Abstract
Nucleic acids have emerged as powerful biological and nanotechnological tools. In biological and nanotechnological experiments, methods of extracting and purifying nucleic acids from various types of cells and their storage are critical for obtaining reproducible experimental results. In nanotechnological experiments, methods for regulating the conformational polymorphism of nucleic acids and increasing sequence selectivity for base pairing of nucleic acids are important for developing nucleic acid-based nanomaterials. However, dearth of media that foster favourable behaviour of nucleic acids has been a bottleneck for promoting the biology and nanotechnology using the nucleic acids. Ionic liquids (ILs) are solvents that may be potentially used for controlling the properties of the nucleic acids. Here, we review researches regarding the behaviour of nucleic acids in ILs. The efficiency of extraction and purification of nucleic acids from biological samples is increased by IL addition. Moreover, nucleic acids in ILs show long-term stability, which maintains their structures and enhances nuclease resistance. Nucleic acids in ILs can be used directly in polymerase chain reaction and gene expression analysis with high efficiency. Moreover, the stabilities of the nucleic acids for duplex, triplex, and quadruplex (G-quadruplex and i-motif) structures change drastically with IL cation-nucleic acid interactions. Highly sensitive DNA sensors have been developed based on the unique changes in the stability of nucleic acids in ILs. The behaviours of nucleic acids in ILs detailed here should be useful in the design of nucleic acids to use as biological and nanotechnological tools.
Keywords: Nucleic acids, Stability, Extraction, Storing, Biosensor, Ionic liquids
Introduction
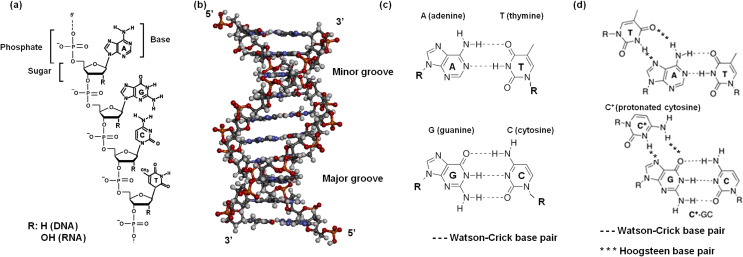
Nucleic acids (DNA and RNA) contain a negatively charged sugar-phosphate backbone, and their bases possess aromatic rings, which have the donor and acceptor sites used for hydrogen bonding (Fig. 1a) (Saenger 1984). The DNA forms a duplex in cells via base pair formation (Fig. 1b, c) and carries genetic information. Extraction, purification, and storage of nucleic acids are required for biological experiments (Hyde and Read 1993). For example, the quantification of transcript and protein levels of target genes is essential for understanding gene regulatory mechanisms and developing drugs that regulate gene expression. Traditionally, extraction, purification, and storage of nucleic acid are based on complicated liquid-liquid extraction techniques that require the use of high toxic reagents such as phenol and chloroform (Tan and Yiap 2009). Therefore, simpler methods for the extraction, purification, and storage of nucleic acids using sustainable reagents are required using an environmentally preferable reagents.
Fig. 1.
a Chemical structure and b ordered structure (duplex) for nucleic acids. The chemical structure of c base pairs in a duplex and d triplets in a triplex
Furthermore, nucleic acids have emerged as powerful nanotechnological tools because of their sequence selectivity and conformational polymorphism. Single strands of nucleic acids recognize and bind to other nucleic acid sequences via highly specific base recognition interactions such as Watson-Crick base pairing (Fig. 1c), and Watson-Crick and Hoogsteen base pairing for triplets (Fig. 1d). Binding of complementary DNA sequences via Watson-Crick base pairing has been applied for developing sequence sensing methods such as DNA microarrays for gene expression analyses (Heller 2002), chips for hybridization-based gene sequencing and phylogenetic studies (Fedrigo and Naylor 2004; Morishima et al. 2004), single nucleotide polymorphism (SNP) detection microarrays (Ott and Hoh 2003), and transcriptome arrays (Mitchell et al. 2017). Moreover, nanoscale self-assembly systems based on duplex formation are useful as functional molecule arrays (Chen and Seeman 1991) and in systems such as molecular transport devices (Phadke 2001), molecular computing devices (switches, logical units, and programmable molecular systems for massively parallel computing) (Phadke 2001), and molecular motors (Yurke et al. 2000) that depend on structural transitions of nucleic acids in response to chemical stimuli.
Although DNA is reasonably stable in an aqueous solution, non-physiological temperature, pH, and ionic strength denature and disrupt the DNA helix. Conventionally, DNA is stored under refrigeration for short- and long-term applications, and the effect of storage temperature on DNA stability has been discussed extensively (Legoff et al. 2006). Nucleic acids are not stable in an aqueous solution at ambient temperatures for long periods (for several days to 1 month) (Armand et al. 2009; Fujita and Ohno 2012) because of degradation by contaminating nucleases (Sasaki et al. 2007) and inherent chemical instability, which is a bottleneck in developing nucleic acid-based bio/nanotechnological tools (Yilmaz et al. 2008). Furthermore, aqueous solutions cannot be used in small-volume technologies as small volumes of water vapourize immediately under open air conditions or at high temperature (Armand et al. 2009). Therefore, solvents that circumvent the limitations of aqueous buffers are required for developing functional nanodevices.
Ionic liquids (ILs) are of interest as solvents in nanotechnology because they are “green” solvents suitable for a wide range of chemical reactions. Certain remarkable features of ILs have made them attractive alternatives to water in a variety of applications, including electrochemistry, separation science, chemical synthesis, and material science (Fujita et al. 2007; Fujita and Ohno 2010; Trincao et al. 2004; Welton 1999). Room temperature ILs provide favourable environments for reactions, because ILs are non-volatile at vapour pressure close to zero (Earle and Seddon 2000). Unfortunately, ILs act as denaturing agents for biomolecules in general as the latter possess negative and positive charges, resulting in non-specific binding to ILs. Nucleic acids are favourable for electrostatic interaction as well as the formation of hydrogen bonds and van der Waals interactions. Moreover, hydration and cosolute binding also affect the stability and structure of nucleic acids (Nakano et al. 2014c). Since ILs can be custom designed to change the anion-cation combination, they act as good solvents for nucleic acids from perspective of IL-nucleic acid interactions (Benedetto and Ballon 2016a, b). This review covers researches on the structure and stability of nucleic acids in ILs and highlights the advantages and unique properties of these solvents. We expect that IL properties will enable novel applications of nucleic acids in biological and nanotechnological fields.
