Abstract
Oncogenic transcription factor FOXQ1 has been implicated in promotion of multiple transformed phenotypes in carcinoma cells. Recently, we have characterized FOXQ1 as a melanoma tumor suppressor that acts via repression of N-cadherin gene, and invasion and metastasis. Here we report that FOXQ1 induces differentiation in normal and transformed melanocytic cells at least partially via direct transcriptional activation of MITF gene, melanocytic lineage-specific regulator of differentiation. Importantly, we demonstrate that pigmentation induced in cultured melanocytic cells and in mice by activation of cAMP/CREB1 pathway depends in large part on FOXQ1. Moreover, our data reveal that FOXQ1 acts as a critical mediator of BRAFV600E-dependent regulation of MITF levels, thus providing a novel link between two major signal transduction pathways controlling MITF and differentiation in melanocytic cells.
Introduction
Metastatic melanoma is one of the most deadly forms of skin cancer with rising incidence [1, 2]. Exposure to harmful ultraviolet radiation from the sun is a major extrinsic risk factor [3, 4]. Melanocytes, found in the basal epidermal layer of the human skin, produce melanin, which offers protection to both melanocytes and keratinocytes from ultraviolet radiation [5, 6]. Baseline epidermal melanin content and pigmentary response to ultraviolet exposure are major determinants of skin type, the most important factor determining melanoma and non-melanoma skin cancer risk [3, 6]. Therefore, molecular mechanisms underlying melanin production in response to external stimuli are critical for skin cancer prevention and may provide therapeutic targets and insight into individual skin cancer susceptibility.
A tanning response is initiated by keratinocytes which sense ultraviolet radiation induced DNA damage in a p53-dependent manner [7] and promote the secretion of ɑ-melanocyte stimulating hormone (α-MSH), which in turn activates cyclic AMP (cAMP) signaling via Gs-coupled melanocortin 1 receptor [8]. Subsequently, cAMP via the cAMP responsive element binding protein (CREB1) activates the expression of micropthalmia-associated transcription factor MITF, a melanocytic lineage-specific transcription factor that regulates expression of several pigmentation enzymes and melanosome components, including tyrosinase (TYR), dopachrome tautomerase, silver, etc. [8, 9]. In addition, several other signaling pathways, including RAF/MAPK [10–13], have been implicated in the regulation of MITF expression [11]. MITF has a critical role not only in melanocyte differentiation and survival but in melanoma progression [11, 14].
Forkhead transcription factor Forkhead box Q1 (FOXQ1) has been characterized as a major activator of epithelial-to-mesenchymal transition (EMT), invasion, and metastasis in cells derived from several carcinomas including breast, colon, ovarian, and lung, where its levels also correlate with poor prognosis [15–20]. Mechanistically, FOXQ1 has been shown to exert oncogenic activity in large part by promoting EMT, via direct regulation of E/N cadherin switch [18, 19]. On the contrary, we have recently demonstrated that levels of FOXQ1 decreased during melanoma progression and in melanoma cells as compared with melanocytes [21]. Moreover, unlike in carcinoma cells, in melanoma cells FOXQ1 suppresses EMT-like processes [21]. Mechanistically, opposite functions of FOXQ1 in carcinoma and melanoma cells were largely due to its ability to transactivate (in carcinoma) or trans-repress (in melanoma) expression of the N-cadherin gene (CDH2).
Here we report that FOXQ1 is also a potent inducer of differentiation and an important mediator of two major signal transduction pathways, namely cAMP/CREB1 and RAF/MAPK, in melanocytic cells.
Results
FOXQ1 induces melanocyte differentiation through direct binding to MITF
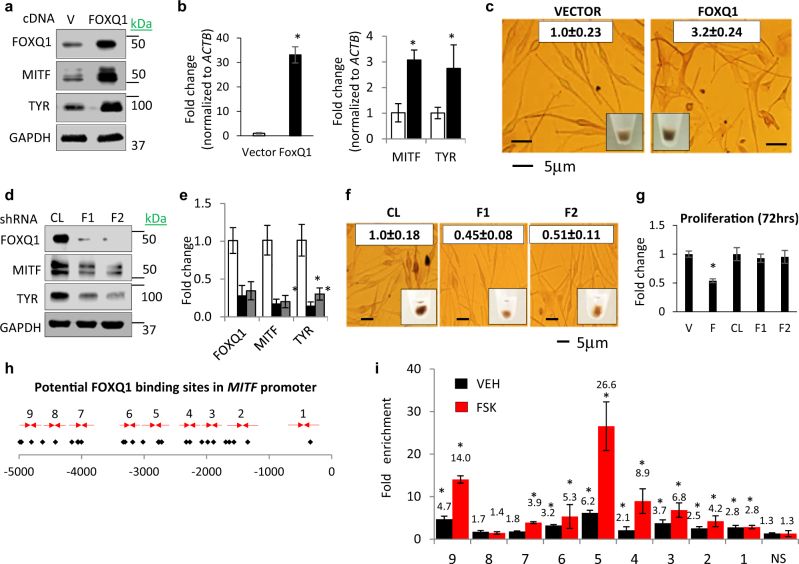
To evaluate differentiation-associated phenotypes induced by FOXQ1 we set up experiments to manipulate its levels in human and mouse melanocytic cells. To this end, we first transduced FOXQ1 cDNA in normal human melanocytes (NHMs), and normal and transformed mouse melanocytic cells (Melan-a and B16, respectively). FOXQ1 upregulated mRNA and protein levels of MITF, MITF direct target gene TYR, and induced differentiation evidenced by visible hyperpigmentation (Fig. 1a-c and Supplementary Figures S1a-c). Conversely, depletion of FOXQ1 in NHMs, Melan-a, and B16 suppressed pigmentation and decreased MITF and TYR levels (Fig. 1d-f and Supplementary Figures S1d-f). In addition, overexpression of FOXQ1 decreased proliferation of NHM, whereas FOXQ1 depletion did not significantly affect it (Fig. 1g).
