Abstract
Telomeres integrity is indispensable for chromosomal stability by preventing chromosome erosion and end-to-end fusions. During meiosis, telomeres attach to the inner nuclear envelope and cluster into a highly crowded microenvironment at the bouquet stage, which requires specific mechanisms to protect the telomeres from fusion. Here, we demonstrate that germ cell-specific knockout of a shelterin complex subunit, Trf1, results in arrest of spermatocytes at two different stages. The obliterated telomere-nuclear envelope attachment in Trf1-deficient spermatocytes impairs homologue synapsis and recombination, resulting in a pachytene-like arrest, while the meiotic division arrest might stem from chromosome end-to-end fusion due to the failure of recruiting meiosis specific telomere associated proteins. Further investigations uncovered that TRF1 could directly interact with Speedy A, and Speedy A might work as a scaffold protein to further recruit Cdk2, thus protecting telomeres from fusion at this stage. Together, our results reveal a novel mechanism of TRF1, Speedy A, and Cdk2 in protecting telomere from fusion in a highly crowded microenvironment during meiosis.
Introduction
Telomeres are nucleoprotein structures located at the end of chromosomes and telomere integrity is essential for genome stability and cell survival. [1] Telomeres become progressively shorter during mitotic cycles or due to environmental stresses. When telomeres are shortened and become dysfunctional, cellular senescence is triggered and tissue/organ aging might be accelerated. [2] Dysfunctional telomeres also tend to form chromosomal end-to-end fusion, eventually causing genomic instability, cell cycle arrest, and finally cell death. [3] Protection of telomere fusion is mainly regulated by the telomerase complex and the shelterin complex, which includes six subunits (TRF1, TRF2, RAP1, TIN2, TPP1, and POT1). [4–11] TRF1 and TRF2 directly bind to double-stranded telomeric DNA and prevent the activation of both ATM- and ATR-dependent DNA damage signaling pathways. [12, 13] Mutations in telomere related genes are associated with many diseases, including Hoyeraal-Hreidarsson syndrome, dyskeratosis congenita, pulmonary fibrosis, and cancer. [14–16]
Telomeres are attached to the nuclear matrix and scattered throughout the nucleus in somatic cells. [17–19] In contrast in germ cells, telomeres display an unprecedented behavior during meiotic prophase I where they attach to the inner nuclear envelope (NE) and cluster to form a structure which resembles a bouquet of flowers, termed the bouquet stage. [17, 20, 21] This phenomenon is conserved in diverse eukaryote species and required for homologous chromosome pairing, synapsis, and proper resolution of recombination events. [22, 23] During the bouquet stage, telomeres are in close proximity to each other, thus creating a crowded microenvironment. Since the prerequisite for telomere fusion is that two telomeres are in close proximity to each other to allow the reaction to occur, telomeres are vulnerable to unwanted end-to-end fusions at the bouquet stage. In this crowded microenvironment, protection mechanisms must have been evolved to protect them fusing with each other.
The attachment of telomeres to the NE is dependent on the transmembrane linker of nucleoskeleton and cytoskeleton (LINC)-complex, which contains SUN1, SUN2, KASH5, and adaptor proteins like TERB1, TERB2, MAJIN, Speedy A, and TRF1. [24–29] Since TRF1 is a core subunit of the shelterin complex and also involved in anchoring telomeres to the NE by interacting with TERB1, [28] we selected TRF1 as the starting point to investigate the telomere fusion protection mechanism at the bouquet stage. We knocked out Trf1 in germ cells by crossing Trf1Flox/Flox mice [13] with Stra8-Cre transgenic mice. [30] We found that Trf1 is essential for spermatogenesis since the knockout of Trf1 in male germ cells led to arrest at two stages: the pachytene-like and the meiotic division stages. It is the defective attachment of telomeres to NE that results in the pachytene-like arrest, whereas the meiotic division arrest stems from chromosome end-to-end fusions in Trf1 knockout spermatocytes. Further investigations uncovered that Speedy A and cyclin-dependent kinase 2 (Cdk2) but not SUN1 were involved in protection from telomere fusion at the bouquet stage. Thus, in addition to their roles in tethering telomeres to NE, our work defines novel functions for TRF1, Speedy A, and Cdk2 in protecting telomeres from fusion in a crowded microenvironment during meiosis.
Results
TRF1 is essential for spermatogenesis
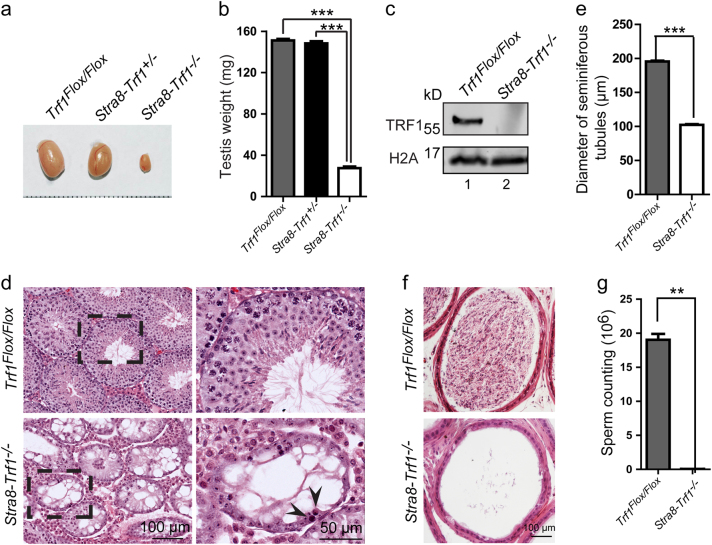
To study the molecular mechanism of protection from telomere fusion at the bouquet stage, the core subunit of the shelterin complex, Trf1, was knocked out in germ cells by crossing mice with a floxed Trf1 allele to Stra8-Cre mice, which express Cre recombinase during early-stage spermatogenesis beginning on day 3 after birth. [30] Germ cell-specific Trf1 knockout mice will be referred to as Stra8-Trf1−/−. We found that the Stra8-Trf1−/− male mice were completely infertile. The size and weight of their testes were significantly reduced compared with that of Trf1Flox/Flox and Stra8-Trf1+/− mice (Fig. 1a-b) due to efficient Trf1 deletion (Fig. 1c). Further histological examination revealed that Trf1-deficient testes lacked post-meiotic cells (Fig. 1d) and the diameter of Trf1-deficient seminiferous tubules was decreased significantly (Fig. 1e). Few spermatozoa could be detected in the cauda epididymis and the total number of spermatozoa was dramatically reduced in the Stra8-Trf1−/− mice compared with that of the control group, indicating that the disruption of Trf1 severely impairs spermatogenesis (Fig. 1f-g). The amount of degenerated cells with highly condensed nuclei found in the seminiferous epithelium of Stra8-Trf1−/− mice (Fig. 1d), which are resembling the characteristic of apoptotic cells, let us speculate that the reduction of post-meiotic cells in Trf1-deficient testes is due to apoptosis. To verify this possibility, we performed TUNEL assays and found that the number of TUNEL positive cells per seminiferous tubule in the Stra8-Trf1−/− mice was significantly increased compared with that of the control group (Supplementary Figure 1), suggesting that spermatocytes in Trf1-deficient testes are eliminated through apoptosis. Altogether, these results indicate that germ cell-specific disruption of Trf1 results in spermatogenetic failure and that TRF1 is essential for spermatogenesis.
Fig. 1. TRF1 is required for spermatogenesis.
a The Stra8-Trf1−/− mice testes were smaller than those of the Trf1Flox/Flox and Stra8-Trf1+/− mice (10-week-old, and the same to the below). b Quantification of testis weight of the Trf1Flox/Flox and Stra8-Trf1+/− mice. Testis weight: Trf1Flox/Flox (grey bar, 151.20 ± 1.42 mg), Stra8-Trf1+/− (black bar, 148.40 ± 1.96 mg), Stra8-Trf1−/− (white bar, 27.40 ± 1.69 mg). Data are presented as mean ± SEM. ***P < 0.001. c The TRF1 protein levels were reduced in the testes of the Stra8-Trf1−/− mice. Histone H2A was used as the loading control. d PAS-hematoxylin analysis of the seminiferous tubules of Trf1Flox/Flox and Stra8-Trf1−/− mice. Arrows indicate apoptotic cells. e The diameter of the seminiferous tubules in Stra8-Trf1−/− mice was smaller than that of the Trf1Flox/Flox mice. Trf1Flox/Flox (gray bar, 195.50 ± 4.49 μm), Stra8-Trf1−/− (white bar, 102.40 ± 3.62 μm). Data are presented as mean ± SEM. ***P < 0.001. f Histological analysis of the caudal epididymides from 10-week-old Trf1Flox/Flox and Stra8-Trf1−/− mice. g The total number of sperm in the cauda epididymis was significantly decreased in the Stra8-Trf1−/− mice. Trf1Flox/Flox (grey bar, 19.03 ± 1.22 × 106), Stra8-Trf1−/− (white bar, 0.06 ± 0.11 × 106). Data are presented as mean ± SEM. **P < 0.01.
