Abstract
Amyloid aggregates are composed of protein fibrils with a dominant β-sheet structure, are water-insoluble, and are involved in the pathogenesis of many neurodegenerative diseases. Development of pharmaceuticals to treat these diseases and the design of recovery agents for amyloid-type inclusion bodies require the successful suppression and dissolution of such aggregates. Since ionic liquids (ILs) are composed of both a cation and anion and are known to suppress protein aggregation and to dissolve water-insoluble compounds such as cellulose; they may also have potential use as suppression/dissolution agents for amyloid aggregates. In the following review, we present the suppression and dissolution effects of ILs on amyloid aggregates so far reported. The protein–IL affinity (the ability of ILs to interact with amyloid proteins) was found to be the biochemical basis for ILs’ suppression of amyloid formation, and the hydrogen-bonding basicity of ILs might be the basis for their ability to dissolve amyloid aggregates. These findings present the potential of ILs to serve as novel pharmaceuticals to treat neurodegenerative diseases and as recovery agents for various amyloid aggregates.
Keywords: Amyloid aggregates, Suppression, Dissolution, Ionic liquids, Protein–IL interaction, Solution structure
Introduction
Amyloid aggregates are composed of protein fibrils with a dominant β-sheet structure and are formed by ordered aggregation of misfolded proteins. Such aggregates are involved in the pathogenesis of many neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases, and are transported to amyloid-type inclusion bodies in response to the overexpression of recombinant proteins (De Groot et al. 2009; Singh et al. 2015). It has been suggested that organic compounds with low formula weights, such as guanidine salts and dimethyl sulfoxide (DMSO), can suppress the formation of amyloid aggregates by dissolving them (Singh et al. 2015; Meersman and Dobson 2006). Electrostatic interactions between protein side chains, such as salt bridges, can influence the amyloid stability by the addition of organic compounds (Shammas et al. 2011). Besides, the basicity of the –S=O group in DMSO may disrupt the intermolecular hydrogen bonding (H-bonding) (Hirota-Nakaoka et al. 2003). Amyloid stability is thought to be effectively controlled by the application of compounds that destabilize these intermolecular interactions such as H-bonding and electrostatic/hydrophobic interactions between proteins, thereby aiding in the suppression and dissolution of amyloid aggregates.
Ionic liquids (ILs) comprising a cation and an anion have shown unique physical and chemical properties, such as negligible vapor pressure and high miscibility with other liquids (Hayes et al. 2015). ILs can be varied to produce solvents with different chaotropic and kosmotropic properties, and these properties are critical in determining the aqueous solubility and protein stability of the solute (Zhang and Cremer 2006). Thus, aqueous mixtures of proteins and ILs play an important role in protein engineering applications, such as protein storage media and biocatalysts, and in the pharmaceutical and biomedical sciences (Greaves and Drummond 2015; Benedetto and Ballone 2015a, b; Egorova et al. 2017). An intriguing feature of aqueous IL solutions is that some ILs cause protein unfolding (Tietze et al. 2013), while others positively influence protein refolding (Summers and Flowers II 2000; Takekiyo et al. 2012). Also intriguing is that the dissolution and regeneration of cellulose and aggregated recombinant proteins have been demonstrated using ILs (Hauru et al. 2012; Fujita et al. 2016). These features imply that ILs may destabilize amyloid aggregates, as the formation and stability of these aggregates are dependent upon the same intramolecular forces present in amyloid aggregates (H-bonding and electrostatic/hydrophobic interactions).
In the following review, we present current information regarding the suppression/dissolution effects of aqueous ILs on amyloid aggregates in solution, giving consideration to protein–IL interactions and solution properties of this media.
Interactions between ionic liquids and proteins
Protein–IL interactions have been the subject of intensive study, primarily the use of ILs as solvents for biochemical reactions such as protein folding/unfolding and protein aggregation (Zhang and Cremer 2006; Patel et al. 2014; Benedetto and Ballone 2015a, b). In fact, protein–IL interactions involving ethylammonium nitrate (EAN) and 1-ethyl-3-methylimidazolium chloride demonstrated a positive effect on protein renaturation (Summers and Flowers II 2000; Tischer et al. 2014). To understand the mechanisms of protein structural changes in aqueous IL solutions, we must first understand protein–IL interactions.
The interactions between human serum albumin and 1-butyl-3-methylimidazolium tetrafluoroborate ([bmim][BF4]) have a denaturing effect, and protein–choline dihydrogen phosphate ([Chl][dhp]) interactions have a stabilizing effect (Akdogan et al. 2011). When bovine serum albumin is combined in aqueous solution with imidazolium-based ILs, these cations localize around the hydrophobic residues such as Phe and Leu, and the NO3− anion strongly interacting with the protein, by entering the protein interior and disrupting its α-helical structure (Shu et al. 2011). Imidazolium-based ILs with longer alkyl chains have been demonstrated to preferentially interact with hydrophobic moieties, resulting in serious distortions in the protein conformation (Silva et al. 2014). Recently, interaction sites of protein with ILs in concentrated solution have been reported (Takekiyo et al. 2015, 2016; Yoshimura et al. 2016). For instance, [bmim][SCN] interacts with proteins, primarily at Lys and Arg residues, and a higher quantity of these residues induced the suppression of intermolecular β-sheet formation due to the specific amino acid residue–IL interaction (Takekiyo et al. 2015). The interaction sites of strong kosmotropic ions such SCN− anion with proteins in concentrated aqueous media are consistent with those in the crystalline state (Hamiaux et al. 1999; Vaney et al. 2001), because the protein–kosmotropic ion interactions are stronger than those of the chaotropic ions.