Fig. 1.
FOXQ1 induces MITF-dependent differentiation. a, b NHM transduced with empty vector (VECTOR) or FOXQ1-expressing vector (FOXQ1) were probed in immunoblotting with indicated antibodies (left) or in Q-RT-PCR (right). FOXQ1/ACTB, MITF/ACTB, and TYR/ACTB signal ratios are shown. c Cells described in a were imaged as adherent cells or as cell pellets followed by quantification of total melanin content (white boxes). d, e Cells transduced with control (CL) or FOXQ1 (F1, F2) shRNAs were probed in immunoblotting with the indicated antibodies (left panel) or in Q-RT-PCR (right panel). FOXQ1/ACTB, MITF/ACTB, and TYR/ACTB signal ratios are shown. f NHM described in d, e) were imaged as adherent cells or as cell pellets followed by quantification of total melanin content (white boxes). g NHM expressing empty vector (V), FOXQ1 cDNA f, or control (CL) or FOXQ1 shRNAs (F1, F2) were counted for 72 h starting 48 h post infection. The cell numbers at 72 h were normalized by those before plating and by the ratio of these numbers in vector or control cells. h Human MITF promoter. Shown are FOXQ1-binding sites (diamonds) and PCR primers (arrows). i Q-PCR signals in reactions with DNA precipitated with FOXQ1-specific antibodies from untreated and FSK-treated NHM cells were normalized by corresponding signals in DNA precipitated with IgG antibodies and by signals obtained with MITF nonspecific control primers (NS). All data represent mean ± SEM. Statistical significance was assessed using two-tailed Student’s t-tests. A p < 0.05 (*) was considered significant
Sequence analysis of human MITF promoter revealed several potential FOXQ1 binding sites (Fig. 1h). Promoter regions encompassing some of these sites were significantly enriched in material precipitated with FOXQ1-specific but not IgG antibodies in chromatin immunoprecipitation (ChIP) assay performed in NHM cells treated with either vehicle or Forskolin (FSK, an activator of adenylate cyclase and an established inducer of MITF [22]) (Fig. 1i).
Therefore, these data demonstrate that FOXQ1 induces differentiation in melanocytic cells via direct transcriptional regulation of the MITF gene.
FOXQ1 mediates cAMP/CREB1-dependent differentiation in cultured melanocytic cells and in mouse skin
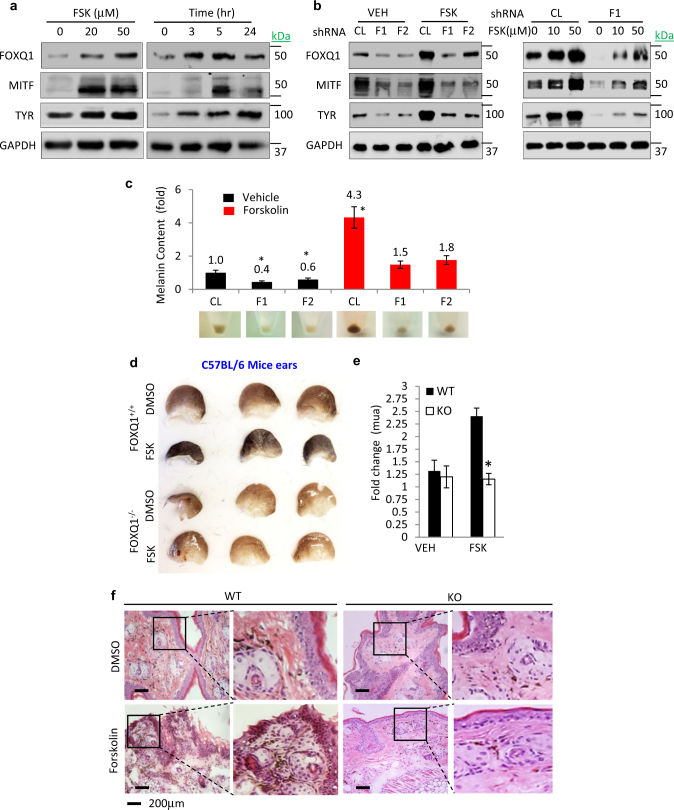
The cAMP/CREB1 pathway has a major role in induction of pigmentation in melanocytic cells [23]. Treatment of NHM, Melan-a, and B16 cells with FSK significantly upregulated FOXQ1 levels in a dose- and time-dependent manner (Fig. 2a and Supplementary Figure S2a). Importantly, depletion of FOXQ1 significantly suppressed FSK-induced upregulation of MITF and pigmentation in these cells (Fig. 2b,c and Supplementary Figures S2b-g).
