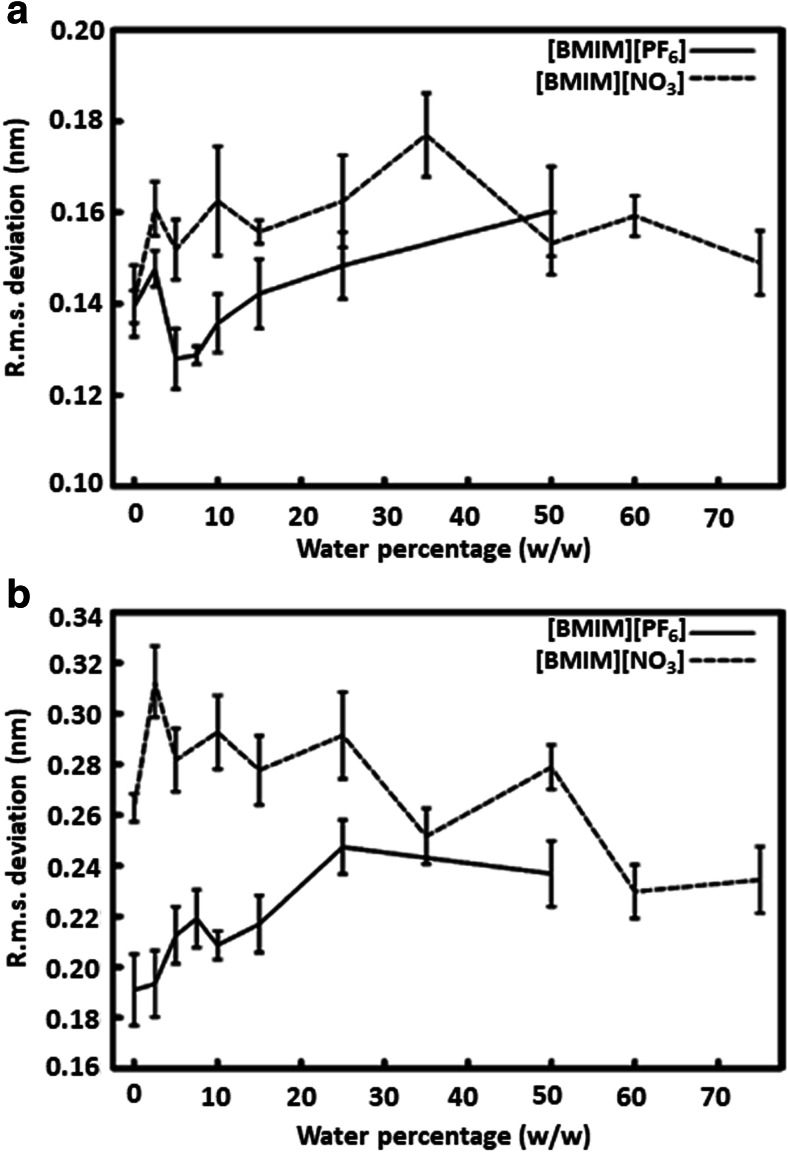
The use of ILs as suitable solvents for proteins (Summers and Flowers 2000) and as appropriate media for enzyme catalyzed reactions (Cull et al. 2000; Madeira Lau et al. 2000) has been established nearly two decades ago. The anion species present in the IL mixture play a critical role in the activity and stability of enzymes dissolved in these IL mixtures (Kaar et al. 2003). The enzymes lipase and protease have been found to be active in ILs containing the anion [BF4]−, [PF6]−, and [Tf2N]− while the activity has been found to be lost in ILs containing the anion [NO3]− ,[CF3CO2]− and [CF3SO3]− (Moon et al. 2006). The first MD study carried out by Micaêlo and Soares on the enzyme serine protease cutinase from Fusarium solani pisi gives insight on this anion dependence for the enzyme activity (Micaêlo and Soares 2008). It was found that the native structure of the enzyme remained more stable in [BMIM][PF6] compared to [BMIM][NO3] at different water contents. This was verified by the root mean square deviation (RMSD) values for the enzyme at different water content as shown in Fig. 3. The IL with [PF6]− can be seen to be more effective in stabilizing the enzyme at higher temperature. The higher affinity of [NO3]− toward the protein main chain has been attributed as the cause for this behavior (Micaêlo and Soares 2008).
Fig. 3.
Average rmsd values of the Cα atoms from the X-ray structure of cutinase in [bmim][PF6] and [bmim][NO3] at a 298 K and b 343 K. Reprinted with permission from ref. Micaêlo and Soares 2008. Copyright 2008 American Chemical Society
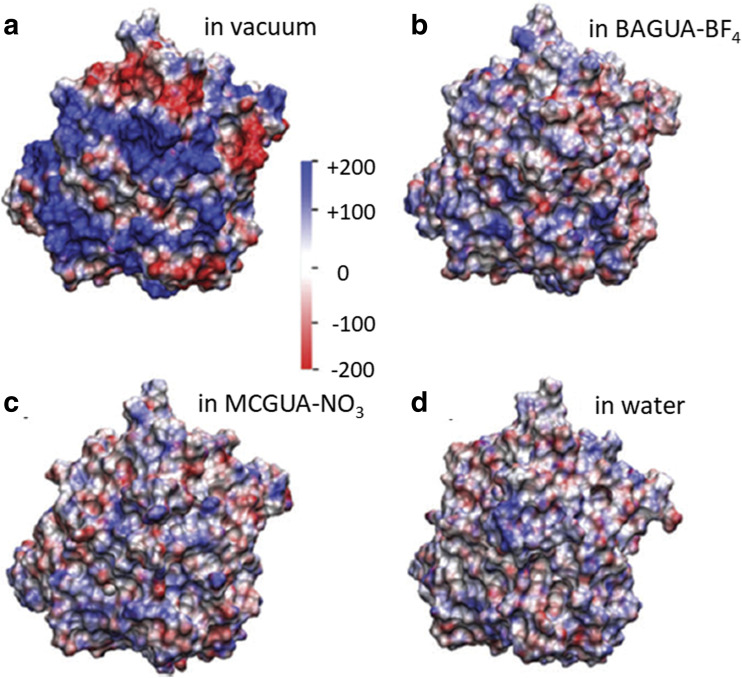
However, the use of these large hydrophobic anions in the ILs is hindered by the poor solubility of proteins and enzymes in these ILs (Madeira Lau et al. 2004). The different interactions shown by cations and anions with the amino acids can alter the electrostatic potential of the protein surface. This has been shown in the study by Wu and co-workers (Klahn et al. 2011) through MD simulation. Figure 4 shows the comparison of surface potential for the enzyme candidaantarctica lipase B (CAL-B) in tetra-methyl guanidinium nitrate (MCGUA-NO3), in butyl-pentamethyl guanidinium tetrafluoroborate (BAGUA-BF4), and in water. It can be seen that the capacity to screen the charges on the enzyme for IL cations and anions are weaker compared to water, due to the lower dielectric constant of these ILs which renders these liquids medium polarity (Wakai et al. 2005). The slightly higher charge screening in case of MCGUA-NO3 can be explained by taking into account the higher density for this IL which causes more accumulation of ions near the enzyme surface. The authors argue that these behaviors contribute toward the solubility and stability of proteins and enzymes in the IL mixtures (Klahn et al. 2011).
Fig. 4.
Electrostatic potential calculated from the equilibrated MD simulation on the surface of CAL-B in a vacuum, b BAGUA-BF4, c MCGUA-NO3, and d in water. The unit for the electrostatic potential is in kV e. The protein has been kept in the same orientation in all four cases. The electrostatic potential calculation in b to d has involved consideration of the entire solvent. A decreasing pattern for the electrostatic potential can be seen from a to d. Reprinted with permission from Klahn et al. 2011. Copyright 2011 Royal Society of Chemistry
Currently, with a large variety of ions available that can combine to give IL solutions, obtaining a general behavior of ILs with proteins is not straight forward. Several key factors are responsible for the stability and activity of proteins and enzymes in the IL solutions. Details of these factors have been summarized in the reviews by Naushad and co-workers (Naushad et al. 2012) and Gao and co-workers (Gao et al. 2015) Here, we briefly discuss some of these factors.