The Trf1-deficient spermatocytes arrest at two different stages.
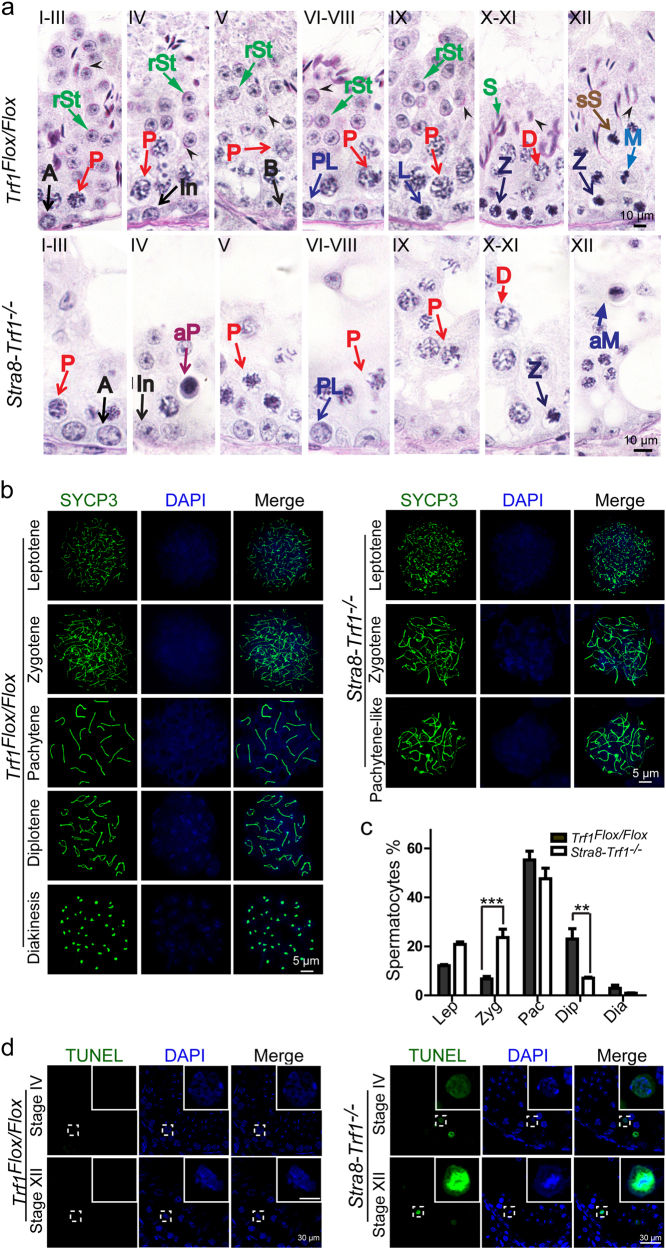
Spermatogenesis is a process in the seminiferous epithelium where various generations of germ cells undergo a series of developmental steps, including spermatogonial mitosis, spermatocytic meiosis, and spermiogenesis. [31, 32] Spermatogenesis can be subdivided into 12 stages in mouse testes using Periodic Acid Schiff (PAS) and hematoxylin staining of sections. [33] To identify which stages of spermatogenesis were affected after Trf1 deletion, sections were stained with PAS and hematoxylin and we determined that spermatogenesis of Trf1-deficient mice was blocked at stages IV and XII (Fig. 2a). Since early spermatocytes gradually develop into mid-pachytene spermatocytes at stage III–V and spermatocytes in either the first or the second meiotic division are present in stage XII, [33] our results indicate that the Stra8-Trf1−/− spermatocytes might be arrested at the pachytene and meiotic division stages.
Fig. 2. Trf1-deficient spermatocytes arrest at two stages.
a Seminiferous tubules paraffin sections from Trf1Flox/Flox and Stra8-Trf1−/− testis were stained with PAS-hematoxylin. A type A spermatogonia, In intermediate spermatogonia, B type B spermatogonia, PL preleptotene spermatocytes, L leptotene spermatocytes, Z zygotene spermatocytes, P pachytene spermatocytes, aP apoptotic pachytene spermatocytes, M meiotic divisions, aM abnormal meiotic divisions, sS secondary spermatocytes, rSt round spermatids, S spermatids. b Spermatocyte stages in pre-metaphase in Trf1Flox/Flox and Stra8-Trf1−/− spermatocytes. Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and DAPI (blue). c Meiotic stage frequencies in Trf1Flox/Flox and Stra8-Trf1−/− testes. Lep (Leptotene): Trf1Flox/Flox (gray bar, 12.07 ± 0.94%), Stra8-Trf1−/− (white bar, 20.85 ± 1.30%). Zyg (Zygotene): Trf1Flox/Flox (gray bar, 6.71 ± 1.34%), Stra8-Trf1−/− (white bar, 23.63 ± 6.42%). Pac (Pachytene): Trf1Flox/Flox (gray bar, 55.31 ± 2.50%), Stra8-Trf1−/− (white bar, 47.60 ± 2.75%). Dip (Diplotene): Trf1Flox/Flox (gray bar, 23.00 ± 2.71%), Stra8-Trf1−/− (white bar, 7.00 ± 0.89%). Dia (Diakinesis): Trf1Flox/Flox (gray bar, 2.88 ± 1.49%), Stra8-Trf1−/− (white bar, 0.89 ± 0.52%). Data are presented as mean ± SEM. **P < 0.01, ***P < 0.001. d Representative TUNEL results in Trf1Flox/Flox and Stra8-Trf1−/− testes. Paraffin sections from Trf1Flox/Flox and Stra8-Trf1−/− testes were stained with TUNEL (green) and DAPI (blue) to determine apoptotic cells in stage IV and stage XII tubules together with pachytene and metaphase spermatocytes.
To confirm this hypothesis, we identified the various stages of meiotic prophase I by staining for a component of the synaptonemal complex, SYCP3. [34] All meiotic prophase I stages could be identified in the spermatocyte nuclei of Trf1Flox/Flox testes, whereas only spermatocytes from the leptotene to the pachytene-like stages were observed in the Stra8-Trf1−/− mice testes (Fig. 2b). Further quantification of the meiotic prophase I stages in testes indicated that the proportion of the zygotene cells was significantly increased and that of diplotene cells was decreased in the Trf1-deficient testes (Fig. 2c). The percentage of spermatocytes in the pachytene stage was not accumulated in Stra8-Trf1−/− mouse testes (Fig. 2c), most likely because the pachytene checkpoint-arrested spermatocytes undergoing apoptosis. [35] To further confirm this possibility, TUNEL staining was performed, and TUNEL-positive signal was detected in the pachytene-like spermatocytes in testes from Stra8-Trf1−/− mice (Fig. 2d), with the percentages of TUNEL positive tubules and spermatocytes in stage IV significantly higher than that of the control (Supplementary Figure 1d-e), suggesting that the deletion of Trf1 leads to a pachytene-like arrest and results in spermatocyte death.
Since germ cells of Trf1-deficient mice also displayed stage XII arrest (Fig. 2a) and some diplotene and diakinesis spermatocytes could be identified in the Stra8-Trf1−/− mouse testes, we speculated that some Trf1-deficient spermatocytes could bypass the pachytene checkpoint due to the variation of TRF1 expression, TRF1 protein half-life, or other reasons. We found that some of the germ cells were then blocked at the meiotic division stage. To determine the fate of the meiotic division stage-arrested germ cells, we performed TUNEL staining and found that some spermatocytes undergoing meiotic division were TUNEL positive (Fig. 2d), with both the percentages of TUNEL positive tubules and spermatocytes in stage XII significantly higher than that of the control (Supplementary Figure 1f-g). Thus, Trf1 disruption leads to most spermatocytes arresting at the pachytene-like stage but some of them bypass the pachytene checkpoint and are later blocked at the meiotic division stage.