DNA structure and stability in ILs
The effect of ILs on nucleic acid structures has been investigated in several studies. Circular dichroism (CD) is used for monitoring nucleic acid structure in most cases. MacFarlane et al. measured the CD spectrum of long DNA duplexes from salmon testes in hydrated choline dihydrogen phosphate (choline dhp) (Vijayaraghavan et al. 2010) and observed that they formed the B-form structure in both choline dhp and aqueous solution. It is also reported that the choline dhp does not change the structures of quadruplexes (G-quadruplexes and i-motifs) (Fujita and Ohno 2012; Tateishi-Karimata et al. 2015a).
In an aqueous solution, cations bind to the phosphate groups of DNA to shield the negative charge. Moreover, cations can also bind non-specifically to several parts of DNA, such as bases and sugars at high cation concentration. In general, the non-specific binding induces aggregation or destabilization of DNA structures. However, certain ILs that show specific interactions with DNA in addition to electrostatic interactions change DNA stability in a unique manner.
Quantitative thermodynamic analyses demonstrated that A–T base pairs (the dash indicates the Watson-Crick base pair) are more stable than G–C base pairs in a solution containing 4 M choline dhp (80 wt% choline dhp) (Tateishi-Karimata and Sugimoto 2012). This is the reverse of the base pair stabilities of DNA duplexes in a buffered NaCl solution because A–T and G–C base pairs have two and three hydrogen bonds, respectively. The stability differences were due to differences in enthalpy contributions. Molecular dynamic (MD) simulations and NMR experimental results also revealed that choline ions bind to A–T-rich regions in B-form DNA duplexes in the minor groove and stabilize the A–T base pairs via hydrogen bonds (Fig. 2(a)) (Marusic et al. 2015; Nakano et al. 2014b), whereas the G–C base pairs are destabilized because of specific binding of choline ions to guanine in single-stranded DNA, which inhibits G–C base pairing (Tateishi-Karimata and Sugimoto 2012). Several experimental in vitro and MD in silico studies also attempted to explore the mechanism underlying this behaviour. Chandran et al. investigated interactions of IL cations with DNA base pairs in detail using MD and experiments (Chandran et al. 2012). The results of MD simulations further suggest that, apart from the electrostatic association of IL cations with the DNA backbone, groove binding of IL cations through hydrophobic and polar interactions contributes significantly to DNA stability (Chandran et al. 2012). They also confirmed using fluorescent dye displacement assay the intrusion of IL molecules into the DNA minor groove. Portella et al. showed using MD simulations that single-stranded DNA with G–C bases possesses more preferable solvation energy than A–T base pairs in IL solution and the solvation differences affect the DNA stability (Portella et al. 2014). Overall, the general behaviour of ILs towards duplexes arises from its specific interactions with the grooves in nucleic acids.
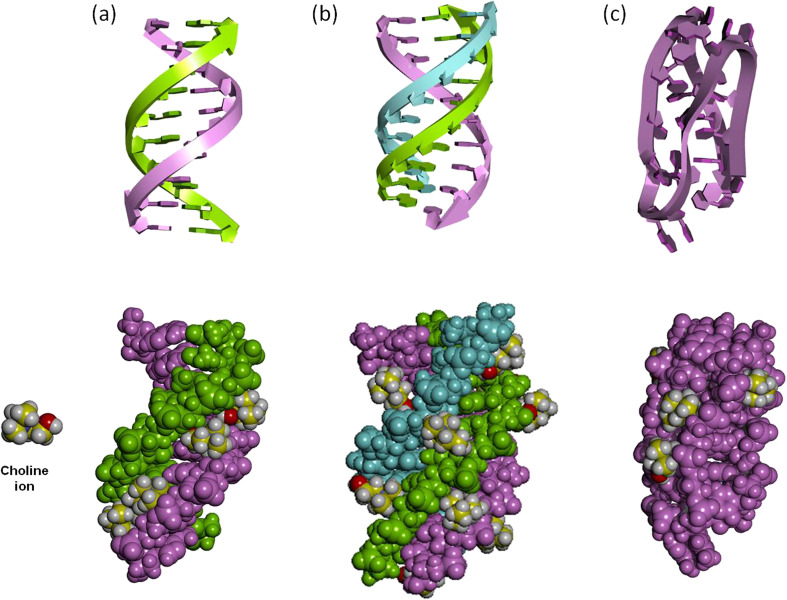
Fig. 2.
Interaction of choline ions with DNA structures of (a) duplex, (b) triplex, and (c) i-motif. Upper figures: DNA structures drawn by ribbon models. Lower figures: Interaction of choline ions with DNA structures drawn by van der Waals models. Carbon, oxygen, nitrogen, and hydrogen atoms in choline ions are shown in yellow, red, blue, and white, respectively
Moreover, we observed that a DNA triplex was significantly stabilized in choline dhp compared to an aqueous buffer at neutral pH, although triplexes are not formed at neutral pH in an aqueous solution (Tateishi-Karimata et al. 2014). Interestingly, the stability of Hoogsteen base pairs was comparable to that of Watson-Crick base pairs in hydrated IL. MD simulations of a DNA triplex in the presence of choline ions revealed that the DNA triplex was stabilized because of the binding of choline ions around the third strand in the grooves (Fig. 2(b)) (Nakano et al. 2014a). Fujita et al. reported that stable G-quadruplexes were formed in choline dhp, in which choline cations existed the core of the quadruplex structure similar to that observed for Na+ or K+ ions (Fujita and Ohno 2012). Interestingly, thermodynamic analyses and molecular dynamic calculations demonstrated that i-motifs in choline dhp were more stable than G-quadruplexes as choline ions bound to loop regions and grooves of the i-motifs (Fig. 2(c)) (Tateishi-Karimata et al. 2015a). Usually, i-motifs are not formed at physiological pH; however, choline ion binding dramatically induced the formation of i-motif at physiological pH. These results indicated that the stabilities of nucleic acids are decidedly difference relative to those in the aqueous solution; therefore, ILs are good solvent to control the stability of nucleic acids.
