Abstract
Background
Lasting changes in gene expression in brain reward regions, including nucleus accumbens (NAc), contribute to persistent functional changes in the addicted brain. We and others have demonstrated that altered expression of several candidate transcription factors in NAc regulates drug responses. A recent large-scale genome-wide study from our group predicted E2F3 as a prominent upstream regulator of cocaine-induced changes in gene expression and alternative splicing.
Methods
We studied the expression of two E2F3 isoforms – E2F3a and E2F3b – in mouse NAc after repeated cocaine administration, and assayed the effects of overexpression or depletion of E2F3 isoforms in NAc on cocaine behavioral responses. We then performed RNA-seq to investigate the effect of E2F3a overexpression in this region on gene expression and alternative splicing, performed qChIP at downstream targets in NAc following E2F3a overexpression or repeated cocaine exposure. Sample sizes vary between experiments and are noted in the text.
Results
We show that E2F3a, but not E2F3b, overexpression in mouse NAc regulates cocaine-induced locomotor and place conditioning behavior. Furthermore, we demonstrate that E2F3a overexpression substantially recapitulates genome-wide transcriptional profiles and alternative splicing induced by cocaine. We further validate direct binding of E2F3a at key target genes following cocaine exposure.
Conclusions
This study establishes E2F3a as a novel transcriptional regulator of cocaine action in NAc. The findings reveal a crucial role for E2F3a in the regulation of cocaine-elicited behavioral states. Moreover, the importance of this role is bolstered by the extensive recapitulation of cocaine’s transcriptional effects in NAc upon expression of E2F3a.
Keywords: E2F, RNA-sequencing, ChIP, gene expression, addiction, splicing
Introduction
Casual drug use transitions into compulsive drug-taking in part due to epigenetic mechanisms in nucleus accumbens (NAc). Lasting changes in NAc gene expression underlie many persistent neurophysiological changes in the addicted brain (1, 2). Transcription factors are key upstream regulators of these molecular responses. Research by our group and others has demonstrated that alterations in the expression or activity of several candidate transcription factors (e.g., ΔFosB, CREB, NFκB, SRF, and Egr3), coupled with chromatin remodeling, in NAc act to either potentiate or inhibit drug responses (1–12).
To further elucidate transcriptional mechanisms of cocaine action, we performed ChIP-and RNA-seq experiments on mouse NAc after repeated investigator-administered cocaine (13). Analyses of these unbiased, genome-wide datasets suggested E2F3 as a novel upstream regulator of cocaine-induced gene expression and alternative splicing (Figure S1)(13). No prior work has implicated E2F3 in cocaine’s effects.
The E2f3 gene encodes two proteins, E2F3a and E2F3b, which arise through alternative splicing of mutually exclusive first exons (Figure 1A)(14). E2F3a and E2F3b retain the same DNA binding, nuclear localization, and transactivation domains but E2F3b lacks an N-terminus ubiquitin-targeting domain (Figure 1B). To date, E2F proteins have only been studied in the context of proliferating tissue, in which E2F3a typically acts as a gene activator and E2F3b as a repressor (15, 16). Given these opposing roles in dividing cells, we hypothesized that the two isoforms may play different roles in NAc in the regulation of cocaine action as well.
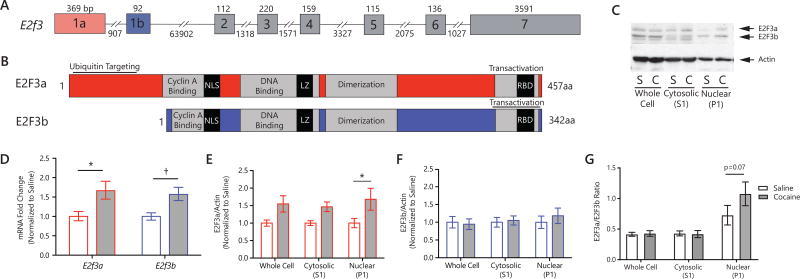
Figure 1. E2F3 regulation in NAc by cocaine.
(A) Structure of E2f3 gene detailing 8 exons, including exons 1a and 1b which are mutually exclusively spliced to form either E2f3a or E2f3b mRNA. (B) Structure of E2F3a and E2F3b proteins. Note that all domains are conserved in both isoforms except the ubiquitin-targeting domain at the N-terminus present in E2F3a only. (C) Western blot of E2F3a and E2F3b in whole cells, cytosolic (S1) and crude nuclear (P1) fractions. N=8–10 (D) qPCR quantification showing mRNA fold change in NAc normalized to saline (following ΔΔCt method using Gapdh as a control). Cocaine specifically increased E2f3a (main effect of treatment was F1,20=12.99, p=0.0018; post-hoc: p=0.0207) but not E2f3b mRNA. N=6 (E–G) Quantification of protein levels in NAc by Western blot for E2F3a/b in whole cell, cytosolic, and crude nuclear fractions, showing (E) E2F3a levels in NAc (main effect of drug: F1,37=16.59, p=0.0002; post-hoc, crude nuclear: p=0.0165); (F) E2F3b levels (no main effect of drug: F1,36=0.01, p=0.9113); (G) E2F3a/E2F3b ratio (main effect of cell compartment: F2,40=11.24, p=0.0001, post-hoc, crude nuclear: p=0.0955) Graphs show mean ± SEM. *p<0.05; †: not significant, p<0.1 (See also Figures S1 and S2)
We first tested the effect of cocaine on expression of E2f3 isoforms, and found selective induction of E2f3a in NAc, with no effect on E2F3b. Using viral-mediated gene manipulation, we established that E2F3a in NAc is both necessary and sufficient for cocaine-mediated locomotor and place conditioning responses. Next, we utilized RNA-seq to compare differential gene expression and alternative splicing after either cocaine administration or E2F3a overexpression, and qChIP to examine functional binding of E2F3 at target genes implicated by RNA-seq. Results of these findings establish a novel role for E2F3a in cocaine action.
Methods and Materials
(See also Supplementary Methods and Materials)
Animals
C57BL/6J male mice (The Jackson Laboratory), 7–8 weeks old, 25–30g, were used in these experiments. All experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Mount Sinai.
Repeated cocaine injections
For RNA-qPCR, immunoblotting, and qChIP assays, mice were injected intraperitoneally (I.P.) with saline or cocaine (20 mg/kg) once daily for seven days in home cages. Mice were euthanized 24 hr after the final cocaine injection. For RNA-seq, the same procedure commenced 48 hr after stereotactic surgery (17).
Viral-mediated gene transfer
Stereotactic injection into NAc of 0.5 µL of HSV (18, 19) was used to neuronally express HSV-GFP or -GFP plus E2F3a, E2F3b, or a miRNA (miR) targeting E2f3a, E2f3b, or LacZ mRNA. miRs were designed using Invitrogen BLOCK-iT Pol II kit (Table S1).
Tissue collection
Mice were euthanized by cervical dislocation and 14- or 15-gauge NAc punches were dissected for cocaine or viral studies, respectively, then frozen on dry ice. Single animal samples were used in all RNA-qPCR, immunoblotting, and RNA-seq experiments, while animals were pooled for qChIP experiments as described below.
Subcellular fractionation, immunoblotting, and antibodies
NAc punches were fractionated into P1 (crude nuclear) and S1 (cytosolic) components as published(20). Protein concentrations were determined by DC protein assay (Bio-Rad). Immunoblotting was carried out using established protocols (20).
Immunohistochemistry
Immunohistochemistry was used to assess cellular expression of E2F3 using a rabbit-anti-E2F3 antibody (abcam ab50917).