Germ cell-specific Trf1 knockout impairs the attachment of telomeres to the nuclear membrane
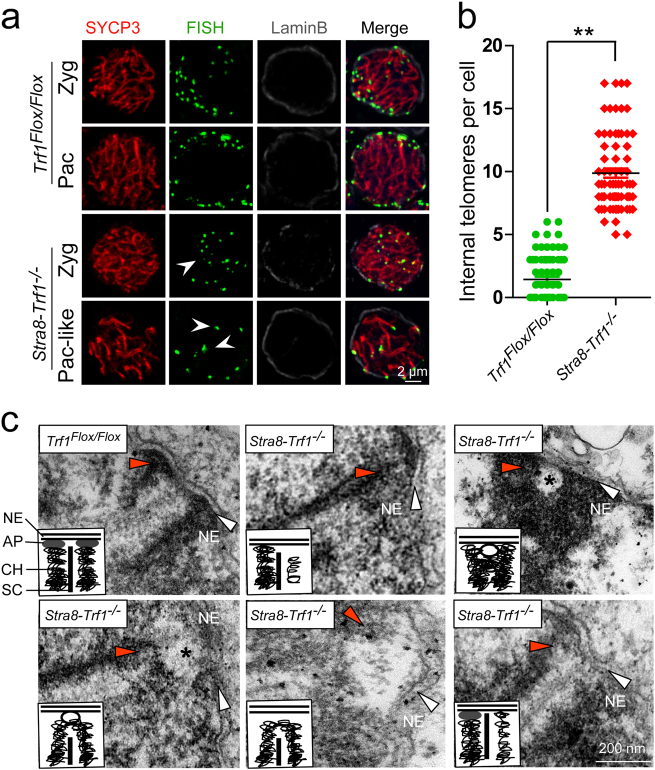
Since it has been reported that TERB1 is involved in anchoring telomeres to the NE by interacting with TRF1, [28] we speculated that knocking out Trf1 might impair NE attachment. To verify this hypothesis, we performed immunofluorescence staining of the NE marker Lamin B and telomere FISH. In Trf1Flox/Flox spermatocytes, most of the telomeres were attached to the NE at the zygotene and pachytene stage, whereas several telomeres were still detached from the NE in the Stra8-Trf1−/− spermatocytes (Fig. 3a-b). Electron microscopy analysis indicated that the telomeres either did not reach the NE, did not form an attachment plate (AP), or were bound to a membrane vesicle just before reaching the NE (Fig. 3c), which is similar to the reported phenomenon in other telomere-NE attachment-deficient mice. [24–29, 36, 37] Thus, the disruption of Trf1 seems to affect the telomere-NE attachment during meiosis.
Fig. 3. TRF1 is required for the attachment of telomeres to the nuclear envelope in spermatocytes.
a Increased detachment of telomeres from NE after Trf1 deletion. An increased number of telomeres were distributed at the inner site of nucleus in Stra8-Trf1−/− spermatocytes compared with the control group. Paraffin sections from Trf1Flox/Flox and Stra8-Trf1−/− testes were immunostained with antibodies against SYCP3 (red), Tel-FISH (green), and Lamin B (gray). An increased number of telomere (FISH) signals was observed at the inner site of the nucleus and did not co-localize with the NE (Lamin B) signals. White arrowheads indicate the inner telomeres. Zyg zygotene spermatocytes, Pac pachytene spermatocytes. b The number of internal telomeres in Stra8-Trf1−/− spermatocytes was increased compared with that of the control group. Zygotene to pachytene spermatocytes were counted for wild-type, whereas pachytene-like spermatocytes were counted for the Stra8-Trf1−/− spermatocytes. Trf1Flox/Flox, 1.17 ± 1.38; Stra8-Trf1−/−, 9.56 ± 1.62. The numbers of internal telomere (FISH) foci are presented as mean ± SEM. **P < 0.01. c An increased number of telomeres were detached from NE in the Trf1-deficient spermatocytes. Electron micrographs showing telomeres (red arrowheads) and inner NE (white arrows) in the Trf1Flox/Flox pachytene and Stra8-Trf1−/− pachytene-like nuclei; asterisks indicate membrane vesicles. Schematic illustrations represent the structures. AP attachment plate, CH chromatin, NE nuclear envelope, SC synaptonemal complex.
TRF1 is essential for homologue synapsis and recombination
During meiotic prophase I, telomeres associate with the LINC complex to attach to the nuclear envelope and the disruption of this attachment might affect homologous chromosome synapsis and recombination, thereby leading to pachytene-like arrest. [24, 26–29, 36, 37] To investigate homologous chromosomes synapsis, we stained chromosome spreads with antibodies against SYCP3 and SYCP1, a key component of the synaptonemal complex [38] and found that the SYCP1 signals were not continuous with SYCP3 and many regions of homologous chromosomes failed to synapse in the Trf1-deficient spermatocytes (Supplementary Figure 2), suggesting the knockout of Trf1 impairs homologue synapsis.
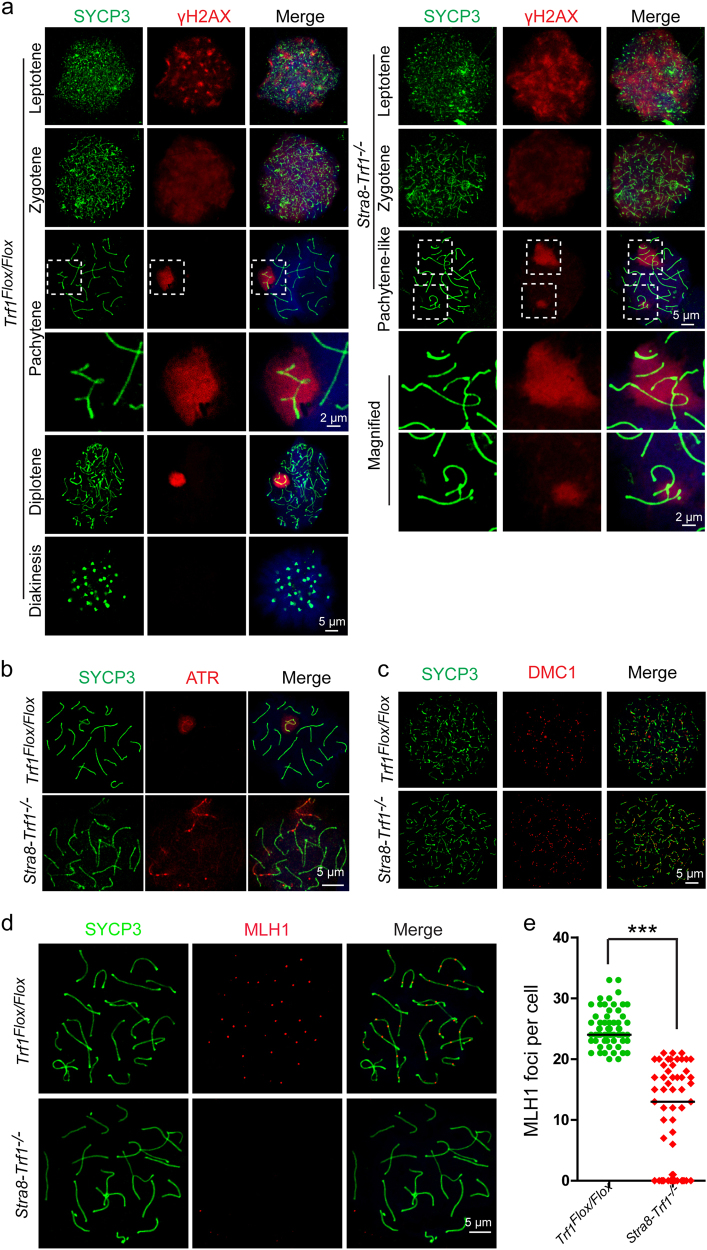
Chromosome synapsis and recombination are facilitated by the introduction of DNA double-strand breaks (DSBs), [39] which can be monitored by detecting γH2AX-positive foci at the leptotene and zygotene stages and these are decreased on autosomes after DSB repaired at the pachytene stage (Fig. 4a). [40] In the Stra8-Trf1−/− spermatocytes, the γH2AX signal was detected at leptotene and zygotene stages, which was similar to Trf1Flox/Flox spermatocytes, indicating the normal production of DSBs (Fig. 4a). However, in the Trf1-deficient spermatocytes, the γH2AX signal was still diffused on the autosomes at the pachytene-like stage, rather than accumulated only on the XY bodies (Fig. 4a). Consistent with the γH2AX staining, ATR, which is integral to the regulation of programmed DSB formation, [41, 42] was also detected on the autosomes of Stra8-Trf1−/− spermatocytes (Fig. 4b). These results indicate that the programmed DSBs are properly introduced, but DSB repair is impaired in the Trf1-deficient spermatocytes. Since the DSB repair in meiosis is mainly dependent on homologous recombination, we stained for the meiosis specific recombination related protein, DMC1 [39] in Stra8-Trf1−/− spermatocytes. The number of DMC1 foci in the Trf1-deficient spermatocytes was dramatically increased compared with that of the control group (Fig. 4c), indicating impaired homologous recombination. The late-recombination marker MLH1 normally appeared at the designated crossing-over sites, [43] and MLH1 immunofluorescence in Trf1-deficient spermatocytes indicated that the number of MLH1 foci was significantly reduced compared with that of the control group (Fig. 4d-e), suggesting the crossover formation was also impaired. Altogether, TRF1 is required for homologue synapsis and recombination, and the disruption of Trf1 might affect the telomere-nuclear envelope attachment, lead to impaired homologue synapsis, recombination and crossover formation, finally resulting in a pachytene-like arrest.
