Abstract
Background
Prostate cancer (PCa), the most common cancer and second leading cause of cancer death in American men, presents the clinical challenge of distinguishing between indolent and aggressive tumors for proper treatment. PCa presents significant alterations in metabolic pathways that can potentially be measured using techniques like mass spectrometry (MS) or mass spectrometry imaging (MSI) and used to characterize PCa aggressiveness. MS quantifies metabolomic, proteomic, and lipidomic profiles of biological systems that can be further visualized for their spatial distributions through MSI.
Methods
PubMed was queried for all publications relating to MS and MSI in human prostate cancer from April 2007 to April 2017. With the goal of reviewing the utility of MSI in diagnosis and prognostication of human PCa, MSI articles that reported investigations of PCa-specific metabolites or metabolites indicating PCa aggressiveness were selected for inclusion. Articles were included that covered MS and MSI principles, limitations, and applications in PCa.
Results
We identified nine key studies on MSI in intact human prostate tissue specimens that determined metabolites which could either differentiate between benign and malignant prostate tissue or indicate prostate cancer aggressiveness. These MSI-detected biomarkers show promise in reliably identifying PCa and determining disease aggressiveness.
Conclusions
MSI represents an innovative technique with the ability to interrogate cancer biomarkers in relation to tissue pathologies and investigate tumor aggressiveness. We propose MSI as a powerful adjuvant histopathology imaging tool for prostate tissue evaluations, where clinical translation of this ex vivo technique could make possible the use of MSI for personalized medicine in diagnosis and prognosis of prostate cancer. Moreover, the knowledge provided from this technique can majorly contribute to the understanding of molecular pathogenesis of PCa and other malignant diseases.
Introduction
The American Cancer Society declares prostate cancer (PCa) as the most common cancer in males, representing 19% of all diagnosed cancer cases in American men, with an estimated number of 26,730 deaths in the United States alone in 2017, PCa the second leading cause of cancer death1.
While the ability of serum prostate-specific antigen (PSA) level to detect early stage disease is evident, its introduction for annual testing in the 1980’s also led to the discovery of many predominantly slow growing prostate tumors that would not have become life-threatening during a patient’s life2–5. However, a considerable percentage of patients harbor aggressive disease that requires early and appropriate treatment consisting of surgery, radiation, chemotherapy, or hormonal therapy. Unfortunately, at present, the clinical ability to identify these patients with aggressive disease and differentiating them from patients harboring indolent tumors is often limited. Thus, the urge to treat cancer is often challenged by the concern of avoiding over-treatment in the PCa clinic6–8.
At present, histopathological examination can reliably distinguish benign from malignant lesions using the Gleason Score9,10 or Prognostic Grade Group system11, but its ability to distinguish indolent PCa from aggressive is still limited. The development and progression of PCa inevitably alters tissue biochemistry, and inspired by the developments in oncological genomics, proteomics, and lipidomics, mass spectrometry (MS)-based metabolic biomarkers have been investigated for their ability to detect and characterize PCa for better prognostication and individual therapy planning. MS investigations have been further enhanced by the innovation of mass spectrometry imaging (MSI), by which metabolic maps of intact tissues are generated12–18.
Here, while we focus our review on the current literature reports of MSI studies of PCa and the promise of this innovative molecular imaging method to improve PCa diagnosis and characterization, we will start our address with brief reviews in the MS and MSI techniques, and in the major achievements in MS evaluations of PCa.
Methods
For this review, PubMed was queried for all publications relating to MS and MSI in human prostate cancer from April 2007 to April 2017. Titles and abstracts of studies found in the search were reviewed, and relevant studies were advanced to full-text review. With the goal of reviewing the utility of MSI in diagnosis and prognostication of human PCa, MSI investigations were included that described identification of PCa-specific metabolites and metabolites indicating PCa aggressiveness. Additionally, MS articles were included which highlighted principles, applications in PCa, or limitations of the technique, and articles that described MSI principles in overcoming these limitations were selected. In a final step, the reference lists of included papers were screened, and new titles were reviewed for potential inclusion as previously described. Relevant data from the selected studies were summarized in the text and tabulated in Table 1.
Table 1.
Key studies on mass spectrometry imaging in intact human prostate tissue.
| Year | Topic | Main Findings | Ref |
|---|---|---|---|
| 2016 | MALDI-FTICR-MSI for PCa diagnosis | Distinction between cancerous and non-cancerous regions in prostate tissue through the expression of neutral acyl glycerides | 70 |
| 2015 | MALDI-MSI for PCa prognosis | Correlation between decreased expression of lysophosphatidylcholine and biochemical recurrence of PCa after surgical treatment | 82 |
| 2014 | MALDI-MSI for PCa diagnosis | Phosphatidylinositols represent potential biomarkers for PCa diagnosis | 65 |
| 2013 | MALDI-MSI for PCa diagnosis | N-glycan profiling provides a new opportunity to evaluate disease status in PCa patients | 88 |
| 2013 | MALDI-MSI for PCa diagnosis | Biliverdin reductase B serves as a potential biomarker for PCa diagnosis | 67 |
| 2013 | MALDI-MSI for PCa diagnosis and prognosis | Identification of potential biomarkers for PCa diagnosis and prognosis (signals associated with Gleason score, disease stage, Ki-67 and PSA recurrence) | 83 |
| 2010 | DESI-MSI for PCa diagnosis | Cholesterol sulfate represents a potential biomarker for PCa diagnosis | 64 |
| 2009 | MALDI-MSI for PCa diagnosis | The overexpression of MEKK2 can be used to discriminate malignant from benign prostate tissue | 63 |
| 2007 | MALDI-MSI for PCa diagnosis | Differentiation of cancerous and non-cancerous prostate tissue with an overall cross-validation of 88.00%, sensitivity 85.21% and specificity 90.74% | 69 |
Results
Mass spectrometry principles
Mass spectrometry is a chemical analysis technique that measures the mass of molecules by ionizing, separating, and detecting the resulting ions as peaks according to their mass-to-charge ratio. The resulting spectrum plots the relative abundance of each molecular fragment as a peak against its mass-to-charge ratio (m/z), and produces a unique profile for every sample. Similar peak patterns, however, can describe a similar family of molecules. The values of data signatures, or fingerprints, for the analyzed molecules are dependent on the procedures of sample preparation, the selection of the ionization method, and the configuration of the mass analyzer.