Storage of DNA in ILs
Media in which DNA is soluble and stable at a room temperature for long periods without loss of structure has been elusive and hence a bottleneck in DNA nanotechnology. Hydrated ILs, like choline dhp with 20% dissolved water, have been shown to be good solvents for proteins (Fujita et al. 2007) and choline dhp has chaperon-like activity for the protein (Fujita et al. 2016). To determine the chemical stability of DNA in ILs, MacFarlane et al. measured the CD spectra of long DNA from salmon testes in hydrated choline dhp (4 M) over time (Vijayaraghavan et al. 2010) and observed a slight change in the CD spectra intensity after 6 months. DNA in an aqueous solution is substantially denatured after 6 months under these conditions (Vijayaraghavan et al. 2010). This suggests that DNA was stabilized chemically by the hydrated IL medium, as has been observed for proteins (Fujita et al. 2005). Prasad et al. studied DNA from salmon testes in the choline ion-based IL of choline-indole-3-acetate (chol-IAA) (Mukesh et al. 2013). DNA was solubilized in the IL at concentrations of up to 3.5% (wt/wt). No structural degradation of the molecule was observed for the sample solubilized in chol-IAA after 6 months of storage. Senapati et al. investigated the interactions between IL cations and a DNA duplex (CGCGAATTCGCG)2 by MD simulations. Results suggest that in addition to the electrostatic association of cations in hydrated ILs with the DNA backbone, groove binding by IL cations via hydrophobic and polar interactions contributes significantly to DNA stability (Chandran et al. 2012). Interestingly, IL ions disrupt the water cage around DNA, including the spine of hydration in the minor groove (Chandran et al. 2012). This partial dehydration by ILs likely prevents hydrolytic reactions and stabilizes DNA in the long run. Moreover, plasmid DNA (pDNA), which is used in gene therapy and as a DNA vaccine, is susceptible to nuclease-mediated degradation in the extracellular environment. It was observed that enhanced enzymatic degradation resistance of pDNA when stored in a hydrated IL based on choline dhp. The stability of pDNA stored in choline dhp in the presence of DNase was studied using agarose gel electrophoresis, which showed longer shelf life at room temperature over a period of 28 days in choline dhp than in phosphate-buffered saline for 10 min. In addition, the biological activity of pDNA was maintained under such conditions, as demonstrated by the expression of yellow fluorescent protein.
Extraction and purification of DNAs using ILs
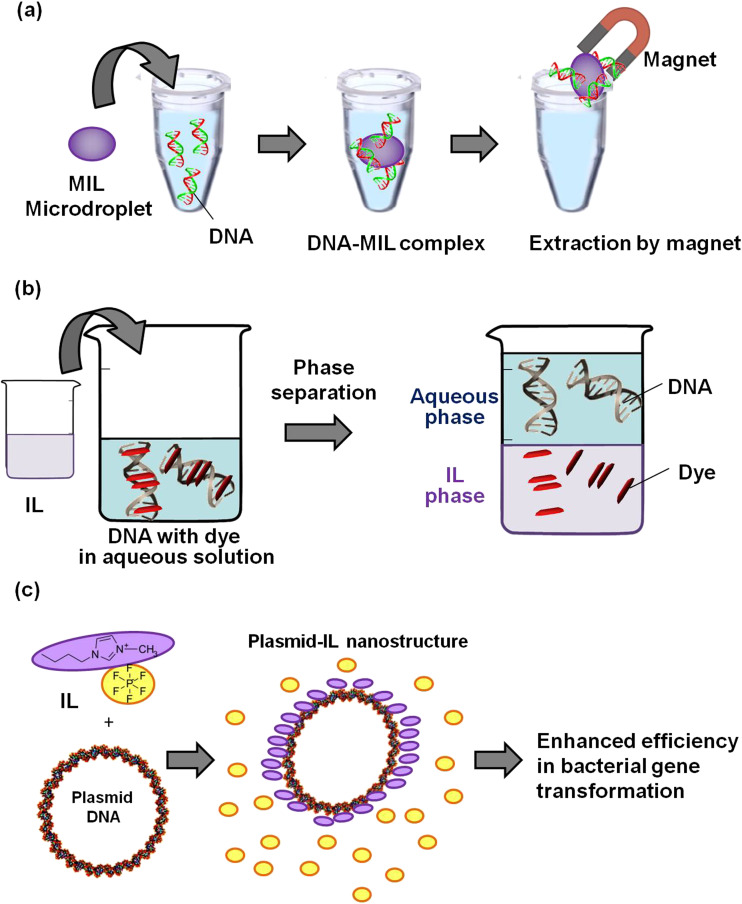
Purification of nucleic acids is essential for biological and bioapplicable studies. Currently, ILs are playing an increasingly important role in separation science, because their physical properties can be varied and their structural design can be tuned to achieve the desired functionality and enhance selectivity, efficiency, and sensitivity for the extraction of nucleic acids. Clark et al. synthesized hydrophobic magnetic ionic liquids (MILs) and used them as solvents for the rapid and efficient extraction of DNA from an aqueous solution (Fig. 3a) (Clark et al. 2015a). High extraction efficiencies were obtained for smaller single-stranded and double-stranded DNA using benzyltrioctylammonium bromotrichloroferrate (III) MIL, whereas higher extraction efficiencies for larger DNA molecules were obtained with dicationic 1,12-di(3hexadecylbenzimidazolium)dodecane bis[(trifluoromethyl)-sulfonyl]imide bromotrichloroferrate(III) MIL. Moreover, several methods for DNA extraction using MILs are also developed and the DNA in MIL is transferred directly to a polymerase chain reaction (PCR) tube for analysis (Clark et al. 2017a, b; 2015b). Shi et al. observed that a bicyclic imidazolium ionic liquid, was highly effective not only for promoting PCR of GC-rich DNA by minimizing non-specific amplification but also for facilitating PCR of normal-GC DNA under mild conditions because of destabilization of DNA duplexes (Shi et al. 2012). Furthermore, most DNA staining dyes commonly used in molecular biology possess large aromatic rings, which can participate in van der Waals interactions. Partitioning of oligonucleotides and DNA staining dyes in few hydrophobic ionic liquids has been studied, where the oligonucleotides remain in the aqueous phase and the DNA staining dyes are extracted in the ionic liquid phase, allowing the separation of these two solutions (Fig. 3b) (Khimji et al. 2013).