Fig. 4. Impaired meiotic recombination in Stra8-Trf1−/− spermatocytes.
a Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and γH2AX (red). γH2AX marked DSBs in leptotene and zygotene spermatocytes. No difference was observed between Trf1Flox/Flox and Stra8-Trf1−/− at these stages. In the Trf1Flox/Flox spermatocytes at the pachytene stage, DSBs were mostly repaired and γH2AX was confined to the XY body, whereas at the pachytene-like stage in Stra8-Trf1−/− spermatocytes, γH2AX remained on some of the autosomal regions. b Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and ATR (red). In the Trf1Flox/Flox spermatocytes at the pachytene stage, DSBs were mostly repaired and ATR was confined to the XY body, while the ATR signal persisted on the autosomes in Stra8-Trf1−/− spermatocytes. c Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and DMC1 (red). The DMC1 signals were dramatically increased in the Stra8-Trf1−/− spermatocytes at the zygotene stage compared with those in the Trf1Flox/Flox spermatocytes. d Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunostained with antibodies against SYCP3 (green) and MLH1 (red). MLH1 is a recombination marker in WT spermatocytes and decreased MLH1 staining was observed in Stra8-Trf1−/− spermatocytes. e MLH1 signals were counted in spreads. Quantification of the MLH1 foci numbers per cell in the Trf1Flox/Flox and Stra8-Trf1−/− spermatocytes. Trf1Flox/Flox, 25.0 ± 1.7; Stra8-Trf1−/−, 10.6 ± 2.8. Data are presented as mean ± SEM. ***P < 0.001.
Impaired telomere fusion protection in Trf1-deficient spermatocytes leads to meiotic division arrest
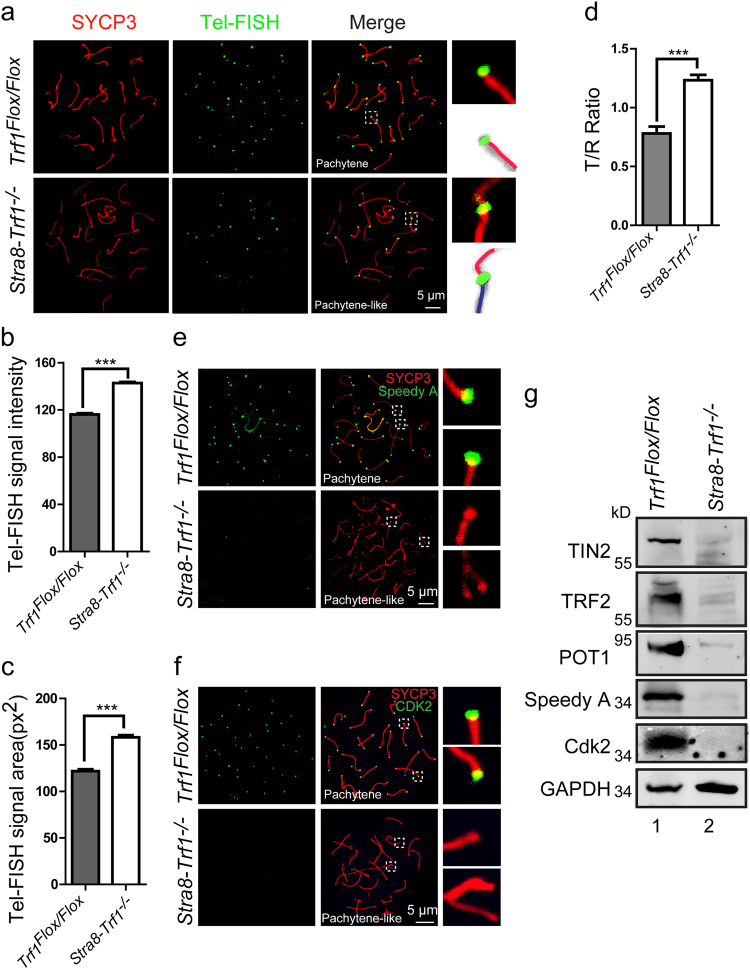
It has been reported that the depletion of shelterin complex subunits, such as TRF1, leads to telomere de-protection and initiation of DNA damage response at the end of chromosomes with telomere fusions and cell cycle checkpoint activation in somatic cells. [5, 12, 15, 16, 44] Thus, it is possible that TRF1 depletion in germ cells could lead to chromosomal end-to-end fusions and result in meiotic division arrest. We noticed fused chromosomes in Stra8-Trf1−/− pachytene-like spermatocytes (Figs. 2b, 4a-b). Therefore, we performed telomeric FISH experiment together with SYCP3 staining in chromosome spreads of spermatocyte nuclei. Indeed we found end-to-end fusions in the Trf1-deficient spermatocytes (Fig. 5a), which might originate from the bouquet stage in Stra8-Trf1−/− spermatocytes (Supplementary Figure 3). In support of the above results, we uncovered that both the Tel-FISH intensity and area in the Trf1-deficient spermatocytes were increased compared to those in the control group (Fig. 5b-c), most likely due to their elongation by telomere fusions (Fig. 5d). These results suggest that germ cell-specific knockout of Trf1 may impair telomere integrity, leading to chromosomal instability with telomere end-to-end fusions, finally resulting in a meiotic division arrest.
Fig. 5. TRF1 is required for preventing telomere fusion at the bouquet stage.
a Telomere fusions occurred after Trf1 depletion. Meiotic chromosome spreads from the Trf1Flox/Flox and Stra8-Trf1−/− testis were immunostained with antibody against SYCP3 (red) and Tel-FISH (green). In the pachytene-like Stra8-Trf1−/− spermatocytes, FISH signal can be found in the middle of the SYCP3 threads, instead of at the ends. Schematic illustrations represent the structures. b and c Telomeric FISH signal intensities and areas per telomere were increased in Stra8-Trf1−/− spermatocytes. Telomeric FISH signal intensities and areas were measured, and the mean Tel-FISH signal intensities and areas were shown. Intensity: Trf1Flox/Flox (gray bar, 116.22 ± 5.42) Stra8-Trf1−/− (white bar, 142.77 ± 5.35). Area: Trf1Flox/Flox (gray bar, 121.98 ± 7.58 px2), Stra8-Trf1−/− (white bar, 158.28 ± 8.87 px2). Data are presented as mean ± SEM. ***P < 0.001. d Telomere length was increased in the Trf1-deficient spermatocytes. Quantitative PCR was performed to measure telomere length and the relative telomere to reference single copy gene (T/R) ratio was calculated to represent the average telomere length in the Trf1Flox/Flox and Stra8-Trf1−/− spermatocytes at pachytene or pachytene-like stage. Trf1Flox/Flox (gray bar, 0.78 ± 0.12), Stra8-Trf1−/− (white bar, 1.23 ± 0.09). Data are presented as mean ± SEM. ***P < 0.001. e and f Speedy A and Cdk2 are not recruited to the telomeres in the Trf1-deficient spermatocytes. Trf1Flox/Flox and Stra8-Trf1−/− chromosome spreads of spermatocytes were immunolabelled with antibodies against SYCP3 (red) and Speedy A (green) or Cdk2 (green). g Selected shelterin and LINC complexes protein levels were decreased after Trf1 depletion. Immunoblotting for shelterin and LINC proteins in Trf1Flox/Flox and Stra8-Trf1−/− spermatocytes. TIN2, TRF2, POT1, Speedy A, and Cdk2 were all decreased to some extent in the Stra8-Trf1−/− spermatocytes. GAPDH was used as loading control.