Quantitative chromatin immunoprecipitation
qChIP was performed on bilateral 14- or 15-gauge NAc punches, pooled from 2 mice, 24 hr after the last drug treatment, or from 3 mice, 72 hr after viral infection. qChIP methodology was modified from previous studies using primers listed in Table S1 (21).
Locomotor activity and conditioned place preference
Locomotor activity and an unbiased place conditioning was assayed as described previously (10, 22). In overexpression experiments, behavioral observations began 2–3 days after surgery, and 5 mg/kg cocaine was used (Figure 2A,D). To allow for sufficient knockdown of E2F3 isoforms, behavioral observation began 4–5 days post-surgery; 7.5 mg/kg cocaine was used (Figure 2F,I).
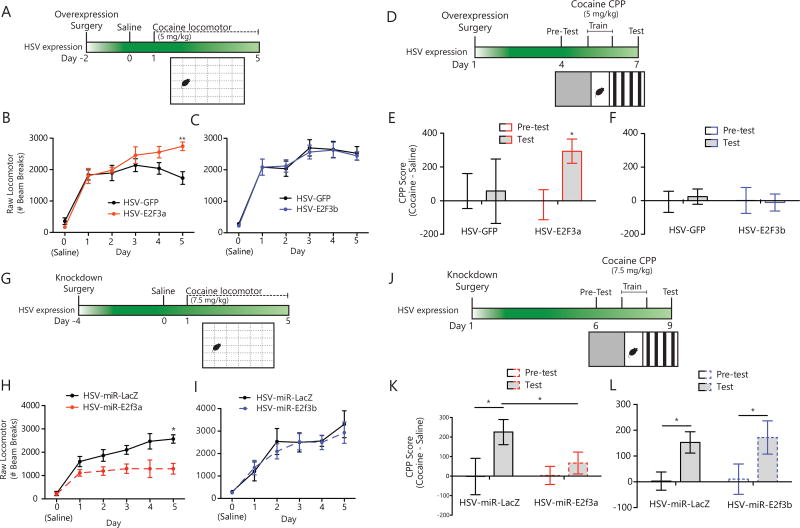
Figure 2. E2F3a expression in NAc regulates behavioral responses to cocaine.
(A) HSV-GFP, HSV-E2F3a, or HSV-E2F3b was infused into NAc. 48 hr post-infusion, mice were tested over 6 days for cocaine (5 mg/kg)-induced locomotor activity. (B) Overexpression of E2F3a in NAc increased locomotor response to cocaine across testing days, with locomotor activity significantly increased above GFP on Day 5 (interaction of virus and day, F5,60=3.78, p=0.0048; post-hoc, day 5: p=0.0019; N=7). (C) Overexpression of E2F3b in NAc had no effect on cocaine-induced locomotor activity compared to GFP (no main effect of virus: F1,85=0.07, p=0.7879; N=10). (D) HSV-GFP or HSV-E2F3a was infused into NAc. 48 hr post-infusion, saline/cocaine (5 mg/kg) pairing began in a conditioned place preference procedure. (E) E2F3a overexpression in NAc increased cocaine preference (E2F3a Test vs. Pre-Test: p=0.0239; N=9). (F) E2F3b overexpression in NAc had no effect on cocaine preference (no main effect of virus: F1,18=0.04179, p=0.8403; N=10). (G) HSV-miR-LacZ, HSV-miR-E2f3a, or HSV-miR-E2f3b was infused into NAc. 96 hr post-infusion, mice were tested over 6 days for cocaine (7.5 mg/kg)-induced locomotor activity. (H) Decreased locomotor responses to cocaine across testing days was seen in mice expressing miR-E2f3a in NAc, with a significant decrease in locomotor activity on Day 5 when compared to miR-LacZ (interaction of virus and day, F5,78=2.74, p=0.0248; post-: p=0.0124; N=7–10). (I) Knockdown of E2F3b had no effect on cocaine-induced locomotor activity compared to miR-LacZ (no main effect of drug: F1,61=0.31, p=0.5815; N=6–10). (J) HSV-miR-LacZ or HSV-miR-E2F3a was infused into NAc. 96 hr post-infusion, saline/cocaine (7.5 mg/kg) pairing began. (K) E2F3a knockdown decreased cocaine preference (miR-E2F3a Test vs. miR-LacZ Test: p=0.0306; miR-LacZ Test vs. Pre-test: p=0.0293; N=10). (L) Knockdown of E2F3b had no effect on cocaine conditioned place preference (no main effect of virus: F1,18=0.04416, p=0.8359; N=10). Graphs show mean ± SEM. *p<0.05. **p<0.01. (See also Figure S3)
RNA isolation and qPCR
RNA isolation, qPCR, and data analysis were performed as described (23). Data were analyzed by comparing C(t) values of treatment condition to control condition with the ΔΔC(t) method (24). Primer sequences are given in Table S1.
RNA-sequencing and library preparation
Tissue was collected 24 hr following the final injection and RNA-seq was performed on samples from individual animals (Figure 3A,B). Library preparation was performed as described previously with minor modifications (13). All samples had a RIN value above 8. Libraries were prepared using the TruSeq mRNA Stranded Sample Prep Kit v2 protocol (Illumina, San Diego, CA).
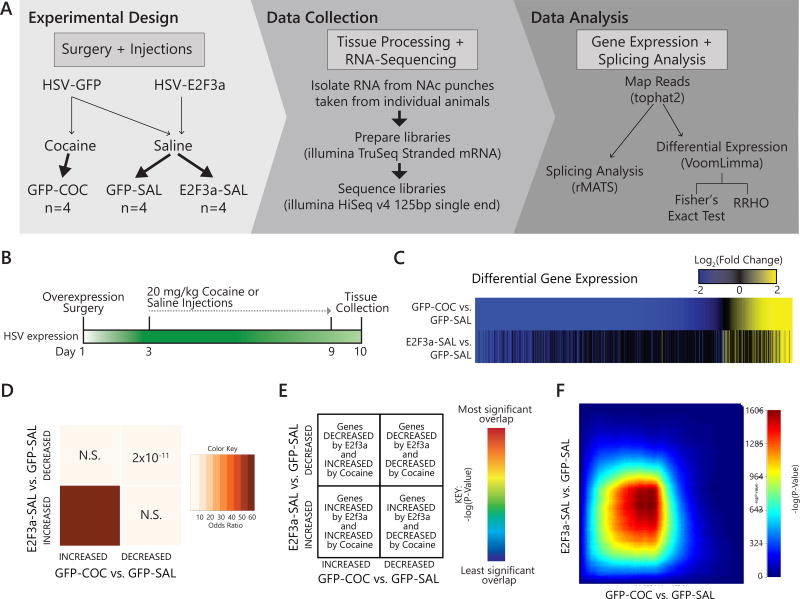
Figure 3. RNA-sequencing identifies E2F3a regulation of gene expression in NAc.