Fig. 2.
FOXQ1 regulates cAMP/CREB1-dependent pigmentation. a NHM were treated with the indicated doses of Forskolin (FSK) for 5 h (left panel) or with 50 μM of FSK for indicated duration (right panel) followed by immunoblotting with the indicated antibodies. b NHM were transduced with control (CL) or FOXQ1 (F1, F2) shRNAs and treated with vehicle (DMSO) or the indicated doses of Forskolin (FSK) for 5 h followed by immunoblotting with the indicated antibodies. c Melanin content in cells described in b. d Representative images of ears from Foxq1+/+ or Foxq1–/– mice (n = 3) treated with either vehicle (DMSO, left ear) or vehicle containing 100μM Forskolin (FSK, right ear). e Pigmentation was quantified using a reflectance spectrometer and represented as fold change in the coefficient of absorption “mua”. f Representative images of H&E staining of mouse ear tissues shown in d. All data represent mean ± SEM. Statistical significance was assessed using two-tailed Student’s t-tests. A p < 0.05 (*) was considered significant
FSK has been widely used to induce pigmentation in the mouse skin [23]. To test the functional involvement of Foxq1 in regulation of pigmentation in in vivo settings, we generated Foxq1 knockout mice. In line with previous reports [24], Foxq1-deficient mice were viable and healthy, exhibiting 100% penetrance of the satin sheen phenotype [24]. Wild type and Foxq1 knockout mice were treated topically with either vehicle (dimethyl sulfoxide (DMSO), left ear) or 100 µM FSK (in DMSO, right ear) twice daily for 10 days. Epidermal pigmentation was detected visually (Fig. 2d) and assessed via reflective spectroscopy (Fig. 2e) and hematoxylin and eosin staining of skin sections (Fig. 2f). We did not detect a significant difference in basal pigmentation between wild-type and knockout mice. However, although treatment with FSK induced robust epidermal pigmentation in wild-type mice, it failed to elicit such a response in Foxq1–/– mice (Fig. 2d-f). We therefore concluded that FOXQ1 has a major role in inducing pigmentation in cultured melanocytic cells and mouse skin.
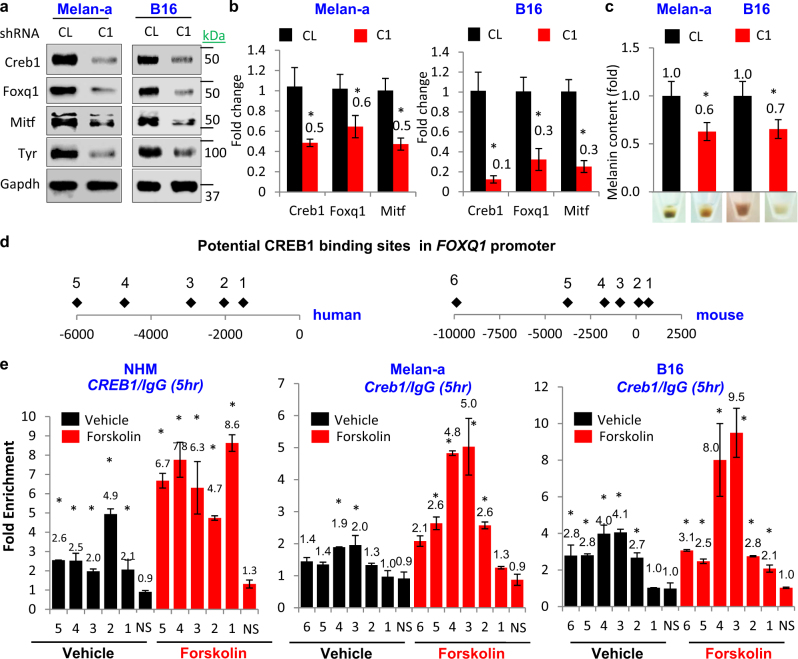
CREB1 is an established effector of FSK-induced phenotypes in melanocytic cells where it functions as a direct transcriptional activator of MITF [25]. On the other hand, data presented in Figs. 1 and 2 suggest that activation of cAMP/CREB1 pathway requires FOXQ1 for upregulation of MITF and melanocytic differentiation. We hypothesized that CREB1 transcriptionally activates FOXQ1, and that such activation is at least in part required for induction of MITF and differentiation by CREB1. Depletion of CREB1 substantially decreased levels of Foxq1 mRNA and protein in Melan-a and B16 cells, concomitant with changes in Mitf, Tyr, and melanin content (Fig. 3a-c). Analysis of the promoter of human and mouse FOXQ1 genes indicated the presence of several conserved CREB1-binding sites (Fig. 3d). ChIP assays in NHM, Melan-a, and B16 cells revealed that several of these sequences were enriched in the material precipitated with CREB1-specific but not IgG antibodies. Treatment with 10 µM FSK substantially increasing CREB1 binding to the FOXQ1 promoter (Fig. 3e).
