Abstract
Aflatoxin B1 (AFB1), which is mainly produced by Aspergillus flavus and Aspergillus parasiticus, is the most toxic and hepatocarcinogenic polyketide known. Chemical fungicides are currently utilized to reduce this fungal contaminant, but they are potentially harmful to human health and the environment. Therefore, natural anti-aflatoxigenic products are used as sustainable alternatives to control food and feed contamination. For example, eugenol, presents in many essential oils, has been identified as an aflatoxin inhibitor. However, its exact mechanism of inhibition is yet to be clarified. In this study, the anti-aflatoxigenic mechanism of eugenol in A. flavus was determined using a comparative transcriptomic approach. Twenty of twenty-nine genes in the aflatoxin biosynthetic pathway were down-regulated by eugenol. The most strongly down-regulated gene was aflMa, followed by aflI, aflJ, aflCa, aflH, aflNa, aflE, aflG, aflM, aflD, and aflP. However, the expression of the regulator gene aflR did not change significantly and the expression of aflS was slightly up-regulated. The down-regulation of the global regulator gene veA resulted in the up-regulation of srrA, and the down-regulation of ap-1 and mtfA. The early developmental regulator brlA was profoundly up-regulated in A. flavus after eugenol treatment. These results suggested a model in which eugenol improves fungal development by up-regulating the expression of brlA by the suppression of veA expression and inhibits aflatoxin production through the suppression of veA expression. Exposure to eugenol also caused dysregulated transcript levels of the G protein-coupled receptors (GPCRs) and oxylipins genes. A Gene Ontology analysis indicated that the genes that were highly responsive to eugenol were mainly enriched in RNA-binding functions, suggesting that post-transcriptional modification plays a pivotal role in aflatoxin biosynthesis. KEGG analysis showed that ribosome biogenesis was the most dysregulated pathway, suggesting that eugenol dysregulates ribosome biogenesis, which then interrupts the biosynthesis of Nor-1, Ver-1, and OmtA, and prevents aflatoxisomes performing their normal function in aflatoxin production. In conclusion, our results indicated that eugenol inhibited AFB1 production by modulating the expression of structural genes in aflatoxin pathway, fungal antioxidant status, post-transcriptional modifications and biosynthesis of backbone enzymes in A. flavus.
Keywords: aflatoxin B1, Aspergillus flavus, oxidative stress, transcriptome, gene regulation, eugenol
Introduction
Aspergillus flavus is a saprotrophic filamentous fungus that occurs widely in agricultural and medical products (Liang et al., 2015). It contaminates many important agro-products including peanut, maize, rice, cottonseed, sunflower seed, herbal material, and feeds (Cleveland et al., 2004; Liang et al., 2015). A. flavus produces many kinds of secondary metabolites including aflatoxin, cyclopiazonic acid, conidial pigment, aflatrem and kojic acid (Bennett and Klich, 2003; Hoffmeister and Keller, 2007). Of them, aflatoxin is the most toxic and hepatocarcinogenic compound (Squire, 1981). As a carcinogen, aflatoxin is estimated to cause up to 28% of the total global cases of hepatocellular carcinoma, the most common form of liver cancer (Wu, 2014; Xing et al., 2017). Moreover, aflatoxin leads to acute intoxication, immune-system disruption and growth impairment in children (Groopman et al., 2008).
Eugenol (4-allyl-2-methoxy phenol), a natural substance used as a food- flavoring agent, was first isolated in 1929 and its commercial production began in 1940 in the United States (da Silva et al., 2018). It is mainly extracted from Syzygium aromaticum, Ocimum tenuiflorum, Pimenta racemosa, Zieria smithii, and Cassia fistula although it can be produced synthetically (Jayashree and Subramanyam, 1999). As an allyl-phenol-type phenylpropanoid, eugenol is a pale yellow oil with clove odor and spicy taste (da Silva et al., 2018). Eugenol can be oxidized in a non-enzymatic manner, such as light via a one-electron pathway to a phenoxyl radical (ArO·), and subsequently to eugenol quinonemthide (QM) by light (Satoh et al., 1998; Choi et al., 2009). It is generally regarded as safe by the Food and Agricultural Organization (Opdyke, 1975), with an acceptable daily intake of up to 2.5 mg/kg body weight in humans (FAO, 1982) based on its non-mutagenic and non-carcinogenic properties [International Agency for Research on Cancer (IARC), 1985]. Eugenol is widely used in the pharmaceutical, food, agricultural and cosmetics industries because it exerts useful antimicrobial and antioxidant effects (da Silva et al., 2018). It also has other biological properties, including antiviral, anti-inflammatory, and anti-cancer effects, and inhibits platelet aggregation. Eugenol has previously been used as an AFB1 inhibitor.
Karapinar (1990) reported that the growth of A. parasiticus NRRL 2999 and A. parasiticus CBS 26027 was inhibited by eugenol at a concentration of 300 μg/mL (~1.83 mmol/L), and the production of aflatoxin particularly by A. parasiticus NRRL 2999 was enhanced by eugenol below 200 μg/mL (~1.22 mmol/L). Jayashree and Subramanyam (1999) found that aflatoxin production in A. parasiticus was inhibited by eugenol in a dose-dependent manner up to a concentration of 0.75 mmol/L without inhibiting fungal growth. They suggested that the anti-aflatoxigenic actions of eugenol were attributable to the inhibition of the ternary steps of aflatoxin biosynthesis, which involve lipid peroxidation and oxygenation. Nam and Kim (2013) demonstrated that eugenol inhibited aflatoxin biosynthesis through disrupting lipid peroxidation by reducing the microsomal activities of cytochorome P450, poly substrate monooxygenase (PSMO), and NADPH-dependent cytochorome C reductase. Using quantitative real-time PCR (q-PCR), Jahanshiri et al. (2015) indicated that eugenol strongly inhibited AFB1 production in A. parssiticus in the range of 15.07–98.0% in a dose-dependent manner. The expressions of major pathway genes such as ver-1 (aflM), nor-1 (aflD), pksA (aflC), omtA (aflP), and aflR were significantly suppressed by eugenol at concentrations of 62.5 and 125 μg/mL (~0.76 mmol/L). In the meantime, Liang et al. (2015) also showed that eugenol (0.8 mM) inhibited AFB1 biosynthesis in A. flavus in Yeast Extract Sucrose (YES) broth by down-regulating the transcript levels of some key biosynthetic genes such as aflP, aflM, and aflD. Using a large-scale qPCR approach, Caceres et al. (2016) found that AFB1 inhibition by eugenol addition at 0.5 mM in a Malt Extract Agar (MEA) medium resulted in a complete inhibition of all but one gene of the AFB1 biosynthesis cluster. This phenomenon was modulated by the down-regulation of aflR and aflS expression and the over-expression of veA and mtfA, which are directly involved in regulating AFB1 cluster. However, the detailed molecular mechanism by which eugenol represses aflatoxin biosynthesis is still largely unknown.
RNA sequencing (RNA-Seq), a high-throughput sequencing technology with a low false-positive rate and high sensitivity, is widely considered a revolutionary tool for transcriptomics studies and has been used to investigate multiple eukaryotic transcriptomes (Wilhelm et al., 2008; Wang et al., 2009; Lin et al., 2013). Compared with the de novo assembly approach, a reference-based method is more accurate and sensitive. The A. flavus genome is a well-annotated genome available in NCBI with accession number AAIH00000000 (http://www.ncbi.nlm.nih.gov/nuccore/AAIH00000000). Transcriptome profiling of A. flavus has been used to investigate the effect of temperature and water activity (aw) on fungal development and aflatoxin biosynthesis (Yu et al., 2011; Zhang et al., 2014; Bai et al., 2015). Lin et al. (2013) profiled transcriptome of A. flavus to explore the inhibitory mechanism of 5-Azacytidine (5-AC) on fungal development and aflatoxin biosynthesis. They found that 5-AC affects fungal development through increasing the expression of brlA by depressing the expression of veA and affects aflatoxin production by suppressing veA expression (Lin et al., 2013).
To reduce aflatoxin contamination in foods, a number of strategies have been developed to either prevent fungal growth or block toxin production (Amaike and Keller, 2011). During planting, atoxigenic biocompetitive A. flavus and/or A. parasiticus strains or yeast are used to prevent fungal infection (Chang et al., 2012). During storage, chemicals, drying, natural products, and microorganisms have been applied to prevent fungal growth and aflatoxin production (Liang et al., 2015; Xing et al., 2017). Currently, chemical-based control remains the common measure used to control post-harvest aflatoxins contamination in a variety of foods. However, the application of chemicals not only increases the risk of toxic residues in foods but also often leads to fungal resistance (Isaac et al., 1999; Hua H. et al., 2014; Hua S. S. et al., 2014). Therefore, much effort has been directed in the recent years toward limiting the use of chemical fungicides in grains and foods. Essential oils from plants, such as phenolic and aldehydic compounds, acetate esters, and alcohols, provide an attractive alternative to inhibit fungal growth and aflatoxins formation because they efficiently eliminate aflatoxin, maintain food quality, are natural resources and are highly volatile (Wright et al., 2000; Bluma and Etcheverry, 2008; Roze et al., 2011).
In this study, the anti-aflatoxigenic mechanism of eugenol was determined with an RNA-Seq approach. A comprehensive view of the A. flavus transcriptome and the differentially expressed genes between eugenol treated and untreated samples were obtained. This study may extend our understanding of the inhibitory pathway of eugenol on aflatoxin biosynthesis and fungal development at the transcriptome level.