Based on these results, protein–IL interaction depends on a variety of factors that induce changes in protein free energy and the cation and anion species of ILs, as well as their position in the Hofmeister series (Zhang and Cremer 2006). These factors play important roles in protein stability. Some previously determined interaction sites in proteins with ILs dissolved in aqueous solution are summarized in Table 1.
Table 1.
Interaction sites of proteins with ionic liquids
| Ionic liquids | Interaction sites | Methods | References |
|---|---|---|---|
| [dBmim][Cl] | Phe, Ala, Leu, Val, and Ser residues | Molecular docking | Shu et al. 2011 |
| [bmim]-based ILs | Phe, Leu, Val, and Ser residues (NO3−: disruption of α-helix) | Molecular docking | Shu et al. 2011 |
| [bmim][SCN] | Lys, Arg, and Glu residues | FTIR | Takekiyo et al. 2015 |
| [bmim][Cl] | Phe, Tyr, and Pro residues | FTIR | Yoshimura et al. 2016 |
| EAN | Ala and Val residues | FTIR | Takekiyo et al. 2016 |
| PAN | Ser, Ala, Lys, Ile, and Val residues | FTIR | Takekiyo et al. 2016 |
Liquid nano-heterogeneity and protein state in ionic liquid solutions
In addition to protein–IL interactions, the effect of ILs on protein stability is also a function of their liquid structure. IL solutions of x > 20 mol% IL often induce formation of unique protein states, such as amyloid-type aggregation and helical structure (Takekiyo et al. 2013; Weaver et al. 2012), and it has been suggested that such unique protein states in IL solutions are strongly related to the unique liquid structure with heterogeneous domains (Takekiyo et al. 2014).
It is known that ILs adopt a nano-heterogeneous structure, with a polar domain arising from the interaction of the IL’s ionic parts, and a nonpolar domain containing the alkyl chains (Hayes et al. 2015). These binary solutions formed IL-aggregated structures that are surrounded by bulk water molecules under water-rich conditions (lower nano-heterogeneity) (Jiang et al. 2007). However, under IL-rich conditions, the water molecules are scattered in the polar domains and self-assemble in the ILs (higher nano-heterogeneity) (Jiang et al. 2007). Moreover, an increase in alkyl chain length results in an increase in nano-heterogeneity (Hayes et al. 2015). It has been suggested that proteins in IL solutions of 70 vol% are hydrated by water molecules surrounded by IL layers (Tietze et al. 2013). Related to this, a decrease in the cationic alkyl chain length of imidazolium-based ILs causes an increase in the protein size at which protein aggregation occurs, suggesting that changes in the alkyl chain length may control the size at which a specific protein forms aggregates under these conditions (Takekiyo et al. 2014). Thus, the relationship between the size of the water domains and the proteins in solution may allow control of amyloid-type aggregation of proteins in concentrated solutions.
Increasing the proportion of ILs to water molecules induces low polarity of the resulting liquid. The ability of aqueous IL solutions to induce helix formation is similar to alcohol denaturation (Takekiyo et al. 2013, 2017a). The effects of alcohols on proteins are shown to arise from their low polarity; low polarity weakens the hydrophobic interactions that stabilize the compact native structure of proteins, but simultaneously strengthens electrostatic interactions, thus stabilizing secondary structures, particularly the α-helix (Shiraki et al. 1995; Luo and Baldwin 1997). The dielectric constant of pure ILs is low (ε = 10~15) (Weingärtner et al. 2001), as is the case for 2,2,2-trifluoroethanol (TFE) (ε = 27.1) (Hong et al. 1999). As such, ILs with low polarity may cause the enhancement of protein intramolecular H-bonds. Thus, the solution property (nano-heterogeneity structure/water hydration and low polarity) acts cooperatively in the helix-forming ability of concentrated IL solutions (Takekiyo et al. 2013, 2017a).
Suppression of amyloid aggregation using ionic liquids
It was known that amyloid stability is affected by the addition of organic compounds, and the interaction between protein and organic compounds can suppress the formation of amyloids (Arora et al. 2004; Shammas et al. 2011). Trimethylamine N-oxide inhibits amyloidogenesis after lysozyme unfolding (Wawer et al. 2014). Besides, indole-3-carbinol interacts with hydrophobic residues such as Trp, Ile, and Ala residues in lysozyme and demonstrates inhibition of amyloidogenesis (Morshedi et al. 2007).
Since ILs also interact with amino acid residues in proteins, it follows that ILs also affect amyloid stability. Some studies have investigated the suppression and promotion of amyloid formation using ILs (Table 2). As for ammonium-based ILs, tetramethylguanidium acetate ([TMG][Ac]) and EAN inhibited lysozyme amyloids, while propylammonium and butylammonium nitrates (PAN and BAN) and 2-methyloxyethylammonium nitrate (MEOAN) slightly promoted amyloid formation (Kalhor et al. 2009; Mangialardo et al. 2012). Triethylammonium-based ILs ([Tea][Y], Y=anion) with H2PO4− and HSO4− anions promote Aβ16-22 amyloid aggregation (Debeljuh et al. 2011a), and triethylammonium mesylate ([Tea][Ms]) suppresses amyloid formation of Aβ16-22 and Aβ1-40 peptides (Debeljuh et al. 2011a, b). Conversely, imidazolium- and pyridinium-based ILs promote amyloid formation (Bae et al. 2011), and the promotion of α-lactalbumin amyloid by [bmim][BF4] was observed in other proteins (Hwang et al. 2009).