Fig. 3.
FOXQ1 is important for cAMP/CREB1-dependent pigmentation. a, b Cells transduced with control shRNA (CL) or CREB1 shRNA (C1) were probed in immunoblotting with the indicated antibodies or in Q-RT-PCR. CREB1/Actb, Foxq1/Actb, and Mitf/Actb signal ratios are shown. c Melanin content and representative cell pellet images of cells described in a, b. d Schematic representation of human and mouse FOXQ1 promoter. Diamonds represent potential CREB1 consensus binding sites. e NHM, Melan-a, and B16 cells treated with vehicle (DMSO) or 10 μM Forskolin were probed in ChIP assay. Shown are ratios of Q-PCR signals in reactions with DNA precipitated with CREB1 or IgG antibodies using primers corresponding to potential CREB1-binding sites in FOXQ1 promoter. NS corresponds to CREB1-nonspecific primer targeting a distal genomic region. All data represent mean ± SEM. Statistical significance was assessed using two-tailed Student’s t-tests. A p < 0.05 (*) was considered significant
In summary, our data identify FOXQ1 as a major mediator of cAMP/CREB1-dependent regulation of MITF and MITF-dependent pigmentation in melanocytic cells in vitro and in vivo.
Suppression of FOXQ1 contributes to BRAFV600E-induced repression of MITF and inhibition of transformation
Ectopic expression of BRAFV600E in normal melanocytic cells downregulates β-catenin and MITF levels [26]. Conversely, BRAFV600E inhibition leads to increased pigmentation in cultured melanoma cells [10] and in some nevi [27]. In addition, in Melan-a cells, BRAFV600E overexpression was reported to cause transformation [12].
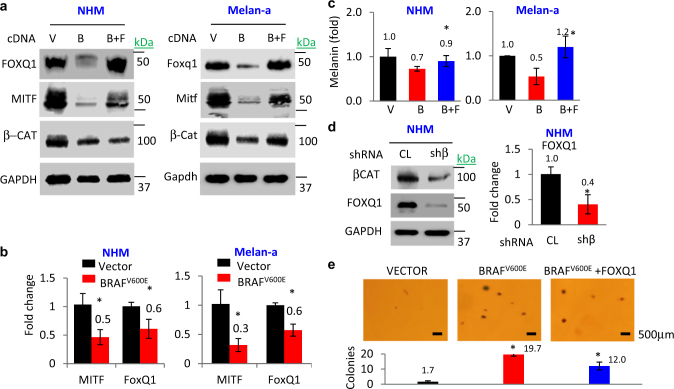
We identified that in addition to suppressing the levels of β-catenin, MITF, and pigmentation (Fig. 4a-c), ectopic expression of BRAFV600E in human and mouse melanocytes (NHM and Melan-a cells, respectively) causes a significant decrease in FOXQ1 mRNA and protein levels (Fig. 4a,b). Consistently, shRNA-mediated depletion of β-catenin also downregulated Foxq1 in Melan-a cells as it has been shown previously in carcinoma cells [28, 29] (Fig. 4d,e).
Fig. 4.
BRAF-β-catenin-FOXQ1 axis controls MITF-dependent phenotypes. a NHM and Melan-a cells were transduced with indicated constructs (V = empty vector, B = BRAFV600E, F = FOXQ1) followed by immunoblotting with the indicated antibodies. b Cells were transduced with the indicated constructs and probed in Q-RT-PCR. (MITF/ACTB (Mitf/Actb) and FOXQ1/ACTB (Foxq1/Actb) signal ratios are shown. c Total melanin content in cells described in a. d NHMs transduced with the indicated constructs were probed in immunoblotting with the indicated antibodies (left panel) and in Q-RT-PCR (right panel). FOXQ1/ACTB signal ratios are shown. e Melan-a cells were transduced with the indicated constructs and assayed for the ability to form colonies in soft agar. Shown are average numbers of colonies per view field (n = 5). All data represent mean ± SEM. Statistical significance was assessed using two-tailed Student’s t-tests. A p < 0.05 (*) was considered significant
To determine the functional role of FOXQ1 in regulation of MITF and colony formation by the BRAFV600E–β-catenin axis, we restored FOXQ1 levels in BRAFV600E-expressing Melan-a cells approximately to its levels in control cells (Fig. 4a). Such restoration strongly reverted BRAFV600E-dependent downregulation of MITF and associated pigmentation in NHM and Melan-a cells (Fig. 4a,c). In addition, restoration of FOXQ1 levels in BRAFV600E-overexpressing Melan-a cells inhibited BRAFV600E-induced transformation evidenced by colony formation in semi-solid agar (Fig. 4e). These data argue that FOXQ1 is a major mediator of BRAFV600E–β-catenin signaling in melanocytic cells.