Mass spectrometry ionization and analysis methods
Two of the most frequently used MS ionization techniques are electrospray ionization (ESI) and matrix assisted laser desorption ionization (MALDI).
Initially, the use of MS was been restricted to gases, volatiles, and thermally stable molecules due to the limitations of early ionization techniques. However, the development of electrospray ionization (ESI) in 1984 by John B. Fenn and colleagues19, and the proposal of MALDI in 198520 with demonstration by Koichi Tanaka and colleagues in 198721, made it possible to analyze heavy and non-volatile molecules such as nucleic acids, proteins, and metabolites. For their respective contributions to the field, Fenn and Tanaka shared the Nobel Prize in Chemistry in 2002 with Kurt Wüthrich for developments in NMR that enabled direct analysis of biomolecules. Today, ESI and MALDI have become increasingly important techniques in the laboratories for structural analyses or quantitative measurements of metabolites in biological samples.
ESI produces ions using an electrospray in which a high voltage is applied to a liquid, creating a fine aerosol. The process of transferring ionic species from solution into the gas phase involves three steps: dispersal of a fine spray of charged droplets, solvent evaporation, and ion ejection from the highly charged droplets. With the aid of elevated temperature or nitrogen drying gas, the charged droplets are reduced in size by evaporation of the solvent, leading to increased charge density and decreased droplet radius. When the electric field strength within the charged droplet reaches a critical point, ions at the surface of the droplets are ejected into the gaseous phase. Finally, the ions are accelerated down a pressure and electric potential gradient toward the mass analyzer.
In MALDI, the sample is mixed with a UV-absorbing crystalline matrix material and spotted onto a target plate. The plate is placed in a vacuum and hit with a UV laser, and the matrix absorbs the irradiation – simultaneously volatilizing and ionizing the sample.
The sensitivity of a mass spectrometer relies on the structure of its mass analyzer; both time-of-flight (TOF)22 and quadrupole mass filters (Q)23 are commonly used mass analyzer structures. Furthermore, they can be combined to produce a high-resolution mass spectrometer (Q-TOF)24. Achieving high mass resolution and accuracy can also be accomplished by means of Fourier transform mass spectrometers, such as Fourier Transform Ion Cyclotron Resonance (FTICR)25.
Mass spectrometry applications in prostate cancer studies
Metabolomic studies of prostate cancer have been accomplished by MS to examine prostate cell lines26–28, animal prostate models29–31 and biological fluids including urine, prostatic fluid, semen and blood16,32–36. As this review focuses on prostate cancer diagnosis and prognosis in prostate tissue samples, three major MS studies investigating metabolomics in prostate tissues are noted.
In 2009, Chinnaiyan and colleagues reported a study of 42 prostate samples of adjacent benign (16) and malignant (12 localized and 14 metastatic PCa) tissues, along with 110 matched specimens of urine and plasma from biopsy-positive PCa patients (59) and biopsy-negative subjects (51)37. A total of 1,126 metabolites were quantified using high-throughput gas chromatography (GC)- and liquid chromatography (LC)-MS37. Among the 626 tissue-specific metabolites, 60 metabolites were exclusively found in malignant samples. Another 50 metabolites were increased and 37 were decreased in localized prostate cancer compared to benign prostate tissue samples. A total of 124 metabolites were increased and 107 metabolites were decreased in all biospecimen samples from patients with metastatic compared to patients with localized disease. In general, an upregulation in amino acid metabolism and nitrogen breakdown pathways during tumor metastasis was revealed. Particularly, the study identified sarcosine, an N-methyl derivative of glycine, as a significant metabolite that increased with PCa progression and metastasis and, most importantly, could be detected both in tissue and, non-invasively, in urine samples of PCa patients.
Recently, combined metabolomic, transcriptomic, and immunohistochemistry studies have been reported on malignant and matched adjacent non-malignant tissue samples from 106 patients with PCa38. Metabolomic analyses were performed by GC-MS and LC-MS and identified a set of 254 metabolites. Among them, 173 showed a capacity to differentiate malignant from non-malignant samples. Furthermore, analysis of a subset of metabolites indicated that twelve were up-regulated and four down-regulated in comparison to increased Gleason score. Specifically, positive correlations with pantothenic acid, and negative correlations with maltose, fructose-6-phosphate, gluconic acid and cholesterol, were reported. The study further observed metabolite changes in transmembrane protease serine 2 (TMPRSS2)-ERG translocation-positive PCa, with 55 metabolites increased and 17 decreased when compared to ERG-negative carcinomas. TMPRSS2-ERG gene fusion represents the predominant molecular subtype in PCa, and various studies have shown that TMPRSS2-ERG positivity is often associated with higher disease-related mortality39,40. In accordance with this report, the study reported a negative correlation between ERG positivity and maltotriose and gluconic acid quantified from MS, with the absence of these metabolites correlated with a greater chance of PCa recurrence17. Although previously correlated with PCa, for the first time the study showed the polyamines spermine and putrescine to be negatively correlated with presence of the poor-prognosis TMPRSS2-ERG fusion gene. In contrast, fatty acid levels (e.g. cerebronic acid, 2-hydroxybehenic acid and tricosanoic acid) were increased in ERG-positive PCa samples. These results clearly demonstrate the dysregulation of fatty acid, sphingolipid, and polyamine metabolism in PCa.