Materials and methods
Natural compound, fungal strain, and culture conditions
Natural eugenol (99% purity) extracted from clove buds was purchased from Xue-Song Company (Jiangxi, China) and dissolved in ethanol. The stock solution was stored at 4°C until use. The A. flavus strain YC-15 (Table S1) used in this study (Liang et al., 2015) was maintained in the dark on PDA medium (200 g boiled potato, 20 g dextrose, 20 g agar, 1 L) at 4°C. The conidia from a 7-day-old PDA culture were washed with 0.01% Tween-20 solution and counted with a hemocytometer. A suspension of 5 × 107 conidia/mL was prepared. YES broth (20 g yeast extract, 150 g sucrose, 0.5 g MgSO4·7H2O, 1 L) was inoculated with the conidia at a final concentration of 106 conidia/mL. The eugenol stock solution was diluted with ethanol to a concentration of 80 mM, and 500 μL of the diluted eugenol stock was added to 50 mL of YES broth, producing a final eugenol concentration of 0.80 mM. The control cultures were treated similarly but without eugenol. Each culture was incubated at 28°C in the dark for 5 days. The mycelia of A. flavus were then harvested. Each treatment was performed in triplicate.
Preparation of cDNA and illumina sequencing
The cDNA preparation and Illumina sequencing were performed according to Zhang et al. (2014) with some modifications. A. flavus mycelia were harvested from YES broth for isolation of RNA. Total RNA was isolated using a Fungal RNA Kit (Omega, Norcross, USA) and genomic DNA was digested using RNA-free DNase I (Thermo Fisher Scientific, CA, USA). An Agilent 2100 Bioanalyzer and Nano Drop 2000 spectrophotometer were used to evaluate the integrity and concentration of RNA. The mRNA was isolated using oligo (dT)-attached magnetic beads. The isolated mRNA was mixed with fragmentation buffer and cleaved into small fragments (380 ± 50 bp) using divalent cations under elevated temperatures. The cDNA was synthesized using these cleaved RNA fragments as templates. After purification, these short cDNA fragments were subjected to an end-repair process with the addition of a single “A” base, and were then ligated to sequencing adaptors using the Illumina TruSeq DNA sample preparation kit. PCR amplification was performed using the qualified fragments as templates. Lastly, the libraries were sequenced using an Illumina HiSeq 4000 system.
Clean reads and normalized gene expression levels
The reads were filtered according to Zhang et al. (2014) with minor modifications. The raw reads were filtered by removing read adaptors, artificial reads, and other low quality reads. After filtering, the clean reads were obtained and used for the subsequent analysis. The clean reads were mapped to the reference genome and assembled according to Lin et al. (2013) with some modifications. Using the programs TopHat 1.31 and Bowtie, the clean reads were mapped to the A. flavus genome, the EST sequencing and rRNA sequencing (Yu et al., 2004; Langmead et al., 2009; Trapnell et al., 2009). Using program Cufflinks, transcripts were assembled (Trapnell et al., 2010). The FPKM (Fragments Per Kb of exon per Million reads) method was used to calculate and normalize the expression levels of genes (Mortazavi et al., 2008).
Identification and analysis of differentially expressed genes
The differentially expressed genes were identified and analyzed according to Lin et al. (2013) with minor modifications. The normalized gene expression levels in A. flavus treated with eugenol and untreated sample were directly compared. The p-value was then used to identify the differentially expressed genes. FDR (False Discovery Rate) control was used to correct p-value and FDR ≤ 0.05 was chosen. Finally, GO functional enrichment analysis and KEGG pathway enrichment analysis were performed using the FungiFun program (Kanehisa et al., 2008; Priebe et al., 2011).
Reverse transcription (RT)-PCR conditions and q-PCR analysis of aflatoxin biosynthesis genes
First-strand cDNA was obtained by RT-PCR using the Takara RNA Kit (AMV) ver. 3.0 (Takara Bio Inc., Japan) according to the manufacturer's instructions. All PCR primers were designed based on the A. flavus NRRL 3357 genomic sequence (GenBank accession number EQ963478A). Primers pair sequences of 18S, aflR, aflS (aflJ), aflA (fas-2), aflC (pksA), aflD (nor-1), aflE (norA), aflF (norB), aflG (avnA), aflH (adhA), aflI (avfA), aflJ (estA), aflK (vbs), aflL (verB), aflM (ver-1), aflN (verA), aflO (omtB), aflP (omtA), aflQ (ordA), aflU (cypA), aflX (ordB), and aflT were adapted from Liu et al. (2017) and the primer sequences of aflB (fas-1), aflCa (hypC), aflLa (hypB), aflMa (hypE), aflNa (hypD), aflV (cypX), aflW (moxY), and aflY (hypA) were adapted from Caceres et al. (2016). All 29 genes encoding aflatoxin biosynthesis were analyzed. Real-time PCR was performed on an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). MicroAmp optical 96-well plates were prepared for PCR with each well containing a total volume of 20 μL: 10 μL of SYBR Green Real-time PCR Master Mix (Applied Biosystems) used as the fluorescent dye, 2 μL of cDNA and 1 μL of each primer. The q-PCR steps were performed as previously described by Liu et al. (2017).
Availability of RNA-seq data
The raw RNA-seq data of A. flavus discussed in this work have been deposited in the NCBI Sequence Read Archive under the accession number of SRP132641.
Results
RNA-seq data
RNA sequencing of eugenol treated and untreated A. flavus YC-15 generated a total of 16.99 Gb of valid data and 64.65 million read pairs (the average length is 150 bp). Of these, 53.87 million passed purity filtering standards, of which approximately 29.63 million (55.00%) were uniquely mapped to the genome of A. flavus. Among all the 107.73 million reads, 64.21 million (59.60%) were mapped to the A. flavus genome and only 0.02% of reads were aligned to rRNA genes. The overall transcription levels of the genes were quantified with FPKM values. The results showed that 11,941 (88.55%) of the 13,485 gene models in the A. flavus genome database were expressed at least once in one of the six samples. In the control and eugenol-treated groups, 10,932 (81.07%) and 10,826 (80.28%) genes were expressed, respectively.
Identification and functional analysis of differentially expressed genes
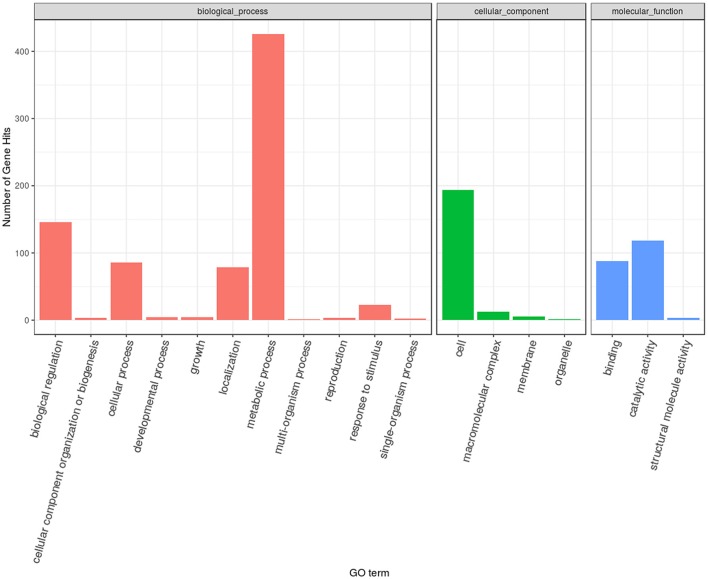
Based on the FPKM values, 735 differentially expressed genes were identified (with FDR ≤ 0.05, log2Ratio ≥ 1 or ≤1) between the eugenol and control groups. Among these, 271 gene displayed up-regulation and 464 genes displayed down-regulation after eugenol exposure. These differentially expressed genes were subjected to GO functional enrichment analysis. The results showed that these genes were mainly involved in RNA binding, hydrolase activity, pyrophosphatase activity, nucleoside-triphosphatase activity, structural molecular activity, transferase activity, methyltransferase activity, helicase activity, or macromolecular complex binding, or were structural constituents of the ribosome (Table 1, Figure 1). KEGG metabolic pathway enrichment analysis indicated that these genes were mainly involved in ribosome biogenesis, the ribosome, RNA transport, pyrimidine metabolism, and RNA polymerase (Table 2).
Table 1.
GO functional enrichment analysis of differentially expressed genes when A. flavus was treated with eugenol.
| GO ID | TERM (Molecular function) | p-Value | q-Value | List hits | List size |
|---|---|---|---|---|---|
| GO:0003723 | RNA binding | 2.56E-22 | 6.86E-20 | 99 | 800 |
| GO:0016818 | Hydrolase activity, acting on acid anhydrides | 3.54E-03 | 3.38E-02 | 61 | 800 |
| GO:0016462 | Pyrophosphatase activity | 4.87E-03 | 4.04E-02 | 60 | 800 |
| GO:0017111 | Nucleoside-triphosphatase activity | 2.99E-03 | 2.96E-02 | 59 | 800 |
| GO:0005198 | Structural molecule activity | 2.08E-03 | 2.42E-02 | 38 | 800 |
| GO:0016741 | Transferase activity, transferring one-carbon | 2.75E-05 | 5.25E-04 | 36 | 800 |
| GO:0008168 | Methyltransferase activity | 2.35E-05 | 4.83E-04 | 34 | 800 |
| GO:0003735 | Structural constituent of ribosome | 1.78E-05 | 3.97E-04 | 33 | 800 |
| GO:0004386 | Helicase activity | 7.41E-10 | 6.60E-08 | 29 | 800 |
| GO:0044877 | Macromolecular complex binding | 4.76E-03 | 4.04E-02 | 25 | 800 |
Figure 1.
The gene ontology annotation of differential expression genes.