Discussion
Epidermal melanin content and melanogenesis in response to ultraviolet stimulus are amongst the most important determinants of skin cancer susceptibility [30–32]. Although skin cancers are the highest incidence human malignancies and melanoma is one of the malignancies associated with most active life years lost [32] benign alterations of pigmentation (solar lentigines, melasma, post-inflammatory hyperpigmentation, etc.) poses an important psychosocial problem for many people. Our current work establishes FOXQ1 as a crucial transcriptional regulator of exogenous stress induced melanogenesis and establishes FOXQ1 as novel target for skin cancer prevention and treatment of cutaneous hyperpigmentation.
A member of forkhead box protein family, FOXQ1, is a transcription factor that has been reported to regulate differentiation in several cell types[18, 33]. Recently, an increasing number of studies have demonstrated that FOXQ1 is significantly associated with the pathogenesis of tumors. Here we describe FOXQ1 as a regulator of melanin production and tanning response via direct regulation of MITF in response to the complex interplay of upstream signaling pathways including α-MSH/cAMP/CREB1 and RAF/MAPK.
MITF has a complex role in melanoma biology, apart from its established role in melanocytic differentiation [11, 14]. Depending on its levels MITF has been described as oncogene or tumor a suppressor. In order to integrate these conflicting reports a ‘rheostat model’ of MITF functioning has been proposed where terminal differentiation and inhibition of proliferation is induced by high levels of MITF; cells with intermediate MITF levels proliferate rapidly but possess reduced invasive potential; low MITF levels decrease proliferation but increase invasive properties of a cell.
Because of its central role in normal and transformed melanocytic cells, several signal transduction programs including BRAFV600E/MAPK- and cAMP/CREB1-dependent pathways converge on MITF. By demonstrating that MITF is a direct transcriptional target of FOXQ1, we were interested in identifying the role of FOXQ1 in these pathways.
Physiologically, melanogenesis is stimulated by ultraviolet radiation [34]-induced DNA damage and resultant α-MSH release [35], which induce the cAMP/CREB1 pathway. cAMP-elevating agents such as FSK have been widely used to study cAMP/CREB1-dependent melanogenesis [23]. Our data position FOXQ1 as a direct CREB1-downstream target and a rate-limiting mediator of FSK-induced melanogenesis, as depletion of FOXQ1 severely suppresses both FSK- and CREB1-induced pigmentation and MITF activation. Interestingly, CREB1 itself has been shown to bind to the MITF promoter and activate its transcription [11]. This suggests a potential functional cooperation between FOXQ1 and CREB1 similar to that between SOX10 and its target gene PAX3. Similar to FOXQ1, SOX10 also directly activates MITF [36] and is also required for full-scale activation of MITF by PAX3 [37]. On the other hand, cooperation between FOXQ1 and CREB1 does not exclude functional involvement in MITF regulation of other CREB1-dependent targets.
Although inactivation of several upstream regulators of MITF including SOX10 [36, 38], PAX3 [36, 39], and YY1 [40] among others [11] results in coat-color phenotypes, Foxq1-null mice retain baseline pigmentation. This observation is consistent with other studies demonstrating that mice deficient of CREB1 or another MITF activator PGC1α also demonstrate “no coat-color” phenotype, underlining the complexity of basal versus induced pigmentation in mouse skin [14, 41]. These findings, taken together with our results, indicate that FOXQ1 is a crucial regulator of adaptive, DNA damage signaling induced melanogenesis. As its deficiency alters hyperpigmentation but not baseline pigmentation, Foxq1 can be considered as an ideal therapeutic target.
Activated BRAFV600E in normal human and mouse melanocytes has been shown to decrease β-catenin and consequently MITF levels [26, 42]. We confirmed these findings and further demonstrated that BRAFV600E causes transcriptional repression of FOXQ1, a direct target of β-catenin [28, 29]. On the other hand, restoration of FOXQ1 levels in BRAFV600E melanocytes while not affecting nuclear β-catenin, upregulated MITF close to its levels in vector melanocytes. These data argue that suppression of FOXQ1 is required for transcriptional repression of MITF by the BRAFV600E–β-catenin axis.
In summary, our data discovered FOXQ1 is a critical mediator of two major signal transduction pathways in melanocytic cells.
Materials and methods
Cell culture
Populations of NHMs were purchased from Invitrogen and maintained in Medium 254 (Invitrogen) supplemented with human melanocyte growth supplement (Invitrogen). B16-F0 cells were purchased from ATCC. HMLER cells were a gift from Dr. Robert A Weinberg (Whitehead Institute). Melan-a mouse melanocytes were grown at 37 °C (10% CO2) in RPMI media containing 12-O-Tetradecanoylphorbol-13-acetate.