In a recently published study, mass spectrometry-based metabolomic analysis was performed in frozen and matched formalin-fixed paraffin embedded (FFPE) human prostate cancer tissue. A total of 352 metabolites were profiled, and physical-chemical characteristics of metabolites measured were related to their preservation or loss following fixation and embedding. Although FFPE tissue sections showed a decrease of metabolites with functional groups (e.g. carboxamide), global metabolomic profiles obtained from FFPE collections still proved useful for the prediction of biological states and discovery of biomarkers in tissue specimens, which offers great opportunity concerning retrospective tissue analysis41.
Recent technology developments have also enabled metabolomic MS evaluation of PCa metabolites and histopathology from the same specimen, especially important given the heterogeneity of PCa pathologies. A new preparation method uses methanol to simultaneously extract metabolites for MS measurement and fix the tissue for histopathological evaluation42. Pathology analyses, including H&E and immunohistochemistry, as well as metabolomic analyses, were successfully carried out with MS in this fashion42. In 96 samples, a total of 260 metabolites in the alcohol extracts were detected, and among them 82 metabolites were increased in PCa containing biopsies compared to histologically benign prostate biopsies.
However, mass spectrometry itself cannot directly attribute the measured MS values to each individual pathology, but the development of mass spectrometry imaging (MSI) has overcome this barrier.
Mass spectrometry imaging principles
As an extension of MS, MSI enables molecular information to be spatially mapped onto the solid biospecimen from which it originated (Figure 1). Histopathology analysis of an adjacent piece of tissue allows metabolites to be localized onto their tissue pathologies of origin. MALDI-MSI, which opened up the histopathology-based MSI field, still remains a first line approach in MSI-based diagnostics. As described above, MALDI occurs in a vacuum and requires sample preparation, in that a tissue section needs to be covered with a UV-absorbing crystalline matrix material. It offers the widest mass range allowing the imaging of small molecules, metabolites, lipids, and proteins, unlike immunohistochemistry, while also remaining the only approach capable of imaging proteins with molecular weights up to 300 kDa43–48. Moreover, MALDI MSI now offers spatial resolution as high as 5 μm, and can routinely image at 100 μm49–51.
Figure 1.

Schematic of mass spectrometry imaging technique, where focused beam irradiation generates a spectral read-out for each micron-scale area of the biospecimen89.
Usually separate pieces of tissue are used for MSI and histopathology analysis, but a histology-compatible tissue preparation method was developed by Agar and colleagues in 200752. The matrix solution fixation (MSF) method allows simultaneous tissue fixation and matrix deposition by incorporating MALDI matrix into solvents that preserve tissue integrity. Solvent fixation treatment of frozen tissue sections usually causes solubilization and displacement of proteins, lipids, and cellular membrane structures. MSF, however, preserves tissue integrity through maintenance of cellular structure and the general ensemble of biomolecules, causing subcellular disruption of only 50 to 300 nm. This has been achieved by a combination of cold solvent tissue fixation protocols with matrix deposition. The matrix can be removed after imaging, enabling histology and MSI of the same tissue section. The method is compatible with common MALDI matrixes and solutions commonly used for histology fixation, and it exhibits spectral quality and spatial resolution similar to matrix deposition techniques used in the current literature17,29. Furthermore, this method is shown to afford flexibility in optimizing crystallization parameters, so that a variety of biomolecules can be imaged52.
Many technical approaches for surface-based ambient ionization processes have been described in the literature in the past years, and a few have been commercialized, including desorption electrospray ionization (DESI) MSI. This technique involves directing a focused beam of charged droplets at a sample, such as thin-layered tissue on a glass slide. As the solvent pools, it forms a thin film on the surface that extracts molecular components from the sample below. The extracted liquid splashes up into the mass analyzer as subsequent solvent impacts the surface. Ambient interface methods can be used to take a snapshot of the sample’s molecular profile or to collect spectra at various points across a sample53. This ambient ionization method enables analysis with basically no preparation procedures. However, this advantage comes at the expense of its lower sensitivity, due to the lack of sample preparation and enrichment procedures; thus, molecules of interest can be lost amidst the signal produced by more abundant but possibly irrelevant compounds. DESI is limited by lower spatial resolution than can be achieved by MALDI (around 200 μm), although values of 40 μm have been reported on non-biological tissues54,55. While it is also limited to the analysis of a smaller mass range, its simple, fast application makes it suitable for clinical translation and able to complement other ionization approaches.
The above mentioned protocols may facilitate clinical acceptance of MS, as verification of tissue histopathology in conjunction with MS analysis is the only way to ensure the accurate interpretation of MS metabolomics. Additionally, the technical advancements of MALDI and DESI MSI, including the development of effective sample preparation procedures, faster and more accurate analysis, and high resolution, encourage mass spectrometry imaging towards further research use and clinical implementation56–62.
MSI identification of prostate cancer-specific metabolites
MSI investigation of tissue specimens represents an innovative field in PCa research. The following studies have focused on either the diagnosis or prognosis of PCa by considering either single metabolites or metabolomic profiles quantified from MALDI or DESI MSI and have reported differences in expression profiles between benign and malignant human prostate tissue63–70. A number of metabolites have been identified as potential diagnostic markers for PCa by means of MSI, such as mitogen-activated protein kinase/extracellular signal-regulated kinase, cholesterol sulfate, phosphatidylinositols, and biliverdin reductase B63–65,67. An overview of the most recent studies of MSI on human PCa whole tissue specimens is presented in Table 1.