(A) Schematic of experimental design, data collection, and analysis. (B) Animals underwent stereotactic surgery to infuse HSV-GFP or HSV-E2F3a into NAc. After 48 hr, cocaine (20 mg/kg) or saline I.P. injections commenced, and tissue was collected 24 hr after the final injection for RNA-seq. (C) Union heat map shows fold change of all genes significantly differentially expressed (FC>1.3, p<0.05) in either comparison for GFP-COC vs. GFP-SAL (top panel) or E2F3a-SAL vs. GFP-SAL (bottom panel) rank ordered by fold change in the GFP-COC vs. GFP-SAL comparison. (D) Matrix summarizes enrichment for genes across the two comparisons using Fisher’s exact test, corrected for multiple comparisons. Darker colors indicate increasing Odds Ratio (OR). P value is noted in each quadrant. Significant overlap was found between genes upregulated (OR=62.77, p=9 × 10−22) or downregulated (OR=5.84, p=2 × 10−11) in both comparisons. (E) Schematic illustrating rank-rank hypergeometric overlap (RRHO) map interpretation(29). (F) RRHO maps compare threshold-free differential expression between the two gene lists. Each pixel represents the overlap between the transcriptome of the two manipulations (cocaine administration or E2F3a overexpression) with the significance of overlap (−log10(P-Value) of a hypergeometric test) color coded, with warmer colors indicated increasing significance. E2F3a-SAL vs. GFP-SAL and GFP-COC vs. GFP-SAL showed a robust overlap in upregulation signature. (See also Figure S4 and Tables S2 and S3)
Bioinformatic data analysis
Differential expression analysis
Sequencing reads were aligned to the mouse mm10 reference transcriptome using Tophat2 and counted against Gencode vM4 annotation using the HTSeq Python package (25, 26). Raw counts per gene for each sample were normalized by sequencing depth and mRNA length to generate reads per kb per million mapped reads (RPKMs). All RNA-seq processing steps were performed in parallel using an in-house pipeline, SPEctRA(27). Subsequently, pair-wise differential expression comparisons were made using Voom Limma on filtered gene lists meeting the criterion that the majority samples within at least one experimental condition have RPKMs of ≥1.0 (28). A nominal significance threshold of p<0.05 and Fold Change>1.3 was used.
Rank Rank Hypergeometric Overlap (RRHO)
To evaluate the similarity of differential expression between cocaine-treatment and E2F3a overexpression, RRHO was conducted on the full differential expression lists (i.e., genome-wide lists with no significance thresholds applied). Gene lists were ranked by the −log10(p-value) multiplied by the sign of the fold change (29).
Differential splicing analysis
Alternative splicing events were analyzed using RNA-seq multivariate analysis of transcript splicing (rMATS)(30). In order to measure the differential splicing occurring in each of these events, one exon is determined to be constitutive and the other to be alternatively spliced (noted for each type of splicing event in Figure 4A). The measurement of differential alternative splicing is the Inclusion Level Difference (ILD), or the difference in read counts between the case (GFP-COC or E2F3a-SAL) and control (GFP-SAL) groups for the alternatively spliced exon. A negative ILD represents a gain of the alternatively spliced exon, and a positive ILD represents a loss. Comparisons of differentially spliced exons were considered significant at p < 0.05. Individual splicing events (gene by gene) were visualized using rmats2sashimiplot (http://www.mimg.ucla.edu/faculty/xing/rmats2sashimiplot/).
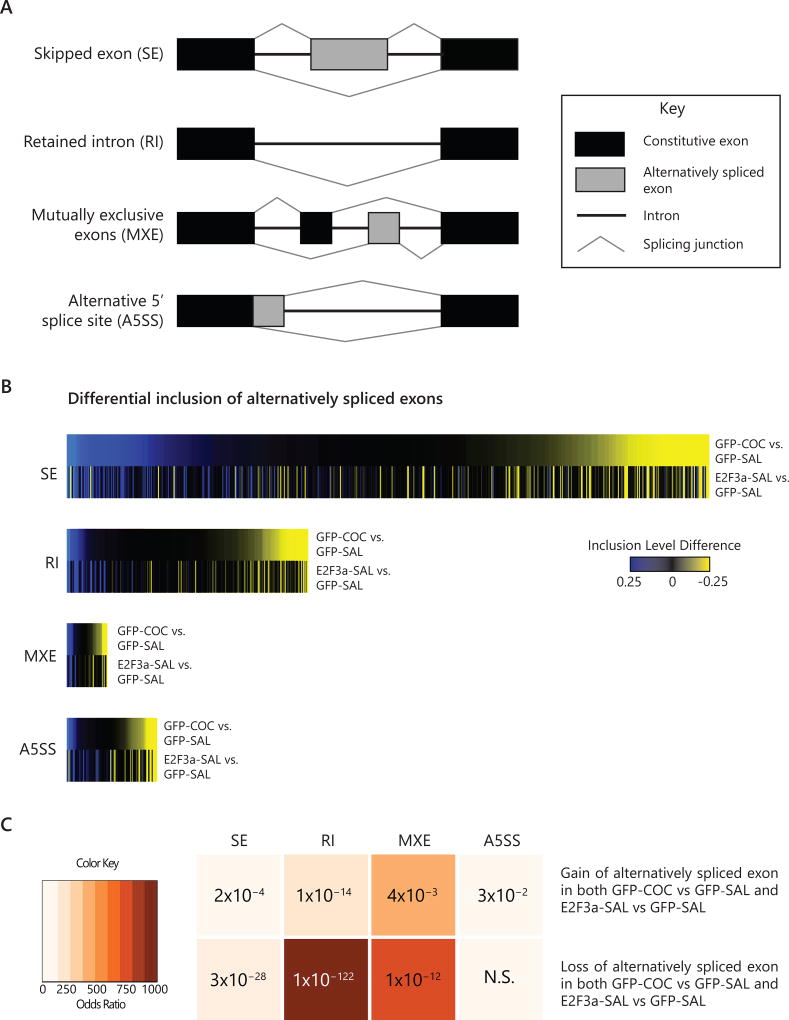
Figure 4. E2F3a regulation of alternative splicing in NAc.
(A) Schematic diagram of four types of alternative splicing events detected using rMATS analysis: skipped exons (SE), retained introns (RI), mutually exclusive exons (MXE), and alternative 5’ splice site (A5SS). (B) Based on the RNA-seq data shown in Figure 3, union heat maps show Inclusion Level Difference (ILD) of all significant differential inclusion events of alternatively spliced exons (p<0.05) in either comparison for GFP-COC vs. GFP-SAL (top panel) or E2F3a-SAL vs. GFP-SAL (bottom panel) rank ordered by ILD in the GFP-COC vs. GFP-SAL comparison for SE, RI, MXE, and A5SS scaled according to number of differentially spliced exons. (C) Matrix for Fisher’s exact test for enrichment of E2F3a-induced alternatively spliced genes in cocaine-induced alternatively spliced genes. Darker colors indicate increasing Odds Ratio (OR). P value is noted in each quadrant. (See also Table S4)
Enrichment analyses
Enrichment between gene lists was analyzed using the GeneOverlap R Package, which utilizes Fisher’s Exact Tests (FETs) to determine whether enrichment is significant (p < 0.05), and reports an odds ratio to denote the level of enrichment (31).
Statistics
All analyses were performed in Prism (GraphPad). In qPCR and immnoblotting analyses, control groups were normalized to 1 by dividing each sample’s value by the control group mean. Then SEM was calculated from the normalized sample values. Student’s t tests were used for all pairwise comparisons; one- or two-way ANOVAs were used for all multiple comparisons, followed by Bonferroni post hoc tests where appropriate. For behavioral analyses, a repeated measures design was used.