Fig. 3.
Biological application of nucleic acids in ILs. a Efficient extraction of DNA using magnetic ionic liquids (MILs), b separation of DNA and its staining dyes, and c functional nanostructure formations by DNA and IL cation
Moreover, bacterial gene transformation is one of the key techniques used in molecular biology and gene cloning, wherein bacteria receive a new genetic trait from fragments of foreign DNA. Generally, such transformation occurs naturally in few prokaryotic systems; however, most of the bacterial cells require artificial transformation because of the slow diffusion of hydrophilic DNA across the hydrophobic lipid bilayer membrane and the slight electrostatic repulsion between anionic DNA and the anionic headgroups of the bilayer membrane. Soni et al. observed that the electrostatic interaction between the negatively charged phosphate groups of pDNA and the cationic part of the hydrophobic ionic liquid 1-butyl-3-methylimidazolium hexafluorophosphate initiates spontaneous self-assembly to form functional nanostructures consisting of DNA and IL (Fig. 3c) (Soni et al. 2015). These functional nanostructures were promising synthetic non-viral vectors for efficient bacterial pGFP transformation. Soni et al. revealed that the electrostatic interaction between negatively charged phosphate oxygen and cationic ions of the IL tends to initiate the self-assembly process. Thermogravimetric analysis of the DNA-IL functional nanostructures showed that these nanostructures consist of about 16 wt% ionic liquid, which may stabilize the pDNA and eventually enhance transformation efficiency. In contrast, toxic effects of 1-octyl-3-methylimidazolium chloride, 1-octyl-3-methylimidazolium bromide, and 1-octyl-3-methylimidazolium tetrafluoroborate in soil on Vicia faba seedlings were assessed. The level of reactive oxygen species (ROS) was increased after exposure to the three ILs, which resulted in lipid peroxidation, DNA damage, and oxidative damage in the cells of the V. faba seedlings (Liu et al. 2015). Therefore, we should assess safety and effectiveness for the ILs before practical use.
DNA nanotechnology in ILs
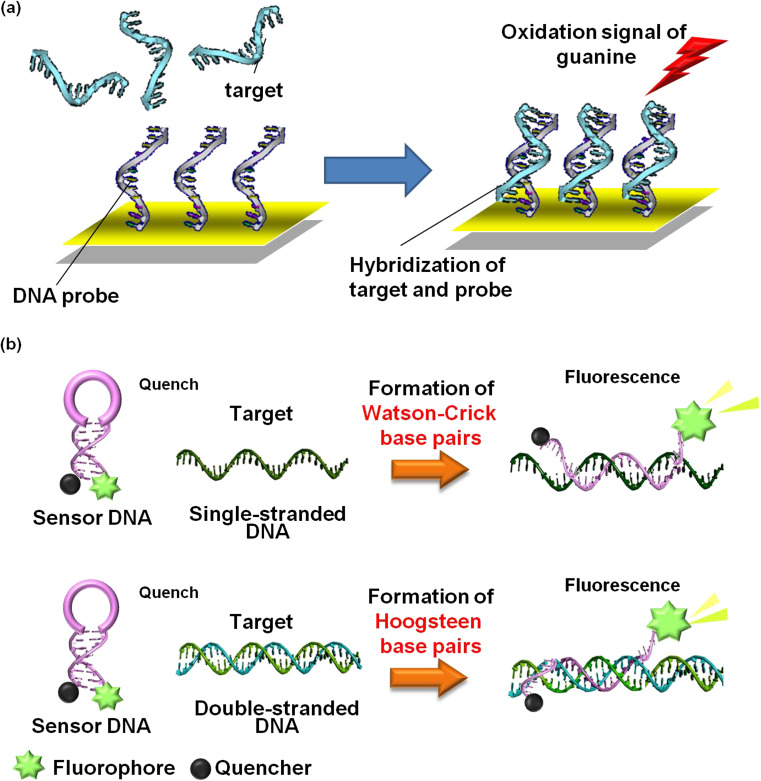
Nucleic acid analysis using biosensing strategies is an attractive topic in many fields including gene analysis, clinical disease diagnosis, biological, environmental, and pharmaceutical and forensic applications. Owing to their unique properties such as low measurable vapour pressure, high thermal stability and conductivity, good solvating properties, non-volatility, low toxicity, and biocompatibility, ILs have been used in different fields including development of electrochemical biosensors. A chronocoulometric DNA sensor based on polyaniline nanotube and IL ([C12mim][PF6])-doped screen-printed electrode were developed (Ren et al. 2010). The target DNA sequences were specifically recognized efficiently by the chronocoulometric DNA sensor. Eksin et al. developed the chitosan/ionic liquid-modified pencil graphite electrodes for enhanced electrochemical monitoring of nucleic acids and interactions of an anticancer drug, mitomycin C with calf thymus double-stranded DNA (Erdem et al. 2014). She et al. investigated the hydrophobic IL-mediated enhancement of hydroquinone detection on carbon paste electrode (She et al. 2010). The IL of 1-octyl-3-methylimidazolium hexafluorophosphate as a hydrophobic conductive pasting binder showed better electrochemical performance compared with the commonly employed binder (She et al. 2010). Moreover, it was also developed that an electrochemical aptasensor for high-sensitivity determination of carcinoembryonic antigen based on l DNAzyme-assisted signal amplification and graphene quantum dot-ionic liquid-nafion composite film (Huang et al. 2018). Sengiz et al. developed electrochemical DNA biosensors using IL (1-butyl-3-methylimidazolium hexafluorophosphate)-disposable pencil graphite electrodes (PGEs) (Sengiz et al. 2015). The IL-PGEs were used for electrochemical monitoring of sequence-selective DNA hybridization for Microcystis spp. without using any external hybridization indicator. The electrochemical biosensor showed high sensitivity and selectivity for the target sequence (Fig. 4a) (Sengiz et al. 2015). MD simulations suggested that the binding of ILs can restrain dynamic conformation and lower the on-site energy of DNA base. Confined movement among the adjacent base pairs was highly related to the increase of electronic coupling among base pairs, which may lead DNA to a charge transport-facilitated state (Meng et al. 2018). Therefore, ILs are useful to enhance the sensitivity of electrochemical DNA sensors by improvement of the electrochemical performance.
Fig. 4.