Antibodies and other reagents
The following antibodis were used: FoxQ1 (K-12) sc-47597, (C-16), MITF (Clone 5, ThermoFisher, MA5-14146), TYR (C-19) sc-7833, GAPDH (ThermoFisher, PA1-987), β-Catenin (CST, D10A8-8480), and CREB1 (CST, 48H2-9197). FSK (LC Labs) was prepared fresh in DMSO, 5-Aza-2′-deoxycytidine (A3656 Sigma).
Plasmid constructs
FOXQ1 open reading frame was PCR amplified from NHM cDNA and cloned into a lentiviral expression vector pLV-puro in frame with an N-terminus flag-tag. CREB1 cDNA was purchased from DNASU repository (DNASU, HsCD00441528). shRNA targeting CREB1 was purchased from OriGene (RC210577). shRNAs targeting human and mouse FOXQ1, MITF, pLV-puro, and pLKO-1.puro lentiviral control vectors were purchased from Sigma-Aldrich (St. Louis, MO). pLKO.1 puro shRNA β-catenin was a gift from Bob Weinberg (Addgene plasmid #18803). pLenti-CMV-Puro-LUC was purchased from Addgene (w168-1-7477).
Cell proliferation analysis
Cells (5 × 103 cells) were seeded onto 96-well pates and grown for 24–72 h followed by fixation and staining in 0.5% Methylene blue in water/methanol (50 : 50) for ~ 1 h. The plates were rinsed using ddH20 and placed in an inverted position overnight to dry. The stain was eluted in 200 μl of 1% SDS in PBS and the optical density was determined at 650 nm using a fluorimeter (SpectraMax).
Chromatin immunoprecipitation
Cultured cells (~ 5 × 107) were fixed with 1% formaldehyde at room temperature for 15 min, followed by addition of glycine (0.125 M) for 5 min. Next, cells were washed with ice-cold phosphate buffered saline, pelleted, and re-suspended in ice-cold cell lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.1, containing protease inhibitors) (EZ-ChIP kit, Millipore, CA, USA). The samples were then incubated on ice for ~ 20 min, followed by sonication (12 × for 30 s each at 30 s intervals) with a Microson (Misonix Inc., USA). Next, the samples were centrifuged at 15,000 r.p.m. at 4 °C for 10 min. A control aliquot (whole-cell extract) was saved and supernatants were diluted 10-fold in ChIP dilution buffer (16.7 mM Tris-HCl (pH 8.1), 167 mM NaCl, 1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA protease inhibitor) (EZ-ChIP kit, Millipore). A 2 h incubation on a rotating platform at 4 °C with protein A/G beads was used to remove nonspecific background.
The pre-cleared chromatin material was incubated with anti-FLAG-conjugated beads (~ 25 μl slurry per 1 × 106 cells) on a rotating platform overnight at 4 °C, followed by magnetic separation. The beads were sequentially washed and the precipitated material was de-crosslinked and treated according to the EZ-ChIP kit instructions (Millipore). The DNA was recovered by column purification and quantified using QUBIT DNA HS assay kit (Invitrogen).
Colony formation assay
A 0.5% agarose gel was prepared, autoclaved, and poured onto six-well plates to form a semi-solid feeder layer. Cells (~ 5 × 105) were embedded into 0.3% agarose gel, poured on top of the feeder layer. The top layer was then covered with ~ 2 ml of complete media and replenished every 2 days for 4–6 weeks. Colonies were observed using standard light microscopy and counted in five random fields per well at × 200 magnification.
Reverse-transcription PCR analysis
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA was prepared using cDNA reverse transcription kit (Invitrogen). Quantitative reverse-transcription PCR was performed using 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using SYBr GreenMaster Mix (Invitrogen).
Immunoblotting
Polyvinylidene difluoride membranes were developed using alkaline phosphatase-conjugated secondary antibodies and signals were detected and visualized using the Alpha-Innotech FluorChem HD2 imaging system (Alpha Innotech) and quantified using ImageQuant software (GE Healthcare Life Sciences).
Generation of Foxq1 knockout mice
Two Foxq1tm1(KOMP)Vlcg mouse embryonic stem cell clones were obtained from the Velocigene (Regeneron) branch of the Knockout Mouse Project (KOMP). Prior genotyping and chromosome analysis was done by KOMP. Both clones were thawed and expanded as per KOMP protocols in VGB6 media, genotyped, and subsequently injected into B6(Cg)-Tyrc-2J/J (Jackson Lab catalog #000058) embryos to create chimeras by the transgenic core facility at Roswell Park Cancer Institute. Embryos were transferred into CD-1 pseudo-pregnant foster mice and one male chimeric mouse was born. Quantitative PCR analysis of his sperm performed by UC Davis showed that 50% of the sperm contained lacZ. Subsequently, a large scale in vitro fertilization (IVF) was performed to generate Foxq1 knockout mice.