Results
Repeated cocaine increases E2f3a mRNA and E2F3a nuclear protein levels in NAc
We probed isoform-specific expression of E2f3 mRNA in NAc following 7 days of 20 mg/kg cocaine (24 hr post final injection). We observed a cocaine-induced increase in E2f3a mRNA levels (main effect of treatment: F1,20=12.99, p=0.0018; post hoc, E2f3a: p=0.0207; E2f3b: p=0.0512; Figure 1D) and a non-significant trend for induction of E2f3b. We next investigated the effects of repeated cocaine on E2F3a and E2F3b protein levels and subcellular localization. We observed an increase in E2F3a after cocaine specifically in NAc crude nuclear fractions (P1; main effect of treatment: F1,37=16.59, p=0.0002; post hoc, nuclear fraction: p=0.0165; Figure 1C,E). In contrast, we observed no change in E2F3b protein levels despite the trend for regulation of its mRNA (no main effect of treatment: F1,36=0.01, p=0.9113; Figure 1C,F). There was more E2F3b protein than E2F3a in the cytosol and whole cell lysate (Figure 1G). In contrast, in the nucleus, where the two proteins regulate gene expression, the two isoforms are expressed to a similar degree. Repeated cocaine increased (trend) the nuclear ratio of E2F3a to E2F3b (main effect of cell compartment: F2,40=11.24, p=0.0001, post hoc, nuclear fraction: p=0.07; Figure 1G). We also found, by immunohistochemistry, that E2F3 protein is found predominantly in post-mitotic neurons in NAc (Figure S2).
E2F3a expression regulates behavioral responses to cocaine
Based on our findings that cocaine selectively regulates E2F3a, we hypothesized that E2F3a, but not E2F3b, would control behavioral responses to cocaine. We examined the effect of HSV-mediated overexpression of either E2F3a or E2F3b in NAc on locomotor sensitization (Figure 2A). Neither E2F3a nor E2F3b overexpression altered baseline locomotor activity (Day 0, Figure 2B,C), habituation to the locomotor chamber (data not shown), or the behavioral response to acute cocaine (Day 1, Fig 2B,C). However, overexpression of E2F3a increased locomotor activity after repeated exposure to a low dose of cocaine (5 mg/kg) that does not elicit increased locomotor responses in control animals (Figure 2A–C)(32). Locomotor activity was significantly increased above GFP control animals on Day 5, indicating an increased sensitization phenotype (interaction of virus and day: F5,60=3.78, p=0.0048; post hoc, Day 5: p=0.0019, Figure 2B). In contrast, E2F3b–overexpressing mice did not differ from GFP controls, supporting the isoform-specific role of E2F3a in regulating cocaine action (no main effect of virus F1,85=0.07, p=0.7879; Figure 2C).
We also tested whether E2F3a enhanced the associative effects of cocaine as measured by conditioned place preference (CPP; Figure 2D). Overexpression of E2F3a, but not E2F3b, increased preference for the cocaine-paired chamber when compared to GFP controls (5 mg/kg; E2F3a Test vs. Pre-Test: p=0.0239; Figure 2E & F). These results, together with the locomotor findings, suggest that E2F3a, specifically, is sufficient to mediate cocaine-elicited behavioral phenotypes in an isoform-specific manner.
We next tested whether E2F3a was also necessary for cocaine-elicited behavior by expressing microRNAs (miRs) to selectively knockdown either E2f3a or E2f3b; a miR targeting LacZ mRNA was used to control for nonspecific off-target miR effects. To allow for sufficient expression of the miRs as well as degradation of existing endogenous E2F3a or E2F3b protein, behavioral testing was conducted 4 days post viral-infusion (Figure 2G)(33). In a separate cohort of mice, we confirmed that knockdown or overexpression of one isoform did not affect levels of the other (Figure S3A,B,C).
Knockdown of either E2F3 isoform did not alter baseline locomotor activity or habituation to the chamber (Day 0, Figure 2H,I). Locomotor activity of the miR-LacZ control group increased in response to a dose of cocaine (7.5 mg/kg), previously shown to increase locomotor responses(17). This effect was prevented by knockdown of E2F3a (interaction of virus and day: F5,78=2.74, p=0.0248). This decreased locomotor activity was significant only on Day 5 (p=0.0124), suggesting that E2F3a is required for locomotor sensitization to cocaine but not its acute locomotor effects (Figure 2H). Knocking down E2F3b had no effect on locomotor responses to cocaine compared to miR-LacZ (F1,61=0.31, p=0.5815; Figure 2I).
Knocking down E2F3a, but not E2F3b, in NAc also blunted cocaine CPP (7.5 mg/kg; miR-E2f3a Test vs. miR-LacZ Test: p=0.0306; miR-LacZ Test vs. Pre-test: p=0.0293; Figure 2J,K,L). Together, these observations confirmed that E2F3a, but not E2F3b, is required for both the place conditioning and locomotor effects of cocaine.
Viral-mediated E2F3a overexpression in NAc recapitulates cocaine-induced transcriptomic profiles
To identify transcriptional mechanisms underlying the role of E2F3a in mediating cocaine-elicited behaviors, we overexpressed E2F3a in NAc and performed RNA-seq. We infused HSV vectors expressing GFP or E2F3a into NAc 48 hr before injecting animals with either 20 mg/kg cocaine or saline for 7 days (Figure 3A & B). To directly compare cocaine-elicited versus E2F3a-overexpression-induced transcriptome-wide alterations, we used the GFP-SAL group as a shared control for both the GFP-COC and E2F3a-SAL groups.
Cocaine induced more differentially expressed genes (DEGs; GFP-COC vs. GFP-SAL: 1030 genes) than did overexpression of E2F3a (E2F3a-SAL vs. GFP-SAL: 86 genes, Fig S4), however, the direction of regulation in these DEGs was similar in both comparisons (Figure 3C, Table S2). Similarity was confirmed by a Fisher’s exact test (FET) comparing the DEG heat maps (Figure 3C), which revealed significant enrichment of upregulated DEGs (Odds Ratio, OR=62.77, p=9 × 10−22) and downregulated DEGs (OR=5.84, p=1 × 10−11) between E2F3a-SAL vs. GFP-SAL and GFP-COC vs. GFP-SAL (Figure 3D). Interestingly, the overlap in upregulated genes was more robust.
We utilized RRHO to extend our analysis in a threshold-free manner (Figure 3E)(18, 29). We observed robust overlap of genes upregulated by both E2F3a and cocaine (max –log(p-value)=1606; Figure 3F, Table S3). This genome-wide result mirrored the highly significant overlap found in upregulated DEGs by FET. RNA-seq also confirmed that E2F3a overexpression did not alter expression of other E2F isoforms expressed in NAc (Table S2).
Viral-mediated E2F3a overexpression in NAc recapitulates cocaine-induced alternative splicing patterns
Next, we employed RNA-seq multivariate analysis of transcript splicing (rMATS) to evaluate a predicted role for E2F3a in cocaine-induced changes in alternative splicing (13, 30). Four types of alternative splicing events were detected (illustrated in Figure 4A): skipped exon (SE), retained intron (RI), mutually exclusive exons (MXE), and alternative 5’ splice site (A5SS). We observed similar overall numbers of differential alternative splicing events elicited by cocaine and E2F3a overexpression (GFP-COC vs. GFP-SAL: 482 SE, 164 RI, 32 MXE, 55 A5SS; E2F3a-SAL vs GFP-SAL: 295 SE, 131 RI, 19 MXE, 51 A5SS; Table S4). Furthermore, for each type of splicing event, cocaine and E2F3a overexpression elicited similar alternative splicing patterns, as measured by Inclusion Level Difference (ILD, detailed in Experimental Procedure; Figure 4B). Statistical comparison by FET revealed that E2F3a overexpression and cocaine elicited significantly overlapping gain in all splicing event types and loss in all but A5SS of alternatively spliced exons (p-values listed in Figure 4C). Together, these data show that E2F3a drives the same types of alternative splicing in NAc in similar directions as cocaine in a robustly overlapping population of genes.