Table 2.
KEGG metabolic pathway enrichment analysis of differentially expressed genes when A. flavus was treated with eugenol.
| ID | TERM (Molecular function) | p-Value | q-Value | List hits | List size |
|---|---|---|---|---|---|
| Afv03008 | Ribosome biogenesis in eukaryotes | 1.63E-16 | 1.39E-14 | 36 | 313 |
| Afv03010 | Ribosome | 7.74E-09 | 3.30E-07 | 26 | 313 |
| Afv03013 | RNA transport | 1.44E-03 | 2.45E-02 | 23 | 313 |
| Afv00240 | Pyrimidine metabolism | 9.68E-04 | 2.06E-02 | 17 | 313 |
| Afv03020 | RNA polymerase | 1.86E-04 | 5.30E-03 | 11 | 313 |
Analysis of gene expression in the pigment, aflatrem, aflatoxin, and cyclopiazonic acid secondary metabolite pathways in A. flavus treated with eugenol
Using the SMURF program and the website of the Center for Integrated Fungal Research (Lin et al., 2013), 55 secondary metabolite pathways of A. flavus were identified and analyzed. The transcription levels in most of these pathways were not significantly affected by eugenol. The transcription levels of the genes involved in the biosynthesis of conidial pigment (#10), aflatrem (#15), aflatoxin (#54), and cyclopiazonic acid (#55) are showed in Table 3. In pathway #10, AFLA_016120 encoding an O-methyltransferase family protein and AFLA_016130 were down-regulated. In pathway #15, most of cluster genes were expressed at a very low level. In pathway #55, AFLA_139490 encoding a hybrid PKS/NRPS enzyme and AFLA_139470 encoding a FAD dependent oxidoreductase were up-regulated, whereas AFLA_139460 encoding a MFS multidrug transporter was down-regulated. In previous studies, we found that aflatoxin biosynthesis was repressed in A. flavus treated with eugenol (Liang et al., 2015). However, in the present study, most genes in pathway #54 were expressed at high levels with only slight changes after eugenol exposure. Of 29 genes in the aflatoxin biosynthetic cluster, 19 genes' transcription were down-regulated to varying degrees, including the key structural genes aflI, aflJ, aflH, aflE, aflG, aflM, aflD, aflP, and afL (Table 3). Most surprising of all, the expression of the regulator gene aflR did not change significantly and the expression of aflS was slightly up-regulated (Table 3).
Table 3.
Transcriptional activity of genes in the biosynthesis of conidial pigment (#10), aflatrem (#15), aflatoxin (#54), and cyclopiazonic acid (#55).
| Cluster ID | Gene ID (AFLA_x) | Untreated (FPKM) | D125 (FPKM) | Log | Annotated_gene_function |
|---|---|---|---|---|---|
| #10 | 016120 | 10.58 | 3.02 | −1.81 | O-methyltransferase family protein |
| #10 | 016130 | 13.25 | 6.45 | −1.04 | Hypothetical protein |
| #10 | 016140 | 14.10 | 17.20 | 0.29 | Conidial pigment biosynthesis scytalone dehydratase Arp1 |
| #15 | 045450 | 37.27 | 64.22 | 0.79 | Ankyrin repeat-containing protein, putative |
| #15 | 045460 | 1.16 | 5.26 | 2.18 | Hypothetical protein |
| #15 | 045470 | 0.10 | 0 | / | Nonsense-mediated mRNA decay protein, putative |
| #15 | 045480 | 0.32 | 1.27 | 2.00 | Conserved hypothetical protein |
| #15 | 045490 | 0.03 | 0.16 | 2.18 | Dimethylallyl tryptophan synthase, putative |
| #15 | 045500 | 0.55 | 0.57 | 0.05 | Cytochrome P450, putative |
| #15 | 045510 | 0.12 | 0 | / | Integral membrane protein |
| #15 | 045520 | 0 | 0 | / | Integral membrane protein |
| #15 | 045530 | 0.23 | 0 | / | Conserved hypothetical protein |
| #15 | 045540 | 0 | 0.06 | / | Cytochrome P450, putative |
| #15 | 045550 | 1.24 | 1.12 | −0.14 | Hypothetical protein AFLA_045550 |
| #15 | 045560 | 1.94 | 1.89 | −0.04 | Carboxylic acid transport protein |
| #15 | 045570 | 1.55 | 4.59 | 1.57 | Acetyl xylan esterase, putative |
| #54 | 139100 | 2.96 | 2.43 | −0.28 | aflYe/orf/Ser -Thr protein phosphatase family protein |
| #54 | 139110 | 2.38 | 2.74 | 0.20 | aflYd/sugR/sugar regulator |
| #54 | 139120 | 1.85 | 1.93 | 0.06 | aflYc/glcA/glucosidase |
| #54 | 139130 | 1.78 | 2.49 | 0.49 | aflYb/hxtA/putative hexose transporter |
| #54 | 139140 | 5.53 | 7.09 | 0.36 | aflYa/nadA/NADH oxidase |
| #54 | 139150 | 101.03 | 92.37 | −0.13 | aflY/hypA/hypP/hypothetical protein |
| #54 | 139160 | 117.97 | 96.36 | −0.29 | AflX/ordB/monooxygenase |
| #54 | 139170 | 49.07 | 43.95 | −0.16 | aflW/moxY/monooxygenase/oxidase |
| #54 | 139180 | 48.91 | 53.16 | 0.12 | aflV/cypX/cytochrome P450 monooxygenase |
| #54 | 139190 | 112.43 | 140.97 | 0.33 | aflK/vbs/VERB synthase |
| #54 | 139200 | 12.91 | 14.38 | 0.16 | aflQ/ordA/ord-1/oxidoreductase/cytochrome P450 monooxigenase |
| #54 | 139210 | 92.70 | 72.78 | −0.35 | aflP/omtA/omt-1/O-methyltransferase A |
| #54 | 139220 | 187.22 | 164.37 | −0.19 | aflO/omtB/dmtA/O-methyltransferase B |
| #54 | 139230 | 15.55 | 9.20 | −0.76 | aflI/avfA/cytochrome P450 monooxygenase |
| #54 | 139240 | 108.16 | 87.23 | −0.31 | aflLa/hypB/hypothetical protein |
| #54 | 139250 | 92.87 | 75.90 | −0.29 | aflL/verB/desaturase/P450 monooxygenase |
| #54 | 139260 | 48.69 | 35.41 | −0.46 | aflG/avnA/ord-1/cytochrome P450 monooxygenase |
| #54 | 139270 | 572.31 | 386.33 | −0.57 | aflNa/hypD/hypothetical protein |
| #54 | 139280 | 34.04 | 34.80 | 0.03 | aflN/verA/monooxygenase |
| #54 | 139290 | 136.29 | 75.22 | −0.86 | aflMa/hypE/hypothetical protein |
| #54 | 139300 | 496.53 | 375.07 | −0.40 | aflM/ver-1/dehydrogenase/ketoreductase |
| #54 | 139310 | 180.51 | 122.95 | −0.55 | aflE/norA/aad/adh-2/NOR reductase |
| #54 | 139320 | 132.91 | 78.55 | −0.76 | aflJ/estA/esterase |
| #54 | 139330 | 192.79 | 128.47 | −0.59 | aflH/adhA/short chain alcohol dehydrogenase |
| #54 | 139340 | 177.63 | 200.51 | 0.17 | aflS/pathway regulator |
| #54 | 139360 | 64.90 | 69.27 | 0.09 | aflR/apa-2/afl-2/transcription activator |
| #54 | 139370 | 35.31 | 38.25 | 0.12 | aflB/fas-1/fatty acid synthase beta subunit |
| #54 | 139380 | 19.45 | 17.79 | −0.13 | aflA/fas-2/hexA/fatty acid synthase alpha subunit |
| #54 | 139390 | 231.56 | 180.60 | −0.36 | aflD/nor-1/reductase |
| #54 | 139400 | 84.14 | 51.61 | −0.71 | aflCa/hypC/hypothetical protein |
| #54 | 139410 | 37.55 | 38.39 | 0.03 | aflC/pksA/pksL1/polyketide synthase |
| #54 | 139420 | 100.86 | 99.84 | −0.01 | aflT/aflT/transmembrane protein |
| #54 | 139430 | 20.76 | 26.04 | 0.33 | aflU/cypa/P450 monooxygenase |
| #54 | 139440 | 14.48 | 15.48 | 0.10 | aflF/norB/dehydrogenase |
| #55 | 139460 | 1293.63 | 1017.37 | −0.35 | MFS multidrug transporter, putative |
| #55 | 139470 | 215.54 | 296.72 | 0.46 | FAD dependent oxidoreductase, putative |
| #55 | 139480 | 243.62 | 278.43 | 0.19 | tryptophan dimethylallyltransferase |
| #55 | 139490 | 9.14 | 17.52 | 0.94 | Hybrid PKS/NRPS enzyme, putative |
Confirmation analysis of gene expression involved in aflatoxin biosynthesis
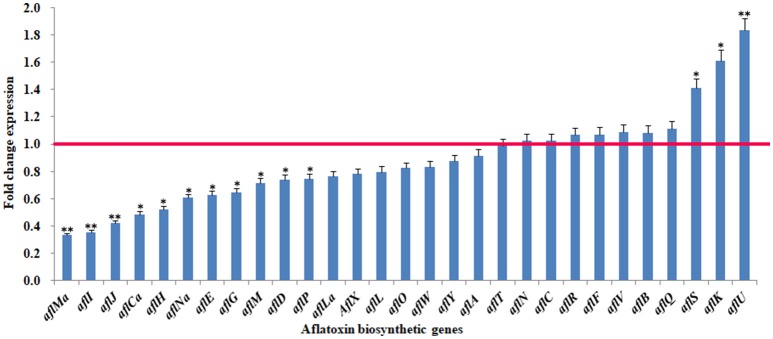
To confirm the observed changes in aflatoxin biosynthetic gene expression, two regulator genes (aflS and aflR) and all the structural genes (aflI, aflJ, aflH, aflE, etc.) were further analyzed by quantitative real-time PCR (q-PCR). The results showed that the transcription of 19 genes was down-regulated to varying degrees after eugenol exposure (Figure 2), which was consistent with the RNA-seq data. For example, aflMa was the most strongly down-regulated gene in A. flavus treated with eugenol in both the RNA-Seq and q-PCR analyses.