One of the crucial aspects of proteins in ILs is the interactions of the constituent ions with the amino acids. The effect of ions on proteins in an aqueous media is a well-known subject. It is known that the presence of ions above 0.05 M can exhibit ion-specific behavior on the proteins (Kunz et al. 2004, Collins and Washabaugh 2009). The observation of salt-induced precipitation of hen egg proteins by Hofmeister (1888) have given rise to the anion series that predicts the effect of ions on protein behavior, known as the Hofmeister series. A similar series is available for the cations as well (Collins and Washabaugh 2009). Further developments in this field have used the activity of enzymes also as criteria to characterize these ions (Kunz et al. 2004; Collins and Washabaugh 2009). Similar systemization has been done for the ions that constitute IL mixtures to determine the effect of these ions on the stability and activity of proteins and enzymes (Constantinescu et al. 2007). The study by Constantinescu et al. (2007) used the melting temperature (Tm) of ribonuclease A to characterize the IL cations and anions in aqueous IL solution. IL cations can be ordered as K+ > Na+ > [C1, 1, 1, 1N]+ > Li+ > [C2, 2, 2, 2N]+ ≈ [ emim]+ > [bmpyrr]+ > [bmim]+ ≈ [C3, 3, 3, 3N]+ > [C6mim]+ ≈ [C4, 4, 4, 4N]+ in terms of the decreasing Tm of ribonuclease A. The pattern indicated that the decrease in Tm is larger with the increase in hydrophobicity of the cation (Constantinescu et al. 2007). Similarly, the anions were arranged in the following manner: [SO4]2− > [HPO4]2− > Cl− > [EtOSO3]− > [BF4]− ≈ Br− > [MeOSO3]−>[TfO]− > [SCN]− ≈ [N(CN)2]− > [Tf2N]−. However, the order of the anions is not straight forward and does not strictly follow the above trend.
The anions with their ability to form H-bonds with the amino acids of the proteins provide a unique behavior in modulating the stability of proteins. These characteristics have been observed in the study of activity and stability of lysozyme in the presence of [emim][BF4], [emim][Tf2N], and [emim][Cl] (Noritomi et al. 2011). It was found that the stability and activity are lowest in case of [emim][Cl] solution. The reason for this could be attributed to the ability of Cl− to make H-bonds with the –OH groups of the protein and subsequent breaking of its structure. A similar observation was found in the case of α-amylase from Bacillus amyloliquefaciens and Bacillus lichiniform where lower stability and activity have been observed with [emim][Cl] and [hmim][Cl] (Dabirmanesh et al. 2011).
The other key factor responsible for protein stability and activity in ILs is the length of the alkyl chain attached with the IL cations. Here, we discuss a few examples of this alkyl chain effect on proteins. The stability and kinetics of horseradish peroxidase have been studied in the presence of imidazolium-based ILs with various chain length ([emim][Cl], [bmim][Cl], and [hmim][Cl]) where the [hmim][Cl] was specifically found to reduce the enzyme activity. (Machado et al. 2014) However, these behaviors are not universal. In some cases, the activity and stability were also seen to reduce in case of reduction of alkyl chain length of imidazolium-based cation in the presence of the same anion (Lou and Zong 2006). It was concluded that the variation in viscosity caused by altering the alkyl chain length was an important factor for the stability and activity of proteins (Gao et al. 2015).
The other factors that govern the protein properties in ILs are the amphiphilicity, polarity, viscosity, and a few other properties of ILs (Gao et al. 2015). While these properties originate from the nature of the IL under consideration, water also plays an important role in altering protein behavior in IL-water solution as discussed in the next section.
Water in protein-IL mixture
The solubility of proteins is low in organic solvents and in neat ILs (Kimizuka and Nakashima 2001; Klibanov 2001; Kragl et al. 2002). There are only a few reports of proteins dissolved in anhydrous ILs (Madeira Lau et al. 2000; Kimizuka and Nakashima 2001; Yang et al. 2008; Lin Huang et al. 2011; Lozano et al. 2015). Also, the absence of water in the ILs has resulted in the unstable behavior of enzymes. Different strategies were employed to counteract this behavior which included solid-support immobilization (Lozano et al. 2015) and protein-polymer surfactant nanoconstruct formation (Brogan and Hallett 2016). However, due to the hygroscopic nature of ILs, a completely anhydrous IL solution is very difficult to obtain. Hence, water can be viewed as an integral part of the protein-IL mixture, the properties of which could affect the stability and activity of proteins. Although it is believed that the IL ions remove the water molecules from the surface of the proteins, the study by Micaelo and Soares (Micaêlo and Soares 2008) on a model protein in the presence of two different ILs [bmim][[PF6] and [bmim][NO3] showed the presence of a small fraction of water molecules in the solvation shell of protein. The number of water molecules was found to be higher in case of [bmim][[PF6] compared to [bmim][NO3], which also resulted in higher stability in the case of [bmim][[PF6].(Micaêlo and Soares 2008) The addition of ILs in aqueous solution of proteins was also known to decrease the hydrodynamic radius of proteins. The study by Ghosh et al. (2015) showed that the hydrodynamic radius for lysozyme reduced from 18 Å in pure water to 11 Å in 1.5 M [pmim][Br] solution. For the solvation of the proteins, the cations were found to be more preferred with the solvation extending up to 30 Å from the protein surface (Ghosh et al. 2015). However, there are several reports that provide evidence for the lower stability of proteins in the presence of higher water content. Eckstein et al. (2002) found the activity of α-chymotrypsin to be higher in [bmim] [(CF3SO2)2N] compared to the organic solvents only when the water activity remained low. Similarly, the study by Persson and Bornscheuer (2003) found enhanced enzyme stability in [bmim][PF6] compared to methyl-tert-butylether and n-hexane at low water activity. These observations point to the fact that the modification in the protein stability and activity in IL solutions for proteins originates from the direct interaction of IL ions rather than the changes in hydration shell structure. This observation has been further confirmed by the MD study by Shao (2013) with investigation of structural properties of a protein in the presence of [bmim][Cl] at different concentrations. The study found that the bmim+ ions interact with the negatively charged amino acids of the proteins, while the alkyl chains interact with the non-polar amino acids. The Cl−ions were mostly expelled to the aqueous phase. Also, the removal of water from the hydration shell protects the backbone H-bonds, which further enhances the protein stability (Shao 2013).
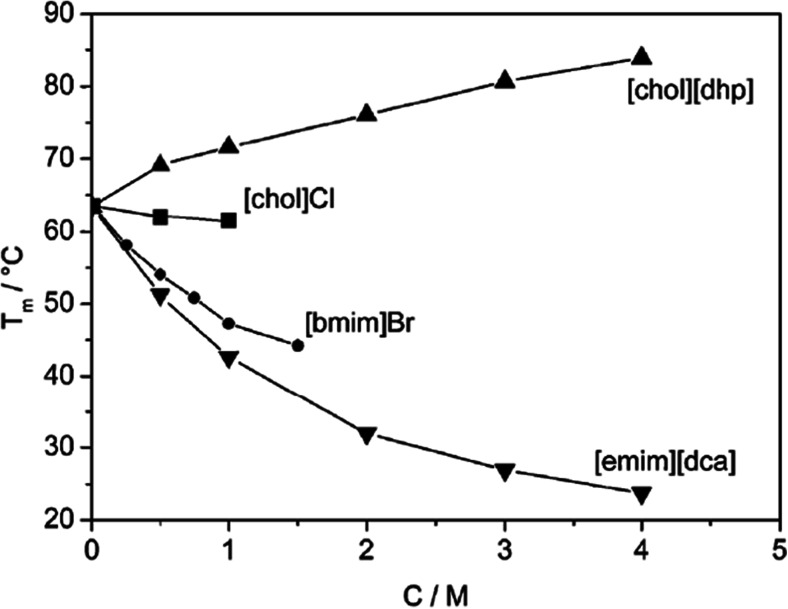
However, explanation for the stability of proteins and enzymes in IL solutions may not always follow a fixed pattern. With the different ways the amino acids of the proteins and IL cations and anions interact with each other and the numerous possibilities to combine these ions to form the ILs make it very difficult to predict the general behavior for a given protein dissolved in an IL a priori. The concentration variation therefore for these ILs can have different behavior for the same protein. This can be explained from the study by Constatinescu et al. (2010) through Fig. 5, where the Tm measurement at different concentrations of the ILs such as choline dihydrogen phosphate ([chol][dhp]), [chol][Cl], 1-ethyl-3-methyl imidazolium dicyanamide ([emim][dca]) and [bmim][Br] for the protein RNaseA have been shown. The figure shows that except for [chol][dhp], all other ILs show denaturation tendency toward the protein. For the case of [chol][dhp], although the hydrophobic organic choline cation is known to impose denaturation, the presence of a strong stabilizing agent such as or [dhp]− can overpower the effect of choline to provide stabilization against denaturation, thus resulting in increase in Tm. Therefore, the stabilization seen for proteins (Fujita et al. 2005; Constatinescu et al. 2010) in the presence of [chol][dhp] mainly originates from the effect of the anion. Further, a number of recent studies have shown that the behavior of anions is often more critical in determining the stability of proteins in IL solutions (Tarannum et al. 2016; Borrell et al. 2017; Diddens et al. 2017). Therefore, it is evident that a better understanding on the general IL anions-protein interactions is crucial for the development of better materials in this field.