Nanotechnological of nucleic acids in ILs. a DNA sensor based on electrochemical detection of target hybridization and b DNA sensor based on molecular beacon systems
On the other hands, using properties of ILs, the DNA sequence selectivity is much improved via the DNA and IL interactions (Tateishi-Karimata et al. 2015b; Tateishi-Karimata and Sugimoto 2016). Systems for sensing specific DNA sequences, particularly single nucleotide polymorphisms (SNPs), are important in medicine and nanobiotechnology (Brown et al. 1990; de Wind and Hays 2001; Ellis and Minton 2003; Erdogan et al. 2001; Krieg et al. 2004; Thoma et al. 2005). Traditional methods for sensing DNA sequences using molecular beacons, DNA microarrays, or in situ hybridisation are based on the formation of A–T and G–C Watson–Crick base pairs between the target sequence and the probe DNA (Drummond et al. 2003; Francois et al. 1988; Lee et al. 2010; Schena et al. 1995). In general, duplexes of fully matched Watson-Crick of A–T and G–C base pairs are more stable than those with mismatched base pairs; therefore, an optimally designed sensor DNA should discriminate a completely complementary target sequence from the one containing mismatches. However, the use of these sensors is compromised by thermodynamically stable mismatches such as the G–T mismatch (Allawi and SantaLucia Jr. 1997; Leonard et al. 1990; Peyret et al. 1999), although approximately 30% of the SNPs result in G–T mismatches between the target and probe (Gu et al. 1991; Mokry et al. 2010), and the formation of thermodynamically stable mismatched duplexes significantly decreases probe sensitivity. We have demonstrated that the choline dhp significantly alters DNA stability using thermodynamic analyses and molecular dynamic simulations (Nakano et al. 2014a; Tateishi-Karimata et al. 2014). Based on our findings, we considered developing new DNA sensors using DNA-choline dhp interactions and determined the stabilities of matched and mismatched Watson-Crick and Hoogsteen base pairs in the choline dhp (Fig. 4b). Mismatched base pairs were significantly more destabilized in choline dhp than in an aqueous buffer (Tateishi-Karimata et al. 2015b). Furthermore, a molecular beacon that forms a triplex with a conserved HIV-1 sequence in choline dhp detected the target duplex via Hoogsteen base pair formation at concentrations as low as 1 pmol 10 μL−1(Tateishi-Karimata et al. 2015b). Notably, the sequence selectivity in choline dhp was 10,000-fold higher than that in an aqueous buffer. Moreover, the molecular beacon was protected from a contaminating nuclease in choline dhp, and the DNA strands in aqueous solution were not sufficiently stable for practical use (Tateishi-Karimata et al. 2015b). Ricci et al. reported a complicated electrochemical sensor for the detection of PCR-amplified HIV-1 targets with triplex-forming oligonucleotides with a detection limit of 10 nM (Patterson et al. 2010). The present simpler sensing system exhibited comparable sensitivity. Moreover, traditional methods for sensing of DNA sequence, including DNA microarrays, Southern blots, and in situ hybridization, are based on the formation of Watson-Crick base pairs and require the generation of single-stranded DNA prior to analysis. To simplify target detection, a number of approaches have been developed in which double-stranded targets are detected directly (Kuhn et al. 2002; Marletta et al. 1981). As these systems use intercalating dyes and groove-binding ligands, these approaches lack sequence specificity and are prone to false-positive detections (Ihmels et al. 2005; Persil and Hud 2007). The triplex is a promising recognition motif for sequence-specific sensing of double-stranded DNA targets (Kuhn et al. 2002; Marletta et al. 1981). Based on our finding that Hoogsteen base pairs are significantly stabilized with choline dhp, sequence-specific sensing of double-stranded DNAs will be possible without the requirement of duplex denaturation or complicated instrumentation (Tateishi-Karimata et al. 2015b).
As a new nanotechnology, it is developed the ion conductive DNA films using IL (Nishimura and Ohno 2005). Ion conductive and flexible DNA film was prepared by direct mixing of DNA with salt-containing PEO and neutralization of DNA bases with an acid. The DNA bases were neutralized with several acids to convert them into ionic liquid. Addition of further ionic liquid (ethylimidazolium tetrafluoroborate (EtImBF4)) allowed the preparation of highly ion conductive films. Moreover, an ionic liquid domain was successfully prepared outside double-stranded DNA by fixing 1-alkyl-3-methylimidazolium cations on the phosphate groups of DNA (Nishimura and Ohno 2005). These reports will usher a new field on the use of DNA as biomass.
Liu et al. studied DNA-linked gold nanoparticles (AuNPs) in choline dhp and in ILs containing propylammonium nitrate, ethylammonium nitrate, methylammonium nitrate, and dimethylammonium nitrate (Menhaj and Liu 2012). DNA-functionalized AuNPs possess high density of negative charges and may acquire new physical properties in ILs. The authors showed that ILs transit from salts to increase or decrease DNA duplex stability depending on IL concentration, with more hydrophobic cations destabilizing DNA at lower IL concentrations. The onset of this transition depends on the structure of ILs. This trend is opposite to that observed with molecular solvents (e.g., ethanol, dimethyl sulphoxide (DMSO), acetonitrile, and dimethyl formamide), which destabilize DNA at low solvent concentrations (Marchand et al. 2013). Specific DNA base pairing is disrupted at high DMSO concentrations, and AuNPs are held together by non-specific interactions. DNA base pairs are maintained in other tested molecular solvents, although strong non-specific interactions are also present. Several ILs can release protons and thus drastically change pH, which also changes the melting temperature of DNA. Liu et al. also demonstrated the feasibility of using ILs as solvents for DNA-functionalized nanomaterials.
Conclusions
Employing IL-DNA interactions can achieve simply and easily purification and solve the problem of long-term storing nucleic acids. Natural DNAs are not chemically stable in solution at ambient temperatures for long periods. It was reported that DNA has long-term stability in hydrated ILs. The nuclease-mediated degradation of DNA is also inhibited in ILs because electrostatic interactions in a solution containing high salt concentration distribute the protein folding. Thus, ILs should be as a chemical DNA stabilizer and nuclease inhibitor. The biological experiments generally use the enzymes such as RNA and DNA polymerase which might deactivate in the high concentration of ILs. To accelerate expansion of biological application using ILs, new ingenuity for keep the activity of specific enzymes in ILs is essential in the future.