Figure 2.
Fold change expression of genes belonging to the cluster responsible for aflatoxin biosynthesis in response to eugenol at 0.80 mM. Red line represents control expression level. *p-value < 0.05; **p-value < 0.01.
Analysis of fungal development-related gene expression in A. flavus treated with eugenol
From the gene expression pattern data, we found that the expression levels of some genes involved in conidiophore development were dysregulated after exposure to eugenol (Table 4). The veA gene (AFLA_066460), encoding a global regulator, was down-regulated with its FPKM value decreasing from 312.87 to 204.64. The transcription of the conidia-specific hydrophobin gene RodA (AFLA_098380) was down-regulated with its FPKM decreasing from 12.35 to 5.42. However, the transcription of the conidial hydrophobin gene RodB (AFLA_014260) was up-regulated with its FPKM increasing from 2.51 to 17.48. The transcription of the C2H2 type conidiation transcription factor BrlA (AFLA_082850) was up-regulated with it FPKM increasing from 2.00 to 7.08. In addition, the transcription levels of the transcription factor AbaA (AFLA_029620), the development regulator FlbA (AFLA_134030), the APSES transcription factor StuA (AFLA_046990), and the transcription factor Medusa (AFLA_136410) showed a mild up-regulation.
Table 4.
Transcriptional activity of genes involved in A. flavus development.
| Gene ID (AFLA_x) | Untreated (FPKM) | D125 (FPKM) | Log | Annotated_gene_function |
|---|---|---|---|---|
| 066460 | 312.87 | 204.64 | −0.61 | Developmental regulator AflYf/VeA |
| 033290 | 31.74 | 29.12 | −0.12 | Regulator of secondary metabolism LaeA |
| 014260 | 2.51 | 17.48 | 2.80 | Conidial hydrophobin RodB/HypB |
| 098380 | 12.35 | 5.42 | −1.19 | Conidial hydrophobin RodA/RolA |
| 081490 | 36.78 | 23.90 | −0.62 | Nucleoside diphosphatase Gda1 |
| 018340 | 46.12 | 54.38 | 0.21 | G-protein complex alpha subunit GpaA/FadA |
| 046990 | 148.93 | 234.07 | 0.65 | APSES transcription factor StuA |
| 136410 | 89.17 | 135.16 | 0.60 | Transcriptional regulator Medusa |
| 029620 | 1.02 | 1.69 | 0.73 | Transcription factor AbaA |
| 082850 | 2.00 | 7.08 | 1.82 | C2H2 type conidiation transcription factor BrlA |
| 039530 | 9.85 | 11.78 | 0.26 | FluG family protein |
| 071090 | 893.92 | 758.74 | −0.24 | GPT-binding protein EsdC |
| 101920 | 2.17 | 3.35 | 0.63 | Extracellular developmental signal biosynthesis protein FluG |
| 020210 | 86.15 | 85.76 | −0.01 | Sexual development transcription factor NsdD |
| 026900 | 10.56 | 12.68 | 0.26 | Developmental regulator VosA |
| 052030 | 7.22 | 8.03 | 0.15 | Developmental regulatory protein WetA |
| 131490 | 37.64 | 41.06 | 0.13 | Conserved hypothetical protein |
| 134030 | 10.32 | 16.26 | 0.66 | Development regulator FlbA |
| 137320 | 104.86 | 92.80 | −0.18 | C2H2 conidiation transcription factor FlbC |
Analysis of the expression of genes involved in fungal oxidative stress in A. flavus treated with eugenol
The transcription levels of oxidative stress-related genes are shown in Table 5. Among the 47 relevant genes, 23 genes were significantly modulated by eugenol. The expression of dioxygenase-encoding ppoC, the transcriptions factor genes msnA, srrA and pacC, the cellular receptor gprC, gprF, gprK, gprM, and gprS, the MAP kinase genes bck1, ste11, and sskB and the superoxide dismutase gene sod1 were up-regulated. Conversely, the addition of eugenol induced the down-regulation of 10 genes encoding for (i) oxylipins ppoB (ii) GPCRs (gprA, gprD, gprG, gprO, and gprB) (iii) protein kinase (sakA and maf1) (iiii) catalase cat2 (iiiii) dehydrogenase gfdB.
Table 5.
Transcriptional activity of MAPK pathway, Oxylipins, and GPCRs genes in A. flavus.
| Gene ID (AFLA_x) | Gene | Untreated (FPKM) | D125 (FPKM) | Log | Annotated_gene_function |
|---|---|---|---|---|---|
| 062500 | Maf1 | 66.38 | 51.64 | −0.36 | Mitogen-activated protein kinase |
| 083380 | Pbs2 | 35.91 | 39.53 | 0.14 | MAP kinase kinase |
| 103480 | Ste7 | 9.60 | 11.75 | 0.29 | MAP kinase kinase |
| 048880 | Ste11 | 13.04 | 16.99 | 0.38 | MAP kinase kinase kinase |
| 035530 | Ste20 | 52.79 | 52.11 | −0.02 | Serine/threonine kinase |
| 021030 | / | 25.45 | 23.47 | −0.12 | Serine/threonine protein kinase |
| 052570 | mpkA | 44.75 | 43.77 | −0.03 | MAP kinase |
| 051240 | Mkk2 | 100.19 | 108.85 | 0.12 | MAP kinase kinase |
| 034170 | Fus3 | 85.07 | 89.43 | 0.07 | MAP kinase |
| 031560 | bck1 | 12.93 | 19.95 | 0.63 | MAP kinase kinase kinase |
| 100250 | cat | 0.40 | 0.38 | −0.07 | Catalase Cat |
| 090690 | Cat1 | 180.19 | 167.61 | −0.10 | Mycelial catalase |
| 122110 | Cat2 | 20.23 | 12.20 | −0.73 | Bifunctional catalase-peroxidase |
| 056170 | catA | 612.92 | 648.71 | 0.08 | Spore-specific catalase |
| 099000 | sod1 | 98.10 | 125.45 | 0.35 | Cu, Zn superoxide dismutase SOD1 |
| 033420 | mnSOD | 1175.47 | 1008.01 | −0.22 | Mn superoxide dismutase |
| 031340 | atfA | 227.96 | 214.80 | −0.09 | bZIP transcription factor |
| 094010 | atfB | 212.65 | 230.11 | 0.11 | bZIP transcription factor |
| 129340 | ap-1 | 158.11 | 135.16 | −0.23 | bZIP transcription factor AP-1 |
| 110650 | msnA | 80.07 | 188.50 | 1.24 | C2H2 transcription factor |
| 091490 | mtfA | 34.54 | 28.89 | −0.26 | C2H2 finger domain protein |
| 030580 | pacC | 129.52 | 165.07 | 0.35 | C2H2 transcription factor |
| 034540 | srrA | 45.97 | 72.01 | 0.65 | Stress response transcription factor |
| 062210 | sskA | 22.66 | 23.11 | 0.03 | Response regulator |
| 068590 | sskB | 14.76 | 18.52 | 0.33 | MAP kinase kinase kinase |
| 061090 | sakA | 2.05 | 1.23 | −0.74 | MAP kinase |
| 026790 | ppoA | 22.17 | 26.58 | 0.26 | Fatty acid oxygenase |
| 120760 | ppoB | 0.49 | 0.03 | −4.03 | Fatty acid oxygenase |
| 030430 | ppoC | 1.04 | 5.19 | 2.32 | Fatty acid oxygenase |
| 002850 | AfPXG | 395.51 | 335.03 | −0.24 | Calcium binding protein Caleosin/Peroxygenase |
| 025100 | gpdA | 7357.45 | 7758.80 | 0.08 | Glyceraldehyde 3-phosphate dehydrogenase |
| 046760 | gfdB | 320.94 | 253.48 | −0.34 | Glycerol 3-phosphate dehydrogenase |
| 060740 | gprA | 11.82 | 5.36 | −1.14 | STE3 GPCR (S. cerevisiae pheromone receptor) |
| 061620 | gprB | 7.64 | 6.13 | −0.32 | STE3 GPCR (S. cerevisiae pheromone receptor) |
| 074150 | gprC | 2.56 | 8.76 | 1.77 | Git3; Git3_C (S. pombe glucose sensor) |
| 135680 | gprD | 10.67 | 5.34 | −1.00 | Git3; Git3_C (S. pombe glucose sensor) |
| 006880 | gprF | 45.33 | 68.78 | 0.60 | PQ loop repeat (S. pombe nitrogen sensor) |
| 067770 | gprG | 40.30 | 22.59 | −0.84 | PQ loop repeat (S. pombe nitrogen sensor) |
| 006920 | gprH | 0.78 | 0.72 | −0.12 | Secretin family (signal through cAMP pathways) |
| 127870 | gprJ | 50.86 | 61.23 | 0.27 | Vacuolar membrane PQ loop repeat protein |
| 009790 | gprK | 0.25 | 0.42 | 0.75 | RGS domain (regulator of G protein signaling) |
| 075000 | gprM | 3.33 | 5.43 | 0.71 | Conserved hypothetical protein |
| 032130 | gprO | 40.06 | 27.13 | −0.56 | Hemolysin III related (broad range of ligands) |
| 088190 | gprP | 33.21 | 31.68 | −0.07 | Hemolysin III related (broad range of ligands) |
| 023070 | gprR | 32.65 | 30.65 | −0.09 | RGS domain (regulator of G protein signaling) |
| 006320 | gprS | 12.95 | 16.26 | 0.33 | PQ loop repeat protein |
| 117970 | nopA | 5388.07 | 4310.21 | −0.32 | Bacteriorhodopsin-like (photoreactive) |
Discussion
To guarantee the health of human beings and food safety, chemical fungicides have been gradually limited in the food chain. In recent years, a number of essential oils from plants have been widely used in food industries as alternatives to chemical fungicides. The inhibitory effects of essential oils, such as citral, cinnamon, clove, litsea, eucalyptus, ginger, anise, spearmint, and camphor oils on fungal growth and toxin production have been reported by many researchers (Velluti et al., 2003; Liang et al., 2015). In our earlier study, of these natural plant-based compounds, 0.80 mM eugenol significantly reduced AFB1 production with inhibitory rate 95.4%, but with no effect on fungal growth (Liang et al., 2015). In this study, the mechanism by which eugenol dysregulates A. flavus growth and aflatoxin production was studied using an RNA-seq analysis.