Fig. 5.
Concentration dependence of Tm of RNase A for the ILs [chol][dhp], [chol]Cl, [bmim]Br, and [emim][dca], respectively. The protein concentration taken is 0.36 mM and pH is 7.0 for the medium. Reprinted with permission from Constatinescu et al. 2010. Copyright 2010 Royal Society of Chemistry
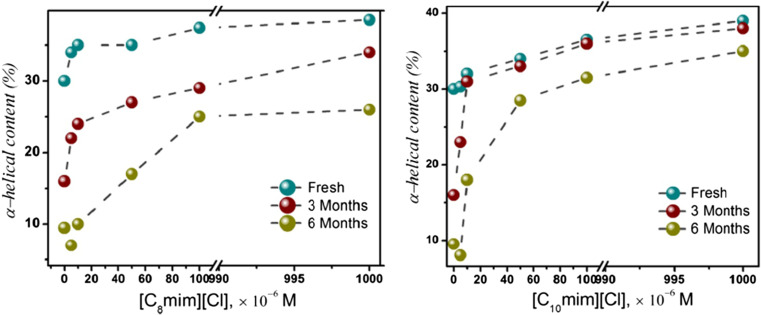
An important aspect for ILs to be useful in the biotechnological applications is not only their ability to stabilize the proteins or enzymes, but also to sustain the stability for a long period of time. Although there have been reports of ILs being able to provide stability for proteins at higher concentration (> 1 M) (Byrne et al. 2007; Fujita et al. 2007), this stability does not guarantee long-term stability (Bihari et al. 2010). However, usage of such high concentrations is not practical because it can hamper the reusability of the protein and can also have an effect on the environment. Further, the ILs at high concentrations has been shown to affect the tertiary structure of the proteins in some cases (Shimojo et al. 2006; Bihari et al. 2010). Therefore, it is desirable to have ILs that can provide long-term stability even at low concentrations. In this regard, the study by Singh et al. (2018) shows an interesting finding by providing evidence of long-term stabilization at room temperature for horse heart cytochrome c (h-cyt c) by employing long-chain imidazolium ILs at low concentrations (1 mM). This study finds that the reason for this stems from the interactions of amino acids with the ILs which makes the loop regions rigid and reduces mobility. The long-term storage capacity of these ILs can be verified by far-UV circular dichroism measurement which gives an idea about the α-helical content in the protein at different time intervals and at different concentrations of the ILs as shown in Fig. 6. It was found that in the case of [C8mim][Cl] and [C10mim][Cl], although the concentrations up to 50 × 10−6 M was not able to protect the secondary structure, the higher concentrations used were capable of protecting the structure to some extent even up to 6 months at room temperature (Fig. 6). The behavior has been explained by considering the interactions between the hydrophobic long chains of the ILs with the hydrophobic amino acids around the heme pocket of h-cyt c (Singh et al. 2018).
Fig. 6.
UV CD spectra showing the percentage of α-helical content in horse heart cytochrome c at 18 × 10−6 M concentration with [C8mim][Cl] and [C10mim][Cl] for fresh sample and the samples stored for 3 and 6 months at room temperature. Reprinted with permission from Singh et al. 2018. Copyright 2018 American Chemical Society
Although the structural stability of proteins and enzymes are a key aspect for ILs for their usage in storage and application purpose, the loss of activity of enzymes in some cases is an issue related to the addition of ILs in aqueous solution of proteins. This has been shown by Jaeger and Pfaendtner (2013) for xylanase where the loss of enzyme activity has been reported due to the dampening of dynamic motions and kinetic trapping of IL cation in the binding pocket.
However, the behavior of IL cations and anions in the aqueous medium of proteins remains an active area of research. The structural changes caused by addition of water in IL solution have implication in the protein behavior in aqueous IL solutions. This can be seen from the difference in behavior shown by proteins to form globular structure or amorphous aggregate depending on the size of the protein in the aqueous IL medium (Takekiyo et al. 2014). It was found that due to the formation of water clusters in the IL mixture, the non-aggregated proteins with radius of gyration below 20 Å interact with these water molecules. On the contrary, the larger proteins with a radius of gyration above 20 Å are not sufficiently hydrated which results in an enhanced protein-protein interaction (Takekiyo et al. 2014). The behavior of different ions also has an effect on whether the ions are present in a neat state or in aqueous medium. For example, while the study by Constantinescu et al. (2007) found the [Tf2N]− ion to be most destabilizing among all the ions studied, it has been found that neat [Tf2N]−-based salts stabilize the enzymes (De Diego et al. 2005). With the advancement in techniques to dissolve proteins in neat IL solutions, the interactions of IL cations and anions with proteins may involve more attention for its application purposes. The reason is that although the Hofmeister series proposed for the IL ions are fairly general when the aqueous mixture of ILs is used, the effect of ions in the cases of neat ILs on proteins is complicated (Zhao 2005). Hence, further studies are required to distinguish the interactions of IL ions with proteins solvated in an aqueous IL mixture and also in neat solutions.
ILs and nucleic acids
Although ILs’ applications on proteins have received more attention, nucleic acid-IL interaction has started drawing interest due to the broadening in the applications of nucleic acids. Recent advancements in the biochemical applications of DNA find its use in molecular computing devices (Burd et al. 2001), in catalysis (Boersma et al. 2010; Park and Sugiyama 2010), and several other areas (Yurke et al. 2000; Kutzler and Weiner 2008). The broadening of the applications of nucleic acid-based materials requires suitable solvents other than water. With the ability to render long-term stability for biomolecules, studies on ILs and nucleic acids provided a larger scope to understand the various aspects of IL ion interactions with DNA, RNA, and G-quadruplex. In the following section, we will discuss some of the recent studies that focus on the mechanism of DNA stabilization in IL solutions and whether there is any role of water in this process.
Interaction of DNA and ILs
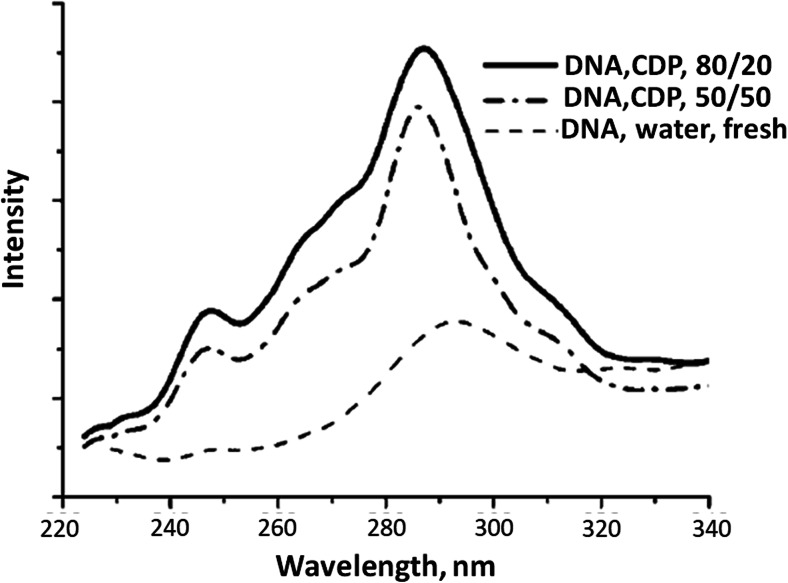
The need for long-term storage of DNA arises for its application in biotechnology and forensic science. Stability of DNA in aqueous medium gets hampered due to the slow hydrolytic reactions that take place in the presence of water (Lindahl and Nyberg 1972). The study by MacFarlane and co-workers (Vijayaraghavan et al. 2010) showed the structural and chemical stability of DNA for up to 1 year in ILs through spectroscopic measurements as demonstrated in Fig. 7. The figure shows that for both concentrations of [chol][dhp] solvent, the characteristic fluorescence emission of DNA at 287 nm corresponding to native DNA structure is seen even after 6 months of the sample preparation at room temperature (Vijayaraghavan et al. 2010). This indicated the usefulness of ILs for the storage purpose of nucleic acids. Therefore, elucidating the nature of interactions taking place between IL ions and DNA base pairs has drawn a large interest.