Table 2.
Amyloid forming ability of ionic liquids
| Proteins | Effect of amyloid formation | Methods | References |
|---|---|---|---|
| Amyloid β (16-22) (Aβ16-22) | [Tea][HSO4] ~ [Tea][H2PO4] > [Tea][Tfac] > [Tea][La] > [Tea][Tf] > [Tea][Ms] | ThT, TEM, and CD | Debeljuh et al. 2011a |
| Amyloid β (1-40) (Aβ1-40) | [Tea][Ms]: suppression | TEM and CD | Debeljuh et al. 2011b |
| α-Lactalbmin | [bmim][BF4] > [bmim][PF6] > [bmim][TFSI] > [bmim][CH3SO4] | ThT and TEM | Bae et al. 2011 |
| α-Synuclein | [bmim][TFSI] > [bmim][BF4] | ThT and TEM | Bae et al. 2010 |
| [bmim][TFSI] > [emim][TFSI] > [hmim][TFSI] > [Bzmim][TFSI] > [eDmim][TFSI] > [omim][FSI] | ThT and TEM | Hwang et al. 2009 | |
| [bpyr][BF4] > [omim][FSI] ~ [Phemim][FSI] ~ [bDmim][BF4] ~ [bmim][BF4] ~ [bmim][PF6] | ThT and TEM | Hwang et al. 2009 | |
| Trypsin | [bmim][BF4]: promotion | ThT and TEM | Bae et al. 2011 |
| Insulin | [bmim][SCN] > PAN ~ EAN | FTIR and CR | Takekiyo et al. 2016 |
| [bmim][BF4]: promotion | ThT and TEM | Bae et al. 2011 | |
| Lysozyme | EAN and [Tea][Ms]: suppression | SEM, CD, and CR | Byrne and Angell 2009 |
| PAN ~ BAN > MEOAN | ThT, TEM. CD, and ANS | Kalhor et al. 2009 | |
| [TMG][Ac]: suppression | Raman | Mangialardo et al. 2012 | |
| [bmim][BF4]: promotion | ThT and TEM | Bae et al. 2011 |
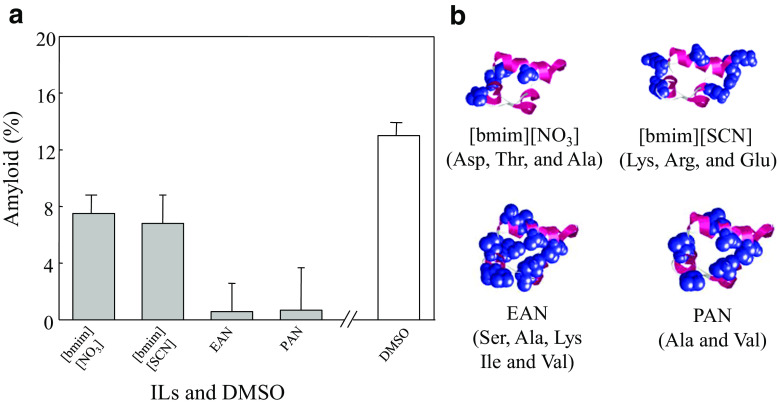
Importantly, investigating the potential use of ILs for suppression of amyloid formation involves elucidation of the mechanisms thereof. It has been suggested that the suppression or promotion of amyloid formation by ILs depends on the specific IL and on the nanostructure of the IL itself (Debeljuh et al. 2011a). Recently, ILs exhibited strong suppression of insulin amyloid formation (Fig. 1a), and this suppression was due to the insulin–IL interactions (Fig. 1b) (Takekiyo et al. 2016). Moreover, the insulin–IL interaction and nano-heterogeneity structure of the aqueous IL solution protect the amyloid formation by heating.
Fig. 1.
a Insulin amyloid contents in aqueous solution with ILs (gray bars) and DMSO (white bar) with x = 20 mol%IL determined by FTIR spectra. b Interaction sites of ILs with insulin. The figure was reproduced from Takekiyo et al. (2016)
As per the results of the insulin amyloid suppression, ILs tend to interact with the amino acid residues through electrostatic and hydrophobic interactions, and the interaction sites of ILs with amino acid residues are dependent upon the specific cationic and anionic species of IL (e.g., Fig. 1b). Therefore, IL nanostructure and the affinity between the ILs and specific amino acid residues both play important roles in the selection of ILs for their suppression and promotion effects on amyloid formation.
Ionic liquid-induced dissolution of amyloid aggregates
Some small compounds such as DMSO and guanidine hydrochloride have been previously used to dissolve amyloid aggregates (Table 3). Since ILs have the ability to dissolve cellulose (Hauru et al. 2012), which has a H-bonding structure similar to that of amyloid aggregates, this ability may be related to the advantage of the reconstruction technique of amyloid aggregates. However, there are few studies reporting the dissolution of amyloid aggregates using IL; one such study reports that ammonium-based ILs such as EAN demonstrated the ability to dissolve lysozyme amyloid (Byrne and Angell 2009).