The expression of 19 of 29 genes in the aflatoxin biosynthetic pathway cluster was down-regulated when A. flavus was treated with eugenol. However, the expression of none of these genes was completely inhibited. The most strongly down-regulated gene was aflMa, followed by aflI, aflJ, aflCa, aflH, aflNa, aflE, aflG, aflM, aflD, aflP, and aflLa. These observed changes in aflatoxin biosynthetic gene transcript levels were confirmed with q-PCR. AflD (nor-1) and aflE (norA) both encode reductases that are involved in the conversion of NOR (norsolorinic acid) to AVN (averantin). AflG (avnA), encoding a cytochrome P450 monooxygenase, converts AVN to HAVN (5′-hydroxy-averantin). AflH (adhA) encodes an alcohol dehydrogenase which is involved in the conversion of HAVN to AVF (averufin). AflI (avfA) encodes an oxidase that converts AVF to VHA (versiconal hemiacetal acetate). AflJ (estA) encodes an esterase that is necessary for the conversion of VHA to VAL (versiconal) (Yu et al., 2004; Cleveland et al., 2009). Our results indicated that the pathway from NOR to VAL was repressed in A. flavus treated with eugenol. AflL (verB) encodes a P450 monooxygenase/desaturase which converts VERB (versicolorin B) to VERA (versicolorin A). AflM (ver-1) encodes a dehydrogenase that can convert VERA to DMST (demethylsterigmatocystin). AflO (omtB) encodes an O-methyltransferase and converts DMST to ST (sterigmatocystin). AflP (omtA) encodes another O-methyltransferase and converts ST to OMST (O-methylsterigmatocystin) (Yu et al., 2004). In the present study, the transcription of aflL, aflM, aflO, and aflP was all down-regulated by eugenol, suggesting that the aflatoxin biosynthetic pathway from VERB to OMST was also repressed in A. flavus treated with eugenol. We obtained similar results in a previous study with q-PCR, and found that the transcription of aflP, aflM, and aflD was also down-regulated by 0.80 mM eugenol (Liang et al., 2015). AflMa (hypE) encodes an enzyme (HypE) which, together with AflE, may be involved in the final two steps in aflatoxin biosynthesis. AflCa (hypC) encodes an oxidase which catalyzes the oxidation of norsolorinic acid anthrone. AflNa (hypD) encodes an integral membrane protein that inhibits A. flavus growth and aflatoxin biosynthesis. AflLa (hypB) encodes an oxidase that is assumed to be involved in one of the oxidation steps in the conversion of OMST to aflatoxin (Wei et al., 2014). These results suggested that eugenol inhibited aflatoxin biosynthesis in A. flavus treated with eugenol by down-regulating the expression of several structural genes.
Most surprising of all, the transcription regulator gene aflR in the cluster did not show significant differential expression after treatment with eugenol, while the transcription regulator gene aflS showed a slight up-regulation (Table 3). The results are similar with the findings of Lin et al. (2013). They found that the transcription regulator genes aflR and aflS showed no significant differential expression after treatment with 5-Azacytidine (5-Ac), an inhibitor of aflatoxin production and development in A. flavus. However, the expressions of aflQ, aflI, and aflLa were totally or almost totally inhibited by 5-Ac. Aflatoxins are produced optimally at 28–30°C and production significantly decreases as temperature approach 37°C, the optimum temperature for fungal growth. OBrian et al. (2007) found that transcript levels of aflR and aflS did not change significantly between 28 and 37°C, while all the structural genes were much lowly expressed at 37°C relative to 28°C. A. flavus exhibits decreased conidiation and aflatoxin biosynthesis under water activity (aw) 0.93 compared to that under 0.99 aw. Zhang et al. (2014) found that transcript levels of aflR and aflS both showed no significant differential expression between two water activities using RNA-seq approach, while the expression of 16 aflatoxin producing-related genes decreased obviously when aw decreased. There are five potential explanations for the decreased transcription of several aflatoxin biosynthesis structural genes while the aflR did not change significantly and aflS was slightly up-regulated after treatment with eugenol: (a) some other transcription regulators may be involved in the down-regulating of these structural genes; (b) post-transcriptional regulation influences the expression of these structural genes; (c) less AFLR is produced after treatment with eugenol and translation process may be involved in the modulation of these structural genes; (d) AFLR is nonfunctional with eugenol exposure; (e) AFLR and AFLS are unable to interact with eugenol exposure.
RNA-binding proteins, which binding to the double or single stranded RNA in cells, participate in the formation of ribonucleoprotein complexes. However, most RNA-binding proteins exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs) because most mature RNA is exported rapidly from the nucleus. RNA-binding proteins have crucial roles in various cellular processes, including cellular function, transport and localization. In particular, they play a major role in the post-transcriptional regulation of RNAs, including their polyadenylation, splicing, mRNA stabilization, localization and translation (Glisovic et al., 2008). Bai et al. (2015) investigated the effects of temperature on transcripts and the corresponding proteins levels using transcriptome-proteome correlation analysis and found that the correlation between protein concentrations and transcript levels was low in A. flavus. Therefore, they proposed that the post-transcriptional regulation process may be involved in aflatoxin biosynthesis (Bai et al., 2015). In the present study, GO functional enrichment analysis showed that RNA binding was the most dysregulated function in A. flavus treated with eugenol, suggesting that post-transcriptional regulation process may be involved in the inhibition of aflatoxin biosynthesis by eugenol.
Ribosome biogenesis is the process by which ribosomes are constructed. In eukaryotes, it takes place in both cytoplasm and the nucleolus. Ribosome biogenesis is intimately associated with many cellular activities including growth, division and secondary metabolism and its process is very tightly regulated (Thomson et al., 2013). The vesicle-vacuole was involved in the conversion of sterigmatocystin (ST) to aflatoxin B1 and compartmentalizing of aflatoxin in A. parasiticus (Chanda et al., 2009). The free ribosomes in the cytoplasm are the sites where the three key enzymes Nor-1, Ver-1, and OmtA are synthesized (Chanda et al., 2009). After synthesis, these three enzymes are packaged into transport vesicles and transported to vacuoles via the cytoplasm-to-vacuole targeting pathway (Chanda et al., 2009). Beside the above pathway, the latest research found that the aflatoxin biosynthesis, exporting, and secretion also occur via cytoplasmic lipid droplets and their associated proteins, oleosins and caleosins (Hanano et al., 2018). Therefore, the free ribosomes also play a critical role in the aflatoxin biosynthesis. In the present study, KEGG metabolic pathway enrichment analysis indicated that ribosome biogenesis was the most dysregulated metabolic pathway in A. flavus treated with eugenol. The result suggested that eugenol dysregulated ribosome biogenesis, which then prevented the synthesis of Nor-1, Ver-1, and OmtA, and the subsequent formation of aflatoxin.
The excess reactive oxygen species (ROS) induces damages to DNA, proteins or lipids, and then causes alteration of cellular functions (Montibus et al., 2013). In Aspergilli, the stress signal transduction is activated by G Protein-Coupled Receptors (GPCRs) and oxylipins (Caceres et al., 2017). These cellular receptors also play an important role in secondary metabolite production (Yu and Keller, 2005). Several compounds inhibit the biosynthesis of aflatoxin in fungi by reducing the ROS levels via the activation of the antioxidant system (Reverberi et al., 2005; Grintzalis et al., 2014; Sun et al., 2015). The transcript levels of oxidative stress-related genes were presented in Table 5. After the addition of eugenol, 10 GPCRs, and two oxylipins genes were significantly regulated. Affeldt et al. (2014) reported that gprK deletion resulted in more aflatoxin in A. flavus treated with inhibitor methyl jasmonte. In this study, the up-regulation of gprK expression was associated with the reduction of AFB1 after eugenol exposure. Caceres et al. (2017) also found that over expressed gprK was associated with AFB1 inhibition by piperine. Oxylipins pathway are known to play important roles in aflatoxin biosynthesis, exporting, fungal development and seed infection. The recent publications show that the oxylipins pathway includes four genes, ppoA, ppoB, ppoC, and afPXG, in A. flavus (Tsitsigiannis and Keller, 2007; Affeldt et al., 2012; Hanano et al., 2015, 2018). AfPXG, the A. flavus caleosin with peroxygenase activity, is associated with the membrane of lipid droplets and mediates fungal development, aflatoxin accumulation, secretion, and seed infection (Hanano et al., 2015, 2018). Among the oxylipins genes, ppoB was the most impacted gene after eugenol exposure in this study. The deletion of ppoB induced more ST which is a precursor of AFB1 (Tsitsigiannis and Keller, 2006), implying a negative correlation between the up-regulation of ppoB and aflatoxin biosynthesis. Caceres et al. (2017) also found that the over expressed ppoB levels was associated with AFB1 inhibition by piperine. However, in the present study, reduced ppoB expression was associated with AFB1 inhibition by eugenol. All these results suggest that the function of GPCRs and oxylipins in aflatoxin production is complicated.