Measurement of melanin content in mice skin
The optical measurement of the skin were conducted with a quantitative optical diffuse reflectance spectroscopy system Zenascope PC1 (Zenalux Biomedical, Durham, NC, www.zenalux.com). The system was validated in a number of in vitro, pre-clinical, and clinical applications to date, including quantification of chemotherapy drug content in tissue in vivo. The principle of operation has been described in a number of publications [43–46]. Briefly, a broadband halogen light source, an optical fiber probe, and a spectrometer are used to collect diffuse reflectance signals from the tissue. The resulting spectra are used as target spectra in an inverse Monte Carlo algorithm, which converts the relative, measured diffuse reflectance spectra into quantitative optical properties (reduced scattering and absorption coefficients). The depth of penetration is about 2–3 mm. Thus, the data are collected primarily from the skin and underlying dermis. Total melanin content in the ear skin was measured as a function of the coefficient of absorption (mua) at 475 nm. Mice were anesthetized and the probe was pressed against the tested area. Multiple readings were taken at three distinct spots and then averaged to yield the mean ± SEM. Data are reported as fold change in the coefficient of absorption (mua).
Statistical analysis
Each experiment was performed at least two times with consistent results. Statistical significance was determined using Student’s t-test. A two-tailed p-value was considered significant for all analyses. Normal distribution was confirmed using normal probability plot (GraphPad Prism 6.0, Graphpad Software, Inc., San Diego, CA, USA), variance was also assessed using GraphPad Prism 6.0 both within and between groups and were approximately the same.
Electronic supplementary material
Acknowledgements
We are grateful to Dr. Smiraglia for critical reading of the manuscript, to the Pathology Resource Network, the Clinical Data Network, and the transgenic shared core facility (funded by NCI P30CA16056) at Roswell Park Cancer Institute. This work has been supported by the following NCI grants: CA190533, CA193981, CA202162, and partially supported by the Program for Basic Research of Russian State Academies of Sciences for 2013−2020.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G Melino
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0066-y) contains supplementary material, which is available to authorized users.
References
- 1.Schadendorf D, Fisher DE, Garbe C, Gershenwald JE, Grob JJ, Halpern A, et al. Melanoma. Nat Rev Dis Prim. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 2.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–58. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 3.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A, et al. Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun. 2012;3:884. doi: 10.1038/ncomms1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi N, Nakagawa A, Muramatsu T, Yamashina Y, Shirai T, Hashimoto MW, et al. Supranuclear melanin caps reduce ultraviolet induced DNA photoproducts in human epidermis. J Invest Dermatol. 1998;110:806–10. doi: 10.1046/j.1523-1747.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi Y, Takahashi K, Zmudzka BZ, Kornhauser A, Miller SA, Tadokoro T, et al. Human skin responses to UV radiation: pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. FASEB J. 2006;20:1486–8. doi: 10.1096/fj.06-5725fje. [DOI] [PubMed] [Google Scholar]
- 7.Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–64. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Weng QY, Fisher DE. UV signaling pathways within the skin. J Invest Dermatol. 2014;134:2080–5. doi: 10.1038/jid.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Orazio J, Fisher DE. Central role for cAMP signaling in pigmentation and UV resistance. Cell Cycle. 2011;10:8–9. doi: 10.4161/cc.10.1.14292. [DOI] [PubMed] [Google Scholar]
- 10.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 2015;72:1249–60. doi: 10.1007/s00018-014-1791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF–stimulated melanocyte and melanoma cell proliferation. J Cell Biol. 2005;170:703–8. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–28. [PubMed] [Google Scholar]
- 15.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS ONE. 2012;7:e39937. doi: 10.1371/journal.pone.0039937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao M, Shih IeM, Wang TL. The role of forkhead box Q1 transcription factor in ovarian epithelial carcinomas. Int J Mol Sci. 2012;13:13881–93. doi: 10.3390/ijms131113881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–63. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M. Forkhead box Q1: A key player in the pathogenesis of tumors (Review) Int J Oncol. 2016;49:51–8. doi: 10.3892/ijo.2016.3517. [DOI] [PubMed] [Google Scholar]