To investigate the protection mechanism of TRF1 in preventing telomere fusion during meiosis, we stained for other shelterin complex subunits. Surprisingly, the protein levels of the shelterin complex proteins TRF2, TIN2, and POT1 were dramatically reduced in the Stra8-Trf1−/− mouse testes but TRF2 was still localized on telomeres, even when fusion occurred (Fig. 5g and Supplementary Figure 4a), which suggests that telomere might be protected during meiosis through a meiosis-specific mechanism. Recently, several LINC complex proteins and their adaptor proteins have been identified, such as Speedy A, Cdk2, TERB1, MAJIN, and SUN1. [24, 26, 28, 29, 36, 37] We screened potential effectors of telomere fusion at the bouquet stage, and found that TRF2, RAP1, and SUN1 were still localized on telomeres, but Speedy A and Cdk2 were not recruited to the telomeres in Trf1-deficient spermatocytes (Fig. 5e-f and Supplementary Figures 4a-c). In addition, the protein levels of Speedy A and Cdk2 were substantially decreased in the Stra8-Trf1−/− mouse testes compared to those of the control groups (Fig. 5g). These results indicate that the disruption of Trf1 impairs the localization of the meiotic telomere-related proteins, Speedy A and Cdk2, on telomeres, which might be indispensable for preventing telomere fusion during meiosis.
Direct physical interaction between TRF1 and Speedy A
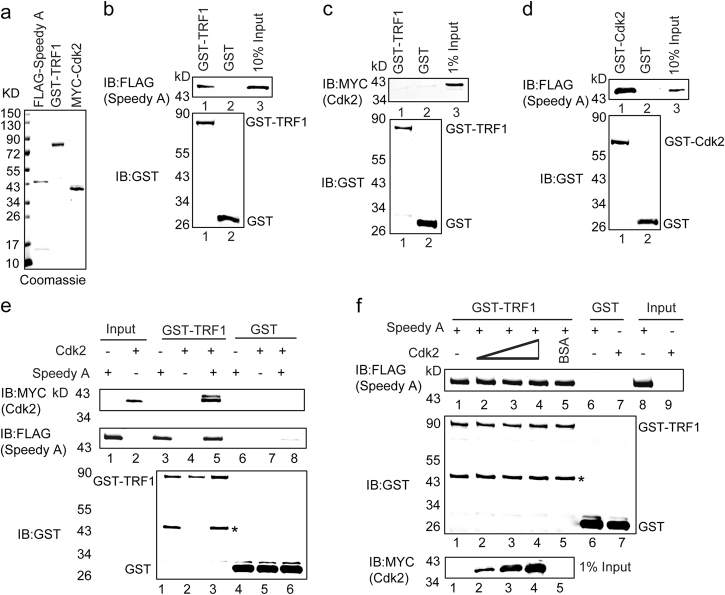
To further investigate the relationship between TRF1, Speedy A and Cdk2, we purified TRF1, Speedy A and Cdk2 proteins (Fig. 6a) and performed the GST pull-down experiments. Our results indicated that TRF1 could directly interact with Speedy A, but not Cdk2 (Fig. 6b-c). We further confirmed the known interaction between Cdk2 and Speedy A, and found that GST-Cdk2 could directly bind to Speedy A (Fig. 6d). Since previous reports indicated that Speedy A was necessary for Cdk2’s localization on telomeres during meiosis, [36] we speculated Speedy A might work as a scaffold protein to facilitate the indirect interaction between TRF1 and Cdk2. To test this possibility, GST-TRF1 was used to pull-down Cdk2 in the presence or absence of Speedy A. The GST pull-down experiments suggested that in the presence of Speedy A, Cdk2 could be pulled down by TRF1 (Fig. 6e). To further demonstrate that Speedy A was a scaffold protein, we did a competition experiment by adding excess Speedy A or Cdk2. When we increased the levels of Cdk2 in the reaction, the binding of Speedy A to TRF1 was unchanged, suggesting that TRF1 and Cdk2 might bind Speedy A at different non-overlapping sites (Fig. 6f). Thus, TRF1 interacts directly with Speedy A, and Speedy A binds to Cdk2, thereby recruiting Cdk2 to the telomeres.
Fig. 6. Direct physical interaction between TRF1 and Speedy A.
a Coomassie blue-stained gels showing the purification of relevant proteins. b Direct physical interaction between TRF1 and Speedy A were determined by GST pull-down assays. GST-TRF1 or GST conjugated sepharose beads were used to pull down His-FLAG-Speedy A. Bound proteins were detected by immunoblotting analysis with anti-FLAG and anti-GST antibodies. c No direct physical interaction between TRF1 and Cdk2, which was determined by GST pull-down assays. GST-TRF1 or GST conjugated sepharose beads were used to pull down His-MYC-Cdk2. Bound proteins were detected by immunoblotting analysis with anti-MYC and anti-GST antibodies. d Direct physical interaction between CDK2 and Speedy A was determined by GST pull-down assays. GST-CDK2 or GST conjugated sepharose beads were used to pull down purified His-FLAG-Speedy A. Bound proteins were detected by immunoblotting analysis with anti-FLAG and anti-GST antibodies. e Speedy A-mediated interaction between TRF1 and Cdk2. GST-TRF1 or GST conjugated sepharose beads were used to pull down His-FLAG-Speedy A and (or) His-MYC-Cdk2. Bound proteins were detected by immunoblotting analysis with anti-FLAG or anti-MYC antibody. Asterisks indicate the bands of Speedy A. Input for His-FLAG-Speedy A and His-MYC-Cdk2 were 10% and 1%, respectively. f TRF1 and Cdk2 do not compete for interaction with Speedy A. GST-TRF1 or GST conjugated sepharose beads were used to pull down His-FLAG-Speedy A in the presence of increasing amounts of His-MYC-Cdk2. Bound proteins were detected by immunoblotting analysis with anti-FLAG and anti-MYC antibodies. Asterisks indicate signals of Speedy A.
Speedy A and Cdk2 are required for protecting telomere fusion at the bouquet stage
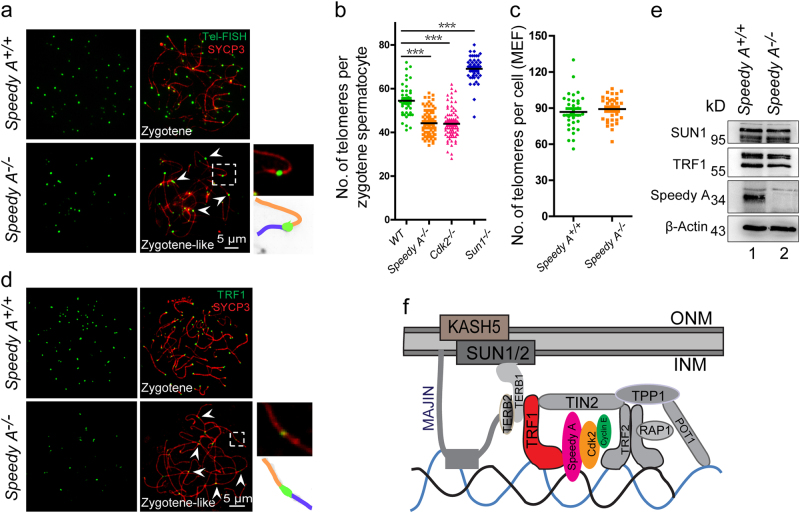
To investigate whether Speedy A and Cdk2 are also required for preventing telomere fusion during meiosis, we performed telomere-FISH and SYCP3 staining of Speedy A, Cdk2, and Sun1-deficient spermatocytes and found some fused telomeres in Speedy A−/− and Cdk2−/− spermatocytes but not in Sun1−/− spermatocytes (Fig. 7a, Supplementary Figures 5a and 5c). Consistent with the telomere fusion results, we found that the total number of telomeres in the Speedy A−/− and Cdk2−/− spermatocytes was significantly decreased compared with their control groups (Fig. 7b), but we did not observe this phenomenon in somatic cells since the total number of telomeres in Speedy A−/− mouse embryonic fibroblasts (MEFs) was similar to that of the control (Fig. 7c). In contrast to Speedy A−/− and Cdk2−/−, the total number of telomeres in Sun1−/− spermatocytes was normal at pachytene-like stage, but was increased at zygotene stage, presumably due to homologous chromosome synapsis failure (Fig. 7b). These results are similar to those findings in the Trf1-deficient spermatocytes, indicating that Speedy A and Cdk2 are required for preventing telomere fusion during meiosis, and telomere fusion in Trf1-deficient spermatocytes might be caused by the absence of Speedy A and Cdk2 on telomeres.