Table 3.
Representative dissolution agent for various amyloids
| Protein | Dissolution agent | Methods | References |
|---|---|---|---|
| β-Microglobulin | Guanidine hydrochloride | CD, ThT, and fluorescence | Narimoto et al. 2004 |
|
Dimethyl sulfoxide (DMSO), Hexafluoroisopropanol and 2,2,2-Trifluoroethanol (TFE) |
CD, light scattering, fluorescence, and EM | Hirota-Nakaoka et al. 2003 | |
| Insulin | DMSO | ThT, AFM, DLS, and FTIR | Loksztejn and Dzwolak 2009 |
| Guanidine thiocyanate | FTIR and TEM | Shammas et al. 2011 | |
| Amyloid β (25-35) (Aβ25-35) | DMSO | NMR, AFM, and UV-Vis | Ippel et al. 2002 |
| Transthyretin (10-19) | TFE | CD and EM | MacPhee and Dobson 2000 |
To further evaluate the amyloid-dissolving ability of ILs, preliminary, we have investigated the dissolution of insulin amyloid aggregates in aqueous solutions with four ILs and DMSO (x = 20 mol%IL or DMSO) using FTIR spectroscopy (Takekiyo et al. unpublished data). Studied ILs were able to dissolve insulin amyloid, and the dissolved amyloid formed an α-helical structure (Fig. 2a). The dissolution ability rank order of ILs for insulin amyloid was [bmim][NO3] ≥ [bmim][SCN] ≥ DMSO = PAN ≥ EAN; this dissolution ability was more strongly dependent on the cationic species of the IL than the anionic species. Imidazolium-based ILs demonstrated a stronger dissolving ability than DMSO and ammonium-based ILs. As mentioned previously, the amyloid-dissolving ability of DMSO is due to its basicity (Hirota-Nakaoka et al. 2003). Differences in dissolution abilities between ILs and DMSO appear to depend upon the basicity of the liquid. It is worth noting that the ability of ILs to dissolve cellulose is dependent upon the H-bonding basicity in the Kamlet-Taft (KT) parameters, a set of factors reflecting the solvent properties, measuring overall polarity by separately considering the H-bond acidity (α), H-bond basicity (β), and polarizability (π*) of the solvent (Hauru et al. 2012). Comparing the β-value in KT parameters of pure DMSO and ILs, the β-value of DMSO is 0.76 (Kamlet et al. 1983), while the β-values of studied imidazolium-based ILs range from 0.66 to 0.95 (Lungwitz et al. 2010). The β-values of imidazolium-based ILs are almost the same or slightly higher than DMSO, and those of EAN (β = 0.46) and PAN (β = 0.52) (Greaves and Drummond 2008) are lower than DMSO. The difference in the dissolution ability of ILs and DMSO for insulin amyloid may correlate with the H-bonding basicity (β-value).
Fig. 2.
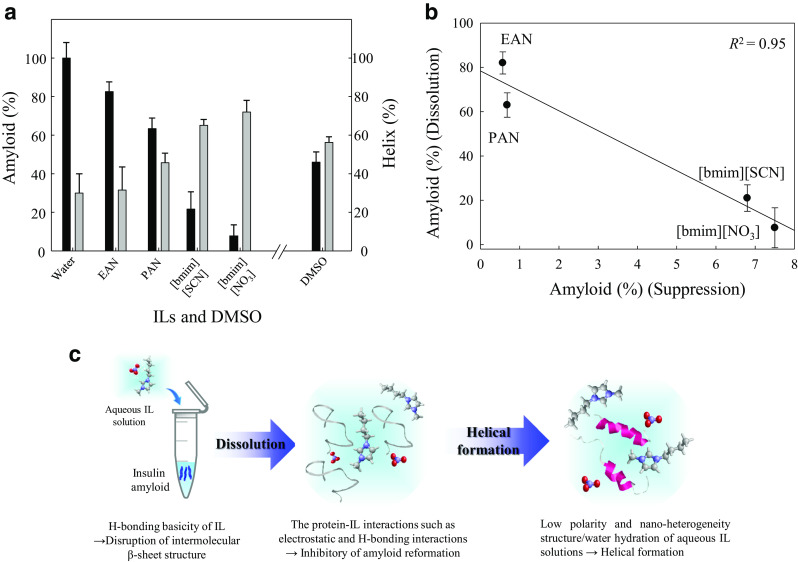
a Amyloid (black bars) and helix (gray bars) contents of dissolved insulin amyloids in the aqueous solutions with ILs and DMSO with x = 20 mol%IL determined by FTIR spectra. b Amyloid contents of the dissolution and suppression of insulin amyloid by the addition of ILs at x = 20. The correlation coefficients (R2) determined by the least-squares analysis. c Scheme of dissolution mechanism of insulin amyloid by the addition of aqueous IL solutions at x = 20
The relationship between the suppression/dissolution abilities of ILs for insulin amyloid is intriguing. Remarkably, ILs with low suppression ability showed high dissolution ability (Fig. 2b). Entirely, imidazolium-based ILs have good suppression/dissolution abilities for insulin amyloid when compared to ammonium-based ILs.