VeA, a global regulator, bridges VelB, and LaeA to form the velvet complex regulating fungal development and secondary metabolism such as aflatoxin (Lin et al., 2013). In addition, it is also involved in the oxidative stress response in A. flavus because it modulates the high osmolarity glycerol (HOG) signaling pathway genes (Duran et al., 2007; Caceres et al., 2017). In A. flavus, veA deletion resulted in the complete inhibition of aflR, aflD, aflM, and aflP expression, and the consequent absence of aflatoxin (Duran et al., 2007). In this study, the decreased expression of veA was also associated with reduced aflM, aflD, and aflP expression and the consequent reduction of aflatoxin production when eugenol treatment. The deletion of veA induced the down-regulation of oxidative stress-related genes such as srrA, msnA, and atfA (Baidya et al., 2014). However, msnA and srrA were up-regulated by eugenol in the present study. These results imply that other regulator factors are also involved in the anti-aflatoxigenic mechanism of eugenol.
In A. parasiticus, the bZIP transcription factors SrrA, AtfB, MsnA, and AP-1 were demonstrated as co-regulators of aflatoxin biosynthesis and oxidative stress (Hong et al., 2013; Caceres et al., 2017). In the present study, we found that the genes encoding these proteins play important roles in the anti-aflatoxigenic mechanism of eugenol. Among these genes, the msnA gene was the most highly up-regulated gene by eugenol. MsnA, encoding a C2H2-type zinc-finger regulator, plays a critical role in fungal growth, aflatoxin and kojic acid biosynthesis, and the oxidative stress response. In A. flavus and A. parasiciticus, msnA deletion resulted in retarded colony growth, slightly increased production of aflatoxin and elevated the production of kojic acid (Chang et al., 2010). In this study, a good correlation between the up-regulation of msnA and the decrease of aflatoxin in A. flavus treated with eugenol. This confirmed that the transcription factor MsnA down-regulated the production of AFB1. Caceres et al. (2016) also found that the msnA gene was up-regulated by 1.9 times after eugenol exposure using a large-scale q-PCR approach. In the present study, atfB and srrA were also up-regulated by eugenol. However, ap-1 was down-regulated. These results suggested that the increased transcription level of the bZIP transcription factor genes msnA, atfB, and srrA is directly involved in the reduced production of aflatoxin induced by eugnol.
The antioxidant-related genes such as genes encoding superoxide dismutases (SODs) and catalase (CAT), are also modulated by the bZIP transcription factors and are involved in the cellular defense against oxidative stress (Caceres et al., 2017). In the present study, the expression of sod1 was up-regulated while cat2 and mnSOD were down-regulated. Many publications have reported that several inhibitors can inhibit aflatoxin production by modulating the antioxidant activities of the fungus (Caceres et al., 2017). However, the effect of inhibitors on the enzymatic defense depends on the type of aflatoxin inhibitor. For example, dithiothreitol, dimethyl sulfoxide, and β-glucans from Lentinula edodes resulted in AFB1 decrease accompanied with a rising in CAT activity (Reverberi et al., 2005; Grintzalis et al., 2014; Caceres et al., 2017). Conversely, ascorbic acid and cinnamaldehyde greatly reduced the production of AFB1 with a rising in SOD activity (Grintzalis et al., 2014; Sun et al., 2015; Caceres et al., 2017). These results indicate that eugenol promotes SOD activity as part of the mechanism of action occurring during AFB1 inhibition.
As a global regulator, VeA is also a critical element coordinating fungal development. VeA trans-regulates the expression of brlA gene which encodes an early regulator of fungal development by modulating the α/β transcript ratio (Kato et al., 2003; Lin et al., 2013). Interestingly, the brlA gene was evidently up-regulated by eugenol in the present study (Table 4). Then the over-expression of brlA gene will activate fungal conidiation and growth. Similarly, Lin et al. (2013) found that brlA gene was up-regulated in A. flavus treated with 5-Azacytidine (5-AC). Calvo et al. (2004) found that brlA gene was highly expressed in the veA deletion strain of A. nidulans. The activation brlA is an essential step of conidiation in Aspergillus (Adams et al., 1988). The BrlA protein includes two C2H2 zinc finger motifs and controls early developmental regulatory genes including abaA, rodA, and yA (Clutterbuck, 1969; Boylan et al., 1987; Chang and Timberlake, 1993; Timberlake and Clutterbuck, 1994). BrlA activates AbaA which plays a critical role in proper differentiation and action of phialides (Sewall et al., 1990; Andrianopoulos and Timberlake, 1994). In this study, the transcription of brlA in A. flavus treated with eugenol was up-regulated, resulting in the up-regulation of abaA and subsequent activation of fungal conidiation and development.
The flbA gene, encoding a development regulator, plays an important role in the expression of nsdD and esdC (Han et al., 2001, 2008). The expression of nsdD and esdC was inhibited by activating FadA and SfaD, directly or indirectly (Han et al., 2008). In this study, the expression of flbA and fadA gene was up-regulated whereas that of esdC gene was slightly down-regulated. Taken together, brlA and flbA were up-regulated by the repression of the veA gene in A. flavus treated with eugenol. The increased FlbA activated FadA and SfaD. Therefore, activated FlbA induced asexual development and sexual development through the up-regulation of brlA gene and the down-regulation of esdC gene, respectively.
Eugenol can be biotransformed by some microbial enzymes. A few eugenol-converting enzymes have been reported, including vanillyl-alcohol oxidase from Penicillium simplicissimum (de Jong et al., 1992), 4-thylphenol methylenehydroxylase from Pseudomonas putida DJ1 (Reeve et al., 1989), eugenol dehydrogenase from Pseudomonas fluorescens E118 (Furukawa et al., 1998), laccase from fungi (Qi et al., 2015), eugenol oxidase from Rhodococcus sp. strain RHA1 (Jin et al., 2007), and lipase from Candida antarctica and Staphylococcus aureus (Horchani et al., 2010; Chiaradia et al., 2012). In A. flavus, there are many similar enzyme genes as well as, such as alcohol dehydrogenase (AFLA_004360, AFLA_005070, AFLA_008880, AFLA_010050, AFLA_024270, AFLA_024700, AFLA_038770, and AFLA_128700), laccase (AFLA_000890 and AFLA_123160), and lipase (AFLA_016150, AFLA_020170, AFLA_057690, and AFLA_058010). In this study, these genes were up-regulated in A. flavus treated with eugenol (Table S2). The results mean that eugenol may be converted by A. flavus. Our previous study showed aflD and aflM were up-regulated at 6–7 d while they were down-regulated at 1–5 d, suggesting that eugenol might be converted to other compounds having lower antiaflatoxigenic activities (Liang et al., 2015).
To decipher the molecular mechanism of action on the aflatoxin production and fungal growth in A. flavus treated with eugenol, we proposed a hypothetical gene modulation mode of action (Figure 3).
Figure 3.
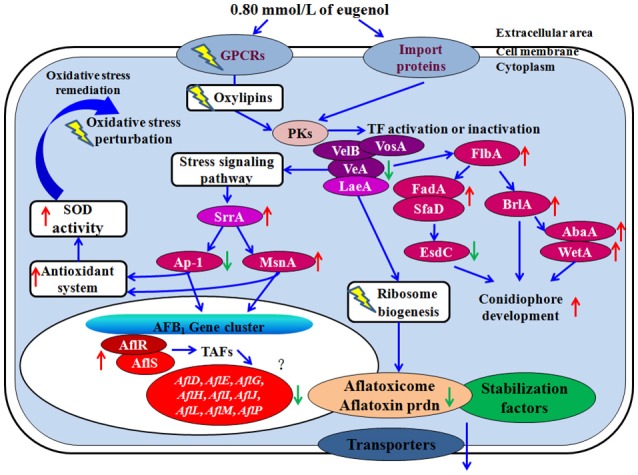
Hypothetical mechanism of action of eugenol. Eugenol perturbs cellular signaling pathway by modulating GPCRs and oxylipins expression levels. Simultaneously, decreased levels of veA might make fungus less tolerant to oxidative stress response which could trigger an activation of several genes involved in the stress signaling pathway such as stress response transcription factor srrA, C2H2 transcription factor msnA, and down-regulate bZIP transcription factor ap-1. Final targets of these modulators correspond to fugal antioxidant system consisting in genes coding for catalasess and superoxide dismutase defenses. The down regulation of genes belonging to the AFB1 cluster may then be a final sequence of the repressive modulation caused by the over expression of OSR transcription factors. Dysregulation of ribosome biogenesis prevents the biosynthesis of Nor-1, Ver-1, and OmtA, and the aflatoxisomes from performing their normal function in aflatoxin formation. For conidiophores development, the growth signaling from the activated fadA and sfaD should be improved by flbA. Upon the activation of them, flbA causes asexual development by the activation of the brlA gene and repression of esdC gene. Up- or down-regulation of gene upon eugenol addition is represented by red and green arrow. PKs, protein kenase; TF, transcription factor.