MALDI-MSI was used to evaluate protein expression in PCa tissue versus benign from 75 prostatectomy cases, and six metabolites were increased in PCa containing-tissues. The overexpression of one of these metabolites – identified as a fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase (MEKK2) – in PCa tissues allowed for a reliable discrimination between malignant and benign tissue specimens with a sensitivity of 90.3% and a specificity of 86.4% (AUC = 0.96)63. MEKK2 is a member of the serine/threonine protein kinase family and is known for its crucial role in relaying cell surface signals through various downstream mitogen-activated protein kinase (MAPK) signaling pathways. The MAPK signaling pathway regulates growth factor-stimulated cell proliferation, differentiation, survival, and death. Dysregulation of this pathway is implicated in diverse diseases and cancers, and compounds inhibiting steps in MAPK signaling pathway are considered as potential drugs for cancer treatment66,71–74. Thus, the presentation of the overexpression of MEKK2 in PCa tissue may not only be useful for disease diagnosis but also for the design of individualized therapy, as well as for monitoring the effect of therapies.
MALDI-MSI has also been applied to malignant and benign epithelium as well as stromal areas of prostatectomy specimens to identify characteristic molecules for each pathological component. A cross-validation analysis, with 13 prostatectomy cases in the testing set and 10 in the validation set, presented high discriminatory ability for two peaks. One such peak, biliverdin reductase B (BVR), is a cytoprotective and growth promoting protein that is overexpressed in PCa, suggesting this enzyme could serve as a potential biomarker for human PCa67. It catalyzes the reaction to bilirubin, which is a potent antioxidant in human cells. Furthermore, BVR activates the expression of genes involved in cell growth, differentiation, and survival through the MAPK- and phosphatidylinositol 3-kinase (PI3K) pathway. This growth-promoting function of BVR as well as its increased expression in tumor cells classifies this enzyme as a tumor promoter75. Recent studies have indicated that many of biliverdin reductase’s growth promoting functions can effectively be suppressed by inhibitors of BVR activity, which could offer a novel approach in PCa treatment76.
To consider an entire cancer metabolomic profile, another MALDI-MSI study created a biomarker algorithm consisting of 22 different metabolites based on protein expression profiles in benign (n=11) and malignant (n=11) human prostate tissues. An overall cross-validation (88.00%), sensitivity (85.21%) and specificity (90.74%) was achieved for the distinction between benign and malignant prostate tissue69. Such remarkable sensitivity and specificity in distinguishing between benign tissue and PCa has also been shown in a high-resolution (HR)-MALDI-MSI study of 14 samples that identified increases of 26 metabolites in malignant compared to benign tissue (including phosphatidylinositols, three phosphatidylethanolamines and three phosphatidic acids). These metabolites together formulate a biomarker algorithm that presented a sensitivity of 87.5% and a specificity of 91.7% for PCa diagnosis65.
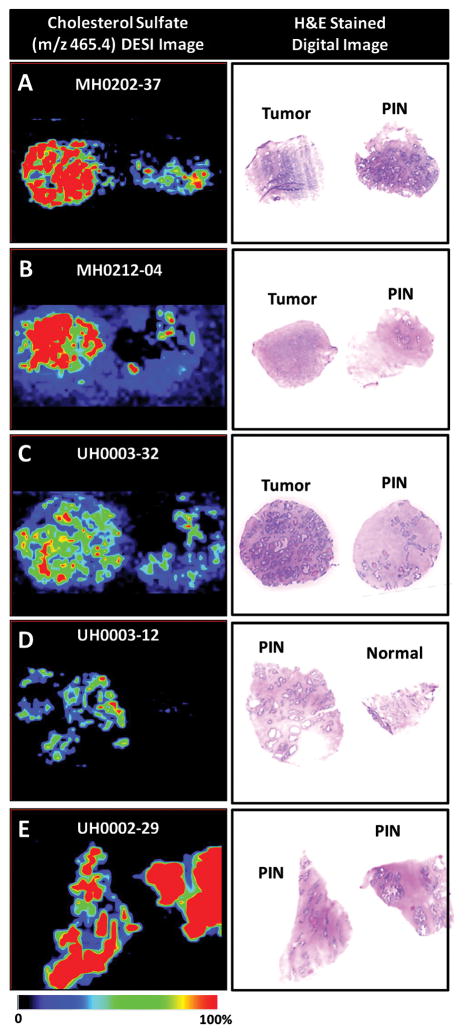
Using DESI-MSI, cholesterol sulfate was identified as a biomarker for PCa diagnosis, as it was almost exclusively found in cancerous and precancerous lesions64. Therefore, cholesterol sulfate might not only be useful to differentiate benign and cancerous tissue but also for the detection of precancerous lesions within histologically normal tissue (Figure 2). Cholesterol sulfate is known for its stabilizing and regulatory role as a cell membrane component and has been reported as a potential tumor marker in various types of cancers77,78. Since the biological process responsible for the expression of cholesterol sulfate in human PCa is unknown, future studies will have to address this issue.
Figure 2.
DESI-MS ion image of m/z 465.4, cholesterol sulfate, in the negative ion mode and H&E stained image of prostate samples (A) MH0202-37, cancer and adjacent normal tissue with PIN, (B) MH0212-04, cancer and normal tissue with PIN, (C) UH003-32, cancer and normal tissue with PIN, (D) UH0003-12, PIN and normal tissue, and (E) UH0002-29, PIN detected within normal regions of tissue64.