Fig. 7.
Fluorescence spectra showing the characteristic peak at 287 nm corresponding to stable DNA sample in hydrated [chol][dhp] IL stored at room temperature for 6 months. The more prominent peak for high IL concentration indicates its ability to store DNA sample for longer periods. Reprinted with permission from Vijayaraghavan et al. 2010. Copyright 2010 Wiley
Because of the negative charge of DNA, IL-DNA interaction is mainly dominated by IL cations. The MD and experimental study by Chandran et al. (2012) have dealt with detailed interactions of IL cations and DNA base pairs. The study found that the IL cations not only bind to the negative phosphate group atoms, they also bind to the grooves in the DNA. Also, the prevention of hydrolytic reactions due to the removal of water molecules from the hydration shell by IL cations was argued to be one of the reasons for the long-term stability of DNA (Chandran et al. 2012). Although DNA is known to undergo transition from its most common B-form in water to the A-form due to dehydration (Franklin and Gosling 1953; Ivanov et al. 1974), it has been found that the DNA retains its native B-form in the presence of ILs (Ding et al. 2010). These properties of ILs make them a suitable choice for DNA-based technologies such as DNA translocation (de Zoysa et al. 2009; Kulkarni and Mukherjee 2016) for genome sequencing purpose.
An interesting feature of DNA stability in ILs is the evidence of a stronger interaction between AT base pairs compared to CG bases. Melting temperature study for double stranded DNA carried out in choline dihydrogen phosphate has revealed that the stability of DNA increases with the increase in AT content in the DNA compared to the DNA dissolved in NaCl solution (Tateishi-Karimata and Sugimoto 2012). Several experimental and simulation studies attempted to explore the mechanism underlying this behavior (Ding et al. 2010; Jumbri et al. 2014; Nakano et al. 2014; Portella et al. 2014). The general conclusion obtained from these studies points to the stronger electrostatic interactions between AT bases and the positively charged IL cations. Furthermore, free energy calculation studies carried out by Portella et al. (2014) found that the single strand DNA with GC bases has more preferable solvation energy compared to AT-DNA in IL solution. Overall, the general behavior of ILs with DNA is somewhat similar to proteins, in which case the water in the solvation shell has been found to get replaced by IL ions. Similar stability of G-quadruplex also have been reported recently in the solution of ILs (Fujita and Ohno 2012; Satpathi et al. 2016), with IL cation being found to occupy the core of the quadruplex structure similar to the generally observed Na+ or K+ ions (Satpathi et al. 2016).
Role of water in DNA-IL solution
Due to the difference in the chemical nature of DNA base pairs, the interaction of water is different for AT and CG base pairs. In general, the CG base pairs are known to be more stable in aqueous media owing to its three H-bonds compared to the two H-bonded AT base pair. Moreover, stronger stacking interaction (Parker et al. 2013) and secondary H-bonds also impart the stability (Jorgensen and Pranata 1990). The chemical structure and the local topography of CG bases cause higher water affinity for these bases. This is reinstated by the highest DNA-water interaction energy difference for CG base pairs for transition from B-DNA to A-DNA compared to other base pairs (Kulkarni and Mukherjee 2013). Therefore, aqueous IL solution will modulate different base pairs differently. In this context, we would discuss a recent finding from a study carried out by us to highlight the importance of water in the context of DNA in aqueous IL solutions (Saha et al. 2016).
We have recently studied the mean residence time (MRT) of IL cations near DNA base pairs. MRT is the time required for a solute to escape the solvation shell of a biomolecule. Our previous study on MRT calculation of water in aqueous solution near DNA base pairs (Saha et al. 2015) showed that the MRT values were the highest for the base pairs located in the middle of the 12-base pair DNA. In this case, the chemical nature and the local topography of the base pair have lesser significance. We concluded that the reason for this originates from the diffusion of water molecules along the DNA before escaping the solvation shell. In our MRT study with ILs (Saha et al. 2016), the residence time of IL cations were checked near the middle four base pair of two 12-base pair DNA with variation in bases: in one, the middle four base pairs had AATT and in the other, we had CGCG base pairs. For five different IL combinations, we found that the MRT values of IL cations near CG base pairs were significantly higher compared to the AT base pairs for the 50 wt% IL solution. The values for the MRTs are given in Table 1. This observation was surprising because AT base pairs have stronger interaction with IL cations compared to CG base pairs (Portella et al. 2014).
Table 1.
The mean residence time (MRT) values for IL cations near different bases in the two DNA sequences considered as reported in Ref Saha et al. 2016
| Cations MRTs (ns) | ||||||
|---|---|---|---|---|---|---|
| Ionic liquids | Major groove | Minor groove | Phosphate | |||
| AATT | CGCG | AATT | CGCG | AATT | CGCG | |
| [bmim][Cl] | 6.8 | 77.0 | 8.2 | 50.9 | 7.2 | 25.3 |
| [choline][Cl] | 17.9 | 2.3 ×104 | 20.2 | 2 ×104 | 13.4 | 20.8 |
| [BPYR][Cl] | 4.9 | 79.9 | 6.2 | 99.2 | 5.7 | 61.3 |
| [BPYR][PF6] | 3.5 | 8.1 | 4.1 | 11.1 | 3.6 | 8.1 |
| [BPYR][BF4] | 3.7 | 7.1 | 4.4 | 9.7 | 4.4 | 9.2 |
The reason for this was traced to the higher affinity of water to the CG base pair. We found that water also had higher MRT values near CG base pairs. The reason for this was attributed to the higher interaction energy (Coulomb + vdW) between phosphate atoms of CG base pairs and IL cations. Also, we observed that the CG base pair system was denser compared to the AT base pairs. The higher density of water near the CG base pair could result in a trapped state for water and IL cations which can render lower configuration entropy for these; thus, making the IL cations thermodynamically more unstable. Hence, this study sheds light on the importance of water for studying DNA in aqueous IL solution.
Discussion and conclusions
The healthy progress in the development of ILs provides enormous opportunity for investigating the interaction between IL and biomolecules. Although the commonly used ILs have been thoroughly studied, many of these ILs are not biodegradable and possess high toxicity. (Jordan and Gathergood 2015) Therefore, the progress in the field of ILs and biomolecules could shift to ILs based on the different amino acids and other biomolecules (Tao et al. 2006; Plaquevent et al. 2008; Socha et al. 2014). These developments in this field are likely to keep the experimental and theoretical researchers involved for a significant amount of time in the future. Hence, it is necessary to understand clearly the existing phenomena observed when biomolecules are dissolved in ILs.
The need for better understanding of IL behavior with biomolecules also stems from the various application points of view for proteins, enzymes, and other biomaterials. One of the major usages of ILs and their aqueous solutions has been involved in the field extraction and separation of proteins from biological fluids (Du et al. 2007). The activity shown by several enzymes in these solutions has shown potential in the field of biofuel cells and for bioinspired catalysis (Armand et al. 2009). A recent review by Sivapragasam et al. (2016) discusses various technological advances for ILs required for proteins, enzymes, and DNA-based applications.
In this contribution for the Biophysical reviews, we have focused on the general behavior of biomolecules in ILs and its aqueous solutions and the role of water in these interactions. Due to the vast nature of this field, we have been able to mention only a small fraction of the research work in IL that has come up recently. Also, the field of deep eutectic solvents has been omitted from the discussion in this review since this topic alone can have its own review article. Our survey of the literature indicates that although there has been a plethora of studies concerning the structure and dynamics of ILs in the presence of water, these effects have not been explored a lot for the case of biomolecules dissolved in ILs. Numerous examples have been shown in literature that indicates structural changes in ILs in the presence of water due to the presence of alkyl chains in most IL cations. These effects are likely to affect the behavior of biomolecules as discussed for the few examples here.
Further development in this area can address some of the important questions for the advancement of ILs applications. These are: (i) Is it possible to obtain a balance between storage ability and dampening of protein dynamics in an aqueous IL mixture so that the proteins can retain their functions? (ii) Can the ILs’ effect on biomolecules be modified in such ways that they can be used for in vivo application? (iii) Can the dynamics of water near different amino acids have a role to play in modulating the behavior of proteins in these solutions as seen for DNA? All these questions may require better understanding of ILs and biomolecules in the presence of water. We expect that in the coming years, with advancements in experimental and computational methods, these goals will be soon addressed.
Acknowledgments
This work was funded by DST SERB (EMR/2016/001069).