E2F3 regulates genes that contribute to RRHO upregulation signature
To confirm that expression changes detected in by RNA-seq were specific to E2F3a, we assayed expression of select genes in a separate cohort following overexpression of E2F3a or E2F3b in NAc (Figure 5A,F). We chose our targets from the intersection of genes that led to the initial prediction of E2F3 as an upstream regulator of cocaine-elicited molecular responses and the list of most significantly co-upregulated genes in our RRHO analysis (−log10(p-value)>1.3; Figure 3F)(13). Representative expression patterns of two such genes from the RNA-seq data are indicated in Figure 5: Fgfr1, fibroblast growth factor receptor 1 (E2F3a-SAL vs GFP-SAL: log2(FC)=0.15) and Tle2, transducin-like enhancer of split 2 (E2F3a-SAL vs GFP-SAL: log2(FC)=0.12). We found by qPCR that both Fgfr1 and Tle2 were significantly upregulated by E2F3a in this second cohort, but were unaffected by E2F3b overexpression (Figure 5B,G). Furthermore, miR-mediated knockdown of E2F3a in NAc reversed the induction of these genes by cocaine, showing that E2F3a is required for their upregulation (Figure 5C,H).
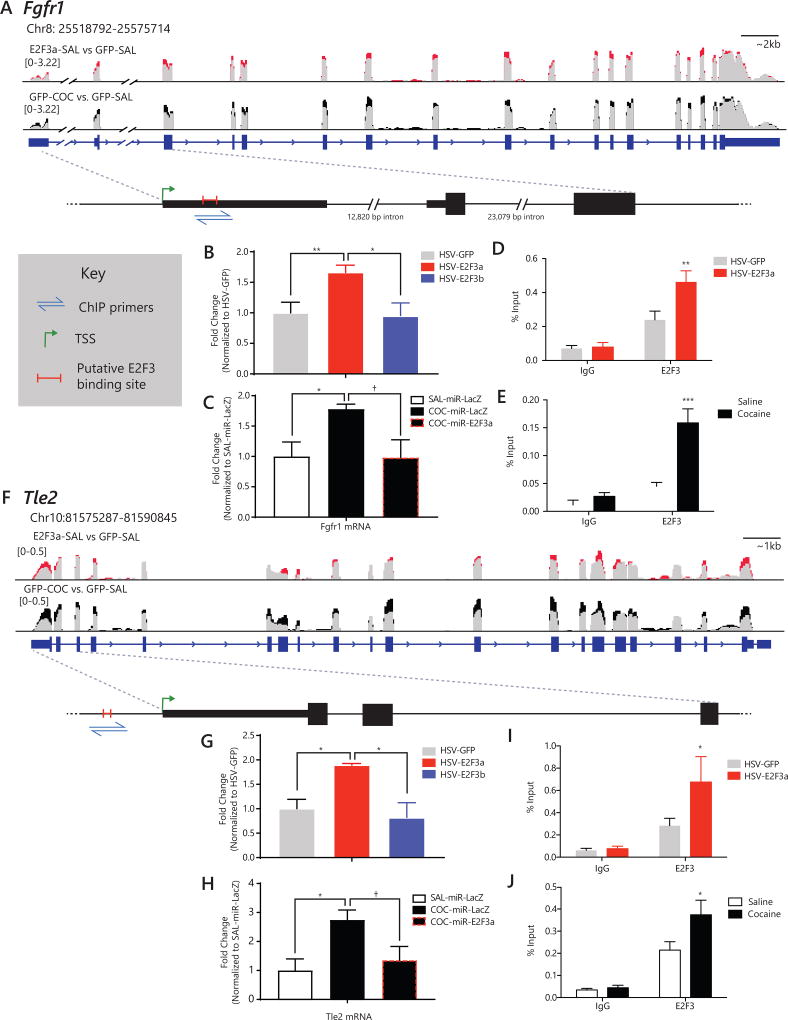
Figure 5. E2F3a regulates expression of Fgfr1 and Tle2 in NAc via direct binding at promoters.
(A & F) Wiggle plots depicting expression profile of genes showing increased expression after E2F3a overexpression (red) or cocaine exposure (black) when compared to control conditions (gray). Fragments per kilobase of transcript per million mapped reads (FPKM) are graphed for the exons. Gene diagram is given below. Red lines in cut out regions depict the site of putative E2F3 binding, illustrated along with the primer set designed to amplify that region. (A) Fgfr1’s putative E2F3 binding site is located within Exon 1. (B) qPCR for Fgfr1 mRNA following expression of HSV-GFP, -E2F3a, or -E2F3b. E2F3a overexpression increases Fgfr1 expression (F2,16=5.721, p=0.0134; post hoc: HSV-GFP vs. HSV-E2F3a, p<0.05; HSV-E2F3a vs. HSV-E2F3b, p<0.05; N=6–7). (C) qPCR for Fgfr1 mRNA following cocaine treatment and expression of either a miRNA targeting E2f3a or LacZ. Cocaine increased Fgfr1 expression, and knockdown of E2F3a blocked the induction by cocaine (F2,11=4.852, p=0.0309; post-hoc: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.0485; COC-miR-LacZ vs. COC-miR-E2f3a, p=0.057; N=6–7). (D) qChIP for E2F3 following HSV-E2F3a infusion into NAc showed increased E2F3 binding at the putative E2F3 binding site in the Fgfr1 promoter (interaction of virus and antibody: F1,18=6.91, p=0.0171; HSV-E2F3a vs. HSV-GFP for E2F3 binding by post-hoc: p=0.0062; N=5–6) (E) qChIP for E2F3 following cocaine (7 days 20 mg/kg; 24 hr) showed increased E2F3 binding at the same putative binding site in the Fgfr1 promoter (interaction of treatment and antibody: F1,18=10.37, p=0.0047; Cocaine vs. Saline for E2F3 binding post-hoc: p=0.0005; N=5–6). (F) Tle2’s putative E2F3 binding site is located approximately 250 bp upstream of the TSS. (G) qPCR for Tle2 mRNA following expression of HSV-GFP, -E2F3a, or -E2F3b. E2F3a overexpression increases Tle2 expression (F2,16=4.918, p=0.0218; post hoc: HSV-GFP vs. HSV-E2F3a, p=0.0436; HSV-GFP vs. HSV-E2F3b, p=0.0257; N=6–7). (H) qPCR for Tle2 mRNA following cocaine treatment and either a miRNA targeting E2f3a or LacZ. Cocaine increased Tle2 expression, and knockdown of E2F3a blocked the induction by cocaine (F2,11=5.072, p 0.0275; post-hoc: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.0336; COC-miR-LacZ vs. COC-miR-E2f3a, p=0.0731; N=4–5). (I) qChIP for E2F3 following HSV-E2F3a infusion into NAc showed increased E2F3 binding at the putative E2F3 binding site in the Tle2 promoter (main effect of antibody was observed: F1,20=15.72, p=0.0008; HSV-E2F3a vs. HSV-GFP for E2F3 binding post-hoc: p=0.023; N=6). (J) qChIP for E2F3 following cocaine (7 days 20 mg/kg; 24 hr) showed increased E2F3 binding at the same putative binding site in the Tle2 promoter (main effect of antibody was observed: F1,17=36.92, p<0.0001; Cocaine vs. saline for E2F3 binding post-hoc: p=0.0313; N=4–6). Graphs show mean SEM. †: not significant, p<0.1. *p<0.05. **p<0.01. ***p<0.001 (See also Figure S5)
We performed qChIP for E2F3 to determine whether these changes could be due to increased direct binding of E2F3a at the genes’ promoters. Because isoform-selective antibodies are unavailable, we first overexpressed E2F3a and tested binding at putative binding motifs in target genes (Figure 5A,F). E2F3a overexpression increased E2F3 binding above GFP-overexpression and negative IgG control levels at Fgfr1 (interaction of virus and antibody: F1,18=6.91, p=0.0171; post-hoc, E2F3: p=0.0062; Figure 5D) and Tle2 (main effect of antibody: F1,20=15.72, p=0.0008; post-hoc, E2F3: p=0.023; Figure 5I). There was no E2F3 binding with or without overexpression at two non-predicted genes lacking an E2F consensus sequences, confirming specificity (Figure S5A,B).