Finally, we propose the dissolution mechanism of insulin amyloid in aqueous IL solutions (x = 20 mol%IL) (Fig. 2c). The addition of aqueous IL solutions to insulin amyloid induced the disruption of intermolecular H-bonding by the H-bonding basicity of ILs, and the dissolved insulin–IL interactions such as electrostatic interaction and H-bonding suppressed amyloid reformation (Takekiyo et al. 2016). Moreover, since the low polarity and nano-heterogeneity structure/water hydration of aqueous IL solutions enhance the intramolecular H-bonding in proteins (Takekiyo et al. 2013, 2017a), dissolved insulin in aqueous IL solutions forms a helical structure. However, although ILs have the ability to dissolve amyloid aggregates from insulin and lysozyme (Byrne and Angell 2009), detailed information for other amyloids is still unclear. It is well-known that the β-sheet structures constructing amyloid aggregates may be either parallel or antiparallel structures (Zou et al. 2013). Therefore, the differences in the amyloid structure may result in different dissolution abilities when different ILs are used. Further systematic investigation of how various amyloid aggregates are affected by ILs will allow further understanding of how ILs may be used to dissolve amyloid aggregates.
Conclusions and perspectives
In this review, we summarized the potential use of ILs for the suppression/dissolution agents of amyloid aggregates. The protein–IL affinity, such as protein–IL interactions (contribution to inhibitory of protein aggregation), and low polarity and nano-heterogeneity structure/water hydration (contribution to helical formation), was found to be the primary biochemical basis for the suppression of amyloid formation. The H-bonding basicity of ILs plays a role in the dissolution ability for insulin amyloids. In addition to these factors, IL solutions easily form a glassy state, which facilitates the cryopreservation of biomolecules at 77 K (Yoshimura et al. 2011). Related to this, ILs are already known for their potential use in cryopreservation/refolding of proteins (Yoshimura et al. 2016; Takekiyo et al. 2017b). Some techniques for removing IL from aqueous protein solutions, such as column chromatography and dialysis, have been reported (Fujita et al. 2016; Takekiyo et al. 2017b). Our understanding of long-term preservation agents that also dissolve aggregated proteins stems from the import and export of valuable proteins, and the challenges of reduced protein function resulting from the preparation of large-scale stock solutions. Aqueous IL solutions may therefore have the potential to increase the suppression/dissolution of amyloid aggregates and also to improve the cryopreservation of dissolved proteins.
Finally, the toxicity of ILs toward biological systems, such as biomolecules and cells, is under intense study (Pham et al. 2010; Kudlak et al. 2015). The toxicity of ILs appears to be dependent on the constructing ions and target biological system. Together with the development of nontoxic ILs for use with protein systems, the basic information of suppression/dissolution/cryopreservation abilities of ILs for target proteins will yield great potential for additional developments in pharmaceutical science, cryobiology, and protein engineering (Fig. 3).
Fig. 3.
Advantages of ILs for potential application in biological engineering and pharmaceutical sciences
Acknowledgements
The authors would like to thank Ms. Yuka Ishikawa, Ms. Erika Yamaguchi, and Prof. Hiroshi Abe at National Defense Academy for fruitful discussions and experimental supports.
Abbreviation
- [emim]
1-Ethyl-3-methylimidazolium
- [bmim]
1-Butyl-3-methylimidazolium
- [hmim]
1-Hexyl-3-methylimidazolium
- [omim]
1-Methyl-3-octylimidazolium
- [Bzmim]
1-Benzyl-3-methylimidazolium
- [eDmim]
1-Ethyl-2,3-dimethylimidazolium
- [bpyr]
N-Butyl-3-methyl-pyridinium
- [Phemim]
1-Phenylpropyl-3-methylimidazolium
- [dBmim]
1,3-Dibutylimidazolium
- [bDmim]
1-Butyl-2,3-dimethylimidazolium
- [TMG]
Tetramethylguanidium
- [Tea]
Triethylammonium
- EAN
Ethylammonium nitrate
- PAN
Propylammonium nitrate
- BAN
Butylammonium nitrate
- MEOAN
2-Methyloxyethylammonium nitrate
- [Ac]
Acetate
- [H2PO4]
Phosphate
- [HSO4]
Hydrogen sulfate
- [Tfac]
Trifluoroacetate
- [La]
Lactate
- [Tf]
Triflate
- [Ms]
Mesylate
- [BF4]
Tetrafluoroborate
- [PF6]
Hexafluorophosphate
- [TFSI]
Bis(trifluoromethanesulfonyl)imide
- [FSI]
Trifluoromethanesulfonate
- [CH3SO4]
Methylsulfonate
- [SCN]
Thiocyanate
- [NO3]
Nitrate
- FTIR
Fourier transform infrared
- ThT
Thioflavin T
- CD
Circular dichroism
- CR
Congo red
- ANS
8-Anilinonaphthalene-1-sulfonic acid
- TEM
Transmission electronic microscopy
- SEM
Scanning electron microscope
- EM
Electron microscope
- AFM
Atomic force microscope
- DLS
Dynamic light scattering
Compliance with ethical standards
Conflict of interest
Takahiro Takekiyo declares that he has no conflict of interest. Yukihiro Yoshimura declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on “Ionic Liquids and Biomolecules” edited by Antonio Benedetto and Hans-Joachim Galla.