Abbreviations
- emim
1-Ethyl-3-methylimidazolium
- bmim
1-Butyl-3-methylimidazolium
- pmim
1-Pentyl-3-methylimidazolium
- hmim
1-Hexyl-3-methylimidazolium
- omim
1-Octyl-3-methylimidazolium
- BPYR
1-butylpyridinium
Compliance with ethical standards
Conflict of interest
Debasis Saha declares that he has no conflict of interest. Arnab Mukherjee declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Ionic Liquids and Biomolecules’ edited by Antonio Benedetto and Hans-Joachim Galla
References
- Anthony JL, Maginn EJ, Brennecke JF. Solution thermodynamics of imidazolium-based ionic liquids and water. J Phys Chem B. 2001;105:10942–10949. doi: 10.1021/jp0112368. [DOI] [Google Scholar]
- Araque JC, Daly RP, Margulis CJ. A link between structure, diffusion and rotations of hydrogen bonding tracers in ionic liquids. J Chem Phys. 2016;144:204504. doi: 10.1063/1.4951012. [DOI] [PubMed] [Google Scholar]
- Araque JC, Yadav SK, Shadeck M, Maroncelli M, Margulis CJ. How is diffusion of neutral and charged tracers related to the structure and dynamics of a room-temperature ionic liquid? Large deviations from Stokes–Einstein behavior explained. J Phys Chem B. 2015;119:7015–7029. doi: 10.1021/acs.jpcb.5b01093. [DOI] [PubMed] [Google Scholar]
- Armand M, Endres F, MacFarlane DR, Ohno H, Scrosati B. Ionic-liquid materials for the electrochemical challenges of the future. Nat Mater. 2009;8:621–629. doi: 10.1038/nmat2448. [DOI] [PubMed] [Google Scholar]
- Attri P, Venkatesu P, Kumar A. Activity and stability of [small alpha]-chymotrypsin in biocompatible ionic liquids: enzyme refolding by triethyl ammonium acetate. Phys Chem Chem Phys. 2011;13:2788–2796. doi: 10.1039/C0CP01291B. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Ballone P. Room temperature ionic liquids meet biomolecules: a microscopic view of structure and dynamics. ACS Sustain Chem Eng. 2016;4:392–412. doi: 10.1021/acssuschemeng.5b01385. [DOI] [Google Scholar]
- Bihari M, Russell TP, Hoagland DA. Dissolution and dissolved state of cytochrome c in a neat, hydrophilic ionic liquid. Biomacromolecules. 2010;11:2944–2948. doi: 10.1021/bm100735z. [DOI] [PubMed] [Google Scholar]
- Blesic M, Marques MH, Plechkova NV, Seddon KR, Rebelo LPN, Lopes A. Self-aggregation of ionic liquids: micelle formation in aqueous solution. Green Chem. 2007;9:481–490. doi: 10.1039/B615406A. [DOI] [Google Scholar]
- Boersma AJ, Megens RP, Feringa BL, Roelfes G. DNA-based asymmetric catalysis. Chem Soc Rev. 2010;39:2083–2092. doi: 10.1039/B811349C. [DOI] [PubMed] [Google Scholar]
- Borrell KL, Cancglin C, Stinger BL, et al. An experimental and molecular dynamics study of red fluorescent protein mCherry in novel aqueous amino acid ionic liquids. J Phys Chem B. 2017;121:4823–4832. doi: 10.1021/acs.jpcb.7b03582. [DOI] [PubMed] [Google Scholar]
- Bowers J, Butts CP, Martin PJ, Vergara-Gutierrez MC, Heenan RK. Aggregation behavior of aqueous solutions of ionic liquids. Langmuir. 2004;20:2191–2198. doi: 10.1021/la035940m. [DOI] [PubMed] [Google Scholar]
- Brogan APS, Hallett JP. Solubilizing and stabilizing proteins in anhydrous ionic liquids through formation of protein–polymer surfactant nanoconstructs. J Am Chem Soc. 2016;138:4494–4501. doi: 10.1021/jacs.5b13425. [DOI] [PubMed] [Google Scholar]
- Bruce DW, Cabry CP, Lopes JNC, et al. Nanosegregation and structuring in the bulk and at the surface of ionic-liquid mixtures. J Phys Chem B. 2017;121:6002–6020. doi: 10.1021/acs.jpcb.7b01654. [DOI] [PubMed] [Google Scholar]
- Burd VN, Bantleon R, van Pee K-H. Oxidation of indole and indole derivatives catalyzed by nonheme chloroperoxidases. Appl Biochem Microbiol. 2001;37:248–250. doi: 10.1023/a:1010220916145. [DOI] [Google Scholar]
- Byrne N, Wang L-M, Belieres J-P, Angell CA (2007) Reversible folding-unfolding, aggregation protection, and multi-year stabilization, in high concentration protein solutions, using ionic liquids. Chem Commun:2714–2716. 10.1039/B618943A [DOI] [PubMed]
- Cammarata L, Kazarian SG, Salter PA, Welton T. Molecular states of water in room temperature ionic liquids. Phys Chem Chem Phys. 2001;3:5192–5200. doi: 10.1039/B106900D. [DOI] [Google Scholar]
- Chandran A, Ghoshdastidar D, Senapati S. Groove binding mechanism of ionic liquids: a key factor in long-term stability of DNA in hydrated ionic liquids? J Am Chem Soc. 2012;134:20330–20339. doi: 10.1021/ja304519d. [DOI] [PubMed] [Google Scholar]
- Chatel G, MacFarlane DR. Ionic liquids and ultrasound in combination: synergies and challenges. Chem Soc Rev. 2014;43:8132–8149. doi: 10.1039/C4CS00193A. [DOI] [PubMed] [Google Scholar]
- Collins KD, Washabaugh MW. The Hofmeister effect and the behaviour of water at interfaces. Q Rev Biophys. 2009;18:323–422. doi: 10.1017/S0033583500005369. [DOI] [PubMed] [Google Scholar]
- Constantinescu D, Weingärtner H, Herrmann C. Protein denaturation by ionic liquids and the Hofmeister Series: a case study of aqueous solutions of ribonuclease A. Angew Chem Int Ed. 2007;46:8887–8889. doi: 10.1002/anie.200702295. [DOI] [PubMed] [Google Scholar]
- Constatinescu D, Herrmann C, Weingartner H. Patterns of protein unfolding and protein aggregation in ionic liquids. Phys Chem Chem Phys. 2010;12:1756–1763. doi: 10.1039/B921037G. [DOI] [PubMed] [Google Scholar]
- Cull SG, Holbrey JD, Vargas-Mora V, Seddon KR, Lye GJ. Room-temperature ionic liquids as replacements for organic solvents in multiphase bioprocess operations. Biotechnol Bioeng. 2000;69:227–233. doi: 10.1002/(SICI)1097-0290(20000720)69:2<227::AID-BIT12>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Dabirmanesh B, Daneshjou S, Sepahi AA, et al. Effect of ionic liquids on the structure, stability and activity of two related α-amylases. Int J Biol Macromol. 2011;48:93–97. doi: 10.1016/j.ijbiomac.2010.10.001. [DOI] [PubMed] [Google Scholar]
- De Diego T, Lozano P, Gmouh S, Vaultier M, Iborra JL. Understanding structure−stability relationships of Candida antartica lipase B in ionic liquids. Biomacromolecules. 2005;6:1457–1464. doi: 10.1021/bm049259q. [DOI] [PubMed] [Google Scholar]
- de Zoysa RSS, Jayawardhana DA, Zhao Q, Wang D, Armstrong DW, Guan X. Slowing DNA translocation through nanopores using a solution containing organic salts. J Phys Chem B. 2009;113:13332–13336. doi: 10.1021/jp9040293. [DOI] [PubMed] [Google Scholar]
- Diddens D, Lesch V, Heuer A, Smiatek J. Aqueous ionic liquids and their influence on peptide conformations: denaturation and dehydration mechanisms. Phys Chem Chem Phys. 2017;19:20430–20440. doi: 10.1039/C7CP02897K. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhang L, Xie J, Guo R. Binding characteristics and molecular mechanism of interaction between ionic liquid and DNA. J Phys Chem B. 2010;114:2033–2043. doi: 10.1021/jp9104757. [DOI] [PubMed] [Google Scholar]
- Dominguez de Marıá P, editor. Ionic liquids in biotransformations and organocatalysis: solvents and beyond. Hoboken: John Wiley & Sons, Inc; 2012. [Google Scholar]
- Domínguez-Pérez M, Tomé LIN, Freire MG, Marrucho IM, Cabeza O, Coutinho JAP. (Extraction of biomolecules using) aqueous biphasic systems formed by ionic liquids and aminoacids. Sep Purif Technol. 2010;72:85–91. doi: 10.1016/j.seppur.2010.01.008. [DOI] [Google Scholar]