Fig. 7. TRF1-mediated Speedy A and Cdk2 recruitment are required for preventing telomere fusion at the bouquet stage.
a Telomere fusions were detected in Speedy A−/− chromosome spreads of spermatocytes. Speedy A+/+ and Speedy A−/− spreads were immunolabelled with antibodies against SYCP3 (red) and telomere FISH (green). Chromosome fusions are indicated by arrows and a schematic illustrations of the structures is shown. b The telomere numbers in WT, Speedy A−/−, Cdk2−/−, and Sun1−/− knockout mice. The telomere numbers were decreased significantly in Speedy A−/− and Cdk2−/− spermatocytes, but increased in Sun1−/− spermatocytes. The numbers of telomere (FISH) foci are presented as mean ± SEM. ***P < 0.001. c Quantification of Tel-FISH signals in Speedy A+/+ and Speedy A−/− MEFs. Telomere numbers were counted and no significant changes were detected after Speedy A knockout. Speedy A+/+, 90.25 ± 3.84; Speedy A−/−, 94.13 ± 3.23. Data are presented as mean ± SEM. d TRF1 is still localized on telomeres in Speedy A−/− spermatocytes. Speedy A+/+ and Speedy A−/− metaphase spreads were immunostained with Telomere FISH (red) and DAPI (blue). e Immunoblotting for telomere related proteins in Speedy A−/− spermatocytes. SUN1 and TRF1 in Speedy A−/− spermatocytes remained unchanged; β-Actin was used as the loading control. f A model of the meiosis specific telomere fusion protection complex, with TRF1, Speedy A, and Cdk2 as the key players to protect telomeres from fusion.
Since the disruption of Trf1, Speedy A, and Cdk2 could cause telomere fusion during meiosis and Trf1 depletion resulted in the exclusion of Speedy A and Cdk2 from telomeres, TRF1 might be an upstream telomeric recruiter for Speedy A and Cdk2. To test this hypothesis, we stained for TRF1 in Speedy A−/− and Cdk2−/− spermatocytes and found that TRF1 was still localized on telomeres albeit the TRF1 signal was decreased compared to the control (Fig. 7d and Supplementary Figure 5b). The lower TRF1 signaling might be due to structural changes of the telomeres because the deletion of Speedy A or Cdk2 leads to telomere fusions in spermatocytes. Immunoblotting analysis of telomeric complex components indicated that SUN1 and TRF1 levels were not changed in Speedy A−/− spermatocytes (Fig. 7e), thus suggesting that TRF1 and Speedy A may be central for the mechanism of protecting telomeres from fusion at the bouquet stage.
Discussion
The cell interior is densely crowded with thousands of macromolecules such as proteins, DNA, RNA, and other molecules, where biological macromolecules have to function in crowded cellular environment. [45, 46] Chromatin, a huge macromolecule, usually distributes all over the nucleus in most somatic cells. [47] In contrast in germ cells, chromatin packs into a relative small space, and chromosome ends cluster in close proximity at the bouquet stage during meiosis, [17, 20, 21] thus creating a crowded microenvironment for telomeres. It is not known how to protect the telomere from fusion in this special microenvironment. Here, we demonstrate that the disruption of the shelterin component Trf1 drives telomere fusion during meiosis (Fig. 5a) and TRF1 is required for the telomeric localization of Speedy A and Cdk2 (Fig. 5e-f). Further analysis indicated that the absence of Speedy A and Cdk2 led to severe telomere fusions in spermatocytes (Fig. 7a and Supplementary Figure 5a). Thus, TRF1, Speedy A, and Cdk2 are found to be involved in protecting telomeres from fusion in this crowded microenvironment (Fig. 7f).
TRF1 is important for the functional telomere structure and it can directly bind double-stranded telomeres by connecting with TRF2 through TIN2. [4, 15] Depletion of TRF1 affects the telomeric association of TRF2, [48] which has a crucial role in chromosome end protection. [12] Similarly, cyclin E deletion causes TRF2 depletion from the telomeres and aberrant telomere structures. [49] Speedy A was dissociated from telomeres in Trf1-deficient spermatocytes and TRF1 could directly bind to Speedy A, suggesting that TRF1 might recruit Speedy A to telomeres during meiosis. Speedy A then serves as a scaffold protein to further recruit Cdk2 to telomeres. Recently, a “telomere localization domain” of Speedy A was identified and it is sufficient for Speedy A to co-localize with TRF1 on telomeres. [36] The C-terminal RINGO domain of Speedy A is also necessary for Cdk2’s localization to telomeres during meiosis. [26, 36] In addition, it has been reported that the formation of cyclin E/Cdk2 complex is necessary for the telomeric localization of Cdk2 and protection of the telomeres from end-to-end fusions. [49, 50] We found that cyclin E levels were increased in Trf1-deficient spermatocytes and cyclin E was still recruited to chromosomes at the pachytene stage (Supplementary Figure 6), suggesting that cyclin E itself is not sufficient to recruit Cdk2 to the telomeres. Thus, we propose that Speedy A may act as a scaffold protein for TRF1 and Cdk2, thus recruiting Cdk2 to telomeres. On the other hand, the RINGO domain of Speedy A interacts with Cdk2 and promotes Cdk2 activation to protect telomere from fusion during meiosis.
Telomere-NE attachment is essential for homologous chromosome synapsis and recombination and many key proteins have been identified for this attachment such as TERB1, TERB2, MAJIN, Speedy A, Cdk2, SUN1, SUN2, and KASH5. [24, 26, 28, 29, 36, 37] TRF1 can interact with TERB1 to form the chimeric complex, TRF1-TERB1/2-MAJIN, which is indispensable for telomere-NE attachment. [28, 29] During mid prophase, telomere cap exchange is achieved by removing the shelterin complex, including TRF1, from the membrane-anchored telomeres, which is dependent on CDK activity. [29] However, TRF1 is still localized near the membrane-anchored telomere region, indicating additional roles of TRF1 during meiosis. Our results demonstrate that in addition to the telomere-NE attachment, TRF1, Speedy A, and Cdk2 are required for protecting telomeres from fusion at the bouquet stage during meiosis. However, the disruption of Sun1, Terb1, Terb2 and Majin do not affect telomere stability because telomere fusions were not observed in Sun1−/− spermatocytes (Supplementary Figure 5c), and the number of telomere does not decrease in Terb1 and Terb2 deficient spermatocytes. [28, 29] Considering that SUN1-dependent chromatin mobility is important for fusion of dysfunctional telomeres in somatic cells, [51] it is possible that the absence of SUN1 or other LINC complex components do not cause telomere fusion during meiosis. Thus, the functions of LINC components and their adaptor proteins in preventing telomere fusion and telomere-NE attachment are not the same process. Whereas SUN1 only participates in telomere-NE attachments, TRF1, Speedy A, and Cdk2 are involved in both of these two processes (Figs. 5 and 7, Supplementary Figures 5a-b). Therefore TRF1, Speedy A, and Cdk2 are key players of the meiosis-specific mechanism in protecting telomeres from fusion at bouquet stage.
Materials and methods
Animals’ experiments
The Trf1Flox/Flox mice (C57BL/6J) were a gift from Prof. Maria A. Blasco from the Spanish National Cancer Centre (CNIO). [13] The Trf1Flox/Flox Stra8-Cre mice were bred from intercrosses of Trf1Flox/Flox mice and Stra8-Cre mice [30]. The Speedy A−/− and Cdk2−/− mice have been reported previously. [36, 52] The Sun1−/− mice were purchased from Jackson Laboratory, numbered 012715-B6; 129S6-Sun1tm1Mhan/J (Bar Harbor, ME). All animal studies were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences.
Tissue collection and histological analysis
For histological examination, at least three adult mice for each genotype were analyzed. The testes were dissected and fixed with Bouin’s fixative for up to 24 h. Next, the testes were dehydrated using graded ethanol and embedded in paraffin. 5 μm sections were collected and covered on glass slides. After deparaffinization, sections were stained with H&E for histological analysis, or stained with Periodic Acid Schiff (PAS)-hematoxylin for determining the seminiferous epithelia cycle stages.
Epididymal sperm count
The cauda epididymis was isolated from 10-week-old mice. Sperm were released from the cauda epididymis and incubated at 37 °C for 30 min under 5% CO2. The sperm solution was diluted and sperm number was counted with a hemocytometer.