References
- Akdogan Y, Junk MJN, Hinderberger D. Effect of ionic liquids on the solution structure of human serum albumin. Biomaclomolecues. 2011;12:1072–1079. doi: 10.1021/bm1014156. [DOI] [PubMed] [Google Scholar]
- Arora A, Ha C, Park CB. Inhibition of insulin amyloid formation by small stress molecules. FEBS Lett. 2004;564:121–125. doi: 10.1016/S0014-5793(04)00326-6. [DOI] [PubMed] [Google Scholar]
- Bae SY, Kim S, Hwang H, Kim H-K, Yoon HC, Kim JH, Lee BY, Kim TD. Amyloid formation and disaggregation of α-synuclein and its tandem repeat (α-TR) Biochem Biophys Res Commum. 2010;490:531–536. doi: 10.1016/j.bbrc.2010.08.088. [DOI] [PubMed] [Google Scholar]
- Bae SY, Kim S, Lee BY, Kim KK, Kim TD. Amyloid formation using 1-butyl-3-methylimidazolium-based ionic liquids. Anal Biochem. 2011;419:354–356. doi: 10.1016/j.ab.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Ballone P. Room temperature ionic liquids interacting with bio-molecules: an overview of experimental and computational studies. Philos Mag. 2015;96(7–9):870–894. [Google Scholar]
- Benedetto A, Ballone P. Room temperature ionic liquids meet biomolecules: a microscopic view of structure and dynamics. ACS Sustain Chem Eng. 2015;4(2):392–412. [Google Scholar]
- Byrne N, Angell CA. Formation and dissolution of hen egg white lysozyme amyloid fibrils in protic ionic liquids. Chem Commum. 2009;9:1046–1048. doi: 10.1039/b817590j. [DOI] [PubMed] [Google Scholar]
- De Groot NS, Sabate R, Ventura S. Amyloid in bacterial inclusion bodies. Trend Biochem Sci. 2009;34:408–416. doi: 10.1016/j.tibs.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Debeljuh N, Barrow CJ, Byrne N. The impact of ionic liquids on amyloid fibrilization of Aβ16-22: tuning the rate of fibrilization using a reverse Hofmeister strategy. Phys Chem Chem Phys. 2011;13:16534–16536. doi: 10.1039/c1cp22256b. [DOI] [PubMed] [Google Scholar]
- Debeljuh N, Barrow CJ, Hemderson L, Byrne N. Structure inducing ionic liquids—enhancement of alpha helicity in the Abeta(1–40) peptide from Altzheimser’s disease. Chem Commum. 2011;47:6371–6373. doi: 10.1039/c1cc10377f. [DOI] [PubMed] [Google Scholar]
- Egorova KS, Gordeev EG, Ananikov VP. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem Rev. 2017;117:7132–7189. doi: 10.1021/acs.chemrev.6b00562. [DOI] [PubMed] [Google Scholar]
- Fujita K, Kajiyama M, Liu Y, Nakamura N, Ohno H. Hydrated ionic liquids as a liquid chaperon for refolding of aggregated recombinant protein expressed in Escherichia coli. Chem Commum. 2016;52:13491–13494. doi: 10.1039/c6cc06999a. [DOI] [PubMed] [Google Scholar]
- Greaves TL, Drummond CJ. Protic ionic liquids: properties and applications. Chem Rev. 2008;108:206–237. doi: 10.1021/cr068040u. [DOI] [PubMed] [Google Scholar]
- Greaves TL, Drummond CJ. Protic ionic liquids: evolving structure–property relationships and expanding applications. Chem Rev. 2015;115:11379–11448. doi: 10.1021/acs.chemrev.5b00158. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Prangé T, Riés-Kautt M, Ducruix A, Lafont S, Astier JP, Veesler S. The decameric structure of bovine pancreatic trypsin inhibitor (BPTI) crystallized from thiocyanate at 2.7Å resolution. Acta Crystallogr D. 1999;55:1037–1113. doi: 10.1107/S0907444998008725. [DOI] [PubMed] [Google Scholar]
- Hauru LKJ, Hummel M, King AWT, Kipeläinen I, Sixta H. Role of solvent parameters in the regeneration of cellulose from ionic liquid solutions. Biomacromolecules. 2012;13:2896–2905. doi: 10.1021/bm300912y. [DOI] [PubMed] [Google Scholar]
- Hayes R, Warr GG, Atkin R. Structure and nanostructure in ionic liquids. Chem Rev. 2015;115:6537–6426. doi: 10.1021/cr500411q. [DOI] [PubMed] [Google Scholar]
- Hirota-Nakaoka N, Hasegawa K, Naiki H, Goto Y. Dissolution of β2-microglobulin amyloid fibrils by dimethylsulfoxide. J Biochem. 2003;134:159–164. doi: 10.1093/jb/mvg124. [DOI] [PubMed] [Google Scholar]
- Hong D-P, Hoshino M, Kuboi R, Goto Y. Clustering of fluorine-substituted alcohols as a factor responsible for their marked effects on proteins and peptides. J Am Chem Soc. 1999;121:8427–8433. [Google Scholar]