- 19.Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71:3076–86. doi: 10.1158/0008-5472.CAN-10-2787. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–301. doi: 10.1158/0008-5472.CAN-10-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagati A, Bianchi-Smiraglia A, Moparthy S, Kolesnikova K, Fink EE, Lipchick BC, et al. Melanoma suppressor functions of the carcinoma oncogene FOXQ1. Cell Rep. 2017;20:2820–32. doi: 10.1016/j.celrep.2017.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khaled M, Levy C, Fisher DE. Control of melanocyte differentiation by a MITF-PDE4D3 homeostatic circuit. Genes Dev. 2010;24:2276–81. doi: 10.1101/gad.1937710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–4. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 24.Hong HK, Noveroske JK, Headon DJ, Liu T, Sy MS, Justice MJ, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genes (New Y, NY: 2000) 2001;29:163–71. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- 25.Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne JP, et al. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J Cell Biol. 1998;142:827–35. doi: 10.1083/jcb.142.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawlikowski JS, McBryan T, van Tuyn J, Drotar ME, Hewitt RN, Maier AB, et al. Wnt signaling potentiates nevogenesis. Proc Natl Acad Sci Usa. 2013;110:16009–14. doi: 10.1073/pnas.1303491110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen PR, Bedikian AY, Kim KB. Appearance of new vemurafenib-associated melanocytic nevi on normal-appearing skin: case series and a review of changing or new pigmented lesions in patients with metastatic malignant melanoma after initiating treatment with vemurafenib. J Clin Aesthet Dermatol. 2013;6:27–37. [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen J, Bentz S, Sengstag T, Shastri VP, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS ONE. 2013;8:e60051. doi: 10.1371/journal.pone.0060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, et al. FOXQ1 mediates the crosstalk between TGF-beta and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16:1099–109. doi: 10.1080/15384047.2015.1047568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bliss JM, Ford D, Swerdlow AJ, Armstrong BK, Cristofolini M, Elwood JM, et al. Risk of cutaneous melanoma associated with pigmentation characteristics and freckling: systematic overview of 10 case-control studies. The International Melanoma Analysis Group (IMAGE) Int J Cancer. 1995;62:367–76. doi: 10.1002/ijc.2910620402. [DOI] [PubMed] [Google Scholar]
- 31.Pho LN, Leachman SA. Genetics of pigmentation and melanoma predisposition. G Ital di Dermatol e Venereol. 2010;145:37–45. [PubMed] [Google Scholar]
- 32.Vogan K. Cancer genetics: pigmentation and skin-cancer risk. Nat Rev Genet. 2008;9:502-. doi: 10.1038/nrg2409. [DOI] [Google Scholar]
- 33.Feuerborn A, Srivastava PK, Kuffer S, Grandy WA, Sijmonsma TP, Gretz N, et al. The Forkhead factor FoxQ1 influences epithelial differentiation. J Cell Physiol. 2011;226:710–9. doi: 10.1002/jcp.22385. [DOI] [PubMed] [Google Scholar]
- 34.Ortonne JP. The effects of ultraviolet exposure on skin melanin pigmentation. J Int Med Res. 1990;18:8C–17C. doi: 10.1177/03000605900180S103. [DOI] [PubMed] [Google Scholar]
- 35.Hunt G, Todd C, Cresswell JE, Thody AJ. Alpha-melanocyte stimulating hormone and its analogue Nle4DPhe7 alpha-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J Cell Sci. 1994;107:205–11. doi: 10.1242/jcs.107.1.205. [DOI] [PubMed] [Google Scholar]
- 36.Hou L, Arnheiter H, Pavan WJ. Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc Natl Acad Sci Usa. 2006;103:9081–5. doi: 10.1073/pnas.0603114103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, et al. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–17. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 38.Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem. 2000;275:37978–83. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- 39.Eccles MR, He S, Ahn A, Slobbe LJ, Jeffs AR, Yoon HS, et al. MITF and PAX3 play distinct roles in melanoma cell migration; outline of a “Genetic Switch” theory involving MITF and PAX3 in proliferative and invasive phenotypes of melanoma. Front Oncol. 2013;3:229. doi: 10.3389/fonc.2013.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Song JS, Bell RJ, Tran TN, Haq R, Liu H, et al. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 2012;8:e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoag J, Haq R, Zhang M, Liu L, Rowe GC, Jiang A, et al. PGC-1 coactivators regulate MITF and the tanning response. Mol Cell. 2013;49:145–57. doi: 10.1016/j.molcel.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biechele TL, Kulikauskas RM, Toroni RA, Lucero OM, Swift RD, James RG, et al. Wnt/beta-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal. 2012;5:ra3. doi: 10.1126/scisignal.2002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bender JE, Vishwanath K, Moore LK, Brown JQ, Chang V, Palmer GM, et al. A robust Monte Carlo model for the extraction of biological absorption and scattering in vivo. IEEE Trans Biomed Eng. 2009;56:960–8. doi: 10.1109/TBME.2008.2005994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown JQ, Wilke LG, Geradts J, Kennedy SA, Palmer GM, Ramanujam N. Quantitative optical spectroscopy: a robust tool for direct measurement of breast cancer vascular oxygenation and total hemoglobin content in vivo. Cancer Res. 2009;69:2919–26. doi: 10.1158/0008-5472.CAN-08-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer GM, Boruta RJ, Viglianti BL, Lan L, Spasojevic I, Dewhirst MW. Non-invasive monitoring of intra-tumor drug concentration and therapeutic response using optical spectroscopy. J Control Release. 2010;142:457–64. doi: 10.1016/j.jconrel.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer GM, Zhu C, Breslin TM, Xu F, Gilchrist KW, Ramanujam N. Monte Carlo-based inverse model for calculating tissue optical properties. Part II: application to breast cancer diagnosis. Appl Opt. 2006;45:1072–8. doi: 10.1364/AO.45.001072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.