In a recently published work, endogenous compounds of cancerous and non-cancerous regions in three human prostate cancer tissue specimens were detected and imaged using matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry imaging (MALDI FTICR MSI)70. The authors combined two different MALDI matrices (quercetin and 9-aminoacridine) and used the newly developed technique of matrix coating assisted by an electric field (MCAEF)79,80. A total of 1091 metabolites were identified, of which 250 and 217 were exclusively found in either the cancerous or the non-cancerous regions, respectively. Of these, 152 metabolites showed differentiating distributions between these two regions of tissue, such as an increased energy charge and a lower expression of neutral acyl glycerides in the cancerous regions. Sixty-two acyl-glycerides were detectable only in non-cancerous regions, which can be interpreted as a result of known low glycolysis in prostate cancer cells and compensatory increased fatty acid beta-oxidation leading to a depletion of these acyl-glycerides in cancerous tissue regions70,81.
MSI prognostication of prostate cancer aggressiveness
Considering the limitations of histology to reliably determine PCa aggressiveness, the ability of MSI to predict and characterize prostate cancer aggressiveness in terms of biochemical recurrence was evaluated. The relationship between the decrease in the expression of lysophosphatidylcholine and the increase in biochemical recurrence potential of PCa after prostatectomy was evaluated with a study of 31 PCa patients82. Reduced expression of lysophosphatidylcholine was an individual predictor and potential prognostic marker of biochemical recurrence (Kaplan-Meier, p=0.027)82.
MALDI-MSI measurement of tissue microarrays of 729 human PCa specimens identified four signals associated with a low Gleason score, early disease stage, and low proliferation marker Ki-67, one signal associated with high Ki-67, and one signal associated with a prolonged time to PSA recurrence83. While these signals might serve as biomarkers for the differential diagnosis of PCa, two additional signals were associated with the overexpression of erythroblast transformation-specific related gene (ERG) and five signals associated with ERG negativity83. This discovery is of particular importance, since ERG can fuse with TMPRSS2 to form an oncogenic fusion gene especially common in hormone-refractory PCa. Therefore, MSI as an imaging tool may identify TMPRSS2-ERG-positive PCa to ensure that affected patients receive individually targeted therapy.
Conclusion and Future Directions
This review highlights the potential significance of MSI in enabling diagnosis and further prognosis of human PCa thus far seen in literature. The potential biomarkers detected during MSI analyses provide a metabolic overview of disease, and at the same time, affirm the potential of metabolomics to improve disease characterization when compared with a single metabolite. Following the development and improvement of various MSI methodologies, applications of the techniques have successfully distinguished PCa from histologically benign prostate tissue, and aggressive from indolent PCa. MSI also offers the valuable ability to map biomarkers onto prostate tissue pathologies, therefore complementing histology, immunohistopathology, and molecular pathology for disease diagnosis and patient prognostication. While the physical principles governing MSI make it an ex vivo technique, improvements in speed, precision, and sensitivity of the technique make feasible MSI clinical use to provide patients with individualized PCa treatment.
Continuing progress in MSI will lead to a higher accuracy in identifying PCa and determining PCa aggressiveness. We consider the next step in the development of MSI to be of three-dimensionality. Working towards this aim, an atmospheric-pressure ion source for MS has recently been developed84, with laser ablation electrospray ionization (LAESI)-MS as an innovative approach for depth profiling. In combination with lateral imaging, this new technology will enable three-dimensional molecular imaging with MS84.
Another huge advance for the field would be the combination of MSI analysis of PCa-containing tissue samples with traditional histopathological examination to develop a computer-based program that can identify different types and aggressiveness of PCa. This knowledge could also be applied to tissues from all over the body to evaluate other diseases, and it has already been done in breast and brain tissue85–88. Thus, MSI can contribute majorly to understanding of molecular pathogenesis of not just PCa and but to other malignant diseases as well.
Acknowledgments
This work was supported by PHS NIH grants R01CA115746, and R01CA115746-08S1 to LLC, and R01 CA201469 and P41EB015898 to NYRA, and by MGH Martinos Center for Biomedical Imaging.
Footnotes
Conflict of interest
NYA is co-founder of BayesianDx and scientific advisor to inviCRO. The other authors declare no conflicts of interest.