Conclusions
In the present study, we have proposed a mechanism to explain the transcription modulation behind the inhibitory function of eugenol on AFB1 using an RNA-seq analysis. Based on the results in previous publications and this study, we concluded that (i) the reduction of aflatoxin biosynthesis is due to the down-regulation of most aflatoxin pathway structural genes in A. flavus treated with eugenol, (ii) eugenol's transcription modulation mechanism includes the down-regulation of the global regulator veA accompanied with the up-regulation of the oxidative stress-related transcription factor genes msnA and srrA, and down-regulation of ap-1 and mtfA, (iii) eugenol induces dysregulated transcription of GPCRs and oxylipins genes, (iiii) post-transcription modification and backbone enzymes biosynthesis may be involved in the inhibition of AFB1 production by eugenol.
Author contributions
FX, YL conceived and designed the experiments. CL, PW, MZ, LM performed the experiments. FX analyzed the data. FX wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the financial support of National Key Research and Development Program of China (2016YFD0400105), National Natural Science Foundation of China (31571938), Fundamental Research Funds for Central Non-profit Scientific Institution (S2016JC02), and National Peanut Industrial Technology System (CARS-13). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01116/full#supplementary-material
References
- Adams T. H., Boylan M. T., Timberlake W. E. (1988). BrlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362. 10.1016/0092-8674(88)90198-5 [DOI] [PubMed] [Google Scholar]
- Affeldt K. J., Brodhagen M., Keller N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins 4, 695–717. 10.3390/toxins4090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affeldt K. J., Carrig J., Amare M. G., Keller N. (2014). Global survey of canonical Aspergillus flavus G protein-coupled receptors. MBio 5, 1501–1514. 10.1128/mBio.01501-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaike S., Keller N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. 10.1146/annurev-phyto-072910-095221 [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A., Timberlake W. E. (1994). The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 14, 2503–2515. 10.1128/MCB.14.4.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Wang S., Zhong H., Yang Q., Zhang F., Zhuang Z., et al. (2015). Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature. Sci. Rep. 5:14582. 10.1038/srep14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidya S., Duran R. M., Lohmar J. M., Harris-Coward P. Y., Cary J. W., Hong S.-Y., et al. (2014). VeA is associated with the response to oxidative stress in the aflatoxin producer Aspergillus flavus. Am. Soc. Microbiol. 13, 1095–1103. 10.1128/EC.00099-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. W., Klich M. (2003). Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. 10.1128/CMR.16.3.497-516.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluma R. V., Etcheverry M. G. (2008). Application of essential oils in maize grain: impact on Aspergillus section Flavi growth parameters and aflatoxin accumulation. Food Microbiol. 25, 324–334. 10.1016/j.fm.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Boylan M. T., Mirabito P. M., Willett C. E., Zimmerman C. R., Timberlake W. E. (1987). Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell Biol. 7, 3113–3118. 10.1128/MCB.7.9.3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres I., Khoury R. E., Bailly S., Oswald I. P., Puel O., Bailly J. D. (2017). Piperine inhibits aflatoxin B1 production in Aspergillus flavus by modulating fungal oxidative stress response. Fungal Genet. Biol. 107, 77–85. 10.1016/j.fgb.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Caceres I., Khoury R. E., Medina A., Lippi Y., Naylies C., Atoui A., et al. (2016). Deciphering the anti-aflatoxinogenic properties of eugenol using a large-scale q-PCR approach. Toxins 8:123. 10.3390/toxins8050123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., Bok J., Brooks W., Keller N. P. (2004). VeA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70, 4733–4739. 10.1128/AEM.70.8.4733-4739.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda A., Roze L. V., Kang S., Artymovich K. A., Hicks G. R., Raikhel N. V., et al. (2009). A key role for vesicles in fungal secondary. Proc. Natl. Acad. Sci. U.S.A. 106, 19533–19538. 10.1073/pnas.0907416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Timberlake W. E. (1993). Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. K., Abbas H. K., Weaver M. A., Ehrlich K. C., Scharfenstein L. L., Cotty P. J. (2012). Identification of genetic defects in the atoxigenic biocontrol strain Aspergillus flavus K49 reveals the presence of a competitive recombinant group in field populations. Int. J. Food Microbiol. 154, 192–196. 10.1016/j.ijfoodmicro.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Chang P.-K., Scharfenstein L. L., Luo M., Mahoney N., Molyneux R. J., Yu J., et al. (2010). Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 3, 82–104. 10.3390/toxins3010082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradia V., Paroul N., Cansian R. L., Júnior C. V., Detofol M. R., Lerin L. A., et al. (2012). Synthesis of eugenol esters by lipase-catalyzed reaction in solvent-free system. Appl. Biochem. Biotechnol. 168, 742–751. 10.1007/s12010-012-9814-5 [DOI] [PubMed] [Google Scholar]
- Choi M.-J., Soottiantawat A., Nuchuchua O., Min S.-G., Ruktanonchai U. (2009). Physical and light oxidative properties of eugenol encapsulated by molecular inclusion and emulsion-diffusion method. Food Res. Int. 42, 148–156. 10.1016/j.foodres.2008.09.011 [DOI] [Google Scholar]
- Cleveland T. E., Yu J., Fedorova N., Bhatnagar D., Payne G. A., Nieman W. C., et al. (2009). Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27, 151–157. 10.1016/j.tibtech.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Cleveland T. E., Yu J., Bhatnagar D., Chen Z. Y., Brown R. L., Chang P. K. (2004). Progress in elucidating the molecular basis of the host plant Aspergillus flavus interaction, a basis for devising strategies to reduce aflatoxin contamination in crops. Toxin Rev. 23, 345–380. 10.1081/TXR-200027892 [DOI] [Google Scholar]
- Clutterbuck A. J. (1969). A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva F. F. M., Monte F. J. Q., de Lemos T. L. G., do Nascimento P. G. G., de Medeiros Costa A. K., de Paiva L. M. M. (2018). Eugenol derivatives: synthesis, characterization, and evaluation of antibacterial and antioxidant activities. Chem. Cent. J. 12:34 10.1186/s13065-018-0407-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong E., van Berkel W. J. H., van der Zwan R. P., de Bont J. A. M. (1992). Purification and characterization of vanillyl-alcohol oxidase from Penicillinum simplicissimum: a novel aromatic alcohol oxidase containing covalently bound FAD. Eur. J. Biochem. 208, 651–657. 10.1111/j.1432-1033.1992.tb17231.x [DOI] [PubMed] [Google Scholar]
- Duran R. M., Cary J. W., Calvo A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. 10.1007/s00253-006-0581-5 [DOI] [PubMed] [Google Scholar]
- FAO (1982). Evaluation of Certain Food Additives and Contaminants. Technical Report Series No. 20. FAO/WHO Expert Committee on Food Additives.