Antibodies
The FLAG (1:2000, M20008L), GST (1:2000, M20007L), MYC (1:1000, M20002M) antibodies were purchased from Abmart (Shanghai, China). Mouse anti-TRF1 (ab10579), rabbit antibodies against SYCP3 (150292) and rabbit anti-SUN1 (ab103021) were purchased from Abcam (Cambridge, MA). Mouse antibodies against γH2AX (05-636) were purchased from Merck Millipore (Darmstadt, Germany). Mouse antibodies against MLH1 (51-1327GR) were purchased from BD Pharmingen (SanDiego, CA). Rabbit antibodies against SYCP1 (NB300-228c) and rabbit anti-TRF2 (NB110-57130) were purchased from Novus Biologicals (Littleton, CO). Mouse antibodies against SYCP3 (SC-74569), goat antibodies against ATR (SC-1187), rabbit antibodies against DMC1 (SC-22768) were purchased from Santa Cruz Biotechnology (Dallas, TX). Mouse anti-RAP1 (A300-306A) was purchased from Bethyl (Montgomery, TX). Mouse antibodies against Cyclin E (MA5-14336) were purchased from Thermo fisher (Rockford, USA). Rabbit anti-Speedy A was generated as previously reported. [36] Goat FITC-conjugated secondary antibodies for rabbit, donkey FITC-conjugated secondary antibodies for mouse, rabbit TRITC-conjugated secondary antibodies for goat, and goat TRITC-conjugated secondary antibodies for mouse IgGs were purchased from Zhong Shan Jin Qiao (Beijing, China). Alexa Fluor®680-conjugated goat secondary antibodies for mouse (A21057) and Alexa Fluor® 680-conjugated rabbit secondary antibodies for goat IgGs (A21088) for immunoblotting were purchased from Invitrogen (Carlsbad, CA), IRDye® 800CW-conjugated goat secondary antibodies for rabbit (926-32211) for immunoblotting was purchased from LI-COR (Lincoln, NE).
Immunoblotting
To prepare protein extracts, the testis albuginea was peeled and the testis or the isolated spermatocytes was suspended in cold RIPA buffer (R0010 Solarbio) supplemented with a protein inhibitor cocktail (Roche Diagnostics, 04693116001, Rotkreuz, Switzerland) and 1 mM phenylmethylsulfonyl fluoride (PMSF, 0754, Amresco). After homogenization and transient sonication, cell extracts were incubated on ice for 30 min. The samples were then centrifuged at 12,000×g for 20 min at 4 °C. The supernatant was transferred to a new tube for immunoblotting. Protein samples were separated via SDS-PAGE and electro-transferred to a nitrocellulose membrane. After incubation with primary and secondary antibodies, the membrane was scanned using an ODYSSEY Sa Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Immunofluorescence and TUNEL assay
The chromosome spreads of spermatocytes were washed with PBS for 3 times and blocked with 5% bovine serum albumin (AP0027, Amresco, Solon, OH). Primary antibodies were incubated at 4 °C overnight, followed by incubation with the secondary antibodies. The nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI). The images were taken immediately using an LSM 780 microscope (Zeiss, Oberkochen, Germany) or a TCS SP8 microscope (Leica, Wetzlar, Germany). 5-μm sections mounted on glass slides were first deparaffinized and then boiled for 15 min in sodium citrate buffer for antigen retrieval. After washing with PBS, sections were blocked and followed by antibody incubation as described above. To detect apoptotic cells in testis, we used the terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assay kit (In Situ Cell Death Detection Kit; Roche, 11684795910) according to the manufacturer’s instructions. [53] Briefly, sections of the testes were deparaffinized and boiled for 15 min in sodium citrate buffer for antigen retrieval. After treated with H2O2 for 10 min at room temperature and sodium citrate for 2 min on ice, the slides were rinsed twice with PBS, the TUNEL reaction mixture was added and incubated in a humidified atmosphere for 60 min at 37 °C in the dark, following by immunofluorescence staining as detailed above.
Immuno-FISH assay
The immuno-FISH assay was carried out based on a FISH protocol with minor modifications. [54] In brief, spermatocyte spreads were treated with DNase free RNase A (V900498, SIGMA, 100 g/ml) at 37 °C for 30 min and then dehydrated with 70, 85, and 100% alcohol. After denaturation at 85 °C for 10 min, they were hybridized for 2 h at 37 °C with fluorescein isothiocyanate (FITC)-labeled Tel (CCCTAA)3 PNA probe (F1009 PNA BIO). The slices were washed sequentially in 2X saline sodium citrate (SSC) with 0.1% Tween (twice) at 65 °C and with 2X SSC (twice) at room temperature for 5 min each time. The preparations were then co-labeled with antibodies.
Genomic DNA extraction and qPCR assay to measure telomere length
The average telomere length was measured from total genomic DNA of pachytene and pachytene-like spermatocytes in Trf1Flox/Flox and Stra8-Trf1−/− mice. Genomic DNA used for average telomere length measurement was extracted following a previous report. [55] Briefly, cells were harvested and lysed in fresh lysis buffer (10 mM Tris-HCL pH 8.0, 0.1 M EDTA, 0.5% SDS, 20 ug/ml RNase) for 3 h and gently inverted every 30 min to release the RNA. Then 200 ug/ml Proteinase K (P6556-100MG, SIGMA) was added and incubated the mixture at 55 °C overnight. After that, phenol:chloroform:isoamyl alcohol = 25:24:1 was used to extract the total genomic DNA, followed by ethanol precipitation. The air-dried DNA was dissolved in 30 µl TE buffer for 2 h in a 37 °C water bath. Purity and integrity of the genomic DNA was tested with NanoDrop and agarose gel electrophoresis. The OD260/OD280 ratio was between 1.60 to 1.90. Telomere length was measured by the qPCR method as previously described. [56, 57]
Protein purification and GST pull-down assay
For purification of GST-TRF1, GST-Cdk2, His-FLAG-Speedy A, and His-MYC-Cdk2, cDNA encoding mouse TRF1, Cdk2 (variant 1) and Speedy A were cloned into pGEX-4t-1 and modified pET28a (one MYC or FLAG tag was first cloned in to the vector) respectively. Briefly, the plasmid was transformed into BL21 (DE3) cells and grown in Terrific Broth at 37 °C. When the optical density reached 1.0, they were transferred to the low temperature (16 °C) shaker, and induced with 0.25 mM isopropyl-D-thiogalactoside (IPTG) for 16 h. After that, cells were harvested and then resuspended in lysis buffer (20 mM Tris, pH 7.4, 500 mM NaCl, 10 mM imidazole, 10% glycerol for hexahistidine-tagged fusion protein; 50 mM Tris, pH 7.4, 500 mM NaCl, 2 mM MgCl2, 5% glycerol for GST-tagged fusion proteins) supplemented with 1 mM PMSF. Sonication was used to lyse the bacteria. Then we collected the supernatant by high-speed centrifugation, and incubated them with Ni Sepharose 6 Fast Flow (GE Healthcare, Marlborough, MA) or Glutathione Sepharose 4B (GE Healthcare, Marlborough, MA) for 2 h at 4 °C. The beads were washed, and the protein was eluted using the lysis buffer supplemented with 250 mM imidazole or 10 mM glutathione. The pull-down assays were performed as described previously [58]. Briefly, GST-TRF1 and GST-Cdk2 were diluted in a buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 2 mM MgCl2, 0.1% BSA and incubated with Glutathione Sepharose 4B agarose beads (GE Healthcare, Marlborough, MA) at 4 °C for 2 h, followed by further incubation for 2 h at 4 °C with FLAG-Speedy A or MYC-Cdk2 or both of them. The beads were washed three times with high-salt buffer, and then subjected to immunoblotting analysis with anti-GST, anti-MYC and anti-FLAG antibodies.
Transmission electron microscopy
Adult mouse testis was dissected and fixed with 2.5% (vol/vol) glutaraldehyde in 0.2 M cacodylate buffer (50 mM cacodylate pH 7.2, 50 mM KCl, 2.5 mM MgCl2) overnight. After washing in cacodylate buffer, the tissues were cut into small pieces of approximately 1 mm3 and immersed in 1% OsO4 in 0.2 M cacodylate buffer for 2 h at 4 °C. Next, the samples were washed and submerged in 0.5% uranyl acetate overnight. Dehydrated through a graded ethanol series and embedded in resin (Low Viscosity Embedding Media Spurr’s Kit, EMS, 14300). Ultrathin sections were cut on an ultramicrotome and mounted on copper grids. Then the sections were stained with uranyl acetate and lead citrate for 10 min and observed using a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan).
Chromosome spreads of spermatocyte
Spermatocyte surface spreading was conducted according to the drying-down technique as previously described. [59] Briefly, testes were dissected and the tubules were washed in phosphate-buffered saline (PBS) pH 7.4 at room temperature. Next, the tubules were submerged in a hypotonic extraction buffer (30 mM Tris pH 8.2, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM DTT and 0.5 mM PMSF) for 30–45 min. Subsequently, the tubules were torn into pieces in 100 mM sucrose pH 8.2 on a clean glass slide and then pipetted gently to make a suspension. The cell suspensions were loaded on slides containing 1% paraformaldehyde (PFA) pH 9.2 and 0.15% Triton X-100. The slides were dried for at least 2 h in a closed box with high humidity. Finally, the slides were washed with 0.4% Photoflo (Kodak, 1464510, Rochester, NY) for 10 min and immunostained with antibodies according to the standard protocols mentioned above.