- Du Z, Yu Y-L, Wang J-H. Extraction of proteins from biological fluids by use of an ionic liquid/aqueous two-phase system. Chem Eur J. 2007;13:2130–2137. doi: 10.1002/chem.200601234. [DOI] [PubMed] [Google Scholar]
- Dupont J, Scholten JD. On the structural and surface properties of transition-metal nanoparticles in ionic liquids. Chem Soc Rev. 2010;39:1780–1804. doi: 10.1039/B822551F. [DOI] [PubMed] [Google Scholar]
- Earle Martyn J, Seddon Kenneth R. Ionic liquids. Green solvents for the future. Pure Appl Chem. 2000;72:1391. doi: 10.1351/pac200072071391. [DOI] [Google Scholar]
- Eckstein M, Sesing M, Kragl U, Adlercreutz P. At low water activity α-chymotrypsin is more active in an ionic liquid than in non-ionic organic solvents. Biotechnol Lett. 2002;24:867–872. doi: 10.1023/a:1015564608261. [DOI] [Google Scholar]
- Egorova KS, Gordeev EG, Ananikov VP. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem Rev. 2017;117:7132–7189. doi: 10.1021/acs.chemrev.6b00562. [DOI] [PubMed] [Google Scholar]
- Erbeldinger M, Mesiano AJ, Russell AJ. Enzymatic catalysis of formation of Z-aspartame in ionic liquid—an alternative to enzymatic catalysis in organic solvents. Biotechnol Prog. 2000;16:1129–1131. doi: 10.1021/bp000094g. [DOI] [PubMed] [Google Scholar]
- Fedorov MV, Kornyshev AA. Towards understanding the structure and capacitance of electrical double layer in ionic liquids. Electrochim Acta. 2008;53:6835–6840. doi: 10.1016/j.electacta.2008.02.065. [DOI] [Google Scholar]
- Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- Freire MG, Neves CMSS, Marrucho IM, Canongia Lopes JN, Rebelo LPN, Coutinho JAP. High-performance extraction of alkaloids using aqueous two-phase systems with ionic liquids. Green Chem. 2010;12:1715–1718. doi: 10.1039/C0GC00179A. [DOI] [Google Scholar]
- Fujita K, MacFarlane DR, Forsyth M (2005) Protein solubilising and stabilising ionic liquids. Chem Commun:4804–4806. 10.1039/B508238B [DOI] [PubMed]
- Fujita K, MacFarlane DR, Forsyth M, et al. Solubility and stability of cytochrome c in hydrated ionic liquids: effect of Oxo acid residues and kosmotropicity. Biomacromolecules. 2007;8:2080–2086. doi: 10.1021/bm070041o. [DOI] [PubMed] [Google Scholar]
- Fujita K, Ohno H. Stable G-quadruplex structure in a hydrated ion pair: cholinium cation and dihydrogen phosphate anion. Chem Commun. 2012;48:5751–5753. doi: 10.1039/C2CC30554B. [DOI] [PubMed] [Google Scholar]
- Gao W-W, Zhang F-X, Zhang G-X, Zhou C-H. Key factors affecting the activity and stability of enzymes in ionic liquids and novel applications in biocatalysis. Biochem Eng J. 2015;99:67–84. doi: 10.1016/j.bej.2015.03.005. [DOI] [Google Scholar]
- Ghosh S, Parui S, Jana B, Bhattacharyya K. Ionic liquid induced dehydration and domain closure in lysozyme: FCS and MD simulation. J Chem Phys. 2015;143:125103. doi: 10.1063/1.4931974. [DOI] [PubMed] [Google Scholar]
- Hayes R, Warr GG, Atkin R. Structure and nanostructure in ionic liquids. Chem Rev. 2015;115:6357–6426. doi: 10.1021/cr500411q. [DOI] [PubMed] [Google Scholar]
- He Y, Li Z, Simone P, Lodge TP. Self-assembly of block copolymer micelles in an ionic liquid. J Am Chem Soc. 2006;128:2745–2750. doi: 10.1021/ja058091t. [DOI] [PubMed] [Google Scholar]
- Hofmeister F. Zur Lehre von der Wirkung der Salze. Arch Exp Pathol Pharmakol. 1888;24:247–260. doi: 10.1007/BF01918191. [DOI] [Google Scholar]
- Huddleston JG, Visser AE, Reichert WM, Willauer HD, Broker GA, Rogers RD. Characterization and comparison of hydrophilic and hydrophobic room temperature ionic liquids incorporating the imidazolium cation. Green Chem. 2001;3:156–164. doi: 10.1039/B103275P. [DOI] [Google Scholar]
- Itoh T. Ionic liquids as tool to improve enzymatic organic synthesis. Chem Rev. 2017;117:10567–10607. doi: 10.1021/acs.chemrev.7b00158. [DOI] [PubMed] [Google Scholar]
- Ivanov VI, Minchenkova LE, Minyat EE, Frank-Kamenetskii MD, Schyolkina AK. The B̄ to Ā transition of DNA in solution. J Mol Biol. 1974;87:817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- Jaeger VW, Pfaendtner J. Structure, dynamics, and activity of xylanase solvated in binary mixtures of ionic liquid and water. ACS Chem Biol. 2013;8:1179–1186. doi: 10.1021/cb3006837. [DOI] [PubMed] [Google Scholar]
- Jens S. Aqueous ionic liquids and their effects on protein structures: an overview on recent theoretical and experimental results. J Phys Condens Matter. 2017;29:233001. doi: 10.1088/1361-648X/aa6c9d. [DOI] [PubMed] [Google Scholar]
- Jeong S, Ha SH, Han S-H, et al. Elucidation of molecular interactions between lipid membranes and ionic liquids using model cell membranes. Soft Matter. 2012;8:5501–5506. doi: 10.1039/C2SM25223F. [DOI] [Google Scholar]
- Jha I, Venkatesu P. Endeavour to simplify the frustrated concept of protein-ammonium family ionic liquid interactions. Phys Chem Chem Phys. 2015;17:20466–20484. doi: 10.1039/C5CP01735A. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang Y, Voth GA. Molecular dynamics simulation of nanostructural organization in ionic liquid/water mixtures. J Phys Chem B. 2007;111:4812–4818. doi: 10.1021/jp067142l. [DOI] [PubMed] [Google Scholar]
- Jordan A, Gathergood N. Biodegradation of ionic liquids—a critical review. Chem Soc Rev. 2015;44:8200–8237. doi: 10.1039/C5CS00444F. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Pranata J. Importance of secondary interactions in triply hydrogen bonded complexes: guanine-cytosine vs uracil-2,6-diaminopyridine. J Am Chem Soc. 1990;112:2008–2010. doi: 10.1021/ja00161a061. [DOI] [Google Scholar]
- Jumbri K, Abdul Rahman MB, Abdulmalek E, Ahmad H, Micaelo NM. An insight into structure and stability of DNA in ionic liquids from molecular dynamics simulation and experimental studies. Phys Chem Chem Phys. 2014;16:14036–14046. doi: 10.1039/C4CP01159G. [DOI] [PubMed] [Google Scholar]
- Kaar JL, Jesionowski AM, Berberich JA, Moulton R, Russell AJ. Impact of ionic liquid physical properties on lipase activity and stability. J Am Chem Soc. 2003;125:4125–4131. doi: 10.1021/ja028557x. [DOI] [PubMed] [Google Scholar]
- Kaintz A, Baker G, Benesi A, Maroncelli M. Solute diffusion in ionic liquids, NMR measurements and comparisons to conventional solvents. J Phys Chem B. 2013;117:11697–11708. doi: 10.1021/jp405393d. [DOI] [PubMed] [Google Scholar]
- Kim K-W, Song B, Choi M-Y, Kim M-J. Biocatalysis in ionic liquids: markedly enhanced enantioselectivity of lipase. Org Lett. 2001;3:1507–1509. doi: 10.1021/ol015824f. [DOI] [PubMed] [Google Scholar]
- Kimizuka N, Nakashima T. Spontaneous self-assembly of glycolipid bilayer membranes in sugar-philic ionic liquids and formation of ionogels. Langmuir. 2001;17:6759–6761. doi: 10.1021/la015523e. [DOI] [Google Scholar]
- Klahn M, Lim GS, Seduraman A, Wu P. On the different roles of anions and cations in the solvation of enzymes in ionic liquids. Phys Chem Chem Phys. 2011;13:1649–1662. doi: 10.1039/C0CP01509A. [DOI] [PubMed] [Google Scholar]