Authors’ Contributions: AK LV, and TF: literature research and manuscript preparation; PH and NA: review; LC: funding, manuscript preparation, and review.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Institute NC. SEER Stat Fact Sheets: Prostate. National Cancer Institute; Bethesda, MD: 2011. [Google Scholar]
- 3.Lu-Yao GL, Albertsen PC, Moore DF, Shih W, Lin Y, DiPaola RS, et al. Outcomes of localized prostate cancer following conservative management. JAMA. 2009;302(11):1202–1209. doi: 10.1001/jama.2009.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albertsen PC, Moore DF, Shih W, Lin Y, Li H, Lu-Yao GL. Impact of comorbidity on survival among men with localized prostate cancer. J Clin Oncol. 2011;29(10):1335–1341. doi: 10.1200/JCO.2010.31.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, Schroder FH, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95(12):868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 6.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J Natl Cancer Inst. 2009;101(19):1325–1329. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch HG, Black WC. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102(9):605–613. doi: 10.1093/jnci/djq099. [DOI] [PubMed] [Google Scholar]
- 9.Zelefsky M, Eastham J, Sartor A. Cancer of the prostate. In: DeVita VJ, Lawrence T, Rosenberg S, editors. Cancer: Principles and Practice of Oncology. 9. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia, USA: 2011. pp. 1220–1271. [Google Scholar]
- 10.Chan TY, Partin AW, Walsh PC, Epstein JI. Prognostic significance of Gleason score 3+4 versus Gleason score 4+3 tumor at radical prostatectomy. Urology. 2000;56(5):823–827. doi: 10.1016/s0090-4295(00)00753-6. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. The American journal of surgical pathology. 2016;40(2):244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 12.Kerian KS, Jarmusch AK, Pirro V, Koch MO, Masterson TA, Cheng L, et al. Differentiation of prostate cancer from normal tissue in radical prostatectomy specimens by desorption electrospray ionization and touch spray ionization mass spectrometry. Analyst. 2015;140(4):1090–1098. doi: 10.1039/c4an02039a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garbis SD, Tyritzis SI, Roumeliotis T, Zerefos P, Giannopoulou EG, Vlahou A, et al. Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (iTRAQ) with two-dimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2008;7(8):3146–3158. doi: 10.1021/pr800060r. [DOI] [PubMed] [Google Scholar]
- 14.Sarafanov AG, Todorov TI, Kajdacsy-Balla A, Gray MA, Macias V, Centeno JA. Analysis of iron, zinc, selenium and cadmium in paraffin-embedded prostate tissue specimens using inductively coupled plasma mass-spectrometry. J Trace Elem Med Biol. 2008;22(4):305–314. doi: 10.1016/j.jtemb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 15.McDunn JE, Li Z, Adam KP, Neri BP, Wolfert RL, Milburn MV, et al. Metabolomic signatures of aggressive prostate cancer. Prostate. 2013;73(14):1547–1560. doi: 10.1002/pros.22704. [DOI] [PubMed] [Google Scholar]
- 16.Saylor PJ, Karoly ED, Smith MR. Prospective study of changes in the metabolomic profiles of men during their first three months of androgen deprivation therapy for prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(13):3677–3685. doi: 10.1158/1078-0432.CCR-11-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung K, Reszka R, Kamlage B, Bethan B, Stephan C, Lein M, et al. Tissue metabolite profiling identifies differentiating and prognostic biomarkers for prostate carcinoma. Int J Cancer. 2013;133(12):2914–2924. doi: 10.1002/ijc.28303. [DOI] [PubMed] [Google Scholar]
- 18.Kami K, Fujimori T, Sato H, Sato M, Yamamoto H, Ohashi Y, et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics. 2013;9(2):444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 20.Karas M, Bachmann D, Hillenkamp F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Analytical chemistry. 1985;57(14):2935–2939. [Google Scholar]
- 21.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, et al. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid communications in mass spectrometry. 1988;2(8):151–153. [Google Scholar]
- 22.Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole–time-of-flight mass spectrometry. Journal of Mass Spectrometry. 2001;36(8):849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 23.Yost R, Enke C. Selected ion fragmentation with a tandem quadrupole mass spectrometer. Journal of the American Chemical Society. 1978;100(7):2274–2275. [Google Scholar]
- 24.Shevchenko A, Loboda A, Shevchenko A, Ens W, Standing KG. MALDI quadrupole time-of-flight mass spectrometry: a powerful tool for proteomic research. Analytical chemistry. 2000;72(9):2132–2141. doi: 10.1021/ac9913659. [DOI] [PubMed] [Google Scholar]
- 25.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev. 1998;17(1):1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Lodi A, Ronen SM. Magnetic resonance spectroscopy detectable metabolomic fingerprint of response to antineoplastic treatment. PloS one. 2011;6(10):e26155. doi: 10.1371/journal.pone.0026155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teahan O, Bevan CL, Waxman J, Keun HC. Metabolic signatures of malignant progression in prostate epithelial cells. Int J Biochem Cell Biol. 2011;43(7):1002–1009. doi: 10.1016/j.biocel.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Burch TC, Isaac G, Booher CL, Rhim JS, Rainville P, Langridge J, et al. Comparative Metabolomic and Lipidomic Analysis of Phenotype Stratified Prostate Cells. PLoS One. 2015;10(8):e0134206. doi: 10.1371/journal.pone.0134206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teichert F, Verschoyle RD, Greaves P, Edwards RE, Teahan O, Jones DJ, et al. Metabolic profiling of transgenic adenocarcinoma of mouse prostate (TRAMP) tissue by 1H-NMR analysis: evidence for unusual phospholipid metabolism. The Prostate. 2008;68(10):1035–1047. doi: 10.1002/pros.20761. [DOI] [PubMed] [Google Scholar]