Repeated cocaine increased E2F3 binding at these same genetic loci. E2F3 binding was increased compared to saline and IgG at the Fgfr1 promoter (F1,18=10.37, p=0.0047; post-hoc, E2F3: p=0.0005; Figure 5E) and Tle2 promoter (main effect of antibody: F1,17=36.92, p<0.0001; post-hoc, E2F3: p=0.0313; Figure 5J). qChIP performed at two non-predicted genes, and regions of Fgfr1 and Tle2 that lack E2F3 binding sites, confirmed absence of E2F3 binding both at baseline and after cocaine administration (Figure S5C–F). These data indicate that the expression differences revealed by RNA-seq at specific target genes following E2F3a overexpression or cocaine administration are mediated by E2F3’s direct binding to the genes of interest.
E2F3a regulates alternative splicing
Because cocaine administration and E2F3a overexpression similarly affected alternative splicing profiles, we investigated the molecular basis of these results. Transcription factors are proposed to bind DNA and recruit splicing factors to affect the balance of spliced isoforms(34). We therefore performed qChIP at alternatively spliced targets, selected with criteria described above. We pursued Tle2 and polypyrimidine tract binding protein 1, Ptbp1, as E2F3a-mediated alternative splicing targets. Both genes contain E2F3 consensus sites proximal to the alternatively spliced exons (Figure 6A,E). We visualized differential splicing using SashimiPlot in conjunction with rMATs, and found that both splicing events illustrated are SEs (Figure 6B,H). Cocaine treatment and E2F3a overexpression led to a loss of the alternatively spliced exon in Tle2 in NAc as compared to GFP controls (GFP-COC vs. GFP-SAL: ILD=0.089, p=0.0348; E2F3a-SAL vs. GFP-SAL: ILD=0.094, p=0.0305). Conversely, both cocaine and E2F3a overexpression induced the alternatively spliced exon in Ptbp1 (GFP-COC vs. GFP-SAL: ILD=-0.095, p=0.0072; E2F3a-SAL vs. GFP-SAL: ILD=-0.07, p=0.0089).
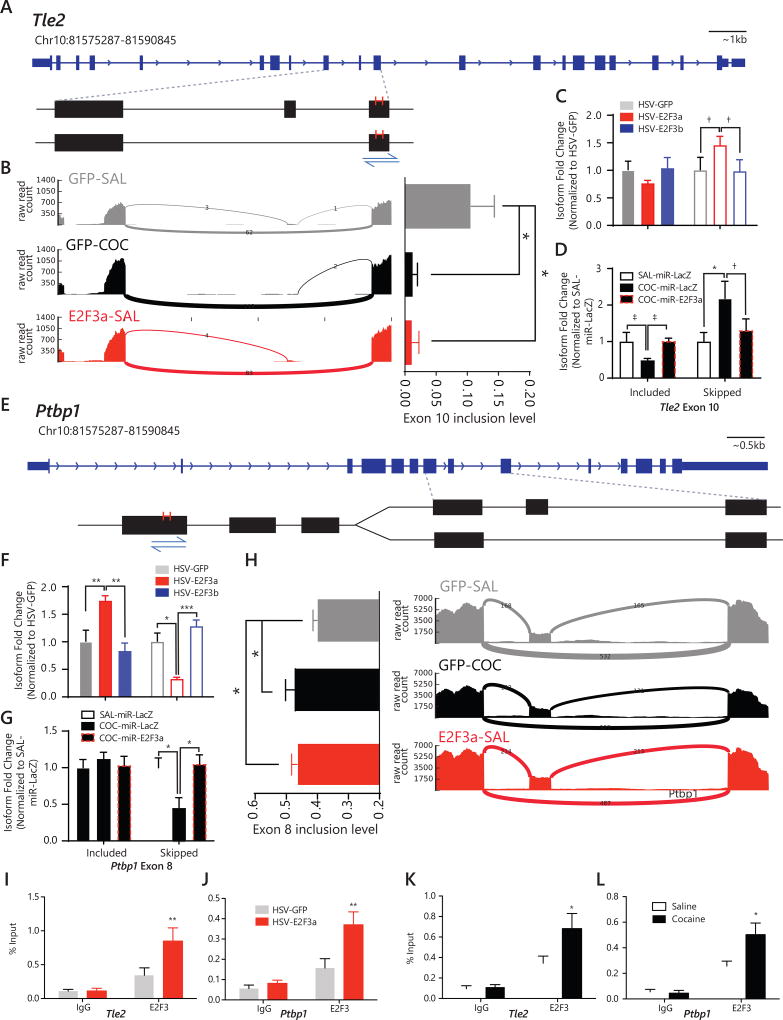
Figure 6. E2F3a regulates alternative splicing of Tle2 and Ptbp1 in NAc via direct binding near splicing sites.
(A & E) Gene diagrams and SashimiPlots – the magnified portion of the genomic loci – illustrate locations of alternative splicing event within the selected genes. Putative E2F3 binding sites and primer sets are noted with red line and blue arrows, respectively. (A) Tle2’s alternative splicing event involves the skipping of Exon 10. The putative E2F3 binding site is located in Exon 11. (B) The SashimiPlots for Tle2 show that there are more junctions between the two constitutive (outer) exons in both GFP-COC and E2F3a-SAL than in GFP-SAL, while there are more junctions between the constitutive exons and the alternatively spliced (center) exon in GFP-SAL than in the other 2 groups. Bar graph illustrates quantification of inclusion level of Exon 10 for each sample (GFP-COC vs. GFP-SAL: inclusion level difference (ILD)=0.089, p=0.0348; E2F3a-SAL vs. GFP-SAL: ILD=0.094, p=0.0305). (C) qPCR for the Tle2 transcripts that either include Exon 10 or skip Exon 10 following expression of HSV-GFP, -E2F3a, or -E2F3b (No main effect of virus: F2,26=0.4596, p=0.6366; N=5–6). (D) qPCR for the Tle2 transcripts that either include Exon 10 or skip Exon 10 following cocaine treatment and expression of either a miRNA targeting E2f3a or LacZ (interaction of virus and isoform: F2,22=7.028, p=0.0044; post-hoc, Included Exon: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.1996; COC-miR-LacZ vs.COC-miR-E2f3a, p=0.2051; Skipped Exon: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.0206; COC-miR-LacZ vs.COC-miR-E2f3a, p=0.0998; N=4–5). (E) Ptbp1’s alternative splicing event involves the skipping of Exon 8. The putative E2F3 binding site is located in Exon 4. (F) qPCR for the Ptbp1 transcripts that either include Exon 8 or skip Exon 8 following expression of HSV-GFP, -E2F3a, or -E2F3b (interaction of virus and isoform: F2,31=22.56, p<0.0001; post-hoc, Included Exon: HSV-E2F3a vs. HSV-GFP, p=0.0023; HSV-E2F3b vs. HSV-E2F3a, p=0.0032; Skipped Exon: HSV-E2F3a vs. HSV-GFP, p=0.0221; HSV-E2F3b vs. HSV-E2F3a, p=0.0002; N=6–7). (G) qPCR for the Ptbp1 transcripts that either include Exon 8 or skip Exon 8 following cocaine treatment and expression of either a miRNA targeting E2f3a or LacZ (interaction of virus and isoform: F2,30=4.488, p=0.0197; post-hoc, Included Exon: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.8870; COC-miR-LacZ vs.COC-miR-E2f3a, p=0.9472; Skipped Exon: SAL-miR-LacZ vs. COC-miR-LacZ, p=0.0131; COC-miR-LacZ vs.COC-miR-E2f3a, p=0.0084; N=6). (H) The SashimiPlots for Ptbp1 show that cocaine exposure and E2F3a overexpression both decrease skipping of Exon 8. Bar graph illustrates quantification of inclusion of Exon 8 level for each sample (GFP-COC vs. GFP-SAL: ILD=-0.095, p=0.0072; E2F3a-SAL vs. GFP-SAL: ILD=-0.07, p=0.0089). (I) qChIP for E2F3 following HSV-E2F3a infusion into NAc showed increased E2F3 binding at the putative E2F3 binding site in Tle2 Exon 11 (interaction of virus and antibody: F1,20=5.53, p=0.0291; HSV-E2F3a vs. HSV-GFP for E2F3 binding post-hoc: p=0.0074; N=6). (J) qChIP for E2F3 following HSV-E2F3a infusion into NAc showed increased E2F3 binding at the putative E2F3 binding site in Ptbp1 Exon 4 (interaction of virus and antibody: F1,18=6.42, p=0.0208; HSV-E2F3a vs. HSV-GFP for E2F3 binding post-hoc: p=0.0046; N=5–6). (K) qChIP for E2F3 following cocaine (7 days 20 mg/kg; 24 hr) showed increased E2F3 binding at the same putative binding site in Tle2 Exon 11 (main effect of antibody was observed: F1,17=19.76, p=0.0004; Cocaine vs. saline for E2F3 binding post-hoc: p=0.0365; N=5–6). (L) qChIP for E2F3 following cocaine (7 days 20 mg/kg; 24 hr) showed increased E2F3 binding at the same putative binding site in Ptbp1 Exon 4 (a main effect of treatment: F1,16=5.63, p=0.0306; main effect of antibody was observed: F1,16=24.22, p=0.0002; Cocaine vs. saline for E2F3 binding post-hoc: p=0.0305; N=5). Graphs show mean SEM. †: not significant, p<0.1. *p<0.05. **p<0.01. (See also Figure S5)