- Klibanov AM. Improving enzymes by using them in organic solvents. Nature. 2001;409:241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- Kohno Y, Ohno H. Ionic liquid/water mixtures: from hostility to conciliation. Chem Commun. 2012;48:7119–7130. doi: 10.1039/C2CC31638B. [DOI] [PubMed] [Google Scholar]
- Kowsari MH, Alavi S, Ashrafizaadeh M, Najafi B (2008) Molecular dynamics simulation of imidazolium-based ionic liquids. I. Dynamics and diffusion coefficient. J Chem Phys 129:224508. doi: 10.1063/1.3035978 [DOI] [PubMed]
- Kragl U, Eckstein M, Kaftzik N. Enzyme catalysis in ionic liquids. Curr Opin Biotechnol. 2002;13:565–571. doi: 10.1016/S0958-1669(02)00353-1. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Mukherjee A. Sequence dependent free energy profiles of localized B- to A-form transition of DNA in water. J Chem Phys. 2013;139:155102. doi: 10.1063/1.4825175. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Mukherjee A. Ionic liquid prolongs DNA translocation through graphene nanopores. RSC Adv. 2016;6:46019–46029. doi: 10.1039/C6RA07017E. [DOI] [Google Scholar]
- Kunz W, Lo Nostro P, Ninham BW. The present state of affairs with Hofmeister effects. Curr Opin Colloid Interface Sci. 2004;9:1–18. doi: 10.1016/j.cocis.2004.05.004. [DOI] [Google Scholar]
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G, Watson PR. Surface tension measurements of N-Alkylimidazolium ionic liquids. Langmuir. 2001;17:6138–6141. doi: 10.1021/la010629v. [DOI] [Google Scholar]
- Lee SH, NgocDoan TT, HoHa S, Chang W-J, Koo Y-M. Influence of ionic liquids as additives on sol−gel immobilized lipase. J Mol Catal B Enzym. 2007;47:129–134. doi: 10.1016/j.molcatb.2007.05.002. [DOI] [Google Scholar]
- Lei Z, Dai C, Chen B. Gas solubility in ionic liquids. Chem Rev. 2014;114:1289–1326. doi: 10.1021/cr300497a. [DOI] [PubMed] [Google Scholar]
- Lin Huang J, Noss ME, Schmidt KM, Murray L, Bunagan MR. The effect of neat ionic liquid on the folding of short peptides. Chem Commun. 2011;47:8007–8009. doi: 10.1039/C1CC11527H. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhou G, He H, Zhang X, Wang J, Zhang S. Rodlike micelle structure and formation of ionic liquid in aqueous solution by molecular simulation. Ind Eng Chem Res. 2015;54:1681–1688. doi: 10.1021/ie503109z. [DOI] [Google Scholar]
- Lou W-Y, Zong M-H. Efficient kinetic resolution of (R,S)-1-trimethylsilylethanol via lipase-mediated enantioselective acylation in ionic liquids. Chirality. 2006;18:814–821. doi: 10.1002/chir.20307. [DOI] [PubMed] [Google Scholar]
- Lozano P, Bernal JM, Garcia-Verdugo E, et al. Sponge-like ionic liquids: a new platform for green biocatalytic chemical processes. Green Chem. 2015;17:3706–3717. doi: 10.1039/C5GC00894H. [DOI] [Google Scholar]
- Machado MF, Queirós RP, Santos MD, Fidalgo LG, Delgadillo I, Saraiva JA. Effect of ionic liquids alkyl chain length on horseradish peroxidase thermal inactivation kinetics and activity recovery after inactivation. World J Microbiol Biotechnol. 2014;30:487–494. doi: 10.1007/s11274-013-1466-2. [DOI] [PubMed] [Google Scholar]
- Madeira Lau R, Sorgedrager MJ, Carrea G, van Rantwijk F, Secundo F, Sheldon RA. Dissolution of Candida antarctica lipase B in ionic liquids: effects on structure and activity. Green Chem. 2004;6:483–487. doi: 10.1039/B405693K. [DOI] [Google Scholar]
- Madeira Lau R, Van Rantwijk F, Seddon KR, Sheldon RA. Lipase-catalyzed reactions in ionic liquids. Org Lett. 2000;2:4189–4191. doi: 10.1021/ol006732d. [DOI] [PubMed] [Google Scholar]
- Méndez-Morales T, Carrete J, Cabeza Ó, Gallego LJ, Varela LM. Molecular dynamics simulation of the structure and dynamics of water–1-Alkyl-3-methylimidazolium ionic liquid mixtures. J Phys Chem B. 2011;115:6995–7008. doi: 10.1021/jp202692g. [DOI] [PubMed] [Google Scholar]
- Micaêlo NM, Soares CM. Protein structure and dynamics in ionic liquids. Insights from molecular dynamics simulation studies. J Phys Chem B. 2008;112:2566–2572. doi: 10.1021/jp0766050. [DOI] [PubMed] [Google Scholar]
- Moon YH, Lee SM, Ha SH, Koo Y-M. Enzyme-catalyzed reactions in ionic liquids. Korean J Chem Eng. 2006;23:247–263. doi: 10.1007/BF02705724. [DOI] [Google Scholar]
- Nakano M, Tateishi-Karimata H, Tanaka S, Sugimoto N. Choline ion interactions with DNA atoms explain unique stabilization of A–T Base pairs in DNA duplexes: a microscopic view. J Phys Chem B. 2014;118:379–389. doi: 10.1021/jp406647b. [DOI] [PubMed] [Google Scholar]
- Naushad M, Alothman ZA, Khan AB, Ali M. Effect of ionic liquid on activity, stability, and structure of enzymes: a review. Int J Biol Macromol. 2012;51:555–560. doi: 10.1016/j.ijbiomac.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Noritomi H, Minamisawa K, Kamiya R, Kato S. Thermal stability of proteins in the presence of aprotic ionic liquids. J Biomed Sci Eng. 2011;4:94–99. doi: 10.4236/jbise.2011.42013. [DOI] [Google Scholar]
- Park S, Sugiyama H. DNA-based hybrid catalysts for asymmetric organic synthesis. Angew Chem Int Ed. 2010;49:3870–3878. doi: 10.1002/anie.200905382. [DOI] [PubMed] [Google Scholar]
- Parker TM, Hohenstein EG, Parrish RM, Hud NV, Sherrill CD. Quantum-mechanical analysis of the energetic contributions to π stacking in nucleic acids versus rise, twist, and slide. J Am Chem Soc. 2013;135:1306–1316. doi: 10.1021/ja3063309. [DOI] [PubMed] [Google Scholar]
- Pârvulescu VI, Hardacre C. Catalysis in ionic liquids. Chem Rev. 2007;107:2615–2665. doi: 10.1021/cr050948h. [DOI] [PubMed] [Google Scholar]
- Persson M, Bornscheuer UT. Increased stability of an esterase from Bacillus stearothermophilus in ionic liquids as compared to organic solvents. J Mol Catal B Enzym. 2003;22:21–27. doi: 10.1016/S1381-1177(02)00294-1. [DOI] [Google Scholar]
- Plaquevent J-C, Levillain J, Guillen F, Malhiac C, Gaumont A-C. Ionic liquids: new targets and media for α-amino acid and peptide chemistry. Chem Rev. 2008;108:5035–5060. doi: 10.1021/cr068218c. [DOI] [PubMed] [Google Scholar]
- Portella G, Germann MW, Hud NV, Orozco M. MD and NMR analyses of choline and TMA binding to duplex DNA: on the origins of aberrant sequence-dependent stability by alkyl cations in aqueous and water-free solvents. J Am Chem Soc. 2014;136:3075–3086. doi: 10.1021/ja410698u. [DOI] [PubMed] [Google Scholar]
- Price DL, Borodin O, González MA, et al. Relaxation in a prototype ionic liquid: influence of water on the dynamics. J Phys Chem Lett. 2017;8:715–719. doi: 10.1021/acs.jpclett.6b02871. [DOI] [PubMed] [Google Scholar]
- Rivera-Rubero S, Baldelli S. Influence of water on the surface of hydrophilic and hydrophobic room-temperature ionic liquids. J Am Chem Soc. 2004;126:11788–11789. doi: 10.1021/ja0464894. [DOI] [PubMed] [Google Scholar]
- Saha D, Kulkarni M, Mukherjee A. Water modulates the ultraslow dynamics of hydrated ionic liquids near CG rich DNA: consequences for DNA stability. Phys Chem Chem Phys. 2016;18:32107–32115. doi: 10.1039/C6CP05959G. [DOI] [PubMed] [Google Scholar]
- Saha D, Supekar S, Mukherjee A. Distribution of residence time of water around DNA base pairs: governing factors and the origin of heterogeneity. J Phys Chem B. 2015;119:11371–11381. doi: 10.1021/acs.jpcb.5b03553. [DOI] [PubMed] [Google Scholar]