- Hwang H, Choi H, Kim H-K, Jo DH, Kim TD. Ionic liquids promote amyloid formation from α-synuclein. Anal Biochem. 2009;386:293–295. doi: 10.1016/j.ab.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Ippel JH, Olofsson A, Scheucher J, Lundgren E, Wijmenga SS. Probing solvent accessibility of amyloid fibrils by solution NMR spectroscopy. Proc Natl Acad Sci U S A. 2002;99:8648–8653. doi: 10.1073/pnas.132098999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang Y, Voth GA. Molecular dynamics simulation of nanostructural organization in ionic liquid/water mixtures. J Phys Chem B. 2007;111:4812–4818. doi: 10.1021/jp067142l. [DOI] [PubMed] [Google Scholar]
- Kalhor HR, KKamizi M, Akbari J, Heydari A. Inhibition of amyloid formation by ionic liquids: ionic liquid affecting intermediate oligomers. Biomacromolecules. 2009;10:2468–2475. doi: 10.1021/bm900428q. [DOI] [PubMed] [Google Scholar]
- Kamlet MJ, Luis J, Abboud M, Abraham MH, Taft RW. Liner solvation energy relationships. 23. A comprehensive collection of the solvatochromic parameters, π*, α, and β, and some methods for simplifying the generalized solvatochromic equation. J Org Chem. 1983;48:2877–2887. [Google Scholar]
- Kudlak B, Owczarek K, Namieśnik J. Selected issues related to the toxicity of ionic liquids and deep eutectic solvents—a review. Environ Sci Pollut Res. 2015;22:11975–11992. doi: 10.1007/s11356-015-4794-y. [DOI] [PubMed] [Google Scholar]
- Loksztejn A, Dzwolak W. Noncooperative dimethyl sulfoxide-induced dissection of insulin fibrils: toward soluble buiding blocks of amyloid. Biochemistry. 2009;48:4846–4851. doi: 10.1021/bi900394b. [DOI] [PubMed] [Google Scholar]
- Lungwitz R, Strehmel V, Spange S. The dipolarity/polarisability of 1-alkyl-3-methylimidazolium ionic liquids as a function of anion structure and the alkyl chain length. New J Chem. 2010;34:1135–1140. [Google Scholar]
- Luo P, Baldwin RL. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- MacPhee CE, Dobson CM. Chemical dissection and reassembly of amyloid fibrils formed by a peptide fragment of tranthyretin. J Mol Biol. 2000;297:1203–1215. doi: 10.1006/jmbi.2000.3600. [DOI] [PubMed] [Google Scholar]
- Mangialardo S, Gontrani L, Caminiti R, Potorino P. Role of ionic liquids in protein refolding: native/fibrillar versus treated lysozyme. RSC Adv. 2012;2:12329–12336. [Google Scholar]
- Meersman F, Dobson CM. Probing the pressure–temperature stability of amyloid fibrils provides new insight into their molecular properties. Biochim Biophys Acta. 2006;1764:452–460. doi: 10.1016/j.bbapap.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Morshedi D, Rezaei-Ghaleh N, Ebraham-Habibi A, Ahmadian S, Nemat-Gorgani M. Inhibition of amyloid fibrillation of lysozyme by indole derivatives—possible mechanism of action. FEBS J. 2007;274:6415–6425. doi: 10.1111/j.1742-4658.2007.06158.x. [DOI] [PubMed] [Google Scholar]
- Narimoto T, Sakurai K, Okamoto A, Chatani E, Hoshino M, Hasegawa K, Naiki H, Goto Y. Conformational stability of amyloid fibrils of β2-microglobulin probed by guanidine-hydrochloride-induced unfolding. FEBS Lett. 2004;576:313–319. doi: 10.1016/j.febslet.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Patel R, Kumari M, Khan AB. Recent advances in the applications of ionic liquids in protein stability and activity: a review. Appl Biochem Biotechnol. 2014;172:3701–3720. doi: 10.1007/s12010-014-0813-6. [DOI] [PubMed] [Google Scholar]
- Pham TPT, Cho C-W, Yun Y-S. Environmental fate and toxicity of ionic liquids: a review. Water Res. 2010;44:352–372. doi: 10.1016/j.watres.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Shammas SL, Knowles TPJ, Baldwin AJ, MacPhee CE, Welland ME, Dobson CM, Devliln GL. Pertubation of the stability of amyloid fibrils through alternation of electrostatic interactions. Biophys J. 2011;100:2783–2791. doi: 10.1016/j.bpj.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki K, Nishikawa K, Goto Y. Trifluoroethanol-induced stabilization of the α-helical structure of β-lactoglobulin: implication for non-hierarchical protein folding. J Mol Biol. 1995;245:180–194. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- Shu Y, Liu M, Chen S, Chen X, Wang J. New insight into molecular interactions of imidazolium ionic liquids with bovine serum albumin. J Phys Chem B. 2011;115:12306–12314. doi: 10.1021/jp2071925. [DOI] [PubMed] [Google Scholar]
- Silva M, Figueiredo M, Cabrita E. Epitope mapping of imidazolium cations in ionic liquid–protein interactions unveils the balance between hydrophobicity and electrostatics towards protein destabilization. Phys Chem Chem Phys. 2014;26:23394–23403. doi: 10.1039/c4cp03534h. [DOI] [PubMed] [Google Scholar]