In a separate cohort, we confirmed that Tle2 transcripts that do not contain Exon 10 are increased in NAc after E2F3a overexpression, but not E2F3b overexpression (Figure 6C). Additionally, knockdown of E2f3a blocked the increase in Exon 10-”skipping” Tle2 transcripts following cocaine exposure (Figure 6D). Similarly, for the Exon 8-containing Ptbp1 transcripts, E2F3a, but not E2F3b, overexpression decreased levels of the Exon 8-”skipping” Ptbp1 transcripts, while E2F3a knockdown blocked cocaine’s effects (Figure 6F,G).
Finally, we found that both cocaine and E2F3a overexpression increased E2F3 binding at consensus sequences near alternative splicing sites. Repeated cocaine increased E2F3 binding compared to saline at the putative E2F3 binding site in Tle2 Exon 11 (main effect of antibody was observed by two-way ANOVA: F1,17=19.76, p=0.0004; post-hoc, E2F3: p=0.0365; Figure 6K), and in Ptbp1 Exon 4 (main effect of treatment: F1,16=5.63, p=0.0306; post-hoc, E2F3: p=0.0305; Figure 6L). There was no E2F3 binding at a control region of Tle2 or Ptbp1 before or following cocaine treatment (Figure S5G). E2F3a overexpression also increased E2F3 binding compared to GFP control at the putative binding motifs in Exon 11 of Tle2 (interaction of virus and antibody: F1,20=5.53, p=0.0291; post-hoc, E2F3: p=0.0074; Figure 6I), and Exon 4 of Ptbp1 (interaction of virus and antibody: F1,18=6.42, p=0.0208; post-hoc, E2F3: p=0.0046; Figure 6J). Together, these data suggest that E2F3a mediates alternative splicing induced by cocaine.
Discussion
Our findings provide direct evidence for the role of E2F3a as a key regulator of cocaine-elicited molecular actions via transcriptional regulation and splicing mechanisms. We identified E2f3 as a novel target of cocaine. Repeated cocaine exposure increased E2f3a mRNA expression in NAc, and increased E2F3a protein levels in the nucleus. This positions E2F3a to bind DNA and regulate the observed changes in gene expression and RNA processing. The isoform-specific role of E2F3a was further confirmed by our behavioral results in which overexpression of E2F3a, but not E2F3b, in NAc enhanced cocaine locomotor and place conditioning behaviors. Conversely, these behaviors were blunted by E2F3a knockdown in NAc, further demonstrating the critical role of E2F3a in regulating cocaine-elicited behaviors. RNA-seq following overexpression of E2F3a in NAc revealed that E2F3a recapitulates a substantial portion of cocaine-induced patterns of gene regulation, specifically gene induction and alternative splicing. Strikingly, qChIP uncovered enhanced E2F3 binding at multiple target genes of interest in NAc following either repeated cocaine exposure or E2F3a overexpression. This strongly suggests that direct binding of the transcription factor to target genes orchestrates the observed expression and splicing changes.
Up to now, most studies have focused on E2Fs in proliferating cell populations as the proteins are known cell-cycle regulators, and there is little knowledge of E2F targets in healthy adult brain tissue(35). Here, we identified thousands of putative E2F3a targets in NAc and specifically probed and confirmed three target genes via qChIP: Fgfr1, Ptbp1 and Tle2.
Fgfr1 and E2f3 have previously been identified in genome-wide screens of human cancers, but the directionality of regulation has not been established (36, 37). Moreover, the FGF system has been implicated in cocaine-elicited molecular mechanisms in rats. Specifically, Fgfr1 mRNA is increased in limbic regions of rats bred for increased locomotor sensitivity to cocaine and enhanced novelty-seeking (38). Fgf2 mRNA is increased in rat striatum by cocaine administration, and a single neonatal infusion of FGF2 increases locomotor sensitivity to cocaine (39, 40). Importantly, Fgf2 is also among the group of genes most significantly co-upregulated by cocaine or E2F3a (Figure 3F, Table S3) and has putative E2F binding motifs present within its promoter (Genomatix). Thus, the FGF system is likely one pathway recruited by cocaine via E2F3a-dependent mechanisms to regulate behavioral responses.
Ptbp1 is a ubiquitously expressed heterogeneous nuclear ribonucleoprotein (hnRNP) known to play a role in pre-mRNA splicing and the regulation of alternative splicing (41, 42). The alternative splicing event induced by both E2F3a overexpression and cocaine administration translates into a 26 amino acid insertion between PTB’s 3rd and 4th RNA binding domains, which has been shown to increase alternative splicing activity in vitro (43). Thus, Ptbp1 may be an important player in the downstream alternative splicing profile orchestrated by cocaine via E2F3a, a possibility which warrants further examination in future studies.
Finally, Tle2 is a transcriptional corepressor that binds to many transcription factors. This gene is particularly interesting as we found it to be both transcriptionally regulated and alternatively spliced by both E2F3a overexpression and cocaine administration – a type of regulation not previously observed for a cocaine-induced transcription factor. Tle2 is one of only 58 genes in our dataset whose expression and splicing were both altered upon repeated cocaine administration or E2F3a overexpression. Other genes on this list (e.g., Camk2a, Nrxn1, Mecp2) may be similarly regulated by E2F3a actions after cocaine.