Another important property of ILs is their specific interaction with DNAs. In general, five interactions play major roles in determining the stabilities and structures of DNAs: hydrogen bonding, base stacking, conformational entropy, hydration, and cation binding (Saenger 1984). The interactions of hydration and cation binding depend upon surrounding conditions (Nakano et al. 2014c; Saha and Mukherjee 2018). The hydration and cation binding should be altered by the addition of IL (Tateishi-Karimata and Sugimoto 2014). These properties of ILs should enhance the function of DNA nanodevices because DNA structure and stability changes have the potential to be used in advanced materials such as biocircuits, biodevices, and biosensors. Moreover, systems for sensing specific DNA sequences which we showed here are important in medicine and nanobiosensing. Thus, information in this review should facilitate development of new DNA materials.
Acknowledgements
We thank Prof. Shigenari Tanaka and Dr. Miki Nakano for useful comments on molecular dynamic simulation.
Funding information
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS), especially a Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (JSPS KAKENHI Grant No. JP17H06351); MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014-2019), Japan; the Hirao Taro Foundation of Konan Gakuen for Academic Research; the Okazaki Kazuo Foundation of Konan Gakuen for Advanced Scientific Research; the Chubei Itoh Foundation; and Kawanishi Memorial ShinMaywa Education Foundation.
Compliance with ethical standards
Conflict of interest
Hisae Tateishi-Karimata declares that she has no conflict of interest. Naoki Sugimoto declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on “Ionic Liquids and Biomolecules” edited by Antonio Benedetto and Hans-Joachim Galla.
References
- Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- Armand M, Endres F, MacFarlane DR, Ohno H, Scrosati B. Ionic-liquid materials for the electrochemical challenges of the future. Nat Mater. 2009;8:621–629. doi: 10.1038/nmat2448. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Ballon P (2016a) Room Temperature Ionic Liquids Meet Biomolecules: A Microscopic View of Structure and Dynamics. ACS Sustainable Chem. Eng. 4 :392–412. 10.1021/acssuschemeng.5b01385
- Benedetto A, Ballon P (2016b) Room temperature ionic liquids interacting with bio-molecules: an overview of experimental and computational studies. Philosophical Magazine 96 :870–894.
- Brown T, Leonard GA, Booth ED, Kneale G. Influence of pH on the conformation and stability of mismatch base-pairs in DNA. J Mol Biol. 1990;212:437–440. doi: 10.1016/0022-2836(90)90320-L. [DOI] [PubMed] [Google Scholar]
- Chandran A, Ghoshdastidar D, Senapati S. Groove binding mechanism of ionic liquids: a key factor in long-term stability of DNA in hydrated ionic liquids? J Am Chem Soc. 2012;134:20330–20339. doi: 10.1021/ja304519d. [DOI] [PubMed] [Google Scholar]
- Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- Clark KD, Nacham O, Yu H, Li T, Yamsek MM, Ronning DR, Anderson JL. Extraction of DNA by magnetic ionic liquids: tunable solvents for rapid and selective DNA analysis. Anal Chem. 2015;87:1552–1559. doi: 10.1021/ac504260t. [DOI] [PubMed] [Google Scholar]
- Clark KD, Yamsek MM, Nacham O, Anderson JL. Magnetic ionic liquids as PCR-compatible solvents for DNA extraction from biological samples. Chem Commun (Camb) 2015;51:16771–16773. doi: 10.1039/c5cc07253k. [DOI] [PubMed] [Google Scholar]
- Clark KD, Purslow JA, Pierson SA, Nacham O, Anderson JL. Rapid preconcentration of viable bacteria using magnetic ionic liquids for PCR amplification and culture-based diagnostics. Anal Bioanal Chem. 2017;409:4983–4991. doi: 10.1007/s00216-017-0439-y. [DOI] [PubMed] [Google Scholar]
- Clark KD, Varona M, Anderson JL. Ion-tagged oligonucleotides coupled with a magnetic liquid support for the sequence-specific capture of DNA. Angew Chem Int Ed Engl. 2017;56:7630–7633. doi: 10.1002/anie.201703299. [DOI] [PubMed] [Google Scholar]
- de Wind N, Hays JB. Mismatch repair: praying for genome stability. Curr Biol. 2001;11:R545–R548. doi: 10.1016/S0960-9822(01)00338-4. [DOI] [PubMed] [Google Scholar]
- Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- Earle MJ, Seddon KR. Ionic liquids. Green solvents for the future. Pure Appl Chem. 2000;72:1391–1398. doi: 10.1351/pac200072071391. [DOI] [Google Scholar]
- Ellis RJ, Minton AP. Cell biology: join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- Erdem A, Muti M, Mese F, Eksin E. Chitosan-ionic liquid modified single-use sensor for electrochemical monitoring of sequence-selective DNA hybridization. Colloids Surf B Biointerfaces. 2014;114:261–268. doi: 10.1016/j.colsurfb.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Erdogan F, Kirchner R, Mann W, Ropers HH, Nuber UA. Detection of mitochondrial single nucleotide polymorphisms using a primer elongation reaction on oligonucleotide microarrays. Nucleic Acids Res. 2001;29:E36. doi: 10.1093/nar/29.7.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedrigo O, Naylor G. A gene-specific DNA sequencing chip for exploring molecular evolutionary change. Nucleic Acids Res. 2004;32:1208–1213. doi: 10.1093/nar/gkh210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois JC, Saison-Behmoaras T, Helene C. Sequence-specific recognition of the major groove of DNA by oligodeoxynucleotides via triple helix formation. Footprinting studies. Nucleic Acids Res. 1988;16:11431–11440. doi: 10.1093/nar/16.24.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Ohno H. Enzymatic activity and thermal stability of metallo proteins in hydrated ionic liquids. Biopolymers. 2010;93:1093–1099. doi: 10.1002/bip.21526. [DOI] [PubMed] [Google Scholar]