- Furukawa H., Wieser M., Morita H., Sugio T., Nagasawa T. (1998). Purification and characterization of eugenol dehydrogenase from Pseudomonas fluorescens E118. Arch. Microbiol. 171, 37–43. 10.1007/s002030050675 [DOI] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986. 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintzalis K., Vernardis S. I., Klapa M. I., Georgiou C. D. (2014). Role of oxidative stress in sclerotial differentiation and aflatoxin B1 biosynthesis in Aspergillus flavus. Appl. Environ. Microbiol. 80, 5561–5571. 10.1128/AEM.01282-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groopman J. D., Kensler T. W., Wild C. P. (2008). Protective interventions to prevent aflatoxin-induced carcinogenesis in developing countries. Annu. Rev. Public Health 29, 187–203. 10.1146/annurev.publhealth.29.020907.090859 [DOI] [PubMed] [Google Scholar]
- Han K.-H., Han K.-Y., Yu J.-H., Chae K.-S., Jahng K.-Y., Han D.-M. (2001). The nsdD gene encodes a putative GATA type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41, 299–309. 10.1046/j.1365-2958.2001.02472.x [DOI] [PubMed] [Google Scholar]
- Han K.-H., Kin J.-H., Moon H., Kim S., Lee S. S., Han D. M., et al. (2008). The Aspergillus nidulans esdC (early sexual development) gene is necessary for sexual development and is controlled by veA and a heterotrimeric G protein. Fungal Genet. Biol. 45, 310–318. 10.1016/j.fgb.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Hanano A., Alkara M., Almousally I., Shaban M., Rahman F., Hassan M., et al. (2018). The peroxygenase activity of the Aspergillus flavus caleosin, AfPXG, modulates the biosynthesis of aflatoxins and their trafficking and extracellular secretion via lipid droplets. Front. Microbiol. 9:158. 10.3389/fmicb.2018.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano A., Almousally I., Shaban M., Blee E. (2015). A caleosin-like protein with peroxygenase activity mediates Aspergillus flavus development, aflatoxin accumulation, and seed infection. Appl. Envrion. Microbiol. 81, 6129–6144. 10.1128/AEM.00867-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister D., Keller N. P. (2007). Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24, 393–416. 10.1039/B603084J [DOI] [PubMed] [Google Scholar]
- Hong S. Y., Roze L. V., Wee J., Linz J. E. (2013). Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in Aspergilli. Microbiologyopen 2, 144–160. 10.1002/mbo3.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horchani H., Salem N. B., Zarai Z., Sayari A., Gargour Y., Chaâbouni M. (2010). Enzymatic synthesis of eugenol benzoate by immobilized Staphylococcus aureus lipase: optimization using response surface methodology and determination of antioxidant activity. Bioresour. Technol. 101, 2809–2817. 10.1016/j.biortech.2009.10.082 [DOI] [PubMed] [Google Scholar]
- Hua S. S., Beck J. J., Sarreal S. B. L., Gee W. (2014). The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 30, 71–78. 10.1007/s12550-014-0189-z [DOI] [PubMed] [Google Scholar]
- Hua H., Xing F., Selvaraj J. N., Wang Y., Zhao Y., Zhou L., et al. (2014). Inhibitory effect of essential oils on Aspergillus ochraceus growth and ochratoxin A production. PLoS ONE 9:e108285. 10.1371/journal.pone.0108285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) (1985). Summaries & Evaluations-Eugenol. 36, 75. [Google Scholar]
- Isaac S. (1999). What is the mode of action of fungicides and how do fungi develop resistance? Mycologist 13, 38–39. [Google Scholar]
- Jahanshiri Z., Shams-Ghahfarokhi M., Allameh A., Razzaghi-Abyaneh M. (2015). Inhibitory effect of eugenol on aflatoxin B1 production in Aspergillus parasiticus by downregulating the expression of major genes in the toxin biosynthetic pathway. World J. Microbiol. Biotechnol. 31, 1071–1078. 10.1007/s11274-015-1857-7 [DOI] [PubMed] [Google Scholar]
- Jayashree T., Subramanyam C. (1999). Antiaflatoxigenic activity of eugenol is due to inhibition of lipid peroxidation. Lett. Appl. Microbiol. 28, 179–183. 10.1046/j.1365-2672.1999.00512.x [DOI] [PubMed] [Google Scholar]
- Jin J., Mazon H., van den Heuvel R. H. H., Janssen D. B., Fraaije M. W. (2007). Discovery of a eugenol oxidase from Rhodococcus sp. strain RHA1. FEBS J. 274, 2311–2321. 10.1111/j.1742-4658.2007.05767.x [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapinar M. (1990). Inhibitory effects of anethole and eugenol on the growth and toxin production of Aspergillus parasiticus. Int. J. Food Microbiol. 10, 193–199. 10.1016/0168-1605(90)90066-E [DOI] [PubMed] [Google Scholar]
- Kato N., Brooks W., Calvo A. M. (2003). The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryotic Cell 2, 1178–1186. 10.1128/EC.2.6.1178-1186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Xing F., Selvaraj J. N., Liu X., Wang L., Hua H., et al. (2015). Inhibitory effect of cinnamaldehyde, citral, and eugenol on aflatoxin biosynthetic gene expression and aflatoxin B1 biosynthesis in Aspergillus flavus. J. Food Sci. 80, M2917–M2924. 10.1111/1750-3841.13144 [DOI] [PubMed] [Google Scholar]
- Lin J., Zhao X., Zhi Q., Zhao M., He Z. (2013). Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 56, 78–86. 10.1016/j.fgb.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Liu X., Guan X., Xing F., Lv C., Dai X., Liu Y. (2017). Effect of water activity and temperature on the growth of Aspergillus flavus, the expression of aflatoxin biosynthetic genes and aflatoxin production in shelled peanuts. Food Control 82, 325–332. 10.1016/j.foodcont.2017.07.012 [DOI] [Google Scholar]
- Montibus M., Pinson-Gadais L., Richard-Forget F., Barreau C., Ponts N. (2013). Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 43, 1–14. 10.3109/1040841X.2013.829416 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Nam H., Kim M. M. (2013). Eugenol with antioxidant activity inhibits MMP-9 related to metastasis in human fibrosarcoma cells. Food Chem. Toxicol. 55, 106–112. 10.1016/j.fct.2012.12.050 [DOI] [PubMed] [Google Scholar]
- OBrian G. R., Georgianna D. R., Wilkinson J. R., Yu J., Abbas H. K., Bhatnagar D., et al. (2007). The effect of elevated temperature on gene transcription and aflatoxin biosynthesis. Mycologia 99, 232–239. 10.1080/15572536.2007.11832583 [DOI] [PubMed] [Google Scholar]
- Opdyke D. L. J. (1975). Monographs on fragrance raw materials: eugenol. Food Cosm. Toxicol. 13, 545–547. 10.1016/0015-6264(75)90011-5 [DOI] [PubMed] [Google Scholar]
- Priebe S., Linde J., Albrecht D., Guthke R., Brakhage A. A. (2011). FungiFun: a webbased application for functional categorization of fungal genes and proteins. Fungal Genet. Biol. 48, 353–358. 10.1016/j.fgb.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Qi Y.-B., Wang X.-L., Shi T., Liu S., Xu Z.-H., Li X., et al. (2015). Multicomponent kinetic analysis and theoretical studies on the phenolic intermediates in the oxidation of eugenol and isoeugenol catalyzed by laccase. Phys. Chem. Chem. Phys. 17, 29597–29607. 10.1039/C5CP03475B [DOI] [PubMed] [Google Scholar]
- Reeve C. D., Carver M. A., Hopper D. J. (1989). The purification and characterization of 4-ethylphenol methylenehydroxylase, a flavocytochrome from Pseudomonas putida DJ1. Biochem. J. 263, 431–437. 10.1042/bj2630431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi M., Fabbri A. A., Zjalic S., Ricelli A., Punelli F., Fanelli C. (2005). Antioxidant enzymes stimulation in Aspergillus parasiticus by Lentinula edodes inhibits aflatoxin production. Appl. Microbiol. Biotechnol. 69, 207–215. 10.1007/s00253-005-1979-1 [DOI] [PubMed] [Google Scholar]
- Roze L. V., Koptina A. V., Laivenieks M., Beaudry R. M., Jones D. A., Kanarsky A. V., et al. (2011). Willow volatiles influence growth, development, and secondary metabolism in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 92, 359–370. 10.1007/s00253-011-3339-7 [DOI] [PubMed] [Google Scholar]
- Satoh K., Sakagami H., Yokoe I., Kochi M. (1998). Interaction between eugenol-related compounds and radicals. Anticancer Res. 18, 425–428. [PubMed] [Google Scholar]
- Sewall T. C., Mims C. W., Timberlake W. E. (1990). AbaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 2, 731–739. 10.1105/tpc.2.8.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire R. A. (1981). Ranking animal carcinogens: a proposed regulatory approach. Science 214, 877–880. 10.1126/science.7302565 [DOI] [PubMed] [Google Scholar]
- Sun Q., Shang B., Wang L., Lu Z., Liu Y. (2015). Cinnamaldehyde inhibits fungal growth and aflatoxin B1 biosynthesis by modulating the oxidative stress response of Aspergillus flavus. Appl. Microbiol. Biotechnol. 100, 1355–1364. 10.1007/s00253-015-7159-z [DOI] [PubMed] [Google Scholar]
- Thomson E., Ferreira-Cerca S., Hurt E. (2013). Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126, 4815–4821. 10.1242/jcs.111948 [DOI] [PubMed] [Google Scholar]
- Timberlake W. E., Clutterbuck A. J. (1994). Genetic regulation of conidiation. Prog. Indust. Microbiol. 29, 383–427. [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Keller N. P. (2006). Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol. Microbiol. 59, 882–892. 10.1111/j.1365-2958.2005.05000.x [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Keller N. P. (2007). Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15, 109–118. 10.1016/j.tim.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Velluti A., Sanchis V., Ramos A., Egido J., Marin S. (2003). Inhibitory effect of cinnamon, clove, lemongrass, oregano and palmarose essential oils on growth and fumonisin B1 production by Fusarium proliferatum in maize grain. Int. J. Food Microbiol. 89, 145–154. 10.1016/S0168-1605(03)00116-8 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D., Zhou L., Selvaraj J. N., Zhang C., Xing F., Zhao Y., et al. (2014). Molecular characterization of atoxigenic Aspergillus flavus isolates collected in China. J. Microbiol. 52, 559–565. 10.1007/s12275-014-3629-8 [DOI] [PubMed] [Google Scholar]
- Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., et al. (2008). Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243. 10.1038/nature07002 [DOI] [PubMed] [Google Scholar]
- Wright M. S., Greene-McDowelle D. M., Zeringue H. J., Bhatnagar D., Cleveland T. E. (2000). Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis. Toxicon 38, 1215–1223. 10.1016/S0041-0101(99)00221-4 [DOI] [PubMed] [Google Scholar]
- Wu F. (2014). Perspective: time to face the fungal threat. Nature 516, S7–S7. 10.1038/516S7a [DOI] [PubMed] [Google Scholar]
- Xing F., Wang L., Liu X., Selvaraj J. N., Wang Y., Zhao Y., et al. (2017). Aflatoxin B1 inhibition in Aspergillus flavus by Aspergillus niger through down regulating expression of major biosynthetic genes and AFB1 degradation atoxigenic A. falvus. Int. J. Food Microbiol. 256, 1–10. 10.1016/j.ijfoodmicro.2017.05.013 [DOI] [PubMed] [Google Scholar]
- Yu J., Chang P. K., Ehrlich K. C., Cary J. W., Bhatnagar D., Cleveland T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. 10.1128/AEM.70.3.1253-1262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Fedorova N. D., Montalbano B. G., Bhatnagar D., Cleveland T. E., Bennett J. W., et al. (2011). Tightcontrol of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 322, 145–149. 10.1111/j.1574-6968.2011.02345.x [DOI] [PubMed] [Google Scholar]
- Yu J.-H., Keller N. (2005). Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43, 437–458. 10.1146/annurev.phyto.43.040204.140214 [DOI] [PubMed] [Google Scholar]
- Zhang F., Guo Z., Zhong H., Wang S., Yang W., Liu Y., et al. (2014). RNA-Seq-Based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 6, 3187–3207. 10.3390/toxins6113187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.