- Satpathi S, Kulkarni M, Mukherjee A, Hazra P. Ionic liquid induced G-quadruplex formation and stabilization: spectroscopic and simulation studies. Phys Chem Chem Phys. 2016;18:29740–29746. doi: 10.1039/C6CP05732B. [DOI] [PubMed] [Google Scholar]
- Schröder C. Proteins in ionic liquids: current status of experiments and simulations. Top Curr Chem. 2017;375:25. doi: 10.1007/s41061-017-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q. On the influence of hydrated imidazolium-based ionic liquid on protein structure stability: a molecular dynamics simulation study. J Chem Phys. 2013;139:115102. doi: 10.1063/1.4821588. [DOI] [PubMed] [Google Scholar]
- Sharma A, Ghorai PK. Effect of water on structure and dynamics of [BMIM][PF6] ionic liquid: an all-atom molecular dynamics simulation investigation. J Chem Phys. 2016;144:114505. doi: 10.1063/1.4944083. [DOI] [PubMed] [Google Scholar]
- Sheldon R (2001) Catalytic reactions in ionic liquids. Chem Commun:2399–2407. 10.1039/B107270F [DOI] [PubMed]
- Shimojo K, Kamiya N, Tani F, Naganawa H, Naruta Y, Goto M. Extractive solubilization, structural change, and functional conversion of cytochrome c in ionic liquids via crown ether complexation. Anal Chem. 2006;78:7735–7742. doi: 10.1021/ac0612877. [DOI] [PubMed] [Google Scholar]
- Singh UK, Kumari M, Khan SH, Bohidar HB, Patel R. Mechanism and dynamics of long-term stability of cytochrome c conferred by long-chain imidazolium ionic liquids at low concentration. ACS Sustain Chem Eng. 2018;6:803–815. doi: 10.1021/acssuschemeng.7b03168. [DOI] [Google Scholar]
- Sivapragasam M, Moniruzzaman M, Goto M. Recent advances in exploiting ionic liquids for biomolecules: solubility, stability and applications. Biotechnol J. 2016;11:1000–1013. doi: 10.1002/biot.201500603. [DOI] [PubMed] [Google Scholar]
- Socha AM, Parthasarathi R, Shi J, et al. Efficient biomass pretreatment using ionic liquids derived from lignin and hemicellulose. Proc Natl Acad Sci U S A. 2014;111:E3587–E3595. doi: 10.1073/pnas.1405685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers CA, Flowers RA. Protein renaturation by the liquid organic salt ethylammonium nitrate. Protein Sci. 2000;9:2001–2008. doi: 10.1110/ps.9.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekiyo T, Fukudome K, Yamazaki K, Abe H, Yoshimura Y. Protein aggregation and partial globular state in aqueous 1-alkyl-3-methylimidazolium nitrate solutions. Chem Phys Lett. 2014;602:22–27. doi: 10.1016/j.cplett.2014.03.089. [DOI] [Google Scholar]
- Tang S, Baker GA, Zhao H. Ether- and alcohol-functionalized task-specific ionic liquids: attractive properties and applications. Chem Soc Rev. 2012;41:4030–4066. doi: 10.1039/C2CS15362A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao G-h, He L, Liu W-s, et al. Preparation, characterization and application of amino acid-based green ionic liquids. Green Chem. 2006;8:639–646. doi: 10.1039/B600813E. [DOI] [Google Scholar]
- Tarannum A, Muvva C, Mehta A, Raghava Rao J, Fathima NN. Role of preferential ions of ammonium ionic liquid in destabilization of collagen. J Phys Chem B. 2016;120:6515–6524. doi: 10.1021/acs.jpcb.6b02723. [DOI] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Sugimoto N. A–T base pairs are more stable than G–C base pairs in a hydrated ionic liquid. Angew Chem Int Ed. 2012;51:1416–1419. doi: 10.1002/anie.201106423. [DOI] [PubMed] [Google Scholar]
- Tomé LIN, Catambas VR, Teles ARR, Freire MG, Marrucho IM, Coutinho JAP. Tryptophan extraction using hydrophobic ionic liquids. Sep Purif Technol. 2010;72:167–173. doi: 10.1016/j.seppur.2010.02.002. [DOI] [Google Scholar]
- Torriero AA. Electrochemistry in ionic liquids. Berlin-Heidelberg: Springer-Verlag; 2015. [Google Scholar]
- Torriero AA. Electrochemistry in ionic liquids. Berlin-Heidelberg: Springer-Verlag; 2015. [Google Scholar]
- Triolo A, Russina O, Bleif H-J, Di Cola E. Nanoscale segregation in room temperature ionic liquids. J Phys Chem B. 2007;111:4641–4644. doi: 10.1021/jp067705t. [DOI] [PubMed] [Google Scholar]
- van Rantwijk F, Sheldon RA. Biocatalysis in ionic liquids. Chem Rev. 2007;107:2757–2785. doi: 10.1021/cr050946x. [DOI] [PubMed] [Google Scholar]
- Ventura SPM, e Silva FA, Quental MV, Mondal D, Freire MG, Coutinho JAP. Ionic-liquid-mediated extraction and separation processes for bioactive compounds: past, present, and future trends. Chem Rev. 2017;117:6984–7052. doi: 10.1021/acs.chemrev.6b00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura SPM, Sousa SG, Freire MG, Serafim LS, Lima ÁS, Coutinho JAP. Design of ionic liquids for lipase purification. J Chromatogr B. 2011;879:2679–2687. doi: 10.1016/j.jchromb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Vicent-Luna JM, Romero-Enrique JM, Calero S, Anta JA. Micelle formation in aqueous solutions of room temperature ionic liquids: a molecular dynamics study. J Phys Chem B. 2017;121:8348–8358. doi: 10.1021/acs.jpcb.7b05552. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan R, Izgorodin A, Ganesh V, Surianarayanan M, MacFarlane DR. Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew Chem Int Ed. 2010;49:1631–1633. doi: 10.1002/anie.200906610. [DOI] [PubMed] [Google Scholar]
- Wakai C, Oleinikova A, Ott M, Weingärtner H. How polar are ionic liquids? Determination of the static dielectric constant of an imidazolium-based ionic liquid by microwave dielectric spectroscopy. J Phys Chem B. 2005;109:17028–17030. doi: 10.1021/jp053946+. [DOI] [PubMed] [Google Scholar]
- Wasserscheid P, Keim W. Ionic liquids—new “solutions” for transition metal catalysis. Angew Chem Int Ed. 2000;39:3772–3789. doi: 10.1002/1521-3773(20001103)39:21<3772::AID-ANIE3772>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Weingärtner H. Understanding ionic liquids at the molecular level: facts, problems, and controversies. Angew Chem Int Ed. 2008;47:654–670. doi: 10.1002/anie.200604951. [DOI] [PubMed] [Google Scholar]
- Welton T. Room-temperature ionic liquids. Solvents for synthesis and catalysis. Chem Rev. 1999;99:2071–2084. doi: 10.1021/cr980032t. [DOI] [PubMed] [Google Scholar]
- Wilkes JS, Zaworotko MJ (1992) Air and water stable 1-ethyl-3-methylimidazolium based ionic liquids. J Chem Soc Chem Commun :965–967. doi: 10.1039/C39920000965
- Yang Z, Yue Y-J, Xing M. Tyrosinase activity in ionic liquids. Biotechnol Lett. 2008;30:153–158. doi: 10.1007/s10529-007-9505-4. [DOI] [PubMed] [Google Scholar]
- Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Zein El Abedin S, Endres F. Electrodeposition of metals and semiconductors in air- and water-stable ionic liquids. ChemPhysChem. 2006;7:58–61. doi: 10.1002/cphc.200500288. [DOI] [PubMed] [Google Scholar]
- Zhao H. Effect of ions and other compatible solutes on enzyme activity, and its implication for biocatalysis using ionic liquids. J Mol Catal B Enzym. 2005;37:16–25. doi: 10.1016/j.molcatb.2005.08.007. [DOI] [Google Scholar]
- Zhao H, Xia S, Ma P. Use of ionic liquids as ‘green’ solvents for extractions. J Chem Technol Biotechnol. 2005;80:1089–1096. doi: 10.1002/jctb.1333. [DOI] [Google Scholar]