- Fujita K, Ohno H. Stable G-quadruplex structure in a hydrated ion pair: cholinium cation and dihydrogen phosphate anion. Chem Commun (Camb) 2012;48:5751–5753. doi: 10.1039/c2cc30554b. [DOI] [PubMed] [Google Scholar]
- Fujita K, MacFarlane DR, Forsyth M (2005) Protein solubilising and stabilising ionic liquids. Chem Commun (Camb):4804–4806 doi: 10.1039/b508238b [DOI] [PubMed]
- Fujita K, MacFarlane DR, Forsyth M, Yoshizawa-Fujita M, Murata K, Nakamura N, Ohno H. Solubility and stability of cytochrome c in hydrated ionic liquids: effect of oxo acid residues and kosmotropicity. Biomacromolecules. 2007;8:2080–2086. doi: 10.1021/bm070041o. [DOI] [PubMed] [Google Scholar]
- Fujita K, Kajiyama M, Liu Y, Nakamura N, Ohno H. Hydrated ionic liquids as a liquid chaperon for refolding of aggregated recombinant protein expressed in Escherichia coli. Chem Commun (Camb) 2016;52:13491–13494. doi: 10.1039/c6cc06999a. [DOI] [PubMed] [Google Scholar]
- Gu XF, Lee JS, Delfau MH, Grandchamp B. PCR detection of a G/T polymorphism at exon 10 of the porphobilinogen deaminase gene (PBG-D) Nucleic Acids Res. 1991;19:1966. doi: 10.1093/nar/19.8.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- Huang JY, et al. A high-sensitivity electrochemical aptasensor of carcinoembryonic antigen based on graphene quantum dots-ionic liquid-nafion nanomatrix and DNAzyme-assisted signal amplification strategy. Biosens Bioelectron. 2018;99:28–33. doi: 10.1016/j.bios.2017.07.036. [DOI] [PubMed] [Google Scholar]
- Hyde JE, Read M. The extraction and purification of DNA and RNA from in vitro cultures of the malaria parasite Plasmodium falciparum. Methods Mol Biol. 1993;21:133–143. doi: 10.1385/0-89603-239-6:133. [DOI] [PubMed] [Google Scholar]
- Ihmels H, et al. Anthryl-substituted heterocycles as acid-sensitive fluorescence probes. J Org Chem. 2005;70:3929–3938. doi: 10.1021/jo047841z. [DOI] [PubMed] [Google Scholar]
- Khimji I, Doan K, Bruggeman K, Huang PJ, Vajha P, Liu J. Extraction of DNA staining dyes from DNA using hydrophobic ionic liquids. Chem Commun (Camb) 2013;49:4537–4539. doi: 10.1039/c3cc41364k. [DOI] [PubMed] [Google Scholar]
- Krieg A, Laib S, Ruckstuhl T, Seeger S. Fast detection of single nucleotide polymorphisms (SNPs) by primer elongation with monitoring of supercritical-angle fluorescence. Chembiochem. 2004;5:1680–1685. doi: 10.1002/cbic.200400044. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J Am Chem Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- Lee JB, Campolongo MJ, Kahn JS, Roh YH, Hartman MR, Luo D. DNA-based nanostructures for molecular sensing. Nano. 2010;2:188–197. doi: 10.1039/b9nr00142e. [DOI] [PubMed] [Google Scholar]
- Legoff J, et al. Influence of storage temperature on the stability of HIV-1 RNA and HSV-2 DNA in cervicovaginal secretions collected by vaginal washing. J Virol Methods. 2006;138:196–200. doi: 10.1016/j.jviromet.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Leonard GA, Booth ED, Brown T. Structural and thermodynamic studies on the adenine.guanine mismatch in B-DNA. Nucleic Acids Res. 1990;18:5617–5623. doi: 10.1093/nar/18.19.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhu L, Wang J, Wang J, Zhang J, Sun X, Zhang C. Biochemical toxicity and DNA damage of imidazolium-based ionic liquid with different anions in soil on Vicia faba seedlings. Sci Rep. 2015;5:18444. doi: 10.1038/srep18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand A, Ferreira R, Tateishi-Karimata H, Miyoshi D, Sugimoto N, Gabelica V. Sequence and solvent effects on telomeric DNA bimolecular G-quadruplex folding kinetics. J Phys Chem B. 2013;117:12391–12401. doi: 10.1021/jp406857s. [DOI] [PubMed] [Google Scholar]
- Marletta MA, Srere PA, Walsh C. Stereochemical outcome of processing of fluorinated substrates by ATP citrate lyase and malate synthase. Biochemistry. 1981;20:3719–3723. doi: 10.1021/bi00516a008. [DOI] [PubMed] [Google Scholar]
- Marusic M, Tateishi-Karimata H, Sugimoto N, Plavec J. Structural foundation for DNA behavior in hydrated ionic liquid: an NMR study. Biochimie. 2015;108:169–177. doi: 10.1016/j.biochi.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Meng Z, Kubar T, Mu Y, Shao F (2018) A MD-QM theoretical study of DNA-mediated charge transport in hydrated ionic liquids. J Chem Theory Comput. 10.1021/acs.jctc.7b01201 [DOI] [PubMed]
- Menhaj AB, Liu J. Exploring the thermal stability of DNA-linked gold nanoparticles in ionic liquids and molecular solvents. Chem Sci. 2012;3:3216–3220. doi: 10.1039/c2sc20565c. [DOI] [Google Scholar]
- Mitchell KA, Zingone A, Toulabi L, Boeckelman J, Ryan BM. Comparative transcriptome profiling reveals coding and noncoding RNA differences in NSCLC from African Americans and European Americans. Clin Cancer Res. 2017;23:7412–7425. doi: 10.1158/1078-0432.CCR-17-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M, Feitsma H, Nijman IJ, de Bruijn E, van der Zaag PJ, Guryev V, Cuppen E. Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Res. 2010;38:e116. doi: 10.1093/nar/gkq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima C, Chung M, Ng KW, Brambilla DJ, Gretch DR. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J Clin Microbiol. 2004;42:421–425. doi: 10.1128/JCM.42.1.421-425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukesh C, Mondal D, Sharma M, Prasad K. Rapid dissolution of DNA in a novel bio-based ionic liquid with long-term structural and chemical stability: successful recycling of the ionic liquid for reuse in the process. Chem Commun (Camb) 2013;49:6849–6851. doi: 10.1039/c3cc42829j. [DOI] [PubMed] [Google Scholar]
- Nakano M, Tateishi-Karimata H, Tanaka S, Sugimoto N. Affinity of molecular ions for DNA structures is determined by solvent-accessible surface area. J Phys Chem B. 2014;118:9583–9594. doi: 10.1021/jp505107g. [DOI] [PubMed] [Google Scholar]