- Singh A, Upadhay V, Upadhay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using dissolution process. Microb Cell Factories. 2015;14(1–10):41. doi: 10.1186/s12934-015-0222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers CA, Flowers RA., II Protein renaturation by the liquid organic salt ethylammonium nitrate. Protein Sci. 2000;9:2001–2008. doi: 10.1110/ps.9.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekiyo T, Yamazaki K, Yamaguchi E, Abe E, Yoshimura Y. High ionic liquid concentration-induced structural change of protein in aqueous solution: a case study of lysozyme. J Phys Chem B. 2012;116:11092–11097. doi: 10.1021/jp3057064. [DOI] [PubMed] [Google Scholar]
- Takekiyo T, Koyama Y, Yamazaki K, Abe H, Yoshimura Y. Ionic liquid-induced formation of α-helical structure of β-lactoglobulin. J Phys Chem B. 2013;117:10142–10148. doi: 10.1021/jp405834n. [DOI] [PubMed] [Google Scholar]
- Takekiyo T, Fukudome K, Yamazaki K, Abe H, Yoshimura Y. Protein aggregation and partial globular states of proteins in aqueous 1-alkyl-3-methylimidazolium nitrate solutions. Chem Phys Lett. 2014;602:22–27. [Google Scholar]
- Takekiyo T, Yamaguchi E, Yoshida K, Kato M, Yamaguchi T, Yoshimura Y. Interaction site between the protein aggregates and thiocyanate ion in aqueous solution: a case study of 1-butyl-3-methylimidazolium thiocyanate. J Phys Chem B. 2015;119:6536–6544. doi: 10.1021/acs.jpcb.5b01650. [DOI] [PubMed] [Google Scholar]
- Takekiyo T, Yamaguchi E, Abe H, Yoshimura Y. Suppression effect on the formation of insulin amyloid by the use of ionic liquid. ACS Sustain Chem Eng. 2016;4:422–428. [Google Scholar]
- Takekiyo T, Yoshida K, Funahashi Y, Nagata S, Abe H, Yamaguchi T, Yoshimura Y. Helix-forming ability for proteins of alkylammonium nitrate. J Mol Liquids. 2017;243:584–590. [Google Scholar]
- Takekiyo T, Ishikawa Y, Yoshimura Y. Cryopreservation of proteins using ionic liquids: a case study of cytochrome c. J Phys Chem B. 2017;121:7614–7260. doi: 10.1021/acs.jpcb.7b05158. [DOI] [PubMed] [Google Scholar]
- Tietze AA, Bordusa F, Giernoth R, Imhof D, Lenzer T, Mrestani-Klaus C, Neudorf I, Oum K, Reith D, Stark A. On the nature of interactions between ionic liquids and small amino-acid-based biomolecules. ChemPhysChem. 2013;14:4044–4064. doi: 10.1002/cphc.201300736. [DOI] [PubMed] [Google Scholar]
- Tischer A, Pultke H, Topf A, Auton M, Lange C, Lilie H. The effects of N-ethyl-N’-methyl imidazolium chloride on the solubility, stability and aggregation of tc-rPA. FEBS J. 2014;281:1738–1749. doi: 10.1111/febs.12736. [DOI] [PubMed] [Google Scholar]
- Vaney MC, Broutin I, Retailleau P, Douangamath A, Lafont S, Hamiaux C, Prangé T, Ducruix A, Riès-Kautt M. Structural effects on monovalent anions on polymorphic lysozyme crystals. Acta Crystallogr D. 2001;57:929–940. doi: 10.1107/s0907444901004504. [DOI] [PubMed] [Google Scholar]
- Wawer J, Krakowiak J, Szociński M, Lustig Z, Olszewski M, Szostak M. Inhibition of amyloid fibril formation of hen egg white lysozyme by trimethylamine N-oxide at low pH. Int J Biol Macromol. 2014;70:214–221. doi: 10.1016/j.ijbiomac.2014.06.057. [DOI] [PubMed] [Google Scholar]
- Weaver KD, Vrikkis RM, Van Vorst MP, Trullinger J, Vijayaraghavan R, Foureau DM, McKillop IH, MacFarlane DR, Kruger JK, Elliott GD. Structure and function of proteins in hydrated choline dihydrogen phosphate ionic liquid. Phys Chem Chem Phys. 2012;14:790–801. doi: 10.1039/c1cp22965f. [DOI] [PubMed] [Google Scholar]
- Weingärtner H, Knocks A, Schrader W, Kaatze U. Dielectric spectroscopy of the room temperature molten salt ethylammonium nitrate. J Phys Chem A. 2001;105:8646–8650. [Google Scholar]
- Yoshimura Y, Kimura H, Okamoto C, Miyashita T, Imai Y, Abe H. Glass transition behaviour of ionic liquid, 1-butyl-3-methylimidazolium tetrafluoroborate–H2O mixed solutions. J Chem Thermodyn. 2011;43:410–412. [Google Scholar]
- Yoshimura Y, Takekiyo T, Mori T. Structural study of lysozyme in two ionic liquids at cryogenic temperature. Chme Phys Lett. 2016;664:44–49. [Google Scholar]
- Zhang Y, Cremer PS. Interactions between macromolecules and ions: the Hofmeister series. Curr Opinin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Zou Y, Li Y, Hao W, Hu X, Ma G. Parallel β-sheet fibril and antiparallel β-sheet oligomer: new insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J Phys Chem B. 2013;117:4003–4013. doi: 10.1021/jp4003559. [DOI] [PubMed] [Google Scholar]