These particular genes provide examples of the E2F3a regulation identified in our dataset. More broadly, our rich dataset paints a picture of the complex genome-wide orchestration by E2F3a of transcriptional and alternative splicing mechanisms in NAc that regulate responses to cocaine. In the future, it will be important to probe the specific role of E2F3a in the two main neuronal subtypes in NAc, D1-type and D2-type medium spiny neurons. These two cell types play different roles in the behavioral and physiological expression of cocaine responses. In the current study, the genome-wide overlap seen in the molecular signature of cocaine and E2F3a overexpression was based on analysis of crude NAc extracts, although E2F3a overexpression was targeted with our HSV vectors to neurons only, and immunohistochemistry confirms a predominantly neuronal localization of E2F3 in this brain region. In any event, the present dataset will prove useful in guiding further exploration of the cell type-specific role of E2F3a in mediating cocaine’s effects in NAc.
Further work is also needed to understand the regulatory and functional implications of the selective role for E2F3a, and not E2F3b, in cocaine action. The two protein isoforms are highly similar (Figure 1A). Thus, it will be interesting to learn how cocaine selectively controls levels of E2F3a in NAc, and how E2F3a, despite similar DNA binding properties as E2F3b, selectively influences cocaine responses. It would also be interesting to perform ChIP-seq for E2F3a in this brain region, although this would require the development of selective antibodies suitable for this approach.
The transcriptomic patterns induced by cocaine that drive downstream behavioral effects rely on a suite of transcription factors that act to regulate expression of the genome in concert. We have now identified a new key player in E2F3a, which orchestrates both transcriptional and splicing profiles leading to locomotor sensitization and enhanced place conditioning. Our study reveals that E2F3a is capable of recapitulating much of cocaine-induced gene upregulation and a large portion of the alternative splicing changes in NAc, indicating that E2F3a is among the most critical upstream effectors of cocaine-elicited molecular regulation. These actions of E2F3a likely contribute to the long-lasting behavioral plasticity that underlies the addicted state. By identifying the critical role of E2F3a in mechanisms of cocaine action, our findings may open the door for novel therapeutic development to target this important molecular regulator or its downstream effectors.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute on Drug Abuse.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Raw and processed RNA-seq gene expression data are available via the Gene Expression Omnibus data (accession number GEO:GSE85936)
Financial Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
Author Contributions (CRediT Taxonomy)
Conceptualization, H.M.C. and E.J.N.; Methodology, H.M.C. and E.J.N.; Software, I.P. and L.S.; Formal Analysis, H.M.C. and I.P.; Investigation, H.M.C, E.A.H., R.C.B., C.K.L., C.J.P., D.M.W. and M.C.; Resources, R.L.N.; Writing – Original Draft, H.M.C.; Writing – Review & Editing, H.M.C., E.A.H., C.K.L., C.J.P., D.M.W., R.C.B. and E.J.N.; Supervision, L.S., R.C.B. and E.J.N.; Funding Acquisition, E.J.N.
References
- 1.Kalivas PW, Volkow ND. The neural basis of addiction: A pathology of motivation and choice. Am J Psychiatry. 2005:162. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 2.Nestler EJ. Review. Transcriptional mechanisms of addiction: role of DeltaFosB. Philos Trans R Soc Lond B Biol Sci. 2008;363:3245–55. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, Lobo MK. Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci. 2015;35:7927–37. doi: 10.1523/JNEUROSCI.0548-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, et al. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–37. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vialou V, Feng J, Robison AJ, Ku SM, Ferguson D, Scobie KN, et al. Serum response factor and cAMP response element binding protein are both required for cocaine induction of ΔFosB. J Neurosci. 2012;32:7577–84. doi: 10.1523/JNEUROSCI.1381-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–74. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brami-Cherrier K, Valjent E, Hervé D, Darragh J, Corvol J-C, Pages C, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–54. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Choi K-H, Renthal W, Tsankova NM, Theobald DEH, Truong H-T, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–14. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–48. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–29. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–50. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn J-H, Maroteaux M, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453:879–84. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Armanious MK, Thomas MJ, Cress WD. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19:3422–33. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 15.Julian LM, Vandenbosch R, Pakenham CA, Andrusiak MG, Nguyen AP, McClellan KA, et al. Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell. 2013;12:440–52. doi: 10.1016/j.stem.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–32. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cates HM, Thibault M, Pfau M, Heller E, Eagle A, Gajewski P, et al. Threonine 149 phosphorylation enhances ΔFosB transcriptional activity to control psychomotor responses to cocaine. J Neurosci. 2014;34:11461–11469. doi: 10.1523/JNEUROSCI.1611-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahr BA, Neve RL, Sharp J, Geller AI, Lynch G. Rapid and stable gene expression in hippocampal slice cultures from a defective HSV-1 vector. Brain Res Mol Brain Res. 1994;26:277–85. doi: 10.1016/0169-328x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 20.Cahill ME, Bagot RC, Gancarz AM, Walker DM, Sun H, Wang Z-J, et al. Bidirectional Synaptic Structural Plasticity after Chronic Cocaine Administration Occurs through Rap1 Small GTPase Signaling. Neuron. 2016;89:566–582. doi: 10.1016/j.neuron.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson D, Shao N, Heller E, Feng J, Neve R, Kim H-D, et al. SIRT1-FOXO3a regulate cocaine actions in the nucleus accumbens. J Neurosci. 2015;35:3100–11. doi: 10.1523/JNEUROSCI.4012-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purushothaman I, Shen L. SPEctRA - A Scalable Pipleline for RNA-seq Analysis 2016 [Google Scholar]
- 28.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111:E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L. GeneOverlap - R package for testing and visualizing gene overlaps [Google Scholar]
- 32.Grueter BA, Robison AJ, Neve RL, Nestler EJ, Malenka RC. ΔFosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc Natl Acad Sci U S A. 2013;110:1923–8. doi: 10.1073/pnas.1221742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci. 2014 doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.KORNBLIHTT AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swiss Va, Casaccia P. Cell-context specific role of the E2F/Rb pathway in development and disease. Glia. 2010;58:377–90. doi: 10.1002/glia.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–51. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 37.Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003;63:2872–80. [PubMed] [Google Scholar]
- 38.Turner CA, Flagel SB, Clinton SM, Akil H, Watson SJ. Cocaine interacts with the novelty-seeking trait to modulate FGFR1 gene expression in the rat. Neurosci Lett. 2008;446:105–7. doi: 10.1016/j.neulet.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fumagalli F, Pasquale L, Racagni G, Riva MA. Dynamic regulation of fibroblast growth factor 2 (FGF-2) gene expression in the rat brain following single and repeated cocaine administration. J Neurochem. 2006;96:996–1004. doi: 10.1111/j.1471-4159.2005.03627.x. [DOI] [PubMed] [Google Scholar]
- 40.Clinton SM, Turner CA, Flagel SB, Simpson DN, Watson SJ, Akil H. Neonatal fibroblast growth factor treatment enhances cocaine sensitization. Pharmacol Biochem Behav. 2012;103:6–17. doi: 10.1016/j.pbb.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB Bound to RNA: Specific Binding and Implications for Splicing Regulation. Science (80- ) 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 42.Romanelli MG, Lorenzi P, Morandi C. Organization of the human gene encoding heterogeneous nuclear ribonucleoprotein type I (hnRNP I) and characterization of hnRNP I related pseudogene. Gene. 2000;255:267–72. doi: 10.1016/s0378-1119(00)00331-0. [DOI] [PubMed] [Google Scholar]
- 43.Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–32. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.