- Nakano M, Tateishi-Karimata H, Tanaka S, Sugimoto N. Choline ion interactions with DNA atoms explain unique stabilization of A-T base pairs in DNA duplexes: a microscopic view. J Phys Chem B. 2014;118:379–389. doi: 10.1021/jp406647b. [DOI] [PubMed] [Google Scholar]
- Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev. 2014;114:2733–2758. doi: 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Ohno H. Design of successive ion conduction paths in DNA films with ionic liquids. J Mater Chem. 2005;12:2299–2304. doi: 10.1016/j.biomaterials.2005.02.005. [DOI] [Google Scholar]
- Ott J, Hoh J. Set association analysis of SNP case-control and microarray data. J Comput Biol. 2003;10:569–574. doi: 10.1089/10665270360688192. [DOI] [PubMed] [Google Scholar]
- Patterson A, Caprio F, Vallee-Belisle A, Moscone D, Plaxco KW, Palleschi G, Ricci F. Using triplex-forming oligonucleotide probes for the reagentless, electrochemical detection of double-stranded DNA. Anal Chem. 2010;82:9109–9115. doi: 10.1021/ac1024528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persil O, Hud NV. Harnessing DNA intercalation. Trends Biotechnol. 2007;25:433–436. doi: 10.1016/j.tibtech.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- Phadke RS. Biomolecular electronics in the twenty-first century. Appl Biochem Biotechnol. 2001;96:269–276. doi: 10.1385/ABAB:96:1-3:279. [DOI] [PubMed] [Google Scholar]
- Portella G, Germann MW, Hud NV, Orozco M. MD and NMR analyses of choline and TMA binding to duplex DNA: on the origins of aberrant sequence-dependent stability by alkyl cations in aqueous and water-free solvents. J Am Chem Soc. 2014;136:3075–3086. doi: 10.1021/ja410698u. [DOI] [PubMed] [Google Scholar]
- Ren R, Leng C, Zhang S. A chronocoulometric DNA sensor based on screen-printed electrode doped with ionic liquid and polyaniline nanotubes. Biosens Bioelectron. 2010;25:2089–2094. doi: 10.1016/j.bios.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acid structure. New York: Springer-Verlag; 1984. [Google Scholar]
- Saha D, Mukherjee A (2018) Effect of water and ionic liquids on biomolecules. Biophys Rev. 10.1007/s12551-018-0399-2 [DOI] [PMC free article] [PubMed]
- Sasaki Y, Miyoshi D, Sugimoto N. Regulation of DNA nucleases by molecular crowding. Nucleic Acids Res. 2007;35:4086–4093. doi: 10.1093/nar/gkm445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sengiz C, Congur G, Erdem A. Development of ionic liquid modified disposable graphite electrodes for label-free electrochemical detection of DNA hybridization related to Microcystis spp. Sensors (Basel) 2015;15:22737–22749. doi: 10.3390/s150922737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Y, Tang Y, Liu H, He P. Electrochemical determination of hydroquinone using hydrophobic ionic liquid-type carbon paste electrodes. Chem Cent J. 2010;4:17. doi: 10.1186/1752-153X-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Liu YL, Lai PY, Tseng MC, Tseng MJ, Li Y, Chu YH. Ionic liquids promote PCR amplification of DNA. Chem Commun (Camb) 2012;48:5325–5327. doi: 10.1039/c2cc31740k. [DOI] [PubMed] [Google Scholar]
- Soni SK, Sarkar S, Mirzadeh N, Selvakannan PR, Bhargava SK. Self-assembled functional nanostructure of plasmid DNA with ionic liquid [Bmim][PF(6)]: enhanced efficiency in bacterial gene transformation. Langmuir. 2015;31:4722–4732. doi: 10.1021/acs.langmuir.5b00402. [DOI] [PubMed] [Google Scholar]
- Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol. 2009;2009:574398. doi: 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Sugimoto N. A-T base pairs are more stable than G-C base pairs in a hydrated ionic liquid. Angew Chem Int Ed Engl. 2012;51:1416–1419. doi: 10.1002/anie.201106423. [DOI] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Sugimoto N. Structure, stability and behaviour of nucleic acids in ionic liquids. Nucleic Acids Res. 2014;42:8831–8844. doi: 10.1093/nar/gku499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Sugimoto N. Expansion of the DNA alphabet beyond natural DNA. Recognition Chembiochem. 2016;17:1301–1303. doi: 10.1002/cbic.201600181. [DOI] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Nakano M, Sugimoto N. Comparable stability of Hoogsteen and Watson-Crick base pairs in ionic liquid choline dihydrogen phosphate. Sci Rep. 2014;4:3593. doi: 10.1038/srep03593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Nakano M, Pramanik S, Tanaka S, Sugimoto N. i-Motifs are more stable than G-quadruplexes in a hydrated ionic liquid. Chem Commun (Camb) 2015;51:6909–6912. doi: 10.1039/c5cc00666j. [DOI] [PubMed] [Google Scholar]
- Tateishi-Karimata H, Pramanik S, Sugimoto N. DNA sensor’s selectivity enhancement and protection from contaminating nucleases due to a hydrated ionic liquid. Analyst. 2015;140:4393–4398. doi: 10.1039/c5an00545k. [DOI] [PubMed] [Google Scholar]
- Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Wolfle WT, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Dpo4 is hindered in extending a G.T mismatch by a reverse wobble. Nat Struct Mol Biol. 2004;11:457–462. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan R, Izgorodin A, Ganesh V, Surianarayanan M, DR MF. Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew Chem Int Ed Engl. 2010;49:1631–1633. doi: 10.1002/anie.200906610. [DOI] [PubMed] [Google Scholar]
- Welton T. Room-temperature ionic liquids. Solvents for synthesis and catalysis. Chem Rev. 1999;99:2071–2084. doi: 10.1021/cr980032t. [DOI] [PubMed] [Google Scholar]
- Yilmaz LS, Bergsven LI, Noguera DR. Systematic evaluation of single mismatch stability predictors for fluorescence in situ hybridization. Environ Microbiol. 2008;10:2872–2885. doi: 10.1111/j.1462-2920.2008.01719.x. [DOI] [PubMed] [Google Scholar]
- Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]