- 30.Raina K, Ravichandran K, Rajamanickam S, Huber KM, Serkova NJ, Agarwal R. Inositol Hexaphosphate Inhibits Tumor Growth, Vascularity, and Metabolism in TRAMP Mice: A Multiparametric Magnetic Resonance Study. Cancer prevention research. 2013;6(1):40–50. doi: 10.1158/1940-6207.CAPR-12-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaurand P, Rahman MA, Hunt T, Mobley JA, Gu G, Latham JC, et al. Monitoring mouse prostate development by profiling and imaging mass spectrometry. Mol Cell Proteomics. 2008;7(2):411–423. doi: 10.1074/mcp.M700190-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Liu T, Ma C, Xue R, Deng C, Zeng H, et al. GC/MS-based metabolomic approach to validate the role of urinary sarcosine and target biomarkers for human prostate cancer by microwave-assisted derivatization. Anal Bioanal Chem. 2011;401(2):635–646. doi: 10.1007/s00216-011-5098-9. [DOI] [PubMed] [Google Scholar]
- 33.Fan Y, Murphy TB, Byrne JC, Brennan L, Fitzpatrick JM, Watson RW. Applying random forests to identify biomarker panels in serum 2D-DIGE data for the detection and staging of prostate cancer. Journal of proteome research. 2011;10(3):1361–1373. doi: 10.1021/pr1011069. [DOI] [PubMed] [Google Scholar]
- 34.Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PloS one. 2011;6(9):e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lokhov PG, Balashova EE, Voskresenskaya AA, Trifonova OP, Maslov DL, Archakov AI. Mass spectrometric signatures of the blood plasma metabolome for disease diagnostics. Biomedical reports. 2016;4(1):122–126. doi: 10.3892/br.2015.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giskeodegard GF, Hansen AF, Bertilsson H, Gonzalez SV, Kristiansen KA, Bruheim P, et al. Metabolic markers in blood can separate prostate cancer from benign prostatic hyperplasia. British journal of cancer. 2015;113(12):1712–1719. doi: 10.1038/bjc.2015.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 38.Meller S, Meyer HA, Bethan B, Dietrich D, Maldonado SG, Lein M, et al. Integration of tissue metabolomics, transcriptomics and immunohistochemistry reveals ERG- and gleason score-specific metabolomic alterations in prostate cancer. Oncotarget. 2016;7(2):1421–1438. doi: 10.18632/oncotarget.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 40.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27(3):253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cacciatore S, Zadra G, Bango C, Penney KL, Tyekucheva S, Yanes O, et al. Metabolic Profiling in Formalin-Fixed and Paraffin-Embedded Prostate Cancer Tissues. Mol Cancer Res. 2017 doi: 10.1158/1541-7786.MCR-16-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuster JR, Lance RS, Troyer DA. Molecular preservation by extraction and fixation, mPREF: a method for small molecule biomarker analysis and histology on exactly the same tissue. BMC Clin Pathol. 2011;11:14. doi: 10.1186/1472-6890-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen LH, Gusev AI. Small molecule analysis by MALDI mass spectrometry. Analytical and bioanalytical chemistry. 2002;373(7):571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 44.Franck J, Longuespee R, Wisztorski M, Van Remoortere A, Van Zeijl R, Deelder A, et al. MALDI mass spectrometry imaging of proteins exceeding 30,000 daltons. Medical science monitor : international medical journal of experimental and clinical research. 2010;16(9):Br293–299. [PubMed] [Google Scholar]
- 45.Mainini V, Bovo G, Chinello C, Gianazza E, Grasso M, Cattoretti G, et al. Detection of high molecular weight proteins by MALDI imaging mass spectrometry. Mol Biosyst. 2013;9(6):1101–1107. doi: 10.1039/c2mb25296a. [DOI] [PubMed] [Google Scholar]
- 46.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38(10):1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 47.Svatos A. Mass spectrometric imaging of small molecules. Trends in biotechnology. 2010;28(8):425–434. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 48.van Remoortere A, van Zeijl RJ, van den Oever N, Franck J, Longuespee R, Wisztorski M, et al. MALDI imaging and profiling MS of higher mass proteins from tissue. Journal of the American Society for Mass Spectrometry. 2010;21(11):1922–1929. doi: 10.1016/j.jasms.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36(4):355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 50.Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM. Tissue protein imaging at 1 mum laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal Bioanal Chem. 2015;407(8):2337–2342. doi: 10.1007/s00216-015-8532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogrinc Potocnik N, Porta T, Becker M, Heeren RM, Ellis SR. Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun Mass Spectrom. 2015;29(23):2195–2203. doi: 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- 52.Agar NY, Yang HW, Carroll RS, Black PM, Agar JN. Matrix solution fixation: histology-compatible tissue preparation for MALDI mass spectrometry imaging. Anal Chem. 2007;79(19):7416–7423. doi: 10.1021/ac071460e. [DOI] [PubMed] [Google Scholar]
- 53.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 54.Ifa DR, Wu C, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135(4):669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- 55.Kertesz V, Van Berkel GJ. Improved imaging resolution in desorption electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(17):2639–2644. doi: 10.1002/rcm.3662. [DOI] [PubMed] [Google Scholar]
- 56.Muller L, Kailas A, Jackson SN, Roux A, Barbacci DC, Schultz JA, et al. Lipid imaging within the normal rat kidney using silver nanoparticles by matrix-assisted laser desorption/ionization mass spectrometry. Kidney Int. 2015;88(1):186–192. doi: 10.1038/ki.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson SN, Barbacci D, Egan T, Lewis EK, Schultz JA, Woods AS. MALDI-Ion Mobility Mass Spectrometry of Lipids in Negative Ion Mode. Anal Methods. 2014;6(14):5001–5007. doi: 10.1039/C4AY00320A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J, et al. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42(8):1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caprioli RM. Imaging mass spectrometry: molecular microscopy for enabling a new age of discovery. Proteomics. 2014;14(7–8):807–809. doi: 10.1002/pmic.201300571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norris JL, Caprioli RM. Imaging mass spectrometry: a new tool for pathology in a molecular age. Proteomics Clin Appl. 2013;7(11–12):733–738. doi: 10.1002/prca.201300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7(4):493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 62.