Speedy A−/− MEF isolation and metaphase spread karyotyping
Speedy A−/− mice was generated from intercrossing of Speedy A+/- mice. We obtained mouse embryonic fibroblasts (MEF) from 13.5-day-old embryos as previously described. [60] The cells were incubated at 37 °C with 5% CO2. For karyotype determination of metaphase spreads, 0.1 µg/mL Colchicine was used to synchronize MEF cells at metaphase, following treatment with hypotonic buffer (0.075M KCl) and incubation at room temperature for 20 min. The suspension was centrifuged and the supernatant was discarded. Cell pellets were fixed twice: in methanol/glacial acetic acid (3:1) and methanol/glacial acetic acid (1:1). Fixed cells were dropped onto ice-cold slides and air dried. The slides were stained with telomere-FISH and DAPI to calculate the proportion of fusion.
Statistical analysis
All data are presented as the mean ± SEM. The statistical significance of the differences between the mean values for the various genotypes was measured by Student’s t-tests with a paired, 2-tailed distribution. The data were considered significant when the P-value was less than 0.05 (*), 0.01 (**) or 0.001(***).
Electronic supplementary material
Supporting Information for Dual roles of TRF1 in tethering telomeres to the nuclear envelope and protecting them from fusion during meiosis.doc
Acknowledgements
We thank Prof. Maria A. Blasco for providing the Trf1 floxed mice. We thank Qingyuan Sun, Kui Liu, and Nathan Palmer for critical reading of the manuscript.
Author contributions
LW, ZT, and CL performed most of the experiments and wrote the manuscript. HL performed part of the experiment. PK constructed the Cdk2−/− and SpeedyA−/− knockout mouse and revised the manuscript. ZC and WL supervised the whole project and wrote the manuscript.
Funding
This work was supported by the National key R&D program of China (Grant No. 2016YFA0500901), National Nature Science of China (Grant No. 31471277 and 91649202) to W. Li and the Biomedical Research Council of A*STAR (Agency for Science, Technology and Research, Singapore) to P. Kaldis.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by R.A. Knight
Lina Wang, Zhaowei Tu and Chao Liu contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1038/s41418-017-0037-8) contains supplementary material, which is available to authorized users.
Contributor Information
Zijiang Chen, Email: chenzijiang@hotmail.com.
Wei Li, Email: leways@ioz.ac.cn.
References
- 1.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–43. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 2.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–7. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 2015;522:492–6. doi: 10.1038/nature14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Bio. 2010;11:171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Gene Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 6.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi A, Smith S, Chong L, Elias P, deLange T. TRF1 is a dimer and bends telomeric DNA. Embo J. 1997;16:1785–94. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–9. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 9.Li BB, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060–8. doi: 10.1091/mbc.e03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor MS, Safari A, Xin HW, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006;103:11874–9. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Podell ER, Zaug AJ, Yang YT, Baciu P, Cech TR, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–5. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez P, Thanasoula M, Munoz P, Liao CY, Tejera A, McNees C, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Gene Dev. 2009;23:2060–75. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allegra A, Innao V, Penna G, Gerace D, Allegra AG, Musolino C. Telomerase and telomere biology in hematological diseases: A new therapeutic target. Leuk Res. 2017;56:60–74. doi: 10.1016/j.leukres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Bio. 2017;18:175–86. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez P, Blasco MA. Telomere-driven diseases and telomere-targeting therapies. J Cell Biol. 2017;216:875–87. doi: 10.1083/jcb.201610111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol life Sci: CMLS. 2007;64:117–24. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange T. Human telomeres are attached to the nuclear matrix. Embo J. 1992;11:717–24. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–81. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reig-Viader R, Garcia-Caldes M, Ruiz-Herrera A. Telomere homeostasis in mammalian germ cells: a review. Chromosoma. 2016;125:337–51. doi: 10.1007/s00412-015-0555-4. [DOI] [PubMed] [Google Scholar]
- 21.Link J, Jahn D, Alsheimer M. Structural and functional adaptations of the mammalian nuclear envelope to meet the meiotic requirements. Nucl-Phila. 2015;6:93–101. doi: 10.1080/19491034.2015.1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–32. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- 23.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Bio. 2001;2:621–7. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 24.Ding X, Xu R, Yu JH, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–72. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, et al. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–98. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikolcevic P, Isoda M, Shibuya H, Barrantes ID, Igea A, Suja JA, et al. Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat Commun. 2016;7:11084. doi: 10.1038/ncomms11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viera A, Alsheimer M, Gomez R, Berenguer I, Ortega S, Symonds CE, et al. CDK2 regulates nuclear envelope protein dynamics and telomere attachment in mouse meiotic prophase. J Cell Sci. 2015;128:88–99. doi: 10.1242/jcs.154922. [DOI] [PubMed] [Google Scholar]
- 28.Shibuya H, Ishiguro K, Watanabe Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol. 2014;16:145–56. doi: 10.1038/ncb2896. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya H, Hernandez-Hernandez A, Morimoto A, Negishi L, Hoog C, Watanabe Y. MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell. 2015;163:1252–66. doi: 10.1016/j.cell.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–42. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roosen-Runge EC. The process of spermatogenesis in mammals. Biol Rev Camb Philos Soc. 1962;37:343–77. doi: 10.1111/j.1469-185X.1962.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 32.Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263–77. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 34.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 35.Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/S0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 36.Tu ZW, Bayazit MB, Liu HB, Zhang JJ, Busayavalasa K, Risal S, et al. Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc Natl Acad Sci USA. 2017;114:592–7. doi: 10.1073/pnas.1618465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horn HF, Kim DI, Wright GD, Wong ESM, Stewart CL, Burke B, et al. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol. 2013;202:1023–39. doi: 10.1083/jcb.201304004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. Embo J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–8. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter N, Borner GV, Lichten M, Kleckner N. Gamma-H2AX illuminates meiosis. Nat Genet. 2001;27:236–8. doi: 10.1038/85781. [DOI] [PubMed] [Google Scholar]
- 41.Wang HY, Wang ML, Wang HC, Bocker W, Iliakis G. Complex H2AX phosphorylation patterns by multiple kinases including ATM and DNA-PK in human cells exposed to ionizing radiation and treated with kinase inhibitors. J Cell Physiol. 2005;202:492–502. doi: 10.1002/jcp.20141. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. Embo J. 2005;24:4345–55. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–42. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 44.Bandaria JN, Qin PW, Berk V, Chu S, Yildiz A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell. 2016;164:735–46. doi: 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev. 2014;114:2733–58. doi: 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- 46.Zimmerman SB. Macromolecular crowding effects on macromolecular interactions—some implications for genome structure and function. Biochim Biophys Acta. 1993;1216:175–85. doi: 10.1016/0167-4781(93)90142-Z. [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008;90:1040–51. doi: 10.1016/j.biochi.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442–8. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 49.Manterola M, Sicinski P, Wolgemuth DJ. E-type cyclins modulate telomere integrity in mammalian male meiosis. Chromosoma. 2016;125:253–64. doi: 10.1007/s00412-015-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinerie L. Mammalian E-type cyclins control chromosome pairing, telomere stability and CDK2 localization in male meiosis. Plos Genet. 2014;10:e1004165. doi: 10.1371/journal.pgen.1004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell. 2015;163:880–93. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–85. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 53.Song ZH, Yu HY, Wang P, Mao GK, Liu WX, Li MN, et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015;6:e1589. doi: 10.1038/cddis.2014.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolzan AD, Bianchi MS. Detection of incomplete chromosome elements and interstitial fragments induced by bleomycin in hamster cells using a telomeric PNA probe. Mutat Res-Fund Mol M. 2004;554:1–8. doi: 10.1016/j.mrfmmm.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Wang F, Pan XH, Kalmbach K, Seth-Smith ML, Ye XY, Antumes DMF, et al. Robust measurement of telomere length in single cells. Proc Natl Acad Sci USA. 2013;110:E1906–E1912. doi: 10.1073/pnas.1306639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callicott RJ, Womack JE. Real-time PCR assay for measurement of mouse telomeres. Comp Med. 2006;56:17–22. [PubMed] [Google Scholar]
- 57.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C, Liu WX, Ye YH, Li W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat Commun. 2017;8:14274. doi: 10.1038/ncomms14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/A:1018445520117. [DOI] [PubMed] [Google Scholar]
- 60.Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol/Ed Frederick M Ausubel [Et al] 2001;Chapter 23:Unit23 22. doi: 10.1002/0471142727.mb2302s51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information for Dual roles of TRF1 in tethering telomeres to the nuclear envelope and protecting them from fusion during meiosis.doc