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 63.Cazares LH, Troyer D, Mendrinos S, Lance RA, Nyalwidhe JO, Beydoun HA, et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15(17):5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 64.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, et al. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82(9):3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goto T, Terada N, Inoue T, Nakayama K, Okada Y, Yoshikawa T, et al. The expression profile of phosphatidylinositol in high spatial resolution imaging mass spectrometry as a potential biomarker for prostate cancer. PLoS One. 2014;9(2):e90242. doi: 10.1371/journal.pone.0090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jing Y, Zhang Z, Ma P, An S, Shen Y, Zhu L, et al. Concomitant BET and MAPK blockade for effective treatment of ovarian cancer. Oncotarget. 2016;7(3):2545–2554. doi: 10.18632/oncotarget.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pallua JD, Schaefer G, Seifarth C, Becker M, Meding S, Rauser S, et al. MALDI-MS tissue imaging identification of biliverdin reductase B overexpression in prostate cancer. J Proteomics. 2013;91:500–514. doi: 10.1016/j.jprot.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One. 2014;9(9):e106255. doi: 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwamborn K, Krieg RC, Reska M, Jakse G, Knuechel R, Wellmann A. Identifying prostate carcinoma by MALDI-Imaging. Int J Mol Med. 2007;20(2):155–159. [PubMed] [Google Scholar]
- 70.Wang X, Han J, Hardie DB, Yang J, Pan J, Borchers CH. Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF) Biochim Biophys Acta. 2017;1865(7):755–767. doi: 10.1016/j.bbapap.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 71.Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328(2):353–361. doi: 10.1016/j.canlet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 72.Hanai J, Doro N, Sasaki AT, Kobayashi S, Cantley LC, Seth P, et al. Inhibition of lung cancer growth: ATP citrate lyase knockdown and statin treatment leads to dual blockade of mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT pathways. J Cell Physiol. 2012;227(4):1709–1720. doi: 10.1002/jcp.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang SY, Miah A, Sales KM, Fuller B, Seifalian AM, Winslet M. Inhibition of the p38 MAPK pathway sensitises human colon cancer cells to 5-fluorouracil treatment. Int J Oncol. 2011;38(6):1695–1702. doi: 10.3892/ijo.2011.982. [DOI] [PubMed] [Google Scholar]
- 74.Yong HY, Koh MS, Moon A. The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin Investig Drugs. 2009;18(12):1893–1905. doi: 10.1517/13543780903321490. [DOI] [PubMed] [Google Scholar]
- 75.Gibbs PE, Miralem T, Maines MD. Characterization of the human biliverdin reductase gene structure and regulatory elements: promoter activity is enhanced by hypoxia and suppressed by TNF-alpha-activated NF-kappaB. FASEB J. 2010;24(9):3239–3254. doi: 10.1096/fj.09-144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibbs PE, Miralem T, Maines MD. Biliverdin reductase: a target for cancer therapy? Front Pharmacol. 2015;6:119. doi: 10.3389/fphar.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiguchi K, Iwamori M, Yamanouchi S, Ishiwata I, Saga M, Amemiya A. Coexpression of cholesterol sulfate and cytokeratin as tumor markers in well-differentiated squamous cell carcinoma of the human uterine cervix. Clin Cancer Res. 1998;4(12):2985–2990. [PubMed] [Google Scholar]
- 78.Rearick JI, Stoner GD, George MA, Jetten AM. Cholesterol sulfate accumulation in tumorigenic and nontumorigenic rat esophageal epithelial cells: evidence for defective differentiation control in tumorigenic cells. Cancer Res. 1988;48(18):5289–5295. [PubMed] [Google Scholar]
- 79.Wang X, Han J, Hardie DB, Yang J, Borchers CH. The use of matrix coating assisted by an electric field (MCAEF) to enhance mass spectrometric imaging of human prostate cancer biomarkers. J Mass Spectrom. 2016;51(1):86–95. doi: 10.1002/jms.3728. [DOI] [PubMed] [Google Scholar]
- 80.Guo S, Wang Y, Zhou D, Li Z. Electric Field-Assisted Matrix Coating Method Enhances the Detection of Small Molecule Metabolites for Mass Spectrometry Imaging. Anal Chem. 2015;87(12):5860–5865. doi: 10.1021/ac504761t. [DOI] [PubMed] [Google Scholar]
- 81.Chandler JD, Williams ED, Slavin JL, Best JD, Rogers S. Expression and localization of GLUT1 and GLUT12 in prostate carcinoma. Cancer. 2003;97(8):2035–2042. doi: 10.1002/cncr.11293. [DOI] [PubMed] [Google Scholar]
- 82.Goto T, Terada N, Inoue T, Kobayashi T, Nakayama K, Okada Y, et al. Decreased expression of lysophosphatidylcholine (16:0/OH) in high resolution imaging mass spectrometry independently predicts biochemical recurrence after surgical treatment for prostate cancer. Prostate. 2015;75(16):1821–1830. doi: 10.1002/pros.23088. [DOI] [PubMed] [Google Scholar]
- 83.Steurer S, Borkowski C, Odinga S, Buchholz M, Koop C, Huland H, et al. MALDI mass spectrometric imaging based identification of clinically relevant signals in prostate cancer using large-scale tissue microarrays. Int J Cancer. 2013;133(4):920–928. doi: 10.1002/ijc.28080. [DOI] [PubMed] [Google Scholar]
- 84.Nemes P, Vertes A. Atmospheric-pressure molecular imaging of biological tissues and biofilms by LAESI mass spectrometry. J Vis Exp. 2010;(43) doi: 10.3791/2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ, Richardson AL, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci U S A. 2014;111(42):15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dekker TJ, Balluff BD, Jones EA, Schone CD, Schmitt M, Aubele M, et al. Multicenter matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) identifies proteomic differences in breast-cancer-associated stroma. J Proteome Res. 2014;13(11):4730–4738. doi: 10.1021/pr500253j. [DOI] [PubMed] [Google Scholar]
- 87.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, et al. Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew Chem Int Ed Engl. 2010;49(34):5953–5956. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pol J, Strohalm M, Havlicek V, Volny M. Molecular mass spectrometry imaging in biomedical and life science research. Histochemistry and cell biology. 2010;134(5):423–443. doi: 10.1007/s00418-010-0753-3. [DOI] [PubMed] [Google Scholar]